Abstract
While reviewing chronic lymphocytic leukemia (CLL) bone marrow slides we identified cytoplasmic lipid vacuoles in CLL cells but not in normal B cells. Because lipoprotein lipase (LPL), which catalyzes hydrolysis of triglycerides into free fatty acids (FFAs), is aberrantly expressed in CLL, we investigated whether LPL regulates the oxidative metabolic capacity of CLL cells. We found that unlike normal B cells, CLL cells metabolize FFAs. Because STAT3 is constitutively activated in CLL cells and because we identified putative STAT3 binding sites in the LPL promoter, we sought to determine whether STAT3 drives the aberrant expression of LPL. Transfection of luciferase reporter gene constructs driven by LPL promoter fragments into MM1 cells revealed that STAT3 activates the LPL promoter. In addition, chromatin immunoprecipitation (ChIP) confirmed that STAT3 binds to the LPL promoter. Furthermore, transfection of CLL cells with STAT3-shRNA downregulated LPL transcripts and protein levels, confirming that STAT3 activates the LPL gene. Finally, transfection of CLL cells with LPL-siRNAs decreased the capacity of CLL cells to oxidize FFAs and reduced cell viability.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by the gradual accumulation of mature-appearing lymphocytes1 whose gene expression profile and expression of the cell surface CD27 antigen resemble typical features of memory B cells.2,3 Normally, memory B cells are quiescent, and extracellular stimuli, such as the CD40 ligand, are required to induce their proliferation.4 Unlike memory B cells, approximately 1% of CLL cells proliferate daily.5 However, what energy source CLL cells use and which metabolic pathways they recruit to provide the energy needed for survival and proliferation are not known.
Lipoprotein lipase (LPL), commonly expressed in adipocytes and muscle cells, plays a central role in lipid metabolism.6 LPL catalyzes the hydrolysis of triglycerides into free fatty acids (FFAs) and increases the cellular uptake of lipoproteins in a non-catalytic manner.7 \
LPL transcripts were found in CLL cells but not in normal B lymphocytes,3,8 and high levels of LPL were detected on the cell surfaces and in the cytosol of CLL cells.9 Remarkably, increased levels of LPL-mRNA were correlated with unmutated immunoglobulin heavy-chain variable region genes, aggressive disease, and an unfavorable prognosis.3,8–11 However, what activates LPL in CLL has not been deciphered.
Signal transducer and activator of transcription–3 (STAT3) is a latent cytoplasmic transcription factor that relays cytokine and growth factor signals from the cell membrane to the nucleus.12 In CLL, STAT3 is constitutively phosphorylated on serine 727 residues. Serine pSTAT3 migrates to the nucleus, binds to DNA, activates transcription, and provides CLL cells with a survival advantage.13,14
In several human tumors, signal transduction pathways regulate metabolic pathways, and in some of those tumors, lipid metabolism is altered.15 Since STAT3 was found to modulate lipid synthesis and modify the expression of genes that regulate cellular metabolism16, and because sequence analysis revealed that the LPL promoter harbors γ-interferon activation sequence (GAS)-like elements, known to bind STAT3, we sought to determine whether STAT3 activates the transcription of LPL.
Here we show that the aberrant expression of LPL is driven by STAT3. Unlike normal B-cells the cytoplasm of CLL cells contains lipid-filled vacuoles, and the cells utilize fat as an energy source in an LPL-dependent manner.
Materials and Methods
Fractionation of CLL cells and normal B cells
Peripheral blood (PB) cells were obtained from previously untreated patients with CLL who were followed at The University of Texas MD Anderson Cancer Center Leukemia Clinic after we received Institutional Review Board approval and written informed consent from the patients (Supplemental Table 1). To isolate low-density cells, the patients’ peripheral blood cells were fractionated using Ficoll-Hypaque 1077 (Sigma-Aldrich, St. Louis, MO). More than 90% of the peripheral blood lymphocytes obtained from these patients were CD19+/CD5+, as assessed by flow cytometry (Becton, Dickinson and Company, Franklin Lakes, NJ). PB samples from healthy donors were obtained from the Central Blood Bank as buffy coats. After Ficoll-Hypaque fractionation, the donors’ B cells were isolated using Miltenyi CD19-coated beads according to the manufacturer’s instructions (Miltenyi Biotec, Bergisch Gladbach, Germany).
Cell culture
The fractionated CLL cells were maintained in Dulbecco’s modified Eagle medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (HyClone, Logan, UT). Cells from the human multiple myeloma line MM1 were obtained from the ATCC (Rockville, MD). MM1 cells were maintained in RPMI 1640 (Sigma-Aldrich) supplemented with 10% fetal bovine serum in a humidified atmosphere of 5% CO2 at 37°C. The human renal epithelial carcinoma 293T cells were grown in Dulbecco modified Eagle medium (Sigma-Aldrich) supplemented with 10% fetal calf serum (HyClone). Human umbilical vein endothelial cells (HUVECs) were maintained in vascular endothelial medium (both from Lonza, Walkersville, MD).
Oil Red O staining of bone marrow aspirates from CLL patients
Slides of bone marrow aspirates from the patients with CLL were placed in absolute propylene glycol for 2 minutes and stained in Oil Red O solution for 16 hours, after which the stained slides were transferred to an 85% propylene glycol solution for 1 minute, rinsed in distilled water, counterstained in Mayer’s hematoxylin solution (Sigma-Aldrich) for 15 seconds, rinsed in distilled water, and mounted in a warmed glycerin jelly solution.
Transmission electron microscopy (TEM)
TEM visualization of CLL and normal B cells were done as previously described.17 Briefly, pieces of agarose containing embedded CLL or normal B cells were trimmed into 1- to 2-mm3 cubes using a razor blade. After staining the samples with en bloc with 1% Millipore-filtered uranyl acetate (EMD Millipore, Billerica, MA), they were dehydrated in increasing concentrations of ethanol and then infiltrated and embedded in LX-112 medium. The samples were then polymerized at 60°C for 2 days and cut using a Leica Ultracut microtome (Leica Microsystems, Buffalo Grove, IL), stained with uranyl acetate and lead citrate using a Leica EM stainer, and examined using a JEM 1010 transmission electron microscope (JEOL USA, Inc., Peabody, MA) at an acceleration voltage of 80 kV. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques, Corp., Woburn, MA).
Confocal microscopy
CLL low-density cells were incubated in microtubes in PBS supplemented with 5% bovine serum albumin (Cell Signaling Technology,). After 1 hour of incubation, the cells were washed three times with PBS and then incubated with mouse anti-LPL antibodies (Abcam, Cambridge, MA) or mouse anti-CD19 antibodies (BD Biosciences, San Jose, CA) for 1 hour. After washing three more times in PBS, the cells were incubated with Alexa Fluor 488–labeled anti-mouse antibodies (Invitrogen/Life Technologies, Grand Island, NY) for 1 hour, washed in PBS, re-suspended in a 0.1% solution of Evans blue dye (Sigma-Aldrich) for 5 minutes, and washed in PBS to remove unbound dye. The cells were resuspended in PBS and placed into µ-slide VI0.4 chamber slides (ibidi, LLC, Verona, WI) for microscopic analysis. The slides were viewed using an Olympus FluoView 500 Laser Scanning Confocal Microscope (Olympus America, Center Valley, PA), and images were analyzed using the FluoView software (Olympus America)
Western blot analysis
Western blot analysis was performed as previously described.18 Briefly, Cell lysates were assayed for their protein concentrations using the BCA protein assay reagent (Pierce Chemical). Each set of paired lyasate was adjusted for the same protein concentration. A lysates of CLL cell extract was mixed with 4× Laemmli sample buffer and was then denatured by boiling for 5 minutes. Forty micrograms of lysates were dissolved separated using 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to a nitrocellulose. The transfer was done overnight at 30 V in a cooled (4°C) reservoir. The nitrocellulose membrane was then placed in a Ponceaus S staining to verify equal loading of protein. The membranes were blocked with 5% dried milk dissolved in 50mL of PBS. After blocking the membrane was incubated with the following primary antibodies: monoclonal mouse anti-human STAT3 (BD Biosciences, San Jose, CA; Cat# 610190; in a dilution of 1:2 × 103), monoclonal rabbit anti-human Tyr705 STAT3 antibodies (Cat# 9131, Cell signaling Danvers, MA; in a 1:103 dilution), monoclonal mouse anti-human LPL (Abcam, Cambridge, MA, Cat# 21356; in a dilution of 1:103) and mouse anti-human β-actin (Sigma-Aldrich, St. Louis, MO). After incubation with horseradish peroxidase–conjugated secondary antibodies (GE Healthcare, Buckinghamshire, UK) for 1 hour, blots were visualized with an enhanced chemiluminescence detection system (GE Healthcare).
Densitometry analysis was performed using an Epson Expression 1680 scanner (Epson America, Inc., Long Beach, CA). Densitometry values were normalized by dividing the numerical value of each sample signal by the numerical value of the signal from the corresponding levels of each sample’s density by the density of the corresponding β actin protein, used as a loading control.
Measurement of cellular O2 consumption
Because fatty acid metabolism increases O2 consumption, palmitic acid and oleic acid utilization was assessed by measuring the level of dissolved O2 (dO2) using the SevenGo pro Dissolved Oxygen Meter (Mettler Toledo, Worthington Columbus, OH).
Preliminary experiments designed to test FFA consumption that used palmitic-acid or oleic-acid dissolved in ethanol, determined that the O2 consumption with 80 mM palmitic acid and 2 mM oleic acid each (both from Sigma-Aldrich) is maximal and therefore we used these concentrations in the following experiments. In these experiments we also found that when used in combination palmitic-acid and oleic-acid increase O2 consumption more than each of these components separately (data not shown).
In each experiment we used CLL cells, normal B-cells or HUVECs at a concentration of 2 to 3 × 106 cells/mL. The cells were incubated with a minimum essential medium (MEM) with Hank’s salts and L-glutamine (Life Technologies, Carlsbad, CA) or with phosphate buffered saline medium (Invitrogen) for 48 to 72 hours in tightly sealed T25 tissue culture flasks (Corning, Tewksbury, MA) at 37°C in the presence or absence of palmitic acid or oleic acid. In control experiments CLL cells were incubated in PBS with or without ethanol. The O2 meter probe was placed in the flask and the reading allowed stabilizing. Then, the dO2 level was recorded. The probe was cleaned before it was re-used. Measurements of dO2 were repeated at least three times for every data point. We used the Student t-test when one experimental condition was compared to non-treated (controls) and one-way ANOVA when 2 experimental conditions were compared to controls. Statistical analyses were performed using GraphPad version 5 (San Diego, California, USA).
Transfection of CLL cells with LPL small interfering RNA (siRNA)
Five microliters of siPORT NeoFX agent and 50 pmol of either siRNA targeting LPL (Applied Biosystems, Foster City, CA) or the FAM-labeled siRNA targeting the human glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Life Science Technologies) were each diluted in 50 µL of OPTI-MEM I and then mixed together and incubated at room temperature for 10 minutes. A total of 5 × 106 cells suspended in 0.2 mL of OPTI-MEM I medium containing the siRNA and transfection agent was incubated at room temperature. After 1 hour of incubation, transfections were performed by electroporation (Bio-Rad Laboratories), and then the cells were cultured in complete RPMI 1640 medium. Transfection efficiency was calculated on the basis of the green fluorescent protein (GFP)-conjugated siRNA measured by flow cytometry (Becton, Dickinson and Company).
