SUMMARY
Like many dsDNA viruses, tumor gammaherpesviruses Epstein-Barr virus and Kaposi’s sarcoma-associated-herpesvirus withstand high internal pressure. Bacteriophage HK97 uses covalent chainmail for this purpose, but how this is achieved non-covalently in the much larger gammaherpesvirus capsid is unknown. Our cryoEM structure of a gammaherpesvirus capsid reveals a hierarchy of four levels of organization: (1) Within a hexon capsomer, each monomer of the major capsid protein (MCP), 1378 amino acids and six domains, interacts with its neighboring MCPs at four sites. (2) Neighboring capsomers are linked in pairs by MCP dimerization domains and in groups of three by heterotrimeric triplex proteins. (3) Small (~280 amino acids) HK97-like domains in MCP monomers alternate with triplex heterotrimers to form a belt that encircles each capsomer. (4) 162 belts concatenate to form non-covalent chainmail. The triplex heterotrimer orchestrates all four levels and likely drives maturation to an angular capsid that can withstand pressurization.
Keywords: gammaherpesvirus, cryo electron microscopy, virus, structure, Rhesus monkey rhadinovirus, non-covalent chainmail
INSTRODUCTION
Gammaherpesviruses constitute a group of double-stranded DNA (dsDNA) tumor viruses that collectively form one of the three subfamilies of the Herpesviridae family (Roizman et al., 2007). These viruses are of significant medical relevance, and the two known human gammaherpesviruses, Kaposi’s sarcoma associated herpesvirus (KSHV) and Epstein-Barr virus (EBV), are associated with lymphomas and other malignancies (Chang et al., 1994; Ganem, 2007; Rickinson and Kieff, 2007). Alphaherpesvirus and betaherpesvirus subfamilies include the well-studied herpes simplex virus type 1 (HSV-1) (Roizman et al., 2007) and human cytomegalovirus (HCMV) (Mocarski et al., 2007), respectively. We previously showed at ~25Å resolution that the overall structural arrangement of the capsid of rhesus monkey rhadinovirus (RRV), a model gammaherpesvirus (O'Connor and Kedes, 2007; Orzechowska et al., 2008), is similar to those of alphaherpesviruses and betaherpesviruses (Yu et al., 2003), even though the protein sequence identities across these subfamilies are only ~20%.
Herpesviruses are highly complex, with a dsDNA genome of about 200 kilobases, encoding about 100 genes (Roizman et al., 2007). To house this much dsDNA, herpesvirus capsids are very large, ~1300Å in diameter. Due to the limited depth of focus in current electron microscopes, this large diameter presents a technical challenge to achievement of high resolution structure by cryo electron microscopy (cryoEM) (Leong et al., 2010; Zhang and Zhou, 2011). Indeed, despite recent progress in near-atomic resolution structural studies of smaller viruses (e.g., Jiang et al., 2008; Liu et al., 2010; Veesler et al., 2013; Yu et al., 2008; Zhang et al., 2010; Zhang et al., 2008), the highest resolution structure obtained thus far among herpesviruses is about 9Å for the capsid of HSV-1, an alphaherpesvirus (Zhou et al., 2000). For human gammaherpesvirus capsids, poor sample quantity and quality have further hindered progress (Germi et al., 2012; Wu et al., 2000). Fortunately, RRV provides an excellent source of gammaherpesviruses, as it grows to high titer in rhesus fibroblasts and allows for purification to obtain uniform capsids (Chang et al., 1994; Desrosiers et al., 1997; O'Connor et al., 2003).
The icosahedral capsid shell of herpesviruses is comprised of four abundant proteins (Newcomb et al., 1993; Rixon, 1993; Trus et al., 1995; Zhou et al., 1995). In gammaherpesviruses, these proteins are (1) the major capsid protein (MCP/ORF25), the monomeric subunit of both hexon and penton capsomers, (2) the triplex monomer protein (TRI-1/ORF62), (3) the triplex dimer protein (TRI-2/ORF26), and (4) the small capsomer interacting protein (SCIP/ORF65) (O'Connor et al., 2003; Yu et al., 2003). The limited resolution of the capsid structures of all herpesviruses has hindered our understanding of the molecular interactions among these proteins. These interactions are essential for capsid assembly, in particular for orchestration of the extensive conformational changes required for capsid maturation from the immature spherical shape to the mature angular shape (Newcomb et al., 2000). Bacteriophage HK97, also a double-stranded DNA virus that must withstand very high internal pressure, up to 50 atmospheres (Gelbart and Knobler, 2009), with just one capsid protein of 280 amino acid (a.a.) residues in the mature virus, also undergoes extensive conformational changes, leading to a chainmail topology in its capsid upon maturation (Bamford et al., 2005; Wikoff et al., 2000). The special fold that enables HK97 to build chainmail, the ‘Johnson’ fold, is found in the capsid protein of other dsDNA bacteriophages (Dai et al., 2010; Jiang et al., 2006) and is also evident in the floor region of the MCP in HSV-1 (Baker et al., 2005). However, how this ancient fold and the chainmail strategy are adapted in the assembly and maturation of the much more complex mammalian herpesvirus capsid remains a puzzle in the absence of knowledge about the folds of all capsid proteins at the secondary structure level.
