Abstract
Background Although rare disease patients make up approximately 6–8% of all patients in Europe, it is often difficult to find the necessary expertise for diagnosis and care and the patient numbers needed for rare disease research. The second French National Plan for Rare Diseases highlighted the necessity for better care coordination and epidemiology for rare diseases. A clinical data standard for normalization and exchange of rare disease patient data was proposed. The original methodology used to build the French national minimum data set (F-MDS-RD) common to the 131 expert rare disease centers is presented.
Methods To encourage consensus at a national level for homogeneous data collection at the point of care for rare disease patients, we first identified four national expert groups. We reviewed the scientific literature for rare disease common data elements (CDEs) in order to build the first version of the F-MDS-RD. The French rare disease expert centers validated the data elements (DEs). The resulting F-MDS-RD was reviewed and approved by the National Plan Strategic Committee. It was then represented in an HL7 electronic format to maximize interoperability with electronic health records.
Results The F-MDS-RD is composed of 58 DEs in six categories: patient, family history, encounter, condition, medication, and questionnaire. It is HL7 compatible and can use various ontologies for diagnosis or sign encoding. The F-MDS-RD was aligned with other CDE initiatives for rare diseases, thus facilitating potential interconnections between rare disease registries.
Conclusions The French F-MDS-RD was defined through national consensus. It can foster better care coordination and facilitate determining rare disease patients’ eligibility for research studies, trials, or cohorts. Since other countries will need to develop their own standards for rare disease data collection, they might benefit from the methods presented here.
Keywords: rare diseases, minimum data set, interoperability, national health program, common data elements, metadata
BACKGROUND
There are 6000–7000 rare diseases according to published estimates.1 A rare disease is defined by a prevalence of less than 1/20002 in Europe and of no more than 1/15003 in the USA. In France, the number of patients affected by a rare disease is estimated to be between 3 and 4 million4 and many diseases have less than 100 identified patients. The anticipated number of patients affected by a rare disease is about 30 million in Europe5 and 25 million in North America.6–8 However, the burden of rare diseases is difficult to calculate since epidemiological data for most rare diseases are at best fragmented or more often lacking. Rare diseases have been identified as a public health priority in Europe. Many EU countries have launched national plans to promote rare diseases care and research.9,10 Since 2004, the French authorities together with field experts, patients’ associations, and other stakeholders have implemented two consecutive rare disease national plans. The first plan (2005–2009) fostered the implementation of a network of 131 rare disease centers of expertise distributed throughout French territory11 and focused on groups of diseases (rare renal diseases, rare pulmonary diseases, rare developmental defects, etc). Each center of expertise consisted of one or more medical units mainly located in university hospitals. A complementary network of 501 units was connected to this first set of centers to better cover the different areas closer to patients’ residences. This rare disease network aimed at building a nation-wide continuum of care for these chronic and disabling diseases. To support clinicians’ rare disease care and research activities, an IT infrastructure has been funded by the second national plan for rare diseases (2011–2014).12 This national information system promotes information exchange tools that can be integrated within the current local or national information systems to avoid data re-entry. Rare disease patients are often barely identifiable within hospital information systems because of the lack of standardized rare disease coding, as well as a lack of systematized data collection at a national level such as used for the Global Rare Diseases Patient Registry and Data Repository (GRDR) common data elements (CDEs) of the US initiative.13 A first objective is to identify patients in the rare disease care network to help build a seamless continuum of care across expert centers and reduce overall costs. Making rare disease-associated activity detectable is essential for the expert centers so they can submit claims for relevant funding. A second objective is to assess whether the rare disease network is a good match for the geographic distribution of rare diseases in the country. A third objective is to help identify rare disease patients likely to be eligible for clinical trials or cohort studies. To meet these objectives, the second French National Plan for Rare Diseases fostered the development of a national minimum data set (F-MDS) (as a clinical data standard) for all French rare disease centers and all rare diseases.
RATIONALE FOR A MINIMUM DATA SET FOR RARE DISEASES
A minimum data set approach was first developed in order to enable large studies at controlled costs.14–18 A data set is composed of a group of data elements (DE). It corresponds to a scheme (or data set specification) which stipulates the sequence of inclusion of the DEs, whether they are mandatory, what verification rules should be employed, and the scope of the collection. The French minimum data set for rare diseases (F-MDS-RD) is a minimum set of DEs agreed for mandatory collection and reporting at a national level.19 A CDE is a DE that is commonly used in various data sets. A specific DE (SDE) is a DE specifically defined for a given purpose (eg, a public health indicator).20 More recently, the same approach was used to define core DEs common between databases (electronic health records (EHRs), registries) at a national level.21 Goetz et al.also proposed eyeGENE, a novel approach combining clinical and genetic data.22 The US cancer network infrastructure also proposed the use of CDEs23 to enable large scale clinical trials. In the rare disease field, such an approach was also developed at the level of a given disease or a group of diseases, but not at a national level.24 The US Office of Rare Diseases Research (ORDR) developed a set of CDEs at a national level as a tool for global collection of rare disease patient data for their GRDR repository of data.25 CDEs or minimum data sets must be built following an appropriate methodology in order to obtain wide consensus across rare disease professionals and public stakeholders. Recently, Svensson-Ranallo et al.26 proposed a comprehensive method to build minimum (clinical) data sets. A methodology for the development of cardiovascular clinical data standards was also proposed.27
INTEROPERABILITY AND STANDARDS
Interoperability between health IT systems for care, epidemiology and public health, or research is an important goal that has fostered much research and received significant funding worldwide. While data collection in care settings should ideally permit data reuse for epidemiology, public health, or research, it is still difficult to reuse the data produced in care setting.28 In this context, several standards are available to support the recording of health data in health information systems. They have been developed to standardize either DEs and information models (Health Level Seven (HL7),29 OpenEHR (EN 13606)30), value sets or terminologies (LOINC,31 SNOMED CT32 for clinical terms, Orphanet1 for orphan drugs and rare disease diagnosis, Human Phenotype Ontology (HPO)33 for signs, Online Mendelian Inheritance in Man (OMIM)34 for genes, Human Genome Variation Society (HGVS) Mutnomen,35 and GenATLAS36 for genes and mutations). For clinical research, the Clinical Data Standards Interchange Consortium37 (CDISC) and the Regulated Clinical Research Information Management (RCRIM) technical committee of HL7 are promoting the development of the Biomedical Research Integrated Domain Group (BRIDG) model, a standard38 to support interoperability between CDISC (clinical research) and HL7 (electronic health records). Standards can facilitate interoperability and data quality in registries.39 These standards must be used to build electronic versions of CDEs or national minimum data sets so they can be seamlessly integrated within electronic medical records to serve the purpose of care, epidemiology and public health, and research.
