Abstract
Epithelial-to-Mesenchymal Transition (EMT) has been implicated in models of tumor cell migration, invasion and metastasis. In a search for candidate therapeutic targets to reverse this process, non-tumorigenic MCF10A breast epithelial cells were infected with an arrayed lentiviral kinome shRNA library and screened for either suppression or enhancement of a 26-gene EMT RNA signature. No individual kinase gene knockdown was sufficient to induce EMT. In contrast, grouped epithelial markers were induced by knockdown of multiple kinases, including mitogen activated protein kinase 7 (MAPK7). In breast cancer cells, suppression of MAPK7 increased E-cadherin (CDH1) expression and inhibited cell migration. In an orthotopic mouse model, MAPK7 suppression reduced the generation of circulating tumor cells (CTCs) and the appearance of lung metastases. Together, these observations raise the possibility that targeting kinases that maintain mesenchymal cell properties in cancer cells, such as MAPK7, may lessen tumor invasiveness.
Implications
Suppression of MAPK7 induces epithelial markers, reduces generation of circulating tumor cells and appearance of lung metastases.
Keywords: EMT, MET, MAPK7, Circulating Tumor Cells (CTCs), Metastasis
INTRODUCTION
The interconversion of epithelial and mesenchymal properties underlies key elements of embryonic development, including the generation of cellular lineages essential to formation of the kidney, heart valves and neural crest-derived tissues (1, 2). EMT converts stationary epithelial cells into migratory mesenchymal cells, with loss of apicobasal cell polarity and gain of stem-like features and apoptosis resistance (3, 4). Its aberrant activation in cancer has been linked to increased invasiveness into the bloodstream, culminating in the generation of circulating tumor cells (CTCs), which may initiate distant metastases (1). While its role is less well defined in tumorigenesis, the opposite transformation, Mesenchymal-to-Epithelial Transition (MET), has been postulated to play a role in the subsequent reversion of CTCs to a less migratory and more proliferative state at distant metastatic sites (5). Accumulating evidence for the relevance of EMT in human cancer include the finding of increased mesenchymal signatures in CTCs from patients with metastatic breast cancer (6).
Signaling pathways known to initiate or modulate EMT include TGF-β, Wnt, Notch, EGF, HGF, FGF and HIF (2, 7, 8). These cellular signals lead to the activation of master transcriptional regulators, such as Snai1, Snai2, Twist, Zeb1/2 and Lbx1, which directly or indirectly mediate the repression of epithelial genes and the induction of mesenchymal markers (9). The complexities of the initial signaling pathways and the subsequent transcriptional outputs have limited efforts to target EMT therapeutically. However, the identification of key, “druggable” downstream effectors would enable the testing and validation of EMT as a therapeutic target in suppressing cancer metastasis.
The use of shRNA screens to reveal potential therapeutic targets modulating complex cellular processes is well established (10–13). We recently used a pooled whole genome shRNA library screen to identify 31 distinct genes whose knockdown increases cellular migration in MCF10A breast epithelial cells (14). Functional analysis of these hits revealed that they function through a common effector, the Ser/Thr kinase (RSK), whose inhibition abrogates mammalian cell migration (14). To extend this analysis to an even more complex phenotype such as EMT, we established an RNA expression signature of epithelial versus mesenchymal cell fates, which could be quantified in high throughput using a microarrayed format. Using a kinome shRNA library, we identified mitogen-activated protein kinase 7 (MAPK7, also known as ERK5 and BMK1), an effector of RSK, as a modulator of epithelial cell properties. Knockdown of MAPK7 enhances epithelial cell characteristics (i.e., MET) and suppresses the blood-borne metastatic potential of human breast cancer cells in a mouse model.
MATERIALS AND METHODS
Tissue culture and Plasmids
MCF10A cells were cultured as described (15). MCF10A:Myc cells were generated by transduction with a lentiviral c-Myc encoding construct. A549, PANC1 and MDA-MB-231 (4175 TGL variant) cells were grown according to the American Type Culture Collection (ATCC) recommendations. All cell lines were obtained, immediately expanded and frozen at early passages. When thawed for experimental use, they were never passaged more than 4 months in culture. All cell lines were tested for mycoplasma contamination giving negative results. Lentiviral vectors containing WT-MAPK7, DN-MAPK7 or Myr-MAPK7 were kindly provided by Dr. Zhengui Xia (University of Washington, Seattle). For Boyden chamber cell migration assays, cultures were set up when confluency of 50%–75% was reached as described (14).
Primary shRNA screen
The Broad Institute RNAi Consortium (TRC) kinome collection of lentiviral shRNAs (16) was arrayed for testing. Briefly, MCF10A:Myc cells were infected at a multiplicity of infection (MOI) of 0.3 in 96 well plate and selected using 2 ug/mL puromycin for two days, after which cells were trypsinized and split 1:3 into a new 96 well plate. The panomics multiplex quantigene assays were performed after three days, according to the manufacturer’s protocols.
Immunoblot analysis
Cells were harvested in RIPA buffer (Roche). Cell lysates were cleared by centrifugation at 14,000 rpm for 10 min at 4°C. For immunoblotting analysis, lysates were loaded onto 4–15% SDS-PAGE gels (Bio-Rad), and subsequently transferred onto Immobilon PVDF membrane (Millipore). Proteins were visualized with Western Lightning Plus chemiluminescence kit (Perkin Elmer). Antibodies used were CDH1 (610181; BD Biosciences), SERPINE1 (612024; BD Biosciences), Fibronectin (F3648, Sigma), β-actin (ab6276, Abcam), phospho-ERK1/2 (Thr 202/Tyr 204) (9101, Cell Signaling), total-ERK1/2 (4695, Cell Signaling) and MAPK7 (3372, Cell Signaling).
qRT-PCR and knockdown studies
RNA was extracted using RNeasy Minikit (Qiagen) and cDNA synthesis was performed using SuperScript III reverse transcriptase (Invitrogen). qRT-PCR quantitation with Power SYBR Green PCR Master Mix (Applied Biosystems) was applied. The sequences of the PCR primer pairs are listed in Table S1. All samples were done in triplicate and the relative abundance of transcripts was derived by standardizing the input to the control signal, GAPDH. To test for knockdown of target transcripts, cells were plated in 6-well plates and spin-infected the next day with individual lentiviruses at an approximate MOI of 1–3. Puromycin selection (at 2 ug/mL) was added at 2 days, and after another 2 days, cells were re-plated to 6 cm2 dishes; RNA and lysates were collected for the knockdown confirmation.
