Abstract
Long non-coding RNAs (lncRNAs) have been found to play a role in gene regulation with dysregulated expression in various cancers. The precise role that lncRNA expression plays in the pathogenesis of B-acute lymphoblastic leukemia (B-ALL) is unknown. Therefore, unbiased microarray profiling was performed on human B-ALL specimens and it was determined that lncRNA expression correlates with cytogenetic abnormalities, which was confirmed by RT-qPCR in a large set of B-ALL cases. Importantly, high expression of BALR-2 correlated with poor overall survival and diminished response to prednisone treatment. In line with a function for this lncRNA in regulating cell survival, BALR-2 knockdown led to reduced proliferation, increased apoptosis, and increased sensitivity to prednisolone treatment. Conversely, overexpression of BALR-2 led to increased cell growth and resistance to prednisone treatment. Interestingly, BALR-2 expression was repressed by prednisolone treatment and its knockdown led to upregulation of the glucocorticoid response pathway in both human and mouse B-cells. Together, these findings indicate that BALR-2 plays a functional role in the pathogenesis and/or clinical responsiveness of B-ALL and that altering the levels of particular lncRNAs may provide a future direction for therapeutic development.
Implications
lncRNA expression has the potential to segregate the common subtypes of B-ALL, predict the cytogenetic subtype, and indicate prognosis.
Keywords: lincRNA, acute lymphoblastic leukemia, glucocorticoid receptor pathway
INTRODUCTION
The advent of high-throughput techniques to study gene expression has led to the recognition that nearly 30–50% of the human genome is transcribed, and non-coding RNA represents a significant portion of this transcriptome (1,2). Perhaps the clearest example of functional non-coding RNA is microRNA (miRNA), which are dysregulated in cancer (reviewed in (3)). In oncogenesis, individual miRNAs have been found to act as either tumor suppressor genes or oncogenes, based on our work and that of others. A new addition to the repertoire of non-coding RNAs is long non-coding RNAs (lncRNAs) (4). These RNAs are frequently found in intergenic regions, lack open reading frames, and have been detected in the transcriptome by high-throughput technologies (4,5). Although several classes of non-coding RNA species are being described, lncRNAs are unique in that there are epigenetic marks in their promoter regions (H3K4me3) and along transcribed regions (H3K36me3), identifying them as unique gene structures (6). At the molecular level, lncRNAs regulate gene expression via transcription, repress microRNA activity by competitive binding, regulate splicing and activate transcription (7–12). At the cellular and organismal level, lncRNAs have been implicated in physiological processes, such as maintenance of embryonic stem cells and erythroid development during hematopoiesis (13–18). Extending their putative roles to pathologic conditions, prior studies have examined lncRNA expression in cell lines as well as in epithelial malignancies, finding dysregulated expression (19).
Given that many hematopoietic malignancies result from mutations that cause dysregulation of gene expression, we reasoned that lncRNAs may play a role in the pathogenesis of these malignancies. Recent studies have demonstrated dysregulation of certain lncRNAs in hematopoietic malignancies, but the majority of these studies were relatively limited in their scope (reviewed in(20)). Amongst malignancies arising from hematopoietic precursors, B-acute-lymphoblastic leukemia (B-ALL), a malignancy of precursor B-cells, has previously been shown to harbor mutations and translocations resulting in dysregulated gene expression (21). Much progress has been made in understanding the molecular pathogenesis of B-ALL, including the role of chromosomal translocations. It is now well-recognized that commonly observed recurrent chromosomal abnormalities have distinct pathogenetic and prognostic implications (21,22). However, to date, there has been no understanding of how lncRNAs may participate in the pathogenesis of this disease. Hence, we undertook a comprehensive study examining lncRNA expression in B-ALL, examining correlations with clinicopathologic parameters, and querying the functional consequences of lncRNA expression.
Here, we found that overall lncRNA expression corresponds with specific cytogenetic abnormalities in B-ALL and that a subset of lncRNAs could correctly predict the cytogenetic subtype of B-ALL amongst the three most common abnormalities. We termed these B-ALL associated long RNAs (BALRs). The four most differentially regulated lncRNAs were mapped to their chromosomal locations and transcripts originating from these genomic loci were sequenced. Interestingly, high expression of one lncRNA, BALR-2, was correlated with a poor patient response to prednisone and worse overall survival. Knockdown of BALR-2 caused an increase in apoptosis of B-ALL cell lines alone and in combination with prednisolone. Interestingly, BALR-2 was repressed when human B-ALL cell lines were treated with prednisolone. Gene expression analyses of cells with knockdown of BALR-2 revealed activation of several genes involved in the response of B-cells to glucocorticoid receptor engagement. Lastly, the expression of BALR-2 in B-ALL cell lines with low basal expression of this lncRNA led to resistance to prednisolone treatment and concordant changes in gene expression. Together with our observation that BALR-2 is repressed by prednisolone treatment, our findings indicate a novel role for a non-coding RNA as a modulator of a treatment response. These data represent some of the first insights into long non-coding RNA expression in B-ALL and reveal that they play a role in pathogenesis, disease severity, and measurement/alteration of their levels may be useful in prognosis and/or treatment of this disease, respectively.
MATERIALS AND METHODS
Patients and samples
The patient cohort consisted of 160 children consecutively admitted to the Pediatric Oncologic Department at the University of Padua, Italy, from 2000 to 2008 with the diagnosis of B-ALL. Following analysis for RNA quality, we had a total of 44 patients for the microarray studies (cytogenetic information only) and 93 patients for RT-qPCR studies (comprehensive clinicopathologic information; See Supp Methods and Table 1). For the initial microarray studies, we utilized 20 patients (7 patients t(4;11) MLL rearranged, 6 t(12;21) TEL-AML1 translocated and 7 t(1;19) E2A-PBX translocated). For validation microarrays, an additional 24 samples were hybridized (n=7 patients for each translocation). For independent validation by RT-qPCR, we utilized 93 samples of de novo B-ALL without selection criteria. For each case, we had data on the following clinicopathological parameters: cytogenetic markers, immunophenotype, age, risk stratification by minimal residual disease (23), response to prednisone, occurrence of recurrence/relapse, time to recurrence/relapse, overall survival, and time to death. Peripheral blood mononuclear cells derived from anonymized donors were obtained from the Center for AIDS Research Virology Core Lab at UCLA, and the Flow Cytometry and Bone Marrow Lab at the Department Pathology and Laboratory Medicine. All procedures were approved by the local institutional review boards, and the study was considered exempt from review at UCLA.
