Abstract
Helicobacter pylori, a bacterial pathogen that can infect human stomach causing gastritis, ulcers and cancer, is known to have a high degree of genome/epigenome diversity as the result of mutation and recombination. The bacteria often infect in childhood and persist for the life of the host. One of the reasons of the rapid evolution of H. pylori is that it changes its genome drastically for adaptation to a new host. To investigate microevolution and adaptation of the H. pylori genome, we undertook whole genome sequencing of the same or very similar sequence type in multi-locus sequence typing (MLST) with seven genes in members of the same family consisting of parents and children in Japan. Detection of nucleotide substitutions revealed likely transmission pathways involving children. Nonsynonymous (amino acid changing) mutations were found in virulence-related genes (cag genes, vacA, hcpDX, tnfα, ggt, htrA and the collagenase gene), outer membrane protein (OMP) genes and other cell surface-related protein genes, signal transduction genes and restriction-modification genes. We reconstructed various pathways by which H. pylori can adapt to a new human host, and our results raised the possibility that the mutational changes in virulence-related genes have a role in adaptation to a child host. Changes in restriction-modification genes might remodel the methylome and transcriptome to help adaptation. This study has provided insights into H. pylori transmission and virulence and has implications for basic research as well as clinical practice.
Introduction
The pathogenic epsilon-proteobacterium Helicobacter pylori is a major cause of human gastric diseases [1]. The H. pylori genome sequence exhibits a high degree of diversity even between closely related strains because of the high rate of mutation and recombination [2–6]. The epigenome is also diverse owing to changes in methyltransferase genes through various mechanisms [7–12].
The significance of such extreme genome/epigenome diversity is not understood in detail, although the ability to adapt to the human host has been often suggested. Evidence for effects of DNA modification on gene transcription in bacteria has been limited primarily because of lack of techniques to sensitively and accurately detect methylated DNA bases and to accurately measure transcripts. The Single-Molecule Real-Time (SMRT) sequencing technology now allows methylome decoding at the single base resolution [13, 14]. On the other hand, RNA-seq method in the next-generation sequencers allows accurate and sensitive measurements of transcriptome changes [15]. As a result, there are now an increasing number of reports on relationship between DNA methylation by modification enzymes and transcriptome in bacteria. For example, Type II restriction-modification (RM) systems affect global gene expression in Escherichia coli [16] and in H. pylori [17]. Type III restriction-modification systems affect expression of various genes in Neisseria, Haemophilus and H. pylori [10, 18, 19]. Type I restriction-modification systems affect gene expression in H. pylori [7] and Streptococcus pneumoniae [20]. There is also evidence that Type II modification enzymes affect H. pylori physiology [21–23].
There have been many studies of human familial transmission in attempts to identify transmission pathways. Early studies used electrophoresis-based methods or multi-locus sequence typing (MLST) based on seven conserved genes (~3 kb in total) [24, 25]. Recently, more sensitive whole genome (~1.6 Mb) sequencing was used to analyze the process of familial infection [26–28].
In order to gain insight into H. pylori adaptive evolution in a new human host, especially in a child who was supposed to be newly infected, we used whole genome sequencing of very closely related H. pylori strains from the same family, which are difficult to distinguish by standard MLST analysis [29] and estimated the order of intrafamilial infection and then investigated which genes have experienced amino acid changes.
Results
Families infected with H. pylori of the same or very similar MLST sequence type
In all, 19 H. pylori strains were isolated from five families (one strain per person) visiting a pediatrician in a hospital in Hokkaido, Japan (Table 1). Each family member was infected with H. pylori strains of the same or very similar sequence type as judged by standard MLST analysis (S1 Table): the mother and the offspring in families K-1 and K-2, the parents and offspring in families K-3 and K-4, and the children in family K-5 [29]. The likely direction of familial transmission in families K-1 and K-2 was from mother to offspring. The allele type numbers of some of the seven MLST genes were different in families K-3 and K-4; this is based on a single nucleotide substitution, however, so we regard them as very similar sequences. The MLST results did not make it clear whether the father or the mother was the H. pylori donor to the offspring. In family K-5, transmission was likely from outside the family, which might be independent among children from the same reservoir or might have been followed by inter-child transmission.
Table 1. Strains and mapping results.
| Family | Strain a | Relationship | Age | Mapped nt (> x5) | Mapped % | Variation # b |
|---|---|---|---|---|---|---|
| K-1 | K16 | Father | 39 | 1456954 | 92.8 | 35242 |
| K17 | Mother | 38 | 1456823 | 92.8 | 34456 | |
| K15 | Child | 10 | 1457425 | 92.8 | 34540 | |
| K-2 | K34 | Father | 51 | 1459303 | 92.9 | 35271 |
| K35 | Mother | 46 | 1419326 | 90.4 | 37269 | |
| K36 | Child | 17 | 1419365 | 90.4 | 37305 | |
| K37 | Child | 14 | 1419197 | 90.4 | 37288 | |
| K-3 | K26 | Father | 43 | 1459932 | 93.0 | 35041 |
| K27 | Mother | 28 | 1460893 | 93.0 | 34981 | |
| K28 | Child | 6 | 1458283 | 92.9 | 34968 | |
| K-4 | K29 | Father | 48 | 1379964 | 87.9 | 37440 |
| K30 | Mother | 44 | 1378891 | 87.8 | 37389 | |
| K32 | Child | 14 | 1377440 | 87.7 | 37387 | |
| K33 | Child | 13 | 1378456 | 87.8 | 37422 | |
| K-5 | K21 | Father | 46 | 1462911 | 93.1 | 35064 |
| K22 | Mother | 47 | 1452079 | 92.5 | 34581 | |
| K23 | Child | 17 | 1455829 | 92.7 | 33890 | |
| K24 | Child | 15 | 1457747 | 92.8 | 33882 | |
| K25 | Child | 5 | 1457010 | 92.8 | 33823 |
a Strains with same sequence type within a family are underlined.
b Number of variations compared with F30 genome.
All these strains were shotgun sequenced by an Illumina high-throughput sequencer and mapped onto the known genome sequence of H. pylori strain F30 (Table 1), which belongs to the hspEAsia population (as do most isolates in Japan) [4, 30]. About 90% of nucleotides were mapped with coverage of more than five reads, with detection of ~35,000 nucleotide substitutions per strain. A lower mapping rate in the strains found in family K-4 can be explained by the lack of a cag pathogenicity island in their genomes. Little difference was observed when other hspEAsia genomes (F16, F32 and F57) [4] were used as a reference genome for mapping (S2 Table), so we used the mapping on H. pylori strain F30 for further analysis.
Estimation of transmission pathways from the number of nucleotide substitutions
Nucleotide substitutions between each strain pair within a family were counted and used as the distance between compared strains (S3 Table) to construct a phylogenetic tree for each family (Fig 1). As expected, the number of nucleotide substitutions between strains of distantly related MLST sequence types was greater compared to the same or very similar MLST sequence type by one to three orders of magnitude. From the whole genome sequence information, various evolutionary relations were inferred between multiple strains of the same or very similar MLST sequence type within a family.