RNAi Assays
A pool of four siRNAs (smartpool) targeting GAPDH and Snai2 was used along with a nontargeting (control smartpool) siRNA from Dharmacon. RNAi transfection was performed according to manufacturer’s protocol. Knockdown of GAPDH and Snai2 were performed 48 hrs prior to RNA extraction. cDNA was synthesized and qRT-PCR was performed as described above. All samples were done in triplicate, and the relative abundance of transcripts was derived by standardizing the input to the control signal, β-actin.
Immunofluorescence microscopy
Cells were fixed in EM grade 4% formaldehyde and permeabilized with 0.1% Triton X-100 and staining with primary antibodies CDH1 (BD Biosciences) and Vimentin (VIM, Santa Cruz) was carried out overnight at 4°C. Alexa594-conjugated goat-anti-mouse secondary antibody (Molecular Probes) was used for primary mouse monoclonal antibodies and Alexa-488 conjugated goat-anti-rabbit secondary antibody was used for primary rabbit polyclonal antibodies (Molecular Probes). Nuclei were visualized using DAPI.
Tumor Generation and Analyses
NOD scid gamma (NSG) mice were purchased from Jackson Laboratory. Mice were housed in a pathogen-free environment at the Massachusetts General Hospital and were handled in strict accordance with Good Animal Practice as defined by the Office of Laboratory Animal Welfare, and all animal experiments were done with approval from Massachusetts General Hospital Subcommittee on Research Animal Care. Six week old female mice were injected with 106 MDA-MB-231 (4175 TGL) cells containing sh-NT (non-target), sh-MAPK7-1 or sh-MAPK7-2 in 100 ul of 50:50 Matrigel:PBS into the fourth right abdominal fat pad. Tumor growth was monitored externally using calipers for up to 6 weeks and animals were sacrificed when tumors volume reached approximately 2000 mm3. Tumor volume = ((π × length × width2)/6), where length represents the largest tumor diameter and width represents perpendicular tumor diameter. Primary tumors and organs were harvested, paraffin embedded, sectioned and stained with hematoxylin and eosin (H&E). At necropsy, lungs were inflated with 4% paraformaldehyde to facilitate surface tumor counts. For lung lesion quantitation, five step sections (at 50 um) were subjected to H&E staining in order to quantify the number of lung metastatic lesions. Enumeration was done by number of lesions, regardless of size.
Ki67 Staining and Analysis
DF/HCC Research Pathology Cores System was used for Ki67 staining. For Ki67 scanning analysis, three randomly selected high-power (40× objective; Aperio scanscope) were counted (between 500 and 2000 tumor cells).
CTC capture and analysis
HBCTC-Chips were manufactured on site at the MGH Cancer Center/BioMEMS Resource Facility, and chips were functionalized using biotinylated antibodies as previously described (17). Chips functionalized with 50 μg of NeutrAvidin were coated with a cocktail of 10 μg/mL each of biotinylated antibodies against EpCAM, HER2 and EGFR. Antibodies were purchased from the following vendors: EpCAM (R&D Systems BAF960); HER2 (R&D Systems BAF1129); EGFR (Cetuximab, Lilly, biotinylated in house); and control goat IgG antibody (R&D Systems BAF108). Following blood processing, captured cells on the HBCTC-Chip were fixed with 4% paraformaldehyde and washed with PBS. Fixed cells were then permeabilized with 1% NP40 in PBS, blocked with 2% goat serum/3% BSA, and immunostained with anti-GFP (ab13970, Abcam). Secondary immunofluorescence tagged antibody was used for signal amplification. The secondary antibody was Goat Anti-Chicken (H+L) (A-11039, Molecular Probes). Nuclei were then stained with DAPI and the devices were washed with PBS and stored at 4°C. The devices were imaged under 20× magnification using the BioView Ltd. automated imaging system (Billerica, MA) as well as an automated upright fluorescence microscope (Eclipse 90i, Nikon, Melville, NY). Positive staining for GFP was required for scoring potential CTCs, which were then manually reviewed.
RNA-in situ hybridization
For quantiGene ViewRNA in-situ hybridization (ISH) on FFPE-tissue, a dual-colorimetric RNA-in situ hybridization (RNA-ISH) branched chain assay (Affymetrix Quantigene, Santa Clara, CA) was used to quantify multiple transcripts. 5-micron sections were fixed in 10% formaldehyde (Fisher Scientific, Pittsburgh, PA), deparaffinized, boiled in pre-treatment solution (Affymetrix, Santa Clara, CA) and digested with proteinase K. Sections were hybridized for 3 hours at 40°C with a cocktail of custom designed QuantiGene ViewRNA probes against epithelial (CDH1, EpCAM, KRT5, KRT7, KRT8, KRT18, KRT19; type 1 probes) and mesenchymal markers (FN1, CDH2, SERPINE1; type 6 probes) (Affymetrix, Santa Clara, CA). Bound probes were then amplified per protocol from Affymetrix using PreAmp and Amp molecules. Multiple Label Probe oligonucleotides conjugated to alkaline phosphatase (LP-AP Type 1) were then added and Fast Red Substrate was used to produce signal (red dots). For two color assays, an LP-AP type 6 probe was used with Fast Blue substrate (blue dots) followed by LP-AP type 1 probe with Fast Red Substrate (red dots) to produce dual colorimetric signals. Slides were then counterstained with Hematoxylin. Images were scanned by Aperio scanscope.
ISH Scoring
For ISH scoring, both mesenchymal and epithelial components of the tumor were scored separately for intensity of the staining: staining observed on 2X magnification is scored as 3, on 20X magnification is scored as 2, on 40X magnification is scored as 1 and no staining even on 40X magnification is scored as 0 (Table S2).
