Abstract
Pancreatic cancer is one of the deadliest cancers with poor survival rates and limited therapeutic options. To improve the understanding of this disease's biology, a prerequisite for the generation of novel therapeutics, new platforms for rapid and efficient genetic and therapeutic screening are needed. Therefore, a combined in vitro/in vivo hybrid shRNA-assay was developed using isolated murine primary pancreatic ductal cells (PDCs), in which oncogenic KrasG12D could be activated in vitro by genomic recombination through 4OH-tamoxifen-induced nuclear translocation of Cre-ERT2 expressed under control of the ROSA26 promoter. Further genetic manipulation was achieved through selective and stable RNA interference (RNAi) against the tumor suppressors p16Ink4a (CDKN2A) or p53 (TP53) using lentiviral gene delivery. Treatment of PDCs with 4OH-tamoxifen increased phosphorylation of extracelluar signal-regulated kinase (ERK) downstream of KRAS, and subsequent lentiviral transduction resulted in sustained target gene repression. Double-mutant PDCs were then re-introduced into the pancreata of NOD-SCID-gamma (NSG) mice and monitored for tumor growth. Orthotopic implantation of PDCs carrying the activated KrasG12D-allele and shRNA against p16Ink4a or p53 resulted in tumor growth, metastasis and reduced survival of NSG mice. In contrast, KrasG12D alone was not sufficient to induce tumor growth. Implications: The combinatory in vitro/in vivo approach described in this study allows for rapid and efficient identification of genes involved in carcinogenesis and opens new avenues for the development of therapeutic strategies to improve cancer treatment.
Keywords: Pancreatic cancer, shRNA, tumor suppressor, carcinogenesis, genetic screen
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer related deaths in the US. The five-year survival rate of all patients suffering from PDAC is 6%, and incidence almost equals mortality rate, underscoring the aggressive behavior of this tumor (1). Most patients suffering from PDAC already present with metastasis, causing the majority of pancreatic cancer associated deaths (2). Although several broad based approaches have been undertaken to shed light on the genetics and biology of pancreatic cancer, only few essential driver mutations have been identified so far (3, 4). One of the most common genetic perturbations in pancreatic cancer is an activating mutation of oncogenic Kras, which can be found in more than 90% of PDAC and is thought to represent an initiating event (5). However, single activation of Kras in mice results in pancreatic intraepithelial neoplasia (PanIN), but shows only infrequent development of invasive PDAC (6). Thus, additional genetic events are required for the development of invasive PDAC, including loss of the cell cycle regulator p16Ink4a (part of the Cdkn2a-locus), and of the tumor suppressor genes Trp53 and/or Smad4 (7). Identification of additional genes involved in tumorigenesis will broaden our understanding of pancreatic cancer biology and eventually lead the way to more effective treatments.
Functional characterization of cancer genes can be cumbersome. Although cell culture assays can be easily performed and are highly reproducible, in vitro models lack the features of the tumor microenvironment and, thus, may not be suitable to detect gene activities linked to cancer initiation or progression.
The standard approach for investigating candidate cancer genes requires the generation of transgenic and knockout mice that harbor germline alterations in the gene of interest. Although these strains are invaluable tools in the field of cancer research, their generation, maintenance and analysis can be costly and time consuming. Moreover, many data obtained with these models rely on manipulation of cancer genes during embryogenesis, and thus, do not reflect somatic mutations occurring during an individual's life span.
To obviate these obstacles, we developed an approach in which we combined the ease of in vitro genetic manipulation and the power of in vivo pancreatic cancer studies. In this model, we took advantage of the well-established primary pancreatic ductal cell culture (PDCs) (8, 9). Isolation of PDCs from mice that harbor the lox-stop-lox-KrasG12D-allele and express Cre-ERT2 under control of the ubiquitous ROSA26-promoter allowed us to induce genetic recombination and subsequent activation of KrasG12D in vitro. We hypothesized that additional depletion of the tumor suppressor genes p16Ink4a or Trp53 in the context of Kras-activation in PDCs will lead to accelerated tumor growth and invasive PDAC. Genetic silencing was achieved by lentiviral delivery of shRNA, and orthotopic implantation of these resulted in tumor growth. However, Kras activation on its own was not sufficient to induce tumor growth. The method described in this study will simplify the identification and validation of new cancer genes with high reliability and without the need for tedious mouse models.
