Abstract
Objective Pooling data across biobanks is necessary to increase statistical power, reveal more subtle associations, and synergize the value of data sources. However, searching for desired data elements among the thousands of available elements and harmonizing differences in terminology, data collection, and structure, is arduous and time consuming.
Materials and methods To speed up biobank data pooling we developed BiobankConnect, a system to semi-automatically match desired data elements to available elements by: (1) annotating the desired elements with ontology terms using BioPortal; (2) automatically expanding the query for these elements with synonyms and subclass information using OntoCAT; (3) automatically searching available elements for these expanded terms using Lucene lexical matching; and (4) shortlisting relevant matches sorted by matching score.
Results We evaluated BiobankConnect using human curated matches from EU-BioSHaRE, searching for 32 desired data elements in 7461 available elements from six biobanks. We found 0.75 precision at rank 1 and 0.74 recall at rank 10 compared to a manually curated set of relevant matches. In addition, best matches chosen by BioSHaRE experts ranked first in 63.0% and in the top 10 in 98.4% of cases, indicating that our system has the potential to significantly reduce manual matching work.
Conclusions BiobankConnect provides an easy user interface to significantly speed up the biobank harmonization process. It may also prove useful for other forms of biomedical data integration. All the software can be downloaded as a MOLGENIS open source app from http://www.github.com/molgenis, with a demo available at http://www.biobankconnect.org.
Keywords: Biobank, Harmonization, Data integration, Search
INTRODUCTION
Researchers increasingly need large data sets to uncover the subtle statistical associations between phenotypes and diseases. It is therefore desirable to pool data from multiple biobanks for analysis.1 However, existing biobanks are usually designed specifically to local requirements and not to be similar to other resources. It therefore requires an incredible amount of time and effort to find relevant data elements across many different biobanks and to combine these into one statistically testable data set.2
The process of integrating comparable, but not necessarily identical, data from different biobanks is often referred to as ‘harmonization’1 and can be separated into several steps:3
Research question parameterization: defining data elements of interest based on the research question, for example, to statistically derive a prediction model for the risk of developing diabetes, data elements for well-known risk factors such as age, smoking status, blood pressure, and cholesterol are desired.4
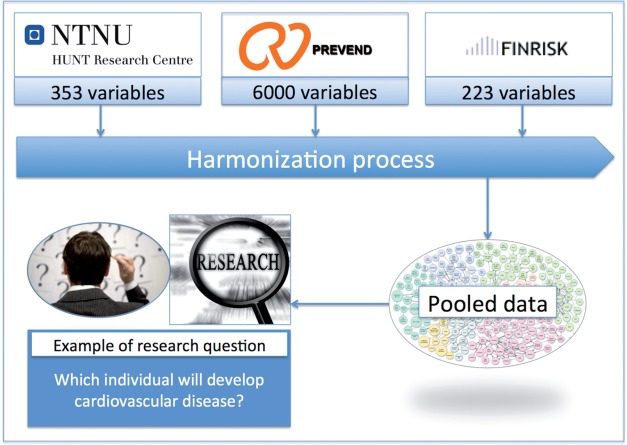
Schema matching: assessing harmonization potential by comparing desired elements within the ‘data dictionaries’ of each biobank. These are usually tab-delimited files that contain all the data elements available in the biobank and their corresponding information such as name, label, and definition (figure 1). The challenges lie in finding the matching elements and deciding whether they are scientifically comparable enough to be used for a pooled analysis.
Data integration: transforming source data into the target schema by creating algorithms based on the matches produced by schema matching that can derive the values of desired data elements from each of the biobanks. For example, pooling the data element ‘body mass index’ from the NCDS biobank cannot be done directly because no such element is available; the alternative elements ‘height in cm’ and ‘weight in kg’ are therefore used to calculate body mass index. During the calculation, the unit for ‘height in cm’ is converted into a unit in meters.
Figure 1:
Harmonization process. Many studies need to pool data in order to reach sufficient statistical power, however matching data elements of interest to the available data elements is a daunting task.
In this paper we describe how we can dramatically reduce the time needed for the second step—schema matching. The current process consists of human experts going through all the data dictionaries manually to identify potentially matching data elements, for example, the Prevend data dictionary which contains 6000 data elements including follow-up studies. Even when researchers are familiar with the content of a data dictionary, it takes multiple iterations to find relevant data elements and decide whether these are a complete, partial, or impossible match. This requires many detailed assessments as to whether the data element ‘is self-reported’ or ‘by a physician,’ ‘is whole life’ or ‘current status only,’ since even a small change in the way information is collected can substantially modify the scientific comparability of elements. This may take 4 h per data element (personal communication from the BioSHaRE project).1,2 Often the desired data element is not available and the best one can do is to identify a proxy element that is strongly related to the element of interest statistically and which can be used as an indirect measure.4 Moreover, the type and definition of potential proxies can vary greatly across biobanks, there are usually hundreds of available data elements in each biobank, and the descriptions use different local terminologies.1 Hypertension, for example, can also be described as ‘high blood pressure’ or ‘increase in blood pressure.’
BACKGROUND
Determining harmonization potential can be generalized as matching data elements from two schemas using unstructured data element descriptions.5 Two major candidate methods to automate this procedure are described in the literature: lexical matching and semantic matching.
