SUMMARY
Circadian clocks and metabolism are inextricably intertwined, where central and hepatic circadian clocks coordinate metabolic events in response to light-dark and sleep-wake cycles. We reveal an additional key element involved in maintaining host circadian rhythms, the gut microbiome. Despite persistence of light-dark signals, germ-free mice fed low or high fat diets exhibit markedly impaired central and hepatic circadian clock gene expression and do not gain weight compared to conventionally-raised counterparts. Examination of gut microbiota in conventionally-raised mice showed differential diurnal variation in microbial structure and function dependent upon dietary composition. Additionally, specific microbial metabolites induced under low or high fat feeding, particularly short chain fatty acids, but not hydrogen sulfide, directly modulate circadian clock gene expression within hepatocytes. These results underscore the ability of microbially-derived metabolites to regulate or modify central and hepatic circadian rhythm and host metabolic function, the latter following intake of a Westernized diet.
INTRODUCTION
Circadian rhythm has a dominant role in determining overall health and physiological homeostasis. The central clock, located within the suprachiasmatic nucleus (SCN) of the brain, is important in coordinating light-dark and sleep-wake cycles with metabolic events that occur within peripheral tissues, including the liver (Huang et al., 2011). Classically, circadian rhythm is maintained through an oscillating autoregulatory transcriptional feedback loop, where the central transcriptional activators of the positive feedback segment are bmal1 and clock. BMAL1/CLOCK induce expression of the feedback loop repressor genes, period (per1-3) and cryptochrome (cry1-2) (Dibner et al., 2010).
Epidemiological studies demonstrate that human subjects with altered sleep cycles, interruption, or sleep quality (i.e., shift work or sleep apnea) exhibit increased appetite, increased susceptibility to diabetes, obesity, and overall metabolic syndrome (Bass and Turek, 2005; Dibner et al., 2010; Huang et al., 2011; Van Cauter et al., 1997). Animal models of circadian clock (CC) disruption show that the consumption of high fat diets adversely alters circadian signaling, suggesting a bi-directional relationship (Kohsaka et al., 2007), however the molecular mechanisms behind this are not clearly understood. Here, using an initial discovery-based approach, we found an additional key element that is pivotal for maintenance of CC, the gut microbial organ or microbiome, an essential mediator for dietary composition entrainment, time of ingestion, and host physiological cues. As a result, gut microbes exhibit their own CC patterns, producing oscillations in key metabolic mediators that are integrated into host circadian rhythm maintaining metabolic homeostasis. Perturbations of gut microbial structure and function caused by diet and host factors adversely affect the delicate balance in CC networks promoting metabolic disturbances that include diet-induced obesity (DIO).
RESULTS
The absence of gut microbes results in differential CC gene expression regardless of dietary challenge
Using Affymetrix microarray, we initially discovered the impact of microbes on liver transcriptomes via comparison of germ-free (GF), conventionalized (CONV), and conventionally-raised specific-pathogen-free (SPF) mice under regular low-fat chow (RC) feeding conditions. As expected, CONV and SPF were similar, however, their hepatic transcriptomes differed significantly from GF confirming previous findings (Bjorkholm et al., 2009; Joyce et al., 2014; Larsson et al., 2012; Mukherji et al., 2013) (Figure S1A). Of the >1000 differentially displayed genes, the majority mapped to metabolic networks; the most significantly altered pathways are highlighted in Figure 1A. Two networks were of particular interest – CC and nuclear receptor (NR) families – which contain many nodal genes that serve important roles in integrating downstream effector responses involved in metabolic regulation (Figure S1B). CC and NR networks in the liver are intimately linked and populated by many genes that appear to be dependent on the presence of microbes. We speculated that GF mice are perhaps metabolically different due to differential expression of these networks, particularly CC, accounting for their resistance to DIO.
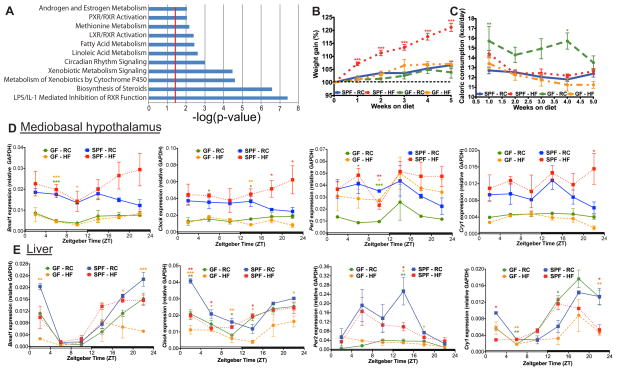
Figure 1. Germ-free mice exhibit differential canonical liver gene transcriptomic signatures and are resistant to diet-induced obesity.
