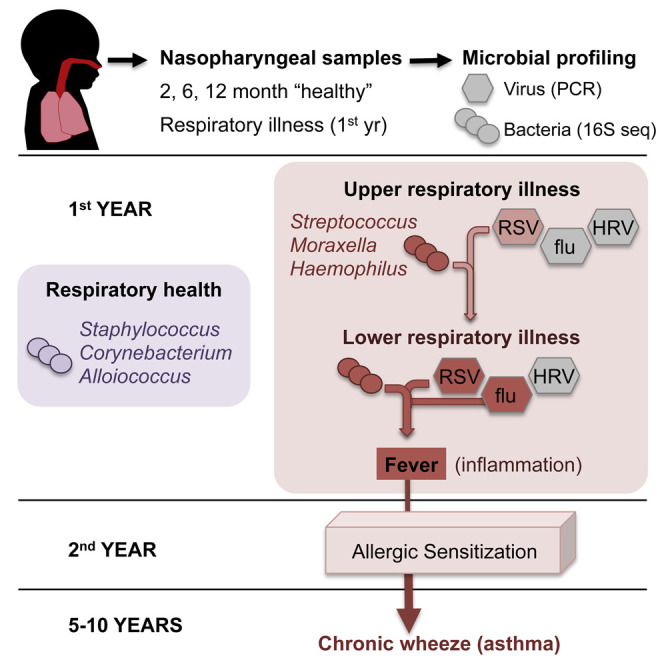
Summary
The nasopharynx (NP) is a reservoir for microbes associated with acute respiratory infections (ARIs). Lung inflammation resulting from ARIs during infancy is linked to asthma development. We examined the NP microbiome during the critical first year of life in a prospective cohort of 234 children, capturing both the viral and bacterial communities and documenting all incidents of ARIs. Most infants were initially colonized with Staphylococcus or Corynebacterium before stable colonization with Alloiococcus or Moraxella. Transient incursions of Streptococcus, Moraxella, or Haemophilus marked virus-associated ARIs. Our data identify the NP microbiome as a determinant for infection spread to the lower airways, severity of accompanying inflammatory symptoms, and risk for future asthma development. Early asymptomatic colonization with Streptococcus was a strong asthma predictor, and antibiotic usage disrupted asymptomatic colonization patterns. In the absence of effective anti-viral therapies, targeting pathogenic bacteria within the NP microbiome could represent a prophylactic approach to asthma.
Graphical Abstract
Highlights
-
•
The nasopharynx microbiome of infants has a simple structure dominated by six genera
-
•
Microbiome composition affects infection severity and pathogen spread to lower airways
-
•
Early asymptomatic colonization with Streptococcus increases risk of asthma
-
•
Antibiotic usage disrupts asymptomatic colonization patterns
Teo et al. characterize bacterial and viral communities within the infant nasopharynx during the first year of life, comparing between asymptomatic colonization and episodes of acute respiratory infections. Microbiome composition affects infection severity and spread to lower airways and risk for future asthma development.
Introduction
The human microbiome is now recognized as playing an important role in the etiology and pathogenesis of myriad diseases (Weinstock, 2012). However, elucidation of these complex roles requires targeted characterization of microbial communities present in relevant spatial niche(s) during critical periods of pathogenesis. The focus of this study is the respiratory tract, in particular the nasopharynx (NP), which is an accessible source of airway microbial communities (Hilty et al., 2010) and serves as a conduit for pathogens associated with lower respiratory illnesses (LRIs) that are responsible for substantial morbidity and mortality worldwide.
Of particular interest is asthma, a multi-factorial disease characterized by airway inflammation and associated smooth muscle hyperplasia. It is now recognized that the hallmark persistent wheeze of asthma is consolidated in childhood and, further, may progress to chronic asthma in adulthood (Holt and Sly, 2012, Sly et al., 2008) and potentially chronic obstructive pulmonary disease (Tai et al., 2014). We and others have previously shown that development of persistent atopic (allergic) wheeze in children is linked to the number of virus-associated febrile and/or wheezy LRIs experienced during infancy (Jackson et al., 2008, Kusel et al., 2007, Kusel et al., 2012, Oddy et al., 2002). The principal virus type of current interest is human rhinoviruses (HRVs), particularly subtype C (HRV-C) (Bochkov and Gern, 2012); however, respiratory syncytial virus (RSV) is also recognized as a major cause of infant LRI (Wu and Hartert, 2011). The relative contributions of these viral pathogens in asthma initiation remain controversial (Stein and Martinez, 2010). Further complicating the picture, recent studies have also implicated bacterial pathogens as potential independent causal factors in infant LRIs and their long-term sequelae. Notably, culture of S. pneumoniae, M. catarrhalis, or H. influenzae from NP samples taken at 1 month of age has been linked to increased risk for subsequent diagnosis of asthma at 5 years of age (Bisgaard et al., 2007). These findings have fuelled debate around the use of antibiotics and vaccine strategies for respiratory illness in children (Penders et al., 2011, Rollins et al., 2010).
Several studies have investigated airway microbiota in children or adults with chronic respiratory illness, including asthma (Bogaert et al., 2011, Hilty et al., 2010, Vissers et al., 2014); however, no study has investigated the airway microbiome during the critical infancy period (0–12 months). In this study, we investigated the NP microbiome during the first year of life using the Childhood Asthma Study (CAS), a prospective cohort of 234 children (Kusel et al., 2006, Kusel et al., 2007, Kusel et al., 2008, Kusel et al., 2012), to elucidate the NP microbiome during respiratory health and illness, its longitudinal dynamics, susceptibility to exogenous factors such as antibiotics, and association with future asthma.
Results
A total of 1,021 NP microbiome profiles were obtained from 234 infants using 16S rRNA gene deep sequencing (see Supplemental Experimental Procedures). These included 487 “healthy” NP samples collected in the absence of respiratory symptoms and 534 “infection” NP samples collected during episodes of acute respiratory illness (ARI) during the first year of life. Three quarters of the infants (n = 177) contributed at least two healthy NP samples at the age of ∼2 months, ∼6 months, and/or ∼12 months. Eighty percent of the infants (n = 186) contributed a healthy sample before experiencing their first ARI. The 534 infection NP samples were from 184 infants who had experienced ≥ 1 ARI within the first year of life. NP samples were analyzed from all (380/381) recorded LRI in this period and a random selection of 20% (154/782) of recorded episodes of upper respiratory illness (URI). The characteristics of the infants are summarized in Table S1.
NP Microbiome Composition
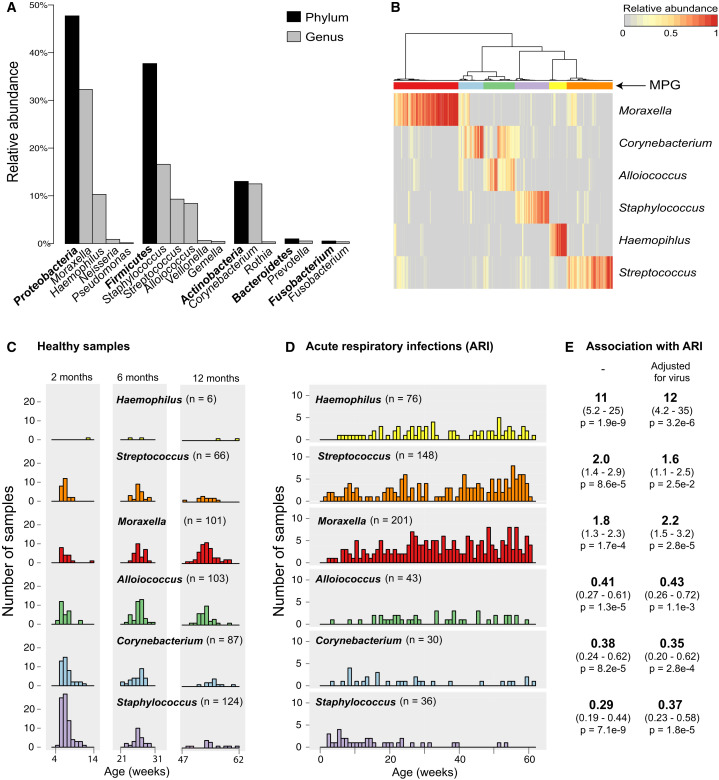
Across all NP samples, > 193 million high-quality 16S rRNA sequences were classified into 14,131 operational taxonomic units (OTUs), of which 1,010 were supported by > 1,000 reads each. The dominant phyla were Proteobacteria (48%), Firmicutes (38%), Actinobacteria (13%), Bacteroidetes (1%), and Fusobacteria (0.5%) (Figure 1 A). The NP microbiomes were dominated by six genera: Moraxella (31.2%), Streptococcus (15.5%), Corynebacterium (13.5%), Staphylococcus (10.3%), Haemophilus (9.7%), and Alloiococcus (8.8%; genus Dolosigranulum in some databases) (Figure 1A). Despite the inclusion of diverse OTUs of these genera in the reference database, our sequences were dominated by one OTU per genus (Figure S1), consistent with culture-based studies reporting NP colonization with the species Moraxella catarrhalis, Streptococcus pneumoniae, Staphylococcus aureus, Haemophilus influenzae, and Alloiococcus otitidis. Hierarchical clustering of NP microbiomes based on relative abundance of the six major genera identified six microbiome profile groups (MPGs, Figure 1B). Each MPG was dominated by one of the six genera, although some samples in the Alloiococcus MPG also had relatively high abundance of Corynebacterium (Figure 1B).