Here, we report a 7.2 Å structure of the RRV capsid by cryoEM and single particle reconstruction. The 1378 a.a. long MCP of RRV is organized into six domains that include one forming a fold homologous to the Johnson fold and another interacting with SCIP. Each triplex heterotrimer is comprised of a TRI-1 monomer with three domains and a dimer of TRI-2 subunits, each with three domains. The structure also reveals how the MCP and triplex domains (1) interact to bind MCP subunits within a capsomer, (2) bind capsomers to each other, (3) form non-covalently linked belts of capsomers that encircle each individual capsomer, and (4) concatenate the belts to form non-covalent chainmail. Two distinct conformations of the Johnson fold are identified, suggesting a role of conformational switching in orchestrating the global architectural changes required for herpesvirus capsid maturation.
RESULTS
Molecular architecture of the RRV capsid
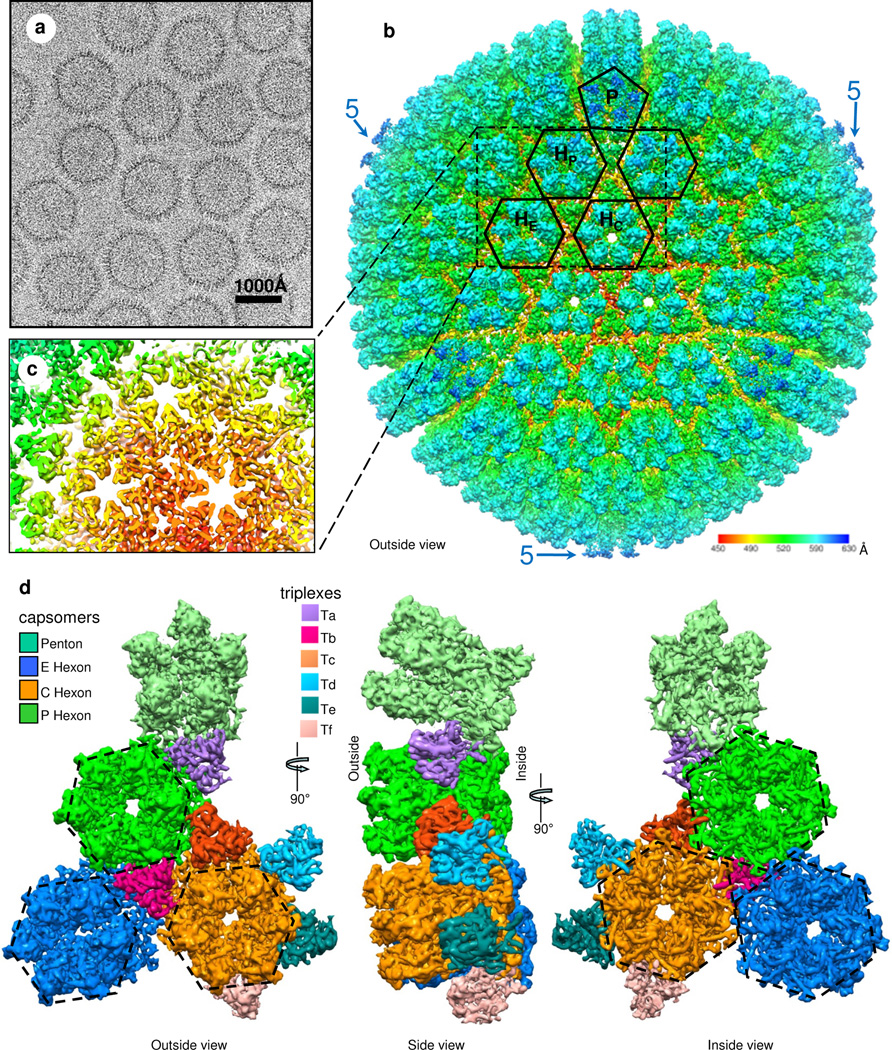
Due to the large size of the RRV capsid, we used higher accelerating voltage (300kV) for cryoEM imaging (Fig. 1a) to partially alleviate the depth-of-field limitation (Zhang and Zhou, 2011; Zhou and Chiu, 2003) and achieved an effective resolution of 7.2 Å based on the 0.143 “gold-standard” Fourier shell correlation (FSC) criterion (Fig. S1a). At this resolution, molecular boundaries (Fig. 1b and Movie S1) and secondary structure elements (Fig. 1c) can be resolved (Nakagawa et al., 2003; Zhang et al., 2003; Zhou et al., 2001), permitting computational segmentation of individual proteins for structural comparison and identification of molecular interactions (Fig. 1d). Indeed, sausage-shaped densities, corresponding to helices, are particularly visible in slices of the capsid shell (Fig. 1c and Fig. S1b,c) and from the inside of the capsid (right panel in Fig. 1d and Movie S2). The validity of the interpretation of α helices and putative β sheets can also be gauged by the excellent agreement of such elements in the upper domain (Fig. S1c) and the Johnson-fold domains between RRV and bacteriophage HK97 (Fig. S1d–f).
Figure 1. Overall structure.