GF and CONV C57Bl/6 liver transcriptomes generated via Affymetrix Mouse Genome 430 2.0 array (n=3 per group). Canonical pathways (A) enriched in GF versus CONV identified by Significant Analysis of Microarray software (FC >1.5 and FDR <5%) followed by submission to Ingenuity Pathway Analysis software. Significant pathways were determined by one-tail Fisher’s Exact test (p-value <0.05, i.e. −log(p-value) >1.3, (red line)). Weight gain (B) and food consumption (C) of SPF and GF mice (n=17–18 per group) fed RC or HF. Data represent mean ± s.e.m. ***p<0.001; **p<0.01; *p<0.05 via one-way ANOVA followed by Dunnett’s post-test relative to SPF-RC control where star color represents treatment exhibiting significance. Diurnal circadian gene expression patterns relative to GAPDH in mediobasal hypothalamus (D) and liver (E) in GF and SPF mice fed RC or HF. ***p<0.001; **p<0.01; *p<0.05 via one-way ANOVA followed by Dunnett’s post-test relative to SPF-RC control - star color indicates treatment exhibiting significance. See also Figure S1A–F and Table S2.
To determine how high-fat diet-induced microbial shifts impact CC specifically in DIO, we conducted studies using age-matched SPF or GF individually-housed male mice maintained under standard 12:12 L/D conditions (Zeitgeber (ZT) time 0 = 6am; lights on and ZT12 = 6pm; lights off) and fed RC or 37.5% (kcal) milk fat diet (HF) that elicits a known impact on gut microbes (Devkota et al., 2012; Huang et al., 2013; Table S1). GF mice fed RC or HF gained significantly less weight compared to SPF counterparts, despite nearly identical, if not increased, caloric consumption (Figure 1B,C). Compared to SPF mice, GF mice fed HF also exhibited reduced gonadal fad pad and liver weight (Figure S1C,D), reflecting resistance to DIO, as previously shown (Rabot et al., 2010). After 5 weeks, liver and mediobasal hypothalamus (MBH) samples were collected every 4 hours (h) over 24h for measurements of CC gene expression (Figure 1E,F and Figure S1C,D). The majority of CC genes in the liver from both SPF and GF mice fed RC or HF exhibited rhythmicity, while fewer CC genes exhibited cosinor expression patterns in the MBH (see Table S2 for CircWave software cosinor output). Despite rhythmicity in CC genes in SPF and GF mice, further analyses at specific time points showed that diet and the presence of diet-induced gut microbiota influence host CC gene expression. Figure 1E shows that HF induced expression of bmal1 and clock in the MBH of SPF mice during the dark phase but not in GF mice, suggesting microbes mediate this induction. Contrary to the MBH responses, HF suppressed bmal1 expression in the liver (Figure 1E, ZT2, 18, and 22), blunting the hepatic CC pattern, as previously shown (Kohsaka et al., 2007; Eckel-Mahan et al., 2013). Similar effects were seen in GF mice yet a strikingly lower level of CC expression was observed compared to SPF, regardless of diet in many CC genes. In the MBH of SPF mice, HF increases per2 and cry1 expression compared to RC. However, in the liver (Figure 1E), HF decreases per2 expression at ZT14 in SPF mice, yet there is a loss of per2 expression on either diet in GF mice. Thus, these data show that HF has opposing roles in modulating CC genes, whereby HF induces bmal1 and clock as well as per2 and cry1 expression in the MBH, but suppresses expression in the liver of SPF mice. Notably, in the case of bmal1 in the MBH, and per2 in the liver, these effects are suppressed in GF mice, implicating microbes as mediators in HF-induced alterations of CC function.
High fat diet alters diurnal patterns of gut microbiota structure and function
Given our results examining host CC gene expression in the absence of gut microbes, a second goal of our studies was to determine the impact of HF on gut microbiota structure/function and subsequent effect on host CC. We first established whether or not gut microbes undergo diurnal oscillations and how this is affected by HF. Fecal pellets were obtained every 6h over 48h via repeat sampling from individual SPF mice and cecal contents were collected from SPF mice fed RC or HF over 24h at time of harvest. Assessment of 16S rRNA V4–V5 region via Illumina MiSeq sequencing showed HF significantly altered microbial community composition as shown by Principal Coordinate Analysis (PCoA) of abundance-weighted beta-diversity in both fecal pellets collected over 48h (Figure 2A) and cecal contents collected over 24h (Figure 2B). Additionally, HF resulted in altered microbial alpha-diversity, indicated by significantly reduced Shannon diversity index as compared to RC (Figure S2A–B) corroborating previous reports (Backhed et al., 2004; Carvhalo et al., 2012; Ridaura et al., 2013; Turnbaugh et al, 2006). Further assessment of beta-diversity (Figure 2C) between HF and RC cecal microbiota showed temporal shifts over 24h that were more evident under RC feeding. Additionally, oscillations of 16S abundance determined by qPCR (Figure 2D), were detected only in cecal contents from RC-fed mice (see Table S5 for CircWave cosinor statistics) suggesting that HF blunts diurnal rhythmicity of luminal gut microbiota abundance. To determine if enteral delivery of nutrients is the sole driver of diurnal patterns in gut microbes, mice were subjected to constant intravenous parenteral nutrition (PN) in absence of any peroral alimentation. Despite significant changes in community structure, diurnal variation was still evident, suggesting that additional host factors contribute to gut microbial chronobiology (Figure S2C,D).