Material and Methods
Isolation of Pancreatic ductal cells
Primary pancreatic ductal cells (PDCs) were isolated from mice carrying the genotype ROSA26Cre-ERT2;Lox-Stop-Lox-KrasG12D (termed Kras-PDCs thereafter) and maintained essentially as described (10). Early passage cells were treated with 4OH-tamoxifen 200 nM (Sigma) or vehicle for 10 days.
Western Blot Analysis
PDCs were collected from collagen by digestion with collagenase type 2 (Worthington) at a final concentration of 1 mg/ml at 37 °C for 15 min. Upon complete digestion, cells were pelleted by centrifugation and washed with ice cold PBS. The final pellet was lysed and protein concentration was normalized using Bradford reagent (Biorad). 50 μg were resolved on a 10% sodium-dodecylsulfate-(SDS)-polyacrylamide gel and transferred to polyvinylidene difluoride membranes (PVDF). Membranes were blocked in PBS containing 0.05% Tween and 3% non-fat dry milk for 1 h at room temperature and incubated with anti-pERK (Cell Signaling 4370) 1:1000, anti-P16INK4a (M-156, Santa-Cruz sc-1207)1:200 or anti-TRP53 (NCL-p53-CM5p, Novocastra, Leica Bisosystems) 1:1000. Membranes were then subsequently incubated with anti-ERK (BD Biosciences 610124) 1:1000 and anti-β-Actin (Sigma-Aldrich, Clone AC-74, A5316) 1:5000. Visualization was performed using IRDye 680 (anti-rabbit) or IRDye 780 (anti-mouse) secondary antibodies on an Odyssey Infrared Imaging System (all LiCor).
RAS activation assay
Detection of activated KRAS was performed essentially as described using a Raf-RBD-pulldown assay (Cytoskeleton) (11). Transfer to PVDF membranes and visualization was conducted as mentioned above using an antibody against KRAS (Merck-Millipore, 1:1000).
Quantitative Real-Time PCR
Total RNA was extracted using the RNeasy kit (Qiagen). Synthesis of cDNA using random hexamers and MMLV-based reverse transcriptase (Life Technologies) was achieved as previously described (12). Quantitative analysis was carried out on StepONEplus real-time PCR system (Applied Biosystems, Life Technologies) and the amount of target gene was normalized to the endogenous reference Ppia (Cyclophilin A) (13).
Murine primers were designed to be intron spanning. The following primers were used: p16Ink4a FW: CCCAACGCCCCGAACT, P16Ink4a RV: GTGAACGTTGCCCATCATCA, Trp53 FW: AGATCCGCGGGCGTAAAC RV: TCTGTAGCATGGGCATCCTTT Ppia FW: ATGGTCAACCCCACCGTGT, Ppia RV: TTCTGCTGTCTTTGGAACTTTGTC.
Lentiviral constructs, virus generation, target cell transduction and selection
Glycerol stocks containing the desired lentiviral constructs were obtained from Open Biosystems (now part of GE Healthcare) and grown according to the manufacturer's instructions. The clone IDs for shRNAp16Ink4a were: TRCN0000077814, target sequence GTGATGATGATGGGCAACGTT, termed shRNAp16Ink4a #1 hereafter, and TRCN0000077813, target sequence CATCAAGACATCGTGCGATAT, termed shRNAp16Ink4a #2 hereafter. The clone IDs for shRNATrp53 were: TRCN0000012362, target sequence CTACAAGAAGTCACAGCACAT, termed shRNATrp53 #1 hereafter, and TRCN0000054551, target sequence AGAGTATTTCACCCTCAAGAT, termed shRNATrp53 #2 herafter. pLKO.1 was used for control. Purified plasmids were tested for integrity prior to transfection using the restriction enzymes BamHI and NdeI.
For virus production 6×106 293T cells in a 10 cm dish were transfected with 12 μg lentiviral construct, 4,2 μg pMD2G-VSVG and 7,8 μg pCMV-dR8.74 using Lipofectamine 2000 (Life Technologies). Twenty-four hours post transfection culture media was changed and virus-containing supernatant was collected 24 h later. Culture media was replaced and collected another 24 h later. Viral supernatant was stored at -80 °C until further use.