Statistical and Bioinformatic Analyses
The computation of the FDR estimates used to construct Figure 1B and Figure S4 contained the following steps: (Step 1) Model and correct for the dependence of each marker on the housekeeping genes, (Step 2) Correct for inter-plate variability, (Step 3) Combine all the hairpins for each gene, (Step 4) Assess statistical significance. The steps are described in detail in the supplemental methods.
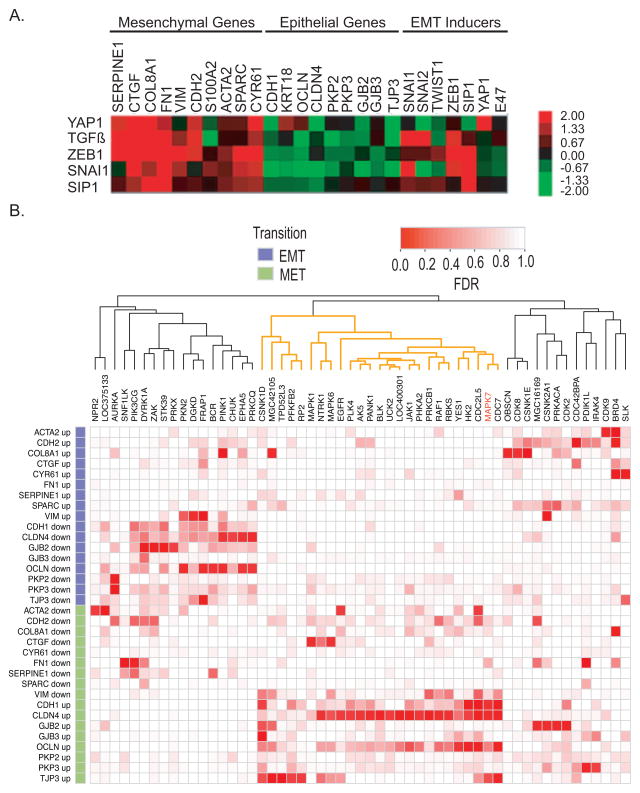
Figure 1. Validation of shRNA kinome screen.
(A) Relative expression levels of EMT signature in response to EMT inducers: YAP1, TGF-β, Zeb1, Snai1 and SIP1 in MCF10A:Myc cells.
(B) Heat map analysis of the arrayed kinome shRNA screening using the EMT gene-set signature. Each row corresponds to a gene and a direction (EMT or MET). Each column corresponds to a gene that was knocked down. Red corresponds to a low False Discovery Rate (FDR) and white to a high FDR using midRSA methodology. RNAi for 752 genes was administered, but only the genes with FDR < 25% for at least one marker and direction are included in the matrix. The columns of the matrix were clustered based on correlation of the FDR values. For details, please see supplemental methods.
RESULTS
Arrayed kinome shRNA screening using an EMT gene expression signature
We defined and optimized an EMT expression signature for microarrayed screening using a highly sensitive and quantitative RNA-based assay (Panomic quantigene, Affymetrix, Santa Clara, CA). Candidate EMT markers were selected from microarray-based expression profiling of immortalized, non-transformed MCF10A human mammary epithelial cells, following ectopic expression of the known EMT inducers Snai1 and Twist (18, 19). We selected a total of 26 markers, including epithelial markers (N=9), mesenchymal markers (N=10) and known regulators of EMT (N=7) (Table 1).
Table 1.
The list of genes utilized for EMT gene-set signature
| Mesenchymal genes | Epithelial genes | EMT inducers | Housekeeping genes |
|---|---|---|---|
| SERPINE1 | CDH1 | SNAI1 | GAPDH |
| CTGF | KRT18 | SNAI2 | RPS20 |
| COL8A1 | OCLN | TWIST1 | RPLPO |
| FN1 | CLDN4 | ZEB1 | PPIB |
| VIM | PKP2 | SIP1 | |
| CDH2 | PKP3 | YAP1 | |
| S100A2 | GJB2 | E47 | |
| ACTA2 | GJB3 | ||
| SPARC | TJP3 | ||
| CYR61 |
MCF10A cells can undergo further specification in either epithelial or mesenchymal direction, making them ideal for manipulations that either enhance or suppress their epithelial properties (20–23). However, these cells spontaneously assume mesenchymal features when grown at low cell density, a characteristic that prohibits a large-scale microarrayed screen (24, 25). Ectopic expression of c-Myc in MCF10A cells is known to suppress this cell density-dependent effect, and we therefore adapted Myc-expressing variants (MCF10A:Myc cells) for the screen (Figure S1A) (26, 27). These cells remain sensitive to EMT-inducing stimuli, including overexpression of Snai1, YAP, SIP1 or Zeb1 genes, as well as treatment with TGF-β (Figure 1A).
The outline of the kinome shRNA screen, performed using an arrayed 96-well plate format (constructs from the RNAi Consortium, Broad Institute), is illustrated in Figure S1B. Since full EMT phenotypes may only become evident after a few days, cells were trypsinized after infection and reseeded to allow continued proliferation. Each plate included negative and positive controls; cells infected with the EMT-inducer YAP are shown, demonstrating robust EMT as measured using the 26 gene-set signature (Figure 1A). The Panomics quantigene assay allowed simultaneous detection and quantitation of the effect of individual RNA hairpins on all the markers in our EMT signature.
We observed that the effect of cell density on EMT was greatly reduced but not abolished by constitutive expression of c-Myc. We therefore established analytic parameters to exclude the effect of cell density on EMT measurements by calculating the cell density within each well from the absolute amount of RNA for four housekeeping genes (GAPDH, RPS10, RPLP0 and PPIB), which were used to standardize the expression of epithelial and mesenchymal markers (see Figures S2 and S3A for details).