Transfection of MM1 cells with LPL promoter fragments and luciferase assay
LPL promoter fragments were transfected into MM1 cells by electroporation as previously described19,20. Each construct included the luciferase reporter gene and the fragment either 143 bp upstream of the transcription start site (TSS) of the LPL gene, a region that includes one putative STAT3 binding site, or 333 bp upstream of the LPL gene TSS, a region that includes two putative STAT3 binding sites. The luciferase activity of unstimulated or interleukin 6–stimulated MM1 cells was assessed 48 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI) and the SIRIUS luminometer V3.1 Berthold Detection Systems (Titertek-Berthold, Pforzheim, Germany). The luciferase activity of each of the human LPL promoter constructs was determined by calculating the constructs’ luciferase activity relative to the activity of the Renilla luciferase produced by the pRL-SV40 control vector.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed according to manufactures of the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) as previously described20. The primers to amplify the human LPL promoter were F’: AGC AAT GAG GTA TGT GTG TAG and R: CTA CAT CAT TAT CAG GGT TAC, which generate a 144-bp product that covers the promoter region flanking 1 to 144 bp upstream of LPL; F’: GAG ATT GAA ACT GAA GCA CTG and R’: TGC CCA TTG CAC TTG ATT GTG, which generate a 83-bp product that covers the promoter region flanking 250 to 333 bp upstream of LPL; and F’: ATT TCT TCA GCA GGG TTT GCC and R’: AAA CTA TGG GAT TCC TAG GGG, which generate a 111-bp product that covers the promoter region flanking 563 to 674 bp upstream of LPL.
Infection of CLL cells with GFP-conjugated lentiviral STAT3 short hairpin RNA (shRNA)
The supernatant of 293T cells, co-transfected with GFP-lentivirus STAT3-shRNA (shRNA) or GFP-lentivirus empty vector and the packaging vectors pCMVdeltaR8.2 was used to infect CLL cells with STAT3-shRNA or the empty vector as previously described14. Briefly, CLL cells (5 × 106/mL) were incubated in 6-well plates (Becton Dickinson, Franklin Lakes, NJ) in 2 mL DMEM supplemented with 10% FCS and transfected with 100 µL of viral supernatant. Polybrene (10 ng/mL) was added to the viral supernatant at a ratio of 1:1000 (v/v). Transfection efficiency was measured after 48 hours and was found to range between 30% and 60% (calculated on the basis of the ratio of propidium iodide (PI)-negative/GFP-positive cells).
RNA purification and quantitative reverse transcription–polymerase chain reaction (qRT-PCR)
RNA was isolated using an RNeasy purification procedure (QIAGEN Inc., Valencia, CA). RNA quality and concentration were analyzed with a NanoDrop spectrophotometer (ND-1000; NanoDrop Products, Wilmington, DE). Five hundred nanograms of total RNA was used in one-step qRT-PCR with the sequence detection system ABI Prism 7700 using TaqMan gene expression assays (all from Applied Biosystems) for STAT3, ROR1, c-Myc, cyclin D1, p21, VEGF-c, RPL30 and LPL according to the manufacturer’s instructions. Samples were run in triplicate, and relative quantification was performed using the comparative CT method.20
Apoptosis assay
The rate of cellular apoptosis was analyzed using double staining with a Cy5-conjugated annexin V kit and propidium iodide (PI; BD Biosciences) according to the manufacturer’s instructions. Briefly, 1×106 cells were washed once with phosphate-buffered saline and resuspended in 200 µL binding buffer with 0.5 µg/mL annexin V-Cy5 and 2 µg/ml PI. After incubation for 10 minutes in the dark at room temperature, the samples were analyzed on a FACSCalibur flow cytometer (BD Biosciences). Cell viability was calculated as the percentage of annexin V positive cells.
Results
Lipid-like vacuoles were detected in CLL cells but not in normal B cells
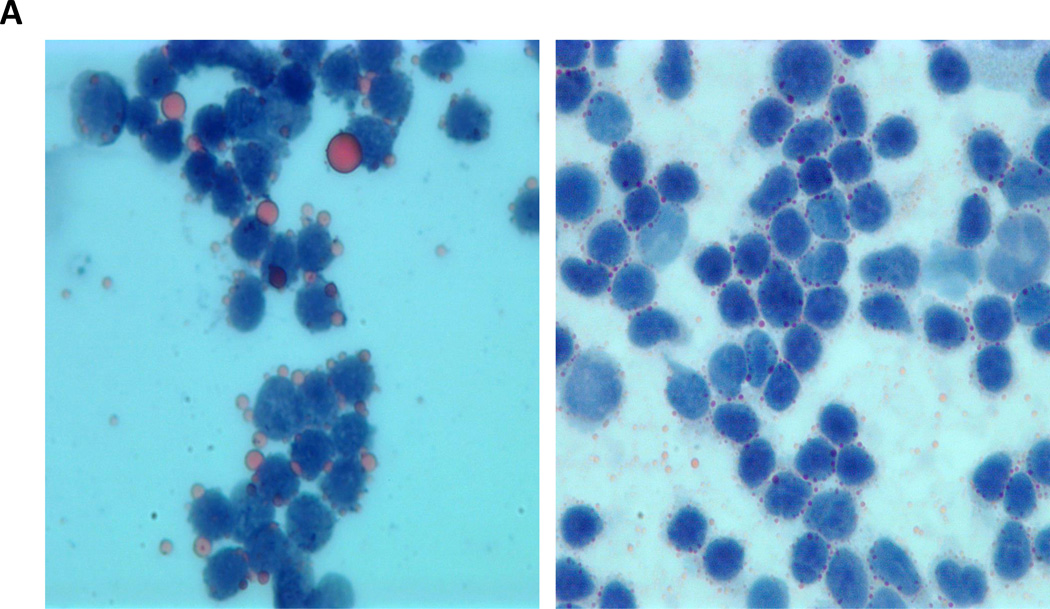
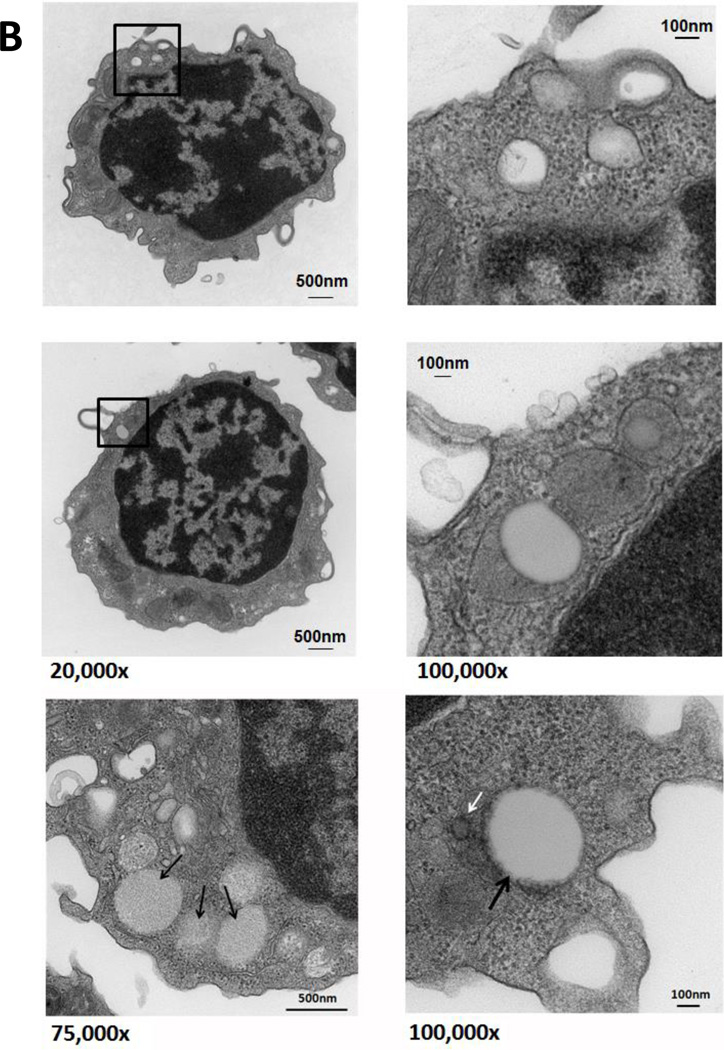
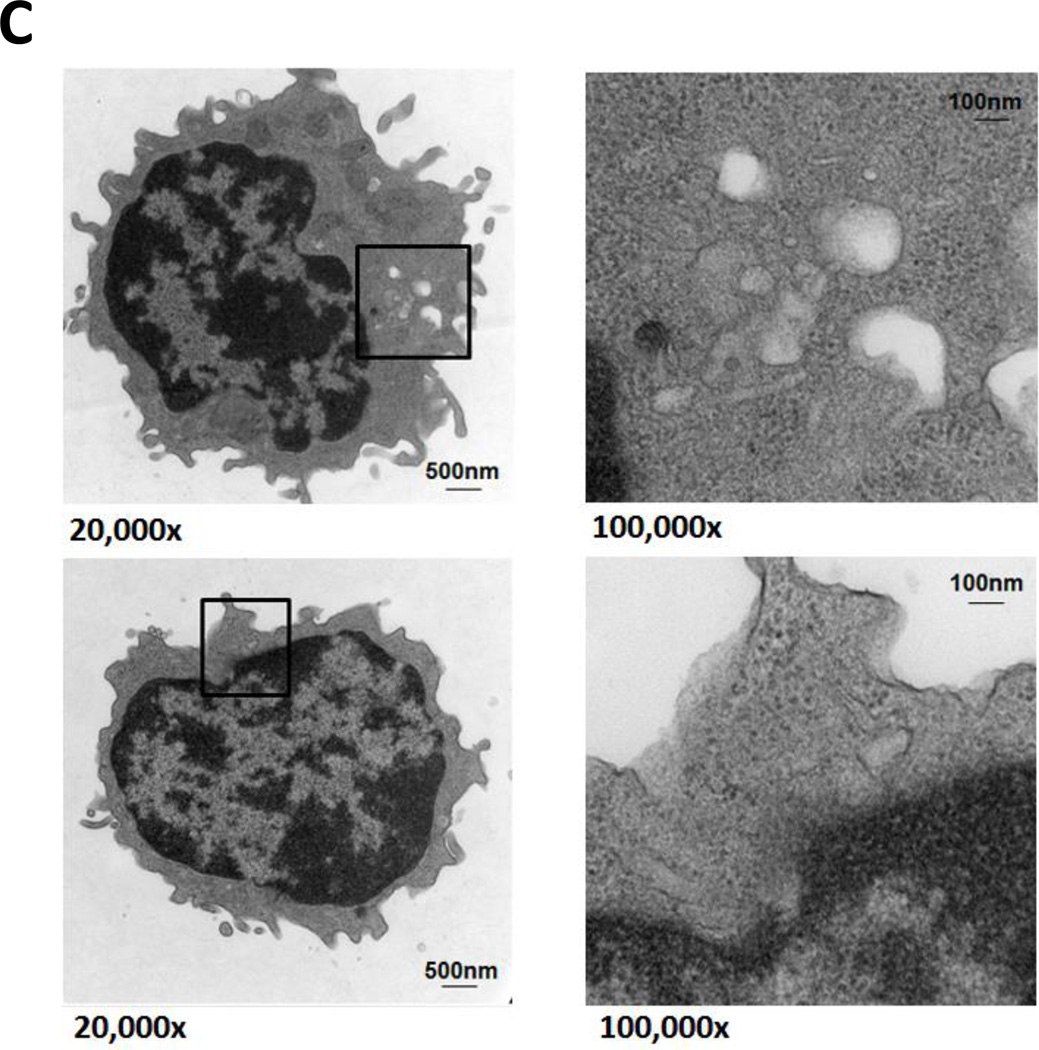
In reviewing bone marrow biopsies of patients with CLL, we identified clear-appearing vacuoles in the cytoplasm of CLL but not normal bone marrow cells. Because clear-appearing inclusion vacuoles are typically those of stored lipids, we stained CLL bone marrow smears from two CLL patients with Oil Red O. As shown in Figure 1A, Oil Red O staining confirmed that lipid deposits are present in CLL bone marrow cells (Figure 1A). To confirm that these lipid deposits were present within the cells, we used TEM and found that the lipid vacuoles visualized by TEM were present in the cytoplasm of 95% of CLL peripheral blood cells (Figure 1B) but not in normal B cells obtained from healthy individuals (Figure 1C).
