Abstract
Background Venous thromboembolisms (VTEs), which include deep vein thrombosis (DVT) and pulmonary embolism (PE), are associated with significant mortality, morbidity, and cost in hospitalized patients. To evaluate the success of preventive measures, accurate and efficient methods for monitoring VTE rates are needed. Therefore, we sought to determine the accuracy of statistical natural language processing (NLP) for identifying DVT and PE from electronic health record data.
Methods We randomly sampled 2000 narrative radiology reports from patients with a suspected DVT/PE in Montreal (Canada) between 2008 and 2012. We manually identified DVT/PE within each report, which served as our reference standard. Using a bag-of-words approach, we trained 10 alternative support vector machine (SVM) models predicting DVT, and 10 predicting PE. SVM training and testing was performed with nested 10-fold cross-validation, and the average accuracy of each model was measured and compared.
Results On manual review, 324 (16.2%) reports were DVT-positive and 154 (7.7%) were PE-positive. The best DVT model achieved an average sensitivity of 0.80 (95% CI 0.76 to 0.85), specificity of 0.98 (98% CI 0.97 to 0.99), positive predictive value (PPV) of 0.89 (95% CI 0.85 to 0.93), and an area under the curve (AUC) of 0.98 (95% CI 0.97 to 0.99). The best PE model achieved sensitivity of 0.79 (95% CI 0.73 to 0.85), specificity of 0.99 (95% CI 0.98 to 0.99), PPV of 0.84 (95% CI 0.75 to 0.92), and AUC of 0.99 (95% CI 0.98 to 1.00).
Conclusions Statistical NLP can accurately identify VTE from narrative radiology reports.
Keywords: support vector machines, automated text classification, deep vein thrombosis, pulmonary embolism, acute care hospital, natural language processing
INTRODUCTION
Venous thromboembolism (VTE), which includes deep vein thrombosis (DVT) and pulmonary embolism (PE), is one of the most common complications of hospitalization.1,2 In the absence of thromboprophylaxis, the incidence of VTE ranges from 10–40% in medical and general surgical populations, to as high as 40–60% in patients who have undergone major orthopedic surgical procedures.1 VTE is also a leading cause of mortality and morbidity. The 30-day VTE case fatality rate in hospitalized patients ranges from 5% to 15%.3 Moreover, it is estimated that 15–50% of VTE patients will experience post-thrombotic syndrome,4,5 and that 4–5% will develop chronic thromboembolic pulmonary hypertension.6,7 VTE is the second most common cause of excess length of hospital stay,8 and each case of hospital-acquired VTE results in an incremental inpatient cost of US $7000–$21 000.8,9 Given the significant mortality, morbidity, and cost associated with VTE, its prevention has been ranked as the most important of 79 strategies aimed at improving patient safety in hospitals.10
Accordingly, regulatory agencies worldwide have launched VTE prevention initiatives that aim to encourage hospitals to assess VTE risk, and to institute appropriate thromboprophylaxis in high-risk patients.11,12 An important requirement to evaluate the success of these initiatives is access to accurate, timely, and efficient methods for monitoring VTE rates. However, at present, there are no such methods.13,14 Although manual chart review is the reference standard in many adverse events studies, it is a time-consuming, resource-intensive, and costly process.13–15 As a consequence, it is an impractical means for routine VTE monitoring. While discharge diagnostic codes have the advantage of being readily available, relatively inexpensive, and easy to use,15,16 previous studies have found that they generally have low to moderate sensitivity and positive predictive value (PPV) for identifying VTEs.17,18 In addition, it can be difficult to determine from discharge diagnostic codes whether a VTE occurred before the patient was hospitalized or during the actual hospitalization.19,20 With the increasing availability of electronic health records (EHRs), a far richer source of clinical information for identifying VTEs is becoming available. Moreover, with the advent of automated methods for encoding and classifying electronic narrative documents, such as natural language processing (NLP), an exciting opportunity has emerged to develop potentially more accurate, timely, and efficient methods for monitoring VTE rates.
NLP refers to automated methods for converting free-text data into computer-understandable format.21 NLP techniques have been divided into two broad categories: symbolic and statistical. Symbolic (or grammatical) techniques use the characteristics of the language (i.e., semantics, syntax, and the relationships among sentences) to interpret a narrative document to the extent necessary for encoding it into one of a set of discrete categories.22 Only a few studies have used symbolic NLP techniques to identify adverse events such as VTEs. While the results of these studies are promising, symbolic NLP techniques were found to have low to moderate sensitivity and PPV for identifying DVT and PE.23–25 This could possibly be attributed to the characteristics of clinical narratives, which are often ungrammatical, composed of short telegraphic phrases, and replete with abbreviations, acronyms, and local dialectal shorthand phrases.26,27
Alternatively, statistical NLP techniques use the frequency distribution of words and phrases to automatically classify a set of documents into one of a discrete set of predefined categories.28 Among the various statistical NLP techniques, support vector machines (SVM), which is a type of supervised machine learning, have been widely used in pattern recognition and classification problems.29,30 SVM models have demonstrated high performance in automatically classifying and detecting diseases,31,32 and are one of the most effective models for automated text classification.33 To our knowledge, no prior study has documented their accuracy in identifying VTEs from narrative clinical documents. We thus sought to determine the accuracy of SVM models for identifying DVTs and PEs.
METHODS
Setting
The study was conducted at the McGill University Health Centre (MUHC), a university health network located in the Canadian province of Quebec. The MUHC is composed of five adult-care hospitals and has more than 800 beds. It serves a population of 1.7 million people (22% of the provincial population), with an annual volume of approximately 865 000 ambulatory visits, 34 000 surgeries, and 38 000 hospitalizations. The research ethics committee of the MUHC approved this study.