Figure 1.
Bacterial Composition of 1,021 Nasopharyngeal Aspirates Collected from 234 Infants during Periods of Respiratory Health and Disease
(A) Frequency of the most abundant phyla and genera (comprising 99.9% of reads).
(B) Clustering of samples into microbiome profile groups (MPGs) based on relative abundance of the six most common genera. Colored bars indicate MPGs, labeled by their dominant genus: Moraxella (red), Corynebacterium (blue), Alloiococcus (green), Staphylococcus (purple), Haemophilus (yellow), and Streptococcus (orange).
(C) Weekly frequencies of MPGs among healthy samples, collected during planned visits at approximately 2, 6, and 12 months of age and following at least 4 weeks without symptoms of acute respiratory infection (ARI).
(D) Weekly frequencies of each MPG among ARI samples.
(E) Odds ratios for association of MPGs with ARI symptoms, adjusted for age, gender, season, number of prior infections, antibiotics intake, mother’s antibiotics intake, delivery mode, and breastfeeding; with and without adjustment for detection of common viruses (RSV, HRV).
NP Microbiome Dynamics
Healthy NP samples collected around 2 months of age were dominated by Staphylococcus (41%) and Corynebacterium (22%) MPGs, but the frequency of these MPGs declined with age (11% and 10%, respectively, at 12 months old) (Figure 1C). In contrast, the prevalence of Alloiococcus and Moraxella MPGs in healthy samples increased with age (14% and 9% at 2 months, 26% and 41% at 12 months, respectively). Analysis of MPG transitions among consecutive healthy NP samples from the same individuals (Figures S3A and S3B) suggested that Staphylococcus carriage was unstable or transient, particularly where an ARI had occurred in the intervening period between sampling. Alloiococcus was a stable colonizer, but less so if an ARI occurred between sampling. Where an ARI occurred in the intervening period between healthy samples, the most common transitions were to the Moraxella MPG from other MPGs or the maintenance of stable colonization with Moraxella (Figure S3B). Almost all Moraxella-colonized infants experienced subsequent ARI before the next healthy sample was taken (explored further below). Haemophilus was very rarely detected in healthy NP microbiomes, while Streptococcus was present in 14% of healthy samples at each sampling time (Figure 1C). Similar age-related patterns were observed among infection samples, with a decline in Staphylococcus and Corynebacterium MPGs and increase in Moraxella, Haemophilus, and Streptococcus MPGs in older children (Figure 1D). Interestingly, males had significantly more Moraxella in healthy samples (OR 1.3 for log abundance; 95% CI 1.1–1.5, p = 0.0014, adjusted for age); no other gender effects were detected.
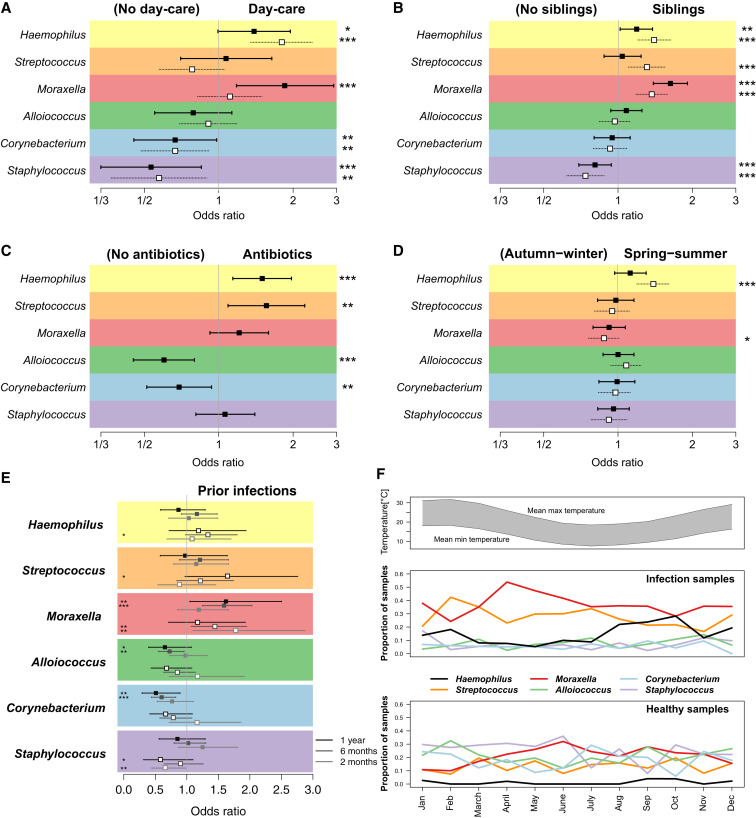
We next assessed the impact of environmental factors on the relative abundance of the common NP microbiome genera. We found no significant effects of delivery mode or breastfeeding on NP colonization at 2 months of age; the latter was unsurprising as nearly all infants (90%) were breastfed for at least 2 months. The abundance of Streptococcus in healthy NP samples was significantly lower among children whose parents reported having furry pets such as dogs or cats in the home (OR 0.84, 95% CI 0.70–1.0, p = 0.046; adjusted for age at NP sampling). No other significant associations were detected for pets. Children attending day care had significantly higher relative abundances of Haemophilus and Moraxella and lower relative abundances of Corynebacterium and Staphylococcus (Figure 2 A) in both healthy and infection samples (note that very few children had commenced day care by 6 months of age, hence the impact of day care attendance was assessed at the 12-month time point only). Co-habiting with siblings was also associated with higher abundances of Haemophilus, Streptococcus, and Moraxella and lower abundance of Staphylococcus during health and ARI (adjusted for age at sampling, Figure 2B). Importantly, among healthy samples, antibiotic usage in the four weeks prior to sampling was associated with higher abundances of Haemophilus, Streptococcus, and Moraxella and lower abundances of Alloiococcus and Corynebacterium (adjusted for age at sampling, Figure 2C). The composition of the healthy NP microbiome was also affected by the number of prior respiratory infections experienced, with higher abundance of Moraxella and lower abundances of Alloiococcus or Corynebacterium in samples following increasing numbers of ARIs (Figure 2E). At the MPG level, ARIs dominated by Haemophilus increased in spring-summer, while those dominated by Moraxella peaked in autumn-winter (Figure 2F). We therefore tested for differences in relative abundance in autumn-winter and spring-summer, adjusting for age and number of prior infections; this confirmed significant seasonal effects on the abundance of Haemophilus (summer associated) and Moraxella (winter associated) among ARI samples and a similar but non-significant trend among healthy NP samples (Figure 2D).
Figure 2.
Impact of Environmental Factors on Relative Abundances of Major Genera of the NP Microbiome
(A–D) Squares, odds ratios; filled squares, healthy samples; empty squares, infection samples; bars, 95% confidence intervals; ∗p < 0.1, ∗∗p < 0.05, ∗∗∗p < 0.01. Associations are estimated using logistic regression and adjusted for age: (A) day care attendance (yes versus no, 12-month samples), (B) co-habiting with siblings, (C) antibiotics intake in the 4 weeks preceding NP sample collection, (D) season (spring-summer versus autumn-winter).
