Abstract
Objective The ShARe/CLEF eHealth 2013 Evaluation Lab Task 1 was organized to evaluate the state of the art on the clinical text in (i) disorder mention identification/recognition based on Unified Medical Language System (UMLS) definition (Task 1a) and (ii) disorder mention normalization to an ontology (Task 1b). Such a community evaluation has not been previously executed. Task 1a included a total of 22 system submissions, and Task 1b included 17. Most of the systems employed a combination of rules and machine learners.
Materials and methods We used a subset of the Shared Annotated Resources (ShARe) corpus of annotated clinical text—199 clinical notes for training and 99 for testing (roughly 180 K words in total). We provided the community with the annotated gold standard training documents to build systems to identify and normalize disorder mentions. The systems were tested on a held-out gold standard test set to measure their performance.
Results For Task 1a, the best-performing system achieved an F1 score of 0.75 (0.80 precision; 0.71 recall). For Task 1b, another system performed best with an accuracy of 0.59.
Discussion Most of the participating systems used a hybrid approach by supplementing machine-learning algorithms with features generated by rules and gazetteers created from the training data and from external resources.
Conclusions The task of disorder normalization is more challenging than that of identification. The ShARe corpus is available to the community as a reference standard for future studies.
Keywords: Natural Language Processing, Disorder Identifciation, Named Entity Recognition, Information Extraction, Word Sense Disambiguation, Clinical Notes
BACKGROUND AND SIGNIFICANCE
Introduction
The clinical narrative within the electronic medical records (EMRs) forming a patient's medical history encapsulates vast amounts of knowledge. Unlocking this information can benefit clinical investigators, caregivers, and patients. The natural language processing (NLP) community has made advances in the past couple of decades in helping represent clinical knowledge.1 The fact that language in clinical reports is usually terse and compressed2,3 complicates its interpretation. Supervised machine-learning techniques are becoming popular, and various corpora annotated with syntactic and semantic information are emerging through various projects such as the Multi-source Integrated Platform for Answering Clinical Questions (MiPACQ),4 Temporal Histories of Your Medical Events (THYME),5–7 Informatics for Integrating Biology and the Bedside (i2b2),8,9 Shared Annotated Resources (ShARe),10 and Strategic Health IT Advanced Research Project: Area 4 (SHARPn).11,12 These resources help advance the development of novel NLP methods, which in turn enable improved tools for analyzing clinical narratives.13 Some examples of applications of these NLP methods are the phenotyping algorithms in Electronic Medical Records and Genomics (eMERGE),14–17 Pharmacogenomics Research Network (PGRN),18–20 and i2b29 projects, all of which make use of information extracted from the clinical narrative.
The tradition of shared tasks in the general NLP domain, such as Computational Natural Language Learning (CoNLL)21 and Semantic Evaluations (SemEval),22 has spread to the biomedical literature domain through BioNLP23 shared tasks and BioCreAtIvE,24 and more recently in the clinical domain through the i2b2 shared tasks.25 Chapman et al.13 highlight the importance of these activities for the clinical language processing community. The ShARe/CLEF 2013 eHealth shared task continued this tradition to accelerate research in this fast-growing and important domain. The focus of this task was disorder mention identification and normalization. A disorder is defined as a span of text that can be mapped to a concept in the Systematized Nomenclature of Medicine—Clinical Terms26 (SNOMED CT) terminology and that belongs to the Disorder semantic group as defined by Bodenreider and McCray.27 The process of normalization involves mapping these disorder mentions to the closest equivalent Unified Medical Language System28 (UMLS) Concept Unique Identifier (CUI) subset of SNOMED CT. This task differs from the other clinical NLP challenges—the i2b2 NLP challenges25—because (i) it uses an ontological definition of disorder (versus a looser and more subjective definition of problem in i2b2) and (ii) it normalizes the mention to an ontology, in this case, SNOMED CT as represented in the UMLS; the normalization task has not been explored previously in clinical NLP challenges.
This journal extension of the ShARe/CLEF eHealth workshop29 describes the gold-standard datasets, presents the task parameters, and provides a detailed analysis of the participating systems in terms of their methodology and performance.
Related work
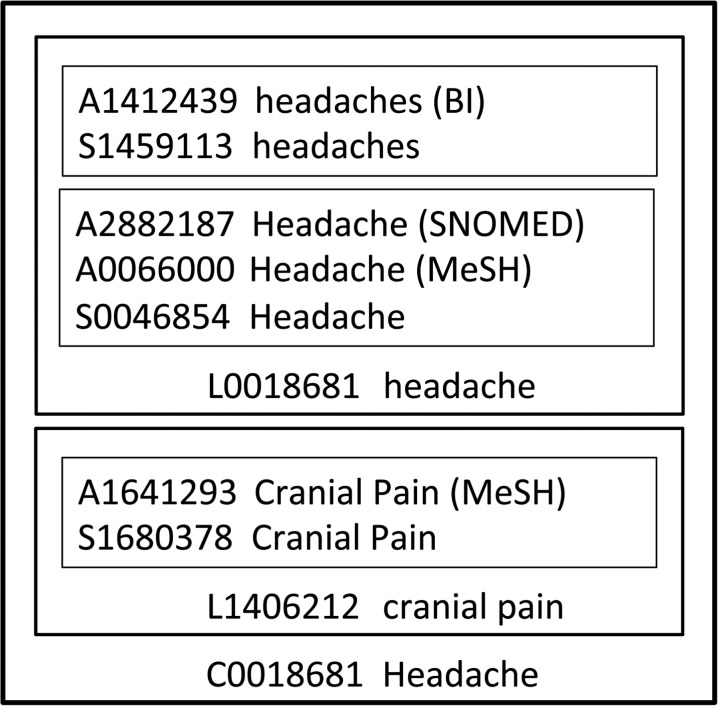
Over the years, the biomedical informatics community has developed rich terminologies such as Medical Subject Headings30 (MeSH) and RxNorm.31 The UMLS28 is an attempt to align existing terminologies with each other. The SNOMED CT26 terminology is an internationally recognized convention for clinical documentation and is part of the UMLS. UMLS defines the terms ‘atom,’ ‘term,’ and ‘concept’ as follows. An atom is defined as the smallest unit of naming in a source, viz, a specific string. A concept represents a single meaning and contains all atoms from any source that express that meaning in any way. A term is a word or collection of words comprising an expression. Thus, ‘Headache,’ and ‘headaches’ would be two strings, each with a unique String Unique Identifier (SUI) and a Atom Unique Identifier (AUI) depending on which terminology they occur in. These would be part of the same term, with a unique Lexical (term) Unique Identifier (LUI), whereas the string ‘Hue’ would be a separate term in the same concept represented with a CUI. Figure1 is an example from the UMLS website which clarifies the four levels of specification—CUI, LUI, SUI, and AUI. Their identifiers start with the letters C, L, S, and A, respectively, followed by seven numbers.