(a) CryoEM image of RRV capsids. (b) Radially color-coded surface representation of the RRV capsid structure at 7Å resolution. One penton (P) and four hexons (two HP, HE, HC) are outlined. Pentons (blue) around the perimeter (‘5’) extend farthest from the center. (c) A slab of density from the dashed rectangular region of (b) containing sausage-shaped densities that correspond to helices. (d) Surface views from outside (left), side (middle) and inside (right) of a region containing a penton, three hexons (E, C and P) and six triplex heterotrimers (Ta, Tb, Tc, Td, Te and Tf). See also Fig. S1, Movie S1 and S2.
The RRV capsid proteins at the innermost radius of the capsid shell (inside view in Fig. 1d) are organized in a T=16 icosahedral lattice (with 12 pentons and 150 hexons that share edges) that is common to the capsids of other herpesviruses. [The putative DNA-translocating portal (Deng et al., 2007) that replaces one of the pentons is not visible here because of icosahedral averaging.] Both penton and hexon capsomers are cylindrical (Side view of Fig. 1d) with central channels (Movie S2), three of which are visible in the center of Figure 1b and in the inside and outside views in Figure 1d. At the outermost radius of the capsid shell, the icosahedrally averaged capsid also contains 320 triangular components (Fig. 1b), each a triplex heterotrimer that interdigitates among the hexons and pentons (outside view in Fig. 1d). An asymmetric unit contains six triplex heterotrimers, Ta through Tf (outside view in Fig. 1d), each spanning half of the height of the hexon and residing at the inner half of the capsid shell (side view in Fig. 1d). Hexagons and pentagons outlining hexons and pentons at the outermost capsid (Fig. 1b) are rotated approximately 30° and 36°, respectively, from their orientations in a standard T=16 lattice. Thus, the radially oriented MCP subunits appear to contribute hexagon/pentagon corners at the radius of the outermost capsid. (See also dashed outlines in the outside view in Fig. 1d.) The difference between the outer lattice and the standard T=16 lattice is similar to the difference between an icosidodecahedron (with 20 triangles and 12 pentagons) and a dodecahedron (with just 12 pentagons).