Microarray data analysis
Agilent SurePrintG3 Human GE 8x60K microarrays (product # G4851A; http://www.genomics.agilent.com/article.jsp?crumbAction=push&pageId=1516) were hybridized at the Caltech microarray core facility. These arrays target 27,958 Entrez Gene RNAs, based on RefSeq Build 36.3, Ensemble Release 52, Unigene Build 216, GenBank (April 2009), as well as 7,419 lincRNAs, based on the initial discovery set from the Broad Institute (4). Data analysis was implemented in the R statistics package (24). The data from two microarray experiments (20 in the initial set and 24 in the validation set) were analyzed independently but following the same protocol. The Agilent feature extraction raw data files were loaded into the R environment and analyzed using the R library of Linear Models for Microarray Data (LIMMA) (25). The raw data were preprocessed for background correction and normalized between arrays using the quantile method, summarized by taking the average of replicates for each gene, and subsequently log2-transformed. Additional microarray data was generated from 4 different cultures of RS4;11 cells with or prednisolone treatment and with a siRNA against BALR-2 (or control vector). Samples were hybridized at the UCLA microarray core facility using Affymetrix HG-U133_Plus_2 microarray. Raw hybridization intensities were summarized and normalized using the RMA method (26) as implemented in Matlab® (The Mathworks Inc.). Differential expression analysis was performed as previously described (27). Functional annotation of differentially expressed genes was retrieved from www.ingenuity.com. All data from the microarray studies have been deposited in the Gene Expression Omnibus (GEO) resource (GEO Series ID GSE65647).
Clinicopathologic data analysis
All data analysis was completed using SPSS software. Clinicopathologic parameters (available for 93 patients) were correlated with the continuous qPCR data obtained for each of the lncRNAs using a Pearson’s Chi-square, and two-tailed T-tests (for dichotomous variables). Next, Kaplan Meier analyses were performed using dichotomized variables as described in supplemental methods.
Vectors, cell culture and flow cytometric analysis
mmu-miR-155 formatted siRNAs were cloned into BamHI and ApaI or XhoI sites in the pHAGE2-CMV-ZsGreen-W vector (28), using the strategy that we have previously described to generate knockdown vectors against protein coding genes (29,30). The BALR-2 sequence was cloned into a bicistronic MSCV vector with PGK driving GFP expression. RS4;11 and MV4;11, (MLL-AF4-translocated; ATCC CRL-1873 and CRL-9591), Reh (TEL-AML1-translocated; CRL-8286), 697 (E2A-PBX1-translocated), Nalm-6, and 70Z/3 (ATCC TIB-158) murine pre-B-cell leukemic cell line, and the HEK 293T cell line (ATCC CRL-11268) were grown as previously described (30). Lentiviruses and MSCV-based retroviral vectors were produced to generate knockdown and overexpression constructs as previously described (28,30). 5.0 × 10^5 cells were spin-infected at 30°C for 90 minutes in the presence polybrene (4 μg/mL). Transduced cell lines were sorted using a BD FACSAriaII cell sorter.
Apoptosis, proliferation, cell cycle and drug response assays
To measure proliferation, cells were plated in 96-well plates with MTS reagent (Promega CellTiter 96) and cells were incubated at 37°, 5% CO2 for 4 hours before absorbance was measured at 490nm. For drug response assays, cells were plated and treated with corresponding chemotherapeutic agent (250 μg/mL prednisolone dissolved in DMSO, 75 μg/mL of dexamethasone dissolved in DMSO, and 100 μg/mL of doxorubicin dissolved in water) for 24 hours, then harvested for RNA isolation or cell proliferation measurement. To measure apoptosis, chemotherapy treated cells were stained with APC tagged Annexin V, propidium iodide to assess the subG0 fraction, or lysed for protein isolation, and analyzed by FACS. For caspase-3 assays, colorimetric reagents were added to 50 μg of protein lysates (Biovision), and absorbance was measured at 405nm. Stably transduced cells were synchronized by serum starvation for 12 hours, replated into fresh media with FBS, collected after 24 hours, fixed with 70% EtOH, and stained with 1X propidium iodide (PI) solution in PBS. The DNA content of the cells was analyzed by flow cytometry using BD FACSDiva software and FlowJo software.
RT-qPCR and Western Blot
RNA collected from human samples was reverse transcribed using iScript reagent (Quanta Biosciences), RNA from cell lines was reverse transcribed using qScript (Quanta Biosciences). Real Time quantitative PCR was performed with the StepOnePlus Real-Time PCR System (Applied Biosystems) using PerfeCTa SYBR Green FastMix reagent (Quanta Biosciences). Primer sequences used are listed in Supp Table 4. For Western Blotting, cells were lysed in RIPA buffer (Boston BioProducts) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific), and Western Blot was performed according to standard procedures. Antibodies were purchased from Cell Signaling: c-Jun (60A8) Rabbit mAb; Bim (C34C5) Rabbit mAb; c-Fos (9F6) Rabbit mAb). Monoclonal anti-beta-ACTIN (AC-15) antibody was purchased from Sigma Aldrich. Fold change values were quantitated using ImageJ software and normalizing to ACTIN levels.
RESULTS
LncRNA expression segregates three common cytogenetic subtypes of B-ALL and can predict the cytogenetic subtype
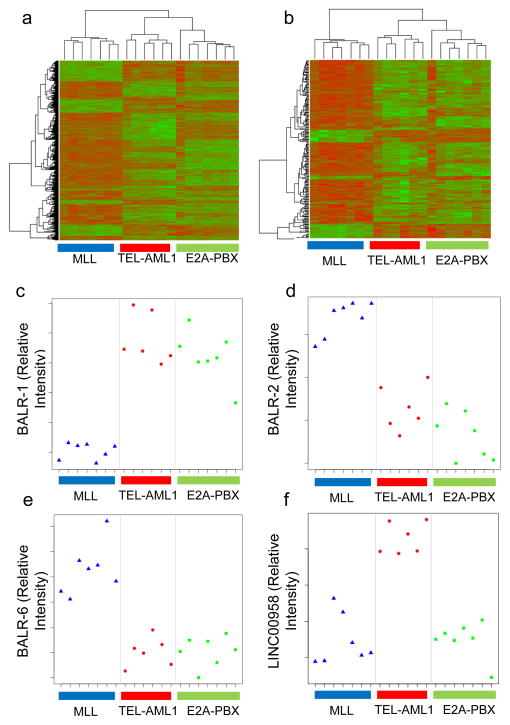
LncRNAs have been ascribed functions in both cancer causation and development of the hematopoietic system. Here, we undertook a microarray study to define patterns of lncRNA expression in different subsets of B-ALL using the Agilent SurePrintG3 Human GE 8x60K platform (4,7,31) (see Supp Fig 1 for experimental pipeline). In our initial sample set, we hybridized 20 cases of B-ALL with three different common translocations. Following correction for multiple hypothesis testing, we performed unsupervised hierarchical clustering with significantly differentially expressed protein-coding genes (p-adj≤0.01), finding that the three cytogenetic subtypes separated into three subsets (Fig 1A). Interestingly, when we focused exclusively on lncRNAs (p-adj≤0.01), lncRNAs were differentially expressed across the 3 different cytogenetic subtypes, although MLL-translocated cases appeared to have the most distinctive lncRNA expression pattern (Fig 1B). Interestingly, lncRNA expression seemed to be highly predictive of the cytogenetic abnormality in B-ALL. Using nearest shrunken centroid analysis, we determined that lncRNA expression is at least equivalent to protein coding gene expression at differentiating these three common cytogenetic abnormalities in an independent set of 24 cases of B-ALL (Supp Fig 2). For instance, one case of ALL became misclassified when 113 protein-coding genes were used to classify the samples, whereas one could use 27 lncRNAs before the same case was misclassified (Supp Fig 2G–H; see Supp Figure legend for complete description of results in Supp Fig 2). We then compared expression of these lncRNAs between B-ALL and normal CD10+CD19+ cells (data not shown), narrowing down the list to 10 ten putative oncogenic and tumor suppressor lncRNAs. These were designated as B-ALL associated long RNAs (BALRs), unless they had already been assigned LINC designations. Four lncRNAs were subsequently selected based on higher expression in leukemic versus normal B-cell precursors in the microarray, and the fact that these lncRNAs were located in well-annotated loci those that showed multiple ESTs or mRNA transcripts and were conserved in human and mouse genomes (Supp Fig 1). These lncRNAs were BALR-1, BALR-2, BALR-6 and LINC00958 which were also the most differentially expressed in the different cytogenetic subtypes (Fig 1C–F). Hence, lncRNAs are differentially regulated in B-ALL with specific translocations.