(E) Impact of prior ARI, estimated using proportional odds ordinal logistic regression and categorized as 0, 1, or ≥ 2 ARI.
(F) Seasonal patterns: top, mean maximum and minimum temperatures in study location (Perth); bottom, monthly proportions of samples in each microbiome profile group (MPG) for infection and healthy samples.
NP Microbial Determinants of ARI Symptoms
The Moraxella, Streptococcus, and Haemophilus MPGs were significantly more frequent in ARI compared to healthy NP samples, even after adjusting for a large set of potential confounders (age, gender, season, number of prior infections, antibiotic intake, mother’s antibiotic intake, delivery mode, and breastfeeding) (Figure 1E). The Staphylococcus, Corynebacterium, and Alloiococcus MPGs were significantly less frequent in ARI (Figure 1E). The rare genus Neisseria was more common in infection samples, especially LRIs (of 28 NP samples with > 5% relative abundance of Neisseria, 4 were from URI and 20 were from LRI, though all were co-colonized with Streptococcus). It was not possible to confirm species from the 16S sequences; however, it is well known that the respiratory pathogens S. pneumoniae, H. influenzae, M. catarrhalis, and N. meningitidis are frequently cultured from respiratory infections in children. We have previously measured IgG to species-specific surface proteins of S. pneumoniae and H. influenzae at 12 months of age in the CAS cohort (Hales et al., 2012). Here we found that H. influenzae-specific IgG was significantly associated with the number of prior ARI samples testing positive for either of the two most common Haemophilus OTUs (Figures S2A–S2C); similar results were obtained for S. pneumoniae-specific IgG antibodies and the dominant Streptococcus OTU (Figures S2D–S2F). Healthy colonization with these genera was not associated with species-specific IgG.
A total of 138 children had ≥ 2 ARI samples profiled, and of these, 97 (70%) had ≥ 2 different MPGs among their ARIs. There was a clear temporal trend, with infections occurring closer in time more likely to be of the same MPG (Figure S3D). Moraxella and Haemophilus MPGs were particularly stable between consecutive infections, i.e., following an ARI in which the Moraxella or Haemophilus MPG was present, the next ARI was more likely to share the same MPG type than expected given the overall frequency of these MPGs among infection samples (Figure S3C).
We further considered the impact of the NP microbiome on infection severity and interactions with viral pathogens. All samples analyzed here were previously screened for a panel of viruses (Kusel et al., 2006). This screen detected viruses in 21% of healthy samples, 68% of URIs, and 69% of LRIs. The most common viruses detected were RSV (11% of ARI) and HRV (40% of ARI) (Figure S4), although subsequent expanded screening and subtyping of HRV in LRI samples suggests this is an underestimate (see below). For all virus groups except adenovirus and coronavirus, virus detection was significantly positively associated with ARI symptoms (i.e., ARI versus healthy samples, Figure S4). The association between ARI and Streptococcus, Haemophilus, and Moraxella MPGs remained after adjusting for detection of virus (OR 7.0, p < 1 × 10−15 for any of these MPGs; individual ORs in Figure 1E), indicating both viruses and bacteria contribute to ARI symptoms. Among the viruses analyzed, only RSV was significantly more frequent in LRIs versus URIs (16% versus 8.3%, OR 2.3; Figure S4, Table 1 ). The illness-associated MPGs Streptococcus, Haemophilus, and Moraxella were significantly associated with LRIs versus URIs (OR > 2), individually and collectively, adjusting for the effect of RSV (Table 1). As a group, the illness-associated MPGs increased in frequency from healthy to URI to LRI samples, regardless of the presence of RSV (Figure S5). Taken together, these analyses indicate that both viruses and bacteria independently contribute to ARI and that bacteria and RSV independently increase the risk of infection spread to the lower airways.
Table 1.
Associations between ARI Symptom Severity and Risk-Associated MPGs
| Symptom Group Comparison | Model |
Streptococcus, Moraxella, or Haemophilus MPG |
RSV | HRV (expanded screen) | ||
|---|---|---|---|---|---|---|
| Streptococcus MPG | Moraxella MPG | Haemophilus MPG | ||||
| LRI versus URI | 1A | OR = 2.2 (1.3–3.7), p = 0.0043 | 2.2 (1.1–4.7), p = 0.034 | – | ||
| 1B | 2.3 (1.2–4.5), p = 0.011 | 2.1 (1.1–3.8), p = 0.017 | 2.1 (1.0–4.6), p = 0.056 | 2.3 (1.1–4.8), p = 0.033 | – | |
| Febrile LRI versus non-febrile LRI | 2A | OR = 3.8 (1.0–14), p = 0.047 | 4.0 (1.6–9.8), p = 0.0031 | 0.45 (0.21–0.95), p = 0.036 | ||
| 2B | 4.3 (1.0–18), p = 0.046 | 3.8 (0.98–15), p = 0.053 | 2.7 (0.53–14), p = 0.23 | 4.0 (1.6–10), p = 0.0031 | 0.46 (0.21–0.98), p = 0.043 | |
| Wheezy LRI versus non-wheezy LRI | 3A | OR = 1.4 (0.60–3.1), p = 0.45 | 0.94 (0.41–2.1), p = 0.88 | 1.2 (0.62–2.2), p = 0.64 | ||
| 3B | 1.1 (0.39–3.0), p = 0.88 | 1.8 (0.74–4.1), p = 0.20 | 1.0 (0.34–3.2), p = 0.93 | 0.95 (0.42–2.2), p = 0.90 | 1.1 (0.60–2.2), p = 0.69 | |
Odds ratio (OR) (95% confidence interval), p value. NP samples taken within a week of antibiotic use were excluded from analysis. The response variable (symptom group comparison) is shown in column 1. Two models (A, B) were fit for each comparison, labeled in column 2. Model A included Streptococcus or Moraxella or Haemophilus MPG as a single covariate; Model B included Streptococcus, Moraxella, and Haemophilus MPGs as separate covariates. Both models also include RSV, HRV, and the potential confounders age, gender, and season. Note: HRV was not included in the LRI versus URI comparison, as enhanced sensitivity re-screening for HRV was performed only in LRI samples.
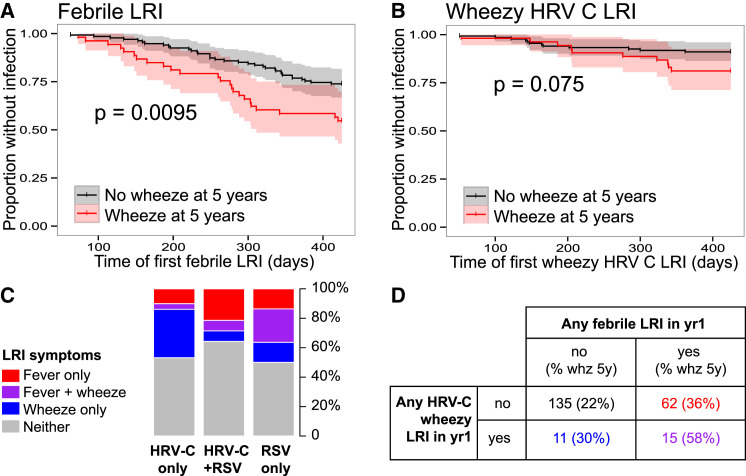
In our earlier studies with this cohort, we observed an association between LRIs (but not URIs) during infancy and risk for wheeze at age 5 years; moreover, this association was restricted to severe LRIs, i.e., those accompanied by fever and/or wheeze (Holt et al., 2010, Kusel et al., 2007, Kusel et al., 2012). Thus we investigated the association of viruses and bacteria with the presence of fever and wheeze symptoms during LRI. To enable more accurate assessment of the role of HRV in this critical sample group, we performed an expanded screen for detection and subtyping of HRV within LRI samples (Supplemental Experimental Procedures). This assay proved more sensitive than the first screen and detected HRV in 66% of LRI samples (twice that in the earlier screen), with equal amounts of HRV-A and HRV-C (Figure S4). During LRI, the presence of any HRV, or HRV-C specifically, was negatively associated with fever (Table 1). The only viruses showing positive associations with fever were RSV (Table 1, Figure 3C, Figure S4) and influenza (8 influenza-positive fever LRI events only, all with illness-associated MPGs, thus not considered further). Among LRIs, the illness-associated MPGs were associated with fever, even after adjusting for RSV, HRV, age, season, and gender (Table 1). Interestingly, Moraxella MPG was also significantly positively associated with fever among RSV-positive LRIs (OR 9.2, 95% CI 1.1–73, p = 0.037; Figure S5), suggesting a possible interaction between Moraxella and RSV whereby their co-presence further enhances risk of fever. Overall, only 10 febrile LRIs (9.5%) could not be explained by the presence of RSV or illness-associated MPGs. Across the cohort, the presence of wheeze during LRI was not significantly associated with any viral or bacterial groups (Table 1), including HRV or HRV-C specifically. Of all ARIs analyzed, there were 61 with concomitant otitis media (OM) affecting 47 infants; however, ARIs with accompanying OM had similar NP microbiome profiles to those of LRIs with no OM diagnosis (Table S2).
