Abstract
Osteolytic bone damage is a major cause of morbidity in several metastatic pathologies. Current therapies using bisphosphonates provide modest improvement, but cytotoxic side effects still occur prompting the need to develop more effective therapies to target aggressive osteoclastogenesis. Increased levels of Receptor Activator of Nuclear Factor Kappa B Ligand (TNFSF11/RANKL), leading to RANKL-RANK signaling, remains the key axis for osteoclast activation and bone resorption. Osteoprotegerin (TNFRSF11B/OPG), a decoy receptor for RANKL is significantly decreased in patients who present with bone lesions. Despite its potential in inhibiting osteoclast activation, OPG also binds to tumor necrosis factor related apoptosis-inducing ligand (TNFSF10/TRAIL), making tumor cells resistant to apoptosis. Towards uncoupling the events of TRAIL binding of OPG and to improve its utility for bone remodeling without inducing tumor resistance to apoptosis, OPG mutants were developed by structural homology modeling based on interactive domain identification and by superimposing models of OPG, TRAIL and its receptor DR5 (TNFRSF10B) to identify regions of OPG for rational design. The OPG mutants were purified and extensively characterized for their ability to decrease osteoclast damage without affecting tumor apoptosis pathway both in vitro and in vivo, confirming their potential in bone remodeling following cancer-induced osteolytic damage.
Keywords: Osteoprotegerin, TRAIL, RANKL, osteoclast, bone
INTRODUCTION
The therapeutic potential of OPG in bone remodeling has been of great interest in the last decade. Human OPG is comprised of 401 amino acids with a molecular weight of approximately 60 kDa. Structurally, OPG consists of four cysteine-rich pseudo-repeats at the N-terminus, two death domains, a heparin binding site at the C-terminus and a 21-amino acid signal peptide (1,2). The importance of OPG as a negative regulator of osteoclastogenesis is evident from studies with transgenic mice where over-expression of OPG has been shown to cause severe osteopetrosis and reduce the number of mature osteoclasts, while OPG gene knockout leads to osteoporosis (3). This understanding led to the use of OPG as a potential therapeutic molecule for various diseases including glucocorticoid-induced osteoporosis, rheumatoid arthritis, vascular calcification, and osteolytic malignancies (1,4–6).
The key regulators of bone remodeling in respect to osteoclast biology belong to the tumor necrosis factor (TNF) superfamily. Activation and maturation of osteoclasts results from the binding of Receptor Activator of Nuclear Factor kappa B Ligand (RANKL) to its receptor RANK, found on osteoclast and osteoclast precursor cells of monocyte lineage (3,7,8). Osteoprotegerin, which is a soluble glycoprotein, is a member of the TNF receptor family and acts as a decoy receptor by binding RANKL with a higher affinity than RANK, thus neutralizing the RANK-RANKL interaction and ultimately osteoclastogenesis (8).
Despite its potential as a possible molecule for therapeutic targeting of osteolytic damage in bone metastasis, OPG also acts as a receptor for the cytotoxic ligand, TRAIL, thereby conferring resistance to tumor cell apoptosis. Thus, while OPG can effectively block osteoclastogenesis, it also promotes tumor cell survival by impairing TRAIL function. To overcome this limitation and to improve the clinical utility of OPG for bone remodeling in osteolytic malignancies, the current study sought to uncouple these two properties of OPG. By using structural homology modeling of OPG, RANKL, and TRAIL, we have identified, developed and validated genetic mutants of OPG, lacking TRAIL binding but preserving RANKL binding. The OPG-mutants were first functionally characterized in vitro to demonstrate selective inhibition of RANKL-mediated osteoclastogenesis, and then their low binding affinity to TRAIL facilitating caspase-3-induced apoptosis of tumor cells upon treatment with TRAIL. Further, in vivo validation of two such OPG mutants, in a preclinical mouse model of a bone-disseminated osteolytic tumor, demonstrated their potential in restoring bone remodeling. The two novel OPG variants will be promising for therapeutic targeting of aggressive osteoclast-induced bone damage and associated morbidity in several osteolytic malignancies.
MATERIALS AND METHODS
Modeling of OPG-TRAIL complex
The structural model of OPG/TRAIL complex was generated by superimposition of human TRAIL monomer (PDB code: 1D4V) onto human RANKL/OPG complex structure (PDB code: 3URF) using human RANKL monomer as a reference structure. The generated model includes binding interface between a monomer of OPG and two monomers of trimeric ligands. Rigid body movement and model analysis were carried out using both UCSF Chimera (9) and Coot (10), and all figures were made with the PyMol program (http://www.PyMol.org).
Cell lines and Reagents
The human osteolytic prostate cancer cell line PC3, expressing firefly luciferase, (a kind gift from Dr. Kenneth J. Pienta, University of Michigan, Ann Arbor, Michigan), was maintained in RPMI-1640 medium (Mediatech Inc. Hendron, VA) supplemented with 10% fetal bovine serum (Mediatech Inc.) and penicillin/streptomycin (Mediatech Inc). The murine macrophage cell line, RAW-264.7 was maintained in DMEM supplemented with 10% FBS, 4 mM L-glutamine, 1% antibiotics and 10% macrophage colony-stimulating factor (M-CSF, a kind gift from Dr. Xu Feng, The University of Alabama at Birmingham). The human melanoma cell line MDA-MB-435 was maintained in 50% DMEM, 50% DMEM F12, 1% penicillin/streptomycin, 10% FBS, and non-essential amino acids. HEK-293 cells were purchased from ATCC and maintained in DMEM supplemented with 10% new born calf serum and 1% penicillin-streptomycin. The proliferation index of PC3 cells was determined by CellTiter 96® AQueous One Solution Cell Proliferation Assay Kit (Promega Corporation, Madison, WI) as recommended by the manufacturer. Recombinant OPG was purchased from R&D Systems and recombinant TRAIL was purchased from Millipore. Purified RANKL was a kind gift from Dr. Xu Feng, UAB. Transfections were performed using Purefection reagent (System Bioscience, Mountain View, CA).
Animal use and care
Athymic nude mice (nu/nu), 6–8 weeks of age, were purchased from Frederick National Laboratory-NCI Frederick, MD or Charles River, CA) and housed in the animal facility at UAB. Animal care and treatments were conducted in accordance with established guidelines and protocols approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Cloning of OPG expression vector
The recombinant OPG used in this study for creating genetic variants was comprised of the ligand-binding domain of human OPG (1-201 amino acids), fused to the Fc domain of human IgG. The OPG-Fc fusion gene was isolated from an adenoviral construct (kindly provided by Dr. Joanne Douglas, University of Alabama at Birmingham) and subcloned into pcDNA3.1 under control of the CMV promoter by directional subcloning using BamH1 restriction site. The recombinant plasmids were verified for proper orientation by restriction digestion, and the open-reading frame by DNA sequencing at the UAB Center for AIDS Research DNA Sequencing Core.