For each kinase gene, 5–10 hairpins were tested individually within the microarrayed format. Two analytic approaches, the midRSA method (28) (Figure 1B) and the Wilcoxon test (Figure S4) were used to assess statistical significance (for details, see supplemental methods). Both approaches generated a cluster of candidate kinase genes whose knockdown directed cells toward an epithelial (MET) fate, but none that enhanced a mesenchymal (EMT) phenotype (Figure 1B, Figure S4). By analyzing individual markers, seven candidate kinase genes whose knockdown triggered increased epithelial gene expression were identified by both midRSA and Wilcoxon tests (MAPK7, CDC7, CDC2L5, CSNK1D, MGC42105, NTRK1 and TPD52L3). Using the mean of pooled MET and EMT markers and a false discovery rate (FDR) threshold of 25%, midRSA analysis identified MAPK7, PDIK1L, EGFR and PAK2 as candidate genes, whereas the Wilcoxon test identified MAPK7, CDC7, CDC2L5, EGFR, PAK2 and LOC402289. Thus, MAPK7 scored consistently as a regulator of MET in both midRSA and Wilcoxon tests, and using both individual and combined marker analysis, while a small set of other kinases were identified by the different analytic methods. Given its known association with an adverse prognosis in cancers of the brain, breast, colon and lymphomas (29–32), we selected MAPK7 for more detailed functional analyses.
Knockdown of MAPK7 induces expression of epithelial markers
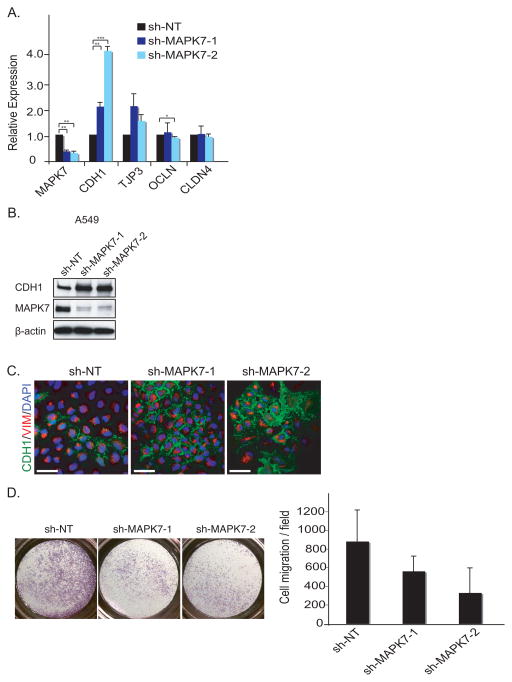
To begin to define its effects on tumorigenic properties, we first validated MAPK7 as a regulator of mesenchymal and epithelial gene expression across different cancer cell lines, selecting cells that like MCF10A cells are in an intermediate state between the two extremes (Figure 2A, Figure S5A). In A549 lung cancer cells (Figure 2A–B) as in MDA-MB-231 breast cancer cells (Figure S5B) and PANC1 pancreatic cancer cells (Figure S5B), knockdown of MAPK7, using two independent shRNA constructs, resulted in increased expression of the epithelial marker CDH1 and a reduction in the mesenchymal marker SERPINE1 (Figure 2B, Figure S5B). The increased expression of CDH1 in A549 cells was accompanied by its relocalization to the cell membrane, consistent with an epithelial phenotype (Figure 2C). Whereas in vitro proliferation of these cells was not affected by MAPK7 knockdown (data not shown), their migration in a Boyden chamber assay was suppressed as these cells shifted from a mesenchymal to a more epithelial phenotype (Figure 2D). Taken together, these observations suggest that MAPK7 regulates cellular mesenchymal features at baseline in these cancer cells, with its knockdown resulting in increased epithelial properties.
Figure 2. Knockdown of MAPK7 induces Mesenchymal-Epithelial Transition (MET).
(A) MET is shown by qRT-PCR upon knockdown of shRNA against non-target (sh-NT) and two independent shRNAs against MAPK7 (sh-MAPK7-1 and sh-MAPK7-2) in A549 cells. Error bars represent SD from three individual experiments. * represents p-value < .05, ** represents p-value < .01 and *** represents p-value < .001.
(B) Knockdown of MAPK7 induces MET. Knockdown of MAPK7 in A549 cells increase CDH1 expression as demonstrated by immunoblot analysis. β-actin is used as loading control.
(C) Knockdown of MAPK7 induces CDH1 expression in A549 cells, demonstrated by immunofluorescence staining. Green = CDH1, red = VIM (Vimentin) and blue = dapi. Scale bar (white) = 50 μm.
(D) Knockdown of MAPK7 in A549 cells reduces cell migration as assayed using a Boyden Chamber. Quantitation of cell migration is derived from five independent fields. Error bars represent SD from three independent experiments.
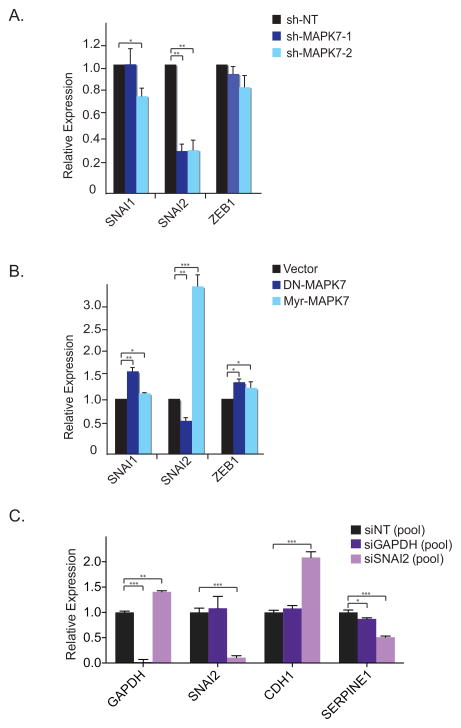
To begin to define potential mechanisms underlying the induction of CDH1 following suppression of MAPK7, we measured the expression of transcriptional repressors of CDH1 that have been implicated as major regulators of EMT. Knockdown of MAPK7 in A549 cells led to markedly reduced expression of Snai2, whereas Snai1 and Zeb1 levels were unaltered (Figure 3A). Lenti-viral transduction of a well-characterized dominant-negative MAPK7 mutant (33) produced similar results (Figure 3B). Moreover, expression of a constitutively activated, myristylated form of MAPK7 (33) dramatically increased Snai2 expression (Figure 3B). siRNA-mediated knockdown of Snai2 itself in A549 cells increased expression of the epithelial marker CDH1 and decreased levels of the mesenchymal marker SERPINE1 (Figure 3C). Thus, baseline expression of MAPK7 in A549 cells regulates Snai2, a well characterized master transcriptional regulator of EMT (34), whose own knockdown phenocopies that of MAPK7 in inducing epithelial cell properties.