Data sources
Data for this study were extracted from three electronic databases at the MUHC and were linked by unit, patient, and hospital admission date. The Discharge Abstract Database provided patient age and sex, and dates of hospital admission and discharge. The Admission, Discharge and Transfer Database was used to identify the unit where the patient was located at the time of the radiological examination. The Radiology Report Database provided data on all radiological examinations that were performed over the study period in patients suspected of having a VTE, including dates when these examinations were performed, a text description of the radiological findings, and the radiologist's interpretation. At the time of this study, no other clinical narratives were available in an electronic format at the MUHC.
Study design and data sample
To determine the accuracy of SVM models in identifying DVTs and PEs, we conducted a validation study. First, we randomly sampled 2000 narrative radiology reports among all radiological examinations that were performed at the MUHC between January 1, 2008 and December 31, 2012 for patients with a suspected DVT or PE. Then, two sets of alternative SVM models were trained and tested: one set predicting DVTs (including DVTs of the lower and upper extremities) and one set predicting PEs. To identify the best performing SVM model for predicting DVT and PE, the accuracy of the models within each set was compared. The decision to develop two sets of SVM models was based on the observation that several narrative reports described the results of a radiological examination (e.g., pulmonary embolus study with distal runoff) that was performed in a patient suspected of having both conditions. While it is possible to train a multi-class SVM model, most work on SVMs and most standard evaluation techniques are designed for binary classification problems.34 As such, we opted for the two-model approach.
SVM model development and validation
To develop and validate the two sets of SVM models, four successive steps were followed: (a) reference standard development, (b) text pre-processing and feature generation, (c) feature selection, and (d) SVM training, testing, and validation.
Reference standard development
First, the 2000 radiology reports were manually coded by a clinical expert (CMR) to identify cases of DVT and PE. During the coding process, each report was assigned two codes: (a) positive or negative for DVT of the lower or upper extremities, and (b) positive or negative for PE.
Positive radiology reports for a DVT were those where a thrombus was identified in the proximal deep veins of the lower extremities (e.g., external iliac, common femoral, deep femoral, or popliteal), in the deep distal veins of the lower extremities (e.g., peroneal and posterior tibial), or in the deep veins of the upper extremities (e.g., brachial, radial, ulnar, axillary, or subclavian). Negative cases included those where no thrombus was identified or where a thrombus was identified in a superficial vein of the lower extremity (e.g., saphenous), in a superficial vein of the upper extremity (e.g., cephalic), or in a perforating vein of the lower extremity but not extending into a deep vein.17 Radiological examinations finding evidence of chronic thrombosis were coded as negative.
Similarly, positive radiology reports for a PE included those where a filling defect was identified in the central, segmental, or subsegmental pulmonary arteries. Radiological reports describing evidence of chronic PE were coded as negative, as were those finding no evidence of the disease. Lastly, a second clinical expert (TE) blindly recoded a 20% random sample of the radiology reports and inter-coder reliability was assessed using the κ statistic, yielding near perfect agreement (κ = 98).
Text preprocessing and feature generation
In preparation for SVM training and validation, the unstructured text data embedded in the narrative radiology reports were transformed into a corpus, which is a database of text documents.35 Then, a series of transformations were applied to each radiology report within the corpus, including: (a) conversion of all words to lower case, and (b) the removal of punctuation marks and superfluous white spaces.33 The transformed corpus was converted into a document-by-term matrix (bag-of-words),36 a structured format holding radiology report IDs as rows, terms as columns, and term frequencies within a given radiology report as matrix elements. The original bag-of-words contained 7370 distinct features (i.e., words or unigrams). To introduce some elements of contextual knowledge, and preserve the local dependencies of each word, bigrams (which are combinations of two consecutive words) were introduced into the bag-of-words as additional features.37 This resulted in a total of 62 416 distinct features (i.e., unigrams and bigrams). Text preprocessing was conducted in R with package tm.38
Lastly, because feature generation is a key determinant of SVM model prediction performances,39 we experimented with several potential enhancements to our proposed feature generation approach, including word stemming (i.e., reducing inflected words to their root form) and using higher order n-grams (e.g., trigrams). These potential enhancements were evaluated as part of the SVM model training and validation.
Feature selection and SVM training and validation
To assess the accuracy of the SVM models for identifying DVT and PE, we used a 10-fold cross-validation approach. To avoid biasing the accuracy of the SVM models by using information from the test sets, we first performed feature selection within each set of k−1 training folds.34,40 We used the Pearson's correlation coefficient (ρ) to identify subsets of features significantly associated with DVT and PE.39 Using a threshold value of |ρ|≥0.10, 118 unigrams and 218 bigrams were associated with DVT, and 301 unigrams and 1242 bigrams were associated with PE. The top 30 most informative features (highest values of ρ) are summarized in table 1. As can be seen from table 1, most of these features are either related to: (a) a relevant anatomical body part (e.g., vein, pulmonary artery, femoral vein, or segmental branches (of the pulmonary arteries)), or (b) pathological manifestations of DVT or PE (e.g., thrombus, emboli, or filling defect).