Site-directed mutagenesis
The OPG mutants (Y49R, N81A, F96A and F107A) were generated based on putative OPG residues identified from structural model of OPG-TRAIL complex. The sequences of primers used for mutagenesis are given in Table. 1. Site-directed mutagenesis was performed using Quickchange XL Kit (Stratagene) and PfuTurbo DNA Polymerase (Stratagene) as per the manufacturer’s instructions with a “touch-down” PCR protocol as follows: After an initial denaturation step at 95°C for two minutes, cycles were performed at 95°C denaturation for 50 seconds, followed by annealing at 65°C, and extension at 72°C for 2 min per kilobase pair of template DNA. The annealing step was reduced by 1°C for 10 cycles until an annealing temperature of 55°C was reached. Eight more cycles were then repeated at an annealing temperature of 55°C for a total of 18 cycles. A final extension step at 72°C was then performed and the reaction mix was digested with restriction enzyme DpnI to remove the template DNA. Five microliters of the reaction mix was then transformed into OneShot Top10 competent cells (Invitrogen) and plated onto agar containing 100 μg/mL ampicillin. Clones were screened for the identification of respective mutations by DNA sequencing. All DNA sequencing was performed at the UAB Center for AIDS Research DNA Sequencing Core.
Table 1.
| Sequences of primers used for site-directed mutagenesis | |
|---|---|
| OPG-Y49R |
|
| OPG-N81A |
|
| OPG-F96A |
|
| OPG-F107A |
|
Expression of recombinant OPG proteins
HEK-293 cells were transfected with either OPGwt or OPGmut (Y49R, N81A, F96A, and Y107A) plasmid constructs individually. Forty-eight hours after transfection, culture media were collected and concentrated using a 30 kDa cut-off centrifugal filter unit (Amicon). The expression of recombinant OPGs in the culture media were analyzed by Western blot using antibodies against OPG (R&D Systems, Inc., Minneappolis MN) and Fc (Sigma-Aldrich, St. Louis, MO).
Osteoclast assay
Primary murine macrophage cells were cultured in 48-well dishes at a density of 2×104 cells per well. Cells were allowed 24 hrs to attach at which point 200 ng of recombinant OPG, OPGwt or OPGmut were added to individual wells in combination with or without 60 ng of purified RANKL. The medium was changed every 48 hours with fresh supplementation of OPG and RANKL. The cultures were maintained for 10 days following which tartrate-resistant acid phosphotase (TRAP) staining was carried out to identify multi-nucleated TRAP-positive osteoclasts. While assessing under light microscope, 10 fields of view were randomly selected to quantify the number of osteoclasts.
Cell proliferation assay
MDA-MB-435 cells were cultured in 96 well dishes at a density of 4×104 cells per well. Twenty-four hours later, 100 ng of recombinant TRAIL was added to each well in combination with 200 ng of recombinant OPG, OPGwt or OPGmut. After forty-eight hours, the cells were fixed in 3.7% paraformaldehyde and stained with 0.05% crystal violet (Sigma) for 30 min and analyzed under a light microscope (100x). Additionally, from replicate cultures, cell proliferation was determined 48 hours later using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) and then colorimetrically measured at 490 nm. The assay was performed three times independently.
TRAIL-mediated apoptosis
To demonstrate further that TRAIL binding to OPGwt and not OPGmut was responsible for apoptosis induction in cancer cells, downstream caspase activity was determined following TRAIL and OPG treatments as above. Briefly, the human osteolytic prostate cancer cell line, PC3 was cultured in 60 mm dishes at 106 cells per dish and allowed to adhere for 24 hours. Combinations of 100 ng of recombinant TRAIL and 200 ng of recombinant OPG, OPGwt or OPGmut were added to the cells and cultured for additional 3 hrs. The cells were then harvested and lysed using cell culture lysis reagent (Promega, Madison, WI). Cell lysates were tested by Western blot analysis using cleaved caspase-3 antibody (Santa Cruz Biotechnology Inc., TX).
Development of genetically-engineered human mesenchymal stem cells for cell therapy
Human mesenchymal stem cells (MSC) were isolated from surgical bone marrow transplant remnants of healthy donors from UAB following an IRB approved protocol. The cells were cultured in DMEM, 10% fetal bovine serum, and 1% penicillin-streptomycin to confluence for cell sorting. Upon FACS analysis for positive MSC surface markers CD44, CD73, CD90 and after verifying the pluripotency by lineage differentiation into adipocytes, chondrocytes and osteoblasts (11), MSC were transfected with pcDNA-OPGwt/mut expression vectors using purefection reagent (Systems Bioscience, Mountain View, CA) as per the manufacturer’s instructions. Media from MSC, transfected with OPGwt or OPGmut (Y49R or F107A) constructs were collected and concentrated using a 30 kDa filter and analyzed by Western blotting using OPG and Fc antibodies.
Therapeutic potential of genetically-engineered MSC expressing OPGmut (Y49R and F107A), in a mouse model of tumor-induced osteolysis in vivo
Athymic nude mice were injected intra-tibially with ~1×105 human osteolytic prostate cancer line, PC3, constitutively expressing firefly luciferase. Twenty-four hours later, non-invasive imaging was performed to confirm implantation of the cells within the tibia and mice were injected with ~3×105 MSC, transfected with either OPGwt or OPGmut (Y49R or F107A) in the same tibia. Similar cohorts of mice were also used to test the therapeutic potential of MSC, engineered to express OPGwt or OPGmut, through systemic administration following implantation of PC3 cells in the tibia. In this group of experiment, 24 hours after implantation of PC3 cells in tibia, 3×105 MSC-OPGwt/mut were systemically administered through tail vein. Mice from both routes of MSC administration were non-invasively imaged for tumor growth based on luciferase expression up to 14 days post MSC therapy and sacrificed for analysis of osteolytic damage and bone remodeling.