Finding a consensus in this context is complex. First, there are numerous rare diseases and a rare disease can be represented in several medical fields. For instance, a disease affecting the kidneys, the eyes, and the central nervous system should be identifiable by nephrologists, ophthalmologists, or neurologists. Second, there are numerous stakeholders: medical and paramedical experts, public authorities, researchers, patients and their associations, and drug manufacturers. Last, the objectives of data collection must be precisely oriented towards specific targets: patient care, monitoring activity, public health, or research. The purpose of our work was to: (i) establish a consistent, interoperable national set of DEs common to all rare diseases; (ii) promote EHR data entry at the bedside; and (iii) facilitate the future development of EU registries by proposing an EU standard for rare disease patient based registries. To set up our F-MDS-RD, we proposed a complete methodology based on a systematic review of the literature as well as design and validation of the DEs by four different groups of experts and decision makers.
METHODOLOGY FOR DEVELOPING A CLINICAL DATA STANDARD
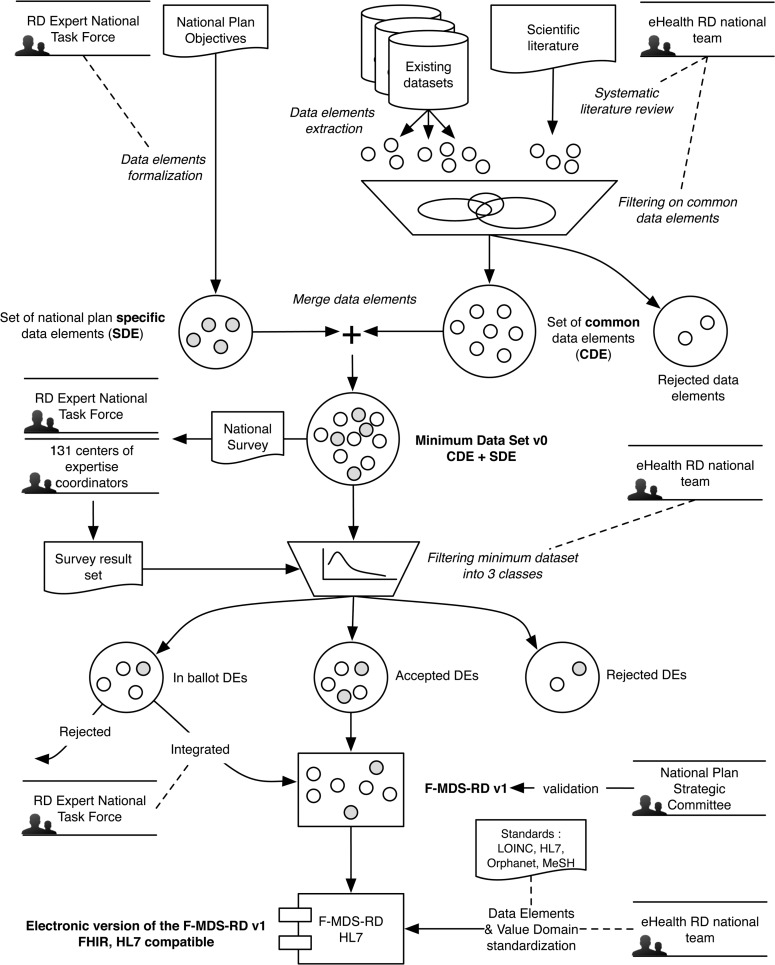
Our approach was based on the following steps: (i) establishment of several groups of experts; (ii) systematic review of the literature; (iii) development of the first draft of the F-MDS-RD by the eHealth team and the rare disease expert national working group; (iv) submission of this draft to the expert panels via a survey in order to formalize F-MDS-RD V.0; (v) validation of this V.0 version by the Rare Diseases Expert National Task Force group and validation of the resultant F-MDS-RD by the National Plan for Rare Diseases Strategic Committee; and (vi) standardization by the national eHealth team using Fast Health Interoperable Resources (FHIR), the latest HL7 data format, to support integration with EHRs and facilitate data reuse. The overall methodology is presented in figure 1.
Figure 1:
The F-MDS-RD methodology diagram. RD, rare diseases.