Table 1:
The top 30* most informative unigrams and bigrams for deep vein thrombosis and pulmonary embolism identification according to the Pearson's correlation statistic
| Deep vein thrombosis |
Pulmonary embolism |
||
|---|---|---|---|
| Unigrams | Bigrams | Unigrams | Bigrams |
| Vein | Length of | Filling | Pulmonary artery |
| Thrombus | The thrombus | Segmental | Filling defect |
| Occlusive | Thrombosis involving | Artery | Lower lobe |
| Peroneal | Popliteal vein | Lobe | Defect in |
| Length | Over a | Pulmonary | Pulmonary emboli |
| Patent | Non occlusive | Defect | A filling |
| Popliteal | Posterior tibial | Subsegmental | There are |
| Over | A length | Strain | With pulmonary |
| Femoral | Is deep | Emboli | Upper lobe |
| Tibial | Peroneal vein | Chest | Main pulmonary |
| Thrombosis | The mid | Lung | Segmental branch |
| Veins | Basilic vein | Main | Defects are |
| Involving | Femoral vein | Branches | Segmental branches |
| Entire | Entire length | Basal | Embolus in |
| Thrombosed | Reminder of | Small | Multiple filling |
*Features were ranked using their Pearson correlation coefficient (ρ) and the top 30 unigrams and bigrams were selected.
Then, using these subsets of features, a nested 10-fold cross-validation was performed within each set of k−1 training folds to identify the optimal value of: (a) the cost parameter C, which controls the trade-off between false positives and false negatives (used with all kernel functions), and (b) the γ parameter, which controls the shape of the hyperplane (used with the radial basis function (RBF) kernel only).34,41 The optimal value of C and γ (when applicable) were then automatically applied to the relevant set of k−1 training folds, and a parametrically optimized multivariate SVM model was trained using either a linear or an RBF kernel.40 Lastly, this optimal SVM model was tested on the remaining kth test fold. During SVM training and validation, repeated radiology reports from the same patient over any given hospitalization (if any) were not allowed to cross the training and test sets to avoid inflating the performances of the SVM models.
Within each test fold, we estimated the sensitivity, specificity, PPV, negative predictive value (NPV), and the area (AUC) under the receiver operating characteristic (ROC) curve. These estimates were then averaged over the 10 folds, and their 95% CIs estimated.42 For each test fold, an ROC curve was generated by using the classification probabilities produced by the SVM models as the discrimination threshold.43 Vertical averaging was used to generate the average ROC curve for each of the two models.43 To account for the asymmetric class sizes (i.e., fewer positive cases of DVT and PE), and to avoid biasing the SVM predictions towards the majority class, each class was weighted by the inverse proportion.34 Then, using this general approach, 10 alternative models were trained and tested. The first eight models were trained using the aforementioned selected subsets of features and were characterized by their kernel function (linear vs RBF), gram type (unigrams only vs unigram and bigrams), and the use or non-use of SVM parameter optimization (tuning vs no tuning). SVM models with no tuning used the default values for the cost (C = 1) and γ (γ = 1/number of features) parameters.34 Two additional SVM models were trained and tested using all available features: (a) all features, linear kernel, uni-bigrams, and tuned parameters, or (b) all features, RBF kernel, uni-bigrams, and tuned parameters. To identify the best performing model, the AUCs of these 10 SVM models were compared using the Friedman test, and pairwise comparisons were performed using Tukey's test with the Bonferroni adjustment for multiple comparisons.34 To determine if potential enhancements to our proposed feature generation approach (i.e., stemming and higher order n-grams) influenced SVM prediction performances, alternative SVM models were trained, tested, and compared using the aforementioned procedures. SVM models were implemented in R using package e1071.44
RESULTS
Overall, 1649 patients contributed 1751 hospitalizations from which the 2000 randomly selected narrative radiology reports were generated. A total of 1544 (88.2%) hospitalizations contributed only one radiology report to the analyses, while 207 (11.8%) hospitalizations contributed between two and five radiology reports (table 2). The other characteristics of these hospitalizations are summarized in table 2.
Table 2:
Characteristics of the 1751 hospitalizations which contributed the 2000 narrative radiology reports
| Hospitalization characteristics | Statistics (n = 1751) |
|---|---|
| Demographic characteristics | |
| Sex | |
| Male, n (%) | 892 (50.9) |
| Female, n (%) | 859 (49.1) |
| Age, mean ± SD | 66.7 ± 16.1 |
| Nursing unit at the time of the examination | |
| Internal medicine, n (%) | 643 (36.7) |
| Surgery, n (%) | 537 (30.7) |
| Intensive care unit, n (%) | 332 (19.0) |
| Other (e.g., geriatrics, neurology, short stay), n (%) | 239 (13.6) |
| Length of hospital stay (days), median (IQR) | 15 (28) |
| Number of radiology reports contributed to the analyses | |
| One radiology report, n (%) | 1544 (88.2) |
| Two radiology reports, n (%) | 173 (9.9) |
| Three to five radiology reports, n (%) | 34 (1.9) |
The typical radiology report had a median length of 110.5 words, ranging from 8 to 727 words. On manual review, 454 narrative radiology reports (22.7%) described a VTE. Of these, 324 reports (16.2%) described a DVT, including 216 DVTs of the lower extremities and 108 DVTs of the upper extremities. In addition, 154 reports (7.7%) described a PE. Notably, 24 reports described 2 simultaneous events, the most prevalent combination being a lower extremity DVT and a PE (n = 19).