Micro-computed tomograph analysis of bone tissue
Superficial computed-tomography scanning of the tibiae was performed on bone tissues using MicroCAT II (Imtek Inc., Knoxville, TN). For the determination of the three-dimensional architecture of trabecular bone, mice were sacrificed, tibiae harvested and then analyzed in an advanced micro-computed tomography instrument (μCT 40, Scanco Medical AG, Wayne, PA). Two scans were performed on each tibia, one for whole tibial bone with 16-μm resolution and one for trabecular analysis with a 6-μm resolution. A three-dimensional reconstruction of the images was created with the region of interest consisting of the trabecular area under the growth plate. The scan of the trabecular bone was performed beginning 25 slices below the growth plate and 100 slices were used for analysis and 3D reconstruction.
Statistical Analysis
Significance levels of the experimental data were determined by student’s t-test and p-values less than 0.05 were considered as statistically significant.
RESULTS
Development of genetically-engineered OPG variants
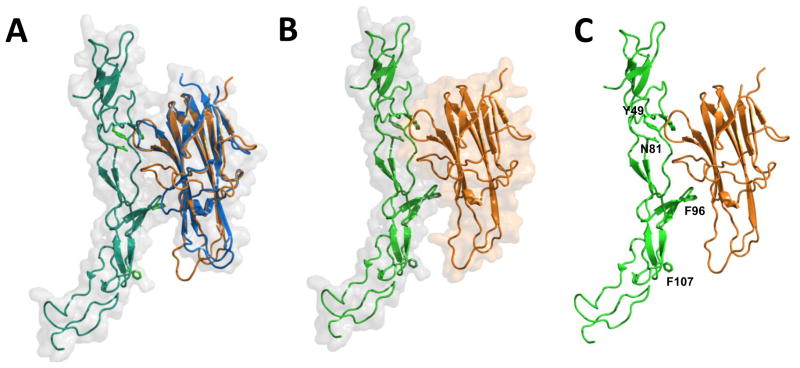
Towards developing OPG variants that lack TRAIL binding, yet retain RANKL binding, the binding interface of OPG with RANKL and TRAIL was structurally aligned. Since the crystal structure of OPG/TRAIL complex was not available, we generated the structural model of OPG/TRAIL complex by rigid body movement to the homologous structure. The RANKL/OPG complex (PDB code: 3URF) was utilized as a reference structure where the structure of TRAIL monomer from TRAIL/DR5 complex (PDB code: 1D4V) was superimposed onto RANKL (rms = 0.735, Fig. 1A). The generated model of OPG/TRAIL complex was then analyzed by using Coot and UCSF Chimera modeling softwares, which further helped in the identification of putative interaction sites between TRAIL, RANKL and OPG (Figs. 1B and 1C). The most striking difference between RANKL and TRAIL was an amino acid loop that showed possible interactions of TRAIL and OPG (Fig. 1A). Analysis of the model indicated that the amino acid loop of TRAIL may interact with OPG in a similar manner as it interacted with DR5. A close-up view of the amino acid loop indicated that residues Y49 and N81 in OPG shared a similar mode of interaction with TRAIL as the respective conserved residues in DR5. However, F107 appeared to be projecting away from the loop. There also appeared to be possible contacts with Q95 and F96 in the 90s loop of OPG. None of these interactions appeared to take place with RANKL. These observations suggest that the OPG binding interface may include amino acid residues Y49, N81, F96, and F107 that potentially interact with TRAIL (Fig. 1C). To test this hypothesis, the above residues were substituted to Y49R, N81A, F96A, and F107A to create individual OPG-mutant open-reading frames (ORF).
Figure 1. Structure model of OPG-TRAIL complex.
Superposition of human TRAIL (orange) with human RANKL (blue)-OPG (green) complex using a RANKL monomer as a reference (A); Surface diagram of TRAIL (orange) and OPG (green) (B); and, putative OPG residues (in green) interacting with TRAIL (C) are shown. Designated amino acid residues (Y49, N81, F96, and F107) were subjected to site-directed mutagenesis. Ribbon and surface diagrams were created with the PyMol program.
Expression purification and characterization of OPG mutants
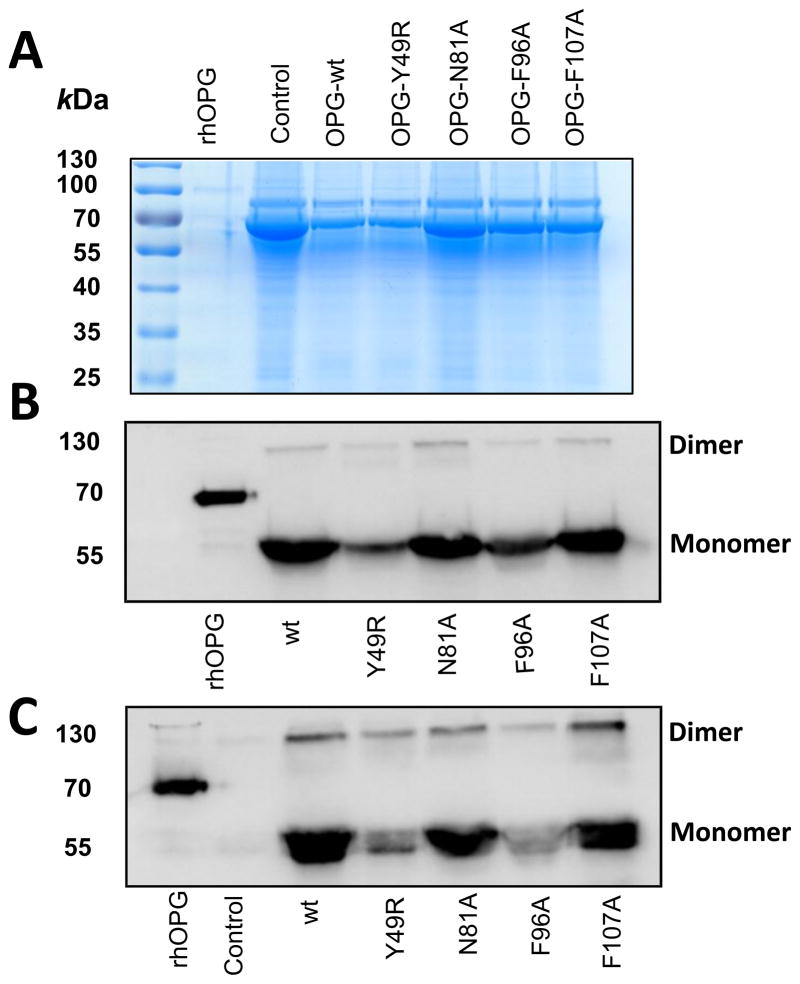
In order to produce OPG mutants in a eukaryotic expression system, the ORF of OPG were subcloned in pcDNA3.1 vector together with the Fc portion of human immunoglobulin molecule as a fusion protein at the C-terminus to enhance the stability of recombinant proteins. Site-directed mutagenesis was performed using this OPG construct and primers corresponding to single point mutations in putative TRAIL-binding sites of OPG (Y49R, N81A, F96A, and F107A). Following verification by sequence analysis of both the DNA strands, plasmids encoding OPGwt and OPGmut were transfected into HEK-293 cells. The supernatants from plasmid transfected cells, containing OPGwt and OPGmut, were concentrated and analyzed by SDS-PAGE to confirm the molecular mass at the expected size. As a measure of loading control for the culture supernatants, identical volume of conditioned media from OPG wt and OPGmut plasmid-transfected HEK-293 cells were resolved in SDS-PAGE and the gel was stained with Coomassie Blue. Results, confirming concentration of proteins between samples is shown in Fig. 2A. Immunoblotting with OPG and Fc antibodies confirmed the specificity of the proteins in monomeric and dimeric configurations (Figs. 2B & C).