Figure 3.
Symptoms of Lower Respiratory Illness during the First Year of Life Are Associated with Viruses Present during the Infection and Predict Chronic Wheeze at 5 Years of Age
(A and B) Kaplan-Meier survival curves for age (days) at (A) first febrile LRI and (B) first HRV-C wheezy LRI, stratified by chronic wheeze status at 5 years. p values shown were estimated using Cox proportional hazards models, adjusted for gender and maternal and paternal history of atopic disease. Shaded areas indicate 95% confidence intervals.
(C) Frequencies of fever and wheeze symptoms during LRI, in which HRV-C and/or RSV were detected. Total numbers are: HRV-C only, n = 79; HRV-C and RSV, n = 14; RSV only, n = 22.
(D) Cross-tabulation of individuals according to their experience of LRI during infancy; percentages in brackets indicate frequency of chronic wheeze at 5 years.
Impact of ARI on Later Chronic Wheeze
We next investigated associations between LRI in the first year of life and subsequent expression of chronic wheeze at age 5 or 10 years. Positive associations were found for two discrete classes of LRI. First, febrile LRI was significantly positively associated with later chronic wheeze and among children who were atopic by 2 years of age (Table 2 ). Furthermore, timing of first febrile LRI appeared to be important, with earlier febrile LRI occurring among children who had chronic wheeze at 5 years (p = 0.0095, Figure 3 A). Second, HRV-C LRI accompanied by wheezing symptoms showed a positive association with later chronic wheeze among all children and particularly strongly so for those who were atopic by 2 years (OR ∼7, Table 2), but not among non-atopics (OR 1, p = 0.95; Table S3). As we found no association between HRV-C and wheeze during LRI across the whole cohort (OR 1.2, 95% CI 0.63–2.2, p = 0.61; adjusting for illness-associated MPGs, RSV, age, season, and gender), we re-examined the association in those who developed atopy by age 2. Among this group of children (n = 65), the presence of HRV-C was significantly positively associated with increased risk of wheeze during LRI (OR 2.7, 95% CI 1.1–7, p = 0.035; adjusting for illness-associated MPGs, RSV, age, season, and gender).
Table 2.
Association between Microbial Events during Infancy and Chronic Wheeze at Age 5 and 10 Years
| Wheeze at 5 Years |
Wheeze at 10 Years |
|||
|---|---|---|---|---|
| All children | Atopics | All children | Atopics | |
| Any febrile LRI | 2.3 (1.2–4.5), p = 0.016∗ | 2.7 (1.1–7), p = 0.034∗ | 2.2 (0.93–5.2), p = 0.071 | 2.7 (0.89–8.7), p = 0.083 |
| Any wheezy LRI | 1.6 (0.79–3), p = 0.2 | 1.3 (0.49–3.3), p = 0.61 | 1.4 (0.58–3.3), p = 0.45 | 1.6 (0.53–4.8), p = 0.4 |
| Any HRV wheezy LRI | 2 (0.93–4.2), p = 0.073 | 2.5 (0.86–7.2), p = 0.092 | 2.1 (0.81–5.4), p = 0.11 | 1.9 (0.55–6.4), p = 0.29 |
| Any HRV-C wheezy LRI | 2.4 (0.93–6.1), p = 0.064 | 7.2 (1.7–35), p = 0.009∗ | 3.5 (1.1–11), p = 0.026∗ | 7.1 (1.6–40), p = 0.014∗ |
| Any HRV-A wheezy LRI | 1.2 (0.42–3.1), p = 0.74 | 0.55 (0.11–2.2), p = 0.43 | 1.4 (0.37–4.7), p = 0.57 | 0.35 (0.02–2.2), p = 0.34 |
| Any risk bacteria LRI | 0.89 (0.45–1.8), p = 0.73 | 0.96 (0.39–2.4), p = 0.93 | 2 (0.82–5.2), p = 0.14 | 1.7 (0.59–5.2), p = 0.34 |
| High-abundance Streptococcus colonization (≤7 weeks) | 3.8 (1.3–12), p = 0.017∗ | 4 (0.88–21), p = 0.077 | 2.7 (0.6–12), p = 0.18 | 3.9 (0.63–28), p = 0.15 |
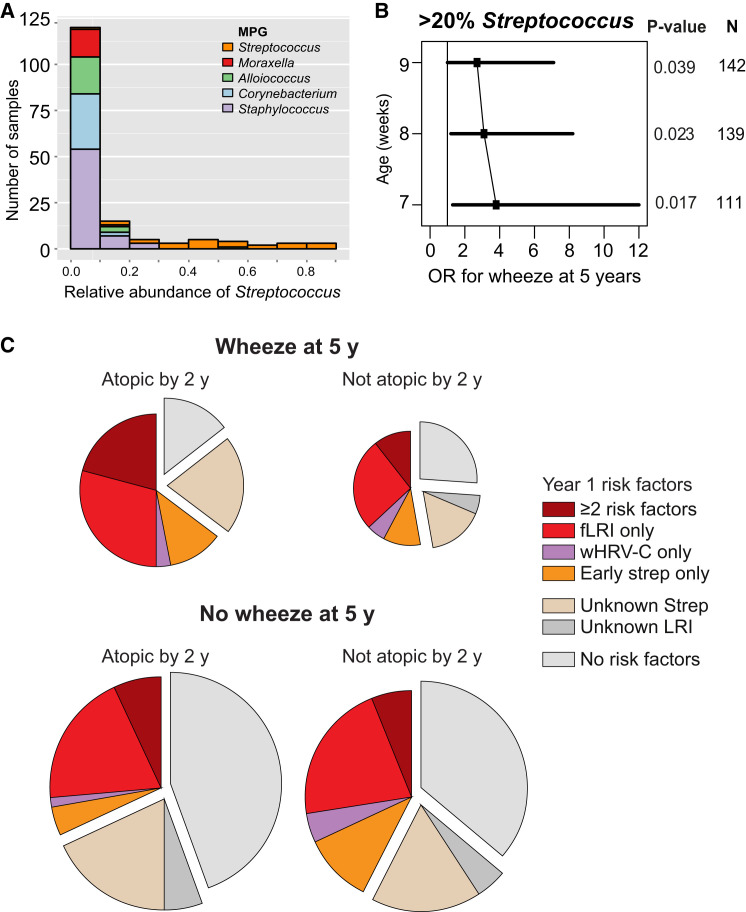
Odds ratio (95% confidence interval), p value; estimated using logistic regression, adjusted for gender and maternal and paternal history of atopic disease; estimated separately for all children and those who were atopic by 2 years of age. LRI, lower respiratory illness, further classified according to microbes and symptoms; any risk bacteria LRI, any LRI with Streptococcus, Moraxella, or Haemophilus MPG. Early Streptococcus colonization was assessed in the first healthy NP sample, collected by 7 weeks of age and prior to any recorded infection; high abundance was classified as > 20% Streptococcus reads (based on distribution in Figure 5A).
Since febrile LRI and HRV-C wheezy LRI were both associated with later chronic wheeze (Table 2), and RSV was associated with febrile LRI (Table 1), we examined the interaction between RSV, HRV-C, LRI symptom severity, and chronic wheeze at age 5. Febrile LRI and wheezy HRV-C LRI in the first year of life appeared to exert independent effects on later chronic wheeze, since only a minority of HRV-C wheezy infections were also febrile (Figure 3C). Further, at the level of individual children, the frequency of chronic wheeze at 5 years was elevated among those with either one of febrile LRI (36%) or wheezy HRV-C LRI (30%) but was greatest among children who experienced both (58%, Figure 3D).