Identifying the expert panels
We first identified several groups of experts or decision makers in order to streamline our workflow and achieve national consensus among all rare disease clinicians and public authorities. We identified four groups assigned with various responsibilities:
The 131 rare disease expert centers represented by their medical coordinator or a representative. This group was responsible for stating their needs in terms of data management as well as selecting the DEs of interest for their group of rare diseases.
A Rare Disease Expert National Task Force composed of rare disease experts, representatives of the centers of expertise for rare diseases, national agencies, the Ministry of Health, researchers, and the national Institute of Health and Medical Research. This group discussed the relevance of the DEs to the specific objectives of the second National Plan for Rare Diseases11
The National Plan for Rare Diseases Strategic Committee is a board of public representatives chaired by the director of the care management department of the Ministry of Health. The board's role was to validate and approve the F-MDS-RD.
A team of eHealth experts to build the necessary tools for the systematic review of the literature, to determine the statistical methods for item selection, and to propose a standardized electronic version of the F-MDS-RD.
Systematic review of the literature and development of F-MDS-RD V.0
We built a preliminary set (V.0) of DEs. We identified DEs used in existing catalogs and the scientific literature, as well as in research projects and rare disease registries. We reviewed the current data catalogs of equivalent systems for rare diseases in order to select a primary set of DEs. We selected sets of CDEs from the US Office of Rare Disease Research,40 from the EPIRARE project,41 and from the CEMARA project.42 We also collected the disease specific data set from the ESID European project.43 We aligned these resources and built a preliminary CDE set for rare diseases.
In parallel, we also conducted a systematic review of the scientific literature. We developed a review tool to scan the PubMed site (http://www.ncbi.nlm.nih.gov/pubmed). We used the Human Rare Diseases Ontology44 in order to build the domain part of the queries. We then associated it to a corpus of keywords related to rare diseases. The ontology of more than 7000 diseases and group of diseases was associated with each of the following keywords: {Data Set, Minimum data set, Data catalogue, Data model, Models of data, Common data elements, Data Elements}. The query was built in the PubMed query language. We first searched for the diseases in Medical Subject Headings (MeSH) to identify relevant MeSH terms. Complementary title and abstract word searches were carried out for disease names not present in MeSH. The results were a set of articles related to data sets in the rare disease field which were then reviewed by a human expert. Information extracted from the relevant articles was used to validate the quality of the preliminary F-MDS-RD developed by the eHealth team.
National survey to review the minimum data set
A national survey instrument was created in order to validate the elements of the F-MDS-RD. It was sent to 160 rare disease experts, 131 coordinators of rare disease centers of expertise, and 29 members of the Rare Disease National Task Force. This survey was conducted through an on-line questionnaire which consisted of 118 items grouped into 12 themes. Questions were aimed at determining whether items should be retained or not, and, if retained, whether they should be mandatory or optional. To analyze the survey results, we used a decision tree that took into account the number of people who responded to each question. We devised a set of decision rules in order to determine whether items should be accepted or rejected:
If an item had more ‘no’ than ‘yes’ responses, the item was discarded;
- If an item had more ‘yes’ than ‘no’ responses, there were two options:
- If the percentage of responses was greater than the median rate of answers for all the questions and the rate of ‘yes’ for this item was above 50%, then the item was retained;
- Otherwise, the item was submitted to the Rare Disease National Task Force for review.
We carried out a sensitivity analysis to assess whether active and less active (defined as having a percentage of item responses lower than the median of responses) participants gave similar answers. Because the coordinators belonged to 18 different medical fields (eg, neuromuscular diseases, developmental anomalies, etc) defined by the national rare disease plan, we assessed whether the distributions of the response rates were balanced between the different fields (Wilcoxon test, Bonferroni correction).
National validation of the F-MDS-RD
We developed a set of DEs common to all rare diseases that were in accordance with the objectives of the second French National Plan for Rare Diseases. The retained DEs were presented and reviewed by the national steering committee of the second French National Plan for Rare Diseases and approved by the Chief Executive of the Care Organization.
An interoperable electronic format of the F-MDS-RD
To support F-MDS-RD integration into the EHR and to facilitate compatibility with other standards, we represented the F-MDS-RD into the latest HL7 standard format, FHIR, which should be easier and faster to implement than its predecessor.45 FHIR is based on modular components called ‘resources’ which can be combined together or extended to meet specific data capture needs. FHIR is backward compatible with HL7 V.2 and V.3. We aligned the F-MDS-RD with standard FHIR components where possible. For the remainder, we defined extensions of existing FHIR resources when necessary. Otherwise the components will be defined as questions of a resource questionnaire to form homogeneous and meaningfully independent data groups. Finally, we checked the concordance of data types and tested the compatibility of value domains. An element might be a ‘codable concept’ if it takes its values from a publicly defined code system like LOINC or from a locally defined code system in a FHIR value set. Some value sets of ‘codable concepts’ are predefined in FHIR. We checked their compliance with the F-MDS-RD value domains and if they did not match, internal value sets had to be created by referencing codes from one or more code systems and/or by defining specific codes.