Figure 3. MAPK7 activates Snai2.
(A) Knockdown of MAPK7 in A549 cells decreases Snai2 expression whereas it has no effect on Snai1 and Zeb1 expression by qRT-PCR. Error bars represent SD from three individual experiments. * represents p-value < .05 and ** represents p-value < .01.
(B) Constitutive active MAPK7 (Myr-MAPK7) increases Snai2 expression, whereas a dominant negative MAPK7 mutant (DN-MAPK7) reduces Snai2 expression in A549 cells as shown by qRT-PCR. Error bars represent SD from three individual experiments. * represents p-value < .05, ** represents p-value < .01 and *** represents p-value < .001.
(C) Smartpool RNAi knockdown (Dharmacon) of Snai2 decreases CDH1 and increases SERPINE1 expression in A549 cells by qRT-PCR. GAPDH serves as knockdown control. Error bars represent SD from three individual experiments. * represents p-value < .05, ** represents p-value < .01 and *** represents p-value < .001.
Suppression of MAPK7 reduces CTCs and number of metastases
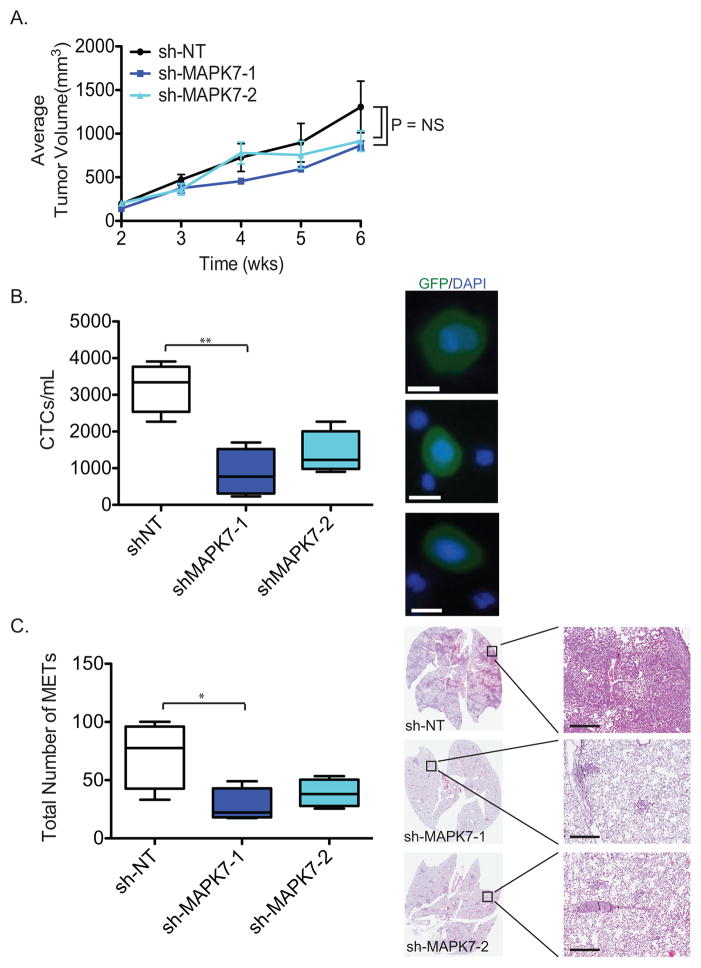
To quantify the effects of MAPK7 knockdown on metastasis, we made use of the highly tumorigenic and metastasis-prone human breast cancer cell line MDA-MB-231 (4175 TGL variant) (35). This triple negative cell line (lacking expression of the estrogen receptor (ER), progesterone receptor (PR) and HER2) has a mesenchymal phenotype, but knockdown of MAPK7 in these cells induces epithelial marker CDH1 (Figure S5B). MAPK7 knockdown does not affect the in vitro proliferation in MDA-MB-231 cells (data not shown). GFP- and luciferase-tagged cells were orthotopically injected into the mammary fat pad of immunosuppressed NSG mice (106 cells/injection), all of which generated tumors within 6 weeks. Suppression of MAPK7 in MDA-MB-231 cells, using two different shRNA constructs (MAPK7-1 and MAPK7-2), did not reduce the size of the primary tumors, compared with non-target (NT) controls (Figure 4A, Figure S6A). At six weeks following orthotopic tumor initiation, mice were sacrificed for quantitation of both CTCs and lung metastasis burden.
Figure 4. Knockdown of MAPK7 decreases circulating tumor cells (CTCs) and lung metastasis.
(A) Knockdown of MAPK7 in MDA-MB-231 (4175 TGL variant) cells does not affect tumor volume in mice. Quantitation of tumor volume (mm3) is shown. Tumor volume data are represented as the mean of 4 tumors in 4 mice for each group (+/− SEM).
(B) Quantitation of GFP+ CTCs from mice bearing sh-MAPK7-1 (N=4) and sh-MAPK7-2 (N=4) knockdown compared to control sh-NT (N=4) in MDA-MB-231 (4175 TGL) cells. Error bars represent SEM. * represents p-value < .05, ** represents p-value < .01 and *** represents p-value < .001. Representative images of CTCs are shown (blue = dapi, green = GFP). Scale bar (white) = 10 μm.