Kras-PDCs were transduced with lentivral constructs as described with only minor modifications (14). Briefly, 4OH-tamoxifen-treated Kras-PDCs were placed in wells of a six-well plate at a cell number of 3×105 cells/well and allowed to adhere on plastic over night. Next day, cells were transduced with viral supernatant containing ploybrene 4 μg/ml (Sigma). Twenty-four hours later PDCs were placed back onto collagen coated wells and were allowed to adhere for another 24 h. Upon complete attachment, cells were selected in the presence of puromycin 8 μg/ml for ten days (termed Kras-shRNA-PDCs hereafter). Successful depletion of target gene mRNA was confirmed by quantitative RTPCR. Two different shRNAs per target gene were tested to reduce off-target effects.
Orthoptopic transplantation and animal procedures
Immunocompromised NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (NOD scid gamma, NSG) were obtained form The Jackson Laboratory. Eight to ten week old animals were anaesthetized using a combination of Medetomidine, Midazolam and Fentanyl. A total of 5×105 Kras-shRNA-PDCs in a volume of 20 μl were injected into the pancreata of NSG-mice as described (12). Briefly, a small left abdominal incision was made and the pancreas was retrieved by gently dislodging the spleen. Tumor cells were injected into the pancreas in an area adjacent to the spleen using a micro liter syringe with a 27 gauge needle. Successful injection was confirmed by an intrapancreatic bleb. The peritoneal layer was sutured with Ethilon 5-0 (Johnson and Johnson) and the cutaneous wound was closed using wound clips. We injected three mice per shRNA. Animals were investigated weekly for tumor growth, development of ascites and weight loss. Animals were euthanized upon palpable local tumor growth > 1cm, development of ascites or loss of body weight > 20%. If none of these occurred, animals were euthanized after a period of 26 weeks. All animal procedures were in agreement with the Government of Upper Bavaria (protocol 55.2-1-54-2532-117-13).
Histology
Mice were euthanized and organs were removed and fixed overnight in 4% paraformaldehyde. Organs were then embedded in paraffin, sectioned at 2.5 μm and mounted on glass slides. Following standard dewaxing and hydration procedures, staining was performed for 30 s in Hematoxylin, followed by a 5 min tap water rinse. Counterstaing was performed in Eosin 30 s, and subsequent dehydration was conducted according to standard procedures. For immunohistochemistry, slides were dewaxed and hydrated as above. Antigen retrieval was perfomed in citrate solution at pH 6.0 for 15 min in a microwave at 600W. The following antibodies were used: anti-P16INK4a (1:100, Santa Cruz, F-12, sc-1661) and anti-TRP53 (1:300, NCL-p53-CM5p, Novocastra, Leica Bisosystems), followed by secondary biotin-conjugated antibodies. Peroxidase conjugated streptavidin was used with 3,3’-diaminobenzidine tetrahydrochloride (DAB, VectorLabs) as a chromogen for detection. Hematoxylin was used for counterstaining. Pictures were then recorded on an AxioImagerA1 microscope with an AxioCam color camera using AxioVision 4.3 software (all Carl Zeiss).
Statistical Analysis
Statistics were performed using graph pad prism. For expression analysis, student's t-test was used. To analyze survival after orthtopic implantation of KrasshRNA-PDCs, Log-Rank (Mantle-Cox) analysis was applied.