Figure 1. LncRNA expression segregates with ALL cytogenetic subtypes.
(A) Hierarchical clustering of differentially regulated protein-coding gene expression data (adjusted p-value after correction for multiple hypothesis testing, p-adj<0.01) in 20 B-ALL samples with three common translocations, namely, t(12; 21), TEL-AML1(n=6); t(1;19) E2A-PBX (n=7); and t(4;11) MLL-AF4 (n=7). Genes that are relatively upregulated appear in green, and those that are relatively downregulated appear in red. (B) Hierarchical clustering of lncRNAs that were differentially expressed (adj. p-value<0.01) showed distinct separation into three subsets of B-ALL, corresponding to the cytogenetic abnormalities. (C–F) Plots of normalized intensity ratios of BALR-1, BALR-2, BALR-6, and LINC00958 in individual cases of B-ALL, respectively.
LncRNA expression is correlated with clinicopathologic parameters in a large set of B-ALL cases
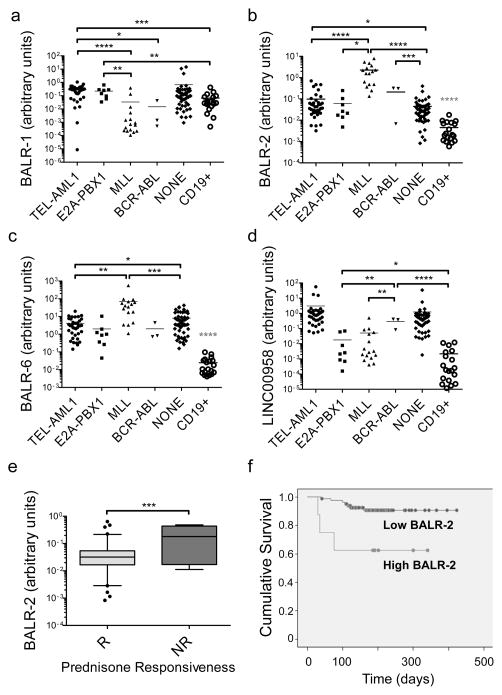
RT-qPCR was used to confirm the expression pattern of four individual lncRNAs in a subset of cases from the microarray cohort as well as in an independent set of B-ALL samples (Fig 2A–D). qPCR primer sets with the most consistent behavior across technical replicates and serial dilutions were chosen (Supp Table 4, qPCR primers). LncRNA expression was most consistently different in the subset of cases with MLL translocations, which is associated with a bad prognosis. For all four lncRNAs, we confirmed that MLL-translocated cases showed significantly different expression levels, compared with cases showing TEL-AML1, E2A-PBX1, or BCR-ABL translocations. Additionally, BALR-2 and BALR-6 expression was significantly upregulated in all subsets of patient samples when compared to normal CD19+ cells (Fig 2B–C).
Figure 2. LncRNAs exhibit differential expression in human B-ALL samples and BALR-2 expression correlates with prednisone response.
(A–D) Expression levels of BALR-1, BALR-2, BALR-6, and LINC00958, respectively, normalized to ACTIN. For these analyses 118 samples were analyzed based on the presence of good quality RNA, including both the discovery and validation cohorts. RT-qPCR was performed with specific primers for these lncRNAs, showing differential expression between the three cytogenetic subtypes of B-ALL used for the initial microarray experiments and an independent cohort of clinical samples. Number of cases used in this analysis: TEL-AML1-translocated, n = 38; E2A-PBX-translocated, n = 8; MLL-translocated, n = 16; BCR-ABL-translocated, n = 3; karyotypically normal cases, n = 53; and CD19+ cells isolated from normal donors, n=19. CD19+ cells showed significantly differential expression of BALR-2 and BALR-6 from all the subsets of B-ALL cases depicted here. (E) Analysis of BALR-2 expression data and response to prednisone treatment shows that BALR-2 expression is significantly higher in B-ALL patients that are not responsive (NR) to prednisone compared to those who do respond to prednisone (R). Number of cases used in this analysis: Responsive, n = 81; Non-Responsive, n = 8. RT-qPCR assays were normalized to ACTIN. Comparisons made using a two-tailed T-test, p<0.05 (*); p<0.005 (**); p<0.0005 (***); p<0.0001 (****). (F) Kaplan Meier survival analysis for two patient groups shows that high BALR-2 expression is associated with poor overall survival (overall survival (OS) high = 62.5% (n = 8), OS low = 91.5% (n = 82), log-Rank test, p=0.005). The two groups were dichotomized based on two step cluster analysis using SPSS software.
To examine the possibility that lncRNA expression is predictive of patient outcomes, we performed statistical analyses on available clinicopathologic parameters associated with these cases. Interestingly, BALR-2 expression was significantly higher in B-ALL patients that were unresponsive to prednisone as opposed to those who had a response to prednisone (Fig 2E, T-test, two-tailed p=0.0003). We dichotomized BALR-2 expression using a cut point based on cluster analysis (Supp Fig 3B), and found that high BALR-2 expression was associated with inferior overall survival (Fig 2F; Kaplan Meier survival analysis; log-Rank test, p=0.005,). Although BALR-2 expression did correlate with translocations, it should be noted that four of the poor-prognosis cases that had high BALR-2 expression did not contain a detectable translocation. When we attempted multivariate logistic regression, BALR-2 was not established as an independent prognostic variable (Supp Table 3). This may be due to insufficient numbers of cases with high BALR-2 expression in our cohort, and requires further study.