Lexical matching
Lexical matching is a method to measure the similarity between two strings. Prior to matching, strings need to be processed by normalization procedures such as lowering case, removing punctuation and blank characters, etc. Two matching algorithms are relevant:3 (1) edit distance techniques using the minimal number of operations that need to be applied to one string in order to get to the other one, such as N-grams and Levenshtein distance; and (2) token-based distance techniques, derived from information retrieval research, for example, vector space models (VSM), which are usually recommended for matching long strings. They treat strings as bags of words, in which each dimension represents a word, with its length representing the number of occurrences of that word. Similarity can be measured using a cosine similarity function that calculates the cosine angles between two vectors representing two different strings. Considering that the descriptions of biobank data elements are usually in the format of unstructured long strings, it was logical to choose a token-based distance matching algorithm over other approaches for our system.
Semantic matching
Semantic matching searches for correspondences using knowledge about the concepts and their relationships.6 In ontologies, some related concepts are connected with a subClassOf (is-a) relationship, which provides the backbone for taxonomic structures. These concepts are considered to be quite similar and could therefore be considered a partial match. For example, matching ‘Parental diabetes mellitus’ with ‘Father diabetes’ cannot be achieved using the lexical matching strategy because the relationship between ‘Father’ and ‘Parent’ cannot be determined by synonyms. However, in an ontology, the fact that ‘Father’ is a subclass of ‘Parent’ is stated explicitly, so matching ‘Father diabetes’ with ‘Parental diabetes mellitus’ becomes possible. Other than the is-a relationship, the concepts could also be connected by associative relationships such as part-of and has-location. However, matching based on these relationships is not useful in this project.3
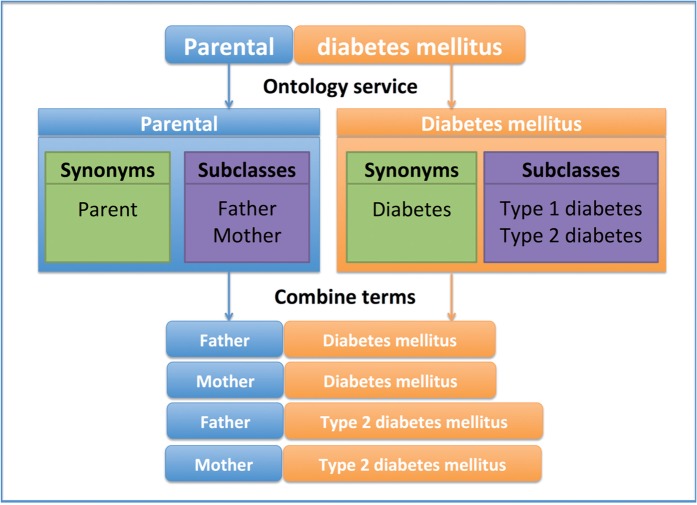
Semantic matching techniques facilitate biobank schema matching because different terminologies are often used to describe equivalent concepts and/or more specialized data elements are available that can be used as a proxy for the desired concept. Query expansion is a useful method to enhance the search for these correspondences by adding semantically similar terms to expand the original query in order to match more data elements.7 Ontologies provide the background knowledge for such query expansion. Normally, synonyms and hyponyms (subclasses) provided by the ontology are used. An example of a query expansion for ‘Parental diabetes mellitus’ is shown in figure 2.
Figure 2:
Example of query expansion. ‘Parental diabetes mellitus’ is annotated with the ontology terms ‘Parental’ and ‘Diabetes mellitus.’ Then the terms are expanded based on synonyms, resulting in three terms for ‘Diabetes mellitus’ and three terms for ‘Parental,’ so all 3 × 3 = 9 combinations are used for the search (only four are shown here).
Existing tools
There are several lexical- and semantic-matching tools that could benefit our system: Díaz-Galiano et al.8 used synonyms for query expansion to improve the performance of the retrieval system. Each query was matched against a set of MeSH terms (concept and synonyms), and as long as the MeSH term could be found in the query, its corresponding set of terms would be appended to the query. A similar approach was used in GoPubMed, where a query was submitted to PubMed and retrieved abstracts were matched against ontology terms in Gene Ontology using a string-matching algorithm based on synonyms.9,10 Rodriguez et al.11 Nilsson and Hjelm,12 and Voorhees13 described similar approaches using ontologies for query expansion to resolve ambiguous terms. Hyponyms (subclasses) were also extracted from ontologies and used to expand queries. The main difference between these projects was the choice of ontologies, implying that the choice depends on the data that need to be dealt with; the data therefore require careful evaluation. Finally, Aleksovski et al.5 described a strategy in which they mapped two lists of unstructured medical terms from two hospitals in Amsterdam. Their strategy best addresses our matching problem. Their process had two major steps: (1) automatically annotating two lists of terms with DICE ontology terms using a string-matching algorithm, which they called ‘ontology term anchoring,’ in order to enrich semantics for both lists; and (2) automatically matching two lists that were annotated with ontology terms using existing ontology matchers such as FOAM and S-Match.14,15
We also searched for tools to manage biobank data dictionaries, and found the Clinical Information Modeling Initiative (CIMI),16 Cancer Data Standards Registry and Repository (caDSR) of common data elements,17 and the Observ-OM phenotype system,18 which all deal with data models not unlike the ‘data schemas’ in our project. But, to our knowledge, there is still little automation support to map non-standard data to these elements, with caDSR coming closest to our needs with UML annotation tools (Semantic Integration Workbench, SIW) that used a simple search for matches by name. We decided to combine elements from these tools in BiobankConnect.