(C) Quantitation of total number of lung metastases (METs) stained by H&E from mice (N=4 for sh-NT, sh-MAPK7-1 and sh-MAPK7-2) bearing primary tumors with either sh-MAPK7-1 or sh-MAPK7-2 knockdown, compared to control in MDA-MB-231 (4175 TGL) cells. Lung metastasis as depicted by H&E staining was enumerated based on number of lung lesions regardless of size. Five step sections (50 microns each) were subjected to H&E staining in order to quantify the number of lung metastatic lesions. Error bars represent SEM. * represents p-value < .05. Representative images of H&E-stained lung metastases are shown from mice for sh-NT, sh-MAPK7-1 or sh-MAPK7-2. Scale bar (black) = 300 μm.
Following blood collection through intracardiac puncture, specimens were processed through a microfluidic device (HBCTC-Chip) whose walls are functionalized with capture antibodies against EpCAM, EGFR and HER2. We have previously shown that the combination of these epithelial and lineage-based antibodies is highly effective in capturing MDA-MB-231 cells spiked into blood specimens (6, 17). Following in-line cell fixation and GFP-staining within the microfluidic device, the number of captured CTCs was scored using multi-Z plane automated fluorescence microscopy (17). Control tumor-bearing mice had a median 3,343 ± 481 CTCs/mL, compared with only 693 ± 331 CTCs/mL for mice whose tumor expressed the sh-MAPK7-1 construct (79% reduction, P=0.006) and 1,228 ± 350 CTCs/mL for those expressing sh-MAPK7-2 (63% reduction, P=0.054) (Figure 4B). Thus, suppression of MAPK7 significantly reduced the number of CTCs generated from primary orthotopic mammary tumors of comparable size. The MDA-MB-231 (4175 TGL variant) cell line has a high rate of lung metastases (35), and indeed, histological analysis of control mice demonstrated abundant lesions (78; median: lesions/50 microns lung sections. For details, see methods) (Figure 4C). Consistent with the reduction in CTCs, histological analysis of lungs from mice whose tumor expressed sh-MAPK7-1 had only 22 metastatic lesions (median: lesions/50 microns lung sections) (71% reduction, P=0.03), while sh-MAPK7-2 had 38 lung lesions (median: lesions/50 microns lung sections) (51% reduction, P=0.075) (Figure 4C).
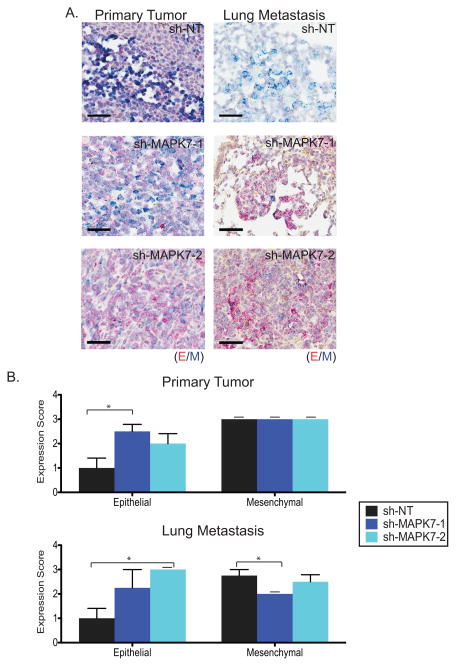
Finally, to correlate the reduction in CTCs and metastatic lesions with epithelial/mesenchymal features of the cancer cells, we used a dual-colorimetric RNA-in situ hybridization (ISH) branched chain assay (Affymetrix Quantigene, Santa Clara, CA) to quantify multiple transcripts in these cells (6). We measured expression of seven pooled epithelial (E) transcripts [keratins (KRT): 5, 7, 8, 18 and 19; EpCAM and CDH1 (cadherin 1)] versus three mesenchymal (M) transcripts [FN1 (fibronectin 1), CDH2 (cadherin 2) and PAI-1 (serpine 1)] by enzyme-colorimetric-based quantitation (6). Increased numbers of E+ cells were detected in primary tumors of mice bearing the MAPK7 knockdown constructs, with a 250% increased score for sh-MAPK7-1-expressing cells (P=0.02), and a 200% increase for sh-MAPK7-2 (P=0.13) (Figure 5A-B, Table S2, see RNA-ISH analysis and scoring in methods). The primary tumor and lung metastases had a comparable epithelial score in control mice, and a similar increase in epithelial transcript expression was evident within metastatic lesions of mice whose tumors harbor sh-MAPK7-1 (225% increased score, P=0.19) or sh-MAPK7-2 (300% increased score, P=0.03) (Figure 5A–B, Table S2). Taken all together, these observations suggest that suppression of MAPK7 in primary tumors derived from a highly aggressive mesenchymal breast cancer model reduces intravascular invasion, leading to fewer CTCs and metastatic lesions, an effect that is correlated with the induction of epithelial features. While knockdown of this MAPK pathway effector may affect tumor cell proliferation in addition to their invasive properties, Ki67 staining of primary tumor cells did not show a difference in proliferative index for tumors with MAPK7 knockdown compared with control (Figure S6B–C).
Figure 5. Knockdown of MAPK7 increases epithelial marker expression in primary tumors and lung metastases.
(A) Representative RNA-ISH images of pooled epithelial (E) (red dots) and mesenchymal (M) (blue dots) markers in primary tumors and lung metastases derived from breast tumor MDA-MB-231 (4175 TGL) cells expressing either sh-NT (N=4), sh-MAPK7-1 (N=4) or sh-MAPK7-2 (N=4). Scale bar (black) = 50 μm. (B) Quantitation of RNA-ISH using pooled E and M markers applied to primary tumors and lung metastases generated from MDA-MB-231 (4175 TGL) cells expressing either sh-NT (N=4), sh-MAPK7-1 (N=4) or sh-MAPK7-2 (N=4). Error bars represent SEM (if dashed line above bar, SEM=0). (Table S2, please see methods for more detail). * represents p-value < .05.