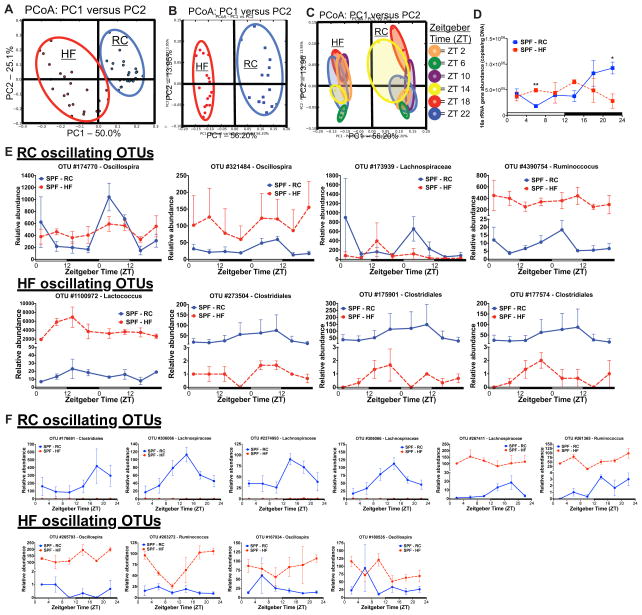
Figure 2. Diurnal gut microbe community structure is altered by high fat feeding.
Principal Coordinate Analysis (PCoA) of weighted UniFrac distances from 16S rRNA amplicon sequences colored by diet in fecal pellets (A) collected via repeat sampling over 48h (n=3 mice/treatment) and in cecal contents (B) collected over 24h (n=2–3 mice/time point) from SPF C57Bl/6 mice fed HF or RC. (C) PCoA analysis of cecal contents colored by Zeitgeber (ZT) time (PC=principle coordinate). (D) 16S rRNA abundance determined via qPCR using universal primers in cecal contents. Data represent mean ± s.e.m. **p<0.01; *p<0.05 via unpaired t-test at each time point. (E) Relative abundance of oscillating 16S rRNA OTUs determined via eJTK_CYCLE in fecal samples collected every 6h over 48h and in cecal contents (F) collected every 4h over 24h at sacrifice. Data represent mean ± s.e.m. See also Figure S2A–E, Table S3 and S5.
Detecting significant oscillatory patterns in 16S rRNA amplicon sequencing data is complicated due to lack of absolute OTU abundance information and limited tools to describe non-linear relationships in relative abundance data (Friedman and Alm, 2012; David, et al. 2014). However, we detected diet-dependent oscillations of gut microbes using an improved version of JTK_CYCLE (Thaiss et al., 2014; Zarrinpar et al., 2014), empirical JTK_CYCLE with asymmetry (Hutchison et al., 2015 in press) (Table S3). HF reduced the number of OTUs exhibiting significant oscillation patterns in both fecal and cecal contents compared to RC. Under RC, the majority of significant oscillating OTUs in both fecal (Figure 2E) and cecal contents (Figure 2F) belonged to the family Lachnospiraceae, which were mostly absent in HF. Despite a significant increase in relative abundance of specific OTUs induced by HF (i.e., Figure 2E,F Ruminococcus, OTU #4390754 and OTU #261365), diurnal oscillations were absent. Thus, the oscillatory nature of specific microbes may be even more important than their relative abundance. Interestingly, we noted a significant oscillation of OTU #1100972 (Lactococcus) in fecal pellets under HF conditions that was not evident amongst microbes from RC. Additionally, we observed significant oscillations in OTUs belonging to the genus Oscillospira under both RC and HF in fecal and cecal contents. Examination of OTU oscillations within cecal contents and fecal pellets collected from the same mouse at the time of harvest over 24h showed that two OTUs maintained oscillations in both compartments under RC that were nearly absent in HF (Figure 2F and Figure S2E). These data, however, did not provide insights into the functional implications of oscillatory microbial patterns that could have more influence on host chronobiology.