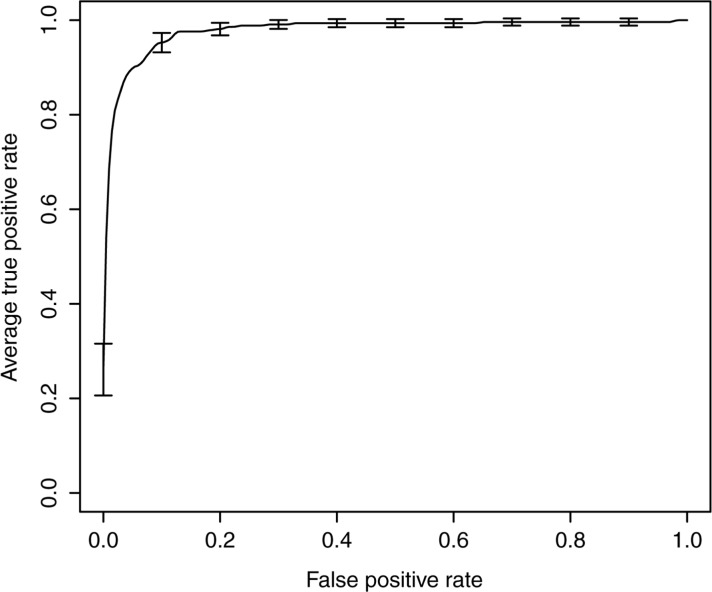
The average sensitivity, specificity, PPV, and NPV of the SVM models for identifying DVTs are shown in table 3. On average, the best performing model correctly identified 80% of the radiology reports describing a true DVT (sensitivity: 0.80 (95% CI 0.76 to 0.85)) and generated 11% of false positives (PPV: 0.89 (95% CI 0.85 to 0.93)). The average AUC of this model was 0.98 (95% CI 0.97 to 0.99), and the associated average ROC curve is presented in figure 1. These performances, which are highlighted in boldface in table 3, were achieved on an SVM model trained on the whole feature set using an RBF kernel for which both the cost and γ parameters were optimized. The performance of this SVM model did not statistically differ from that of similar SVM models trained using a linear kernel function on either the whole feature set or a selected subset of features (table 3). It was also not statistically significantly influenced by feature selection (table 3). Nonetheless, it performed statistically significantly better than any other alternative SVM models (table 3).
Table 3:
Average accuracy estimates of the SVM models for identifying deep vein thrombosis (DVT)
| Average estimates (95% CI)† |
|||||
|---|---|---|---|---|---|
| SVM models | Sensitivity | Specificity | PPV | NPV | AUC |
| Unigram only, linear kernel, no tuning | 0.65 (0.58 to 0.71) | 0.97 (0.96 to 0.98) | 0.82 (0.77 to 0.87) | 0.93 (0.90 to 0.96) | 0.94 (0.92 to 0.96)‡ |
| Unigram only, RBF kernel, no tuning | 0.57 (0.45 to 0.68) | 0.96 (0.95 to 0.97) | 0.72 (0.66 to 0.79) | 0.92 (0.89 to 0.95) | 0.93 (0.91 to 0.96)‡ |
| Unigram only, linear kernel, with tuning | 0.69 (0.64 to 0.73) | 0.97 (0.96 to 0.98) | 0.82 (0.77 to 0.87) | 0.94 (0.91 to 0.97) | 0.95 (0.94 to 0.97)‡ |
| Unigram only, RBF kernel, with tuning | 0.70 (0.65 to 0.74) | 0.97 (0.96 to 0.98) | 0.81 (0.74 to 0.88) | 0.94 (0.92 to 0.97) | 0.95 (0.92 to 0.97)‡ |
| Uni + bigrams, linear kernel, no tuning | 0.67 (0.63 to 0.73) | 0.98 (0.97 to 0.99) | 0.87 (0.82 to 0.93) | 0.94 (0.90 to 0.97) | 0.95 (0.93 to 0.98) |
| Uni + bigrams, RBF kernel, no tuning | 0.59 (0.49 to 0.69) | 0.96 (0.95 to 0.97) | 0.74 (0.66 to 0.82) | 0.92 (0.89 to 0.96) | 0.94 (0.92 to 0.96)‡ |
| Uni + bigrams, linear kernel, tuning | 0.70 (0.63 to 0.77) | 0.98 (0.97 to 0.99) | 0.85 (0.81 to 0.90) | 0.94 (0.91 to 0.97) | 0.96 (0.93 to 0.98) |
| Uni + bigrams, RBF kernel, tuning | 0.77 (0.72 to 0.84) | 0.97 (0.97 to 0.98) | 0.84 (0.79 to 0.89) | 0.96 (0.92 to 0.99) | 0.97 (0.94 to 0.99) |
| Uni + bigrams, linear kernel, tuning, all features. | 0.79 (0.74 to 0.84) | 0.98 (0.97 to 0.99) | 0.89 (0.84 to 0.94) | 0.96 (0.93 to 0.98) | 0.98 (0.97 to 0.99) |
| Uni + bigrams, RBF kernel, tuning, all features | 0.80 (0.76 to 0.85) | 0.98 (0.97 to 0.99) | 0.89 (0.85 to 0.93) | 0.96 (0.93 to 0.99) | 0.98 (0.97 to 0.99)* |
Bold typeface is used to highlight the characteristics of the best performing SVM model. *p<0.001; statistically significant difference in performance compared to alternative SVM models.
†Averages correspond to the mean accuracy estimates obtained after 10 rounds of cross-validation.
‡Statistically significantly different compared to the best performing SVM model (i.e., Uni + bigrams, RBF kernel, tuning, all features).
AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value; RBF, radial basis function kernel; SVM, support vector machine.
Figure 1:
Average receiver operator characteristic (ROC) curve associated with the deep vein thrombosis (DVT) model. The average ROC curve for the DVT model was estimated by vertically averaging the 10 ROC curves generated during 10-fold cross-validation. The best performances were achieved using an SVM model trained on the whole feature set using an RBF kernel for which both the cost and γ parameters were optimized.