Figure 1. Lipid vacuoles are detected in bone marrow CLL cells.
(A) CLL patients’ bone marrow aspirate smears stained with Oil Red O imaged using light microscopy show leukemia cells with stained inclusion bodies. (B) TEM showing numerous lucent vacuoles 100 to 500 nm in diameter (black arrows) scattered in the cytoplasm of a CLL cells. Enlarged micrographs of the areas in the boxes (left panel) are depicted in the right panel. The white arrow points to a 30-nm peroxisome adjacent the nuclear membrane. (C) TEM images of normal B cells. Unlike in CLL cells, lipid vacuoles are not seen in the cytoplasm of normal B lymphocytes. Cell regions suspected to contain vacuoles (black boxes, left panel) were enlarged and, as shown in the right panel, did not contain lipid vacuoles.
LPL was detected on the surface and in the cytosol of CLL cells
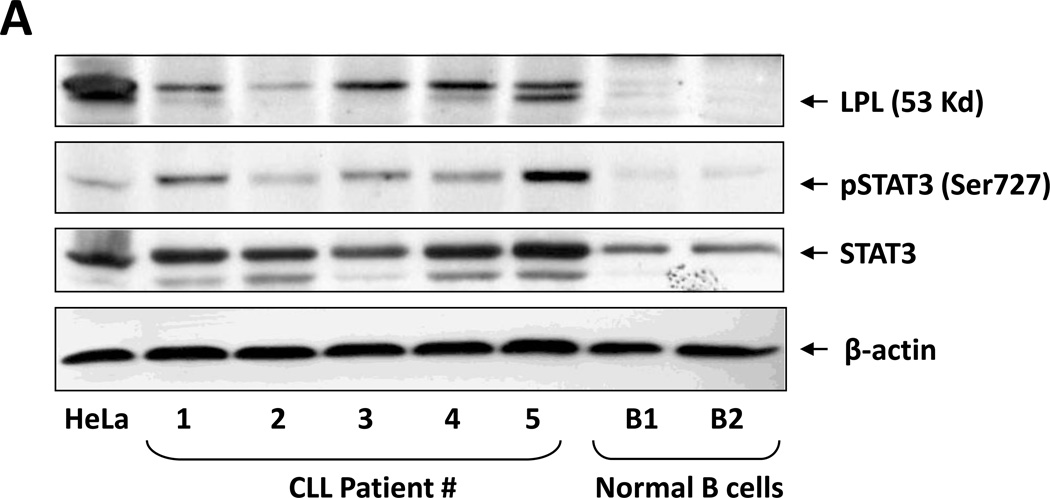
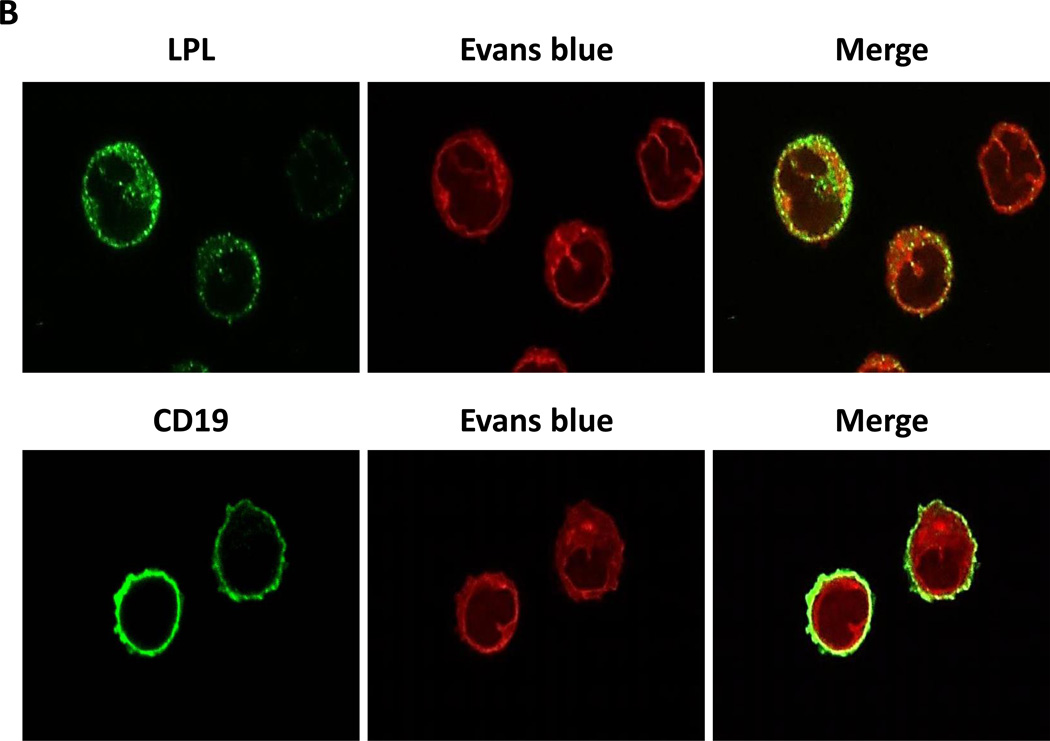
Several reports have suggested that CLL cells, like other lipid vacuole–containing cells (e.g., adipocytes and myocytes21–23), express LPL.3,8,24 To confirm this observation, we performed Western blot analysis on low-density peripheral blood cells from five CLL patients. As shown in Figure 2A, we detected LPL protein in all five CLL patients’ samples but not in normal B cells. Then, using confocal microscopy, we confirmed this observation and found that LPL was present on the surfaces and in the cytoplasm of CLL cells (Figure 2B).
Figure 2. LPL is detected in CLL cells but not in normal B cells.
(A) Western blot analysis of lysates from five CLL cells and two normal B cells detected LPL and pSTAT3 in CLL cells but not in normal B cells. STAT3 was detected in normal B cells but at levels lower than those in CLL cells. (B) Confocal microscopic images (×400) of freshly isolated CLL cells stained with anti-CD19 and anti-LPL antibodies for 1 hour. LPL was detected on cell surfaces and cytoplasm of CLL cells.
FFAs increase the metabolic rate of CLL cells
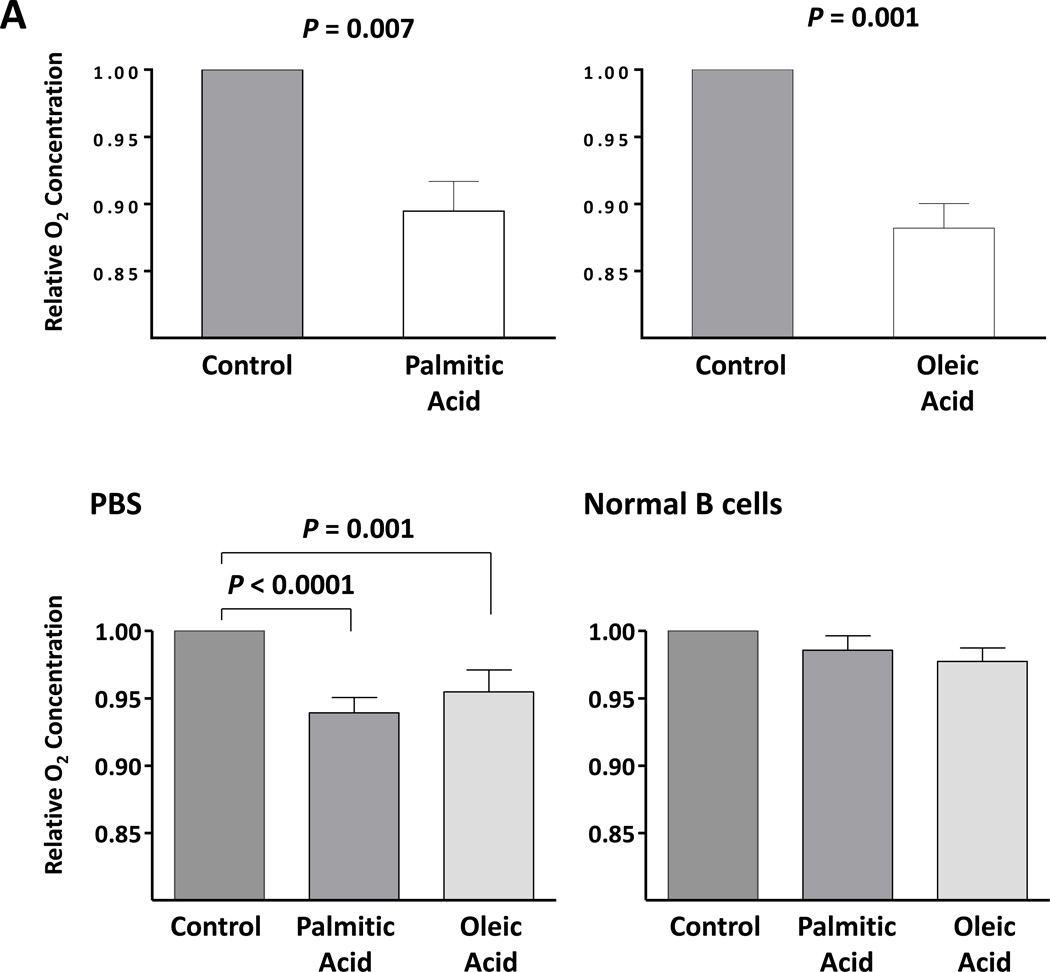
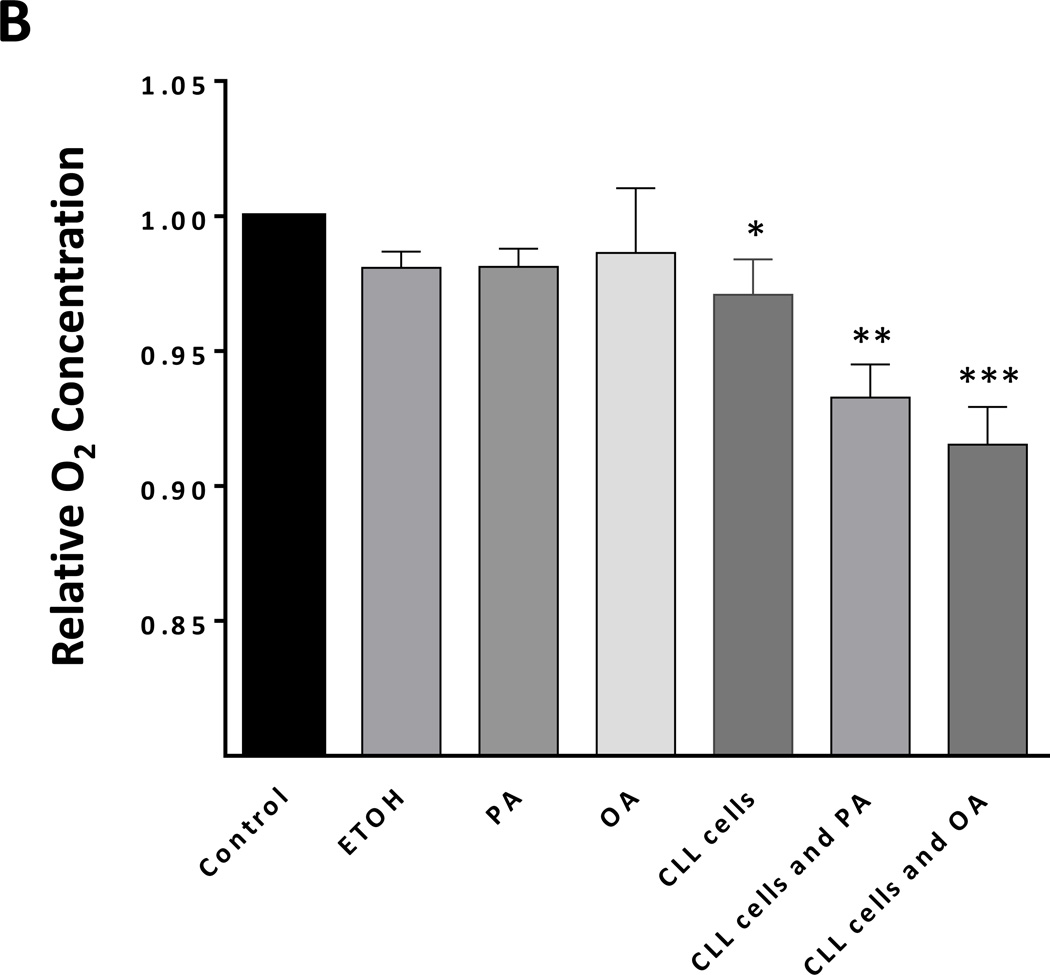
The presence of lipid vacuoles in the cytoplasm of CLL cells suggested that, like adipocytes and myocytes, CLL cells utilize lipids as a cellular energy source. Active metabolism increases O2 consumption and, as a result, decreases the levels of dO2 in the cells’ culture medium. Therefore, to estimate levels of CLL cell metabolism, we measured the levels of dO2 in the medium of the CLL cells incubated in the presence or absence of palmitic acid or oleic acid. After 48 hours of incubation with or without glucose-free media, the levels of dO2 were significantly lower in the presence of palmitic acid or oleic acid than in the absence of either FFA (Figure 3A). In contrast, incubation