RESULTS
A systematic review of existing DEs for rare diseases
The first systematic PubMed search based on title and abstracts returned 2126 articles. The relevance of the results was checked by an expert (PL). Several articles were not retained because they were too specific or because they were out of scope. It should be noted that the expert did not select articles presenting minimum data sets but not describing the methods used for their creation. Moreover, data sets were often very disease specific with most items not applicable to a range of rare diseases, so only a few items were retained, mostly the patient's personal information. We thus obtained a very small set of six relevant articles.46–51
National rare diseases expert survey to review the F-MDS-RD
The response rate of the experts to the survey was 81%. A median of 60% of questions were answered. The survey contained 105 questions on DEs. After application of the decision rules, 54 items were retained and 30 were rejected. The 21 remaining items were submitted to a second round of discussion with the working group. During this second review, some items accepted by the decision rules were rejected. We assessed whether the distributions of the response rates were balanced between the different medical fields (Wilcoxon test, Bonferroni correction). Response rates were not affected by the type of medical field to which the experts belonged.
Towards an electronic standard
We selected five major resources within the FHIR resource pool: the Patient resource to gather administrative and demographic information on a patient, the Family history resource to regroup contextual family information, the Encounter resource to include information about care activities performed on the patient, the Condition resource to regroup the diagnosis, diagnosis history, and confirmation data, and the Medication resource to specify ongoing rare disease treatment. We added two resource questionnaires, one for ante- and neonatal information and one for research data. More than half of the F-MDS-RD DEs were defined as extensions or questionnaire elements, and 17% of value sets remained exclusively internal value sets.
THE F-MDS-RD
The resulting F-MDS-RD is described in online supplementary table 1. It is built from 42 CDEs and 16 specific national DEs assigned to 13 groups: consent, patient identification, personal information, family information, vital status, care pathway, care activity, diagnosis history, diagnosis, diagnosis confirmation, treatment, ante- and neonatal information, and participation in research.
If use of the F-MDS-RD is integrated with use of an EHR, as many as 38 of these 58 DEs are likely to be captured and coded in the EHR as part of usual care. The coding of the medical domain values can be adapted to a country's existing practices. For example, the rare disease diagnosis can be coded in Orphanet, OMIM, or SNOMED CT depending on the context and the required granularity of the coding. In France, diagnostic coding relies on Orphanet codes and is done by clinicians using an application developed in-house.52 France does not have the necessary license to use SNOMED CT at present.
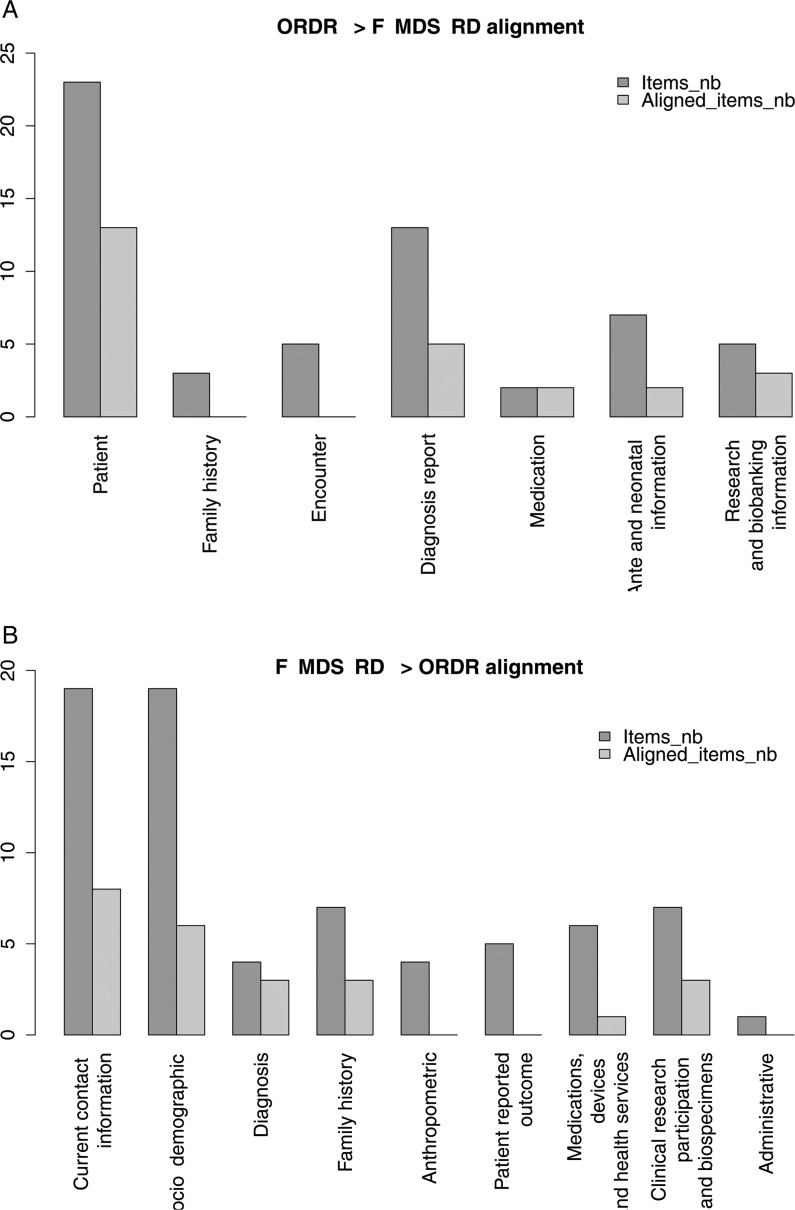
Online supplementary table 1 shows the correspondence between the F-MDS-RD and the US GRDR CDEs25 we considered as a gold standard in our work. Both alignments between GRDR CDEs25 and the F-MDS-RD were assessed.