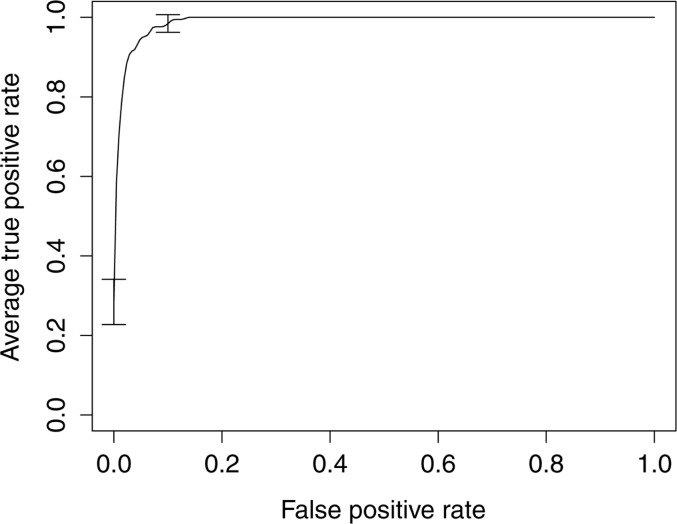
Similarly, on average, the best SVM model predicting PE correctly identified 79% of the true PEs (sensitivity: 0.79 (95% CI 0.73 to 0.85)) and generated 16% of false positives (PPV: 0.84 (95% CI 0.75 to 0.92)) (table 4). This model had an AUC of 0.99 (95% CI 0.98 to 1.00), and the associated average ROC curve is shown in figure 2. These performances were achieved on an SVM model trained on the whole feature set using an RBF kernel for which both the cost and γ parameters were optimized (table 4). The performance of this SVM model did not significantly differ from that of similar SVM models trained using a linear kernel function on either the whole feature set or a selected subset of features (table 4). This model was also not statistically significantly influenced by feature selection (table 4). Nonetheless, this model performed statistically significantly better than any other alternative SVM models (table 4).
Table 4:
Average accuracy estimates of the SVM models for identifying pulmonary embolism (PE)
| Average estimates (95% CI)‡ |
|||||
|---|---|---|---|---|---|
| SVM models | Sensitivity | Specificity | PPV | NPV | AUC |
| Unigram only, linear kernel, no tuning | 0.51 (0.34 to 0.68) | 0.99 (0.98 to 1.00) | 0.75 (0.62 to 0.88) | 0.96 (0.95 to 0.98) | 0.92 (0.87 to 0.96)† |
| Unigram only, RBF kernel, no tuning | 0.39 (0.32 to 0.46) | 0.98 (0.97 to 0.99) | 0.63 (0.45 to 0.81) | 0.95 (0.94 to 0.96) | 0.93 (0.92 to 0.95)† |
| Unigram only, linear kernel, with tuning | 0.53 (0.36 to 0.70) | 0.98 (0.97 to 0.99) | 0.75 (0.65 to 0.85) | 0.96 (0.94 to 0.98) | 0.92 (0.88 to 0.96)† |
| Unigram only, RBF kernel, with tuning | 0.55 (0.42 to 0.67) | 0.98 (0.97 to 0.99) | 0.70 (0.54 to 0.86) | 0.96 (0.95 to 0.98) | 0.95 (0.93 to 0.97)† |
| Uni + bigrams, linear kernel, no tuning | 0.60 (0.45 to 0.75) | 0.99 (0.98 to 1.00) | 0.85 (0.77 to 0.93) | 0.97 (0.95 to 0.98) | 0.95 (0.90 to 1.00) |
| Uni + bigrams, RBF kernel, no tuning | 0.40 (0.33 to 0.47) | 0.98 (0.97 to 0.99) | 0.67 (0.51 to 0.83) | 0.95 (0.94 to 0.96) | 0.95 (0.93 to 0.96)† |
| Uni + bigrams, linear kernel, tuning | 0.61 (0.46 to 0.76) | 0.99 (0.98 to 1.00) | 0.84 (0.76 to 0.92) | 0.97 (0.95 to 0.98) | 0.95 (0.90 to 1.00) |
| Uni + bigrams, RBF kernel, tuning | 0.66 (0.49 to 0.83) | 0.99 (0.98 to 1.00) | 0.80 (0.68 to 0.93) | 0.97 (0.96 to 0.99) | 0.96 (0.92 to 1.00) |
| Uni + bigrams, linear kernel, tuning, all features | 0.78 (0.72 to 0.85) | 0.99 (0.98 to 0.99) | 0.84 (0.76 to 0.91) | 0.98 (0.98 to 0.99) | 0.99 (0.98 to 1.00) |
| Uni + bigrams, RBF kernel, tuning, all features | 0.79 (0.73 to 0.85) | 0.99 (0.98 to 0.99) | 0.84 (0.75 to 0.92) | 0.98 (0.98 to 0.99) | 0.99 (0.98 to 1.00)* |
Bold typeface is used to highlight the characteristics of the best performing SVM model. *p<0.001; statistically significant difference in performance compared to alternative SVM models.
†Statistically significantly different compared to the best performing SVM model (i.e., Uni + bigrams, RBF kernel, tuning, all features).
‡Averages correspond to the mean accuracy estimates obtained after 10 rounds of cross-validation.
AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value; RBF, radial basis function kernel; SVM, support vector machine.
Figure 2:
Average receiver operator characteristic (ROC) curve associated with the pulmonary embolism (PE) model. The average ROC curve for the PE model was estimated by vertically averaging the 10 ROC curves generated during 10-fold cross-validation. The best performances were achieved using an SVM model trained on the whole feature set using an RBF kernel for which both the cost and γ parameters were optimized.