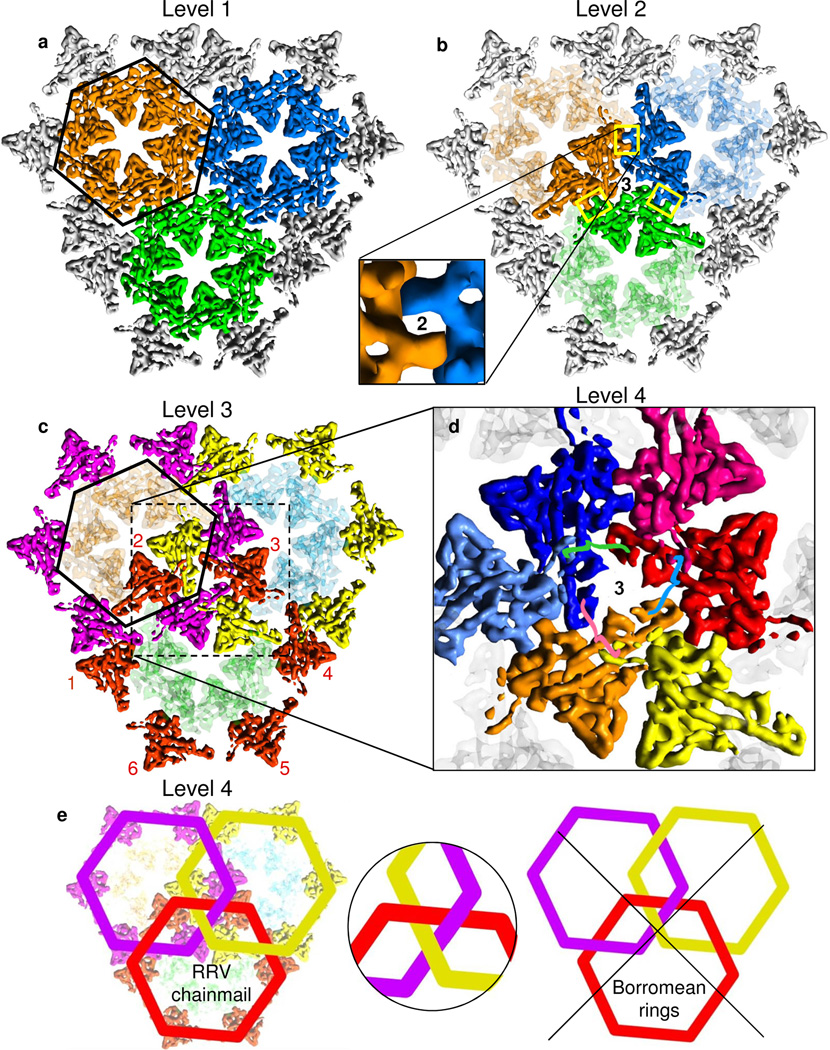
To facilitate the following description of the complex capsid structure, we show a small region of the floor of the capsid shell, a region containing three colored MCP capsomers (orange, blue and green) (Fig. 2a,b). These happen to be three hexons, but they could be two hexons and a penton. At the lowest level of organization (Fig. 2a), the six MCP subunits in a hexon capsomer are held together by (“intra-capsomer”) interactions between neighboring MCP subunits. At the second (“inter-capsomer”) level (Fig. 2b), every pair of neighboring capsomers is linked together by an MCP subunit in one capsomer binding to an MCP subunit in the other. The binding site sits on each and every local 2-fold axis (‘2’ in the inset to Fig. 2b). In addition, three capsomers are “stapled” together by interactions with a triplex heterotrimer at each and every local and global 3-fold position centered among the three capsomers (‘3’ in Fig. 2b). The triplex proteins play another role at the third (“belts”) level by non-covalently connecting belts of MCP subunits (e.g., the yellow, the magenta and the red belts in Figure 2c and Movie S3), six around a central hexon or five around a central penton. Comprising a fourth level, the belts concatenate (Fig. 2d,e). If chainmail is an extended fabric composed of concatenated belts (Fig. 2e), then the capsid of RRV may be described as chainmail.
Figure 2. Four levels of hierarchical organization.
All views are from outside, but only the floor regions of MCP monomers are shown. We use three color schemes to highlight different organizational levels, one for panels (a) and (b) as in Figure 1d, a second for (c), and a third for (d) and Fig. S4. Panels (a) to (c) contain all of the same monomers. Panel (d) is an enlarged version of the center of panel (c). (a) Within a capsomer, each MCP subunit interacts with its neighbors. MCP subunits within each capsomer are marked by the same color (orange, green or blue), and the orange capsomer is enclosed by a hexagon. (b) Binding of neighboring capsomers (bright and pale orange MCP subunits, bright and pale green, bright and pale blue) in groups of two by interactions between MCP dimerization domains (marked by rectangles – see inset) across local two-fold axes (‘2’), binding the orange capsomer with the green, the green with the blue, and the blue with the orange). Also, triplex heterotrimers bind groups of three (orange, green and blue) capsomers at the local 3-fold axis marked by ‘3’. (c) The magenta belt, surrounding the outlined capsomer, is created by a ring of six Johnson folds from six (magenta) MCP subunits – each subunit a member of a different capsomer – joined by triplex heterotrimers. We also show yellow and red belts, the latter with numbered MCP subunits. (d) Formation of non-covalent chainmail by concatenated belts. Each of the three belts is joined by one subunit of the triplex heterotrimer (not shown) centered at the position marked by ‘3’, each joint denoted by a differently colored curly brace to correspond with its (pink, cyan or green) triplex monomer, consistent with the color scheme in Figure 6. See also Fig. S4. (e) Belts (e.g., red crossing over magenta, over yellow, under magenta, and under yellow) in the three-fold symmetric pattern of the RRV capsid do not separate if one belt breaks, thus forming chainmail. By contrast, Borromean rings, with a strictly alternating pattern (e.g., red crossing under magenta, over yellow, under magenta again, over yellow again), do separate if any one ring breaks. Nonetheless, at local three-fold axes, the crossing patterns are the same (inset). See also Movie S3.