Results
Development of an in vitro Kras activation method
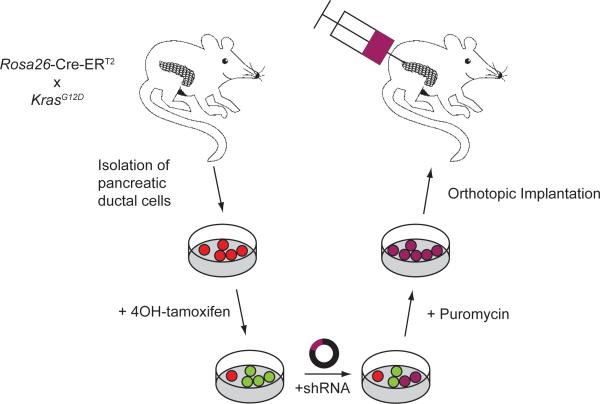
To obtain a strictly genetically defined model without contaminating stromal cells we decided to isolate a purely ductal cell population from the pancreas for further in vitro manipulation. Since Kras is mutated in over 90% of PDAC cases, we chose in vitro activation of LSL-KrasG12D by nuclear translocation of Cre-ERT2 through the application of 4OH-tamoxifen. Genomic re-arrangement and activation of KrasG12D was followed by introduction of a well-defined genetic second hit by the virtue of shRNA against Trp53 or p16Ink4a. These double-mutant PDCs were then re-introduced into the pancreata of NSG mice (Figure 1).
Figure 1.
Approach to generate double-mutant primary pancreatic ductal cells. PDCs were harvested from ROSA26CreERT2;KrasG12D animals and treated with 4OH-tamoxifen to induce recombination. A second genetic hit was subsequently introduced by infection with lentiviral particles containing empty control vector or short hairpin RNAs directed against p16Ink4a or Trp53. Upon selection for viral integration by puromycin, double-mutant PDCs were injected orthotopically into recipient mice (n=3 per lentiviral construct).
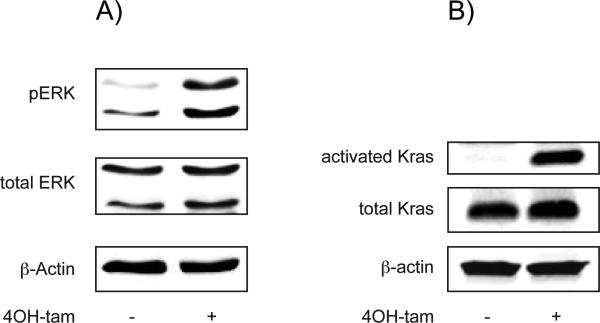
Indirect and direct assessment of Kras-activation by determination of the phosphorylation status of extracelluar signal-regulated kinase (ERK) and by using a Raf-RBD-GST-pulldown assay clearly demonstrated an increase of acitvated Kras in 4OH-tamoxifen treated Kras-PDCs (Figure 2 A and B).
Figure 2.
Validation of KRAS activation in 4-OHtamoxifen treated PDCs.
A) Western blot analysis reveals increased levels of pERK in 4OH-tamoxifen treated PDCs as compared to control as a functional readout for Kras activation.
B) Direct evidence for KRAS-activation by 4OH-tamoxifen by detection of active KRAS using a Raf-RBD-assay, followed by western blot analysis. Note the absence of active endogenous KRAS in non-treated cells and comparable levels of total KRAS.
Stable expression of shRNA leads to long-term gene regulation
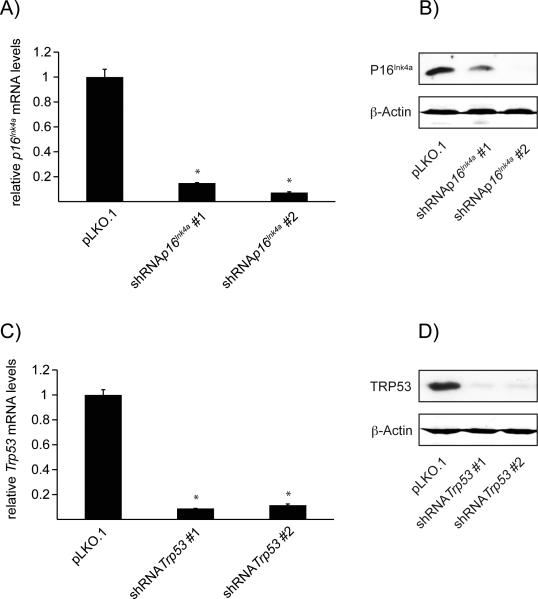
As lentiviral transduction results in stable integration of the lentiviral genome into the host genome, we evaluated for sustained gene silencing after lentiviral infection. To that end, we tested target gene expression ten days after withdrawal of puromycin. Indeed, we observed long-term gene silencing of p16Ink4a and Trp53 in all shRNA constructs used as demonstrated by assessment of target gene expression by qRT-PCR (Figure 3 A and C). Additional western blot analyis confirmed significant reduction of protein expression of P16INK4a and TRP53, respectively. (Figure 3 B and D).