Figure 1:
Unique Identifiers in the Unified Medical Language System (UMLS) Metathesaurus.
Normalizing phrases to standardized terminology is not a new task. However, until recently, corpora that would allow gauging of the state of the art of normalization on clinical narrative have not been publicly available, despite its importance and downstream applications. For example, in ‘Patient diagnosed with RA,’ ‘Patient diagnosed with rheumatoid arthritis,’ and ‘Patient diagnosed with atrophic arthritis,’ the terms RA, rheumatoid arthritis and atrophic arthritis are text spans of type Disease/Disorder (the entity recognition/identification step); all of them are mapped to the same CUI (C0003873) (the normalization step). Therefore, processing clinical text to normalize it against a standard terminology provides a unified representation over the many textual forms of the same concept. This unified representation, in turn, can be the lingua franca across institutions, which can be shared in a similar fashion as in the eMERGE14–17 and PGRN18–20 phenotyping efforts. However, ours is the first formal evaluation of the state-of-the-art systems for entity recognition and normalization of the clinical narrative. This evaluation is the main goal of the presented shared task and the focus of this manuscript.
Existing systems
Much of the research in biomedical informatics has centered on named entity recognition and normalization tasks. Although most methods are rule based, systems have emerged that implement hybrid approaches combining machine learning and rules. The following are some of these systems:
MedLEE32—The Medical Language Extraction and Encoding System (MedLEE) is a rule-based tool for processing clinical text. It was originally designed to work with radiology reports of the chest and has since been extended to handle other types of clinical narratives.
MetaMap33,34—MetaMap is developed by the National Library of Medicine (NLM) to map scholarly biomedical text to the UMLS Metathesaurus. It is highly configurable and uses a knowledge-intensive approach.
cTAKES35—clinical Text Analysis and Knowledge Extraction System (cTAKES) is an open source Apache top-level project for information extraction from clinical narratives. It is a comprehensive platform for performing many clinical information extraction tasks in addition to mapping text to UMLS concepts (eg, syntactic and semantic parsing).
YTEX36—This system is a series of extension modules on top of cTAKES that provides a generalizable framework for mapping clinical phrases to various terminologies, including UMLS and its terminologies.
DNorm37—This system uses a machine-learning approach to compute similarities between mentions and concept names.
Related corpora
A few projects have focused on annotation of disease mentions in biomedical text:
The National Center for Biotechnology Information (NCBI) disease corpus38—comprises about 6900 disease mentions from 793 PubMed abstracts.
Arizona Disease Corpus39 (AZDC)—contains 2784 sentences from the MEDLINE abstracts annotated with disease names and mapped to UMLS CUIs. It includes about 3228 total disease mentions, with 1202 unique diseases, mapped to 686 concepts. The corpus comprises about 80 K tokens.
i2b2/VA corpus8—comprises discharge summaries from various hospitals annotated with, among other things, medical problems as concepts. It includes a total of 394 training reports, 477 test reports, and 877 de-identified, unlabeled reports. This corpus does not provide a mapping of mentions to any standardized terminology/ontology.
Corpus for Disease Names and Adverse Effects40 (DNAE)—includes 400 MEDLINE abstracts generated using the ‘Disease OR Adverse Effect’ query and annotated for disease mentions mapped to UMLS CUIs, totaling 1428 disease and 813 adverse effect annotations.
Multi-source Integrated Platform for Answering Clinical Questions4 (MiPACQ) corpus—comprises about 130 K words of clinical narrative from Mayo Clinic, and is annotated with various layers of syntactic and semantic information as well as UMLS core semantic types such as Disorder, Sign or Symptoms, Procedures, Medications, and Labs.
MATERIALS AND METHODS
In this section, we describe the dataset and the gold standard, which are the bases for the evaluation conducted within the CLEF/ShARe 2013 shared task.
Data
The ShARe corpus comprises annotations over de-identified clinical reports from a US intensive care EMR repository (V 2.5 of the Multiparameter Intelligent Monitoring in Intensive Care (MIMIC) II database).41 The corpus used for the CLEF/ShARe 2013 shared task consists of 298 discharge summaries, electrocardiograms, echocardiograms, and radiology reports, covering a total of about 180 K words. All shared-task participants were required to register, accept a Data Use Agreement (DUA), and obtain a US human subjects training certificate.
Gold standard
Two professional coders (a healthcare professional who has been trained to analyze clinical records and assign standard codes using a classification system) trained for this task annotated each clinical note in a double-blind manner, followed by adjudication. The quality of the annotations was assessed through inter-annotator agreement (IAA). Details of the IAA are presented later in table 2.
Table 2:
Inter-annotator (A1 and A2) and gold standard (GS) agreement as F1 score for the disorder mentions and their normalization to the Unified Medical Language System Concept Unique Identifier (UMLS CUI)
| Disorder | CUI | |||
|---|---|---|---|---|
| Relaxed | Strict | Relaxed | Strict | |
| F1 | F1 | Accuracy | Accuracy | |
| A1–A2 | 0.909 | 0.769 | 0.776 | 0.846 |
| A1–GS | 0.968 | 0.932 | 0.954 | 0.973 |
| A2–GS | 0.937 | 0.826 | 0.806 | 0.863 |
The following are the salient aspects of the guidelines used to annotate the data. The ShARe project guidelines relevant to Task 1 are available at: https://physionet.org/works/ShAReCLEFeHealth2013/files/Task1ShAReGuidelines2013.pdf and https://sites.google.com/site/shareclefehealth/data. The first site is password protected and requires users to obtain a DUA because it also contains the data used for the evaluation.
Annotations represent the most specific disorder span. For example, small bowel obstruction is preferred over bowel obstruction.