METHODS
We implemented a three-step harmonization strategy. First, data elements of interest, which are defined based on the research question, are manually annotated with ontology terms, for example, users can choose from a drop-down menu to annotate a data element of interest such as smoking status or cardiovascular disease. Then, these ontology terms are used to automatically scan the descriptions of the thousands of available data elements from each biobank to find potential matches. Finally, all candidate matches are sorted from ‘best’ to ‘worst’ so researchers can quickly decide on a useful match.
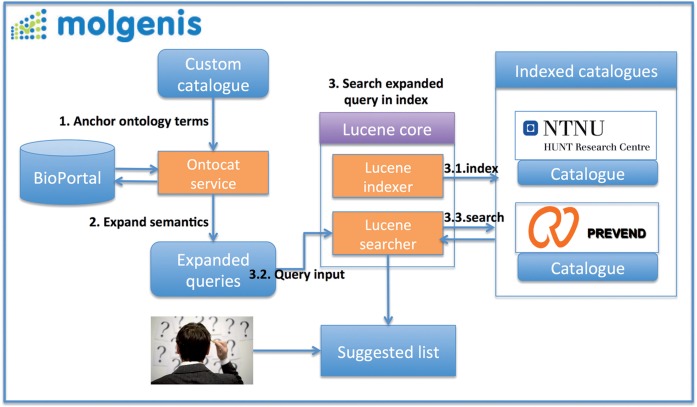
Figure 3 shows an overview of our matching strategy, which can be seen as a simplified version of that of Aleksovski et al.5 The process is implemented on top of the Observ-OM data model for describing the data elements and the MOLGENIS web database software in Java.18,19 Details of each step are described below.
Figure 3:
Overview of BiobankConnect. Data elements of interest (target) are matched to all available data elements (source), based on knowledge from the ontology terms.
Step 1: Manually annotate the search elements with ontology terms
To improve the accuracy of matching, we enable researchers to annotate data elements of interest with ontology terms either automatically or by hand. We added this option because some concepts are described in ontologies with a slightly different label than the desired data elements, something a human expert can quickly resolve. Moreover, there are typically only a few data elements of interest and this manual work is therefore limited. For example, to apply a prediction model for type 2 diabetes, about 10 predictors (data elements of interest) needed to be ontologically annotated.
To access ontologies we use BioPortal,20 an online ontology service with more than 400 ontologies currently available. To carefully select which ontology to use for our test case, we indexed questionnaires (collections of data elements) for 53 studies1 taken from the P3G observatory and matched those against all the ontologies that are available on BioPortal.21 Among these ontologies, we chose NCI Thesaurus and SNOMED Clinical Terms (SNOMED CT) because both are characterized by broad ranges (90 000 and 300 000 concepts, respectively) and matched the most terms in the 53 studies. For medication-related data elements we use the Anatomical Therapeutic Chemical (ATC) Classification System, because ATC codes are commonly used to store information regarding medication usage. Users can use any or all of these ontologies. The ontology terms are retrieved from BioPortal20 using OntoCAT22 software and stored in a local Lucene index.23 Use of this local index solves the problem of slow response when many requests are made over slow internet connections.
Step 2: Automatically expand semantics for search elements
The ontology annotations are used to automatically expand a query for data elements of interest using synonyms and subclasses. This succeeds in many cases where the biobank does not contain a perfectly matched data element by retrieving similar or more specific elements that can be used as proxies. For example, when matching ‘Current use of alcohol,’ the annotated ontology term ‘Alcoholic beverage’ in the NCI ontology lists more specific types of alcoholic beverages, such as ‘beer,’ ‘wine,’ and ‘liquor,’ and biobanks with data elements that are related to any of these beverages can then be matched. A complete query is created based on the expansions of the desired data element definitions using both synonyms and subclasses from the ontology terms. For example, the query ‘Hypertension’ is written as {‘Hypertension’ OR ‘Increased blood pressure’ OR ‘High blood pressure’ OR ‘Hypertensive disorder’ OR ‘HTN’}. Figure 2 shows another example. When the data elements of interest are not annotated with ontology terms, just the labels will be used as the query in the search.
Step 3: Lexical matching of the expanded query
Finally, all data dictionaries are searched via lexical matching and potential matches are shortlisted for manual decision-making. The retrieved data elements are sorted by Lucene VSM scores and then presented as ordered lists of candidate data elements per biobank from which users can decide on a suitable match. An all-to-all comparison of search data elements against all elements from all biobank dictionaries is a computationally expensive task, which took days in our original prototype. To speed up this process, we pre-indexed all the data dictionaries using Lucene.23 Prior to indexing, the sophisticated language pre-processing of Lucene removes ‘stop words’ (such as ‘what’ and ‘where’) from data elements to increase the sensitivity of matching. Lucene also stems terms in data elements so that different variations can be recognized during a search, for example, the stem for ‘smoking’ and ‘smoked’ is ‘smoke.’