Figure 2. Immunoblot analysis of purified OPG-mutants.
Forty-eight hours following transfection of expression plasmids containing the sequences of OPG-wt and OPG-mutants in HEK-293 cells, supernatant media were collected and concentrated using a 30 kDa filter. The concentrated media were then separated in a SDS-PAGE and transferred to nitrocellulose membrane. Loading control was assessed by Coomassie Blue staining (A) and detection of OPG-wt and OPG-mutants was performed using antibodies for IgG (B) and OPG (C).
OPG-mutants inhibit RANKL-induced osteoclastogenesis
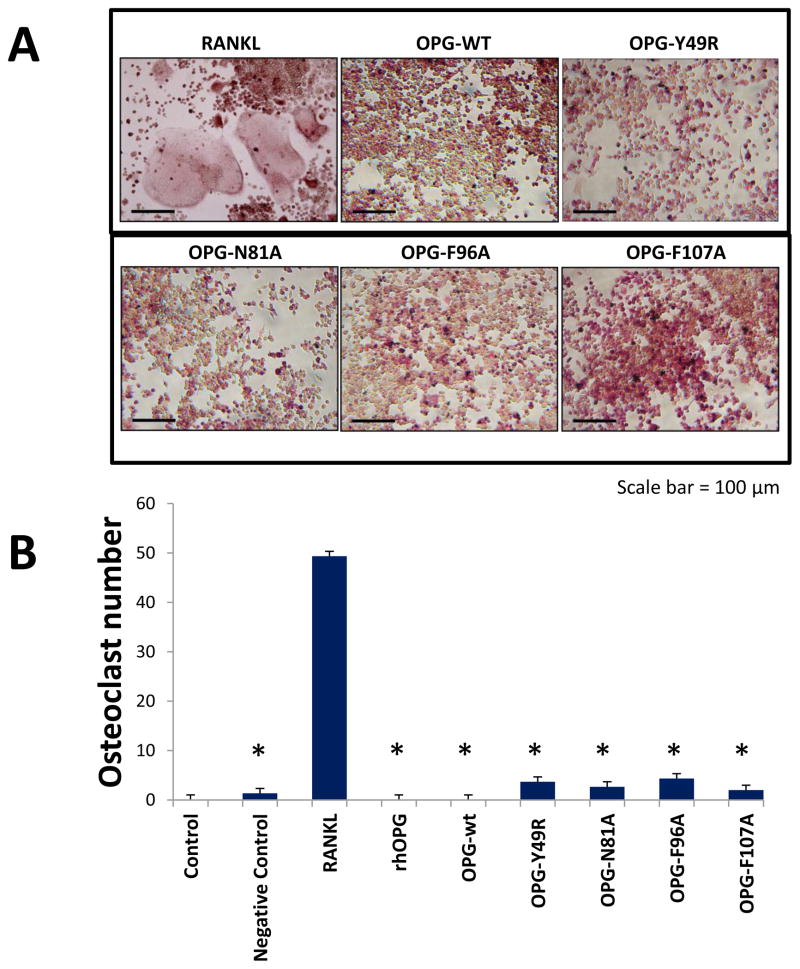
To determine the ability of OPG mutants to inhibit osteoclastogenesis, RAW-264.7 cells were cultured in the presence of RANKL, and OPGwt or OPGmut. Results of tartrate-resistant acid phophatase (TRAP) staining indicated the appearance of large, multinucleated osteoclasts in RAW-264.7 cell cultures with RANKL only (Fig. 3A). Cultures with RANKL and recombinant OPG or OPGwt and RANKL resulted in no osteoclast differentiation, as expected. Interestingly, in cultures containing OPGmut proteins, there was also a significant reduction in the formation of osteoclasts even in the presence of RANKL, confirming the genetically-engineered OPG mutants were functionally active in inhibiting osteoclast formation by retaining RANKL binding affinity. Quantitative analysis of osteoclasts from different culture conditions indicated a statistically significant difference in osteoclast formation between RANKL-treated controls and those treated with recombinant wild-type or mutant OPG, in combination with RANKL (Fig. 3B).
Figure 3. Osteoclast assay to determine retention of RANKL binding of OPG-mutants.
Primary macrophage cells were cultured in the presence of 200 ng of recombinant OPG, OPG-wt or indicated OPG-mutants in the presence or absence of 60 ng of recombinant RANKL. As a positive control, macrophage cells were cultured with RANKL alone. Tartrate-resistant acid phosphatase (TRAP) staining of macrophage cell cultures were performed 7–14 days after cells were cultured in the presence of OPG-wt or OPG-mutants and RANKL (A). Quantitative analysis based on the number osteoclast seen in 10 random fields (B) (*p<0.005 compared to culture with RANKL only).