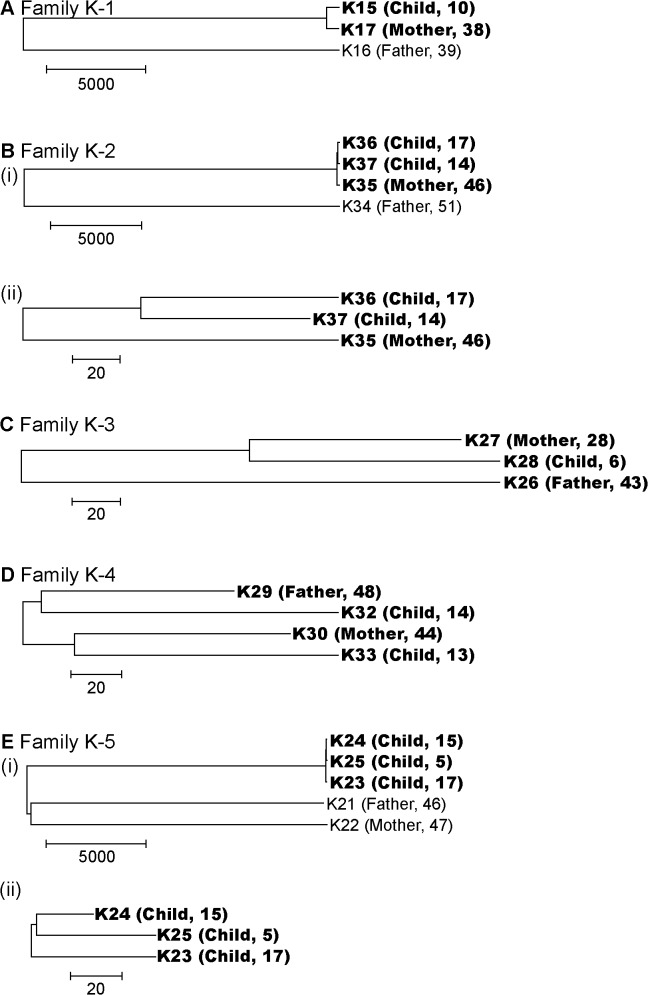
Fig 1. Phylogenetic trees of H. pylori genomes in the families.
The number of nucleotide substitutions was used as the distance matrix. The relation and age of the host is added next to the strain name. (A) Family K-1. (B)(i) Family K-2. (ii) Strains with the same MLST sequence type in Family K-2. (C) Family K-3. (D) Family K-4. (E)(i) Family K-5. (ii) Strains with the same MLST sequence type in Family K-5. The bar indicates the number of nucleotide substitutions.
In family K-2, comparison of three strains with the same MLST sequence type showed the distance between the two strains from children K36 and K37 was, on average, 1.7-fold smaller compared to K35 (from mother) and K36 (from child) and to K35 (from mother) and K37 (from child) (Fig 1B (ii)). The difference was statistically significant (P < 10–5), supporting the hypothesis that the latest infection (transfer) occurred between the hosts of K36 and K37; i.e. transmission between children. (This inference is discussed in Discussion, below.)
A comparison of three strains with the same MLST sequence type in family K-3 showed the distance between K27 (from mother) and K28 (from child) is about half compared to K26 (from father) and K27 (from mother) and to K26 (from father) and K28 (from child) (Fig 1C). The difference is significantly greater compared to assuming a random distribution of substitution mutations (P < 10–5), suggesting the latest infection occurred between the hosts of K27 (mother) and K28 (child); i.e. transmission between mother and child. The likely direction is from mother to child, considering the general pathway of H. pylori infection. (See Discussion, below.)
No significant difference between distances was found for the four strains of very similar MLST sequence type in family K-4 (P > 0.01, Fig 1D).
There were three MLST sequence types corresponding to father, mother and offspring in family K-5 (see the preceding section). No significant difference between distances was detected for the three strains isolated from the offspring (P > 0.01, Fig 1E (ii)).
Detection and classification of strain-specific nucleotide substitutions
All the nucleotide variations in strains with the same or very similar MLST sequence type were compared within each family for the detection of strain-specific nucleotide substitutions. Bases corresponding to the same site were compared, and assigned as a strain-specific substitution when only a single strain had a base different from the other strains in the comparison. In the case of family K-1, there were only two strains with the same sequence type, so we could not distinguish K15-specific from K17-specific substitutions and treated all the substitutions as strain-specific mutations.
We classified base substitutions to single nucleotide variations (SNVs) or clusters of nucleotide polymorphisms (CNPs) in order to estimate the frequency of recombination (Table 2). CNP is defined as a cluster of two or more substitutions separated by <200 bp and flanked by >200 bp of identical sequence on both sides and is considered as a sign of substitution by a recombination event [31]. Substitutions not clustered as CNPs were assigned as SNVs. A large number of CNPs was observed in family K-1, which accounted for about 85% of substitutions. The number of substitutions included in a CNP was, on average, about 11 for those of family K-1 but less than three in all the other strains.
Table 2. Strain specific nucleotide substitutions.
| Strain specific | Substitution | non-synonymous/ | ||||||
|---|---|---|---|---|---|---|---|---|
| Family | Strain | Relationship | age | nucleotide substitution | SNV/CNP | per CNP | synonymous/non-coding | Mutation rate a |
| K-1 | K17 & K15 b | Mother and Child | N/A c | 1213 | 183/95 | 10.8 | 437/695/83 | N/A |
| K-2 | K35 | Mother | 46 | 182 | 154/12 | 2.3 | 96/62/24 | N/A |
| K36 | Child | 17 | 83 | 69/7 | 2.0 | 50/22/11 | 2.6 x 10–6 | |
| K37 | Child | 14 | 71 | 54/7 | 2.4 | 40/20/11 | 2.5 x 10–6 | |
| K-3 | K26 | Father | 43 | 291 | 214/27 | 2.9 | 186/64/41 | N/A |
| K27 | Mother | 28 | 87 | 74/6 | 2.2 | 46/24/17 | N/A | |
| K28 | Child | 6 | 103 | 80/8 | 2.9 | 49/34/20 | 5.2 x 10–6 | |
| K-4 | K29 | Father | 48 | 69 | 64/2 | 2.5 | 39/20/10 | N/A |
| K30 | Mother | 44 | 78 | 74/2 | 2.0 | 41/28/7 | N/A | |
| K32 | Child | 14 | 110 | 89/8 | 2.6 | 77/20/33 | 4.0 x 10–6 | |
| K33 | Child | 13 | 97 | 78/9 | 2.1 | 68/23/6 | 3.8 x 10–6 | |
| K-5 | K23 | Child | 17 | 50 | 37/6 | 2.2 | 19/13/18 | 1.3 x 10–6 |
| K24 | Child | 15 | 17 | 17/0 | N/A | 12/4/1 | 0.7 x 10–6 | |
| K25 | Child | 5 | 46 | 30/6 | 2.7 | 25/10/11 | 3.8 x 10–6 |
a Rate of SNV per site per year.
b K15 and K17 were counted together because we cannot assign strain specific substitutions by comparison of two strains.
c N/A, not applicable.