BALR-2 is a nucleocytoplasmic lncRNA whose expression is regulated by glucocorticoid treatment
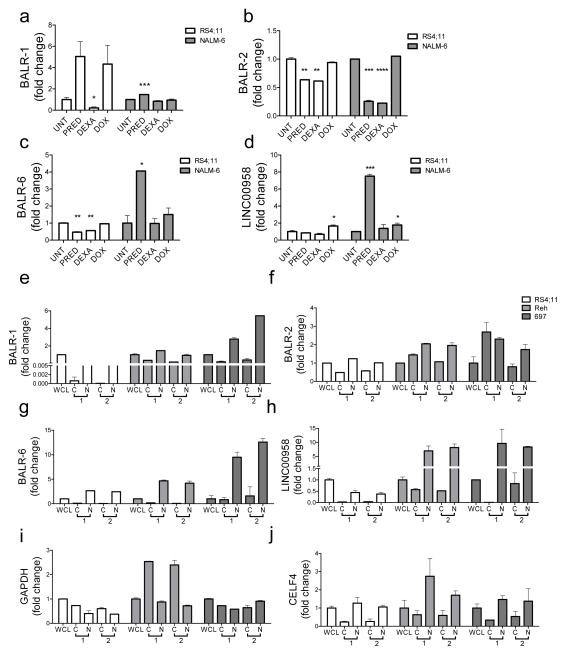
Given the interesting clinicopathologic findings with BALR-2, we began to study whether the expression of lncRNAs was regulated during treatment of B-ALL cells with chemotherapeutic agents. Interestingly, while the expression of these lncRNAs showed some variation with treatment, only BALR-2 was specifically regulated by glucocorticoid treatment (Fig 3A–D). Specifically, BALR-2 was downregulated in two different B-ALL cell lines, RS4;11 and NALM6, following treatment with either prednisolone or dexamethasone, but not with doxorubicin (Fig 3B). This finding is in line with the idea that prednisolone treatment downregulates BALR-2, but in B-ALL with constitutively high BALR-2, the cells become resistant. Next, we characterized the subcellular localization of these four lncRNAs, finding that in contrast to the other three lncRNAs, BALR-2 expression was on the same order of magnitude in nuclear and cytoplasmic fractions; and was in fact higher in the cytoplasm in one set of experiments (Fig 3E–H). This was confirmed in multiple B-ALL cell lines, including RS4;11, REH and 697, which show differing levels of expression of BALR-2 (Fig 3F). As expected GAPDH mRNA is primarily localized to the cytoplasm (Fig 3I) and the nuclear RNA CELF4 is primarily in the nuclear fraction (Fig 3J). Hence, BALR-2 expression appears to be regulated by glucocorticoid treatment and significant amounts of BALR-2 are found in the cytoplasm, making this lncRNA amenable to siRNA mediated knockdown.
Figure 3. LncRNA expression following glucocorticoid treatment and subcellular localization of lncRNA.
(A–D) Expression of BALR-1, BALR-2, BALR-6, and LNC00958 following treatment of RS4;11 and NALM6 cells with prednisolone (PRED), dexamethasone (DEXA), and doxorubicin (DOX). Control cells are denoted as untreated (UNT). (E–H) Subcellular localization of lncRNA transcripts. RT-qPCR data showing expression of BALR-2 (E), BALR-6 (F), BALR-1 (G), and LINC00958 (H) in cytoplasm and nuclear fractionations of from RS4;11 (left), Reh (middle) and 697 (right). All 4 lncRNAs are expressed in the nucleus while BALR-2 is also expressed in the cytosolic fraction. GAPDH (cytoplasmic) (I) and CELF4 (nuclear) (J) mRNAs were used to demonstrate successful fractionation. The data are represented as fold-change values over the relative expression of each gene in the whole cell lysate. Each panel represents two experiments, with overall similar results. Comparisons made using a two-tailed T-test, p<0.05 (*); p<0.005 (**); p<0.0005 (***); p<0.0001 (****).
Knockdown of BALR-2 results in growth inhibition and increased apoptosis
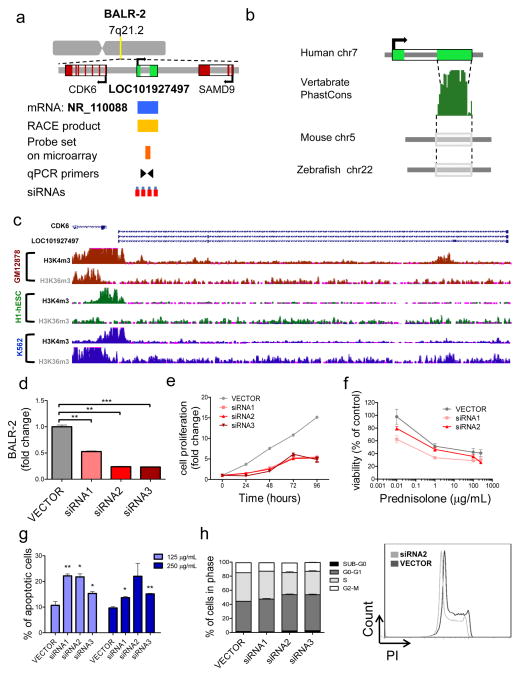
Given the correlation of BALR-2 with overall survival, we first characterized its chromosomal location, 7q21.2. The area surrounding BALR-2 is conserved in mammals and mRNA NR_110088 is localized to this region (Fig 4A). Similar gene diagrams are provided for the other three lncRNAs in Supp Fig 4a–c. To map the transcript, we carried out 5′RACE and 3′RACE using mRNA extracted from B-ALL cell line RS4;11. We confirmed the annotated mRNA and an alternate splice form with a previously unannotated exon (Supp Fig 4D). 3′ RACE confirmed the transcript as being approximately 500 base pairs in length (Supp Table 4). This transcript lacked an open reading frame and translation initiation site, and was predicted to be non-coding (32). Vertebrate PhastCons analysis demonstrated significant conservation within the terminal exon in 91 vertebrate species, suggesting a functional transcript (Fig 4B). Moreover, analysis of H3K4m3 and H3K36m3 revealed that signals along the promoter and gene body are consistent with a transcriptional element (Fig 4C) (33). To define a functional role for this transcript in B-ALL, siRNAs against BALR-2 were cloned into a modified lentiviral vector using miRNA formatted flanking and loop sequences (28,30). These lentiviral constructs were used to transduce the RS4;11 cell line (Fig 4D). RS4;11 cells stably expressing siRNA against BALR-2 showed significantly decreased proliferation, measured by MTS (Fig 4E). Decreased cell viability and increased apoptosis was observed both at baseline (data not shown) and after treatment with prednisolone (Fig 4F–G) (34,35). In line with cell proliferation data, RS4;11 cells treated with siRNAs1-3, revealed an increase in G0-G1-phase, as well as a decrease in S- and G2-M phase, as measured by propidium iodide staining. A significant increase in apoptotic cells (cells in Sub-G0 phase) was observed in cells treated with siRNAs2-3 (Fig 4H).