OPG-mutants do not interfere with TRAIL-induced apoptosis of cancer cells
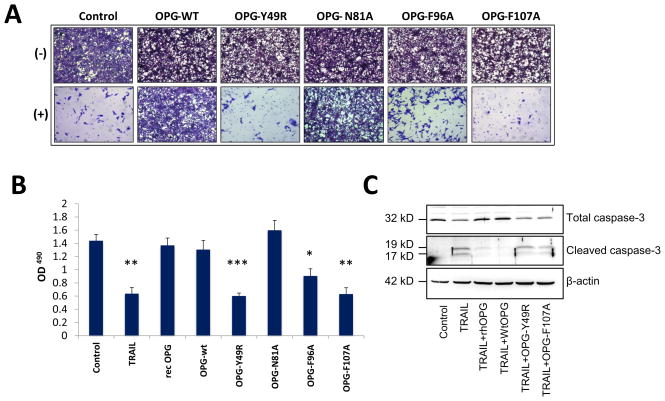
After confirming that the OPG-mutants effectively inhibited osteoclastogenesis, they were assessed for interference with TRAIL-mediated apoptosis of cancer cells. Supernatants containing OPGmut were concentrated using a 30 kDa filter and tested in MDA-MB-435 cells, in the presence or absence of recombinant human TRAIL. MDA-MB-435 cells were used in this study as they express the TRAIL receptor, DR5, and have been shown to be sensitive to TRAIL (12,13). Results of the study indicated that MDA-MB-435 cells underwent cell death when cultured with TRAIL. Conversely OPGwt inhibited cytotoxic effects of TRAIL as proliferation of cancer cells was found to be comparable to untreated cells (Fig. 4A). While analyzing the mutants for blocking TRAIL-induced apoptosis, the mutant N81A appeared to bind to TRAIL, inhibiting its apoptosis-inducing property, as MDA-MB-435 cells demonstrated resistance to TRAIL treatment. However, OPG-mutants Y49R, F96A and F107A showed abolished TRAIL binding with a significant decrease in cell proliferation and increased cell apoptosis (Fig. 4B). The MDA-MB-435 cells, cultured with recombinant OPG, OPG-wt and OPGmut N81A indicated that these two forms of OPG retained intact TRAIL binding, and inhibited TRAIL-mediated apoptosis leading to cell survival (Fig. 4B). To determine the downstream signaling pathway for TRAIL-induced apoptosis, MDA-MB-435 cells were cultured with OPGwt or OPGmut (Y49R or F107A) and TRAIL to determine activation of cleaved caspase-3 for apoptotic cell death. Western blot analysis confirmed that OPG mutants (Y49R and F107A) did not interfere with TRAIL-mediated apoptosis, as shown in Fig. 4C.
Figure 4. Apoptosis assay to determine abrogation of TRAIL binding by OPG-mutants.
MDA-MB-435 cells were cultured in the presence of 100 ng of TRAIL and 200 ng of OPG-wt or OPG-mutants. As a positive control, MDA-MB-435 cells were cultured with TRAIL alone. After 24 hours, MDA-MB-435 cells were either fixed in 3.7% paraformaldehyde and then stained with 0.05% Crystal violet for 30 minutes and viewed using a light microscope (100x) (A) or cultured with 20 μl of the solution 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) for 2 hours and then measured at an absorbance of 490nm (B) (*p<0.05, **p<0.005, ***P<0.001, compared to control). To determine downstream activity of TRAIL function, a human osteolytic cancer cell line, PC3, was cultured in a combination of TRAIL and OPGwt or OPGmut (Y49R or F107A) for 3 hours. Cells lysates were prepared by harvesting the cultures and Western blot analysis was performed for cleaved caspase-3 activity (C).
Osteoprotegerin mutants Y49R and F107A demonstrated bone protection from cancer-induced osteolysis in vivo
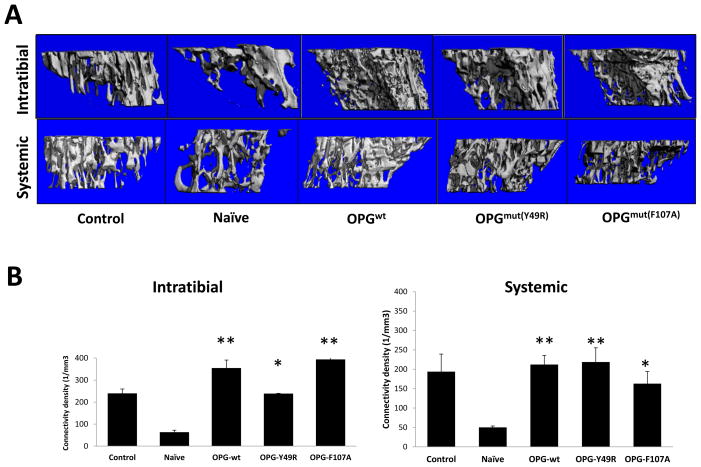
After determining the biological activity of the OPG-mutants in vitro, based on the results from osteoclast and TRAIL assays, we sought to determine the effects of mutant Y49R and F107A (which demonstrated highest activity in RANKL binding and TRAIL-mediated apoptosis) in bone remodeling following cancer-induced osteolysis in vivo. A genetically-engineered stem cell therapy approach was used to deliver the OPG-mutant proteins (Y49R and F107A) in vivo in a therapy model of bone-disseminated human osteolytic prostate cancer in mouse. Cohorts of male athymic nude mice were challenged intra-tibially with a human osteolytic prostate cancer cell line PC3, constitutively-expressing firefly luciferase. Following confirmation of engraftment of the tumor cells by non-invasive imaging, human MSC, transfected with expression vectors encoding OPGwt or OPGmut (Y49R or F107A), were injected in the same tumor microenvironment within the tibia or systemically via tail vein. Mice were sacrificed on day-14 post MSC therapy and tibiae were isolated for micro-computed tomography (Micro-CT) analysis. Results of this analysis confirmed significant bone destruction in the naïve group that received PC3 cells only. However, mice treated with either OPGwt or OPGmut (Y49R or F107A) demonstrated a significant increase in trabecular bone architecture and trabecular connectivity density as compared to the control group both by intratumoral and systemic administration of genetically-engineered MSC, expressing OPGmut thereby providing evidence that the developed OPG mutants Y49R and F107A were therapeutically effective in vivo against cancer induced osteolytic bone damage (Figs. 5A & B).
Figure 5. Micro-CT assessment to determine the potential of OPGmut in inhibiting tumor-induced osteolytic damage.
Nude mice were injected intratibially with ~1×105 osteolytic prostate cancer cell line PC3. Twenty-four hours later, ~3×105 MSC over-expressing OPGwt, OPGmY49R or OPGmF107A were injected into the same tibia for intratumoral administration and by tail vein injection for systemic administration. Mice from both routes of MSC administration were sacrificed 14 days post MSC therapy and tibia were collected for Micro-CT analysis to determine changes in the overall trabecular architecture (A) and connectivity density (B) (*p<0.05; **p<0.01, compared to Naïve).