As a final step, alternative SVM models predicting DVT and PE were trained and tested using potential enhancements to the proposed feature representation (i.e., word stemming and including trigrams). However, none of these approaches were superior to using a bag-of-words representation enhanced with bigram features. As such, they are not detailed any further.
DISCUSSION
In this study, we assessed the accuracy of a statistical NLP technique, based on SVM models, for the purpose of identifying VTEs from electronic narrative radiology reports. We found that DVTs and PEs can be accurately identified from a bag-of-words representation of narrative radiology reports that is augmented with bigram features.
To our knowledge, there have been few studies using NLP techniques for the purpose of identifying VTEs, and most relied on symbolic NLP. For instance, Melton and Hripcsak23 assessed whether symbolic NLP techniques could accurately identify 45 adverse events, including DVTs, from narrative discharge summaries. For DVTs, a PPV of 0.51 was observed. In another study, Murff et al.24 evaluated the accuracy of symbolic NLP techniques for identifying several postoperative complications, including VTEs, from an integrated EHR at six Veterans Health Administration (VHA) medical centers. The sensitivity of the NLP system for identifying VTEs was 0.59 (95% CI 0.44 to 0.72) and the specificity was 0.91 (95% CI 0.90 to 0.92). In a subsequent study based on a larger sample of VHA patients, FitzHenry et al.25 used symbolic NLP techniques to detect nine post-operative complications, including DVT and PE. For DVT, sensitivity was 0.56 (95% CI 0.45 to 0.67), specificity was 0.94 (95% CI 0.93 to 0.95), and PPV was 0.15 (95% CI 0.11 to 0.20). For PE, a sensitivity of 0.80 (95% CI 0.66 to 0.89), a specificity of 0.97 (95% CI 0.96 to 0.98), and a PPV of 0.23 (95% CI 0.17 to 0.30) were observed. Overall, the results of our study add to this emerging body of literature and provide further evidence that automated methods based on NLP techniques can successfully be applied to EHR data for the purpose of identifying adverse events such as DVT and PE.
Interestingly, we were able to achieve relatively good prediction performances for both DVTs and PEs using a bag-of-words representation of the narrative radiology reports that was augmented with bigram features. While these performances compare to those noted in recent statistical text classification studies,45–48 some characteristics of the models validated in this study should be emphasized. First, we found, in contrast to Bejan et al.46 that feature selection did not significantly increase the performances of the SVM models beyond what can be achieved with bigrams and SVM parameter optimization. This observation provides evidence than when computational resources are limited, feature selection, as performed in this study, may provide acceptable results. Second, we observed, consistent with Hsu et al.49 that when SVM parameters are optimized, both the linear and RBF kernel achieve similar performances. Lastly, we observed that several potential enhancements to the feature representation (e.g., using higher order n-grams) did not improve the SVM model predictive performances. Similar results were observed in a recent study where SVM models were used to categorize EHR progress notes pertaining to diabetes.48 It is possible that more complex features, such as higher order n-grams, do not occur in a high enough frequency to significantly improve the performance of an SVM model. Moreover, it is possible that these alternative features do not contain the information needed to discriminate narrative reports from different classes. Recently, Garla and Brandt50 used symbolic NLP techniques to encode semantic relationships between concepts in the narrative reports, and were able to improve the prediction performance of a machine learning clinical text classifier pertaining to obesity. Additional studies are needed to determine if this approach can be successfully replicated in other clinical contexts, including the identification of VTEs. Nonetheless, the combination of symbolic and statistical NLP techniques represents an interesting area of future research, and promises improved performance in adverse event detection.
Automated methods based on statistical or symbolic NLP techniques have a number of advantages over traditional adverse event monitoring methods, such as manual chart review. Because they are automated, they can rapidly scan large numbers of patient records and clinical data with minimal human effort and cost, potentially allowing surveillance of an entire healthcare organization's population rather than just subsamples. In addition to being scalable, another potential benefit of automation is that human resources traditionally assigned to manual adverse event monitoring could be more productively reassigned to the development, implementation, and follow-up of preventive interventions. Moreover, because NLP techniques use electronic data that are available in near-real time, they offer the potential for timely adverse event monitoring and for prompt intervention. This represents an important advantage over automated methods based on discharge diagnostic codes, which are only available several months after discharge.
An important strength of this study is the approach used to fit the SVM models. Indeed, feature selection performed on the whole dataset would have created a positive bias in prediction accuracy by indirectly using information from the test sets. To guard against this, we performed feature selection within each set of k−1 training folds, which ensured independence from the test sets. Similarly, we performed a nested 10-fold cross-validation within each set of k−1 training folds to identify the optimal values for the SVM parameters (C and γ). This approach penalized over-fit models during the parameter search process, therefore increasing generalizability. Lastly, we reported on the average accuracy estimates of the SVM models over the 10 test folds. This reduced the influence that any bias in the distribution of the data between the training and test sets could have on the predictive performance of the SVM models. It also provided more reliable insights on how the SVM models would perform on an independent data set drawn from the same population of radiology reports.
Another strength of this study was to train and validate the SVM models on narrative radiology reports from a network of five adult-care hospitals. Because these hospitals operate for the most part independently, and have their dedicated pool of radiologists, this provided us with a range of reporting practices and styles (as observed during the manual coding process). Moreover, by randomly sampling radiology reports over a 5-year period, we could also capture variations through time, if any, in reporting practices by the radiologists. For these reasons, we are confident that our SVM models are relatively robust to variations in reporting styles.