We therefore performed shotgun metagenomic analysis of cecal contents from RC-fed mice, which failed to reveal broad functional diurnal gene changes (Figure S3A). Nonetheless, a few genes involved in sporulation, protein synthesis, and protein degradation did show significant diurnal variation (Figure S3B). Community function was further assessed using Biolog™, a tetrazolium dye indicator assay, an alternative approach for analyzing functional community metabolic potential (Table S4 outlines substrates and sensitivity assays). Cecal contents from RC or HF tested under anaerobic conditions showed that both diets promote unique carbon substrate utilization signatures (Figure 3A,B). PCoA analysis revealed similar ordination as the 16S rRNA data, where PC1 describes separation by dietary treatment (Figure 3A). Interestingly, communities from HF exhibit tighter clustering within ZT as compared to RC, yet the variability across ZT was reduced (Figure 3B). Fecal samples collected over 48h assessed via Biolog™ revealed that both metabolic potential as well as chemical sensitivity of communities from RC-fed mice exhibit greater oscillatory behavior as compared to HF (Figure 3C; Table S4). Ultimately, this data suggests that not only is the gut microbe metabolic activity dependent on diet, but there is a distinct interaction between diet and time of day, which are both essential factors that modulate microbial function.
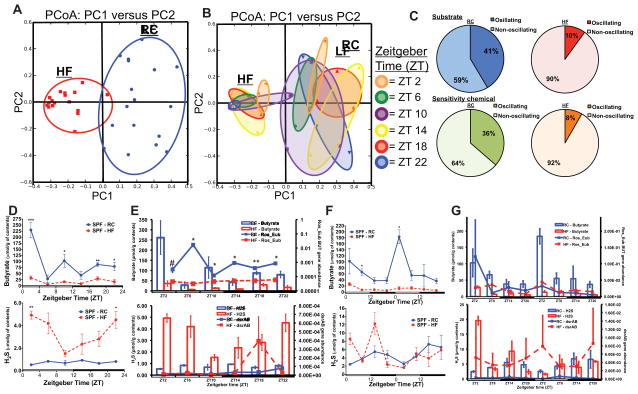
Figure 3. High fat diet shifts diurnal microbial function and metabolite production.
Principle Coordinate Analysis (PCoA) of Biolog™ spectrophotometric analysis of cecal contents from mice fed RC and HF incubated anaerobically and colored by diet (A) and Zeitgeber (ZT) time (B). Percent oscillating vs. non-oscillating substrate and sensitivity chemicals in fecal pellets (C) from HF or RC collected over 48h (n=3/time point). Diurnal cecal butyrate concentration (D, top) and H2S production (bottom) in cecal contents collected from RC or HF (expressed as μmol/g of content) (n=2–3/time point). Data represents mean ± s.e.m. *p<0.05,**p<0.01 determined via unpaired t-test at each time point. (E) Rosburia butyryl-CoA: acetate CoA-transferase (BUT Ros_Eub, top) and dissimilatory sulfite reductase (dsrAB, bottom) gene abundance (right axis, normalized to 16S abundance) in cecal contents from mice fed RC or HF. Butyrate and H2S concentration are presented on the left axis. (F) Fecal butyrate concentration (top) and H2S production (bottom) collected via repeat sampling over 48h from mice fed RC and HF (expressed as μmol/g of content) (n=3/time point. (G) BUT Ros_Eub (top) and dsrAB (bottom) gene abundance (right axis, normalized to 16S abundance) in fecal pellets over 48h. Data represents mean ± s.e.m. #p<0.1, *p<0.05, **p<0.01 determined via unpaired t-test at each time point. See also Figure S3A–D, Table S4 and S5.