Figure 3.
Transduction with shRNA results in long-term gene silencing.
A) Quantitative RT-PCR of p16Ink4a. 4OH-tamoxifen treated PDCs have been transduced with lentiviral particles containing either pLKO.1 empty control vector or shRNA against p16Ink4a. Ten days after 4OH-tamoxifen withdrawal PDCs show significant decrease of target gene mRNA in both experimental groups vs. control. * = p<0.05, n=3.
B) Western blot analysis of P16INK4a expression. 4OH-tamoxifen treated PDCs have been transduced with lentiviral particles containing either pLKO.1 empty control vector or shRNA against p16Ink4a. Ten days after 4OH-tamoxifen withdrawal PDCs show significant decrease of P16INK4a protein expression in both experimental groups vs. control.
C) Quantitative RT-PCR of Trp53. 4OH-tamoxifen treated PDCs have been transduced with lentiviral particles containing either pLKO.1 empty control vector or shRNA against Trp53. Ten days after 4OH-tamoxifen withdrawal PDCs show significant decrease of target gene mRNA in both experimental groups vs. control. * = p<0.05, n=3
D) Western blot analysis of TRP53 expression. 4OH-tamoxifen treated PDCs have been transduced with lentiviral particles containing either pLKO.1 empty control vector or shRNA against Trp53. Ten days after 4OH-tamoxifen withdrawal PDCs show significant decrease of TRP53 protein expression in both experimental groups vs. control.
Depletion of p16Ink4a in Kras-PDCs results in tumorigenesis
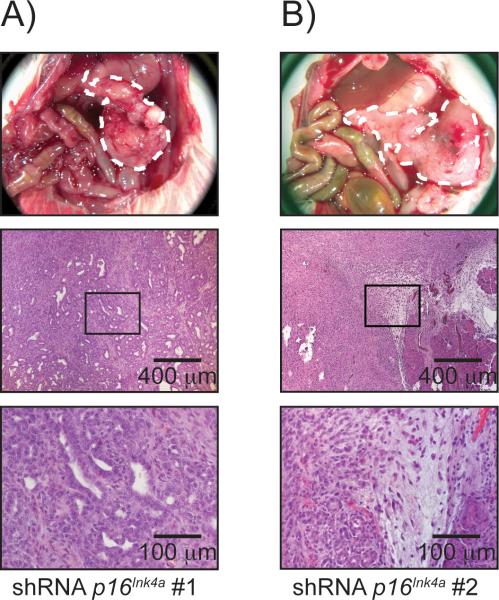
Tumor growth did not occur in animals that received Kras-shRNA-Control-PDCs over a time period of 26 weeks. In particular, the histological examination of control pancreata did not reveal any PanINs or other atypical or premalignant cell formation. By contrast, stable knock down of P16Ink4a resulted in rapid local tumor growth in five out of six animals in total. Interestingly, implantation of KrasshRNA-p16Ink4a #1-PDCs resulted in tumor growth in two out of three animals, whereas Kras-shRNA-p16Ink4a #2-PDCs led to development of pancreatic tumors only in all animals. In addition, the median survival in animals receiving KrasshRNA-p16Ink4a #1-PDCs was 160.0 days and 105.0 days in those receiving Kras-shRNA-p16Ink4a #2-PDCs. However, gross anatomy as well as histological findings did not differ between the two shRNAs against p16Ink4a. Immunohistological staining for P16INK4a did not yield any signal in tumors that developed from Kras-shRNA-p16Ink4a-PDCs, indicating sustained gene silencing by both shRNAs directed against p16Ink4a. PanINs from three month old Ptf1a-Cre; LSL-KrasG12D animals served as positive control (Figure S1 A). Of note, the tumors did not show the classical, desmoplastic architecture typical for PDAC but rather displayed a tumor cell rich growth with only sparse duct formation in vivo and almost no stromal reaction (Figure 4 A and B). Macroscopic liver metastasis did not occur upon depletion of p16Ink4a. However, micro-metastasis could be found in one animal out of five mice that developed tumors upon depletion of p16Ink4a (Figure S2).