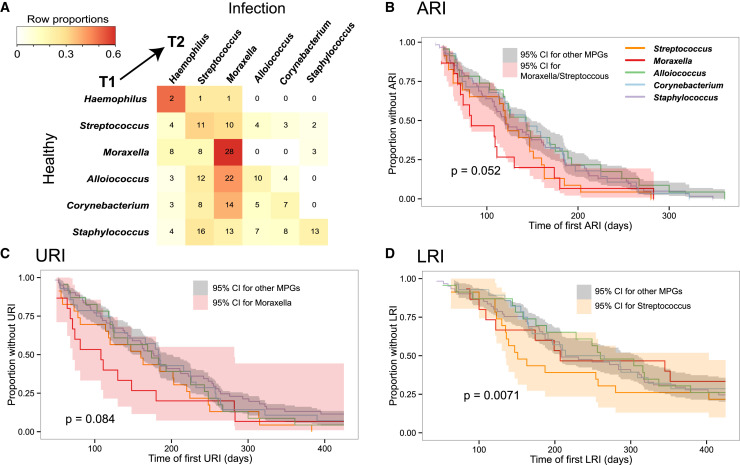
Impact of NP Colonization on ARI
Whereas Streptococcus, Haemophilus, or Moraxella MPGs were detected in healthy NP samples, subsequent infections tended to belong to the same MPG (Figure 4 A). Critically, we investigated whether healthy colonization in early infancy was associated with subsequent episodes of ARI. Since prior infections also affect the healthy NP microbiome (Figure 2E), we restricted these analyses to healthy samples collected at 5–9 weeks of age and prior to each infant’s first reported ARI (n = 160). Using Cox proportional hazards models, infants whose earliest healthy NP sample was of the Moraxella or Streptococcus MPG tended to experience ARI at a younger age than those with other MPGs (Figure 4B; note that Haemophilus MPGs were extremely rare in early healthy samples). When examining URIs and LRIs separately, we found that early Moraxella colonization was associated with earlier first URI, whereas early Streptococcus colonization was strongly associated with earlier first LRI (Figures 4C and 4D). Since our data suggested that Alloiococcus and Moraxella were key stable colonizers of the NP microbiome, we also examined whether Alloiococcus-colonized infants differed from Moraxella-colonized infants (defined as those with ≥ 1 healthy sample of Moraxella MPG and none with Alloiococcus MPG, and vice versa). These groups did not differ in terms of overall numbers of ARI, LRI, or later wheezing phenotypes. However, compared to either Moraxella-colonized infants or those not in either group, Alloiococcus-colonized infants had fewer RSV infections, especially RSV LRIs (OR 0.27, Table S4).
Figure 4.
Impact of Early Colonization on Age at First Respiratory Infection
(A) Microbiome profile group (MPG) transitions between healthy samples (T1) and the next sequenced infection (T2). Cell numbers indicate the number of times the respective transition from T1 to T2 was observed in the dataset; cells are colored to indicate the row proportions as per legend.
(B–D) Kaplan-Meier survival curves for age (days) of first (B) acute respiratory illness (ARI), (C) upper respiratory illness (URI), and (D) lower respiratory illness (LRI), stratified according to the MPG of the first healthy sample (collected by 9 weeks of age and prior to any infection, n = 160). Cox proportional hazards models were adjusted for age, gender, season, virus status in the early healthy sample, and virus status at the first event. Shaded areas indicate 95% confidence intervals.
Early NP Colonization Impacts Later Chronic Wheeze
We next assessed the association between early pre-ARI asymptomatic NP colonization and current wheeze at 5 and 10 years of age, stratified by atopic sensitization by 2 years of age (Supplemental Experimental Procedures). This analysis was restricted to the 160 infants (70%) who had an asymptomatic NP sample taken prior to their first ARI (and ≤ 9 weeks of age). Haemophilus MPG was not detected in pre-ARI healthy samples, and Moraxella MPG showed no evidence of association with future wheeze. The Streptococcus MPG showed a weak association; however, the relative abundance of Streptococcus in these samples was highly skewed, so we divided samples into those with high (> 20%) or low (≤ 20%) relative abundance of Streptococcus reads (Figure 5 A). High Streptococcus abundance in the first pre-ARI healthy NP sample was more frequent in infants who later displayed wheeze at 5 years, and this association was stronger when restricting the analysis to earlier NP samples (Figure 5B, Table 2). The same trend was evident for wheeze at 10 years of age, despite reduced sample size due to loss to follow-up (Table 2). Early Streptococcus colonization was associated with younger age at first LRI (Figure 4D), but not with presence of Streptococcus in the first LRI or with detection of S. pneumoniae antibodies at 12 months of age. No statistically significant associations were observed for future chronic wheeze when aggregating the 7-week healthy Streptococcus, Haemophilus, and Moraxella MPGs into a single predictor (with or without atopy by 2 years). Overall, infants who were atopic by age 2 and developed chronic wheeze at age 5 were twice as likely to have had early Streptococcus colonization, febrile LRI, and/or HRV-C wheezy LRI in the first year of life, compared to those that did not develop chronic wheeze (Figure 5C).
Figure 5.
Predictors of Chronic Wheeze at Age 5
(A) Streptococcus abundance among healthy samples collected by 9 weeks of age, broken down by microbiome profile group (MPG).
(B) Adjusted odds ratio (OR, squares) and 95% confidence intervals (bars) for association between chronic wheeze at age 5 and high (> 20%) abundance of Streptococcus in the first healthy NP sample; p values and sample sizes (n) are indicated; individuals who experienced an infection prior to first healthy NP sample collection were excluded.
(C) Distribution of microbial events during infancy that were identified as risk factors for chronic wheeze at 5 years of age, stratified according to atopic status by age 2. fLRI, febrile LRI; wHRV-C, HRV-C LRI accompanied by wheeze; Strep, > 20% Streptococcus abundance in healthy NP sample taken in by 9 weeks old and prior to any ARI; unknown Strep, no such NP sample available (mainly due to ARI before 9 weeks of age); unknown LRI, incomplete viral/symptom profiling for LRI. Size of pie chart is proportional to the number of infants in each condition.
Discussion
NP Microbiome Composition and Dynamics
Our data provide a detailed prospective characterization of bacterial communities within the human NP microbiome during the first year of life. The NP microbiome was qualitatively simple (Figure 1), dominated by six common genera: Haemophilus, Streptococcus, Moraxella (each more common in ARI), Staphylococcus, Alloiococcus, and Corynebacterium (more common in healthy samples). This is consistent with previous studies of NP microbiome composition in children aged 12–14 months (Biesbroek et al., 2014, Bogaert et al., 2011) and adults (Hilty et al., 2010), although intra- and inter-sample diversity was greater in these older groups, likely due in part to extreme seasonal variation in those study locations. In contrast, our study site in Perth, Australia has a very moderate climate, and we detected only limited seasonal effects (Figure 2D) that were readily adjusted for, enabling us to assess the dynamics of the NP microbiome at different stages of infancy and to examine its association with other factors.
We found that early NP colonization typically involved Staphylococcus or Corynebacterium, which was later replaced by Moraxella or Alloiococcus. Staphylococcus was the dominant colonizing bacteria in the early healthy NP microbiomes, but its presence declined rapidly with age (Figure 1). S. aureus is a common cause of neonatal sepsis, and high rates of S. aureus nasal colonization in the first months of life have been reported (Bisgaard et al., 2007, Bisgaard et al., 2010). Few studies have examined longitudinal colonization; however, a recent study in African children reported S. aureus colonization in 42% of infants at 1 month and 12% at 12 months of age, a trend that was mirrored in maternal colonization rates (Schaumburg et al., 2014) and is strikingly similar to the patterns we observed (41% at 2 months, 11% at 12 months). Staphylococcus and Corynebacterium are both common components of the human skin microbiome, and in our data they showed a comparable temporal pattern, with high rates in the early NP microbiome declining with age (Figure 1). We speculate that infants tend to be colonized initially with skin-dwelling bacteria (acquired from parents and others), which is replaced over time by stable colonization with Moraxella or Alloiococcus and punctuated by transient acquisition of Streptococcus or Haemophilus that is frequently accompanied by ARI symptoms. Our data further suggest the transition to Moraxella is associated with exposure to other children (in the home or at day care) and episodes of ARI, and that colonization with Moraxella, Streptococcus, and Haemophilus are selected for by antibiotic exposure (Figure 2).