Extent of coverage of GRDR CDEs in F-MDS-RD
DEs level: 43% (value domain alignment: 100%) (figure 2A)
Value domain level: 43%.
Figure 2:
Number of common data elements between US Office of Rare Diseases Research (ORDR) (GRDR CDEs) and F-MDS-RD (from US CDE to French CDE). CDE, common data elements; F-MDS-RD, French national minimum data set for rare diseases; GRDR, Global Rare Diseases Patient Registry and Data Repository; nb, numbers.
Extent of coverage of F-MDS-RD in GRDR CDEs
DEs level: 33% (value domain alignment: 96%) (figure 2B)
Value domain level: 32%.
DISCUSSION
The mission of the French rare disease centers is to organize patient care as well as conduct rare disease research. The second French National Plan for Rare Diseases (2011–2014) aims to provide the centers with information technology tools to help improve rare disease care and research. The design and setup of a national information system and databank for rare diseases is thus promoted. The collection of standardized data at a national level and the descriptions of patients’ diseases at an individual level were identified as important steps for fostering rare disease care and clinical and epidemiological research. Two main issues were identified: interoperability to avoid double data entry, and data privacy and security.
The International Rare Diseases Research Consortium (IRDiRC), developed under the 7th EU Framework Program for Research and Technological Development to expand rare disease research, is currently hampered by a lack of data and samples due to the absence of a complete rare disease classification, standard terms of reference, shared ontologies, and harmonized regulatory requirements. The European Union Committee of Experts on Rare Diseases (EUCERD) jointly with EPIRARE41 issued recommendations on rare disease patient recording and data collection in June 2013.9 These recommendations encourage national data collection for rare diseases based on common data sets for all patients and interoperability through the dissemination of agreed CDEs for rare diseases, and guidelines for data sharing. The US ORDR launched the GRDR. The main goals were: to develop a set of CDEs to facilitate data collection in a standardized and meaningful manner; to assist organizations that have no patient registry to establish a registry and to share de-identified data with the GRDR or other databases; and to facilitate harmonization among the many organizations collecting patient information.40 To our knowledge, a description of the methodology used for developing the GRDR CDEs has not been published.
The second French National Plan for Rare Diseases encouraged the setting up of a national minimum data set for rare diseases (F-MDS-RD) for all French rare disease centers of expertise. It was oriented towards enabling homogenous and standardized data collection, while limiting multiple or redundant data entries. This F-MDS-RD had to include CDEs for all rare diseases as well as a set of SDEs that met the national plan objectives. The resulting minimal data set was nationally approved and establishes a first mandatory set of standardized data for all rare diseases.
Compared with the GRDR CDEs, we gather less information. We collect less contact or demographic information and do not collect information on the patient's quality of life or the possibility of transplantation. On the other hand, we collect more information on the structure of care, ante- and neonatal care for patients early in life, rare disease activity, and diagnosis (history and mode of diagnosis confirmation). Harmonization of the F-MDS-RD at an international level is very important for future cooperation among registries. Maintaining updated alignment between coding resources will be an absolute necessity. Orphanet aligns its coding resource to SNOMED CT, OMIM, and ICD, but the coverage is not complete and not easy to maintain: a better continuous and coordinated effort is needed.
Our objective was to push the implementation of the F-MDS-RD in healthcare information systems (EHRs, registries) to facilitate interoperability. We privilege data capture at the ‘bedside’ or in the outpatient clinic by the physicians themselves even though we are aware this is a difficult goal to achieve. Adherence to the system and data collection has to be strong and reliable and bring a real ‘return on investment’ for professionals. The rare disease community is quite motivated to capture data since they are already involved in epidemiological and research programs. Healthcare professionals are very interested in avoiding double data entry when possible as well as having a secure data store for their patient data. Even if data are available from an existing EHR system, they are rarely reusable as it is the case for clinical research.53 In fact, even if the system has a proper coding system (Orphanet codes for instance), the data quality is often not sufficient for research.54 Our experience with these problems is based on use of CEMARA42 which at present gathers data from 250 000 patient records for more than 4000 rare diseases.
It is very important when sharing patient data to strictly comply with national and international regulations on patient privacy and intellectual property. These aspects need to be considered early in the process of setting up a cohort or a registry. Informed consent is required for all patients to be included in the French registry. An assent and a consent process are required for minors and individuals not competent to provide informed consent. Enrolled minors must be re-contacted when they reach adulthood and re-consent obtained. The importance of understanding the content of the consent form is explained as well as the meaning of the participant’s signature.55
A recent successful European project (epSOS56) has led to the issuance of an EU guideline on a minimum and non-complete patient summary data set for electronic exchange in accordance with the cross-border directive 2011/24/EU.57 We now aim to help define a comparable minimum data set for rare diseases at the European level to support development of an EU recommendation for rare diseases as epSOS did for emergency care. The F-MDS-RD only responds to part of the information needs of the French rare disease centers of expertise. In fact, for patient care or research, more data are needed. For this reason, we promote the sharing of information with clinical trials and cohorts studies. We also aim to define specialized minimum data sets for each group of rare diseases in order to continue our efforts to make data interoperable.
ACKNOWLEDGEMENTS
The authors warmly thank the reviewers and the editor for their constructive and stimulating remarks and comments.