Domain organization of MCP monomer
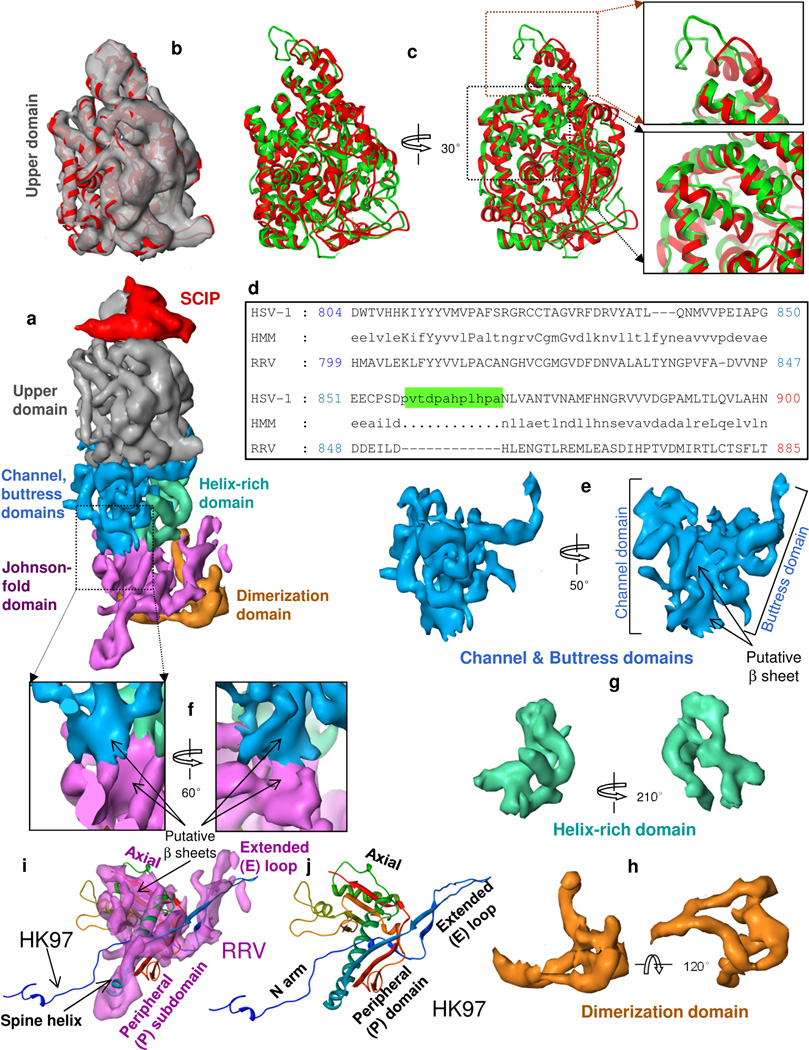
Pentons and hexons contain five and six MCP subunits, respectively. With 7.2 Å resolution, we can see that each subunit contains a radially elongated MCP monomer and a V-shaped SCIP monomer on top (Fig. 3a). Each MCP subunit has six domains distributed in three regions (Fig. 3a and Movie S4): in the upper region the upper domain (Fig. 3b–d), in the middle region the channel, buttress (Fig. 3e, f) and helix-rich (Fig. 3g) domains, and in the floor region the dimerization domain (Fig. 3h) and a domain that contains a fold similar to the gp5 structure of bacteriophage HK97 (Fig. 3i,j) (Wikoff et al., 2000). (Also compare the structures of HK97 and RRV MCP in the superposition of Movie S4 and Supplementary Fig. S1d with the RRV MCP Johnson fold domain in Supplementary Fig. S1e–f.) This Johnson fold was subsequently seen in other viruses (e.g., Baker et al., 2005; Bamford et al., 2005; Dai et al., 2010; Jiang et al., 2006). The dimerization domain (Fig. 3h) extends along the side of the Johnson-fold domain and reaches beneath it (Fig. 3a).