Moraxella was the most common genus in our study population, dominating 21% of all healthy NP microbiomes and 38% of infection samples. Moraxella was represented in our sequence data by a single OTU matching that of M. catarrhalis, a human-restricted, unencapsulated, Gram-negative bacterium previously associated with both commensal NP colonization and pathogenicity in the respiratory tract and inner ear (de Vries et al., 2009). Increased rates of NP colonization with M. catarrhalis have been reported in children following pneumococcal vaccination (Revai et al., 2006); however, the samples in our study predate introduction of this vaccine. In our cohort the presence of Moraxella increased with age, and Moraxella was a particularly stable component of the NP microbiome during both health and disease (Figure S3). These findings are consistent with Moraxella’s known ability to form biofilms (de Vries et al., 2009), which offer protection from antibiotics and promote co-colonization with other common bacteria such as S. pneumoniae and H. influenzae (Verhaegh et al., 2011). Moraxella was more abundant during the cooler months (Figure 2), a seasonal trend consistent with recent reports that cold shock at 26°C stimulates growth, colonization, and expression of virulence-associated traits in M. cattarhalis (Spaniol et al., 2011). These data are consistent with a Dutch culture-based study of M. catarrhalis carriage in young children (Verhaegh et al., 2011), which found increasing prevalence of M. catarrhalis during the first year of life, with a strong seasonal effect (also observed in a Swedish cohort [Gisselsson-Solén et al., 2014]) and a significant positive association with day care attendance and siblings. However, these studies did not examine the stability of Moraxella colonization within individuals over time. Importantly, our data show that the presence of Moraxella contributes to severity of RSV respiratory infections in infants (Figure S5, Table S3), which may be mechanistically related to RSV-Moraxella interactions reported in OM pathogenesis (Brockson et al., 2012).
Alloiococcus was a common and stable component of healthy NP samples and demonstrated enhanced stability in healthy NP microbiomes (Figure S3). Alloiococcus is a Gram-positive bacterium with one named species, A. otitidis, frequently detected in the ear canals of children with OM. Little is known about its mechanisms of colonization; however, the stable colonization patterns we observed may reflect a propensity to form stable biofilms, similar to Moraxella. The Alloiococcus MPG included some samples with high abundances of both Alloiococcus and Corynebacterium, indicating compatibility between these bacteria somewhat reminiscent of that between the Moraxella biofilm and Streptococcus or Haemophilus. Whether Alloiococcus is involved in OM pathology or is merely a healthy component of the microbiome is controversial (Tano et al., 2008); in our data, Alloiococcus was not associated with ARI or OM (Figure 1, Table S2).
Haemophilus was almost exclusively associated with ARI symptoms and was often found in consecutive infections, suggesting it can persist in the nasopharynx for some time (Figure 1, Table 1, Figure S3). Our analyses also suggest Haemophilus is highly transmissible between children and selected for by antibiotic exposure (Figure 2). Although our data include multiple Haemophilus OTUs, these epidemiological patterns are consistent with the human-adapted pathogen H. influenzae, which was supported by antibody data (Figure S2).
Role of the NP Microbiome in Asthma Development
We have previously reported that within the CAS cohort, the frequency during the first year of life of severe (wheezy and/or febrile) LRI was positively associated with subsequent risk for persistent wheeze at 5 and 10 years of age (Holt et al., 2010, Kusel et al., 2007, Kusel et al., 2008), and similar findings have been reported in the U.S.-based COAST cohort (Gern, 2009). In both these study populations, the susceptible subgroup of children were those who developed early allergic sensitization (Gern, 2009, Holt et al., 2010, Holt et al., 2014, Jackson et al., 2008, Kusel et al., 2007, Kusel et al., 2012, Oddy et al., 2002), and a variety of evidence suggests that the underlying mechanism involves synergistic interactions between atopic and anti-viral inflammatory pathways triggered within the infected airway mucosa (Holt and Sly, 2012). Of note, ARIs that remain restricted to the upper respiratory tract, and infections resulting in only mild LRI without wheezing or febrile symptoms, are relatively benign with respect to asthma risk in this cohort (Holt et al., 2010, Kusel et al., 2007, Kusel et al., 2012) (Table S3). Hence cofactors that can enhance the spread and ensuing severity of viral-initiated ARI are potentially central to asthma pathogenesis.
Here we found that two specific classes of LRI, namely febrile LRI and HRV-C-positive wheezy LRI, were independent risk factors for later chronic wheeze, especially among atopics (Table 2, Figure 3). Febrile LRI was common, occurring in one-third of all children and half of atopic chronic wheezers at age 5 (Figure 3 and Figure 5). Notably, the presence of illness-associated bacteria within the NP microbiome at the time of ARI increased both the risk for progression to the lower respiratory tract and the development of fever (Table 1). These mechanisms likely include myriad bacterial-viral interactions (recently reviewed by Vissers et al., 2014). In contrast, neither specific virus groups nor bacterial MPGs were associated with wheezy versus non-wheezy LRI within the overall population (Table 1), and the impact of wheezy LRI on later chronic wheeze was limited to LRI with HRV-C (Table 2), which occurred in just 11% of children. Re-examination of wheeze symptoms during LRI stratified by atopic status identified a positive association between HRV-C and wheeze during LRI among those children who later became sensitized (by 2 years of age), consistent with observations that HRV-C can induce wheeze in high-risk individuals (Lee et al., 2012). However, the presence of wheezy LRI alone, without further stratification by HRV-C detection and later atopic status, was not a significant predictor of later chronic wheeze, while febrile LRI per se was a significant predictor of chronic wheeze at 5 and 10 years, both among atopics and across the whole cohort (Figure 5, Table 2).
Early asymptomatic Streptococcus colonization at ∼2 months of age, which occurred in 14% of children tested at that time, was significantly associated with chronic wheeze at 5 years of age (Figure 5, Table 2). Early Streptococcus colonization was not associated with incidence of infections with Streptococcus MPG, nor with detection of S. pneumoniae-specific antibodies at 12 months, suggesting that the mechanism is independent of innate immune response to Streptococcus. Rather, early Streptococcus colonization was associated with younger age of first LRI, and the level of subsequent asthma risk appears inversely related to age at initial Streptococcus colonization (Figure 4 and Figure 5). Collectively, these observations suggest that developing airway tissues may be maximally susceptible to the long-term effects of Streptococcus-mediated or LRI-mediated damage during the early postnatal period, when lung growth rates are most rapid.
To our knowledge, the only other study reporting the impact of early life NP colonization on wheeze/asthma at pre-school age is the 2007 report on the Copenhagen Prospective Study on Asthma in Childhood (n = 321, children born to mothers with asthma), in which a higher rate of wheeze at 5 years of age was detected among children colonized with S. pneumoniae, M. catarrhalis, and/or H. influenzae at 1 month post-birth (in culture-confirmed colonized, asthma prevalence was 33%, not colonized, 10%, OR 4.57) (Bisgaard et al., 2007). In that study, associations were not reported individually for the three species and colonization was measured as positive or negative by microbiological culture. In our cohort, wheeze at 5 years was not associated with early colonization with Haemophilus or Moraxella; however, Haemophilus was very rare in healthy NP samples, and Moraxella colonization was established later during infancy, which may be related to the warmer climate in Perth; these associations may differ in populations where asymptomatic carriage of these organisms is higher.