DISCUSSION
The suppression of metastasis remains one of the most important and poorly addressed challenges in cancer therapy. Here, we sought to tackle the interconversion between epithelial and mesenchymal cell fates, which is thought to constitute a critical component of epithelial cancer invasion and metastasis. We used an arrayed kinome shRNA library to interrogate non-transformed MCF10A cells for expression of an EMT RNA signature, and confirmed hits using tumorigenic cancer cell lines. We did not uncover an individual kinase whose knockdown was sufficient to trigger EMT in MCF10A cells, suggesting that the maintenance of epithelial phenotypes may be dependent upon multiple pathways. In contrast, the individual knockdown of several kinases resulted in an increased epithelial phenotype. Thus, baseline mesenchymal properties of MCF10A cells appear to be maintained by the activity of these kinases, whose suppression leads to enhanced epithelial gene expression.
Our kinome shRNA screen relied on an arrayed format, with 5 to 10 shRNA constructs per gene, and on the high infectivity of MCF10A cells by lentiviral constructs. MCF10A cells also express both epithelial and mesenchymal markers, making it possible in one screen to test for changes in either direction. We used simple selection criteria, namely EMT versus MET scoring using a 26 gene-set signature, but the analysis of individual markers allowed us to resolve partial MET phenotypes. Among the regulators of MET, we selected one candidate, MAPK7, as a proof of principle, although additional targets identified in this screen may warrant additional investigation. MAPK7 knockdown induced epithelial phenotypes in multiple cancer cell lines, an effect that was correlated in A549 cells with its regulation of the EMT master regulator Snai2. Knockdown of MAPK7 in highly invasive mesenchymal breast cancer cells resulted in increased expression of epithelial cell markers, without affecting cell proliferation in vitro or in vivo. Significantly, it also led to a marked reduction in the generation of CTCs and in the formation of lung metastases.
MAPK7 is known to be activated by stress and growth factor signaling, in part through its upstream mitogen-activated kinase kinase 5 (MEK5) (36). In turn, MAPK7 has been implicated in diverse phenotypes, ranging from resistance to TNF-α (37) and regulation of estrogen receptor expression (38) to migration of epithelial cells in response to Hepatocyte Growth Factor (HGF) and of keratinocytes during wound healing (31, 39, 40). Our observation that MAPK7 mediates inter-conversion between epithelial to mesenchymal phenotypes is consistent with previous reports, which have also linked these effects to its regulation of expression of EMT master regulators (37, 38, 40). Whereas in A549 cells, suppression of MAPK7 induces expression of Snai2, but not other transcriptional regulators of EMT, MAPK7 may also regulate Zeb1 in some other cell types (37).
The role of MAPK signaling, including MAPK7 and its upstream kinases, in enhancing cell proliferation, angiogenesis, invasion and metastasis is well established (32, 36, 41). In mouse models, MAPK7 modulates tumor growth and metastasis by prostate and breast cancer cell lines (31, 38, 42), while clinical studies indicate that deregulated MAPK7 is associated with metastatic risk in prostate, breast and oral cancers (30, 31, 43). Our results extend these observations by showing that even in highly invasive mesenchymal and metastasis-prone breast cancer cells, such as the 4175 TGL variant of MDA-MB-231 cells, knockdown of MAPK7 is capable of increasing tumor epithelialization and suppress intravascular invasion, as measured by the reduced generation of CTCs.
The potentially opposing roles of EMT and MET in tumor invasiveness and CTC generation versus the proliferative potential of metastatic lesions is of considerable interest (4, 44–47). Mesenchymal cells are more migratory and invasive, but have a reduced proliferative rate. They may thus be enriched among tumor cells migrating into the bloodstream, but have a growth disadvantage once distant metastases are established. Indeed in a mouse model of K-Ras-driven skin cancer, transient induction of Twist through topical administration of tamoxifen initiates limited EMT and enhances metastasis, whereas stable expression of Twist facilitates tumor cell dissemination but suppresses proliferation of distant metastases (48). While the risks and benefits of targeting EMT regulators in human cancer remain the subject of debate, the apparent role of MAPK-dependent signals in mediating both EMT and cellular proliferation may present a therapeutic opportunity. In the mouse model described here, suppression of MAPK7 resulted in a reduction in tumor invasiveness, without a corresponding increase in proliferation rate or tumor size by the more epithelial cancer cells. If confirmed in additional tumor models, these observations suggest that MAPK7 may constitute a therapeutic target to modulate cancer-associated mesenchymal transformations.
Supplementary Material
Acknowledgments
We thank Dr. Zhengui Xia (University of Washington, Seattle) for kindly sharing with us the WT-MAPK7, DN-MAPK7 and Myr-MAPK7 plasmids. We thank Laura Libby for expert technical support and members of the Haber, Maheswaran and Ramaswamy laboratories for helpful discussions. This work was supported by the Howard Hughes Medical Institute (D.A.H.), the NIH (grant CA-129933 to D.A.H., NIBIB-EB008047) and the National Foundation for Cancer Research (NFCR).
Footnotes
Authors’ Contribution: S.J., J.Z., G.A.S., M.Y. and D.A.H. conceived and designed the study. S.J., J.Z., G.A.S., M.Y., A.S., K.S.A., M.W.M., R.D., M.J.Z. and B.J.S. conducted experiments; B.S.W. performed statistical analysis; B.S.W., S.R., D.T.T., S.L.S. and T.S. developed analytical and technical tools; S.J., J.Z., G.A.S., M.Y., B.S.W., M.T., S.M. and D.A.H analyzed and interpreted data and wrote the manuscript. All authors reviewed and revised the manuscript.
Competing Financial Interests Statement: The authors declare that actual or potential competing financial interests do not exist.