A disorder mention is a concept in the SNOMED CT portion of the Disorder semantic group.
Negation and temporal modifiers are not considered part of the disorder mention span.
All disorder mentions are annotated—even the ones related to a person other than the patient.
Mentions of disorders that are coreferential/anaphoric are also annotated.
The following are a few examples of disorder mentions from the data.
(E1) The patient was found to have left lower extremity DVT.
Here, lower extremity DVT is marked as the disorder. It corresponds to CUI C0340708 (preferred term: Deep vein thrombosis of lower limb). The span DVT can be mapped to CUI C0149871 (preferred term: Deep Vein Thrombosis), but this mapping would be incorrect because it is part of a more specific disorder in the sentence, namely, lower extremity DVT.
(E2) A tumor was found in the left ovary.
Here tumor … ovary is annotated as a disorder with a discontiguous mention.
(E3) The patient was admitted with low blood pressure.
Some disorders do not have a representation to a CUI as part of the SNOMED CT within the UMLS. However, if the annotators deemed them as important, they annotated them as CUI-less mentions. Here, low blood pressure is a finding that is normalized as a CUI-less disorder.
We constructed the annotation guidelines to require that the disorder be a reasonable synonym of the lexical description of a SNOMED CT disorder. In a few instances, the disorders are abbreviated or shortened in the clinical note. One example is w/r/r, which is an abbreviation for the concepts wheezing (CUI C0043144), rales (CUI C0034642), and ronchi (CUI C0035508). This abbreviation is also sometimes written as r/w/r and r/r/w. Another is gsw for ‘gunshot wound’ and tachy for ‘tachycardia.’
The final gold standard comprises 11 167 disorder mentions split into training and test sets (table 1), which were made available to the participating teams. The fraction of discontiguous mentions is substantial at about 11%, which is significant considering the fact that discontiguous mentions have almost never been part of the annotation schema in most open domain corpora. Only a few of the corpora in the biomedical literature domain exhibit this phenomenon. About 30% of the mentions are CUI-less; that is, they lack a representation in the SNOMED CT portion of the UMLS. One reason is the fact that we do not annotate the UMLS semantic group findings with a CUI, because this semantic group was found to be a noisy, catch-all category, and attempts to consistently annotate against it did not succeed in our preliminary studies. Another reason for CUI-less annotations is that many terms in clinical text still are not represented in standard vocabularies.
Table 1:
Distribution of disorder mentions across the training and test set according to the two criteria—whether they map to a Concept Unique Identifier (CUI) and whether they are contiguous
| Training | Test | |||
|---|---|---|---|---|
| Total disorder mentions | 5816 | 5351 | ||
| CUI-less mentions | 1639 | (28.2%) | 1750 | (32.7%) |
| CUI-ed mentions | 4177 | (71.8%) | 3601 | (67.3%) |
| Contiguous mentions | 5165 | (88.8%) | 4912 | (91.8%) |
| Discontiguous mentions | 651 | (11.2%) | 439 | (8.2%) |
Inter-annotator agreement
Table2 shows the IAA statistics for the data. For the disorders we measure, the agreement in terms of the F1 score as traditional agreement measures such as Cohen's κ and Krippendorf's α are not applicable for measuring agreement for entity mention annotation. We computed agreements between the two annotators as well as between each annotator and the final adjudicated gold standard. The latter is to give a sense of the fraction of corrections made in the adjudication process. The strict criterion considers two mentions correct if they agree in terms of the class and the exact string, whereas the relaxed criterion considers overlapping strings of the same class as correct. The reason for checking the class is as follows. Although we only use the disorder mention in this task, the corpus has been annotated with some other UMLS types as well and therefore, in some instances, the second annotator assigns a different UMLS type to the same character span in the text. If exact boundaries are not taken into account, the IAA agreement score is in the mid-90s.
For normalization to CUIs, we used accuracy to assess agreement. This was so that we could compare it with the same criteria used for evaluating system performance. For the relaxed criterion, all overlapping disorder spans with the same CUI were considered correct. For the strict criterion, only disorder spans with identical spans and the same CUI were considered correct.
Novelty of the ShARe corpus
As source text and annotation scheme, the ShARe corpus is closest to the i2b2/VA 2010 and MiPACQ corpora. Some notable differences exist: (i) in the i2b2 corpus, modifiers such as determiners are included in the mention span; (ii) the i2b2 corpus does not contain discontiguous mentions; (iii) the i2b2 corpus does not provide UMLS semantic typing and normalization to a CUI; and (iv) the MiPACQ corpus does provide UMLS semantic typing but not normalization to a CUI. The i2b2 training corpus contains about 18 550 concepts. The MiPACQ corpus includes about 4206 (14.74% of mentions in the corpus) disorder mentions.
Some other corpora containing annotations of disorder mentions are the NCBI, DNAE, and AZDC corpora. However, they use the scholarly literature from Medline and PubMed as the source text. In addition to marking the disease mentions, the NCBI corpus also categorizes them into four subcategories. The AZDC corpus annotates overlapping disease mentions in case of coordination but does not map discontiguous spans. For example, it annotates the phrase Duchenne and Becker muscular dystrophy as two separate but overlapping disease mentions—Duchenne and Becker muscular dystrophy and Becker muscular dystrophy. The same phrase is marked in whole as a Composite Mention in the NCBI corpus, whereas in the ShARe corpus, it is marked as overlapping discontiguous spans—Duchenne…muscular dystrophy and Becker muscular dystrophy.
STUDY DESIGN
Task description
The ShARe corpus presents a novel opportunity to evaluate performance on the tasks of identification of disorder mentions and normalization to a standardized vocabulary. The 2013 ShARe/CLEF Evaluation Lab42 was composed of two parts:
Task 1a—Identification of the text spans in the document that represent disease mentions.
Task 1b—Mapping the mention to the appropriate SNOMED CT CUI in the UMLS (optional task).
In Tasks 1a and 1b, each participating team was permitted to upload the outputs of up to two systems. Teams were allowed to use additional annotations in their systems; systems that used annotations outside of those provided were evaluated separately. We provided the participants with a training set with gold standard annotations. The participants had approximately 2 months from the date the training data were released (February 15, 2013) to the date the test results had to be uploaded (April 24, 2013). The website to the ShARe/CLEF lab is located at https://sites.google.com/site/shareclefehealth/
Evaluation
We used the following evaluation criteria:
Task 1a—Correctness in identification of the character spans of disorders.