Despite these strengths, several limitations of this study should be acknowledged. First, the SVM models were trained and tested on a single source of data: electronic narrative radiology reports. It is thus possible that their accuracy would have been different if additional sources of clinical data had been included in the models. However, at the time of this study, there was no integrated EHR available at the MUHC, and discharge summaries were still in paper format. As a consequence, no other source of electronic clinical narratives was available to us. While the adoption of integrated EHRs is improving, only a small minority of institutions currently use them. As such, the approach taken in this study probably represents what could be realistically implemented in most hospitals across the USA and Canada. Second, we have focused on a single condition—VTEs. As such, we have not tested our proposed approach on other types of adverse events. Nonetheless, we have provided evidence that SVM models can accurately detect the two clinical manifestations of VTE (i.e., DVTs and PEs) with similar prediction performances. While we believe that the approach taken in this study can be generalized to other types of adverse events, this has yet to be demonstrated.
CONCLUSION
We found that SVM models based on narrative radiology reports can identify VTEs with high accuracy. The SVM models developed and validated in this study have many potential applications. Accurate identification of VTEs could assist hospital quality improvement staff monitor VTE rates, and evaluate the effectiveness of preventive interventions. Future studies should assess if the approach used in this study can be generalized to the detection of other types of adverse events, and if the combination of symbolic and statistical NLP techniques results in higher prediction performances than either method used alone.
CONTRIBUTORS
All listed authors: have made substantial contributions to conception and design, or acquisition of data, or analysis and interpretation of data; have been involved in drafting the manuscript or revising it critically for important intellectual content; and have given final approval of the version to be published.
FUNDING
This study was funded by the Canadian Institutes of Health Research (CIHR).
COMPETING INTERESTS
None.
ETHICS APPROVAL
McGill University Health Centre Research Ethics Committee approved this study.
PROVENANCE AND PEER REVIEW
Not commissioned; externally peer reviewed.
REFERENCES
- 1.Geerts WH, Bergqvist D, Pineo GF, et al. Prevention of venous thromboembolism: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition). Chest 2008;133(6 Suppl):381S–453S. [DOI] [PubMed] [Google Scholar]
- 2.Spyropoulos AC, Hussein M, Lin J, et al. Rates of venous thromboembolism occurrence in medical patients among the insured population. Thromb Haemost 2009;102:951–7. [DOI] [PubMed] [Google Scholar]
- 3.White RH. The epidemiology of venous thromboembolism. Circulation 2003;107(23 Suppl 1):I4–8. [DOI] [PubMed] [Google Scholar]
- 4.Ginsberg JS, Hirsh J, Julian J, et al. Prevention and treatment of postphlebitic syndrome: results of a 3-part study. Arch Intern Med 2001;161:2105–9. [DOI] [PubMed] [Google Scholar]
- 5.Kahn SR, Kearon C, Julian JA, et al. Predictors of the post-thrombotic syndrome during long-term treatment of proximal deep vein thrombosis. J Thromb Haemost 2005;3:718–23. [DOI] [PubMed] [Google Scholar]
- 6.Pengo V, Lensing AW, Prins MH, et al. Incidence of chronic thromboembolic pulmonary hypertension after pulmonary embolism. N Engl J Med 2004;350:2257–64. [DOI] [PubMed] [Google Scholar]
- 7.Korkmaz A, Ozlu T, Ozsu S, et al. Long-term outcomes in acute pulmonary thromboembolism: the incidence of chronic thromboembolic pulmonary hypertension and associated risk factors. Clin Appl Thromb Hemost 2012;18:281–8. [DOI] [PubMed] [Google Scholar]
- 8.Zhan C, Miller MR. Excess length of stay, charges, and mortality attributable to medical injuries during hospitalization. JAMA 2003;290:1868–74. [DOI] [PubMed] [Google Scholar]
- 9.Dobesh PP. Economic burden of venous thromboembolism in hospitalized patients. Pharmacotherapy 2009;29:943–53. [DOI] [PubMed] [Google Scholar]
- 10.Shojania KG, Duncan BW, McDonald KM, et al. Making health care safer: a critical analysis of patient safety practices. Evid Rep Technol Assess (Summer) 2001;(43):i–x, 1–668. [PMC free article] [PubMed] [Google Scholar]
- 11.The Joint Commission. Specifications Manual for National Hospital Inpatient Quality Measures. 2014. http://www.jointcommission.org/specifications_manual_for_national_hospital_inpatient_quality_measures.aspx (accessed 11 Feb 2014).