EVALUATION
To evaluate BiobankConnect, we used schema matching data from the EU-BioSHaRE Healthy Obese Project (HOP).24,25 In this project a team of biobank experts integrated a schema of 32 data elements for pooled analysis across the six biobanks with 7461 data elements available: Prevend (The Netherlands),26 NCDS (UK), HUNT (Norway), MICROS (Italy), KORA (Germany), and FINRISK (Finland). First, we calculated precision and recall metrics by comparing the automatically retrieved ‘relevant matches’ with a human curated match set created by the authors. Second, we evaluated the ordering of the results by assessing the ranks of the best matches that were eventually chosen for use in the pooled analysis of this 'healthy obese' study.
Precision and recall
Finding relevant matches out of all possible matches has, at its base, a binary classification. Binary classification performance can be evaluated using the widely accepted measures of precision (the fraction of retrieved instances that are relevant) and recall (also known as sensitivity, the fraction of relevant instances that are retrieved):27
 |
In order to calculate recall, we classified all possible matches between all the 32 desired and all the available data elements, and marked them as relevant or not for five of our biobanks (we excluded the largest). Out of 41 184 possible matches, 420 were classified as relevant (see online supplementary table 1 for the full data).
Prioritization of matches
While precision and recall are good performance measures, not all the relevant matches will be used for data integration. In practice, human experts will decide to use one or two data elements from the list of relevant matches for their research, for example, out of two data elements, ‘weight at baseline’ and ‘weight at year 1,’ only the first might be chosen because baseline data are preferred. Ideally, these best matches should be at the top of the list of relevant matches.
We were fortunate to have a set of 191 manually selected best matches that were used in the pooled analysis of the HOP. See Fortier et al.1 and Doiron et al.24 for the guidelines used for creating the matches, the qualifications of the experts, and the quality assurance procedures. Using this set, we evaluated the prioritization of matches in the results generated by BiobankConnect (see online supplementary table 2 for the full data).
User interface
The harmonization workflow can be summarized as follows:
Upload a target data dictionary containing data elements of interest
Upload one or more source data dictionaries with the data elements available from the biobanks
Either manually or with the help of the annotation wizard, tag the target data dictionary with ontology terms
Choose the target data dictionary as well as the source biobank dictionaries
Automatically produce the shortlist of candidate matches to choose from.
Figure 4 shows an example of the matched results from BiobankConnect for the search elements ‘Parental diabetes mellitus’ and ‘History of hypertension’ in the Prevend and NCDS biobanks, respectively, sorted by Lucene score from high to low. Figure 4A shows that ‘Parental diabetes mellitus’ was matched successfully by using the information from subclasses of ontology term annotations, for example, ‘father’ or ‘mother’ must be a ‘parent.’
Figure 4:
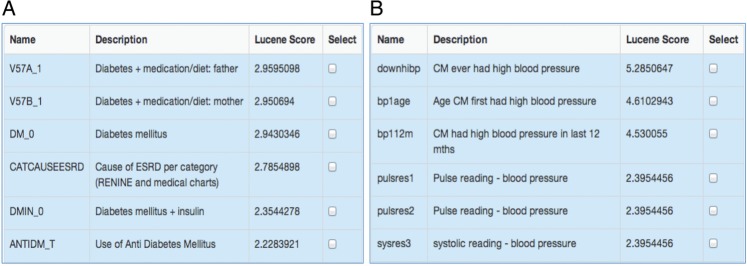
Matching results produced by BiobankConnect. (A) Matching data elements for ‘Parental diabetes mellitus’ in Prevend. The gold standard matches are two data elements, V57A_1 and V57B_1, located in the second and third positions. (B) The matching data element for ‘History of hypertension’ in the NCDS database. The best match in the experts’ opinion is ‘downhibp,’ located in the first position on the candidate list. CM, cohort member.
Figure 4B shows the successful use of synonyms in matching ‘History of hypertension’ in NCDS. Note that the description ‘Ever had high blood pressure’ is quite different from the database term ‘Hypertension,’ which could not be matched automatically by only using string-matching algorithms. However, with the BiobankConnect harmonization method, ‘History of hypertension’ is annotated with the ontology term ‘NCI:Hypertension,’ which has a list of synonyms including ‘High blood pressure,’ and using this knowledge ‘History of hypertension’ was matched with ‘CM [cohort member] ever had high blood pressure’ in NCDS within seconds.
We annotated the data elements with ontology terms (without extensive training or instruction) using a rather simple approach in which as long as any synonyms of the ontology term were similar to the data element description, the ontology term would be used for annotation. For example ‘Parental diabetes mellitus’ was annotated with NCI:parent and NCI:Diabetes Mellitus; the full list of ontology terms and external knowledge annotations for all 32 data elements is given in online supplementary table 3.
RESULTS
Precision and recall of relevant matches
We calculated BiobankConnect's precision and recall for 32 desired data elements across the five biobanks, with a total of 41 184 possible matches, of which 420 were classified as relevant. Overall, we observed an average precision of 0.75 at rank 1 and recall of 0.74, 0.82, and 0.88 at ranks 10, 20, and 50, respectively (see table 1 and figure 5).