The system performance was evaluated against the gold standard using the F1 score of the precision and recall values. There were two variations: (i) strict and (ii) relaxed. In the strict case, a span is counted as correct if it is identical to the gold standard span. In the relaxed case, a span overlapping the gold standard span is also considered correct. The formulae for computing these metrics are as follows:
| 1 |
| 2 |
| 3 |
where Dtp is the number of true positives disorder mentions, Dfp is the number of false positive disorder mentions, and Dfn is the number of false negative disorder mentions.
Task 1b—Correctness in mapping disorders to SNOMED CT codes.
We used accuracy as the performance measure for Task 1b. The reason we chose accuracy was in order to analyze the performance at each of the two stages separately and not let the performance in Task 1a potentially eclipse the performance at the normalization stage. It was defined as follows:
| 4 |
| 5 |
where Dtp is the number of true positive disorder mentions with identical spans as in the gold standard, Ncorrect isthe number of correctly normalized disorder mentions, and Tg is the total number of disorder mentions in the gold standard.
The relaxed accuracy only measures the ability to normalize correct spans. Therefore, obtaining very high values for this measure is possible by simply dropping any mention with a low confidence span. We performed non-parametric statistical significance tests through random shuffling.43
RESULTS
A total of 22 teams competed in Task 1a, and 16 of them also participated in Task 1b. We received working notes from 15 teams describing the methods behind the systems. This task was used as a course project at West Virginia University (WVU). Six students submitted separate runs but wrote a single working note describing the details. More information on the teams can be found in the task overview paper.42
System performance for Tasks 1a and 1b is detailed in tables 3 and 4, respectively. Two different systems performed best across the two subtasks. The best system for Task 1a (UTHealthCCB.A) had an F1 score of 0.75 (0.80 precision, 0.71 recall); the best system for Task 1b (NCBI.2) had an accuracy of 0.59.
Table 3:
Evaluation for Task 1a
| System ({ID}.{run}) | Strict | Relaxed | ||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| No additional annotations | ||||||
| (UTHealthCCB.A).2 | 0.800 | 0.706 | 0.750* | 0.925 | 0.827 | 0.873 |
| (UTHealthCCB.A).1 | 0.831 | 0.663 | 0.737* | 0.954 | 0.774 | 0.854 |
| NCBI.1 | 0.768 | 0.654 | 0.707* | 0.910 | 0.796 | 0.849 |
| NCBI.2 | 0.757 | 0.658 | 0.704* | 0.904 | 0.805 | 0.852 |
| CLEAR.2 | 0.764 | 0.624 | 0.687* | 0.929 | 0.759 | 0.836 |
| (Mayo.A).1 | 0.800 | 0.573 | 0.668* | 0.936 | 0.680 | 0.787 |
| (UCDCSI.A).1 | 0.745 | 0.587 | 0.656 | 0.922 | 0.758 | 0.832 |
| CLEAR.1 | 0.755 | 0.573 | 0.651* | 0.937 | 0.705 | 0.804 |
| (Mayo.B).1 | 0.697 | 0.574 | 0.629* | 0.939 | 0.766 | 0.844 |
| CORAL.2 | 0.796 | 0.487 | 0.604 | 0.909 | 0.554 | 0.688 |
| HealthLanguageLABS.1 | 0.686 | 0.539 | 0.604* | 0.912 | 0.701 | 0.793 |
| LIMSI.2 | 0.814 | 0.473 | 0.598* | 0.964 | 0.563 | 0.711 |
| LIMSI.1 | 0.805 | 0.466 | 0.590 | 0.962 | 0.560 | 0.708 |
| (AEHRC.A).2 | 0.613 | 0.566 | 0.589* | 0.886 | 0.785 | 0.833 |
| (WVU.DG + VJ).1 | 0.614 | 0.505 | 0.554* | 0.885 | 0.731 | 0.801 |
| (WVU.SS + VJ).1 | 0.575 | 0.496 | 0.533 | 0.848 | 0.741 | 0.791 |
| CORAL.1 | 0.584 | 0.446 | 0.505 | 0.942 | 0.601 | 0.734 |
| NIL-UCM.2 | 0.617 | 0.426 | 0.504 | 0.809 | 0.558 | 0.660 |
| KPSCMI.2 | 0.494 | 0.512 | 0.503* | 0.680 | 0.687 | 0.684 |
| NIL-UCM.1 | 0.621 | 0.416 | 0.498 | 0.812 | 0.543 | 0.651 |
| KPSCMI.1 | 0.462 | 0.523 | 0.491* | 0.651 | 0.712 | 0.680 |
| (AEHRC.A).1 | 0.699 | 0.212 | 0.325* | 0.903 | 0.275 | 0.422 |
| (WVU.AJ + VJ).1 | 0.230 | 0.318 | 0.267* | 0.788 | 0.814 | 0.801 |
| UCDCSI.2 | 0.268 | 0.175 | 0.212* | 0.512 | 0.339 | 0.408 |
| SNUBME.2 | 0.191 | 0.137 | 0.160* | 0.381 | 0.271 | 0.317 |
| SNUBME.1 | 0.302 | 0.026 | 0.047 | 0.504 | 0.043 | 0.079 |
| (WVU.FP + VJ).1 | 0.024 | 0.446 | 0.046 | 0.088 | 0.997 | 0.161 |
| Additional annotations | ||||||
| (UCSC.CW + RA).2 | 0.732 | 0.621 | 0.672 | 0.883 | 0.742 | 0.806 |
| (UCSC.CW + RA).1 | 0.730 | 0.615 | 0.668* | 0.887 | 0.739 | 0.806 |
| RelAgent.2 | 0.651 | 0.494 | 0.562* | 0.901 | 0.686 | 0.779 |
| RelAgent.1 | 0.649 | 0.450 | 0.532 | 0.913 | 0.636 | 0.750 |
| (WVU.AL + VJ).1 | 0.492 | 0.558 | 0.523* | 0.740 | 0.840 | 0.787 |
| (THCIB.A).1 | 0.445 | 0.551 | 0.492* | 0.720 | 0.713 | 0.716 |
| (WVU.RK + VJ.1 | 0.397 | 0.465 | 0.428 | 0.717 | 0.814 | 0.762 |
In the Strict F1 score column, * indicates the F1 of the system was significantly better than the one immediately below (random shuffling, p<0.01). The .1 and .2 suffixes represent run number 1 and 2, respectively.