DISCUSSION
Osteoclasts are the primary cells for physiological and pathological bone resorption during bone remodeling, and RANKL is critically involved in the differentiation, activation, and survival of these cells upon binding to its specific receptor RANK and inducing osteoclast differentiation of progenitor macrophages (7,14). Increased expression of RANKL with certain malignancies including breast cancer, prostate cancer and multiple myeloma causes activation of osteoclasts and remains an important mechanism in the formation of osteolytic bone lesions (15–17). The RANKL-RANK signaling axis, has therefore, has become an important target for therapeutic intervention of osteolytic bone pathology. On the basis of the potent inhibitory actions of OPG on osteoclast differentiation and function, the therapeutic application of OPG in treatment of metastatic bone destruction has been recently studied (18–22). The outcome of phase-1 clinical trial using purified OPG in patients with multiple myeloma and breast cancer indicated that OPG was well tolerated and that a single dose suppressed levels of bone resorption markers, comparable to treatment with pamidronate (23). Thus, improving the biological properties of OPG by uncoupling its TRAIL binding ability will provide a crucial advancement in the utility of OPG for treatment of osteolytic malignancies. Results from the present study indicate that the two novel OPG variants, Y49R and F107A indeed show a positive effect in bone remodeling following cancer-induced osteolytic damage without inhibiting TRAIL-induced apoptosis of cancer cells suggesting a strong therapeutic potential.
Osteoprotegerin also binds to TRAIL with similar affinity as with RANKL (24,25), and therefore, OPG treatment raises the concern that it may affect the function of TRAIL. TRAIL is a cytokine expressed on the surface of tumor-infiltrating macrophages that induce apoptosis, specifically in malignant cells via interactions with the death receptors DR4 and DR5 (26). Although a precise mechanism by which TRAIL specifically induces death of transformed cells is not known, a substantial role for TRAIL has been established as a critical effector molecule in tumor immunosurveillance (27–31), and TRAIL deficient mice suffer from increased susceptibility to tumor initiation and metastases (32). These studies highlight the importance of TRAIL and raise the concern that therapeutic administration of OPG might also diminish host immune defense mechanisms against malignant cells. Thus, the newly developed and characterized OPG mutants in this study will have the potential to overcome this limitation and concern.
Osteoprotegerin and RANK are members of the tumor necrosis factor receptor (TNF-R) superfamily, while RANKL and TRAIL are members of the tumor necrosis factor (TNF) superfamily. The crystal structures of RANKL/OPG and TRAIL/DR5 complex have been resolved and the atomic models demonstrate conserved structural features of their respective superfamilies (33–37). Proteins in the TNF-R family adopt the elongated structures characterized by variable numbers of cysteine-rich domains (CRDs) that form a scaffold of disulfide bridges (38) where the DR5 and OPG contains two and four CRDs, respectively. Monomers of both RANKL and TRAIL contain two antiparallel β-pleated sheets that form a β-sandwich as a core scaffold, which interact with adjacent subunits in a head-to-tail fashion to form a bell-shaped homotrimer (33–36). The elongated receptors fit into the grooves of adjacent protomers of the homotrimeric ligands, and this particular mode of interaction was demonstrated by the crystal structure of TRAIL/DR5 and RANKL/OPG (34,35). Due to this conserved mode of interaction between the TNF and TNF-R family members, we hypothesized that OPG also shared a similar mode of interaction with TRAIL. Conservation in residues among family members is an indication of their functional importance; hence it is plausible that the amino acid loop of TRAIL might also interact with the conserved residues in OPG in a similar manner. Since RANKL lacks this elongated loop structure, we predicted that the amino acid loop of TRAIL may provide a unique interaction with OPG and mutations on OPG residues that bind to TRAIL would generate OPG variants that will inhibit osteoclastogenesis but not TRAIL-mediated apoptosis of cancer cells. The crystal structure of OPG in complex with RANKL indicated that CRD2 and CRD3 of OPG play substantial roles in binding of OPG and RANKL, where the binding interface consists of two binding sites: site-1 identifying amino acid loop 50s (H47-L65) in CRD2, and site-2 indentifying amino acid loop 90s (A90-L98) in CRD3 (39). Of the mutants generated and characterized for therapeutic bone remodeling, mutant Y49R falls within binding site-1 and mutant F96R falls within binding site-2. However, mutant F107A does not fall within these binding sites yet resulted in significant abolishment of TRAIL binding. It remains possible that conformational changes in the amino acid F107 to F107A may have altered the binding that may have been retained in full-length OPG protein as opposed to truncated OPG containing CRDs.
Results of the in vitro studies also confirmed that despite mutagenesis at amino acids, based on interactive domain analysis, abolishment of TRAIL binding domains in three of the mutants did not affect binding of RANKL to the OPG mutants. In the osteoclast assay, all recombinant OPG proteins significantly inhibited RANKL-mediated osteoclastogenesis confirming functionality of the mutated proteins. When functionally testing for TRAIL binding activity, the OPG mutant N81A strongly inhibited TRAIL-mediated apoptosis, similar to wild-type OPG indicating that despite the amino acid substitution, the binding affinity of OPG to TRAIL was retained. However, OPG mutants Y49R, F96A and F107A demonstrated a significant decrease in TRAIL binding, which resulted in a significant decrease in cell viability. Further, results of the in vivo studies provided key evidence confirming functional activity of these two OPG mutants in a preclinical mouse model of a bone disseminated osteolytic tumor demonstrated protection from bone destruction in both trabecular architecture and connectivity density.
Realizing the therapeutic potential of OPG in bone remodeling for tumor-induced osteolytic damage, much effort has gone to its clinical utility. In this study, we proposed the use of a novel OPG mutant protein against cancer induced osteolysis while potentially not interfering with TRAILs ability to induce apoptosis in cancer cells. Often times, epithelial carcinomas such as that of breast and prostate metastasize to the spine and long bones causing severe bone damage. With OPG being the native inhibitory protein of osteoclastogenesis, using this protein in conditions where there is severe osteolysis due to an imbalance in the RANKL/RANK/OPG signaling triad can potentially result in increased bone healing and low toxicity. In this regard, soluble RANK and antibodies targeting RANK have shown to be effective (40,41), but toxicity remains an issue and therefore, direct site injection of OPGmut, abolished in TRAIL binding by cell/gene therapy approach whereby low, yet sustained concentration of OPG can be systemically released to inhibit aggressive osteolytic damage. Moreover, the fact that the OPG mutants possess an altered TRAIL binding domain would potentially allow endogenous TRAIL to target transformed cells. Further, exogenous TRAIL therapy can be combined with the proposed OPGmut therapy to diminish both osteolytic burden and tumor cell killing. In many advanced osteolytic malignancies where osteolytic lesions exist throughout the skeleton, a systemic approach will be more beneficial. In this context, it is noteworthy that use MSC is currently being tested in clinical studies (42).