Table 1:
Precision and recall performance
| FINRISK |
Hunt |
KORA |
MICROS |
NCDS |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank | P | R | P | R | P | R | P | R | P | R | P | R |
| 1 | 0.91 | 0.50 | 0.61 | 0.16 | 0.88 | 0.53 | 0.73 | 0.27 | 0.59 | 0.17 | 0.75 | 0.28 |
| 2 | 0.68 | 0.72 | 0.65 | 0.34 | 0.67 | 0.79 | 0.53 | 0.37 | 0.48 | 0.27 | 0.60 | 0.44 |
| 3 | 0.57 | 0.88 | 0.59 | 0.46 | 0.48 | 0.83 | 0.45 | 0.46 | 0.37 | 0.30 | 0.49 | 0.52 |
| 4 | 0.45 | 0.90 | 0.53 | 0.55 | 0.40 | 0.89 | 0.39 | 0.52 | 0.31 | 0.33 | 0.42 | 0.58 |
| 5 | 0.39 | 0.95 | 0.47 | 0.60 | 0.34 | 0.92 | 0.33 | 0.56 | 0.27 | 0.36 | 0.36 | 0.62 |
| 6 | 0.34 | 0.97 | 0.42 | 0.64 | 0.31 | 0.96 | 0.30 | 0.61 | 0.25 | 0.39 | 0.32 | 0.65 |
| 7 | 0.29 | 0.97 | 0.39 | 0.69 | 0.27 | 0.96 | 0.27 | 0.63 | 0.23 | 0.41 | 0.29 | 0.68 |
| 8 | 0.26 | 0.97 | 0.37 | 0.73 | 0.25 | 0.98 | 0.25 | 0.67 | 0.21 | 0.44 | 0.27 | 0.71 |
| 9 | 0.23 | 0.97 | 0.35 | 0.77 | 0.24 | 1.00 | 0.24 | 0.68 | 0.19 | 0.44 | 0.25 | 0.72 |
| 10 | 0.22 | 0.98 | 0.33 | 0.81 | 0.22 | 1.00 | 0.22 | 0.70 | 0.17 | 0.44 | 0.23 | 0.74 |
| 11 | 0.20 | 0.98 | 0.31 | 0.82 | 0.21 | 1.00 | 0.21 | 0.71 | 0.16 | 0.44 | 0.22 | 0.75 |
| 12 | 0.19 | 0.98 | 0.29 | 0.83 | 0.20 | 1.00 | 0.20 | 0.72 | 0.15 | 0.45 | 0.21 | 0.75 |
| 13 | 0.18 | 0.98 | 0.27 | 0.84 | 0.19 | 1.00 | 0.19 | 0.74 | 0.14 | 0.46 | 0.20 | 0.76 |
| 14 | 0.17 | 0.98 | 0.25 | 0.84 | 0.18 | 1.00 | 0.19 | 0.77 | 0.14 | 0.47 | 0.19 | 0.77 |
| 15 | 0.16 | 0.98 | 0.24 | 0.85 | 0.17 | 1.00 | 0.19 | 0.79 | 0.13 | 0.49 | 0.18 | 0.78 |
| 16 | 0.15 | 0.98 | 0.23 | 0.86 | 0.16 | 1.00 | 0.18 | 0.82 | 0.13 | 0.50 | 0.17 | 0.80 |
| 17 | 0.14 | 0.98 | 0.22 | 0.86 | 0.16 | 1.00 | 0.18 | 0.84 | 0.13 | 0.51 | 0.17 | 0.79 |
| 18 | 0.14 | 0.98 | 0.21 | 0.87 | 0.15 | 1.00 | 0.18 | 0.85 | 0.12 | 0.51 | 0.15 | 0.81 |
| 19 | 0.13 | 0.98 | 0.20 | 0.87 | 0.14 | 1.00 | 0.17 | 0.87 | 0.12 | 0.52 | 0.16 | 0.81 |
| 20 | 0.13 | 0.98 | 0.19 | 0.88 | 0.14 | 1.00 | 0.17 | 0.87 | 0.11 | 0.53 | 0.14 | 0.82 |
| 30 | 0.09 | 0.98 | 0.13 | 0.91 | 0.11 | 1.00 | 0.14 | 0.93 | 0.08 | 0.57 | 0.11 | 0.85 |
| 50 | 0.06 | 0.98 | 0.09 | 0.94 | 0.10 | 1.00 | 0.11 | 0.96 | 0.06 | 0.64 | 0.08 | 0.88 |
Calculated per biobank and for total.
P, precision; R, recall.
Figure 5:
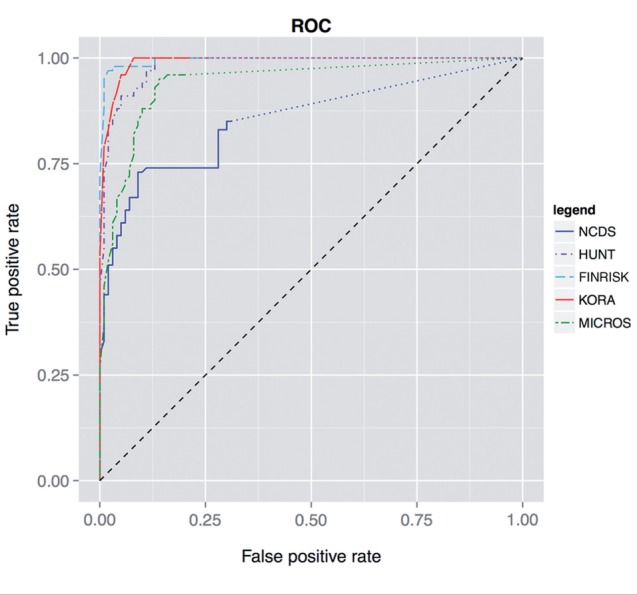
Receiver operating characteristic (ROC) curve. Matching performance for 32 data elements in five different biobanks. Note that BiobankConnect only retrieves a subset of data elements based on the semantic/lexical similarity queries, therefore the ROC curves end before reaching 1.00, 1.00. For the remaining data elements we simulated a line of non-discrimination, indicated by dotted lines.