Gut microbes exhibit diet-dependent diurnal patterns of metabolite production that elicit direct effects on hepatic CC gene expression in vitro and in vivo
To examine specific microbial metabolites that have known impact on host physiology, we determined relative short chain fatty acid (SCFA) concentration in cecal contents by GC-MS-MS (Renom et al., 2001) (Figure 3D–F) as well as proxy H2S production via methylene blue culture assay (Kartha et al., 2012). Since many of the oscillatory OTUs under RC-fed conditions belonged to the family Lachnospiraceae, which is well characterized for its capacity to synthesize butyrate under specific dietary conditions (Vital et al., 2014), we hypothesized that diurnal variation of SCFAs would be observed in RC-fed mice. Indeed, cecal contents from RC-fed mice exhibited a distinct, diurnal pattern of butyrate concentration, while contents from HF-fed mice did not (Figure 3D). Propionate showed similar diurnal patterns, albeit less pronounced, while acetate, although present at lower levels under HF feeding conditions did not (Figure S3C,D). Robust oscillations in fecal butyrate were also observed over 48h which were absent under HF feeding. Quantitative PCR analysis of gene abundance of microbial butyryl-CoA: acetate CoA-transferase (but; Vital et al., 2013; Louis et al., 2010), an enzyme involved in butyrate production, also exhibited diurnal patterns in cecal contents from RC mice, but not in HF mice. The latter most likely results from dramatic changes in community membership and function in response to dietary intake (Figure 3E). In contrast, cecal H2S production from HF mice exhibited diurnal patterns, which was absent in RC counterparts (Figure 3F). Gene abundance of microbial dissimilatory sulfite reductase (dsrAB), a microbial enzyme involved in H2S production shown to be increased by HF feeding, exhibited diurnal oscillations in cecal contents from HF-fed mice that was not seen in samples from RC-fed animals. Conversely, analysis of fecal pellets collected over 48h from HF-fed mice showed loss of rhythmicity in H2S production, yet this rhythmicity became evident in RC controls, although at a significantly reduced level (see Table S5 for CircWave statistics). This dissonance between cecal and fecal samples could be due to HF-induced alterations in luminal transit which confounds comparisons of oscillatory patterns with those occurring in the cecum.
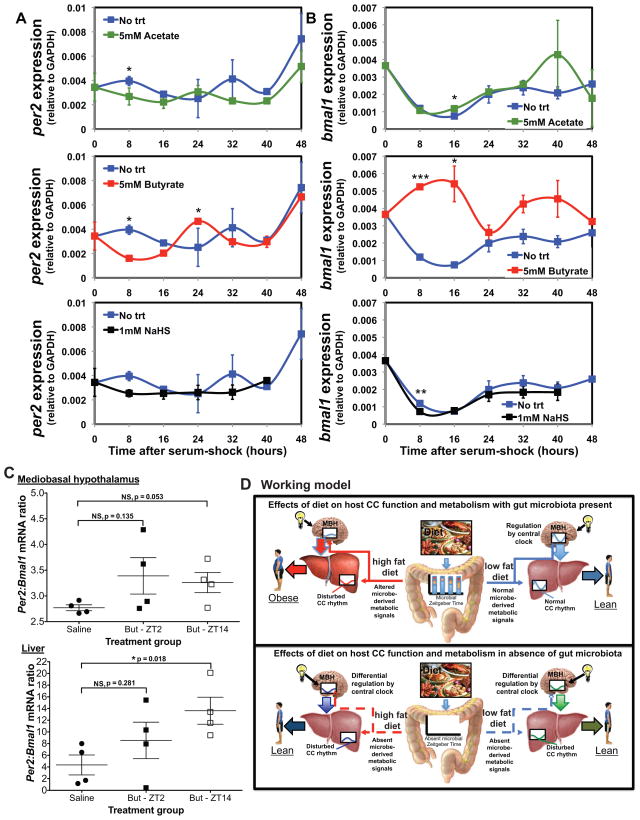
We next determined the direct impact that these particular metabolites have on the cycling of liver CC genes. Hepatic organoids (Huch et al., 2013) (hepanoids) were established in vitro and their CC cell synchronization was initiated via serum shock (Balsalobre et al, 1998). As shown in Figure 4A,B, a nominal diurnal and reciprocating variation in per2 and bmal1 gene expression was evident under basal conditions (no treatment; no trt). In contrast, butyrate, and to a lesser extent, acetate, caused significant shifts in rhythmicity and enhancement in CC gene amplitude, maintaining and accentuating their reciprocating phasic relationships (see Table S5 for CircWave statistics). Exposure to NaHS (an exogenous H2S donor) elicits an opposite effect, causing a blunting and near loss of all phasic changes in both bmal1 and per2 gene expression. To determine if SCFAs elicit direct effects on host CC gene expression in vivo, we intraperitoneally treated GF mice two times daily with either saline twice or saline and butyrate (But, 1000mg/kg) at ZT2 or ZT14 for 5 days followed by harvest of MBH and liver at ZT10 after the final ZT14 treatment. Figure 4C shows peripheral delivery of But does not impact the per2:bmal1 ratio in the MBH, although treatment at ZT14 approaches significance. However, ZT14 But injection significantly increased per2:bmal1 mRNA ratio in the liver compared to saline controls, suggesting peripheral tissues are more sensitive to this SCFA. Together, these results provide a direct link between microbe-derived metabolites and a key host hepatic gene network that could partially explain the observed changes in HF-induced metabolomic shifts.
Figure 4. In vitro and in vivo exposure to diet-induced microbial metabolites alters host circadian gene expression.