P, precision; R, recall.
Table 4:
Evaluation for Task 1b
| System ({ID}.{run}) | Strict | Relaxed |
|---|---|---|
| Accuracy | Accuracy | |
| No additional annotations | ||
| NCBI.2 | 0.589* | 0.895 |
| NCBI.1 | 0.587* | 0.897 |
| (Mayo.A).2 | 0.546* | 0.860 |
| (UTHealthCCB.A).1 | 0.514* | 0.728 |
| (UTHealthCCB.A).2 | 0.506 | 0.717 |
| (Mayo.A).1 | 0.502* | 0.870 |
| KPSCMI.1 | 0.443* | 0.865 |
| CLEAR.2 | 0.440* | 0.704 |
| CORAL.2 | 0.439* | 0.902 |
| CORAL.1 | 0.410* | 0.921 |
| CLEAR.1 | 0.409* | 0.713 |
| NIL-UCM.2 | 0.362 | 0.850 |
| NIL-UCM.1 | 0.362* | 0.871 |
| (AEHRC.A).2 | 0.313* | 0.552 |
| (WVU.SS + VJ).1 | 0.309 | 0.622 |
| (UCDCSI.B).1 | 0.299* | 0.509 |
| (WVU.DG + VJ).1 | 0.241 | 0.477 |
| (AEHRC.A).1 | 0.199* | 0.939 |
| (WVU.AJ + VJ).1 | 0.142 | 0.448 |
| (WVU.FP + VJ).1 | 0.112* | 0.252 |
| (UCDCSI.B).2 | 0.006 | 0.035 |
| Additional annotations | ||
| (UCSC.CW + RA).2 | 0.545* | 0.878 |
| (UCSC.CW + RA).1 | 0.540* | 0.879 |
| (THCIB.A).1 | 0.470* | 0.853 |
| (WVU.AL + VJ).1 | 0.349* | 0.625 |
| (WVU.RK + VJ).1 | 0.247 | 0.531 |
In the Strict Accuracy column, * indicates the accuracy of the system was significantly better than the one immediately below (random shuffling, p<0.01). The .1 and .2 suffixes represent run number 1 and 2, respectively.
The best-performing system by Tang et al.44 combined several resources: (i) systems—MetaMap, cTAKES; (ii) rich features—Brown word clusters,45 terms from UMLS; (iii) algorithms: Conditional Random Fields (CRF) and structured Support Vector Machines (SVM), SVMhmm, term frequency-inverse document frequency (tf-idf); and (iv) feature representation—the most complicated version of the Inside-Outside-Begin (IOB) representation (see 1d in the Discussion section).
Most of the participating systems employed hybrid approaches by supplementing features to a machine-learning algorithm, applying rules and gazetteers extracted from the training data, and using other resources. Only three of the 14 systems were completely rule based. A total of 10 out of the 14 systems incorporated either Apache cTAKES or MetaMap in their methods. Of these, four used both in conjunction and two used them in disjunction. Of the 14 systems, seven used only cTAKES and eight used only MetaMap. Online tables 1 and 2 in the appendix summarize the participating teams and the tasks for which they submitted outputs, along with the various strategies for the selection of classifiers, feature sets, and representations. Columns 3 and 4 list the individual runs that were submitted and what subtask they represent. Online table 3 in the appendix lists all the participants along with the number of runs they submitted for each subtask of Task 1.
DISCUSSION
At first sight, the best results of Task 1a (0.750) seem inferior (by about 0.10 absolute F1 score) to the best reported in the open domain and also some reported in the i2b2 2010 shared task.8 In the case of the open domain, prior art has been reported predominantly on newswire, and as such it is not directly comparable. More recent creations of more diverse corpora have indicated that overall, named entity recognition performance can vary considerably depending on the source of the text.46 The comparison with the i2b2 2010 shared task corpus is also not straightforward, because the ShARe corpus is based on the UMLS as opposed to a more relaxed definition in the i2b2 corpus. The annotation of discontiguous arguments in the ShARe corpus further explains this discrepancy.
The participation by Xia and Zhong47 involved building a baseline using cTAKES and MetaMap individually as well as in combination. The authors found the latter approach is better than the former, so each system seems to solve different aspects of the problem. Of the three rule-based systems, Fan et al.48 used a system based on OpenNLP and employed post-processing to identify discontiguous spans; for the normalization task, the authors utilized a simple list of frequency-ranked CUIs from the training set. Ramanan et al.49 built on a system called Cocoa, which originally contained 30 rules. It was augmented with 70 acronyms and abbreviations and 15 phrases from the training data. The third rule-based system, that of Wang and Akella,50 applied some post-processing rules generated from the frequency of occurrences of patterns in the training data, augmented with regular expressions to handle discontiguous spans. They also created a blacklist of rules to remove the false positives predicted by MetaMap.
Of the machine-learning systems, the most popular classifiers were CRF and SVMs. They typically involved an IOB-style tagger applied to Task 1a. Two systems, which were also the two top performing systems—by Leaman et al.51 and Tang et al.44—used some form of term frequency-inverse document frequency (tf-idf)-based similarity.
One salient characteristic of the 2013 CLEF/ShARe task was the presence of discontiguous mentions. The following are the various discontinuous mention representations and/or techniques used by the participating systems, from simple to more complex:
-
1.Rule-based post-processing using stop-words, intermediate words from training data, and so on, using the standard IOB or more complex variations. This technique used four main approaches as illustrated in table 5:
IO representation—the simplest of the representation, which uses only two classes: one for indicating the word is inside an annotation span and the other used to indicate otherwise.
IOB/IOE representation—a more standard form of representation with B, E, I, and O representing Begin, End, Inside, and Outside classes, respectively. IOB2/IOE2 is a slightly different version of this representation, where B and E are used for marking all begin or end tokens rather than just those that immediately follow or precede another class. The name IOB1 sometimes refers to the IOB representation.
BEISTO representation—a slightly modified version of IOB/IOE in which single tokens are tagged with an S, and tokens between two discontiguous spans are tagged with a T.