Strain-specific nucleotide substitutions were classified into nonsynonymous (changing amino acid), synonymous (retaining amino acid) and substitutions in noncoding regions. The mutation rate was calculated for isolates from children by dividing the number of SNVs (substitutions not included in CNPs) by their age in years, assuming H. pylori infection occurred soon after birth. The rate ranged from 0.7 x 10–6–5.2 x 10–6 per year per site (S4 Table). The value is comparable to those reported for earlier estimations for intra-family evolution [26, 31, 32].
Genes with an amino acid substitution
We investigated which genes had experienced an amino acid substitution using function-based gene grouping with clusters of orthologous groups (COG) [33] (Table 3, S5 Table). The number of genes with at least one nonsynonymous substitution was counted. We found enrichment of amino acid substitutions in COG categories of M (cell wall/membrane/envelope biogenesis) and T (signal transduction) genes in multiple strains. The category M genes included those for outer membrane proteins (OMPs), lipopolysaccharide biosynthesis, lipoprotein-related proteins, penicillin-binding proteins in the cell wall and cell division proteins (S6 Table). Category T genes included spoT (included also in category K) and chemotaxis-related genes (included also in category N) (S6 Table). The genes with nonsynonymous substitutions included those decayed in hspEastAsia, including molybdenum-related genes (mogA and modB), and those decayed in hpEurope, including ackA for acetate formation [4]. The substitution mutations might represent a step in their decay.
Table 3. COG enrichment of genes with strain specific non-synonymous substitutions.
| K-1 | K-2 | K-3 | K-4 | K-5 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COG | Function | F30 a | K15 & K17 b | K35 | K36 | K37 | K26 | K27 | K28 | K29 | K30 | K32 | K33 | K23 | K24 | K25 |
| C | Energy production and conversion | 75 | 8 | 2 | 1 | 3 | 2 | 1 | 2 | 1 | 1 | 3 | 0 | 0 | 0 | 2 |
| D | Cell cycle control and mitosis | 20 | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| E | Amino Acid metabolism and transport | 97 | 9 | 3 | 2 | 0 | 7 | 4 | 2 | 3 | 6 | 1 | 2 | 0 | 0 | 0 |
| F | Nucleotide metabolism and transport | 39 | 3 | 1 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 0 |
| G | Carbohydrate metabolism and transport | 43 | 3 | 2 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| H | Coenzyme metabolis | 77 | 5 | 2 | 2 | 1 | 5 | 1 | 0 | 3 | 1 | 0 | 0 | 0 | 1 | 2 |
| I | Lipid metabolism | 42 | 4 | 2 | 1 | 0 | 4 | 1 | 1 | 1 | 1 | 4 | 1 | 0 | 0 | 0 |
| J | Tranlsation | 122 | 7 | 6 | 1 | 2 | 7 | 3 | 2 | 2 | 0 | 6 | 5 | 1 | 0 | 0 |
| K | Transcription | 33 | 3 | 2 | 2 | 1 | 4 | 0 | 0 | 1 | 1 | 2 | 4 | 0 | 0 | 0 |
| L | Replication and repair | 88 | 9 | 7 | 1 | 2 | 4 | 4 | 3 | 1 | 2 | 6 | 2 | 1 | 1 | 0 |
| M | Cell wall/membrane/envelop biogenesis | 98 | 16 | 8 | 4 | 5 | 20* | 3 | 3 | 4 | 8* | 5 | 5 | 1 | 1 | 3 |
| N | Cell motility | 52 | 5 | 2 | 5 | 2 | 7 | 1 | 1 | 2 | 0 | 3 | 5 | 3 | 0 | 0 |
| O | Post-translational modification | 63 | 3 | 5 | 3 | 0 | 9 | 2 | 3 | 3 | 2 | 4 | 3 | 1 | 1 | 1 |
| P | Inorganic ion transport and metabolism | 62 | 10 | 4 | 1 | 2 | 6 | 2 | 1 | 0 | 4 | 1 | 3 | 1 | 1 | 0 |
| Q | Secondary Structure | 15 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| R | General Functional Prediction only | 131 | 17 | 10 | 3 | 3 | 7 | 4 | 3 | 4 | 2 | 5 | 2 | 1 | 0 | 1 |
| S | Function Unknown | 72 | 7 | 1 | 1 | 0 | 8 | 4 | 2 | 0 | 0 | 5 | 4 | 0 | 0 | 2 |
| T | Signal Transduction | 25 | 5 | 2 | 5* | 1 | 5 | 0 | 1 | 1 | 1 | 3 | 3 | 2 | 0 | 0 |
| U | Intracellular trafficking and secretion | 51 | 1 | 1 | 2 | 2 | 2 | 0 | 2 | 0 | 2 | 2 | 1 | 0 | 1 | 0 |
| Z | Cytoskeleton | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OMP | Outer membrane protein | 46 | 18* | 11* | 3 | 4 | 14* | 3 | 5* | 3 | 4 | 8* | 8* | 3 | 2 | 6* |
| N/A | 355 | 31 | 16 | 9 | 6 | 24 | 6 | 12 | 7 | 3 | 9 | 12 | 4 | 3 | 5 | |
| Sum | 1607 | 148 | 78 | 44 | 36 | 107 | 40 | 38 | 39 | 32 | 62 | 53 | 18 | 12 | 16 | |
a Strain used as reference strain for mapping.
b K15 and K17 were counted together because we cannot assign strain specific substitutions by comparison of two strains.
* Counts were tested by Fischer's exact test, P < 0.01
Genes not assigned to a COG category were also analyzed. Genes annotated as encoding OMPs [34] were the most frequently enriched with the amino acid changing mutations among the strains after transmission, as reported [27]. These included alpAB, hofABDEF, horD, hopADFGINQZ, homBD, babAB and other hypothetical OMPs (Table 4 and S6 Table).
Table 4. Select genes with an amino-acid substitution in H. pylori from children.
| Group | Comment | Gene |
|---|---|---|
| Virulence-related | cagPAI (cag pathogenicity island) | cagA, cagC, cagV, cagW, cagY, cag5 |
| vacA homologs | vacA, vacA paralog, vacA paralog | |
| Helicobacter cysteine-rich protein | hcpD, hcpX | |
| Proteases | htrA, collagenase | |
| Type IV secretion system | virB2, comH | |
| Lewis antigen mimicry | fucT | |
| Others | tnfα (Tumor Necrosis Factor alpha-inducing protein), | |
| ggt (gamma-glutamyltranspeptidase) | ||
| Outer membrane protein | Hop family | hopADFGINQZ |
| Hof family | hofABDEF | |
| Others | alpAB, homBD, babAB, horD | |
| Restriction-modification | Type I | Restriction gene |
| Type IIG | ||
| Type III | Restriction gene, Modification gene |
Various known virulence factors were identified among them (Table 4). The cagACTWY genes reside on the cag pathogenicity island [35]; vacA is known to cause vacuolation in host cells [36] and virB2 and comH are included in Type IV secretion systems, which are used for DNA import from the surrounding environment [37]. Cysteine-rich proteins [38] and HtrA protease [39] are involved in host interaction. The tnfα gene induces tumor necrosis factor alpha in the host [40] and gamma-glutamyltranspeptidase promotes pathogenesis [41–43]. The fucT gene is used for Lewis antigen mimicry and is important for immunity avoidance [44]. We also found amino acid substitutions in several restriction-modification genes (Table 4).