Rank order of final matches compared with expert decisions
We also evaluated BiobankConnect prioritization performance by evaluating the ranks of the best matches from the BioSHaRE project, that is, the position of the match that the human experts chose from the longer lists of relevant matches. The median rank was 1 and the mean rank was 1.85. Table 2 summarizes the frequencies of the ‘best matches’ per rank. The complete list of BioSHaRE best matches and BiobankConnect's suggested matches is given in online supplementary table 1.
Table 2:
Ranking performance
| Rank | P1 (using ontology) | Cumulative P1 | P2 (Lucene matching) | Cumulative P2 |
|---|---|---|---|---|
| 1 | 63.9% (n = 122) | 63.9% (n = 122) | 51.3% (n = 98) | 51.3% (n = 98) |
| 2 | 14.1% (n = 27) | 78.0% (n = 149) | 12.0% (n = 23) | 63.4% (n = 121) |
| 3 | 8.40% (n = 16) | 86.4% (n = 165) | 8.37% (n = 16) | 71.7% (n = 137) |
| 4 | 3.10% (n = 6) | 89.5% (n = 171) | 4.18% (n = 8) | 75.9% (n = 145) |
| 5 | 3.70% (n = 7) | 93.2% (n = 178) | 5.23% (n = 10) | 81.2% (n = 155) |
| 6 | 3.10% (n = 6) | 96.3% (n = 184) | 1.04% (n = 2) | 82.2% (n = 157) |
| 7 | 0.00% (n = 6) | 96.3% (n = 184) | 0.00% (n = 0) | 82.2% (n = 157) |
| 8 | 1.50% (n = 3) | 97.8% (n = 187) | 1.04% (n = 2) | 83.2% (n = 159) |
| 9 | 0.60% (n = 1) | 98.4% (n = 188) | 2.09% (n = 4) | 85.3% (n = 163) |
| 10 | 0.00% (n = 0) | 98.4% (n = 188) | 0.52% (n = 1) | 85.6% (n = 164) |
| ≥10 | 0.00% (n = 0) | 98.4% (n = 188) | 3.66% (n = 7) | 89.5% (n = 171) |
| Not found | 1.60% (n = 3) | 10.5% (n = 20) | ||
| Total | 100% (n = 191) | 100% (n = 191) |
P1,2 shows the rank of 191 expert selected ‘best’ matches within the automatically produced lists of relevant matches, using ontology annotations of the desired data elements or Lucene matching only, respectively. BiobankConnect predicted ‘best’ matches as first choice (rank 1) in 63.9% of cases and within the ‘top 10’ in 98.4% of cases.
Contribution of ontology annotations
We compared the ranking of ‘best’ matches using ontological and Lucene lexical matching with using lexical matching only (see table 2). Out of 191 matches, using ontology annotations led to 17 matches that would otherwise have been missed, 28 large improvements (4.17 ranks on average), and 7 small decreases (1.71 ranks on average), which were significant changes (p = 0.03, Wilcoxon rank-sum test; see online supplementary tables 4 and 5). In particular, the first rank category increased by 12.6% while other ranks hardly changed (between −1.50% and + 2.60%).
DISCUSSION
While the time spent using BiobankConnect is easily calculated, it is difficult to quantify the time spent by human experts on performing the same task. Instead, we can approximate the gain by estimating by how much BiobankConnect reduces the number of data elements that need manual evaluation by an expert. Obviously, in an ideal world the expert would look at each available data element and decide if it is a suitable match for each of the desired data elements. In the worst case, each expert would have to visit on average half of the total data elements before the ‘best’ match is found. This would be a lot of work, so a more realistic comparison is to assume some smart searching strategies. We used Lucene string matching to simulate a best case where the expert would use advanced lexical searches. Table 3 shows the average ranks of best matches per biobank using BiobankConnect (1.8, with 3 missing), Lucene string matching (2.8, with 20 missing) only, and random searching (3730), respectively. This suggests that BiobankConnect reduces the number of data elements that need to be evaluated by a factor of 1.5 to 2000. The string-matching algorithms miss relevant elements due to non-standard descriptions or unexpected data elements that turn out to be valid proxies.
Table 3:
BiobankConnect reduces the amount of data elements that need to be checked
| Biobank (number of elements) |
R1 (via BiobankConnect) | R2 (string matching) | R3 (random search) |
|---|---|---|---|
| KORA (75) | 1.5 | 1.8 | 36 |
| MICROS (119) | 2.0 | 1.3 | 59 |
| FINRISK (223) | 1.5 | 1.9 | 111 |
| Hunt (353) | 2.5 | 4.1 | 174 |
| NCDS (516) | 1.2 | 1.8 | 260 |
| Prevend (6174) | 2.2 | 4.3 | 3109 |
| Average | 1.8 | 2.7 | 3730 |
| Missed elements | 3 | 20 | 0 |
R1,2,3 shows the average rank of the ‘best’ match when searching using BiobankConnect, using Lucene string matching only, and random iteration, respectively.