CONTRIBUTORS
RC designed the overall methodology, conducted the research, and drafted and revised the paper. He is guarantor. MM built the FHIR electronic version of the MDS. AdC conducted the systematic review and alignment analysis with the GRDR CDEs. CM carried out the statistical analysis for item selection. EL revised the draft paper. PL devised the MDS idea, participated in every decision during the project, and drafted and revised the paper. The following coordinators of the 131 centers of expertise selected the data elements and provided medical expertise on rare diseases: Professor Gilles Grateau, Professor Eric Hachullah, Professor Arnaud Jaccard, Professor Isabelle Koné-Paut, Professor Jean-louis Pasquali, Professor Jean-Charles Piette, Professor Pierre Quartier dit Maire, Professor Damien Bonnet, Professor Philippe Charron, Professor Philippe Chevalier, Professor Hervé Le Marrec, Professor Patrick Edery, Professor Didier Lacombe, Professor Sylvie Manouvrier-Hanu, Professor Sylvie Odent, Professor Laurence Olivier-Faivre, Professor Nicole Philip, Professor Pierre Sarda, Professor Alain Verloes, Professor Pascal Joly, Professor Jean Philippe Lacour, Professor Alain Taieb, Professor Jérôme Bertherat, Professor Thierry Brue, Professor Pierre Chatelain, Professor Juliane Leger, Professor Agnès Linglart, Professor Patrice Rodien, Professor Maithé Tauber, Dr Elisabeth Thibaud, Professor Frédéric Gottrand, Professor Olivier Goulet, Professor Emmanuel Jacquemin, Professor Raoul Poupon, Professor Sabine Sarnacki, Professor Dominique Valla, Professor Paul Coppo, Dr Maryse Etienne-Julan, Professor Frédéric Galacteros, Dr Bertrand Godeau, Dr Paquita Nurden, Professor Gérard Socie, Dr Isabelle Thuret, Professor Agnès Veyradier, Dr Nadia Belmatoug, Professor Pierre Brissot, Professor Brigitte Chabrol, Professor Jean-Charles Deybach, Professor Dries Dobbelaere, Professor François Feillet, Professor Philippe Labrune, Professor Pascale de Lonlay, Professor Arnold Munnich, Professor Véronique Paquis-Flucklinger, Dr France Woimant, Professor Odile Boespflug-Tanguy, Professor Dominique Bonneau, Professor Alexis Brice, Professor Hugues Chabriat, Professor Yves Dauvilliers, Professor Vincent Des Portes, Professor Bruno Dubois, Dr Delphine Héron, Professor Jérôme Honnorat, Professor Marc Tardieu, Professor David Adams, Professor Jean-Christophe Antoine, Dr Rémi Bellance, Professor Bruno Eymard, Dr Xavier Ferrer-Monasterio, Professor Bertrand Fontaine, Professor Vincent Meininger, Dr Claude Mignard, Professor André Thévenon, Professor Christine Tranchant, Professor Jean-Michel Vallat, Professor Gabriel Bellon, Professor Annick Clement, Professor Jean-François Cordier, Professor Ti-Tuyet-Ha Trang, Dr Gilles Rault, Professor Joseph Colin, Professor Hélène Dollfus, Professor Jean-Louis Dufier, Professor Christian Hamel, Dr Sandrine Marlin, Professor José Sahel, Professor Dominique Chauveau, Professor Pierre Cochat, Professor Dil Sahali, Professor Rémi Salomon, Dr Martine Le Merrer, Professor Alain Fischer, Professor Dominique Paul Germain, Professor Guillaume Jondeau, Professor Eréa-Noël Garabedian, Professor Marie-Cécile Maniere, Professor Philippe Pellerin, Professor Marie-Paule Vazquez, Dr Andréa Manunta, Professor Laurence Bouillet, Dr Didier Pilliard, Professor Yves Ville, and Professor Pierre Wolkenstein. Mrs Carey Suehs revised the paper. The following rare diseases national group of experts participated in designing national data elements and selecting data elements: Professor Serge Amselem, Dr Ségolène Aymé, Professor Christophe Béroud, Professor Annick Clément, Dr Patrice Dosquet, Mrs Monica Ensini, Mr Philippe Faveaux, Mrs Céline Hubert, Professor Odile Kremp, Mrs Lydie Lemonnier, Professor Nicolas Levy, Dr Laetitia May, Mrs Samantha Parker, Mrs Virginie Picard, and Dr Ana Rath. The National Plan for Rare Diseases Strategic Committee also contributed. Mrs Céline Angin reviewed the paper.
FUNDING
This work was supported by the French Ministry of Health.
COMPETING INTERESTS
None.
PROVENANCE AND PEER REVIEW
Not commissioned; externally peer reviewed.
REFERENCES
- 1.Orphanet knowledge base for rare diseases. http://www.orpha.net (accessed 1 Jan 2014).
- 2.EURORDIS. Eurordis Position Paper on the WHO Report on Priority Medicines for Europe and the World 1. The inconsistency used throughout the Report when addressing the issue of Rare Diseases. 2000. http://www.eurordis.org/IMG/pdf/eurordis_position_WHO__priority_medicines_dec04.pdf
- 3.van Weely S, Leufkens HGM. Orphan diseases. http://www.pharmaceuticalpolicy.nl/Publications/Reports/7.5Orphandiseases.pdf.
- 4.French health ministry information website on rare diseases. http://www.sante.gouv.fr/les-maladies-rares-qu-est-ce-que-c-est.html (accessed 1 Jan 2014).