Expression of per2 (A) and bmal1 (B) relative to GAPDH following addition of 5mM sodium acetate, 5mM sodium butyrate, or 1mM sodium hydrosulfide (NaHS) in hepanoids after serum-shock. Data is represented as mean ± s.e.m (n=3 replicates/treatment/time point). ***p<0.001; **p<0.01; and *p<0.05 determined via unpaired t-test compared to no trt within time point. See also Table S5. Ratio of per2:bmal1 mRNA (C) in MBH and liver of GF mice treated with saline or butyrate (But) at ZT2 or ZT14 for 5 days. Treatments are saline(ZT2)-saline(ZT14), saline(ZT2)-But(ZT14), But(ZT2)-saline(ZT14). Data represent mean ± s.e.m (n=4/treatment). *p<0.05; NS, not significant determined via unpaired t-test. Proposed experimental model (D) diet-induced change in gut microbe metabolic oscillatory patterns alters the balance between food consumption, the central CC, and hepatic regulatory networks of metabolism promoting DIO.
DISCUSSION
Based on our findings, we propose the working hypothesis shown in Fig. 4D. HF Western diets perturb the chronobiology and composition of microbe-dependent metabolites to which the liver is exposed, resulting in disruption of hepatic CC regulatory networks of host metabolism. Under GF conditions, chronobiological cues from gut microbes are absent, resulting in reduced amplitude of circadian gene expression in both central and peripheral tissues even in the presence of light-dark signals. Dysregulated CC function leads to a heightened metabolic state, resulting in leanness even in the presence of HF. However, SPF mice fed HF results in an altered gut microbiota community membership. Here, oscillations of diurnal production of known microbial metabolites are either lost (SCFAs) or gained (H2S), which directly impacts CC gene expression in the liver, shifting the host to a lower metabolic state and DIO. While shifts in time of food intake due to HF feeding may play a dominant role in altering diurnal oscillations of gut microbes as well as the production of microbial metabolites (Thaiss et al., 2014; Zarrinpar et al. 2014), it is clear from our studies with constant, peripheral infusion of nutrients via PN that host factors can also provide cues or substrates (i.e., mucin) that can sustain microbial diurnal patterns.
Recent evidence shows that SPF mice fed HF leads to an overall shift in the oscillating liver metabolome with corresponding changes in oscillations of the liver transcriptome, particularly of transcripts under CC control (Eckel-Mahan et al., 2013). Other work identified diurnal fluctuations in the liver of a specific phospholipid metabolite that influences downstream activation of NRs leading to fatty acid uptake and utlization in peripheral skeletal muscle. Under HF, diurnal production of this metabolite is blunted and plays a role in the development of DIO (Liu et al., 2013). Despite this knowledge, the contribution of HF-induced gut microbiota and the subsequent alterations in the oscillating microbial metabolome which can directly impact host peripheral tissues remains unknown. Our study however highlights the complex and synchronous relationships between dietary consumption/composition, gut microbial function and metabolomes, as well as host metabolic networks that are critical for maintenance of health, yet when perturbed, promote metabolic imbalances. The evidence suggests that gut microbes have co-evolved as host functions became more complex (i.e., host CC rhythms). Our work and the work of others supports the notion that a number of gut microbes and their functions can entrain on timing and dietary composition/intake. We speculate microbial metabolites promoted by a diverse, yet healthy, diet are crucial to orchestrate and maintain proper oscillations of host molecular metabolism in central and peripheral tissues that follow a consistent pattern of diurnal variation, which can be integrated with other entrainment cues (i.e., light dark signals). Additionally, perturbations of diet (HF or PN) profoundly impact oscillations and content of microbially-derived metabolites which elicit feedback on the host CC. The knowledge gained from our study opens opportunities for unique and targeted interventional strategies to treat and prevent DIO, ranging from the manipulation of gut microbial function to pharmacological targeting of host pathways to restore metabolic balance. Finally, our studies provide a scientific basis for many recommendations that have been based on empirical observation, such as the importance of when and what you eat, in addition to lifestyle changes.
EXPERIMENTAL PROCEDURES
Mice
All murine experimental procedures were institutionally approved. Male 8–10 wk-old C57Bl/6J SPF mice were individually housed and acclimated for 2 wks. Age- and gender-matched, individually-housed GF C57Bl/6 mice were bred and maintained in plastic isolators. All mice were maintained on a 12:12 L/D cycle (ZT0 = 6am) in the same vivarium room and fed ad libitum with normal rodent chow (Harlan Teklad 2018S) or 37.5% saturated milk fat diet (Harlan Teklad TD.97222 customized diet, Table S1). Gnotobiotic diets were autoclaved or irradiated and sterility tested prior to and after the experiment. Weekly body weight and food consumption were recorded and after 5 wks, fecal pellets were collected every 6h over 48h (n=3/treatment). Three mice per group were sacrificed via ketamine/xylazine overdose every 4h over 24h. MBH and liver were isolated, snap-frozen in Trizol, and stored at −80C. Cecal contents were snap-frozen and stored at −80C. Male 8–10 wk-old GF C57Bl/6 mice were divided into 3 treatments (n=4/trt) and i.p. injected twice per day with 100uL saline or butyrate (1000mg/kg) for 5 days. Butyrate was administered once daily at ZT2 or ZT14 and MBH and liver were harvested at ZT10 after the final ZT14 injection.