Common and different patterns of evolution in familial transmission
We focused on three families involving children to gain insight into the steps of H. pylori adaptation to a human child host (Table 5and S6 Table). We emphasize that each lineage has a unique pattern of amino acid changing mutations but they all showed a change in cag and other virulence-related genes.
Table 5. Genes with non-synonymous mutations and annotations in H. pylori from children.
| Family | Strain | Group/ COG category | Gene | Annotation | Locus tag of F30 ortholog |
|---|---|---|---|---|---|
| K-5 | K23 | Virulence-related | cagA | Cag pathogenicity island protein | HPF30_0779 |
| Virulence-related | cagC | Cag pathogenicity island protein | HPF30_0780 | ||
| Outer membrane protein | hopQ | Outer membrane protein | HPF30_0214 | ||
| Outer membrane protein | hopI | Outer membrane protein HopI | HPF30_0234 | ||
| Outer membrane protein | alpB | Outer membrane protein AlpB | HPF30_0426 | ||
| J | sfhB | Pseudouridine synthase | HPF30_0951 | ||
| M | ftsI | Cell division protein | HPF30_1449 | ||
| NT | Methyl-accepting chemotaxis protein | HPF30_0728 | |||
| NT | mcpB | Methyl-accepting chemotaxis protein | HPF30_1183 | ||
| NO | flgA | Flagellar basal body P-ring biosynthesis protein FlgA | HPF30_1345 | ||
| P | fecA_2 | Iron dicitrate transport protein | HPF30_0524 | ||
| R | rny | Ribonuclease Y | HPF30_0573 | ||
| hpaA | Flagellar sheath adhesin HpaA | HPF30_0534 | |||
| K24 | Virulence-related | comH | Periplasmic competence protein | HPF30_1399 | |
| Outer membrane protein | alpB | Outer membrane protein AlpB | HPF30_0426 | ||
| Outer membrane protein | hopN-2 | Outer membrane protein HopN2 | HPF30_1068 | ||
| V | hsdR_2 | Type I restriction enzyme R protein | HPF30_0485 | ||
| MU | hefA | Outer membrane protein HefA | HPF30_0721 | ||
| P | Catalase-related peroxidase | HPF30_0836 | |||
| H | hemA | Glutamyl-tRNA reductase | HPF30_1056 | ||
| F | purB | Adenylosuccinate lyase | HPF30_0276 | ||
| K25 | Outer membrane protein | babB | Outer membrane protein | HPF30_0154 | |
| Outer membrane protein | hopL | Outer membrane protein HopL | HPF30_0233 | ||
| Outer membrane protein | alpB | Outer membrane protein AlpB | HPF30_0426 | ||
| Outer membrane protein | alpA | Outer membrane protein AlpA | HPF30_0427 | ||
| Outer membrane protein | babA | Outer membrane protein BabA | HPF30_0441 | ||
| Outer membrane protein | hopF | Outer membrane protein HopF | HPF30_1043 | ||
| Virulence-related | cagC | Cag pathogenicity island protein | HPF30_0780 | ||
| O | pcm | Protein-beta-aspartate methyltransferase | HPF30_0297 | ||
| M | cfa | Cyclopropane fatty acid synthase | HPF30_0349 | ||
| C | atpB | F-ATPase subunit 6 | HPF30_0502 | ||
| H | mogA | Molybdenum cofactor biosynthesis protein | HPF30_0532 | ||
| M | Putative outer membrane protein | HPF30_0673 | |||
| C | ppa | Pyrophosphate phospho-hydrolase | HPF30_0706 | ||
| H | hemA | Glutamyl-tRNA reductase | HPF30_1056 | ||
| M | rfaJ-1 | Putative lipopolysaccharide biosynthesis protein | HPF30_1136 | ||
| R | engB | Probable GTP-binding protein EngB | HPF30_1459 | ||
| hpaA | Flagellar sheath adhesin HpaA | HPF30_0534 | |||
| K-2 | K36 | Outer membrane protein | hopQ | Outer membrane protein | HPF30_0214 |
| Outer membrane protein | alpA | Outer membrane protein AlpA | HPF30_0427 | ||
| Outer membrane protein | hopA | Outer membrane protein HopA | HPF30_1066 | ||
| Virulence-related | fucT | Alpha-(1,3)-fucosyltransferase | HPF30_0677 | ||
| Virulence-related | Tumor necrosis factor alpha-inducing protein | HPF30_0731 | |||
| Virulence-related | cagA | Cag pathogenicity island protein | HPF30_0779 | ||
| Restriction-modification | res-2 | Type III restriction enzyme R protein | HPF30_0033 | ||
| O | groS | Protein Cpn10 | HPF30_0009 | ||
| H | pnuC | Nicotinamide mononucleotide transporter | HPF30_0110 | ||
| K | cobB | Regulatory protein SIR2 homolog | HPF30_0138 | ||
| V | ABC-transporter, ATP-binding domain | HPF30_0187 | |||
| C | bisC-frg | Biotin sulfoxide reductase BisC | HPF30_0341 | ||
| M | capJ | Type 1 capsular polysaccharide biosynthesis protein J | HPF30_0354 | ||
| R | hypA | Probable hydrogenase nickel incorporation protein HypA | HPF30_0464 | ||
| O | tig | PPIase | HPF30_0536 | ||
| TK | spoT | Penta-phosphate guanosine-3'-pyrophosphohydrolase | HPF30_0555 | ||
| FE | prsA | Phosphoribosyl pyrophosphate synthase | HPF30_0592 | ||
| NT | Methyl-accepting chemotaxis protein | HPF30_0728 | |||
| H | bioF | 8-amino-7-oxononanoate synthase | HPF30_0729 | ||
| M | pldA | Phospholipase A1 | HPF30_0822 | ||
| N | fliG | Flagellar motor switch protein G | HPF30_0946 | ||
| I | dxr | 2-C-methyl-D-erythritol 4-phosphate synthase | HPF30_1079 | ||
| M | rfaJ-2 | Putative lipopolysaccharide biosynthesis protein | HPF30_1087 | ||
| M | rfaJ-1 | Putative lipopolysaccharide biosynthesis protein | HPF30_1136 | ||
| P | C(4)-dicarboxylates and tricarboxylates/succinate antiporter | HPF30_1152 | |||
| NT | tlpA | Methyl-accepting chemotaxis protein | HPF30_1179 | ||
| NT | mcpB | Methyl-accepting chemotaxis protein | HPF30_1183 | ||
| NT | Methyl-accepting chemotaxis transducer | HPF30_1218 | |||
| L | topA | DNA topoisomerase I | HPF30_1195 | ||
| U | comB1 | ComB8 competence protein | HPF30_1259 | ||
| U | comB6 | NADH-ubiquinone oxidoreductase subunit | HPF30_1260 | ||
| F | Putative endonuclease, split and separated by inversion, N-terminus part | HPF30_1272 | |||