We wish to improve BiobankConnect and therefore investigated why recall was worse in, for example, the NCDS biobank, and why some best matches were not ranked as top candidates. We discovered that bad matches were often caused by ‘too many matches,’ ‘repeated measurements,’ ‘too specific data elements,’ or ‘complex proxy variable’ (see online supplementary table 6). We discuss these issues and suggest some solutions below:
The issue of ‘too many matches’ resulted in relatively low recall in NCDS. Scrutiny revealed this was caused by a large number of relevant matches for one particular data element. While for most desired data elements, only one to five NCDS data elements were marked as relevant, 58 elements were relevant for ‘EDU_HIGHEST’ because they all cover some aspect of education. However, BiobankConnect only retrieved 11 out of 58, which had a large impact on the calculation of recall.
The issue with ‘repeated measurements’ occurred in Prevend, where data elements were measured multiple times at different time points. For example, for ‘Current quantity of cigarettes smoked,’ two data elements had been manually matched: ‘V29_4’ with the description ‘Numbers of cigarettes per day’ and ‘V28_1’ with the description ‘Cigarettes or fine-cut tobacco in history or present.’ V29_4 was ranked second in the suggested list, whereas V28_1 was ranked eighth because another six data elements had a similar description to V29_4. This search could be improved using ontology annotations that pinpoint the desired time points.
The issue with ‘too specific data element’ occurred when matching ‘Current quantity of spirits/liquor consumed’ in MICROS. For example, descriptions of the manually determined matches were ‘Quantity of schnapps’ and ‘Previous quantity of schnapps,’ in which ‘schnapps’ is an example of spirits/liquor. However, schnapps had not been defined in any of the ontologies on BioPortal, so it was not recognized as a special type of liquor and was therefore not mapped. This could be addressed by improving details in the current ontologies.
The issue with ‘complex proxy variable’ was due to the proxy data elements used in matching being very difficult to find automatically. For example, ‘Fasting status’ and ‘Blood glucose level’ were measured separately in Prevend and, in addition, ‘Fasting status’ was derived from another two data elements: ‘When was the last meal?’ and ‘When was the last drink?’ Similarly, in NCDS, the data element ‘Blood glucose’ was not measured, but a human expert picked a proxy data element ‘Glycated hemoglobin,’ which is known to correlate with plasma glucose. Matching for these data elements could be improved by using a new ontology that defines such complex relationships between biobank data elements.
In the current version of BiobankConnect, data elements are matched based only on the label or short description of the element, which may result in erroneous matching of some elements. However, biobanks contain more information that is not yet being used. For example, the data element ‘Blood pressure’ was recorded in all our biobanks, but the protocols used to measure blood pressure may differ across biobanks. If detailed protocol descriptions could be provided by the biobanks and incorporated into our system, the matches produced by BiobankConnect could be made more accurate. For the categorical data, information on the various categories could also be used to improve the match. Access to individual-level data could also employ statistical characteristics of the data to evaluate the pooling potential by comparing instance-based matching to schema matching. In addition, the use or development of more biobank-oriented ontologies might improve our system's performance. For example, the problem of ‘too many matches’ for education data elements could be alleviated by using a more specific ontology for the education parameters captured in biobanks.
Finally, we would like to be able to keep track of users’ choices because this human expertise could provide important information to train our system and reproduce the findings thus far. For example, where ‘Fasting glucose’ was manually matched with a proxy variable ‘Glycated hemoglobin’ in NCDS, this relationship could be added to suitable ontologies, so that the information can be re-used for query, thereby developing BiobankConnect into a community knowledge base.
CONCLUSION
Within a matter of minutes BiobankConnect is able to find relevant data element matches with 0.75 precision at rank 1 and 0.74 recall at rank 10. The best matches are in the top 10 in 98.4% of cases. BiobankConnect is therefore a useful tool to speed up the harmonization and integration of data across biobanks, with potential for use in other biomedical integration challenges. A demonstration and the open source software are available at http://www.biobankconnect.org.
ACKNOWLEDGEMENTS
We thank Dany Doiron for providing us with the data dictionaries of the KORA, FINRISK, MICROS, and NCDS biobanks and Kirsti Kvaløy for the data dictionary of the HUNT biobank. We thank Jackie Senior for editing the final text.
CONTRIBUTORS
CP, MS, and HH conceived the methods and designed the software. CP, MD, DH, KJvdV, JK and MS implemented and tested the software. CP, HH, and MS drafted the manuscript. All authors read and agreed with the software and the manuscript.
FUNDING
This work was supported by European Union Seventh Framework Programme (FP7/2007–2013) grant 261433 (Biobank Standardisation and Harmonisation for Research Excellence in the European Union—BioSHaRE-EU) and grant 284209 (BioMedBridges), TI Food and Nutrition grant TIFN GH001, and BBMRI-NL grant 184.021.007, a research infrastructure financed by the Netherlands Organization for Scientific Research (NWO).
COMPETING INTERESTS
None.
PROVENANCE AND PEER REVIEW
Not commissioned; externally peer reviewed.