Figure 4. BALR-2 shows a functional role in human B-ALL cell lines.
(A) Map showing the position of BALR-2 in the genome, including the locations of neighboring genes, corresponding annotated mRNA, RACE product confirmation, probe set on microarray, qPCR primers and siRNAs targeting the lncRNA. (B) The Vertebrate PhastCons plot from the UCSC whole-genome alignments to mouse and zebrafish shows conserved regions within the terminal exon, including a region highly conserved among 91 vertebrates. (C) Chip-seq histone modification map from the ENCODE/Broad institute, taken from UCSC genome browser, shows H3K4m3 and H3K36m3 pattern at the BALR-2 locus in three different cell types indicating active promoter and transcription of the lincRNA (D) siRNA-mediated knockdown of BALR-2 in RS4;11 cell line, shown by RT-qPCR (normalized to ACTIN). (E) Reduction of cell proliferation in RS;411 cells stably transduced with siRNAs against BALIR-2, measured by MTS assay. Absorbance was normalized to the 0 hour time point (p-value < 0.05 for all non-zero hour time points). (F) In vitro prednisolone response curves for RS4;11 cell lines transduced with vector control, siRNA1 and siRNA2. Prednisolone response was assessed by MTS assay after 24 hours of plating. Untreated samples were set to 100%. Absorbance readings were normalized to untreated samples. (G) Increased apoptosis measured by AnnexinV staining. RS4;11 cells stably transduced with siRNAs against BALR-2 were treated with 125 μg/mL, and 250 μg/mL prednisolone for 24 hours. (H) Propidium iodide staining of RS4;11 cells treated with siRNAs, showed an increase in G0-G1 cells (p-value < 0.05 for all siRNAs), as well as a decrease in S cells (p-value < 0.05 for all siRNAs), and G2-M cells (p-value <0.05 for siRNAs1-3) The right hand panel shows a representative histogram of propidium iodide staining. Comparisons made using a two-tailed T-test, p<0.05 (*); p<0.005 (**); p<0.0005 (***).
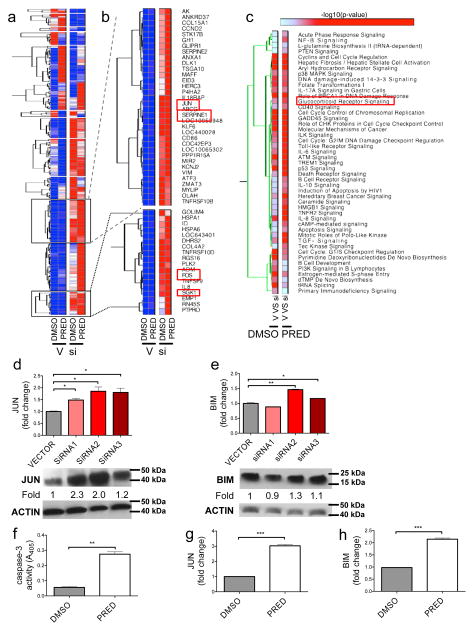
BALR-2 regulates apoptosis by modulating the glucocorticoid receptor signaling pathway
To examine the mechanism by which BALR-2 affects proliferation and apoptosis in BALL cell lines, we examined gene expression in RS4;11 cell lines stably transduced with siRNA2 (which showed the greatest degree of knockdown against BALR-2), with and without prednisolone treatment. Hierarchical clustering analysis identified clusters (Figure 5A–B) of genes that were upregulated in the siRNA group, both with and without prednisolone treatment. Several of these clusters consist of genes involved in the glucocorticoid receptor signaling pathway, confirmed by functional annotation results (2-4 fold increases in FOS, HSPA6, SGK1, IL8, JUN, SERPINE1, CDKN1A and ICAM1 in siRNA group, both with and without prednisone treatment; Fig 5B and 5C). We confirmed knockdown of BALR-2 and upregulation of FOS, JUN, SGK1 and SERPINE1 by RT-qPCR (Supp Fig 5 A–E). When we tried to validate the findings with multiple siRNAs, expression of JUN showed the most consistent changes (Fig 5D). We also observed upregulation of the pro-apoptotic protein BIM, which is a downstream target of JUN and an important mediator of glucocorticoid-induced apoptosis of lymphocytic cells (36) (Fig 5E). Hence, BALR2 knockdown mirrored the effects of prednisolone treatment, which also resulted in induction of apoptosis and increased expression of JUN and BIM (Fig 5F–H).
Figure 5. BALR-2 plays a role in the glucocorticoid response pathway.
(A) Hierarchical gene clustering of microarray data from RS4;11 cells treated with or without siRNA2 against BALR-2 and with or without prednisolone. Abbreviations, V, Vector; si, siRNA 2 against BALR-2; DMSO, Dimethylsulfoxide (used to solubilize prednisolone); PRED, prednisolone. (B) Two clusters of genes significantly over-expressed in siRNA2 treated cells include genes involved in glucocorticoid response (FOS, JUN, SGK1 and SERPINE1) (C) Functional analysis of genes differentially expressed in siRNA2 treated cells shows significant enrichment of various canonical pathways, including Glucocorticoid Receptor Signaling. (D–E) RT-qPCR (top panels), showing upregulation of JUN (D), and BIM (E) in multiple siRNA knockdown lines, normalized to ACTIN, with corresponding Western blot (bottom panels). Fold change values are as quantitated by ImageJ and normalized to ACTIN. (F–H) Prednisolone treatment of RS4;11 cells resulted in induction of apoptosis, as measured by Caspase-3 activity (F) and upregulation of JUN (G) of BIM (H), as measured by RT-qPCR, normalized to ACTIN. Overall, the effects of the siRNA are similar to those induced by prednisolone treatment. Comparisons made using a two-tailed T-test, p<0.05 (*); p<0.005 (**); p<0.0005 (***).
To confirm our findings, we tested additional cell lines that show high-level expression of BALR-2, including Nalm-6, Reh, and MV4;11. The glucocorticoid signaling pathway is operant in Nalm-6 cells, despite this cell line not carrying an MLL translocation (Supp Fig 5F–H). Consistent with our data in RS4;11 cells, knockdown of BALR-2 in Nalm-6 cells resulted in decreased cell proliferation, increased apoptosis, and increased expression of FOS (siRNA1 and 2), JUN (siRNA1), and BIM (siRNA1 and 2) (Supp Fig 6 A–D). Knockdown of BALR-2 in Reh cells by targeting of the BALR-2 splice junction by siRNA (sisplice1), as well as with siRNA3, resulted in increased apoptosis, decreased cell proliferation and significant upregulation of FOS, JUN, and BIM (Supp Fig 6E–J) (37). Moreover, MV-4-11 cells, which carry the t(4;11) translocation showed an increased number of apoptotic cells and increased expression of glucocorticoid response genes, FOS, and JUN, following knockdown of BALR-2 with siRNA3 (Supp Fig 6K–M). Overall, these findings demonstrated parallel effects of prednisolone treatment and BALR-2 knockdown in multiple cell lines, suggesting that BALR-2 is an important regulator of this pathway.