| J | rpsB | 30S ribosomal protein S2 | HPF30_1429 | ||
| K37 | Outer membrane protein | hopQ | Outer membrane protein | HPF30_0214 | |
| Outer membrane protein | hopL | Outer membrane protein HopL | HPF30_0233 | ||
| Outer membrane protein | hofG | Putative outer membrane protein | HPF30_0425 | ||
| Outer membrane protein | alpB | Outer membrane protein AlpB | HPF30_0426 | ||
| Virulence-related | cagA | Cag pathogenicity island protein | HPF30_0779 | ||
| Restriction-modification | res-2 | Type III restriction enzyme R protein | HPF30_0033 | ||
| Restriction-modification | Type IIG restriction-modification enzyme | HPF30_0661 | |||
| F | pyrF | Orotidine 5'-phosphate decarboxylase | HPF30_0005 | ||
| M | tonB | Siderophore-mediated iron transport protein | HPF30_0059 | ||
| J | rpsK | 30S ribosomal protein S11 | HPF30_0105 | ||
| K | rpoB | DNA-directed RNA polymerase subunit beta/beta | HPF30_0196 | ||
| P | Carbonic anhydrase | HPF30_0205 | |||
| M | Putative lipopolysaccharide biosynthesis protein | HPF30_0284 | |||
| C | bisC-frg | Biotin sulfoxide reductase BisC | HPF30_0341 | ||
| C | ackA | Acetokinase | HPF30_0435 | ||
| P | Iron(III) dicitrate transport protein | HPF30_0645 | |||
| L | rnhA | Ribonuclease H | HPF30_0667 | ||
| V | hefC | Cytoplasmic pump protein of the hefABC efflux system HefC | HPF30_0719 | ||
| MU | hefA | Outer membrane protein HefA | HPF30_0721 | ||
| M | pbp-1a | Penicillin-binding protein 1A | HPF30_0730 | ||
| NU | flhA | Flagellar biosynthesis protein FlhA | HPF30_0891 | ||
| J | infB | Translation initiation factor IF-2 | HPF30_0898 | ||
| D | minC | Septum formation inhibitor | HPF30_0903 | ||
| L | ruvB | Holliday junction ATP-dependent DNA helicase RuvB | HPF30_0909 | ||
| R | Oligopeptide permease integral membrane protein | HPF30_1044 | |||
| NT | mcpB | Methyl-accepting chemotaxis protein | HPF30_1183 | ||
| C | Ferrodoxin-like protein | HPF30_1378 | |||
| K-3 | K28 | Outer membrane protein | hopQ | Outer membrane protein | HPF30_0214 |
| Outer membrane protein | alpB | Outer membrane protein AlpB | HPF30_0426 | ||
| Outer membrane protein | hofE | Outer membrane protein HofE | HPF30_0548 | ||
| Outer membrane protein | hofB | Outer membrane protein HofB | HPF30_0932 | ||
| Outer membrane protein | hopN-2 | Outer membrane protein HopN2 | HPF30_1068 | ||
| Virulence-related | cagV | Cag pathogenicity island protein | HPF30_0794 | ||
| Virulence-related | cagW | Cag pathogenicity island protein | HPF30_0795 | ||
| Virulence-related | cagY | Cag pathogenicity island protein | HPF30_0797 | ||
| Virulence-related | cag5 | Cag pathogenicity island protein | HPF30_0800 | ||
| Virulence-related | vacA | Vacuolating cytotoxin A | HPF30_0448 | ||
| Restriction-modification | mod-4 | Putative type III restriction enzyme M protein | HPF30_1273 | ||
| Restriction-modification | Type I restriction enzyme R protein | HPF30_0858 | |||
| Restriction-modification | hsdR_3 | Type I restriction enzyme R protein | HPF30_1407 | ||
| L | dnaG | DNA primase | HPF30_0010 | ||
| R | Lipid A phosphoethanolamine transferase | HPF30_0020 | |||
| O | clpX | ATP-dependent Clp protease ATP-binding subunit ClpX | HPF30_0030 | ||
| I | Isoprenyl transferase | HPF30_0175 | |||
| J | valS | Valyl-tRNA synthetase | HPF30_0237 | ||
| E | nifS_2 | NifS-like protein | HPF30_0338 | ||
| S | truD | tRNA-uridine isomerase D | HPF30_0415 | ||
| C | ackA | Acetokinase | HPF30_0435 | ||
| S | Uncharacterized protein | HPF30_0472 | |||
| M | waaE | Bifunctional protein HldE | HPF30_0475 | ||
| M | amiA | N-acetylmuramoyl-L-alanine amidase | HPF30_0558 | ||
| R | rny | Ribonuclease Y | HPF30_0573 | ||
| L | uvrA | Excinuclease ABC subunit A | HPF30_0629 | ||
| M | Putative outer membrane protein | HPF30_0673 | |||
| P | kefB | Glutathione-regulated potassium-efflux system protein | HPF30_0851 | ||
| O | htrA | Protease DO | HPF30_0869 | ||
| J | infB | Translation initiation factor IF-2 | HPF30_0898 | ||
| NT | tlpA | Methyl-accepting chemotaxis protein | HPF30_1179 | ||
| O | clpA | ATP-dependent C1p protease | HPF30_1264 | ||
| Response regulator | HPF30_0871 |
(i) Family K-2: K36 and K37
K36 had specific nonsynonymous nucleotide substitutions in five motility genes, five signal transduction genes, cagA, tnfα, hcpD, fucT, rfaJ-1/2, and one Type III R (restriction) gene. K37 had these types of mutations in cagA, hcpD, the RNA polymerase subunit gene (rpoB) and two restriction-modification genes (Type IIG and Type III R) as well as four OMP genes.
(ii) Family K-3: K28
Among the 49 genes with at least one nonsynonymous nucleotide substitution specific to the child strain, there are four cag genes (cagV, cagW, cagY and cag5), vacA, htrA, five OMP genes, three restriction-modification genes (two Type I R and one Type III M) and clpX/clpP protease genes (Table 5).
(iii) Family K-5: K23, K24 and K25
Among the three strains isolated from the three children in family K-5, K24 had the fewest (n = 12) genes with an amino acid change, among which are a virulence-related gene (comH), a restriction-modification gene (Type I R), purB for nucleotide metabolism, a peroxidase gene and a heme biosynthesis gene. K23 had nonsynonymous substitutions in cagAC genes, four genes related to motility and chemotaxis and two genes related to signal transduction. K25 had nonsynonymous mutations in cagC, three genes (ppa, rfaJ-1 and cfa) related to lipids, atpB for membrane ATP synthase and pcm for protein repair.