DNA extraction and 16S rRNA amplicon sequencing analysis
Extraction of bacterial DNA from fecal and cecal contents was performed as previously described (Wang et al., 2009). The V4–V5 region of the 16S rRNA encoding gene was amplified using standard protocols (http://www.earthmicrobiome.org/emp-standard-protocols/, Earth Microbiome Project, 2011). Sequencing was performed at the High-Throughput Genome Analysis Core (Institute for Genomics & Systems Biology) at Argonne National Laboratory. Sequences were trimmed and classified using QIIME (Caporaso et al., 2010). OTUs were picked at 97% sequence identity using open reference OTU picking against the Greengenes database (05/13 release) (McDonald et al., 2012). Representative sequences were aligned via PyNAST (Caporaso et al., 2010), taxonomy assigned using the RDP Classifier (Wang et al., 2007), and a phylogenetic tree was built using FastTree (Price et al., 2010). Weighted UniFrac distances were computed to produce a beta-diversity dissimilarity matrix (Lozupone and Knight, 2005). Significant changes in OTU abundance were assessed using Analysis of Variance (ANOVA) and Pearson correlations (Bonferroni correction for multiple tests; α=0.05) (Caporaso et al., 2010). Multivariate statistical tests include Analysis of Similarity (ANOSIM) and Mantel tests (Caporaso et al., 2010). Rhythmic OTUs were detected in cecal contents via eJTK_CYCLE analysis (Hutchison et al., 2015) using a set period of 24h, a phase search every 2h from ZT0 to ZT22, and an asymmetry search every 2h from 2 to 22. Rhythmic OTUs were detected in 48h fecal collections using eJTK_CYLCE by combining time courses from different mice and using a set period of 24h, a phase search at ZT2, ZT8, ZT14, ZT20, and an asymmetry search at 4h, 12h, and 20h. OTUs exhibiting an empirical P-value less than 0.1 were considered significant.
Microbial metabolism measurements
Biolog GenIII plate wells were inoculated with cecal contents or fecal pellets from individual mice resuspended in Tetrazolium indicator dye and incubated anaerobically at 37°C overnight. Absorbance at 590nm was determined and PCoA derived from Bray Curtis dissimilarity of absorbance was generated in QIIME. For the detection of rhythmic substrate utilization in 48h fecal collections, time courses from different mice were combined and eJTK_CYLCE analysis (Hutchison et al., 2015) was performed as previously described. SCFAs were extracted from cecal contents or fecal pellets as previously described (Renom et al., 2001). Fresh samples were collected, weighed, homogenized in water, and centrifuged at 13,000 x g. Supernatants were acidified and Isobutyric acid (3mM) was added (internal standard). SCFAs were extracted using diethyl ether, derivatized using MTBS-TFA, and run on a Varian Saturn 2000 GC-MS-MS. H2S production potential of cecal and fecal microbes was determined via the Agar-trap method (Kartha et al., 2012). Fresh cecal or fecal contents were collected, weighed, transferred into an anaerobic chamber, and resuspended in Bru media based on weight. Samples were inoculated into culture flasks with complete Bru media (containing Hemin and Vit K) with pre-set Zinc agar, sealed, and incubated anaerobically at 37°C. H2S was estimated spectrophotometrically at 670nM via Methylene blue assay after 48h.
Hepanoid culture
Hepanoids were prepared from 8–10 week old SPF C57Bl/6 mice as previously described (Huch et al., 2013). Post differentiation, hepanoids were synchronized (Balsalobre et al., 1998), serum-rich media was removed, and fresh media was added containing sterile-filtered treatments (5mM butyrate, 5mM acetate, or 1mM H2S). Wells were collected every 8h for 48h and snap-frozen in Trizol. RNA was extracted, cDNA synthesized, and qPCR performed as previously described.