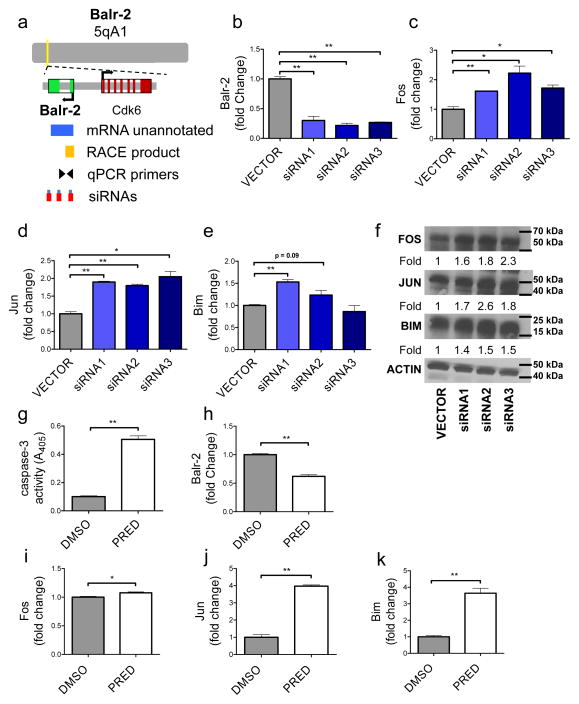
The function of BALR-2 is conserved in human and mouse cells
To assess functional conservation of BALR-2, we mapped and characterized murine Balr-2 to 5qA1, and the murine transcript demonstrates 90% homology to the human sequence (Fig 6A). RACE was performed and two products were identified at this locus (Supp Fig 4E). We generated miRNA-formatted siRNAs (30) against the murine transcript and confirmed decreased expression in the murine pre-B cell line 70Z/3 (Fig 6B). Similar to what we observed in human cell lines, Balr-2 knockdown led to an upregulation of Fos and Jun in all three of the cell lines with upregulation of Bim in two out of three cell lines by RT-qPCR (Fig 6C–E). We further confirmed the upregulation of these proteins by western blot analysis, finding increased expression Fos, Jun and Bim with all three knockdown constructs (Fig 6F). Prednisolone treatment in the murine cells led to downregulation of Balr-2 concomitant with increased apoptosis, and upregulation of Fos, Jun, and Bim (Fig 6G–K), mirroring our observations in the human cells. Our data indicate that BALR-2 plays a key role in regulating the glucocorticoid receptor signaling pathway, thereby regulating the cellular response to prednisolone treatment (A putative schematic mechanism is presented in Supp Fig 7).
Figure 6. The mouse homolog of BALR-2, Balr-2, shows a functional role in mice B-ALL cell lines.
(A) Map showing the position of Balr-2 in the genome, including the locations of neighboring genes, un-annotated mRNA, RACE product confirmation, qPCR primers and siRNAs targeting the mouse lncRNA. (B) siRNA-mediated knockdown of Balr-2 in 70Z/3 mouse cell line, shown by RT-qPCR (normalized to L32). (C–F) Expression of glucocorticoid response genes Fos (C, F), Jun (D, F) and its target Bim (E, F), upon siRNA mediated knockdown of the mouse lncRNA. Expression was analyzed by RT-qPCR (normalized to L32) (C–E), and Western blot (F). Fold change values as quantitated by ImageJ and normalized to Actin. (G–K) Caspase-3 activity, expression of mouse Balr-2 (H), and glucocorticoid response genes Fos, Jun, and Bim, respectively, are altered in 70Z/3 cells upon prednisolone treatment for 6 hours. Comparisons made using a two-tailed T-test, p<0.05 (*); p<0.005 (**).
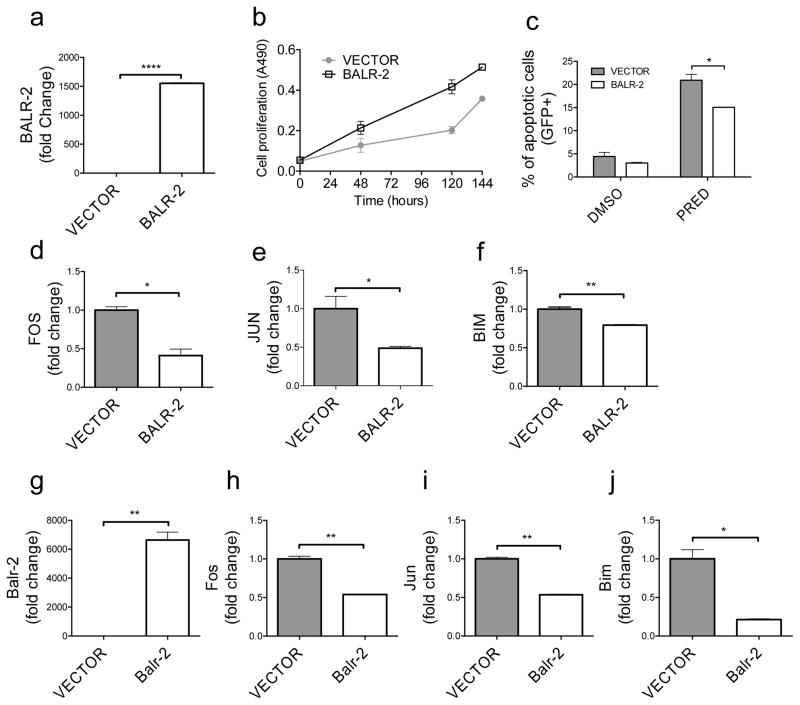
Enforced expression of BALR-2 promotes cell growth and inhibits apoptosis
Given that normal and malignant B-lymphoid cells show markedly differential expression of BALR-2, we wanted to investigate the effects of gain-of-function of BALR-2. We designed and validated a MSCV-based two-promoter vector to constitutively express BALR-2 in E2A-PBX translocated 697 cells, which show very low basal levels of BALR-2 (Fig 7A). In juxtaposition to knockdown, overexpression of BALR-2 resulted in increased growth of the cells, as measured by MTS assay (Fig 7B). Remarkably, overexpression of BALR-2 led to a partial rescue of prednisolone-induced apoptosis in 697 cells (Fig 7C). Baseline levels of apoptosis were also somewhat lower with overexpression, but this finding was not statistically significant. Also in line with its anticipated function, the expression of FOS, JUN, and BIM was reduced (Fig 7D–F). These findings suggest that BALR-2 plays a similar function in primary human B-ALL, leading to prednisone resistance and a poor prognosis. Next, we used the retroviral vector system to overexpress Balr-2 in murine 70Z/3 cells (Fig 7G). This led to similar changes in gene expression as observed in 697 cells. Moreover, the expression changes of Fos, Jun, and Bim were the opposite of those seen with knockdown of Balr-2 (Fig. 7H–J). Hence, knockdown and overexpression of BALR-2 led to opposing phenotypes and gene expression changes in human and mouse cells, consistent with the role we propose here, and once again suggesting a conserved function for this lncRNA.