of normal B cells with palmitic acid or oleic acid did not change the dO2 levels in the culture media of normal B cells (Figure 3A) or HUVEC cells (data not shown), suggesting that CLL cells, but not normal B cells, metabolize FFAs. To determine whether the decrease in dO2 resulted from increased cellular metabolism we performed an additional experiment in which we measured dO2 in the presence or absence of CLL cells. An insignificant decrease in dO2 was observed in the absence of cells. However the addition of CLL cells significantly decreased dO2 and the addition of FFA induced the most significant decrease in dO2 levels, suggesting that increased cellular metabolism induced a major reduction in dO2 levels (Figure 3B).
Figure 3. Palmitic acid and oleic acid increase CLL cells’ metabolism.
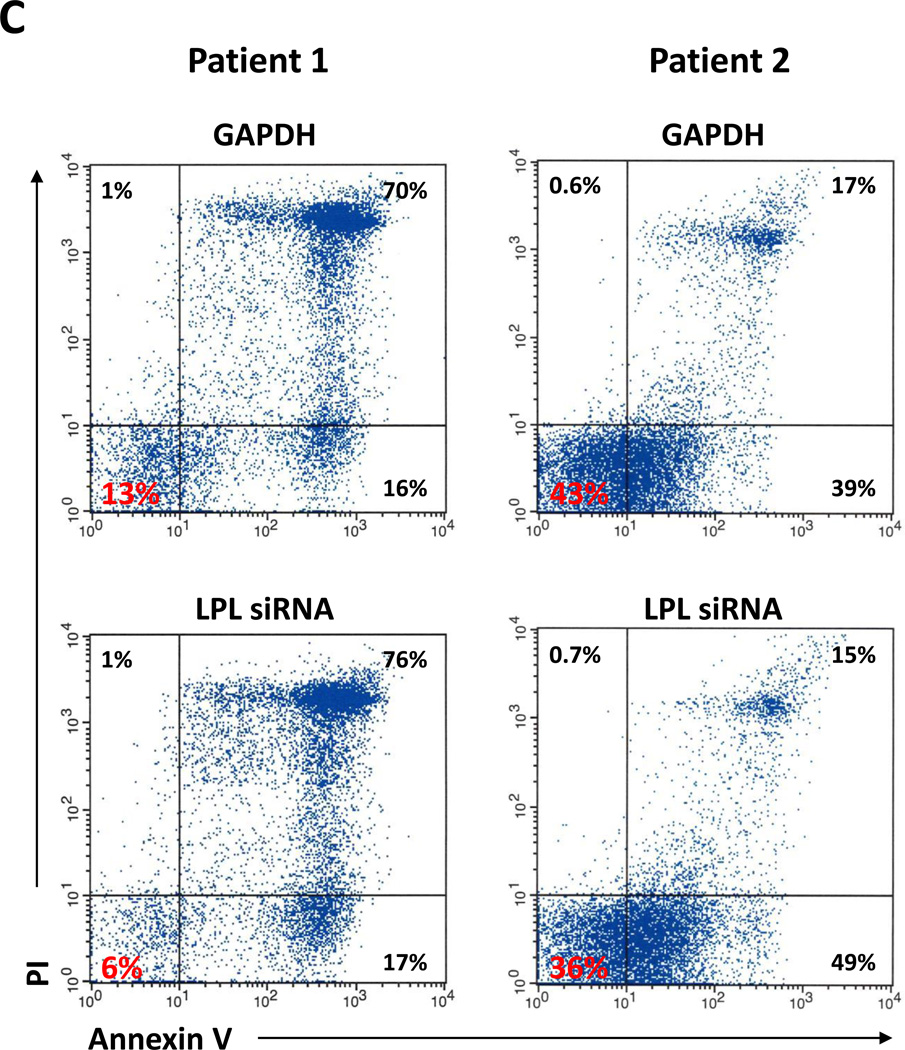
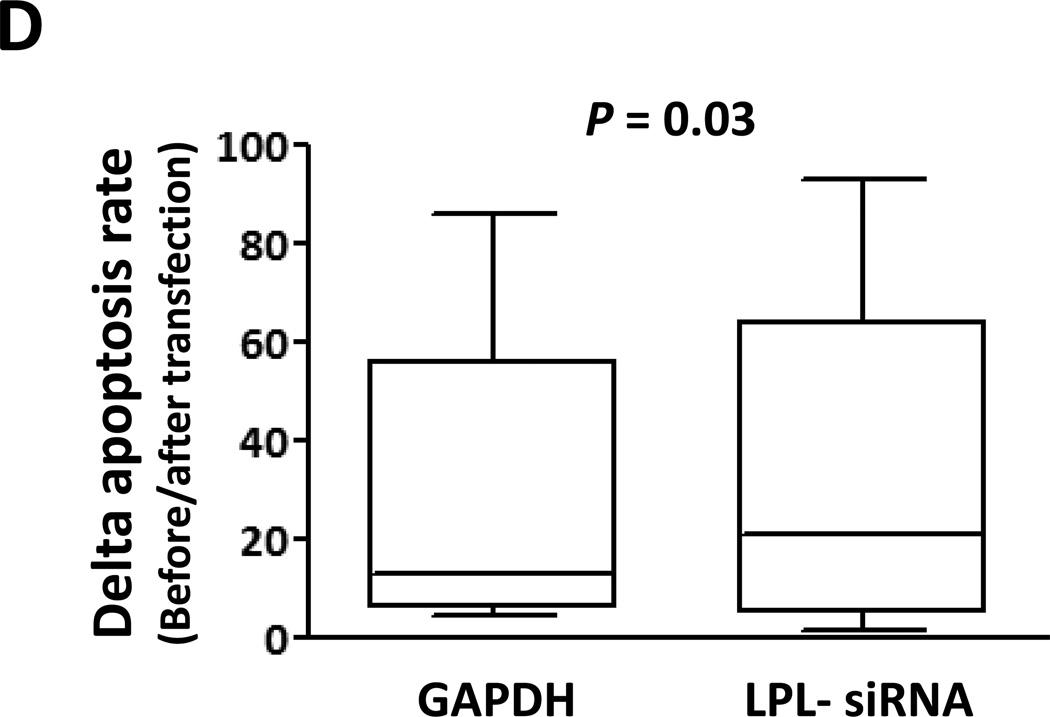
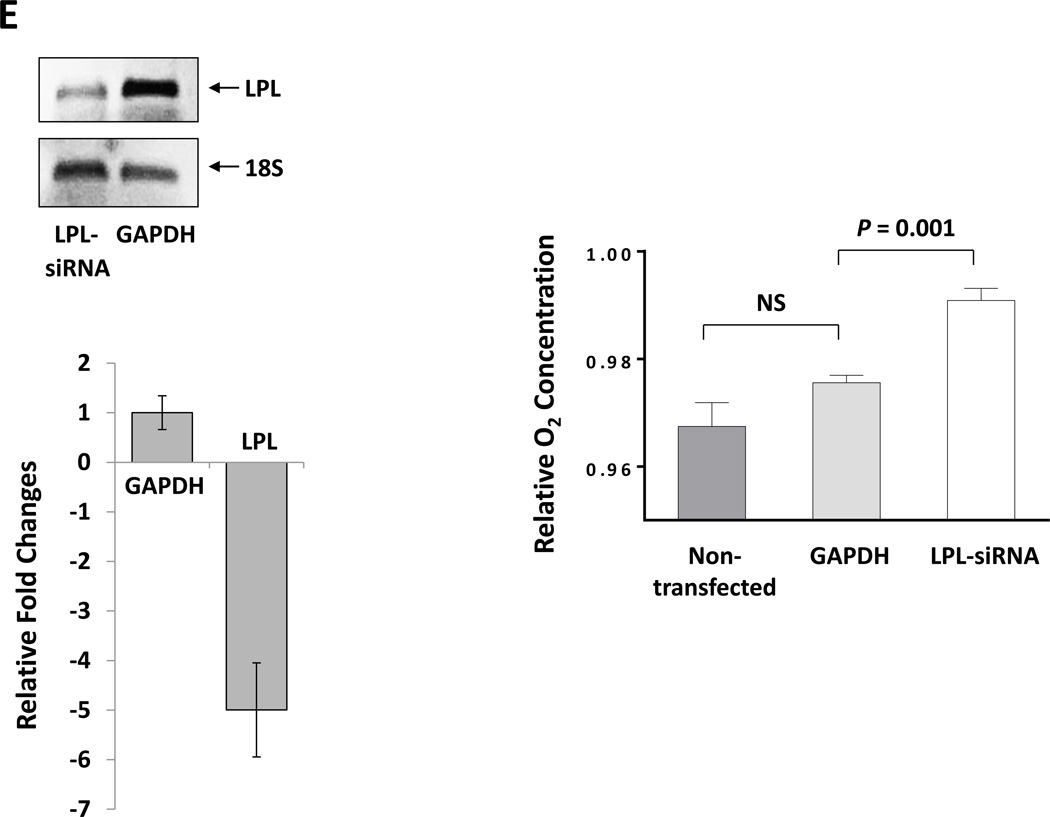
(A) CLL cells were incubated with MEM in the presence or absence of 80 mM palmitic acid (left upper panel) or oleic acid (right upper panel upper) in sealed tissue culture flasks. In a separate experiment CLL cells were incubated with PBS in the presence or absence of FFA (left lower panel). The culture media dO2 concentration was assessed prior to and after 48 hours of incubation. All control cultures contained ethanol at the same concentration as in palmitic acid and oleic acid. To compare dO2 we used the difference in 02 before and after the incubation (in mg/l).The relative difference in dO2 before after adding FFA is depicted after the dO2 in the controls was set to 1. Similar experiments with normal B cells (right lower panel) showed no change in the culture media dO2 concentration. The mean and standard error of the mean from 3 different patients for each condition are represented by the bars. (B) dO2 concentration was measured after 48 hours incubation of PBS (control), ethanol (ETOH; at the concentration present in palmitic acid and oleic acid), palmitic acid (PA) or oleic acid (OA) with or without CLL cells. Marked reduction in dO2 after 48 hours was observed only when CLL cells were added to culture. * P = 0.02, ** P = 0.002, *** P <.0001. (C) Flow cytometry analysis of CLL cells from two patients. Depicted are analyses of CLL cells from 2 different CLL patients transfected with LPL-siRNA or with GAPDH (transfection control). The percentages of viable cells are shown in the left lower quadrant (annexin V /PI-negative) of each panel. (D) Spontaneous apoptosis rate was recorded from CLL cells of 7 CLL patients prior to and 72 hours after transfecting the cells with LPL-siRNA or with GAPDH (transfection control). Significant increase in apoptosis rate in cells transfected with LPL-siRNA