DATA SHARING STATEMENT
All data and software are available as open source from the demo application (http://www.biobankconnect.org) and source code repository (http://github.com/molgenis), respectively.
REFERENCES
- 1.Fortier I, Doiron D, Little J, et al. Is rigorous retrospective harmonization possible? Application of the DataSHaPER approach across 53 large studies. Int J Epidemiol 2011;40:1314–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortier I, Burton PR, Robson PJ, et al. Quality, quantity and harmony: the DataSHaPER approach to integrating data across bioclinical studies. Int J Epidemiol 2010;39:1383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Euzenat J, Shvaiko P. Ontology Matching. 2nd edn. Berlin: Springer, 2013:333 http://www.springer.com/computer/database+management+&+information+retrieval/book/978-3-642-38720-3 [Google Scholar]
- 4.Abbasi A, Corpeleijn E. External validation of the KORA S4/F4 prediction models for the risk of developing type 2 diabetes in older adults: the PREVEND study. Eur J Epidemiol 2012;27:47–52. [DOI] [PubMed] [Google Scholar]
- 5.Aleksovski Z, Klein M, Ten Kate W, et al. Matching unstructured vocabularies using a background ontology. Lect Notes Comput Sci 2006;4248:182–97. [Google Scholar]
- 6.Giunchiglia F, Shvaiko P. Semantic matching. Knowl Eng Rev 2003;18:265–80. [Google Scholar]
- 7.Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Brief Bioinform 2008;9:75–90. [DOI] [PubMed] [Google Scholar]
- 8.Díaz-Galiano MC, Martín-Valdivia MT, Ureña-López LA. Query expansion with a medical ontology to improve a multimodal information retrieval system. Comput Biol Med 2009;39:396–403. [DOI] [PubMed] [Google Scholar]
- 9.Doms A, Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res 2005;33:W783–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez MDB, Hidalgo JMG, Agudo BD. Using WordNet to complement training information in text categorization. Recent Advances in Natural Language Processing II Selected Papers from the Second International Conference on Recent Advances in Natural Language Processing RANLP 1997 March 2527 1997 Stanford CA USA. arXiv:cmp-lg/9709007v1, 1997:16. [Google Scholar]
- 12.Nilsson K, Hjelm H, Oxhammar H. SUiS—cross-language ontology-driven information retrieval in a restricted domain. In: Proceedings of the 15th NODALIDA Conference, 2005:139–45. [Google Scholar]
- 13.Voorhees EM. Using WordNet to disambiguate word senses for text retrieval. Proceedings of the 16th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, SIGIR 93, 1993:171–80. [Google Scholar]
- 14.Ehrig M. Foam—framework for ontology alignment and mapping; results of the ontology alignment initiative. Proceedings of the Workshop on Integrating Ontologies Volume 156, CEUR-WS.org, 2005:72–6. [Google Scholar]
- 15.Giunchiglia F, Autayeu A, Pane J. S-match: an open source framework for matching lightweight ontologies. Semant Web 2012;3:307–17. [Google Scholar]
- 16.Clinical Information Modeling Initiative (CIMI). http://informatics.mayo.edu/CIMI/index.php/Main_Page (accessed 6 Mar 2014).
- 17.Data Standards Registry and Repository (caDSR). http://cbiit.nci.nih.gov/ncip/biomedical-informatics-resources/interoperability-and-semantics/metadata-and-models (accessed 6 Mar 2014). [Google Scholar]
- 18.Swertz MA, Dijkstra M, Adamusiak T, et al. The MOLGENIS toolkit: rapid prototyping of biosoftware at the push of a button. BMC Bioinformatics 2010;11:S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamusiak T, Parkinson H, Muilu J, et al. Observ-OM and Observ-TAB: universal syntax solutions for the integration, search and exchange of phenotype and genotype information. Hum Mutat 2012;33:867–73. [DOI] [PubMed] [Google Scholar]
- 20.Whetzel PL, Shah NH, Noy NF, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res 2009;37:170–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. P3G Observatory. 2005. http://www.p3gobservatory.org
- 22.Adamusiak T, Burdett T, Kurbatova N, et al. OntoCAT––simple ontology search and integration in Java, R and REST/JavaScript. BMC Bioinformatics 2011;12:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Apache Software Foundation. Apache Lucene. Agenda 2006;2009. http://lucene.apache.org/. Date (accessed 27 Oct 2014). [Google Scholar]
- 24.Doiron D, Burton P, Marcon Y, et al. Data harmonization and federated analysis of population-based studies: the BioSHaRE project. Emerg Themes Epidemiol 2013;10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolffenbuttel B. Healthy obese project, 2013. https://www.bioshare.eu/content/healthy-obese-project [Google Scholar]
- 26.Diercks GF, Van Boven AJ, Hillege HL, et al. Microalbuminuria is independently associated with ischaemic electrocardiographic abnormalities in a large non-diabetic population. The PREVEND (Prevention of REnal and Vascular ENdstage Disease) study. Eur Heart J 2000;21:1922–7. [DOI] [PubMed] [Google Scholar]
- 27.Mao M, Peng Y, Spring M. An adaptive ontology mapping approach with neural network based constraint satisfaction. Web Semant Serv Agents World Wide Web 2010;8:14–25. [Google Scholar]