Figure 7. Enforced expression of BALR-2 led to increased cell growth and resistance to apoptosis.
(A) Overexpression of BALR-2 in 697 cells shown by RT-qPCR (normalized to ACTIN). (B) Increased cell proliferation in 697 cells stably transduced with BALR-2, measured by MTS assay. (C) AnnexinV staining showed that 697 cells stably transduced with BALR-2 resulted in reduction of apoptosis upon treatment with 250 μg/mL prednisolone for 24 hours. (D–F) Consistent with the function, the expression of FOS (D), JUN (E), and BIM (F) are downregulated with the overexpression of BALR-2, as measured by RT-qPCR, normalized to ACTIN. (G–J) Overexpression of Balr-2 (G) in mouse 70Z/3 cells rescued the gene expression of glucocorticoid responsive genes Fos (H), Jun (I), and Bim (J), respectively, as measured by RT-qPCR, normalized to L32. Comparisons made using a two-tailed T-test, p<0.05 (*); p<0.005 (**); p<0.0001 (****).
DISCUSSION
In this report, we describe how long non-coding RNAs may play a role in B-ALL pathogenesis. We find that patterns of lncRNA expression are correlated with and can predict certain common chromosomal translocations in B-ALL. We found that expression of one lncRNA, BALR-2, correlated with overall survival and with response to prednisone. To characterize this functionally, we successfully knocked down BALR-2, finding increased apoptosis with chemotherapy. Most interestingly, we demonstrate a putative mechanism for BALR-2 in regulating cell survival in B-ALL- namely, that it is downregulated by glucocorticoid receptor engagement, and that its downregulation results in activation of the glucocorticoid receptor signaling pathway. These findings were conserved in human and murine B-ALL-derived cells, and were achieved with multiple siRNA sequences targeting either the exonic mRNA sequence that we have defined here, or via targeting of exon-intron boundaries (37).
Our results present some of the first clinicopathologic correlations with lncRNA expression data in B-ALL, along with a recent publication that reports dysregulated lncRNA expression in B-ALL (38). Due to the limitations of the microarray platform, we may not have completely profiled all lncRNAs, and future efforts via high throughput RNA sequencing as well as alternate array platforms may yield additional relevant lncRNAs. Nonetheless, we found several correlations between the expression level of a few specific lncRNAs and clinicopathologic parameters. These studies validate the idea that lncRNAs are important players in biological processes and are tied to oncogenesis. Although BALR-2 expression segregated mainly with MLL-translocated cases in the curated data set, there were several cases of B-ALL without translocations that also showed high level expression of BALR-2. We suggest that the expression level of BALR-2 may represent another variable for prognostication in this disease. Although high-risk cytogenetic abnormalities characterize the majority of patients who do not respond well to therapy, we suggest further study of BALR-2 expression, with a view towards prospectively identifying patients who will not respond well to standard therapeutic interventions. This is particularly relevant in the context of responsiveness to prednisone. Although not addressed in this study, lncRNA expression in other types of leukemia may also represent a prognostic and therapeutically relevant variable, and warrants further study.
In addition to the clinical relevance of our findings, the functional studies present a compelling argument for the further study of lncRNA in cancer causation and therapy. Together with the clinicopathologic correlation between high BALR-2 and poor responsiveness to prednisone, our functional studies suggest that BALR-2 expression plays a role in causing resistance to apoptosis. This is supported by both loss-of-function and gain-of-function approaches in human and murine B-cells, presented herein. Although the mechanism is incompletely understood, prednisone is thought to cause apoptosis in lymphoid cells by binding to the glucocorticoid receptor with subsequent activation of the intrinsic apoptosis pathway by repression of anti-apoptotic proteins such as BCL2 or activation of pro-apoptotic BCL2 protein family members, such as BIM (39–41). Here, we find that knockdown of BALR-2 by siRNA caused an increase in expression of the BCL2 family member BIM and increased cell death. These findings are particularly interesting in light of the proposed critical role for BIM in the regulation of the glucocorticoid response in B-ALL. Previous work has demonstrated that BIM is upregulated in leukemic blasts isolated from pediatric B-ALL patients undergoing treatment and that it is downregulated in patients who have poor responses to prednisone (42,43). Hence, BALR-2 may be a determinant of glucocorticoid response in patients, and our study points to therapeutic inhibition of BALR-2 as a possible strategy to overcome resistance to glucocorticoid treatment in B-ALL.
While our data suggest that BALR-2 acts in promoting cell survival via inhibition of genes downstream of the glucocorticoid receptor, such as Fos, Jun, and Bim, the molecular mechanism of its action remains to be defined. Recent work has demonstrated that non-coding RNA can lead to recruitment of transcriptional regulatory complexes to chromosomal loci (7,8,44). Additionally, the expression of certain lncRNAs affects the expression of nearby protein coding genes (37,45). In humans and mice, BALR-2 is located in a chromosomally adjacent region to CDK6. This presents an attractive target as CDK proteins have important roles in cell cycle progression and cancer (46). However, alternate modes of action, including interactions with proteins or other RNAs, have been ascribed to lncRNAs, and BALR-2 may act via any of these mechanisms to regulate the glucocorticoid response pathway (8,10,11,47,48). To comprehensively address the molecular mechanism of BALR-2’s function, future studies are required to study its function as a cis-regulatory element of cell cycle regulation in human cells and in murine model systems, given conservation of its function in human and mouse B-cells.
Future directions will include examination of the prognostic relevance of BALR-2 in BALL, assessment of BALR-2-mediated oncogenesis and delineating the therapeutic utility of BALR-2 knockdown in mouse models of B-ALL. Moreover, a detailed exploration of the molecular mechanisms of action of BALR-2 is warranted. Given some initial studies reporting the involvement of lncRNA in hematopoiesis, another important question is whether BALR-2 and other lncRNAs that are differentially regulated in B-ALL also play a role in normal development of B-lymphocytes (17). Hence, these findings open up a new area of research into the role of lncRNAs in B-cell development and malignancy, and promise to further refine prognostication and therapeutic intervention in B-ALL.