Conclusions and Implications
These findings collectively suggest that bacterial pathogens present in the NP microbiome at the time of upper respiratory viral infections during infancy are significant determinants of risk for the spread of infection to the lower airways and for the resultant expression of inflammatory symptoms marked by fever, and further, they may contribute directly and indirectly to the ensuing risk for development of persistent asthma, which itself is linked to these prior infectious episodes. Prevention of RSV or HRV infections in high-risk children, using immunoprophylaxis or vaccines, has been proposed as a mechanism for preventing the development and/or exacerbation of childhood asthma (Gern, 2009, Wu and Hartert, 2011). Our data suggest that in the absence of effective anti-viral therapies, targeting pathogenic bacteria present within the NP microbiome in this age group could represent an alternative approach toward the same goal. The pneumococcal vaccine, currently recommended from 2 months of age, could play a role; however, the niche created by eliminating vaccine-targeted S. pneumoniae serotypes can be readily filled by other serotypes and other bacteria such as H. influenzae and M. catarrhalis (Biesbroek et al., 2014, Revai et al., 2006). Manipulation of the microbiome by antibiotics may appear attractive; however, the association between antibiotic use in early childhood and subsequent asthma is controversial. In the CAS cohort (Kusel et al., 2008) and others (Semic-Jusufagic et al., 2014), this association has been proposed to arise through confounding and reverse causation, whereby genetic or clinical factors increase the likelihood of both antibiotic prescription (e.g., via genetic susceptibility to viral infection [Semic-Jusufagic et al., 2014]) and asthma development; others have proposed that antibiotic-induced disruption of the gut microbiota could explain the link (Arrieta and Finlay, 2014). Our analyses provide a potential causal pathway linking antibiotics to later asthma, whereby antibiotic use in infants selects for illness-associated bacteria in the NP microbiome, leading to increased risk of febrile LRI and later asthma development. Genetic analysis of the infants in our cohort could potentially help to unravel this in the future. Importantly, further studies are required to prospectively assess the impact of antibiotic use on NP bacterial colonization during health and infection, and the subsequent development of ARI, atopy, and wheeze.
Experimental Procedures
Study Design
This study is an extension of the Childhood Asthma Study (CAS), a birth cohort of 234 infants at high risk of atopy as previously described (Kusel et al., 2007, Kusel et al., 2008). Briefly, healthy NP aspirates (NPAs) were collected from subjects at planned visits at 2, 6, and 12 months of age. NPAs were also collected within 48 hr from the onset of an ARI; these were classified as either lower respiratory infection (LRI, if wheeze or rattly chest detected) or upper respiratory infection (URI, otherwise). NPAs were divided into four aliquots and stored at −80°C. One aliquot was used for bacterial 16S rRNA amplicon sequencing via Illumina MiSeq and another to screen for viruses including RSV, HRV, and other picornaviruses, influenza, parainfluenza, coronavirus, adenovirus, and human metapneumovirus (hMPV). For LRI, a third aliquot was used in an expanded screen for detection and subtyping of HRV. Study ethics were approved by the ethics committees of King Edward Memorial and Princess Margaret Hospitals in Western Australia. Fully informed parental consent was obtained for all subjects. Details of sample and data collection, viral screening, DNA extraction, and sequencing are in Supplemental Experimental Procedures.
16S rRNA Sequence Analysis
Paired end reads were merged using Flash v1.2.7 (Magoč and Salzberg, 2011) with read length 151 base pairs (bp) and expected fragment length 253 bp. Merged sequences were quality filtered as follows: ≤ 3 low-quality bp (Phred score < 3) allowed before trimming, ≥ 189 consecutive high-quality bp with no uncalled bases (Ns) (Bokulich et al., 2013). Thirty-three million sequences were filtered out (13%), leaving 219 million for analysis. These were assigned to OTUs using the closed reference method in QIIME v1.8 (Caporaso et al., 2010) with the Greengenes 99% OTU reference set, version 13_5 (McDonald et al., 2012) (which consists of > 200,000 representative sequences obtained from clustering all sequences from the Greengenes reference database at 99% sequence similarity). Briefly, this method uses UCLUST (Edgar, 2010) to search each sequence against the reference set and assigns it to an OTU based on the best hit at ≥ 99% sequence identity. Sequences not matching the reference database (3%) were excluded from analysis; these sequences could be chimeras, sequencing errors, or novel sequences that are not well characterized in the database. This left > 200,000 taxonomy-assigned sequences on average for each NPA (interquartile range: 108,000–255,000; eight samples with < 1,000 reads). Compared to NPAs, negative control samples (see Supplemental Experimental Procedures) had clearly lower total reads (interquartile range: 1,044–2,346) with a different community structure (Figure S6).
Clustering into MPGs
For each NPA, the relative abundance of each OTU was calculated. Most analyses were summarized at genus level, whereby all OTUs assigned to the same genus were collapsed into a single group for reporting. Samples were assigned to microbiome profile groups (MPGs) based on hierarchical clustering of the relative abundances of the six most common genera (distance metric: 1-Pearson’s correlation; clustering method: Ward’s minimum variance, implemented in R hclust). Previous microbiome studies have reported clinically or environmentally meaningful associations with community profiles or types (e.g., enterotypes) identified using similar clustering approaches (Ravel et al., 2011, Zhou et al., 2014).
Statistical Methods
All statistical analyses were performed using R. ORs were estimated using generalized estimating equations (GEE) logistic regression with unstructured correlation and robust standard errors, to take into account multiple samples from the same subject. Survival analyses were done using Cox proportional hazards models. Potential confounders were adjusted for by including them in the regression models. Full details of each analysis and variable definitions are given in the Supplemental Experimental Procedures.
Acknowledgments
This study was supported by the NHMRC of Australia (Project Grant #1049539; Fellowships #1061409 [K.E.H.] and #1061435 [M.I., co-funded with the Australian Heart Foundation]) and the Victorian Life Sciences Computation Initiative (VLSCI) (#VR0082). S.L.J. is supported in part by a Chair from Asthma UK (CH11SJ) and by Medical Research Council Centre grant G1000758. J.E.G., Y.A.B., and K.G. are supported by NIH grants U19 AI104317 and P01 HL070831.
Published: April 9, 2015
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2015.03.008.
Contributor Information
Kathryn E. Holt, Email: kholt@unimelb.edu.au.
Michael Inouye, Email: minouye@unimelb.edu.au.
Accession Numbers
Short read 16S data for this study have been deposited in NCBI GenBank under accession number SRP056779.