Figure 4.
Loss of p16Ink4a results in PDAC formation
Animals receiving Kras-shRNAControl-PDCs did not develop tumors. Animals injected with Kras-shRNAp16Ink4a-PDCs develop tumors and die due to their tumor burden. A) and B): Gross anatomy shows tumor growth within the anatomical site of injection and histology confirms tumor growth (magnification 50× and 200×).
Loss of Trp53leads to tumor growth
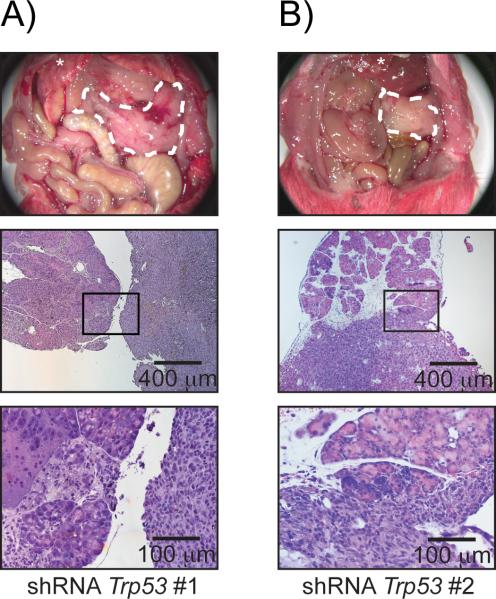
To address the question whether increased tumor growth is specific to the loss of p16Ink4a in the setting of Kras-activation, we asked if depletion of another tumor suppressor, Trp53, would result in tumorigenesis as well. In line with the findings mentioned above, implantation of Kras-shRNA-Trp53-PDCs resulted in tumor development in five out of six animals in total. Two out of three animals receiving Kras-shRNA-Trp53 #1-PDCs and all animals receiving Kras-shRNA-Trp53 #2-PDCs developed pancreatic tumors. The median survival was 140 and 111 days, respectively. Loss of TRP53 was demonstrated by the absence of nuclear staining compared to TRP53 positive PanINs of three month old Ptf1a-Cre; LSL-KrasG12D animals (Figure S1 B), demonstrating downregulation of TRP53 in these tumors. Interestingly, the macroscopic and histologic findings were similar to those seen in animals that have been implanted with Kras-shRNA-p16Ink4a-PDCs, with tumors containing densely arranged tumor cells but almost no stromal reaction (Figure 5 A and B). Macrosopic liver metastasis occurred in three out of five tumor bearing animals and was confirmed by histology (Figure S2).
Figure 5.
Loss of Trp53 results in PDAC formation
Animals receiving Kras-shRNAControl-PDCs did not develop tumors. Animals injected with Kras-shRNATrp53 develop tumors and die due to their tumor burden. A) and B) Gross anatomy shows tumor growth within the anatomical site of injection and histology confirms tumor growth (magnification 50× and 200×).
shRNA mediated gene silencing results in decreased survival
In total, five out of six animals receiving Kras-shRNAp16Ink4a-PDCs or Kras shRNATrp53-PDCs developed tumors. These animals display a significantly shorter survival when compared to animals that received Kras-shRNAControl. However there was no difference between animals implanted with KrasshRNAp16Ink4a-PDCs or Kras-shRNATrp53-PDCs (Figure S3).
Discussion
The data presented here demonstrate a stepwise manipulation of adult pancreatic ductal cells to model PDAC in vivo. First, we report isolation of an already well-defined pancreatic cell population that can be genetically altered by ex vivo recombination events through transient nuclear translocation of Cre-ERT2 (15), thereby activating oncogenic Kras. Second, we show sustained long-term gene silencing in Kras-PDCs using selective shRNAs against the tumor suppressor genes p16Ink4a or Trp53. Third, we clearly demonstrate that in vitro modeling of genetic pathways that have been implicated in pancreatic cancer development and progression to lead to malignant pancreatic tumors in vivo. One main advantage of the system used is the rapid generation of the desired cell line carrying the shRNA against the gene of interest within a few weeks. In addition, cell lines can be produced in parallel and the impact on pancreatic cancer biology of various genes can be studied simultaneously. An overall reduction of time consuming and expensive generation of germline-altered animal models and subsequent breedings and genotyping, not to mention long-term backcrossing, will emerge as a consequence.