- 5.EUCERD. Report on the state of the art of rare disease activities in Europe of the European Union committee of experts on rare diseases PART I: overview on rare diseases. 2012. http://www.eucerd.eu/?post_type=document&p=1378
- 6.U.S. Food and Drug Administration. Developing products for rare diseases & conditions. http://www.fda.gov/forindustry/DevelopingProductsforrareDiseasesConditions/default.htm (accessed 1 May 2014).
- 7.European Commission. Guideline on the format and content of applications for designation as orphan medicinal products and on the transfer of designations from one sponsor to another, 27.03.2014. 2014. http://ec.europa.eu/health/files/orphanmp/2014-03_guideline_rev4_final.pdf
- 8.Wästfelt M, Fadeel B, Henter J-I. A journey of hope: lessons learned from studies on rare diseases and orphan drugs. J Intern Med 2006;260:1–10. [DOI] [PubMed] [Google Scholar]
- 9.EUCERD Core recommendation on rare disease patient registration and data collection. http://www.eucerd.eu/wp-content/uploads/2013/06/EUCERD_Recommendations_RDRegistryDataCollection_adopted.pdf (accessed 1 Jan 2014).
- 10.European projects information website. http://www.europlanproject.eu/ (accessed 1 Jan 2014).
- 11.Evaluation du plan national maladies rares 2005–2008. http://www.sante.gouv.fr/IMG/pdf/hcspr20090317_maladiesRares.pdf (accessed 1 Jan 2014).
- 12.2nd French National Plan for Rare Diseases. http://www.sante.gouv.fr/2eme-plan-national-maladies-rares-pnmr-2011–2014.html (accessed 1 Jan 2014).
- 13.Rubinstein YR, Groft SC, Bartek R, et al. Creating a global rare disease patient registry linked to a rare diseases biorepository database: Rare Disease-HUB (RD-HUB). Contemp Clin Trials 2010;31:394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird SM, Farrar J. Minimum dataset needed for confirmed human H5N1 cases. Lancet 2008;372:696–7. [DOI] [PubMed] [Google Scholar]
- 15.Pheby DF, Etherington DJ. Improving the comparability of cancer registry treatment data and proposals for a new national minimum dataset. J Public Health Med 1994;16:331–40 http://www.ncbi.nlm.nih.gov/pubmed/7999387 (accessed 25 Feb 2013). [PubMed] [Google Scholar]
- 16.Tilyard MW, Munro N, Walker SA, et al. Creating a general practice national minimum data set: present possibility or future plan? N Z Med J 1998;111:317–18 320. http://www.ncbi.nlm.nih.gov/pubmed/9765630 (accessed 8 Feb 2013). [PubMed] [Google Scholar]
- 17.Webster D. A minimum dataset for newborn screening. J Med Screen 1998;5:109 http://www.ncbi.nlm.nih.gov/pubmed/9718531 (accessed 25 Feb 2013). [DOI] [PubMed] [Google Scholar]
- 18.Werley HH, Devine EC, Zorn CR, et al. The nursing minimum data set: abstraction tool for standardized, comparable, essential data. Am J Public Health 1991;81:421–6 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1405031&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National minimum data sets and data set specifications. http://meteor.aihw.gov.au/content/index.phtml/itemId/344846 (accessed 1 Jan 2014).
- 20.National metadata registry. http://meteor.aihw.gov.au/content/index.phtml/itemId/344846 (accessed 1 Jan 2014).
- 21.Häyrinen K, Saranto K. The core data elements of electronic health record in Finland. Stud Health Technol Inform 2005;116:131–6 http://www.ncbi.nlm.nih.gov/pubmed/16160248 (accessed 8 Feb 2013). [PubMed] [Google Scholar]
- 22.EyeGENE–National Ophthalmic Disease Genotyping Network. Insight 34:27 http://www.ncbi.nlm.nih.gov/pubmed/19534233 (accessed 27 Aug 2013). [PubMed] [Google Scholar]
- 23.Winget MD, Baron JA, Spitz MR, et al. Development of common data elements: the experience of and recommendations from the early detection research network. 2003;5056:41–8. [DOI] [PubMed] [Google Scholar]
- 24.Jason L, Unger E, Dimitrakoff J. Minimum data elements for research reports on CFS. Brain Behav Immun 2012;26:401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forrest CB, Bartek RJ, Rubinstein Y, et al. The case for a global rare-diseases registry. Lancet 2011;377:1057–9. [DOI] [PubMed] [Google Scholar]
- 26.Svensson-Ranallo PA, Adam TJ, Sainfort F. A framework and standardized methodology for developing minimum clinical datasets. AMIA Summits Transl Sci Proc 2011;2011:54–8 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3248746&tool=pmcentrez&rendertype=abstract [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson HV, Weintraub WS, Radford MJ, et al. Standardized cardiovascular data for clinical research, registries, and patient care: a report from the Data Standards Workgroup of the National Cardiovascular Research Infrastructure project. J Am Coll Cardiol 2013;61:1835–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Köpcke F, Kraus S, Scholler A, et al. Secondary use of routinely collected patient data in a clinical trial: an evaluation of the effects on patient recruitment and data acquisition. Int J Med Inform 2013;82:185–92. [DOI] [PubMed] [Google Scholar]
- 29.Landgrebe J, Smith B. The HL7 approach to semantic interoperability. Int Conf Biomed Ontol 2011:140–6. [Google Scholar]
- 30.Kohl CD, Garde S, Knaup P. Facilitating secondary use of medical data by using openEHR archetypes. Stud Health Technol Inform 2010;160:1117–21 http://www.ncbi.nlm.nih.gov/pubmed/20841857 (accessed 23 May 2013). [PubMed] [Google Scholar]
- 31.LOINC search website. http://search.loinc.org (accessed 1 Jan 2014).