Discussion
H. pylori is known for sequence diversity between different strains, but strains from the same lineage can be difficult to distinguish by the standard MLST analysis using only seven genes. We undertook whole genome sequencing and distinguished the strain-specific nucleotide substitutions for isolates with the same or very similar MLST sequence type from the same family. On the basis of sequence difference, we revealed the likely pathway of evolution between these strains in some cases. Furthermore, after analyzing the nonsynonymous mutations, we suggested the strategy of H. pylori evolution during infection.
Following the construction of phylogenetic trees based on SNPs in the whole genome sequences. We inferred the direction of transfer between family members and other details (Fig 1). However, a recent study revealed a broad diversity in genome sequences in strains isolated from one specimen from one person [45]. The diversity represented in the above phylogenetic trees might well be accounted for by the diversity of lineages within an individual. In Fig 1B(ii), for example, the mother might have transferred one lineage (the ancestor of K36) to one child and transferred another, well diversified strain (the ancestor of K37) to the other child. In order to clarify transmission pathways accurately, we need genome sequences of multiple strains from one host.
The mutation rate was calculated on the basis of strain-specific SNVs. Most of the value was included with the mutation rate range in earlier calculations from whole genome information [26, 31, 32] and only the K24 strain was a little below the range (S4 Table). This difference might reflect the origin of H. pylori strains. Earlier work using whole genome sequences was with strains from South Africa [26], whereas strains isolated in Japan were used in the present study. Another possibility is recombination between lineages [45]. By contrast, work using 78 genes for analysis gave a much lower value for the mutation rate, using strains derived from USA, UK, Colombia, the Netherlands and South Korea [32]. This result might be related to the large difference in sequence diversity among genes: indeed, different genes can evolve at very different rates [4, 5]. More analysis of whole genomes and individual genes in strains from various regions is required to fully understand the apparent variation of mutation rate.
Genes with an amino acid change might provide insight into the adaptation process. Many of these genes are related to the surface structure of the H. pylori cell, including OMP genes, lipoprotein-related genes and fucT, as found in other work on intrafamilial transmission and intrahost transmission [27, 31]. H. pylori can attach to human gastric epithelial cells through various kinds of adhesion factors, including BabA (HopS), BabB, SabA (HopP), SabB, AlpA and AlpB. Protein BabA is one of the major adhesion molecules associated with severe pathogenesis in H. pylori infection, although babA expression was reported to disappear by six months after infection of Mongolian gerbils with nucleotide changes introducing a stop codon of the gene [46]. AlpA and AlpB were shown to contribute to laminin binding of H. pylori and to induction of inflammatory changes of gastric mucosa [47]. HopQ might be important in initial colonization and long-term persistence of H. pylori in the stomach by modulating the adherence to gastric epithelial cells [48]. Two different alleles of hopQ were shown to be associated significantly with the positivity of other virulence genes, including cagA and vacA [49, 50].
An unexpected finding was the occurrence of amino acid substitutions in many virulence genes other than OMP genes and other surface-related genes. The genes of vacA and cagA are well known as important virulence-related genes of H. pylori. Mutations in these virulence genes were detected in H. pylori isolates of one or two family member(s) in each family but not, in general, in the isolates of all members of a family. One interesting possibility responsible for this observation is that these changes in virulence factors are related to adaptation to children in intrafamilial transmission. A related finding is that CagA is the most reactive antigen recognized by H. pylori-positive sera from children [51].
Amino acid changes in restriction-modification systems were detected in the three families involving children (see the last section in Results). A restriction-modification system consists of DNA methyltransferase, a modification enzyme, and a restriction endonuclease. DNA methyltransferase transfers a methyl group to a specific DNA sequence in the genome, which likely affects global gene expression among others. The restriction enzyme destroys DNA lacking such specific methylation resulting in genetic isolation. Recent work demonstrated the restriction-modification systems in H. pylori frequently change their presence/absence, sequence specificity and expression to remodel the methylome [7, 10]. The mutations mentioned above could be related to adaptation to a new host through such epigenetics-driven adaptive evolution [52]. Substitution of target recognition domains of restriction-modification systems underlying drastic changes in recognition sequence [11, 12], however, cannot be detected, in principle, by the present method based on mapping a genome sequence and SNP analysis.
Mutations were found in earlier work comparing whole genome sequences of closely related strains, especially in OMP genes, which is consistent with the results presented here [27, 28, 31, 53]. Comparison of whole genome sequences of H. pylori isolated from grandfather, son and grandson of a family in England found amino acid changes in OMP genes [28]. Substitution mutations in OMP genes were found in inter-spouse transmission in Australia [27]. A mutation burst was found during the acute phase of H. pylori infection leading to mutational changes in OMPs and cag-related genes in humans and primates [53]. These results are consistent with the present study and we additionally found mutations commonly observed in isolates from children in two categories; i.e. virulence factors other than cag-related and restriction-modification enzymes. This difference might be caused because of isolation in young children, compared to isolation from adults in other studies, but we cannot exclude the possibility it is caused by differences in environment.
We assumed that the individual human hosts have driven the bacterial mutations in the above genes. Our procedures involved culturing the bacteria ex vivo as in almost all the works on bacterial variations. Have the ex vivo steps, as opposed to in vivo steps, induced or selected the mutations we observed here? We think that most of the strain-specific nonsynonymous substitutions were generated in vivo for the following reasons. First, many of the genes with those mutations also show rapid sequence changes in phylogeny giving long branches in their phylogenetic tree [4, 5]. A simplest interpretation is the host adaptation through a nonsynonymous mutation is repeated for many generations to result in the rapid evolutionary rate. Second, comparable mutation rates per year were obtained in this and other studies based on strain culture ex vivo (Table 2). This indicates that the number of mutations is approximately proportional to the years within a human body. This cannot be expected only through mutagenesis and selection in vivo. Third, the strain-specific nonsynonymous mutations are unique to each of the strains. For example, K23, but no other strains, carries such mutations in multiple cag genes and several motility/chemotaxis-related genes among 14 genes with annotation (Table 5). Many strains carry those strain-specific nonsynonymous mutations in OMP genes, but the OMP repertoire is quite diverse among the strains. The difference is not likely a result of mutagenesis and/or selection during ex vivo culture, during which we used a medium of the same recipe, especially, the same batch of horse serum (see Materials and methods). From these considerations, it is, at present, natural to interpret that most of these strain-specific non-synonymous mutations were introduced during the long-term (years of) growth in individual human stomachs although we cannot exclude some contribution of ex vivo growth.
In conclusion, our whole genome decoding of H. pylori strains from family members including children suggested adaptation of these bacteria to a new human host through mutations in virulence-related genes and restriction-modification genes in addition to OMP genes.
Materials and Methods
Ethics statement
This study was undertaken with approval from the Ethics Committees of Kyorin University, Tokyo (No. 537) and Sapporo Kosei General Hospital (H24-104). Written informed consent was obtained from the patients (5 years old or older) and also from their parents when the patients are minor.
Strains
In all, 19 H. pylori strains were obtained from five families during April 2011—December 2012 in Sapporo Kosei General Hospital, Sapporo, Hokkaido, Japan. A single colony was isolated and subcultured on Brucella medium supplemented with 1.5% (w/v) agar and 7% (v/v) horse serum (BHS medium) under microaerobic conditions. The same batch of horse serum was used for the culture to minimize possible variation between cultures. Typing of strains was done initially by seven-gene MLST for all five families [29].