Although the advantage of shRNA has been widely used in screening assays, many of the studies performed so far use either cells from animals that have already undergone embryonic loss of tumor suppressor genes (16, 17), cells with introduction of more than one genetic lesion prior to the transduction with shRNA (17, 18) or use already established and immortalized cancer cell lines (19, 20). The use of cells that are derived from embryonic tissue and/or harbor constitutive activation of oncogenes might lead to interaction with various developmentally activated but otherwise inactive pathways, thus resulting in cell fate decisions and phenotypes that do not occur upon sporadic oncogene activation in somatic cells. This is especially true for pancreatic cancer research as most animal models utilize Pdx1- of Ptf1a-driven Cre, which leads to oncogene activation or tumor suppressor deletion in all functional compartments of the pancreas due to their early promoter activity on days E8.5 and E9.5, respectively (21, 22). Also, sensitizing cells by more than one genetic alteration may lead to over interpretation of a newly identified tumor suppressor gene's impact as cells may be “supersensitized” to only minor oncogenic events. Third, long-term cultured cancer cell lines carry numerous genetic and epigenetic changes and do only partly reflect the cell of origin. In contrast, our model is designed to recapitulate truly somatic oncogene activation as we were able to avoid germline-activation of oncogenic Kras. In addition, one additional genetic hit was sufficient to induce tumor growth. It is important to realize that pancreatic loss p16Ink4a/p19Arf on its own does not result in PanIN or PDAC development in mice (23). Although Trp53-/- animals are prone to develop malignancies, these are mostly lymphomas and soft tissue tumors. The development of epithelial cancers in these animals is rare (reviewed in (24)), and we do not know about any report of development of PDAC upon pancreas specific Trp53-deletion. Thus, our model more closely resembles a truly sequential second-hit carcinogenesis as initially proposed by Knudson in 1971 (25).
In contrast to the human disease and most genetically engineered mouse models of pancreatic cancer, the tumors described in this study lack the classical stromal component and show a more dedifferentiated phenotype. This observation may be due to various reasons. First, we injected ductal cells that have undergone genetic maipulation. However, the induction of a stromal cell response is known to take place early in PDAC development, so that this critical phase might be missed in our model (26). Second, NOD scid gamma mice used in this study are depleted for B- and T-cells, which are also believed to play an important role during the generation of a stromal response (27). Third, injection of a cell suspension might not reflect the hypoxic conditions naturally occurring in a solid tumor, thereby reducing levels of secreted factors that usually foster development of a stromal reaction (28). Intriguingly, tumors described in this study closely resemble those seen in mouse models that a priori lack the stromal compartment (29, 30). However, we argue that orthotopic implantation is preferred over subcutaneous tumor xenograft models as the latter completely lack tumor cell interaction with neighboring cells at the naturally occurring site of origin of PDAC. This might not only be important in tumor initiation processes but also during the course of metastasis.
Because p16Ink4a and Trp53 act non-redundantly through either the control of cell cycle regulation or DNA damage repair mechanisms (31), we argue that our model will be an expandable and powerful tool to screen for new tumor suppressor genes and will broaden our understanding of cancer biology. Moreover, the principle of stepwise in vitro acquisition of genetic hits in primary pancreatic ductal cells may be transferrable to other techniques of gene modulation, including genome editing using CRIPSR/Cas9, as it has already been described for liver cancer (32).
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Institutes of Health (NIH R01DK060694, NIH/NIDDK P30DK050306) and American Cancer Society (RP-10-033-01-CCE) to A.K.R. and Max-Eder-Programm, Deutsche Krebshilfe (#111273) to M.R.