A set of B, I, O, DB, DI, HB, HI tags—none of the above three representations explicitly models discontiguous arguments that share the same head word. This modification uses the standard IOB tags for contiguous disorder mentions, but for discontiguous mentions, it adds four more tags. These tags are variations of the B and I tags, with either a D or an H prefix. H indicates that the word or word sequence is the shared head, and D indicates otherwise. This approach was the most comprehensive one for this task and was implemented by the best-performing system.
2.Training classifiers for ‘discontiguous’ relations by Patrick et al.52 This strategy has an underlying assumption of only one discontiguous mention (without a shared head) in a sentence.
3.Incorporating information from semantic role labels and relation classifiers as in Gung.53 Here the discontiguous mentions are split into separate mentions, and then once the IOB tagging is complete, the discontiguous cases are identified based on information from semantic role labeling and other relations (eg, LocationOf).
Table 5:
Instantiations of the four Inside-Outside-Begin encoding variations for three sentences
| IO | IOB | IOB2 | BIESTO | B, I, O, DB, DI, HB, HI | |
|---|---|---|---|---|---|
| The | O | O | O | O | O |
| aortic | I | I | B | B | DB |
| root | I | I | I | I | DI |
| and | O | O | O | O | O |
| ascending | O | O | O | B | DB |
| aorta | O | O | O | I | DI |
| are | O | O | O | T | O |
| moderately | O | O | O | T | O |
| dilated | I | I | B | E | HB |
| . | O | O | O | O | O |
| The | O | O | O | O | O |
| left | I | I | B | B | DB |
| atrium | I | I | I | I | DI |
| is | O | O | O | T | O |
| moderately | O | O | O | T | O |
| dilated | I | I | B | E | DB |
| . | O | O | O | O | O |
| No | O | O | O | O | O |
| pain | I | I | B | S | B |
| . | O | O | O | O | O |
The words that are part of the disorder mention are in bold along with the respective encodings.
Systems that did not encode explicitly discontiguous spans tried to recover them using some post processing. All except the Tang et al.system ignored the discontiguous spans with shared heads, because they are a very small fraction of the data. Machine-learning systems that do not particularly apply a formulation for discontiguous spans do not seem to perform much differently than the ones with heuristic post-processing. Gung found that utilizing a classifier-based approach to recover discontiguous spans with LibSVM and features representing relation information from cTAKES4 and semantic role information from ClearNLP,54 gave a 2–3 points absolute improvement on the F1 scores. Choosing the right input encoding for the learning algorithms can be difficult, and one could apply techniques such as hill climbing. The utilization of error-correcting codes55 has also been shown to improve performance. None of the systems in this shared task resorted to either technique. For a more thorough analysis of the encodings, the reader is referred to Loper.56
Another interesting observation by Leaman et al.51 was that allowing the annotators to select a discontiguous span that best represents the most specific disease/disorder has possibly had the effect of lowering the amount of term variance in the ShARe corpus with respect to that observed in the NCBI disease corpus. So although discontiguous spans tend to increase the task complexity, they seem to capture the phenomena more precisely.
MetaMap has shown very high precision but low recall, which is one reason the normalization accuracy is in the 90s for the relaxed case for some systems incorporating it. A combination of MetaMap with CRF or SVM tagging, or disease mentions output by cTAKES improves recall without a significant drop in precision, making the combination work much better. As expected, rule-based systems using relatively similar resources tend to have more variation among their results. For example, the reason for an 8-point difference between the performance of the Zuccon et al.57 system and the Xia and Zhong system in the relaxed case, which both employed MetaMap, is unclear. The inclusion of simple nouns when the annotators expected a compound noun significantly reduced YTEX precision. As a result, YTEX performed poorly relative to MetaMap on the strict task. On the other hand, MetaMap tended to include additional text (mostly prepositions and modifiers) that the gold standard did not.
Leaman et al.51 used the 2012AB version of UMLS and obtained the best normalization performance. In addition, the boundary revision approach as a feedback from the normalization system (DNorm) helped their performance. Tokens were added or removed from the left and added to the right (not removed, because doing so tends to delete head words) only if the score improves over a certain threshold. For the MetaMap baseline, the authors fed sentence chunks to MetaMap and received back various terms and their normalized CUIs. These terms were then post-processed and larger overlapping terms were selected. Sometimes MetaMap provides multiple CUIs for the same term, for example, ‘heart failure.’ In such cases, the word sense disambiguation tool built into MetaMap was used to select the best match. On a development subset using just MetaMap, the authors obtained a strict F1 score of 0.44, whereas DNorm, with boundary revisions, obtained a score of 0.66—an increase of almost 22 points.
Liu et al.58 performed an experiment with features based on semantic vectors trained on the Mayo Clinic corpus versus others trained on the MIMIC corpus. Surprisingly, the first approach achieved better performance.
Recently, YTEX has improved on the cTAKES dictionary lookup algorithm by adding a sense disambiguation component that allows the most appropriate concept mapping given the context. It uses the adapted Lesk algorithm59 to compute semantic similarity over a context window, whereas MetaMap implements a series of weighted heuristics to select the appropriate candidate. Osborne et al.60 compare the two systems in their submissions. With MetaMap, they report a score of 0.42, which is similar to that of Leaman et al.51 but on the entire training set.
The 2013 CLEF/ShARe task was a class final project at WVU, and various students teamed to put systems together. They submitted a total of four runs, which span a wide range of performance. These systems used CRF classifiers from the FACTORIE toolkit.61,62 All these systems were outlined in a single overview paper. Given the compressed nature of the description, it is not possible to speculate why their performance spans over such a wide range.
Given the variety of systems and approaches, a thorough error analysis was out of the scope of this paper. Instead, we present a synthesis of the main types of errors. The following are the frequent categories of errors for Task 1a:
False negatives—The recall for these systems is significantly lower than their precision in both the strict and relaxed settings. The difference ranges from 0.10 to 0.18 and is roughly the same across the two evaluation criteria. The fact that 1287 out of a total of 5815 disorder instances in the test data are not seen in the training data partly explains the impact on recall (eg, ‘hyperpigmentation,’ ‘elevated alkaline phosphatase,’ ‘cystic collection’).