was observed. The data are depicted as delta apoptosis rate before and after transfection. To assess the differences in apoptosis rates of cells transfected with LPL-siRNA and GAPDH we used the Student t-test with delta apoptosis as the dependent variable. (E) LPL knockdown abrogates oxygen consumption in the presence of palmitic acid. CLL cells were transfected by electroporation with LPL-siRNA or with FAM-labeled GAPDH or left untreated. LPL-siRNA transfection efficiency in the CLL cells ranged from 35% to 50%. Left upper panel: PCR gel electrophoresis showing reduction in the LPL transcript level after treatment with LPL-siRNA. Left lower panel: RNA expression after LPL-siRNA transfection as shown by qRT-PCR. LPL expression was five times lower in cells transfected with LPL-siRNA than in cells transfected with GAPDH. Right panel: Untreated cells or cells transfected with LPL-siRNA or with GAPDH were incubated in the presence of palmitic acid. After 48 hours, the dO2 concentration was measured. Shown are the means and standard error of the mean of three experiments using cells from three patients. The dO2 level in the culture medium of the LPL siRNA–transfected cells was significantly higher than the dO2 levels in either control. NS, not significant.
LPL provides CLL cells with survival advantage
Although LPL is universally overexpressed in CLL cells, LPL’s mRNA and protein levels were reported to be higher in high-risk CLL9. To test whether LPL provides CLL cells with survival advantage we transfected CLL cells with LPL-shRNA. While a considerable overlap in cell viability between cells treated with LPL-shRNA and controls was observed, LPL knockdown significantly reduced CLL cell viability (Figures 3C and 3D) and abrogated dO2 consumption in the presence of palmitic acid, suggesting that LPL-siRNA reduced the capacity of CLL cells as to oxidize FFA (Figure 3E).
STAT3 activates the LPL promoter in MM1 cells
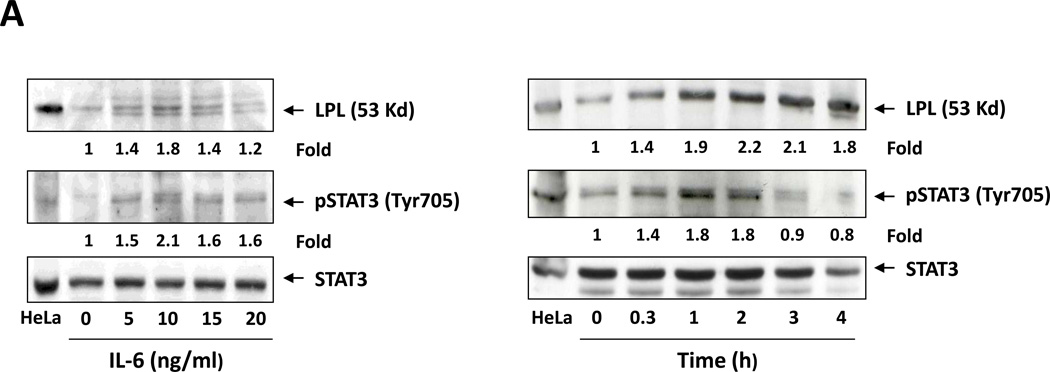
Because STAT3 is constitutively phosphorylated14 and LPL is aberrantly expressed in CLL cells,3,8 we wondered whether STAT3 activates the transcription of LPL. Using the TFSEARCH database,25 we identified two GAS-like elements that are known putative STAT3 binding sites26 within 400 bp upstream of the LPL TSS. To determine whether STAT3 activates the LPL promoter we used MM1 cells. IL-6 induces the phosphorylation of STAT3 in various cell types including MM1 cells14,27,28 thus providing us with a pSTAT3-inducible system. Incubation of MM1 cells with IL-6 induced STAT3 phosphorylation and upregulated LPL protein levels in a dose- and time-dependent manner (Figure 4A).
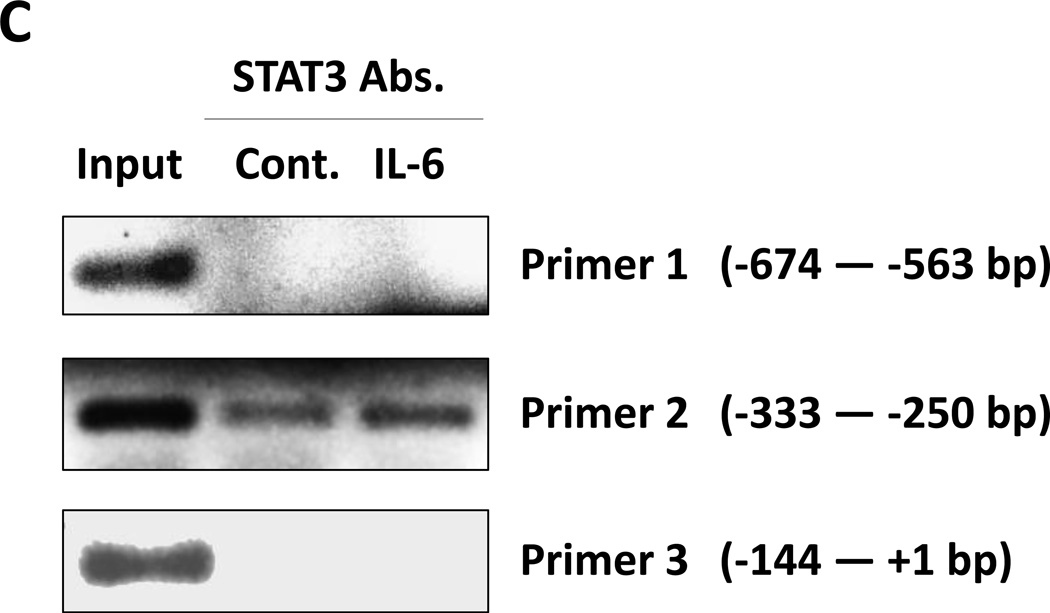
Figure 4. STAT3 activates the LPL promoter in MM1 cells.