Taken together, the current study demonstrates that RANKL-OPG-TRAIL molecular triad may be a valid target to develop novel therapy, particularly for tumor-associated bone destruction. The potential therapeutic application of the variant OPG that effectively inhibits osteoclastogenesis by retaining RANKL binding, while abolishing TRAIL binding, will be beneficial in malignant osteolytic bone pathologies encountered in breast cancer, prostate cancer, and multiple myeloma. The novel variants of OPG presented here may be used in therapeutic applications either as purified protein or by gene-based approaches through cell and gene therapy applications depending on the required duration of the therapy. Both OPG and TRAIL have been tested in preclinical settings and both proteins were well tolerated when used to treat pathological conditions (43,44). Furthermore, as studies have shown cells from human osteolytic malignancies such as U266, RPMI8226, MDA-MB-231 are sensitive to TRAIL (13,45,46) using OPG therapy in combination with TRAIL should result in the protection of bone from aggressive osteoclast damage and increase cancer cell death due to cytotoxic effects of TRAIL.
IMPLICATIONS.
OPG variants were developed that lack TRAIL binding, yet, retain RANKL binding and suggest new possibilities for therapeutic targeting in osteolytic malignancies.
Acknowledgments
Financial support of the National Institutes of Health grants AR050251; AR560948, CA132077, CA133737, P30 AR046031 and P30 AR48311 and DoD grant DoD-BC101411 is gratefully appreciated.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest exist with any of the authors.
AUTHOR CONTRIBUTIONS
Conception and design: JH, JJ, SP
Development of methodology: JH, JJ, JHL, CD, SP
Acquisition of data: JH, JJ, JHL, DC, CL
Analysis and interpretation of data: JH, JJ, SP
Writing, review, and/or revision of the manuscript: JH, SP
Administrative, technical, or material support: SP
Study supervision: SP
References
- 1.Khosla S. Minireview: the OPG/RANKL/RANK system. Endocrinology. 2001;142:5050–55. doi: 10.1210/endo.142.12.8536. [DOI] [PubMed] [Google Scholar]
- 2.Maginn EN, Browne PV, Hayden P, Vandenberghe E, MacDonagh B, Evans P, et al. PBOX-15, a novel microtubule targeting agent, induces apoptosis, upregulates death receptors, and potentiates TRAIL-mediated apoptosis in multiple myeloma cells. Br J Cancer. 2011;104:281–9. doi: 10.1038/sj.bjc.6606035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20:345–57. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 4.Hofbauer LC, Khosla S, Dunstan CR, Lacey DL, Spelsberg TC, Riggs BL. Estrogen stimulates gene expression and protein production of osteoprotegerin in human osteoblastic cells. Endocrinology. 1999;140:4367–70. doi: 10.1210/endo.140.9.7131. [DOI] [PubMed] [Google Scholar]
- 5.Hofbauer LC, Gori F, Riggs BL, et al. Stimulation of osteoprotegerin ligand and inhibition of osteoprotegerin production by glucocorticoids in human osteoblastic lineage cells: potential paracrine mechanisms of glucocorticoid-induced osteoporosis. Endocrinology. 1999;140:4382–9. doi: 10.1210/endo.140.10.7034. [DOI] [PubMed] [Google Scholar]
- 6.Itonaga I, Fujikawa Y, Sabokbar A, Murray DW, Athanasou NA. Rheumatoid arthritis synovial macrophage-osteoclast differentiation is osteoprotegerin ligand-dependent. J Pathol. 2000;192:97–104. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH672>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Yasuda H, Shima N, Nakagawa N, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci U S A. 1998;95:3597–602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonet WS, Lacey DL, Dunstan CR, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–19. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 9.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 10.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 11.Chanda D, Kumar S, Ponnazhagan S. Therapeutic potential of adult bone marrow-derived mesenchymal stem cells in diseases of the skeleton. J Cell Biochem. 2010;111:249–57. doi: 10.1002/jcb.22701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szafran AA, Folks K, Warram J, Chanda D, Wang D, Zinn KR. Death receptor 5 agonist TRA8 in combination with the bisphosphonate zoledronic acid attenuated the growth of breast cancer metastasis. Cancer Biol Ther. 2009;8:1109–16. doi: 10.4161/cbt.8.12.8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L, Huang X, Choi KY, et al. Real-time monitoring of caspase cascade activation in living cells. J Control Release. 2012;163:55–62. doi: 10.1016/j.jconrel.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacey DL, Timms E, Tan HL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–76. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 15.Kitazawa S, Kitazawa R. RANK ligand is a prerequisite for cancer-associated osteolytic lesions. J Pathol. 2002;198:228–36. doi: 10.1002/path.1199. [DOI] [PubMed] [Google Scholar]
- 16.Thomas RJ, Guise TA, Yin JJ, et al. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999;140:4451–58. doi: 10.1210/endo.140.10.7037. [DOI] [PubMed] [Google Scholar]
- 17.Wittrant Y, Theoleyre S, Chipoy C, et al. RANKL/RANK/OPG: new therapeutic targets in bone tumours and associated osteolysis. Biochim Biophys Acta. 2004;1704:49–57. doi: 10.1016/j.bbcan.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Corey E, Brown LG, Kiefer JA, et al. Osteoprotegerin in prostate cancer bone metastasis. Cancer Res. 2005;65:1710–8. doi: 10.1158/0008-5472.CAN-04-2033. [DOI] [PubMed] [Google Scholar]
- 19.Croucher PI, Shipman CM, Lippitt J, et al. Osteoprotegerin inhibits the development of osteolytic bone disease in multiple myeloma. Blood. 2001;98:3534–40. doi: 10.1182/blood.v98.13.3534. [DOI] [PubMed] [Google Scholar]
- 20.Morony S, Capparelli C, Sarosi I, Lacey DL, Dunstan CR, Kostenuik PJ. Osteoprotegerin inhibits osteolysis and decreases skeletal tumor burden in syngeneic and nude mouse models of experimental bone metastasis. Cancer Res. 2001;61:4432–6. [PubMed] [Google Scholar]