Genome sequencing and mapping
After incubation for 48 h under microaerobic conditions at 37°C on BHS medium, the culture of H. pylori (about 5×108 colony-forming units) was collected. A Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) was used according to the manufacturer’s instructions to isolate genomic DNA. A DNA library for genome sequencing was constructed by Nextera XT (Illumina, CA, USA) and sequenced by HiSeq2500 (Illumina, CA, USA).
About 1.4×106 reads (~×200 coverage) with a length of 100 bp in the form of paired ends were selected from each read data (DRA accession no. 002504) and mapped against the genome sequence of H. pylori strain F30 (accession no. NC_017365) by BWA [54]. Nucleotide substitutions were detected by SAMtools software [55] without misalignment filtering to avoid pseudo-negative detection. Lists of nucleotide substitutions were compared by customized Perl scripts for calculation of the distance between strain pairs and for detection of strain-specific nucleotide substitutions. Nucleotide sites with coverage of more than five reads for all the members of a family with the same or very similar MLST sequence type were used for the detection of nucleotide substitution. Classification of nucleotide substitution to nonsynonymous, synonymous or substitutions at noncoding regions were done according to the gene annotation of H. pylori F30 [4]. For the calculation of strain-specific substitutions, the substitutions in strains K17 and K15 in family K-1 were counted together because the family has only two strains with the same MLST sequence type and it is not possible to assign substitutions to either of the strains.
The significance of differences between distances among strains with the same or very similar sequence type was analyzed by generating a matrix assuming the same probability of nucleotide substitution accumulation for all strain pairs. Matrices were constructed 1×106 times and the rank list of standard deviation was compared with the standard deviation of distances in the real data for calculation of the P value.
COG enrichment
COG of genes in the H. pylori F30 genome was annotated by rpsblast [56]. Genes with strain-specific nonsynonymous substitutions were counted and the significance of COG enrichment was tested by Fisher’s exact test. A gene was counted only once even if it had more than two strain-specific nonsynonymous substitutions.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Koji Yahara for discussions.
Data Availability
All the sequence data is available from the DRA database (accession number DRA002504).
Funding Statement
This work was supported, in part, by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (grant nos. 21370001 and 25291080 to I.K., 24790412 to Y.F., 23590518 to S.K. and 24593166 to H.Ys.); and from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (grant nos. 24113506 to I.K. and 24119503 to Y.F.); Health and Labor Sciences Research Grants for Clinical Cancer Research from the Ministry of Health, Labor and Welfare, Japan, grants for Scientific Research (grant no. 20132033 to S.K.) from JSPS; the global COE project of Genome Information Big Bang from MEXT to I.K.; Grant in Promotion of Basic Research Activities for Innovative Biosciences (grant no. 121205003001002100019) from Bio-oriented Technology Research Advance Institution to I.K.; Cooperative Research Grant of the Genome Research for BioResource, NODAI Genome Research Center, Tokyo University of Agriculture; and from the Takeda Science Foundation to Y.F.
References
- 1. Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347(15):1175–86. [DOI] [PubMed] [Google Scholar]
- 2. Bjorkholm B, Sjolund M, Falk PG, Berg OG, Engstrand L, Andersson DI. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori . Proc Natl Acad Sci U S A. 2001;98(25):14607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Falush D, Kraft C, Taylor NS, Correa P, Fox JG, Achtman M, et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci U S A. 2001;98(26):15056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawai M, Furuta Y, Yahara K, Tsuru T, Oshima K, Handa N, et al. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 2011;11:104 10.1186/1471-2180-11-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yahara K, Kawai M, Furuta Y, Takahashi N, Handa N, Tsuru T, et al. Genome-wide survey of mutual homologous recombination in a highly sexual bacterial species. Genome Biol Evol. 2012;4(5):628–40. 10.1093/gbe/evs043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori . Nature. 1999;397(6715):176–80. [DOI] [PubMed] [Google Scholar]
- 7. Furuta Y, Namba-Fukuyo H, Shibata TF, Nishiyama T, Shigenobu S, Suzuki Y, et al. Methylome diversification through changes in DNA methyltransferase sequence specificity. PLoS Genet. 2014;10(4):e1004272 10.1371/journal.pgen.1004272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krebes J, Morgan RD, Bunk B, Sproer C, Luong K, Parusel R, et al. The complex methylome of the human gastric pathogen Helicobacter pylori . Nucleic Acids Res. 2014;42(4):2415–32. 10.1093/nar/gkt1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vale FF, Megraud F, Vitor JM. Geographic distribution of methyltransferases of Helicobacter pylori: evidence of human host population isolation and migration. BMC Microbiol. 2009;9:193 10.1186/1471-2180-9-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Srikhanta YN, Gorrell RJ, Steen JA, Gawthorne JA, Kwok T, Grimmond SM, et al. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori . PLoS One. 2011;6(12):e27569 10.1371/journal.pone.0027569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Furuta Y, Kawai M, Uchiyama I, Kobayashi I. Domain movement within a gene: A novel evolutionary mechanism for protein diversification. PLoS One. 2011;6(4):e18819 10.1371/journal.pone.0018819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Furuta Y, Kobayashi I. Movement of DNA sequence recognition domains between non-orthologous proteins. Nucleic Acids Res. 2012;40(18):9218–32. 10.1093/nar/gks681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, Boitano M, et al. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40(4):e29 10.1093/nar/gkr1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nature methods. 2010;7(6):461–5. 10.1038/nmeth.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinto AC, Melo-Barbosa HP, Miyoshi A, Silva A, Azevedo V. Application of RNA-seq to reveal the transcript profile in bacteria. Genet Mol Res. 2011;10(3):1707–18. [DOI] [PubMed] [Google Scholar]
- 16. Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol. 2012;30(12):1232–9. 10.1038/nbt.2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vitoriano I, Vítor JMB, Oleastro M, Roxo-Rosa M, Vale FF. Proteome variability among Helicobacter pylori isolates clustered according to genomic methylation. J Appl Microbiol. 2013;114(6):1817–32. 10.1111/jam.12187 [DOI] [PubMed] [Google Scholar]
- 18. Srikhanta YN, Dowideit SJ, Edwards JL, Falsetta ML, Wu HJ, Harrison OB, et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria . PLoS Pathog. 2009;5(4):e1000400 10.1371/journal.ppat.1000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci U S A. 2005;102(15):5547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, et al. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun. 2014;5:5055 10.1038/ncomms6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Donahue JP, Israel DA, Torres VJ, Necheva AS, Miller GG. Inactivation of a Helicobacter pylori DNA methyltransferase alters dnaK operon expression following host-cell adherence. FEMS Microbiol Lett. 2002;208(2):295–301. [DOI] [PubMed] [Google Scholar]