- 12.Kahn SR, Morrison DR, Cohen JM, et al. Interventions for implementation of thromboprophylaxis in hospitalized medical and surgical patients at risk for venous thromboembolism. Cochrane Database Syst Rev 2013;7:CD008201. [DOI] [PubMed] [Google Scholar]
- 13.Govindan M, Van Citters AD, Nelson EC, et al. Automated detection of harm in healthcare with information technology: a systematic review. Qual Saf Health Care 2010;19:e11. [DOI] [PubMed] [Google Scholar]
- 14.Murff HJ, Patel VL, Hripcsak G, et al. Detecting adverse events for patient safety research: a review of current methodologies. J Biomed Inform 2003;36:131–43. [DOI] [PubMed] [Google Scholar]
- 15.Klompas M, Yokoe DS. Automated surveillance of health care-associated infections. Clin Infect Dis 2009;48:1268–75. [DOI] [PubMed] [Google Scholar]
- 16.Bates DW, Evans RS, Murff H, et al. Detecting adverse events using information technology. J Am Med Inform Assoc 2003;10:115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson KE, Recktenwald A, Reichley RM, et al. Clinical validation of the AHRQ postoperative venous thromboembolism patient safety indicator. Jt Comm J Qual Patient Saf 2009;35:370–6. [DOI] [PubMed] [Google Scholar]
- 18.White RH, Sadeghi B, Tancredi DJ, et al. How valid is the ICD-9-CM based AHRQ patient safety indicator for postoperative venous thromboembolism? Med Care 2009;47:1237–43. [DOI] [PubMed] [Google Scholar]
- 19.Houchens RL, Elixhauser A, Romano PS. How often are potential patient safety events present on admission? Jt Comm J Qual Patient Saf 2008;34:154–63. [DOI] [PubMed] [Google Scholar]
- 20.Bahl V, Thompson MA, Kau TY, et al. Do the AHRQ patient safety indicators flag conditions that are present at the time of hospital admission? Med Care 2008;46:516–22. [DOI] [PubMed] [Google Scholar]
- 21.Allen J. Natural language understanding Redwood City, CA: Benjamin/Cummings Publishing Company, 1995. [Google Scholar]
- 22.Chapman WW. Natural language processing for biosurveillance. In: Wagner MM, Moore AW, Aryel RM, eds. Handbook of biosurveillance. Burlington, MA: Elsevier Academic Press, 2006:255–71. [Google Scholar]
- 23.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc 2005;12:448–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA 2011;306:848–55. [DOI] [PubMed] [Google Scholar]
- 25.FitzHenry F, Murff HJ, Matheny ME, et al. Exploring the frontier of electronic health record surveillance: the case of postoperative complications. Med Care 2013;51:509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeves RM, Ong FR, Matheny ME, et al. Detecting temporal expressions in medical narratives. Int J Med Inform 2013;82:118–27. [DOI] [PubMed] [Google Scholar]
- 27.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008;47(Suppl 1):128–42. [PubMed] [Google Scholar]
- 28.Mitchell TM. Machine learning Boston, MA: McGraw-Hill, 1997. [Google Scholar]
- 29.Cortes C, Vapnik V. Support vector network. Mach Learn 1995;20:273–97. [Google Scholar]
- 30.Vapnik V. Statistical learning theory New York, NY: Wiley, 1998. [Google Scholar]
- 31.Yu W, Liu T, Valdez R, et al. Application of support vector machine modeling for prediction of common diseases: the case of diabetes and pre-diabetes. BMC Med Inform Decis Mak 2010;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maglogiannis I, Loukis E, Zafiropoulos E, et al. Support Vectors Machine-based identification of heart valve diseases using heart sounds. Comput Methods Programs Biomed 2009;95:47–61. [DOI] [PubMed] [Google Scholar]
- 33.Thorsten J. Learning to classify text using support vector mac hines. Methods, theory and algorithms. Norwell, MA: Kluwer, 2002. [Google Scholar]
- 34.Japkowicz N, Shah M. Evaluating learning algorithms: a classification perspective 1st edn New York, NY: Cambridge University Press, 2011. [Google Scholar]
- 35.Manning C, Schutze H. Foundations of statistical natural language processing 1st edn Cambridge: MIT Press, 2003. [Google Scholar]
- 36.Feinerer I, Hornik K, Meyer D. Text mining infrastructure in R. J Stat Software 2008;25:1–54. [Google Scholar]
- 37.Conway M, Doan S, Kawazoe A, et al. Classifying disease outbreak reports using n-grams and semantic features. Int J Med Inform 2009;78:e47–58. [DOI] [PubMed] [Google Scholar]
- 38.Feinerer I. Tm: Text mining package. R package version 0.5–6. 2012 February 15. http://cran.r-project.org/web/packages/tm/tm.pdf (accessed 14 Apr 2013).
- 39.Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007;23:2507–17. [DOI] [PubMed] [Google Scholar]
- 40.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning. Data mining, inference, and prediction 2nd edn New York, NY: Springer, 2009. [Google Scholar]
- 41.Karatzoglou A, Meyer D, Hornik K. Support vector machines in R. J Stat Software 2012;15:1–28. [Google Scholar]
- 42.Nadeau C, Bengio Y. Inference for the generalization error. Mach Learn 2003;52:239–81. [Google Scholar]
- 43.Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett 2006;27:861–74. [Google Scholar]
- 44.Dimitriadou E, Hornik K, Leisch F, et al. Package ‘e1071’. Vienna, Austria: Technische Universität, Department of Statistics, 14 February 2012. http://cran.r-project.org/web/packages/e1071/e1071.pdf (accessed 14 Apr 2013).
- 45.Garla V, Taylor C, Brandt C. Semi-supervised clinical text classification with Laplacian SVMs: an application to cancer case management. J Biomed Inform 2013;46:869–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bejan CA, Xia F, Vanderwende L, et al. Pneumonia identification using statistical feature selection. J Am Med Inform Assoc 2012;19:817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCart JA, Berndt DJ, Jarman J, et al. Finding falls in ambulatory care clinical documents using statistical text mining. J Am Med Inform Assoc 2013;20:906–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright A, McCoy AB, Henkin S, et al. Use of a support vector machine for categorizing free-text notes: assessment of accuracy across two institutions. J Am Med Inform Assoc 2013;20:887–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu CW, Chang CC, Lin CJ. A practical guide to support vector classification Taipei, Taiwan: Department of Computer Science, National Taiwan University, 2010. http://www.csie.ntu.edu.tw/~cjlin/papers/guide/guide.pdf (accessed 14 Apr 2013). [Google Scholar]
- 50.Garla VN, Brandt C. Ontology-guided feature engineering for clinical text classification. J Biomed Inform 2012;45:992–8. [DOI] [PMC free article] [PubMed] [Google Scholar]