Boundary identification—The other significant contributor to errors was the exact disorder boundary identification. The fact that the F1 measure for the relaxed scores is about 0.11–0.15 points shows that identifying the exact boundaries has an impact on the score. These also include cases of discontiguous disorders in which only one of the spans was identified correctly.
Because Task 1b is directly dependent on the performance on Task 1a, the above errors directly affect its performance. When boundaries were not correctly identified, the likelihood of mapping to a different (more or less specific) CUI increased (eg, discovering DVT instead of lower extremity DVT prompted a wrong CUI mapping).
Interestingly, the higher-performing systems are also the ones with the most intricate set of features and multiple layers of possibly stacked learners. The question arises whether one needs such complex machinery to solve this problem, or whether the excess machinery generates models that overfit the corpus.
In order to get a better idea of the performance ceiling, we had permitted systems to submit runs where they were allowed to use additional data. A few systems ran tasks using additional data. Not all these additional data could be classified as being from external sources. For example, RelAgent used a custom-made dictionary of abbreviations from the training data that we had provided. THCIB considered the use of MetaMap and cTAKES as use of external data.
CONCLUSION
We presented the results from the 2013 CLEF/ShARe shared task on disorder entity identification and normalization. The task evaluated the state of the art of this critical component not previously evaluated on clinical narrative. We observed the use of many interesting variations of existing and novel resources among the system submissions. The majority of the participating systems applied a hybrid machine-learning and rule-based combination. Unfortunately, not all participants submitted working notes, but most of the top-performing systems did submit them, and we could analyze their performance in more detail. Named entity tagging in the clinical domain is a hard task, and the presence of discontiguous spans was a challenging addition to the task. We witnessed the development of some novel approaches—especially the novel variations of the transformation-based learning approaches. Although the discontiguous spans make the span identification task harder, they also seem to allow for a better normalization.
Acknowledgments
We greatly appreciate the hard work and feedback of our program committee members and annotators, including but not limited to David Harris, Glenn Zaramba, Erika Siirala, Qing Zeng, Tyler Forbush, Jianwei Leng, Maricel Angel, Erikka Siirala, Heljä Lundgren-Laine, Jenni Lahdenmaa, Laura Maria Murtola, Marita Ritmala-Castren, Riitta Danielsson-Ojala, Saija Heikkinen, and Sini Koivula.
CONTRIBUTORS
SP, GS, NE, and WWC defined the task. WWC and BRS led the overall task. SP, GS, NE, AV and WWC led the annotation effort. HS co-chaired the lab. BRS and LC developed the annotation infrastructure. BRS, LC, and DM processed and distributed the corpus. DM and WWC led result evaluations. SP and GS drafted the manuscript, and then all co-authors reviewed and contributed to the final version.
FUNDING
This work was partially supported by Shared Annotated Resources (ShARe) project NIH 5R01GM090187, Strategic Health Advanced Research Project: Area 4 project from the US Office of the National Coordinator of Healthcare Technology (SHARP 90TR0002), Temporal Histories of Your Medical Events (THYME) project from NIH (R01LM10090), VAHSR&D HIR 08-374, NICTA (National Information and Communications Technology Australia), funded by the Australian Government as represented by the Department of Broadband, Communications and the Digital Economy and the Australian Research Council through the ICT Centre of Excellence program, and NLM 5T15LM007059.
COMPETING INTERESTS
GS is on the Advisory Board of Wired Informatics, LLC, which provides services and products for clinical NLP applications.
PATIENT CONSENT
Patient consent was waived in all studies due to the use of a retrospective de-identified corpus.
PROVENANCE AND PEER REVIEW
Not commissioned; externally peer reviewed.
DATA SHARING STATEMENT
The data and scripts used in this evaluation are available through PhysioNetWorks (physionet.org) through a data use agreement.
REFERENCES
- 1.Demner-Fushman D, Chapman WW, Mcdonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform 2009;42:760–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oglensky BD, Davidson EJ. Teaching and learning through clinical report-writing genres. Int J Learn 2009;16:139–52. [Google Scholar]
- 3.Zaner RM. Clinical ethics and the necessity of stories. Dordrecht: Springer, 2011. [Google Scholar]
- 4.Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc 2013;20:922–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller T, Bethard S, Dligach D, et al. Discovering temporal narrative containers in clinical text. Proceedings of the 2013 Workshop on Biomedical Natural Language Processing Sofia, Bulgaria. [Google Scholar]
- 6.THYME – Temporal Histories of Your Medical Event. http://thyme.healthnlp.org
- 7.Styler WF, Bethard S, Finan S, et al. Temporal annotation in the clinical domain. Trans Comput Linguist 2014;2:143–54. [PMC free article] [PubMed] [Google Scholar]
- 8.Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.i2b2 – Informatics for Integrating Biology & the Bedside. https://www.i2b2.org/
- 10.Elhadad N, Chapman WW, O'Gorman T, et al. The ShARe Schema for The Syntactic and Semantic Annotation of Clinical Texts. Under Review.