- 32.Wang AY, Sable JH, Spackman KA. The SNOMED clinical terms development process: refinement and analysis of content. Proc AMIA Symp 2002:845–9. [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson PN, Mundlos S. The human phenotype ontology. Clin Genet 2010;77:525–34. [DOI] [PubMed] [Google Scholar]
- 34.Online Mendelian Inheritance in Man. http://www.ncbi.nlm.nih.gov/omim (accessed 1 Jan 2014).
- 35.Laros JFJ, Blavier A, den Dunnen JT, et al. A formalized description of the standard human variant nomenclature in Extended Backus-Naur Form. BMC Bioinformatics 2011;12(Suppl 4):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genatlas. http://genatlas.medecine.univ-paris5.fr (accessed 1 Jan 2014).
- 37.Sinaci AA, Laleci Erturkmen GB. A federated semantic metadata registry framework for enabling interoperability across clinical research and care domains. J Biomed Inform 2013;46:784–94. [DOI] [PubMed] [Google Scholar]
- 38.Fridsma DB, Evans J, Hastak S, et al. The BRIDG project: a technical report. J Am Med Inform Assoc 2008;15:130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higashi T, Nakamura F, Shibata A, et al. The national database of hospital-based cancer registries: a nationwide infrastructure to support evidence-based cancer care and cancer control policy in Japan. Jpn J Clin Oncol 2014;44:2–8. [DOI] [PubMed] [Google Scholar]
- 40.The GRDR CDEs. 2014. https://grdr.ncats.nih.gov/index.php?option=com_content&view=article&id=3&Itemid=5 (accessed 20 Jun 2014).
- 41.EPIRARE project website. http://www.epirare.eu (accessed 1 Jan 2014).
- 42.Landais P, Messiaen C, Rath A, et al. CEMARA an information system for rare diseases. Stud Health Technol Inform 2010;160:481–5 http://www.ncbi.nlm.nih.gov/pubmed/20841733 (accessed 25 Feb 2013). [PubMed] [Google Scholar]
- 43.European Society for Immuno Deficiencies. http://www.esid.org (accessed 1 Jan 2014).
- 44.Dhombres F, Vandenbussche P-Y, Rath A, et al. OntoOrpha: an ontology to support the editing and audit of rare diseases knowledge in Orphanet. In: International Conference of Biomedical Ontology (ICBO 2011) 2011. [Google Scholar]
- 45.Bender D, Sartipi K. HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. In: Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems; IEEE, 2013:326–31. 10.1109/CBMS.2013.6627810 [Google Scholar]
- 46.Buchanan RJ, Wang S, Ju H. Analyses of the minimum data set: comparisons of nursing home residents with multiple sclerosis to other nursing home residents. Mult Scler 2002;8:512–22 http://www.ncbi.nlm.nih.gov/pubmed/12474994 (accessed 10 May 2014). [DOI] [PubMed] [Google Scholar]
- 47.McCormick J, Sims EJ, Green MW, et al. Comparative analysis of Cystic Fibrosis Registry data from the UK with USA, France and Australasia. J Cyst Fibros 2005;4:115–22. [DOI] [PubMed] [Google Scholar]
- 48.Buchanan RJ, Martin RA, Moore L, et al. Nursing home residents with multiple sclerosis and dementia compared to other multiple sclerosis residents. Mult Scler 2005;11:610–16 http://www.ncbi.nlm.nih.gov/pubmed/16193901 (accessed 10 May 2014). [DOI] [PubMed] [Google Scholar]
- 49.O'Donnell JL, Stevanovic VR, Frampton C, et al. Wegener's granulomatosis in New Zealand: evidence for a latitude-dependent incidence gradient. Intern Med J 2007;37:242–6. [DOI] [PubMed] [Google Scholar]
- 50.Adelson PD, Pineda J, Bell MJ, et al. Common data elements for pediatric traumatic brain injury: recommendations from the working group on demographics and clinical assessment. J Neurotrauma 2012;29:639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kerr AM, Nomura Y, Armstrong D, et al. Guidelines for reporting clinical features in cases with MECP2 mutations. Brain Dev 2001;23:208–11 http://www.ncbi.nlm.nih.gov/pubmed/11376997 (accessed 10 May 2014). [DOI] [PubMed] [Google Scholar]
- 52.Linking Open Rare Disease data, an application for RD coding. http://enlord.bndmr.fr (accessed 1 May 2014).
- 53.Dregan A, Toschke MA, Wolfe CD, et al. Utility of electronic patient records in primary care for stroke secondary prevention trials. BMC Public Health 2011;11:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sagreiya H, Altman RB. The utility of general purpose versus specialty clinical databases for research: warfarin dose estimation from extracted clinical variables. J Biomed Inform 2010;43:747–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubinstein YR, Groft SC, Chandros SH, et al. Informed consent process for patient participation in rare disease registries linked to biorepositories. Contemp Clin Trials 2012;33:5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Project epSOS. http://www.epsos.eu (accessed 1 Jan 2014).
- 57.Commission E. guidelines on minimum/non- exhaustive patient summary dataset for electronic exchange in accordance with the cross-border directive 2011/24/eu. 2013. http://ec.europa.eu/health/ehealth/docs/guidelines_patient_summary_en.pdf.