Statistical analysis
Statistical analyses were performed using GraphPad Prism v5.0d via unpaired t test or ANOVA. Dunnett’s post-test was applied to compare to SPF RC control. Samples were removed from host gene expression analyses if GAPDH values were out of range; all time points contain 2 or more mice. CircWave v1.4 software (www.huttlab.nl, www.euclock.org) was used to detect rhythmicity of host CC gene expression, hepanoid CC gene expression, microbial metabolites, and microbial gene expression by applying Fourier-curve fit analyses.
Supplementary Material
Acknowledgments
This work was supported by NIDDK grants DK42086, DK47722, UH3 DK083993, DK097268, the Peter and Carol Goldman Family Research Fund, the Gastrointestinal Research Foundation of Chicago (E.B.C), DDRCC NIDDK P30DK42086 (V.L.), and DARPA D12AP00023 (A.R.D.). S.M.G. is supported by an EPA STAR Graduate Fellowship. A.L.H. is a trainee of the University of Chicago NIH MSTP program (NIGMS T32GM07281). Amplicon sequence data are deposited at figshare (http://dx.doi.org/10.6084/m9.figshare.882928). Metagenomic shotgun sequence data are deposited at MG-RAST (http://metagenomics.anl.gov) under Project ID 5867. Murine microarray data can be found at NCBI Geo (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE53590. We are grateful to S. Owens and J. Marcell for microbial sequencing (ANL). We thank A. Vest, M. Yabes, and M. Bales for GF husbandry (Charles River). We are indebted K. Keegin for initial suggestions (UC, ANL), D. Antonopoulos for critical reagents (microbial pCR4-TOPO primer plasmid standards, ANL), and T. Hasbe, K. Touw, A. Bobe, (UC) and S. Devkota (Harvard University) for assistance with sample collection.
Footnotes
Author contributions: V.L. and E.B.C. were involved in all aspects of these experiments. K.M., E.Y.H., C.M.C., J.F.P., A.F.H., A.N., N.H., E.Z., Y.W., and M.W.M. helped perform experiments. Y.H., B.P., S.M.G. A.L.H., A.R.D., and J.A.G. provided critical feedback and data analysis of host and microbial datasets. B.T., K.A.K., and B.P. provided critical experimental feedback.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERNCES
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15724. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bass J, Turek FW. Sleepless in America: a pathway to obesity and the metabolic syndrome? Arch Intern Med. 2005;165:15–16. doi: 10.1001/archinte.165.1.15. [DOI] [PubMed] [Google Scholar]
- Bjorkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. Intestinal Microbiota Regulate Xenobiotic Metabolism in the Liver. PLoS One. 2009;4:e6958. doi: 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costella EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, Gonzalez A, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulous DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–109. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, Chang EB. Composition of Dietary Fat Source Shapes Gut Microbiota Architecture and Alters Host Inflammatory Mediators in Mouse Adipose Tissue. J Parenter Enteral Nutr. 2013;37:746–754. doi: 10.1177/0148607113486931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison AL, Maienschein-Cline M, Chiang AH, Tabei SMA, Gudjonson H, Bahroos N, Allada R, Dinner AR. Improved statistical methods enable greater sensitivity in rhythm detection for genome-wide data. PLoS Comp Bio. 2015 doi: 10.1371/journal.pcbi.1004094. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CGM. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci USA. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartha RV, Zhou J, Hovde LB, Cheung BW, Schroder H. Enhanced detection of hydrogen sulfide generated in cell culture using an agar trap method. Anal Biochem. 2012;423:102–108. doi: 10.1016/j.ab.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Backhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K, Bhargava P, Gangl MR, Dai L, Hatano B, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502:550–554. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hogenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murkherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabot S, Membrez M, Bruneau A, Gerard P, Harach T, Moser M, Raymond F, Mansourian R, Chou CJ. Germ-free C57Bl/6 mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24:4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- Renom G, Bulois P, Hafraoui S, Colombel JF, Degand PM. Simple gas chromatography analysis of faecal butyrate: Application to patients at risk of pouchitis. Clin Chem Lab Med. 2001;391:15–19. doi: 10.1515/CCLM.2001.005. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss C, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostatis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hoeing JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauter E, Polonsky KS, Scheen AJ. Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr Rev. 1997;18:716–738. doi: 10.1210/edrv.18.5.0317. [DOI] [PubMed] [Google Scholar]
- Vital M, Penton CR, Wang Q, Young VB, Antonopoulos DA, Sogin ML, Morrison HG, Raffals L, Chang EB, Huffnagle GB, et al. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome. 2013;1:8–21. doi: 10.1186/2049-2618-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Gao J, Rizzo M, Harrison T, Tiedje JM. Diet is a major factor governing the fecal butyrate-producing community structure across Mammalia, Aves, and Reptilia. ISME J. 2014:1–12. doi: 10.1038/ismej.2014.179. advanced online publication, Oct 24 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.