- 11.SHARPn: Strategic Health IT Advanced Research Projects. http://sharpn.org
- 12.Pathak J, Bailey KR, Beebe CE, et al. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. J Am Med Inform Assoc 2013;20:e341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman WW, Nadkarni PM, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Informatics Assoc 2011;18:540–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pacheco JA, Avila PC, Thompson JA, et al. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. AMIA Annu Symp Proc 2009;2009:497–501. [PMC free article] [PubMed] [Google Scholar]
- 15.Waudby CJ, Berg RL, Linneman JG, et al. Cataract research using electronic health records. BMC Ophthalmol 2011;11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011;3:79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc 2010;17:568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin C, Karlson EW, Canhao H, et al. Automatic prediction of rheumatoid arthritis disease activity from the electronic medical records. PLoS ONE 2013;8:e69932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Jiang M, Oetjens M, et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc 2011;18:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther 2011;89:379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CoNLL – Computational Natural Language Learning. http://ifarm.nl/signll/conll/
- 22.SemEval – Semantic Evaluations. http://en.wikipedia.org/wiki/SemEval
- 23.BioNLP. http://compbio.ucdenver.edu/BioNLP2013
- 24.BioCreAtIvE. http://www.biocreative.org/
- 25.i2b2 Shared Tasks. https://www.i2b2.org/NLP/Relations/
- 26.SNOMED Clinical Terms (SNOMED CT). http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html
- 27.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. J Biomed Inform 2003;36:414–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UMLS Metathesaurus. http://www.nlm.nih.gov/research/umls
- 29.Pradhan S, Elhadad N, South BR, et al. Task 1: ShARe/CLEF eHealth Evaluation Lab 2013. Proceedings of the ShARe/CLEF Evaluation Lab 2013:1–6 http://www.nicta.com.au/pub?id=7264 [Google Scholar]
- 30.MeSH – Medical Subject Headings. http://www.nlm.nih.gov/mesh/meshhome.html
- 31.RxNorm. https://www.nlm.nih.gov/research/umls/rxnorm/
- 32.Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp 2000:270–4. [PMC free article] [PubMed] [Google Scholar]
- 33.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 34.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010;17:229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garla VN, Brandt C. Knowledge-based biomedical word sense disambiguation: an evaluation and application to clinical document classification. J Am Med Inform Assoc 2013;20:882–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leaman R, Islamaj Dogan R, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics 2013;29:2909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doğan RI, Lu Z. An improved corpus of disease mentions in PubMed citations. Proceedings of the 2012 Workshop on Biomedical Natural Language Processing Montréal, Canada: 2012. 91–9 http://www.aclweb.org/anthology/W12-2411 [Google Scholar]
- 39.Leaman R, Miller C, Gonzalez G. Enabling recognition of diseases in biomedical text with machine learning: corpus and benchmark. Proceedings of the 2009 Symposium on Languages in Biology and Medicine, 2009:82–9. [Google Scholar]
- 40.Gurulingappa H, Klinger R, Hofmann-Apitius M, et al. An empirical evaluation of resources for the identification of diseases and adverse effects in biomedical literature. 2nd Workshop on Building and Evaluating Resources for Biomedical Text Mining (7th edition of the Language Resources and Evaluation Conference) 2010:15–22. [Google Scholar]
- 41.Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II). http://mimic.physionet.org/
- 42.Suominen H, Salanterä S, Velupillai S, et al. Overview of the ShARe/CLEF EHealth Evaluation Lab 2013. Proceedings of the ShARe/CLEF Evaluation Lab, 2013:1–6. [Google Scholar]
- 43.Yeh A. More accurate tests for the statistical significance of result differences. Proceedings of the 18th Conference on Computational Linguistics Morristown, NJ, USA: Association for Computational Linguistics, 2000:947. 10.3115/992730.992783 [Google Scholar]
- 44.Tang B, Wu Y, Jiang M, et al. Recognizing and encoding disorder concepts in clinical text using machine learning and vector space. Proceedings of the ShARe/CLEF Evaluation Lab. 2013. [Google Scholar]
- 45.Brown PF, Pietra VJ Della, DeSouza PV, et al. Class-based n-gram models of natural language. Comput Linguist 1992;18:467–79. [Google Scholar]
- 46.Pradhan S, Moschitti A, Xue N, et al. Towards robust linguistic analysis using OntoNotes. Proceedings of the Seventeenth Conference on Computational Natural Language Learning Sofia, Bulgaria: Association for Computational Linguistics, 2013:143–52 http://www.aclweb.org/anthology/W13-3516 [Google Scholar]
- 47.Xia Y, Zhong X. Combining MetaMap and cTAKES in disorder recognition: THCIB at CLEF eHealth Lab 2013 Task 1. Proceedings of the ShARe/CLEF Evaluation Lab 2013:2–6. [Google Scholar]
- 48.Fan J, Sood N, Huang Y. Disorder concept identification from clinical notes an experience with the ShARe/CLEF 2013 challenge. Proceedings of the ShARe/CLEF Evaluation Lab. 2013. [Google Scholar]
- 49.Ramanan SV, Broido S, Nathan PS. Performance of a multi-class biomedical tagger on clinical records. Proceedings of the ShARe/CLEF Evaluation Lab 2013:1–6. [Google Scholar]
- 50.Wang C, Akella R. ShARe/CLEF Task 1 Working Notes Team UCSC introduction to Task 1. Proceedings of the ShARe/CLEF Evaluation Lab 2013. [Google Scholar]
- 51.Leaman R, Khare R, Lu Z. Disorder normalization in clinical notes with DNorm. Proceedings of the ShARe/CLEF Evaluation Lab 2013. [Google Scholar]
- 52.Patrick JD, Safari L, Ou Y. ShARe/CLEF eHealth 2013 named entity recognition and normalization of disorders challenge. Proceedings of the ShARe/CLEF Evaluation Lab 2013. [Google Scholar]
- 53.Gung J. Using relations for identification and normalization of disorders: team CLEAR in the ShARe/CLEF 2013 eHealth Evaluation Lab. Proceedings of the ShARe/CLEF Evaluation Lab 2013. [Google Scholar]
- 54.The ClearNLP Project. https://code.google.com/p/clearnlp/
- 55.Dietterich TG. Solving multiclass learning problems via error-correcting output codes. J Artif Intell Res 1995;2:263–86. [Google Scholar]
- 56.Loper E. Encoding structured output values [Ph.D. Thesis]. University of Pennsylvania. 2008. [Google Scholar]
- 57.Zuccon G, Holloway A, Koopman B, et al. Identify disorders in health records using conditional random fields and metamap. Proceedings of the ShARe/CLEF Evaluation Lab 2013. [Google Scholar]
- 58.Liu H, Wagholikar K, Jonnalagadda S, et al. Integrated cTAKES for concept mention detection and normalization. Proceedings of the ShARe/CLEF Evaluation Lab 2013. [Google Scholar]
- 59.Lesk ME. Automatic sense disambiguation using machine readable dictionaries: how to tell a pine cone from an ice cream cone. Proceedings of the 5th Annual International Conference on Systems Documentation Toronto: 1986. 24–6. 10.1145/318723.318728 [Google Scholar]
- 60.Osborne JD, Gyawali B, Solorio T. Evaluation of YTEX and MetaMap for clinical concept recognition. Proceedings of the ShARe/CLEF Evaluation Lab 2013. [Google Scholar]
- 61.McCallum A, Schultz K, Singh S. FACTORIE: probabilistic programming via imperatively defined factor graphs. Neural Information Processing Systems (NIPS) 2009:1–9. [Google Scholar]
- 62.FACTORIE Toolkit. http://factorie.cs.umass.edu/