Footnotes
Conflicts of interest: None to declare.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin 2014. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Schneider G, Siveke JT, Eckel F, Schmid RM. Pancreatic cancer: basic and clinical aspects. Gastroenterology. 2005;128:1606–25. doi: 10.1053/j.gastro.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142:730–3. e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 7.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 8.Deramaudt TB, Takaoka M, Upadhyay R, et al. N-cadherin and keratinocyte growth factor receptor mediate the functional interplay between Ki-RASG12V and p53V143A in promoting pancreatic cell migration, invasion, and tissue architecture disruption. Mol Cell Biol. 2006;26:4185–200. doi: 10.1128/MCB.01055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Burstin J, Reichert M, Wescott MP, Rustgi AK. The pancreatic and duodenal homeobox protein PDX-1 regulates the ductal specific keratin 19 through the degradation of MEIS1 and DNA binding. PLoS One. 2010;5:e12311. doi: 10.1371/journal.pone.0012311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiber FS, Deramaudt TB, Brunner TB, et al. Successful growth and characterization of mouse pancreatic ductal cells: functional properties of the Ki-RAS(G12V) oncogene. Gastroenterology. 2004;127:250–60. doi: 10.1053/j.gastro.2004.03.058. [DOI] [PubMed] [Google Scholar]
- 11.Eser S, Reiff N, Messer M, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23:406–20. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 12.von Burstin J, Eser S, Seidler B, et al. Highly sensitive detection of early-stage pancreatic cancer by multimodal near-infrared molecular imaging in living mice. Int J Cancer. 2008;123:2138–47. doi: 10.1002/ijc.23780. [DOI] [PubMed] [Google Scholar]
- 13.Saur D, Vanderwinden J- M, Seidler B, Schmid RM, De Laet M- H, Allescher H- D. Single-nucleotide promoter polymorphism alters transcription of neuronal nitric oxide synthase exon 1c in infantile hypertrophic pyloric stenosis. Proc Natl Acad Sci USA. 2004;101:1662–7. doi: 10.1073/pnas.0305473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reichert M, Takano S, Heeg S, Bakir B, Botta GP, Rustgi AK. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat Protoc. 2013;8:1354–65. doi: 10.1038/nprot.2013.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–7. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 16.Mills JR, Malina A, Lee T, et al. RNAi screening uncovers Dhx9 as a modifier of ABT-737 resistance in an Emu-myc/Bcl-2 mouse model. Blood. 2013;121:3402–12. doi: 10.1182/blood-2012-06-434365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zender L, Xue W, Zuber J, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–64. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iorns E, Ward TM, Dean S, et al. Whole genome in vivo RNAi screening identifies the leukemia inhibitory factor receptor as a novel breast tumor suppressor. Breast Cancer Res Treat. 2012;135:79–91. doi: 10.1007/s10549-012-2068-7. [DOI] [PubMed] [Google Scholar]
- 19.Collins CS, Hong J, Sapinoso L, et al. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci U S A. 2006;103:3775–80. doi: 10.1073/pnas.0600040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Meer R, Yong Song H, Park SH, Abdulkadir SA, Roh M. RNAi screen identifies a synthetic lethal interaction between PIM1 overexpression and PLK1 inhibition. Clin Cancer Res. 2014;20:3211–21. doi: 10.1158/1078-0432.CCR-13-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–9. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 22.Krapp A, Knofler M, Ledermann B, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–63. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguirre AJ, Bardeesy N, Sinha M, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donehower LA. The p53-deficient mouse: a model for basic and applied cancer studies. Semin Cancer Biol. 1996;7:269–78. doi: 10.1006/scbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- 25.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–6. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiraoka N, Onozato K, Kosuge T, Hirohashi S. Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin Cancer Res. 2006;12:5423–34. doi: 10.1158/1078-0432.CCR-06-0369. [DOI] [PubMed] [Google Scholar]
- 28.Tsuzuki Y, Mouta Carreira C, Bockhorn M, Xu L, Jain RK, Fukumura D. Pancreas microenvironment promotes VEGF expression and tumor growth: novel window models for pancreatic tumor angiogenesis and microcirculation. Lab Invest. 2001;81:1439–51. doi: 10.1038/labinvest.3780357. [DOI] [PubMed] [Google Scholar]
- 29.Lee JJ, Perera RM, Wang H, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111:E3091–100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhim AD, Oberstein PE, Thomas DH, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–47. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bardeesy N, Aguirre AJ, Chu GC, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014 doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.