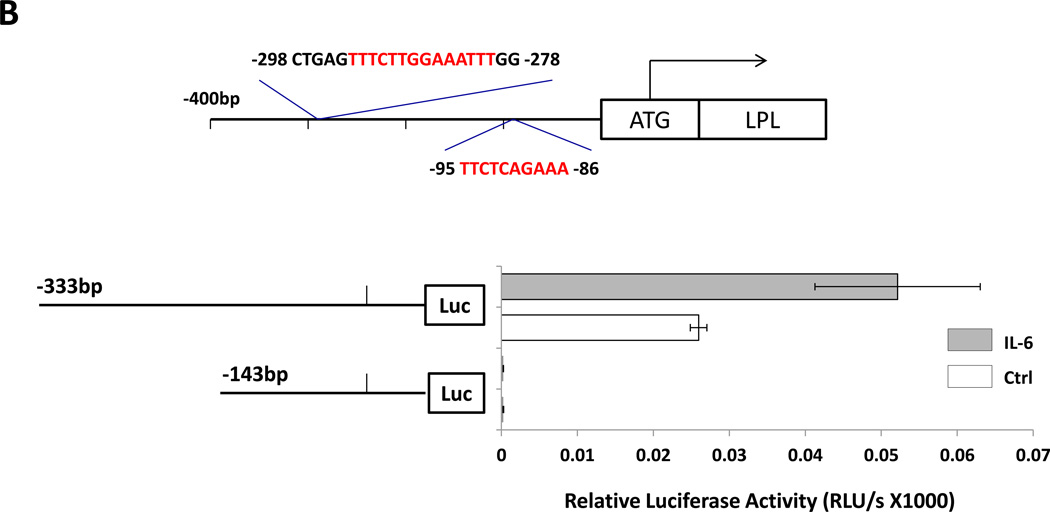
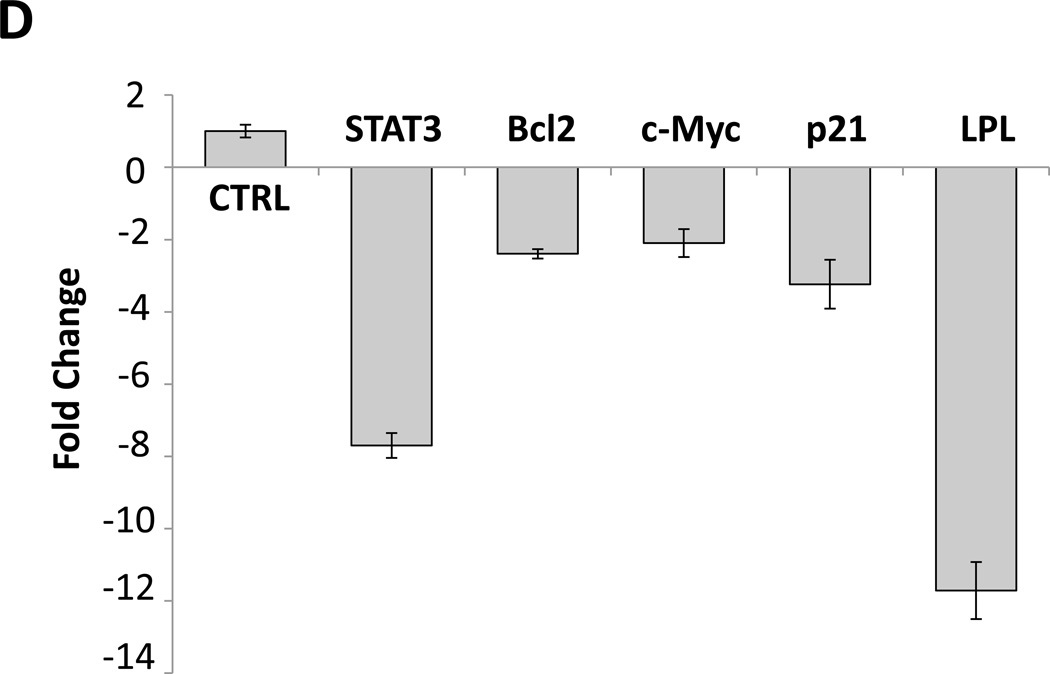
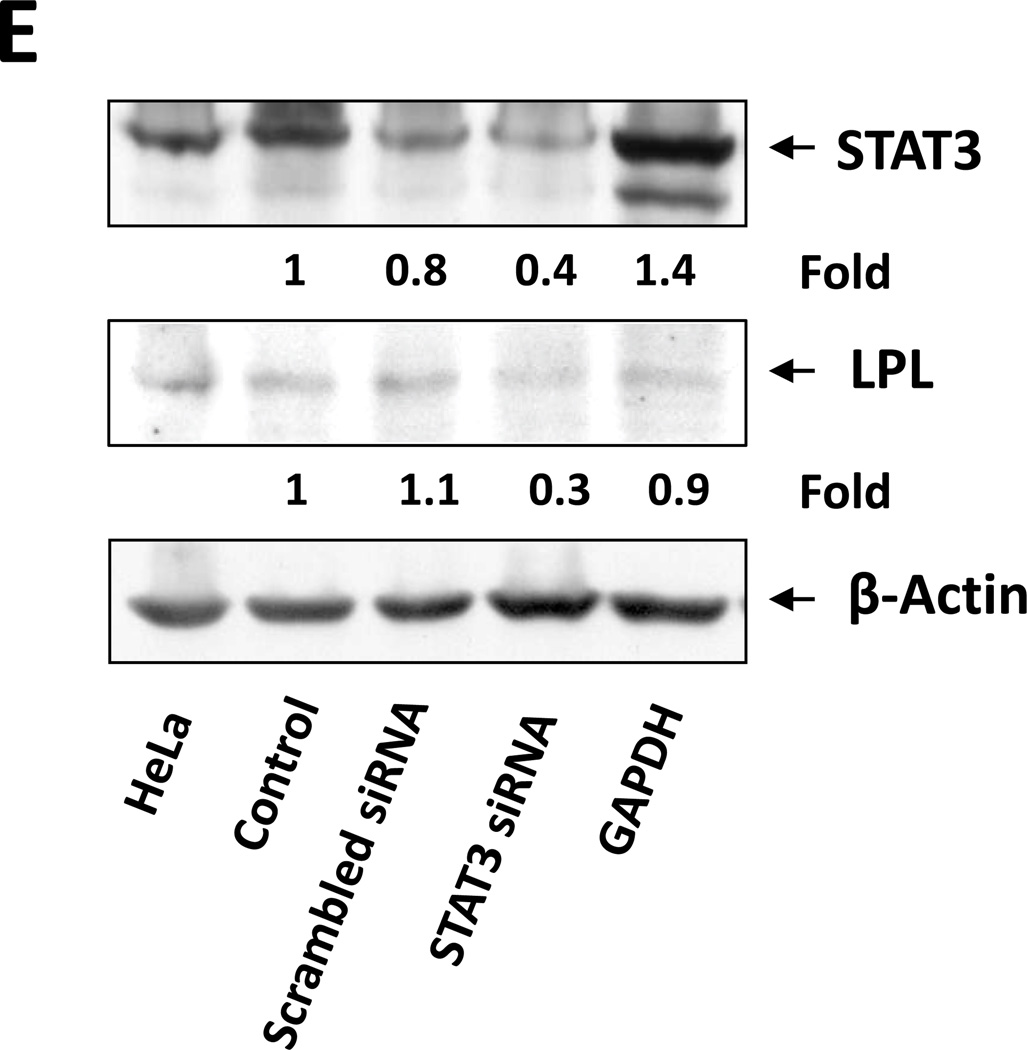
(A) Detection of pSTAT3 and LPL in IL-6–stimulated MM1 cells. MM1 cells were incubated with increasing concentrations of IL-6 (0 to 20 ng/mL for 2 hours (left panel), and with 10 ng/mL of IL-6 for 0 to 4 hours (right panel). Cell lysates underwent Western blot analysis with anti-tyrosine pSTAT3, anti-STAT3, and anti-LPL antibodies. HeLa cells incubated with IL-6 for 4 hours were used as positive controls in each panel. Incubation with IL-6 increased the levels of tyrosine pSTAT3 and LPL in a dose- and time-dependent manner. (B) On the basis of the presence and locations of GAS-like elements in the LPL promoter (upper panel), we transfected MM1 cells with truncated forms of the LPL promoter and the luciferase reporter gene (left lower panel). In the right lower panel, the horizontal bars show the mean ± standard error of the mean for luciferase activity levels of transfected MM1 cells incubated without or with 20 ng/mL IL-6. (C) ChIP assay. DNA obtained from chromatin fragments of MM1 cells incubated without or with IL-6, before (input) or after co-immunoprecipitation with anti-STAT3 antibodies, was analyzed using primers directed at three GAS-like elements located between +1 bp and −674 bp upstream of the LPL gene TSS. DNA co-immunoprecipitated with anti-STAT3 antibodies could be amplified by with primer 2 but not 1 or 3. The amplification of the DNA from IL-6–treated cells was stronger than that from untreated cells ((D) Transfection with STAT3 siRNA. MM1 cells were transfected with STAT3 siRNA using electroporation. Transfection efficiency, calculated by assessing the level of intracellular GFP-conjugated siRNA, was 50%. The cells were incubated with IL-6 for 2 hours, and then RNA was extracted and qRT-PCR performed to determine the levels of the STAT3-regulated genes STAT3, ROR1, c-Myc, cyclin D1, p21, and LPL. The experiment was repeated three times. The means and the standard error of the mean of mRNA levels are depicted. As shown, a 12-fold decrease in the LPL transcript level and 2- to 8-fold decreases in STAT3-regulated transcript levels compared with the control were observed. (E) CLL cells were transfected with STAT3 siRNA, scrambled siRNA, or GAPDH. Cellular protein was extracted and analyzed by Western blot analysis.
To assess the capability of each putative STAT3 binding site to activate the LPL promoter activity we transfected MM1 cells with the luciferase reporter gene driven by fragments of the LPL promoter (Figure 4B, upper panel). As shown in the lower panel of Figure 4B, the promoter activity was detected only in MM1 cells transfected with the 333-bp fragment and not in those transfected with the 143-bp fragment, suggesting that the GAS-like element located between bp −280 and bp −294 but not the one between bp −86 and bp −95 upstream of the TSS is an active STAT3 binding site. Moreover, when MM1 cells transfected with the 333-bp promoter fragment were incubated with IL-6, the promoter activity was markedly increased (Figure 4B).
To decipher this observation, we used a ChIP assay. As shown in Figure 4C, only primer 2, corresponding to the −333 bp to −250 bp sequence, but not primer 1 (downstream of primer 2) or primer 3 (upstream of primer 2), amplified DNA that co-immunoprecipitated with anti-STAT3 antibodies (Figure 4C). To determine whether pSTAT3-LPL promoter binding activates the transcription of LPL, we transfected MM1 cells with STAT3 siRNA, incubated them with IL-6 for 2 hours, and assessed LPL and various other STAT3-regulated genes’ mRNA levels using qRT-PCR. As shown in Figure 4D, STAT3 siRNA downregulated mRNA levels of LPL and the STAT3-regulated genes STAT3, Bcl2, c-Myc, and p21/WAF1. In addition, transfection of MM1 cells with STAT3-siRNA, but not with scrambled siRNA or GAPDH, downregulated STAT3 protein levels by 60% and LPL protein levels by 70% (Figure 4E), suggesting that STAT3 binds to the LPL promoter, activates the LPL gene and induces the production of LPL protein in MM1 cells.
STAT3 activates the LPL promoter in CLL cells
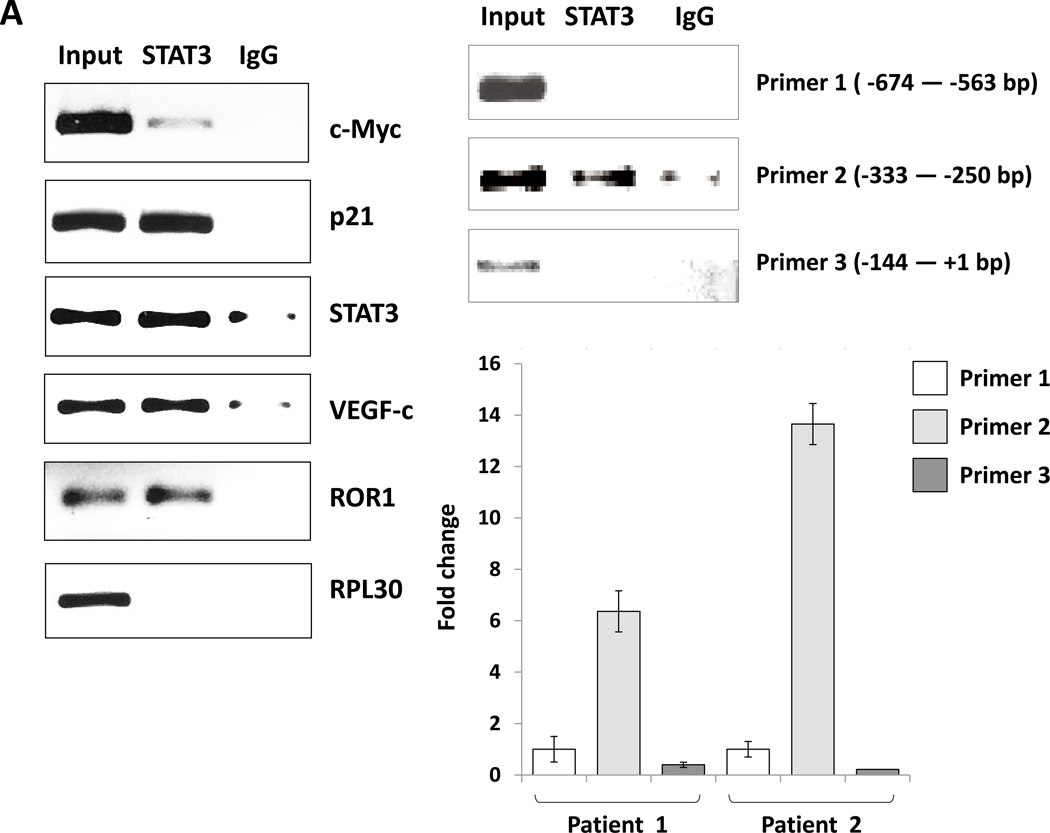
After establishing that STAT3 activates the LPL promoter in MM1 cells, we sought to determine whether STAT3 activates the transcription of LPL in CLL cells. To that end we performed ChIP. As shown in Figure 5A, we found that anti-STAT3 antibodies co-immunoprecipitated DNA of the LPL promoter and of the STAT3-regulated genes STAT3, c-Myc, p21, VEGF-c, and ROR1. As in MM1 cells, primers of the LPL promoter binding site 2, but not site 1 or 3, amplified the STAT3 co-immunoprecipitated DNA (Figure 5A, right panel), suggesting that the GAS-like element 280 bp upstream of the LPL coding region is an active STAT3 binding site in CLL.