Supplementary Material
Acknowledgments
We thank Vijaya Rao and Igor Antoshechkin at the California Institute of Technology Millard and Muriel Jacobs Genetics and Genomics Laboratory for performing the microarray hybridization experiments, Alejandro Balazs at Caltech for lentiviral vector backbones, Gay Crooks and Kathy Sakamoto for cell lines, Subba S Rao for statistical consultation, Ken Dorshkind, Jayanth Kumar and Jorge Contreras for helpful discussions. Also, we thank Tiffany Tran, Parth Patel and Jasmine Gajeton for their technical support. This research was supported by Tumor Biology Training Grant T32 CA009056 from the National Institute of Health (T.R.F.), Eugene V. Cota-Robles Fellowship from UCLA (N.I.R.M.), the Graduate Research Fellowship Program from the National Science Foundation (N.I.R.M.), a Career Development Award K08CA133521 (D.S.R.), the Sidney Kimmel Translational Scholar Award SKF-11-013 (D.S.R), the Irving Feintech Family Foundation/ Tower Cancer Research Foundation Research Grant (D.S.R), the University of California Cancer Research Coordinating Committee (D.S.R) and the Stein-Oppenheimer Endowment Award (D.S.R). Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards CA-16042 and AI-28697, and by the JCCC, the UCLA AIDS Institute, the UCLA Council of Bioscience Resources, and the David Geffen School of Medicine at UCLA.
Footnotes
Conflict of Interest: The authors have no relevant conflicts of interest.
References
- 1.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science (New York, NY. 2002;296(5569):916–9. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (New York, NY. 2007;316(5830):1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nature immunology. 2008;9(8):839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 4.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5[prime]-modified long and short RNAs. Nature. 2009;457(7232):1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell. 2011;147(2):358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012 doi: 10.1038/nature11508. advance online publication. [DOI] [PubMed] [Google Scholar]
- 12.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3[prime] UTRs via Alu elements. Nature. 2011;470(7333):284–88. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenyo A, Rao M, Koneru S, Hallinan W, Shah S, Massey HT, et al. Risk of mortality for ventricular arrhythmia in ambulatory LVAD patients. Journal of cardiovascular electrophysiology. 2012;23(5):515–20. doi: 10.1111/j.1540-8167.2011.02223.x. [DOI] [PubMed] [Google Scholar]
- 14.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Research. 2008;18(9):1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16(2):324–37. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes & Development. 2011;25(24):2573–78. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paralkar VR, Weiss MJ. A new ‘Linc’ between noncoding RNAs and blood development. Genes & Development. 2011;25(24):2555–58. doi: 10.1101/gad.183020.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotech. 2011;29(8):742–49. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garitano-Trojaola A, Agirre X, Prosper F, Fortes P. Long non-coding RNAs in haematological malignancies. Int J Mol Sci. 2013;14(8):15386–422. doi: 10.3390/ijms140815386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122(10):3407–15. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borowitz MJ, Chan JKC. B lymphoblastic leukaemia/lymphoma. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4. Vol. 2. International Agency for Research on Cancer; 2008. pp. 168–75. [Google Scholar]
- 23.Conter V, Bartram CR, Valsecchi MG, Schrauder A, Panzer-Grumayer R, Moricke A, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115(16):3206–14. doi: 10.1182/blood-2009-10-248146. [DOI] [PubMed] [Google Scholar]
- 24.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 25.Smith GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17(6):509–19. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 28.O’Connell RM, Balazs AB, Rao DS, Kivork C, Yang L, Baltimore D. Lentiviral vector delivery of human interleukin-7 (hIL-7) to human immune system (HIS) mice expands T lymphocyte populations. PLoS One. 2010;5(8):e12009. doi: 10.1371/journal.pone.0012009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106(17):7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33(1):48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niazi F, Valadkhan S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3’ UTRs. RNA. 2012;18(4):825–43. doi: 10.1261/rna.029520.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147(7):1628–39. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vangipuram SD, Buck SA, Lyman WD. Wnt pathway activity confers chemoresistance to cancer stem-like cells in a neuroblastoma cell line. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33(6):2173–83. doi: 10.1007/s13277-012-0478-0. [DOI] [PubMed] [Google Scholar]
- 35.Tissing WJE, den Boer ML, Meijerink JPP, Menezes RX, Swagemakers S, van der Spek PJ, et al. Genomewide identification of prednisolone-responsive genes in acute lymphoblastic leukemia cells. Blood. 2007;109(9):3929–35. doi: 10.1182/blood-2006-11-056366. [DOI] [PubMed] [Google Scholar]
- 36.Heidari N, Miller AV, Hicks MA, Marking CB, Harada H. Glucocorticoid-mediated BIM induction and apoptosis are regulated by Runx2 and c-Jun in leukemia cells. Cell death & disease. 2012;3:e349. doi: 10.1038/cddis.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang K, Han BW, Chen ZH, Lin KY, Zeng CW, Li XJ, et al. A distinct set of long non-coding RNAs in childhood MLL-rearranged acute lymphoblastic leukemia: biology and epigenetic target. Human molecular genetics. 2014;23(12):3278–88. doi: 10.1093/hmg/ddu040. [DOI] [PubMed] [Google Scholar]
- 39.Alnemri ES, Fernandes TF, Haldar S, Croce CM, Litwack G. Involvement of BCL-2 in glucocorticoid-induced apoptosis of human pre-B-leukemias. Cancer Res. 1992;52(2):491–5. [PubMed] [Google Scholar]
- 40.Casale F, Addeo R, D’Angelo V, Indolfi P, Poggi V, Morgera C, et al. Determination of the in vivo effects of prednisone on Bcl-2 family protein expression in childhood acute lymphoblastic leukemia. Int J Oncol. 2003;22(1):123–8. doi: 10.3892/ijo.22.1.123. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Malone MH, He H, McColl KS, Distelhorst CW. Microarray analysis uncovers the induction of the proapoptotic BH3-only protein Bim in multiple models of glucocorticoid-induced apoptosis. The Journal of biological chemistry. 2003;278(26):23861–7. doi: 10.1074/jbc.M301843200. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt S, Rainer J, Riml S, Ploner C, Jesacher S, Achmuller C, et al. Identification of glucocorticoid-response genes in children with acute lymphoblastic leukemia. Blood. 2006;107(5):2061–9. doi: 10.1182/blood-2005-07-2853. [DOI] [PubMed] [Google Scholar]
- 43.Bachmann PS, Gorman R, Papa RA, Bardell JE, Ford J, Kees UR, et al. Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res. 2007;67(9):4482–90. doi: 10.1158/0008-5472.CAN-06-4244. [DOI] [PubMed] [Google Scholar]
- 44.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–06. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang B, Arun G, Mao Yuntao S, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 Is Dispensable for Mouse Development but Its Transcription Plays a cis-Regulatory Role in the Adult. Cell Reports. 2012;2(1):111–23. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Placke T, Faber K, Nonami A, Putwain SL, Salih HR, Heidel FH, et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood. 2014;124(1):13–23. doi: 10.1182/blood-2014-02-558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.