- 21.Vanderkerken K, De Leenheer E, Shipman C, et al. Recombinant osteoprotegerin decreases tumor burden and increases survival in a murine model of multiple myeloma. Cancer Res. 2003;63:287–9. [PubMed] [Google Scholar]
- 22.Zhang J, Dai J, Qi Y, et al. Osteoprotegerin inhibits prostate cancer-induced osteoclastogenesis and prevents prostate tumor growth in the bone. J Clin Invest. 2001;107:1235–44. doi: 10.1172/JCI11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Body JJ, Greipp P, Coleman RE, et al. A phase I study of AMGN-0007, a recombinant osteoprotegerin construct, in patients with multiple myeloma or breast carcinoma related bone metastases. Cancer. 2003;97:887–92. doi: 10.1002/cncr.11138. [DOI] [PubMed] [Google Scholar]
- 24.Emery JG, McDonnell P, Burke MB, et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem. 1998;273:14363–7. doi: 10.1074/jbc.273.23.14363. [DOI] [PubMed] [Google Scholar]
- 25.Vitovski S, Phillips JS, Sayers J, Croucher PI. Investigating the interaction between osteoprotegerin and receptor activator of NF-kappaB or tumor necrosis factor-related apoptosis-inducing ligand: evidence for a pivotal role for osteoprotegerin in regulating two distinct pathways. J Biol Chem. 2007;282:31601–9. doi: 10.1074/jbc.M706078200. [DOI] [PubMed] [Google Scholar]
- 26.Griffith TS, Wiley SR, Kubin MZ, Sedger LM, Maliszewski CR, Fanger NA. Monocyte-mediated tumoricidal activity via the tumor necrosis factor-related cytokine, TRAIL. J Exp Med. 1999;189:1343–54. doi: 10.1084/jem.189.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth MJ, Cretney E, Takeda K, et al. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contributes to interferon gamma-dependent natural killer cell protection from tumor metastasis. J Exp Med. 2001;193:661–70. doi: 10.1084/jem.193.6.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda K, Hayakawa Y, Smyth MJ, et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat Med. 2001;7:94–100. doi: 10.1038/83416. [DOI] [PubMed] [Google Scholar]
- 29.Takeda K, Smyth MJ, Cretney E, et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in NK cell-mediated and IFN-gamma-dependent suppression of subcutaneous tumor growth. Cell Immunol. 2001;214:194–200. doi: 10.1006/cimm.2001.1896. [DOI] [PubMed] [Google Scholar]
- 30.Zerafa N, Westwood JA, Cretney E, et al. Cutting edge: TRAIL deficiency accelerates hematological malignancies. J Immunol. 2005;175:5586–90. doi: 10.4049/jimmunol.175.9.5586. [DOI] [PubMed] [Google Scholar]
- 31.Takeda K, Smyth MJ, Cretney E, et al. Critical role for tumor necrosis factor-related apoptosis-inducing ligand in immune surveillance against tumor development. J Exp Med. 2002;195:161–9. doi: 10.1084/jem.20011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cretney E, Takeda K, Yagita H, Glaccum M, Peschon JJ, Smyth MJ. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J Immunol. 2002;168:1356–61. doi: 10.4049/jimmunol.168.3.1356. [DOI] [PubMed] [Google Scholar]
- 33.Cha SS, Kim MS, Choi YH, et al. 2.8 A resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity. Immunity. 1999;11:253–61. doi: 10.1016/s1074-7613(00)80100-4. [DOI] [PubMed] [Google Scholar]
- 34.Cha SS, Sung BJ, Kim YA, et al. Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity. J Biol Chem. 2000;275:31171–7. doi: 10.1074/jbc.M004414200. [DOI] [PubMed] [Google Scholar]
- 35.Hymowitz SG, Christinger HW, Fuh G, et al. Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell. 1999;4:563–71. doi: 10.1016/s1097-2765(00)80207-5. [DOI] [PubMed] [Google Scholar]
- 36.Lam J, Nelson CA, Ross FP, Teitelbaum SL, Fremont DH. Crystal structure of the TRANCE/RANKL cytokine reveals determinants of receptor-ligand specificity. J Clin Invest. 2001;108:971–9. doi: 10.1172/JCI13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mongkolsapaya J, Grimes JM, Chen N, et al. Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat Struct Biol. 1999;6:1048–53. doi: 10.1038/14935. [DOI] [PubMed] [Google Scholar]
- 38.Naismith JH, Devine TQ, Kohno T, Sprang SR. Structures of the extracellular domain of the type I tumor necrosis factor receptor. Structure. 1996;4:1251–62. doi: 10.1016/s0969-2126(96)00134-7. [DOI] [PubMed] [Google Scholar]
- 39.Nelson CA, Warren JT, Wang MW, Teitelbaum SL, Fremont DH. RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor. Structure. 2012;20:1971–82. doi: 10.1016/j.str.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummings SR, San Martin J, McClung MR, et al. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N Engl J Med. 2009;361:756–65. doi: 10.1056/NEJMoa0809493. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Z, Chen C, Zhang J, et al. Safety of denosumab in postmenopausal women with osteoporosis or low bone mineral density: a meta-analysis. Int J Clin Exp Pathol. 2014;7:2113–22. [PMC free article] [PubMed] [Google Scholar]
- 42.Gopal K, Amirhamed HA, Kamarul T. Advances of human bone marrow-derived mesenchymal stem cells in the treatment of cartilage defects: a systematic review. Exp Biol Med (Maywood) 2014;239:663–9. doi: 10.1177/1535370214530364. [DOI] [PubMed] [Google Scholar]
- 43.Zierhut ML, Gastonguay MR, Martin SW, et al. Population PK-PD model for Fc-osteoprotegerin in healthy postmenopausal women. J Pharmacokinet Pharmacodyn. 2008;35:379–9. doi: 10.1007/s10928-008-9093-5. [DOI] [PubMed] [Google Scholar]
- 44.Doi T, Murakami H, Ohtsu A, et al. Phase 1 study of conatumumab, a pro-apoptotic death receptor 5 agonist antibody, in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol. 2011;68:733–7. doi: 10.1007/s00280-010-1544-1. [DOI] [PubMed] [Google Scholar]
- 45.Shipman CM, Croucher PI. Osteoprotegerin is a soluble decoy receptor for tumor necrosis factor-related apoptosis-inducing ligand/Apo2 ligand and can function as a paracrine survival factor for human myeloma cells. Cancer Res. 2003;63:912–6. [PubMed] [Google Scholar]
- 46.Labrinidis A, Diamond P, Martin S, et al. Apo2L/TRAIL inhibits tumor growth and bone destruction in a murine model of multiple myeloma. Clin Cancer Res. 2009;15:1998–2009. doi: 10.1158/1078-0432.CCR-08-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]