References
- 1.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial- mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 3.Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–76. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 4.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunasinghe NP, Wells A, Thompson EW, Hugo HJ. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev. 2012;31:469–78. doi: 10.1007/s10555-012-9377-5. [DOI] [PubMed] [Google Scholar]
- 6.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheel C, Weinberg RA. Phenotypic plasticity and epithelial-mesenchymal transitions in cancer and normal stem cells? Int J Cancer. 2011;129:2310–4. doi: 10.1002/ijc.26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 10.Westbrook TF, Stegmeier F, Elledge SJ. Dissecting cancer pathways and vulnerabilities with RNAi. Cold Spring Harb Symp Quant Biol. 2005;70:435–44. doi: 10.1101/sqb.2005.70.031. [DOI] [PubMed] [Google Scholar]
- 11.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–48. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–64. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smolen GA, Zhang J, Zubrowski MJ, Edelman EJ, Luo B, Yu M, et al. A genome-wide RNAi screen identifies multiple RSK-dependent regulators of cell migration. Genes Dev. 2010;24:2654–65. doi: 10.1101/gad.1989110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 16.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 17.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–7. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim S, Becker A, Zimmer A, Lu J, Buettner R, Kirfel J. SNAI1-mediated epithelial-mesenchymal transition confers chemoresistance and cellular plasticity by regulating genes involved in cell death and stem cell maintenance. PLoS One. 2013;8:e66558. doi: 10.1371/journal.pone.0066558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–84. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 21.Gordon LA, Mulligan KT, Maxwell-Jones H, Adams M, Walker RA, Jones JL. Breast cell invasive potential relates to the myoepithelial phenotype. Int J Cancer. 2003;106:8–16. doi: 10.1002/ijc.11172. [DOI] [PubMed] [Google Scholar]
- 22.Lombaerts M, van Wezel T, Philippo K, Dierssen JW, Zimmerman RM, Oosting J, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94:661–71. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda M, Johnson KR, Wheelock MJ. Cadherin switching: essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J Cell Sci. 2005;118:873–87. doi: 10.1242/jcs.01634. [DOI] [PubMed] [Google Scholar]
- 25.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–97. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 26.Nair R, Roden DL, Teo WS, McFarland A, Junankar S, Ye S, et al. c-Myc and Her2 cooperate to drive a stem-like phenotype with poor prognosis in breast cancer. Oncogene. 2013 doi: 10.1038/onc.2013.368. [DOI] [PubMed] [Google Scholar]
- 27.Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, Tanner SM, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–45. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 28.Konig R, Chiang CY, Tu BP, Yan SF, DeJesus PD, Romero A, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–9. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 29.Carvajal-Vergara X, Tabera S, Montero JC, Esparis-Ogando A, Lopez-Perez R, Mateo G, et al. Multifunctional role of Erk5 in multiple myeloma. Blood. 2005;105:4492–9. doi: 10.1182/blood-2004-08-2985. [DOI] [PubMed] [Google Scholar]
- 30.Montero JC, Ocana A, Abad M, Ortiz-Ruiz MJ, Pandiella A, Esparis-Ogando A. Expression of Erk5 in early stage breast cancer and association with disease free survival identifies this kinase as a potential therapeutic target. PLoS One. 2009;4:e5565. doi: 10.1371/journal.pone.0005565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsay AK, McCracken SR, Soofi M, Fleming J, Yu AX, Ahmad I, et al. ERK5 signalling in prostate cancer promotes an invasive phenotype. Br J Cancer. 2011;104:664–72. doi: 10.1038/sj.bjc.6606062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whyte J, Bergin O, Bianchi A, McNally S, Martin F. Key signalling nodes in mammary gland development and cancer. Mitogen-activated protein kinase signalling in experimental models of breast cancer progression and in mammary gland development. Breast Cancer Res. 2009;11:209. doi: 10.1186/bcr2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Cundiff P, Abel G, Wang Y, Faigle R, Sakagami H, et al. Extracellular signal-regulated kinase (ERK) 5 is necessary and sufficient to specify cortical neuronal fate. Proc Natl Acad Sci U S A. 2006;103:9697–702. doi: 10.1073/pnas.0603373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–61. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 35.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker F, Kato A, Gonez LJ, Hibbs ML, Pouliot N, Levitzki A, et al. Activation of the Ras/mitogen-activated protein kinase pathway by kinase-defective epidermal growth factor receptors results in cell survival but not proliferation. Mol Cell Biol. 1998;18:7192–204. doi: 10.1128/mcb.18.12.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou C, Nitschke AM, Xiong W, Zhang Q, Tang Y, Bloch M, et al. Proteomic analysis of tumor necrosis factor-alpha resistant human breast cancer cells reveals a MEK5/Erk5-mediated epithelial-mesenchymal transition phenotype. Breast Cancer Res. 2008;10:R105. doi: 10.1186/bcr2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antoon JW, Martin EC, Lai R, Salvo VA, Tang Y, Nitzchke AM, et al. MEK5/ERK5 signaling suppresses estrogen receptor expression and promotes hormone-independent tumorigenesis. PLoS One. 2013;8:e69291. doi: 10.1371/journal.pone.0069291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castro NE, Lange CA. Breast tumor kinase and extracellular signal-regulated kinase 5 mediate Met receptor signaling to cell migration in breast cancer cells. Breast Cancer Res. 2010;12:R60. doi: 10.1186/bcr2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnoux V, Nassour M, L’Helgoualc’h A, Hipskind RA, Savagner P. Erk5 controls Slug expression and keratinocyte activation during wound healing. Mol Biol Cell. 2008;19:4738–49. doi: 10.1091/mbc.E07-10-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res. 2006;66:3903–11. doi: 10.1158/0008-5472.CAN-05-4363. [DOI] [PubMed] [Google Scholar]
- 42.Cronan MR, Nakamura K, Johnson NL, Granger DA, Cuevas BD, Wang JG, et al. Defining MAP3 kinases required for MDA-MB-231 cell tumor growth and metastasis. Oncogene. 2012;31:3889–900. doi: 10.1038/onc.2011.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sticht C, Freier K, Knopfle K, Flechtenmacher C, Pungs S, Hofele C, et al. Activation of MAP kinase signaling through ERK5 but not ERK1 expression is associated with lymph node metastases in oral squamous cell carcinoma (OSCC) Neoplasia. 2008;10:462–70. doi: 10.1593/neo.08164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer. 2012;12:425–36. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 45.Ledford H. Cancer theory faces doubts. Nature. 2011;472:273. doi: 10.1038/472273a. [DOI] [PubMed] [Google Scholar]
- 46.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–40. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 47.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 48.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–36. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.