Supplemental Information
References
- Arrieta M.C., Finlay B. The intestinal microbiota and allergic asthma. J. Infect. 2014;69(1):S53–S55. doi: 10.1016/j.jinf.2014.07.015. [DOI] [PubMed] [Google Scholar]
- Biesbroek G., Wang X., Keijser B.J., Eijkemans R.M., Trzciński K., Rots N.Y., Veenhoven R.H., Sanders E.A., Bogaert D. Seven-valent pneumococcal conjugate vaccine and nasopharyngeal microbiota in healthy children. Emerg. Infect. Dis. 2014;20:201–210. doi: 10.3201/eid2002.131220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard H., Hermansen M.N., Buchvald F., Loland L., Halkjaer L.B., Bønnelykke K., Brasholt M., Heltberg A., Vissing N.H., Thorsen S.V. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- Bisgaard H., Hermansen M.N., Bønnelykke K., Stokholm J., Baty F., Skytt N.L., Aniscenko J., Kebadze T., Johnston S.L. Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ. 2010;341:c4978. doi: 10.1136/bmj.c4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov Y.A., Gern J.E. Clinical and molecular features of human rhinovirus C. Microbes Infect. 2012;14:485–494. doi: 10.1016/j.micinf.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert D., Keijser B., Huse S., Rossen J., Veenhoven R., van Gils E., Bruin J., Montijn R., Bonten M., Sanders E. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I., Knight R., Mills D.A., Caporaso J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockson M.E., Novotny L.A., Jurcisek J.A., McGillivary G., Bowers M.R., Bakaletz L.O. Respiratory syncytial virus promotes Moraxella catarrhalis-induced ascending experimental otitis media. PLoS ONE. 2012;7:e40088. doi: 10.1371/journal.pone.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S.P., Bootsma H.J., Hays J.P., Hermans P.W. Molecular aspects of Moraxella catarrhalis pathogenesis. Microbiol. Mol. Biol. Rev. 2009;73:389–406. doi: 10.1128/MMBR.00007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Gern J.E. Rhinovirus and the initiation of asthma. Curr. Opin. Allergy Clin. Immunol. 2009;9:73–78. doi: 10.1097/ACI.0b013e32831f8f1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson-Solén M., Henriksson G., Hermansson A., Melhus A. Risk factors for carriage of AOM pathogens during the first 3 years of life in children with early onset of acute otitis media. Acta Otolaryngol. 2014;134:684–690. doi: 10.3109/00016489.2014.890291. [DOI] [PubMed] [Google Scholar]
- Hales B.J., Chai L.Y., Elliot C.E., Pearce L.J., Zhang G., Heinrich T.K., Smith W.A., Kusel M.M., Holt P.G., Sly P.D., Thomas W.R. Antibacterial antibody responses associated with the development of asthma in house dust mite-sensitised and non-sensitised children. Thorax. 2012;67:321–327. doi: 10.1136/thoraxjnl-2011-200650. [DOI] [PubMed] [Google Scholar]
- Hilty M., Burke C., Pedro H., Cardenas P., Bush A., Bossley C., Davies J., Ervine A., Poulter L., Pachter L. Disordered microbial communities in asthmatic airways. PLoS ONE. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt P.G., Sly P.D. Viral infections and atopy in asthma pathogenesis: new rationales for asthma prevention and treatment. Nat. Med. 2012;18:726–735. doi: 10.1038/nm.2768. [DOI] [PubMed] [Google Scholar]
- Holt P.G., Rowe J., Kusel M., Parsons F., Hollams E.M., Bosco A., McKenna K., Subrata L., de Klerk N., Serralha M. Toward improved prediction of risk for atopy and asthma among preschoolers: a prospective cohort study. J. Allergy Clin. Immunol. 2010;125:653–659, e1, e7. doi: 10.1016/j.jaci.2009.12.018. [DOI] [PubMed] [Google Scholar]
- Holt P.G., Strickland D.H., Hales B.J., Sly P.D. Defective respiratory tract immune surveillance in asthma: a primary causal factor in disease onset and progression. Chest. 2014;145:370–378. doi: 10.1378/chest.13-1341. [DOI] [PubMed] [Google Scholar]
- Jackson D.J., Gangnon R.E., Evans M.D., Roberg K.A., Anderson E.L., Pappas T.E., Printz M.C., Lee W.M., Shult P.A., Reisdorf E. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am. J. Respir. Crit. Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusel M.M., de Klerk N.H., Holt P.G., Kebadze T., Johnston S.L., Sly P.D. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr. Infect. Dis. J. 2006;25:680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- Kusel M.M., de Klerk N.H., Kebadze T., Vohma V., Holt P.G., Johnston S.L., Sly P.D. Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J. Allergy Clin. Immunol. 2007;119:1105–1110. doi: 10.1016/j.jaci.2006.12.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusel M.M., de Klerk N., Holt P.G., Sly P.D. Antibiotic use in the first year of life and risk of atopic disease in early childhood. Clin. Exp. Allergy. 2008;38:1921–1928. doi: 10.1111/j.1365-2222.2008.03138.x. [DOI] [PubMed] [Google Scholar]
- Kusel M.M., Kebadze T., Johnston S.L., Holt P.G., Sly P.D. Febrile respiratory illnesses in infancy and atopy are risk factors for persistent asthma and wheeze. Eur. Respir. J. 2012;39:876–882. doi: 10.1183/09031936.00193310. [DOI] [PubMed] [Google Scholar]
- Lee W.M., Lemanske R.F., Jr., Evans M.D., Vang F., Pappas T., Gangnon R., Jackson D.J., Gern J.E. Human rhinovirus species and season of infection determine illness severity. Am. J. Respir. Crit. Care Med. 2012;186:886–891. doi: 10.1164/rccm.201202-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddy W.H., de Klerk N.H., Sly P.D., Holt P.G. The effects of respiratory infections, atopy, and breastfeeding on childhood asthma. Eur. Respir. J. 2002;19:899–905. doi: 10.1183/09031936.02.00103602. [DOI] [PubMed] [Google Scholar]
- Penders J., Kummeling I., Thijs C. Infant antibiotic use and wheeze and asthma risk: a systematic review and meta-analysis. Eur. Respir. J. 2011;38:295–302. doi: 10.1183/09031936.00105010. [DOI] [PubMed] [Google Scholar]
- Ravel J., Gajer P., Abdo Z., Schneider G.M., Koenig S.S., McCulle S.L., Karlebach S., Gorle R., Russell J., Tacket C.O. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA. 2011;108(1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revai K., McCormick D.P., Patel J., Grady J.J., Saeed K., Chonmaitree T. Effect of pneumococcal conjugate vaccine on nasopharyngeal bacterial colonization during acute otitis media. Pediatrics. 2006;117:1823–1829. doi: 10.1542/peds.2005-1983. [DOI] [PubMed] [Google Scholar]
- Rollins D.R., Beuther D.A., Martin R.J. Update on infection and antibiotics in asthma. Curr. Allergy Asthma Rep. 2010;10:67–73. doi: 10.1007/s11882-009-0086-2. [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Alabi A.S., Mombo-Ngoma G., Kaba H., Zoleko R.M., Diop D.A., Mackanga J.R., Basra A., Gonzalez R., Menendez C. Transmission of Staphylococcus aureus between mothers and infants in an African setting. Clin. Microbiol. Infect. 2014;20:O390–O396. doi: 10.1111/1469-0691.12417. [DOI] [PubMed] [Google Scholar]
- Semic-Jusufagic A., Belgrave D., Pickles A., Telcian A.G., Bakhsoliani E., Sykes A., Simpson A., Johnston S.L., Custovic A. Assessing the association of early life antibiotic prescription with asthma exacerbations, impaired antiviral immunity, and genetic variants in 17q21: a population-based birth cohort study. Lancet Respir Med. 2014;2:621–630. doi: 10.1016/S2213-2600(14)70096-7. [DOI] [PubMed] [Google Scholar]
- Sly P.D., Boner A.L., Björksten B., Bush A., Custovic A., Eigenmann P.A., Gern J.E., Gerritsen J., Hamelmann E., Helms P.J. Early identification of atopy in the prediction of persistent asthma in children. Lancet. 2008;372:1100–1106. doi: 10.1016/S0140-6736(08)61451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaniol V., Troller R., Schaller A., Aebi C. Physiologic cold shock of Moraxella catarrhalis affects the expression of genes involved in the iron acquisition, serum resistance and immune evasion. BMC Microbiol. 2011;11:182. doi: 10.1186/1471-2180-11-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R.T., Martinez F.D. Respiratory syncytial virus and asthma: still no final answer. Thorax. 2010;65:1033–1034. doi: 10.1136/thx.2009.133967. [DOI] [PubMed] [Google Scholar]
- Tai A., Tran H., Roberts M., Clarke N., Wilson J., Robertson C.F. The association between childhood asthma and adult chronic obstructive pulmonary disease. Thorax. 2014;69:805–810. doi: 10.1136/thoraxjnl-2013-204815. [DOI] [PubMed] [Google Scholar]
- Tano K., von Essen R., Eriksson P.O., Sjöstedt A. Alloiococcus otitidis—otitis media pathogen or normal bacterial flora? APMIS. 2008;116:785–790. doi: 10.1111/j.1600-0463.2008.01003.x. [DOI] [PubMed] [Google Scholar]
- Verhaegh S.J., Snippe M.L., Levy F., Verbrugh H.A., Jaddoe V.W., Hofman A., Moll H.A., van Belkum A., Hays J.P. Colonization of healthy children by Moraxella catarrhalis is characterized by genotype heterogeneity, virulence gene diversity and co-colonization with Haemophilus influenzae. Microbiology. 2011;157:169–178. doi: 10.1099/mic.0.042929-0. [DOI] [PubMed] [Google Scholar]
- Vissers M., de Groot R., Ferwerda G. Severe viral respiratory infections: are bugs bugging? Mucosal Immunol. 2014;7:227–238. doi: 10.1038/mi.2013.93. [DOI] [PubMed] [Google Scholar]
- Weinstock G.M. Genomic approaches to studying the human microbiota. Nature. 2012;489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Hartert T.V. Evidence for a causal relationship between respiratory syncytial virus infection and asthma. Expert Rev. Anti Infect. Ther. 2011;9:731–745. doi: 10.1586/eri.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Mihindukulasuriya K.A., Gao H., La Rosa P.S., Wylie K.M., Martin J.C., Kota K., Shannon W.D., Mitreva M., Sodergren E., Weinstock G.M. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014;15:R66. doi: 10.1186/gb-2014-15-5-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.