Figure 5. STAT3 activates the LPL promoter in CLL cells.
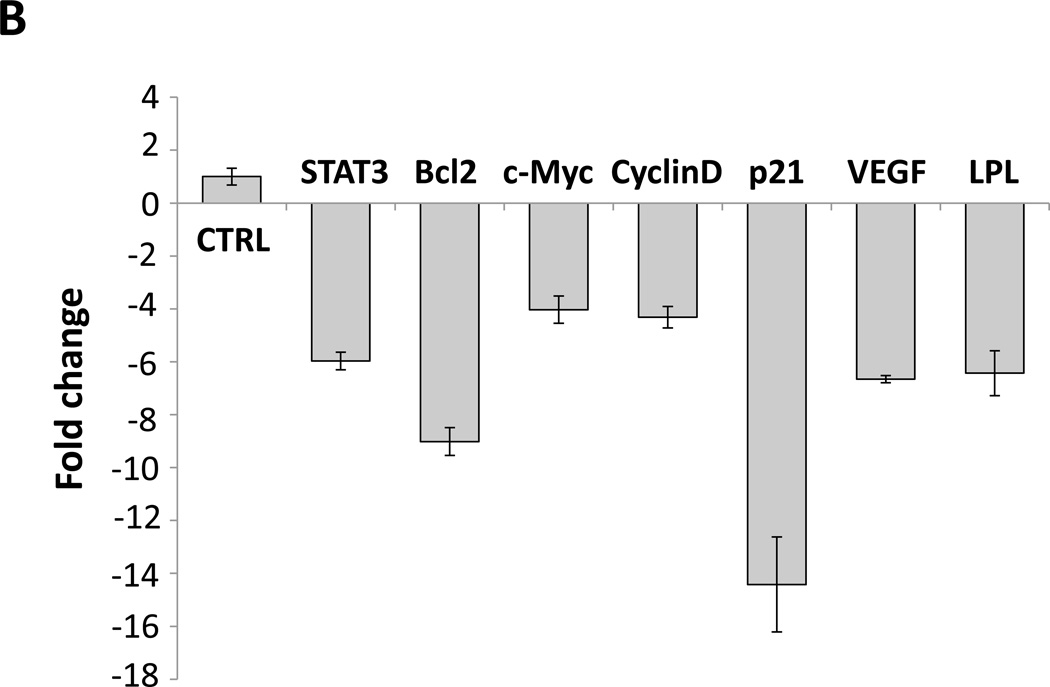
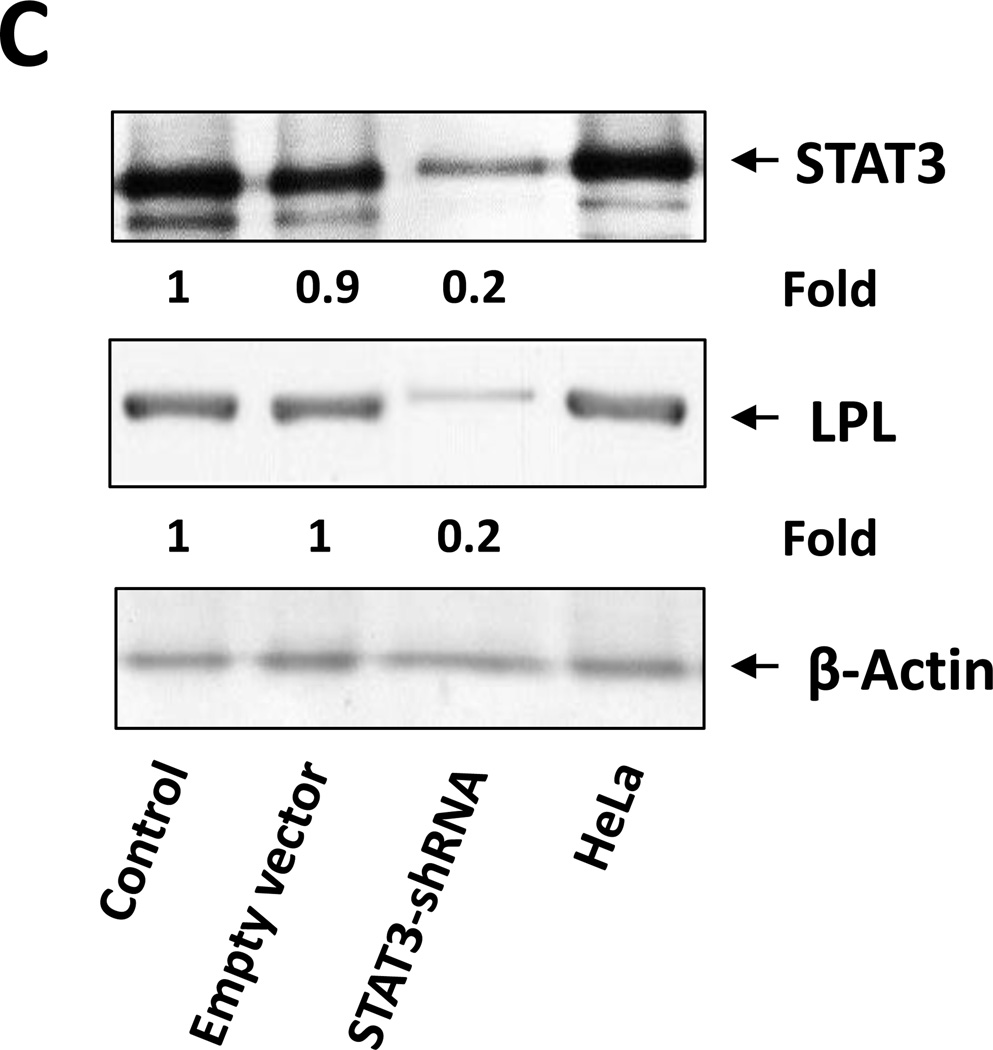
(A) ChIP assay. CLL cell protein extract was incubated without or with anti-STAT3 antibodies, and DNA was extracted from chromatin fragments. As shown in the left panel, anti-STAT3 antibodies co-immunoprecipitated DNA of the STAT3 target genes c-Myc, p21, STAT3, VEGF-c, and ROR1 but not of the ribosomal RPL30 gene (used as negative control). As in Figure 4C, “Input” denotes DNA extracted from non-immunoprecipitated CLL cell chromatin fragments (negative control) IgG is the isotype of the anti-STAT3 antibodies. The right panel upper depicts ChIP of CLL cells. As shown, STAT3 co-immunoprecipitated DNA that was amplified with primers designed to amplify site 2 but not primers designed to amplify site 1 or site 3 of the LPL-promoter regions. The right lower panel depicts results of two separate experiments analyzed using qRT-PCR. Similar to the results depicted in the right upper panel, STAT3 co-immunoprecipitated DNA was significantly amplified with primer 2. (B) CLL cells were transfected with STAT3-shRNA or with an empty vector. Compared with cells transfected with empty vector (CTRL), the cells that were transfected with STAT3-siRNA expressed significantly lower levels of STAT3-regulated genes including LPL. (C) Western blot analysis of CLL cells transfected with STAT3 shRNA or empty vector showed that compared with an empty vector, STAT3 shRNA downregulated STAT3 and LPL protein levels by 80%.
We then sought to determine whether STAT3 activates LPL in CLL cells. We transfected CLL cells with a lentiviral STAT3 shRNA (transfection efficiency, 50%) or with an empty vector and quantitated LPL and STAT3-regulated gene mRNA levels by using relative qRT-PCR. As shown in Figure 5B, transfection with STAT3 shRNA reduced the mRNA levels of LPL and the STAT3-regulated genes STAT3, Bcl2, c-Myc, cyclin D1, p21/WAF1, and VEGF. Furthermore, unlike the empty vector, STAT3 shRNA downregulated STAT3 and LPL protein levels by 80% (Figure 5C). Taken together, our findings suggest that STAT3 binds and activates the LPL promoter and induces the expression and production of LPL.
Discussion
In this study, we found that CLL cells, unlike normal B lymphocytes but similar to adipocytes and myocytes, store lipids in cytoplasmic vacuoles and are capable of metabolizing stored FFAs in an LPL-dependent manner.
Traditionally, CLL has been characterized by an accumulation of immunologically abnormal, long-lived lymphocytes.29 However data generated in the past decade showed that approximately 1% of CLL cells are regenerated daily.5 Various cells use different strategies to provide energy to fuel proliferation.30 One such strategy, used physiologically by adipocytes and myocytes, is the generation of cytoplasmic lipid stores that serve as a readily available energy source. This strategy has been adopted by various cancer cells. For example, the “starry sky” pattern, characteristically found in Burkitt’s lymphoma, is caused by an abundance of intracellular lipid vacuoles used as an energy source in this highly aggressive lymphoma.31 We found that CLL cells, similar to Burkitt’s lymphoma cells but unlike normal B lymphocytes, store lipids in a form of cytoplasmic lipid vacuoles. We could not study lymph node CLL cells. However because CLL cells traffic between the lymph nodes blood and bone marrow1, it is reasonable to assume that lymph node CLL cells carry similar features. Remarkably, it was found that the gene expression profile of CLL cells is skewed towards the expression of genes usually expressed in muscle and fat tissue32. It is therefore likely that to adjust for cellular survival and a proliferation rate higher than in their cell of origin, CLL cells adopted the strategy of mammalian muscle cells and adipocytes, which store intracellular lipids and proliferate at similar rates33.