- 22. Kumar R, Mukhopadhyay AK, Ghosh P, Rao DN. Comparative transcriptomics of H. pylori strains AM5, SS1 and their hpyAVIBM deletion mutants: possible roles of cytosine methylation. PLoS One. 2012;7(8):e42303 10.1371/journal.pone.0042303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takeuchi H, Israel DA, Miller GG, Donahue JP, Krishna U, Gaus K, et al. Characterization of expression of a functionally conserved Helicobacter pylori methyltransferase-encoding gene within inflamed mucosa and during in vitro growth. J Infect Dis. 2002;186(8):1186–9. [DOI] [PubMed] [Google Scholar]
- 24. Burucoa C, Lhomme V, Fauchere JL. Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: experimental results and meta-analysis. J Clin Microbiol. 1999;37(12):4071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parkhill J, Wren BW. Bacterial epidemiology and biology—lessons from genome sequencing. Genome Biol. 2011;12(10):230 10.1186/gb-2011-12-10-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Didelot X, Nell S, Yang I, Woltemate S, van der Merwe S, Suerbaum S. Genomic evolution and transmission of Helicobacter pylori in two South African families. Proc Natl Acad Sci U S A. 2013;110(34):13880–5. 10.1073/pnas.1304681110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Linz B, Windsor HM, Gajewski JP, Hake CM, Drautz DI, Schuster SC, et al. Helicobacter pylori genomic microevolution during naturally occurring transmission between adults. PLoS One. 2013;8(12):e82187 10.1371/journal.pone.0082187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krebes J, Didelot X, Kennemann L, Suerbaum S. Bidirectional genomic exchange between Helicobacter pylori strains from a family in Coventry, United Kingdom. Int J Med Microbiol. 2014;304(8):1135–46. 10.1016/j.ijmm.2014.08.007 [DOI] [PubMed] [Google Scholar]
- 29. Osaki T, Konno M, Yonezawa H, Hojo F, Zaman C, Takahashi M, et al. Analysis of intra-familial transmission of Helicobacter pylori in Japanese families. J Med Microbiol. 2014;64:67–73. 10.1099/jmm.0.080507-0 [DOI] [PubMed] [Google Scholar]
- 30. Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–5. [DOI] [PubMed] [Google Scholar]
- 31. Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, et al. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci U S A. 2011;108(12):5033–8. 10.1073/pnas.1018444108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morelli G, Didelot X, Kusecek B, Schwarz S, Bahlawane C, Falush D, et al. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 2010;6(7):e1001036 10.1371/journal.pgen.1001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278(5338):631–7. [DOI] [PubMed] [Google Scholar]
- 34. Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68(7):4155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42(5):1337–48. [DOI] [PubMed] [Google Scholar]
- 36. Atherton JC, Peek RM Jr., Tham KT, Cover TL, Blaser MJ. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori . Gastroenterology. 1997;112(1):92–9. [DOI] [PubMed] [Google Scholar]
- 37. Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41(2):379–91. [DOI] [PubMed] [Google Scholar]
- 38. Roschitzki B, Schauer S, Mittl PR. Recognition of host proteins by Helicobacter cysteine-rich protein C. Curr Microbiol. 2011;63(3):239–49. 10.1007/s00284-011-9969-2 [DOI] [PubMed] [Google Scholar]
- 39. Hoy B, Lower M, Weydig C, Carra G, Tegtmeyer N, Geppert T, et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO reports. 2010;11(10):798–804. 10.1038/embor.2010.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suganuma M, Watanabe T, Yamaguchi K, Takahashi A, Fujiki H. Human gastric cancer development with TNF-alpha-inducing protein secreted from Helicobacter pylori . Cancer Lett. 2012;322(2):133–8. 10.1016/j.canlet.2012.03.027 [DOI] [PubMed] [Google Scholar]
- 41. Rimbara E, Mori S, Kim H, Shibayama K. Role of gamma-glutamyltranspeptidase in the pathogenesis of Helicobacter pylori infection. Microbiol Immunol. 2013;57(10):665–73. 10.1111/1348-0421.12089 [DOI] [PubMed] [Google Scholar]
- 42. Gong M, Ling SS, Lui SY, Yeoh KG, Ho B. Helicobacter pylori gamma-glutamyl transpeptidase is a pathogenic factor in the development of peptic ulcer disease. Gastroenterology. 2010;139(2):564–73. 10.1053/j.gastro.2010.03.050 [DOI] [PubMed] [Google Scholar]
- 43. Oertli M, Noben M, Engler DB, Semper RP, Reuter S, Maxeiner J, et al. Helicobacter pylori gamma-glutamyl transpeptidase and vacuolating cytotoxin promote gastric persistence and immune tolerance. Proc Natl Acad Sci U S A. 2013;110(8):3047–52. 10.1073/pnas.1211248110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang G, Ge Z, Rasko DA, Taylor DE. Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Mol Microbiol. 2000;36(6):1187–96. [DOI] [PubMed] [Google Scholar]
- 45. Cao Q, Didelot X, Wu Z, Li Z, He L, Li Y, et al. Progressive genomic convergence of two Helicobacter pylori strains during mixed infection of a patient with chronic gastritis. Gut. 2014;64(4):554–61. 10.1136/gutjnl-2014-307345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohno T, Vallstrom A, Rugge M, Ota H, Graham DY, Arnqvist A, et al. Effects of blood group antigen-binding adhesin expression during Helicobacter pylori infection of Mongolian gerbils. J Infect Dis. 2011;203(5):726–35. 10.1093/infdis/jiq090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senkovich OA, Yin J, Ekshyyan V, Conant C, Traylor J, Adegboyega P, et al. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infect Immun. 2011;79(8):3106–16. 10.1128/IAI.01275-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Loh JT, Torres VJ, Algood HM, McClain MS, Cover TL. Helicobacter pylori HopQ outer membrane protein attenuates bacterial adherence to gastric epithelial cells. FEMS Microbiol Lett. 2008;289(1):53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sicinschi LA, Correa P, Bravo LE, Peek RM Jr., Wilson KT, Loh JT, et al. Non-invasive genotyping of Helicobacter pylori cagA, vacA, and hopQ from asymptomatic children. Helicobacter. 2012;17(2):96–106. 10.1111/j.1523-5378.2011.00919.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cao P, Cover TL. Two different families of hopQ alleles in Helicobacter pylori . J Clin Microbiol. 2002;40(12):4504–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Akada J, Okuda M, Hiramoto N, Kitagawa T, Zhang X, Kamei S, et al. Proteomic characterization of Helicobacter pylori CagA antigen recognized by child serum antibodies and its epitope mapping by peptide array. PLoS One. 2014;9(8):e104611 10.1371/journal.pone.0104611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Furuta Y, Kobayashi I. Mobility of DNA sequence recognition domains in DNA methyltransferases suggests epigenetics-driven adaptive evolution. Mobile genetic elements. 2012;2(6):292–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Linz B, Windsor HM, McGraw JJ, Hansen LM, Gajewski JP, Tomsho LP, et al. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat Commun. 2014;5:4165 10.1038/ncomms5165 [DOI] [PubMed] [Google Scholar]
- 54. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37(Database issue):D205–10. 10.1093/nar/gkn845 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All the sequence data is available from the DRA database (accession number DRA002504).