Similar to adipocytes, CLL cells express LPL on their surface and in their cytoplasm and store lipids in cytoplasmic vacuoles. However the functional significance of the expression of LPL in CLL cells is not entirely clear. Overexpression of lipase activity-associated genes34 and increased lipase activity were found in CLL cells34. Conversely, other investigators suggested that LPL is catalytically inactive in CLL cells35 and that LPL knockdown did not affect the survival of CLL cells36. Here we show that LPL provides CLL cells with survival advantage. Transfection of CLL cells with LPL-siRNA increased CLL cell apoptosis rate by an average of 32%. Whether this effect is induced by an abrogation of the catalytic or non-catalytic function of LPL, remains to be determined.
Free fatty acids, the product of triglyceride hydrolysis by LPL, are the fuel used for oxidative phosphorylation, and are also required to prompt the enzymatic machinery needed for their own metabolism. FFAs bind the peroxisome proliferator-activated receptor activated alpha (PPARα) which translocates to the nucleus and induces the transcription enzymes that are needed for fatty acid oxidation37. Remarkably, PPARα was found to be expressed by CLL cells, and CLL cell palmitic acid oxidation rate was similar to the oxidation rate typically found in fat-burning cells38.
Our previous studies suggested that the unique gene signature of CLL is driven in part by constitutively activated STAT3.19,20,39,40 In the current study, we show that STAT3 binds to the LPL promoter and activates the transcription of LPL. Transfection of CLL cells with STAT3-shRNA significantly downregulated LPL mRNA and protein levels, suggesting that the transcription of LPL in CLL cells is STAT3 dependent. STAT3 is not the only transcription factor that induces LPL gene expression. Abreu et al. suggested that in CLL patients with unmutated IgHV microenvironment-induced demethylation contributes to the increased expression of LPL in CLL41. Nonetheless, our data suggest that the survival advantage provided by constitutively activated STAT314 is mediated in part by the contribution of STAT3 to the CLL cells’ metabolism.
Taken together, our findings suggest that CLL cells store lipid vacuoles, produce LPL, and adapt their metabolism to utilize intracellular stored lipids for energy production in an LPL-dependent manner, a process that is driven by STAT3.
Supplementary Material
Implications.
Our study suggests that CLL cells adopt their metabolism to oxidize FFA. Activated STAT3 induces LPL, which catalyzes the hydrolysis of triglycerides into FFA. Therefore, inhibition of STAT3 is likely to prevent the capacity of CLL cells to utilize FFA.
Acknowledgments
We thank Sarah Bronson of the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for editing the manuscript.
This study was supported by a grant from the CLL Global Research Foundation. The University of Texas MD Anderson Cancer Center is supported in part by the National Institutes of Health through a Cancer Center Support Grant (P30CA16672).
Footnotes
Authorship
U.R.: designed the experiments, analyzed the data and drafted the manuscript.
S.G.: performed the western blot experiments, ChIP and drafted parts of the manuscript.
C.B-R.: performed the pathologic classification and the cytochemistry staining.
D.M.H.: performed the confocal and the TEM experiments.
P.L.: performed the ChIP, RNA studies and the luciferase assay experiments.
Z.L., J.Y.W.: performed the western blot experiment.
W.W., J.B., A.F., N.J.: treated the patients and obtained blood samples.
P.J.: obtained the patients’ clinical data.
S.O.: recruited, treated the patients and obtained blood samples.
M.K.: recruited and treated patients and participated in designing the study.
Z.E.: conceived, designed the study and participated in drafting the manuscript.
All authors approved the final manuscript.
Conflict-of-interest disclosure
The authors declare no competing financial interests.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Damle RN, Ghiotto F, Valetto A, et al. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood. 2002;99(11):4087–4093. doi: 10.1182/blood.v99.11.4087. [DOI] [PubMed] [Google Scholar]
- 3.Klein U, Tu Y, Stolovitzky GA, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194(11):1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tangye SG, Avery DT, Deenick EK, Hodgkin PD. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J Immunol. 2003;170(2):686–694. doi: 10.4049/jimmunol.170.2.686. [DOI] [PubMed] [Google Scholar]
- 5.Chiorazzi N. Cell proliferation and death: forgotten features of chronic lymphocytic leukemia B cells. Best Pract Res Clin Haematol. 2007;20(3):399–413. doi: 10.1016/j.beha.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation, and role in disease. J Mol Med (Berl) 2002;80(12):753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg IJ. Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J Lipid Res. 1996;37(4):693–707. [PubMed] [Google Scholar]
- 8.Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194(11):1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintel D, Kienle D, Shehata M, et al. High expression of lipoprotein lipase in poor risk B-cell chronic lymphocytic leukemia. Leukemia. 2005;19(7):1216–1223. doi: 10.1038/sj.leu.2403748. [DOI] [PubMed] [Google Scholar]
- 10.Oppezzo P, Vasconcelos Y, Settegrana C, et al. The LPL/ADAM29 expression ratio is a novel prognosis indicator in chronic lymphocytic leukemia. Blood. 2005;106(2):650–657. doi: 10.1182/blood-2004-08-3344. [DOI] [PubMed] [Google Scholar]
- 11.Kaderi MA, Kanduri M, Buhl AM, et al. LPL is the strongest prognostic factor in a comparative analysis of RNA-based markers in early chronic lymphocytic leukemia. Haematologica. 2011;96(8):1153–1160. doi: 10.3324/haematol.2010.039396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Crowe PJ, Goldstein D, Yang JL. STAT3 inhibition, a novel approach to enhancing targeted therapy in human cancers (review) Int J Oncol. 2012;41(4):1181–1191. doi: 10.3892/ijo.2012.1568. [DOI] [PubMed] [Google Scholar]
- 13.Frank DA, Mahajan S, Ritz J. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest. 1997;100(12):3140–3148. doi: 10.1172/JCI119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazan-Halevy I, Harris D, Liu Z, et al. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood. 2010;115(14):2852–2863. doi: 10.1182/blood-2009-10-230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13(4):227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Ikegami M, Wang Y, Matsuzaki Y, Whitsett JA. Gene expression and biological processes influenced by deletion of Stat3 in pulmonary type II epithelial cells. BMC Genomics. 2007;8:455. doi: 10.1186/1471-2164-8-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bozzola JJ. Conventional specimen preparation techniques for transmission electron microscopy of cultured cells. Methods Mol Biol. 2007;369:1–18. doi: 10.1007/978-1-59745-294-6_1. [DOI] [PubMed] [Google Scholar]
- 18.Ferrajoli A, Faderl S, Van Q, et al. WP1066 disrupts Janus kinase-2 and induces caspase-dependent apoptosis in acute myelogenous leukemia cells. Cancer Res. 2007;67(23):11291–11299. doi: 10.1158/0008-5472.CAN-07-0593. [DOI] [PubMed] [Google Scholar]
- 19.Li P, Grgurevic S, Liu Z, et al. Signal transducer and activator of transcription-3 induces MicroRNA-155 expression in chronic lymphocytic leukemia. PLoS One. 2013;8(6):e64678. doi: 10.1371/journal.pone.0064678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Harris D, Liu Z, Liu J, Keating M, Estrov Z. Stat3 activates the receptor tyrosine kinase like orphan receptor-1 gene in chronic lymphocytic leukemia cells. PLoS One. 2010;5(7):e11859. doi: 10.1371/journal.pone.0011859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzales AM, Orlando RA. Role of adipocyte-derived lipoprotein lipase in adipocyte hypertrophy. Nutr Metab (Lond) 2007;4:22. doi: 10.1186/1743-7075-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;43(12):1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- 23.Pradines-Figueres A, Vannier C, Ailhaud G. Lipoprotein lipase stored in adipocytes and muscle cells is a cryptic enzyme. J Lipid Res. 1990;31(8):1467–1476. [PubMed] [Google Scholar]
- 24.Ruby MA, Goldenson B, Orasanu G, Johnston TP, Plutzky J, Krauss RM. VLDL hydrolysis by LPL activates PPAR-alpha through generation of unbound fatty acids. J Lipid Res. 2010;51(8):2275–2281. doi: 10.1194/jlr.M005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinemeyer T, Wingender E, Reuter I, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26(1):362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296(5573):1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 27.Berishaj M, Gao SP, Ahmed S, et al. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9(3):R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leu CM, Wong FH, Chang C, Huang SF, Hu CP. Interleukin-6 acts as an antiapoptotic factor in human esophageal carcinoma cells through the activation of both STAT3 and mitogen-activated protein kinase pathways. Oncogene. 2003;22(49):7809–7818. doi: 10.1038/sj.onc.1207084. [DOI] [PubMed] [Google Scholar]
- 29.Dameshek W. Chronic lymphocytic leukemia--an accumulative disease of immunolgically incompetent lymphocytes. Blood. 1967;29(4) Suppl:566–584. [PubMed] [Google Scholar]
- 30.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 31.Ambrosio MR, Piccaluga PP, Ponzoni M, et al. The alteration of lipid metabolism in Burkitt lymphoma identifies a novel marker: adipophilin. PLoS One. 2012;7(8):e44315. doi: 10.1371/journal.pone.0044315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilban M, Heintel D, Scharl T, et al. Deregulated expression of fat and muscle genes in B-cell chronic lymphocytic leukemia with high lipoprotein lipase expression. Leukemia. 2006;20(6):1080–1088. doi: 10.1038/sj.leu.2404220. [DOI] [PubMed] [Google Scholar]
- 33.Neese RA, Misell LM, Turner S, et al. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proc Natl Acad Sci U S A. 2002;99(24):15345–15350. doi: 10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallasch CP, Schwamb J, Konigs S, et al. Targeting lipid metabolism by the lipoprotein lipase inhibitor orlistat results in apoptosis of B-cell chronic lymphocytic leukemia cells. Leukemia. 2008;22(3):585–592. doi: 10.1038/sj.leu.2405058. [DOI] [PubMed] [Google Scholar]
- 35.Mansouri M, Sevov M, Fahlgren E, et al. Lipoprotein lipase is differentially expressed in prognostic subsets of chronic lymphocytic leukemia but displays invariably low catalytical activity. Leuk Res. 2010;34(3):301–306. doi: 10.1016/j.leukres.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 36.Porpaczy E, Tauber S, Bilban M, et al. Lipoprotein lipase in chronic lymphocytic leukaemia - strong biomarker with lack of functional significance. Leuk Res. 2013;37(6):631–636. doi: 10.1016/j.leukres.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010;2010 doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spaner DE, Lee E, Shi Y, et al. PPAR-alpha is a therapeutic target for chronic lymphocytic leukemia. Leukemia. 2012 doi: 10.1038/leu.2012.329. [DOI] [PubMed] [Google Scholar]
- 39.Liu Z, Hazan-Halevy I, Harris DM, et al. STAT-3 activates NF-kappaB in chronic lymphocytic leukemia cells. Mol Cancer Res. 2011;9(4):507–515. doi: 10.1158/1541-7786.MCR-10-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badoux X, Bueso-Ramos C, Harris D, et al. Cross-talk between chronic lymphocytic leukemia cells and bone marrow endothelial cells: role of signal transducer and activator of transcription 3. Hum Pathol. 2011;42(12):1989–2000. doi: 10.1016/j.humpath.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abreu C, Moreno P, Palacios F, et al. Methylation status regulates lipoprotein lipase expression in chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54(8):1844–1848. doi: 10.3109/10428194.2013.796057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.