Figure 3. Domain organization of MCP.
(a) Side view of a hexon subunit. Each hexon subunit contains one monomer of SCIP (red) and one monomer of MCP. Each MCP monomer can be divided into six domains: upper domain in the upper region, channel, buttress and helix-rich domains in the middle region, and Johnson-fold and dimerization domains in the floor region. (b) Superposition of the cryoEM density map (semi-transparent gray) and a pseudo-atomic model (red ribbon) of the MCP upper domain (MCPud) of RRV. (c) The pseudo atomic model of the RRV MCPud (red) superimposed with the atomic model of HSV-1 MCPud (green). While most secondary structure elements in RRV and HSV-1 MCPud match (lower inset box), there are differences at the tip (upper inset box), possibly related to binding of SCIP and tegument proteins. (d) Alignment of the amino acid sequence between HSV-1 and RRV MCP in the tip of MCPud. HSV-1 MCP has a string of 12 additional amino acids (green highlighting). HMM: hidden Markov model, used to assist in alignment. (e–h) Surface representations of the channel and buttress domains (e), the helix-rich domain (g) and the dimerization domain (h). (i) Comparison of the surface representation of the Johnson-fold domain of RRV (semitransparent purple) with the atomic model of the Johnson fold of the bacteriophage HK97 (ribbon). (j) Atomic model of the HK97 gp5 protein (i.e., the Johnson fold), colored from N (blue) to C (red) termini. See also Fig. S2 and Movie S4.
The MCP upper domain (MCPud) is rich in helices and loops (Fig 3b). To build a pseudo-atomic model of RRV MCPud, we used these secondary structure elements as constraints for comparative modeling with the crystal structure of HSV-1 MCPud (Bowman et al., 2003) as template. Ramachandran statistics of the MCPud homology model (Fig. S1g,h) indicate model quality on par with the original HSV-1 VP5ud structure. The differences and similarities of the RRV MCPud (red) and the HSV-1 MCPud (green) are clear in the superimposed ribbon models (Fig. 3c). Whereas the interior structures, which are mostly helical, are highly conserved, structures on and near the external surfaces show some differences, especially at the uppermost tip (magnified in the top box at the upper right in Fig. 3c), where the red and green loops do not superimpose. Indeed, a proline-rich, 12 amino-acid segment within the HSV-1 (green) loop (green highlighted portion of a.a. sequence in Fig. 3d) is absent in the RRV (red) loop.
The middle region of MCP encompasses the channel, buttress (Fig. 3e) and the helix-rich domains (Fig. 3g). We divided the middle region into these three domains because they are spatially separated and have distinctive structural roles (e.g., channel and buttress domains forms the channel and provides support, respectively) (Movie S4) and because the conformation of the helix-rich domain differs greatly between MCP in hexon and penton. The channel domain contains a large, twisted, central, putative β-sheet (Fig. 3e). The helix-rich domain of hexon MCP contains three long helices, one of which adopts a different conformation in penton MCP.
For the RRV Johnson-fold domain (Fig. 3i), we have followed the HK97 terminology to describe structural features (named subdomains) that are homologous to domains established in the HK97 gp5 (Wikoff et al., 2000)(Fig. 3j). The RRV Johnson-fold domain contains an extended loop (E loop) and a peripheral (P) subdomain with a 58-Å long ‘spine’ helix that together represent the contribution of each MCP subunit to the belt described in Figure 2c. Based on sequence analysis, we assign the spine helix of RRV MCP to a.a. 147–184 (Fig. S2), the only segment of the MCP sequence that contains a helix of more than 30 amino acids. The spine helix and its connected, putative β sheet in the P subdomain contribute to the long tongue-shaped density in the hexon MCP in Figures 3a and 4a and in the penton MCP in Figure 4b, which are similar but not identical (Fig. 4c). The axial (A) subdomain has a central, putative β-sheet flanked by two helices. The long N-terminal arm (N Arm), which is not clearly resolved in our RRV map, can adopt multiple conformations in HK97 (Gertsman et al., 2009; Veesler et al., 2012).
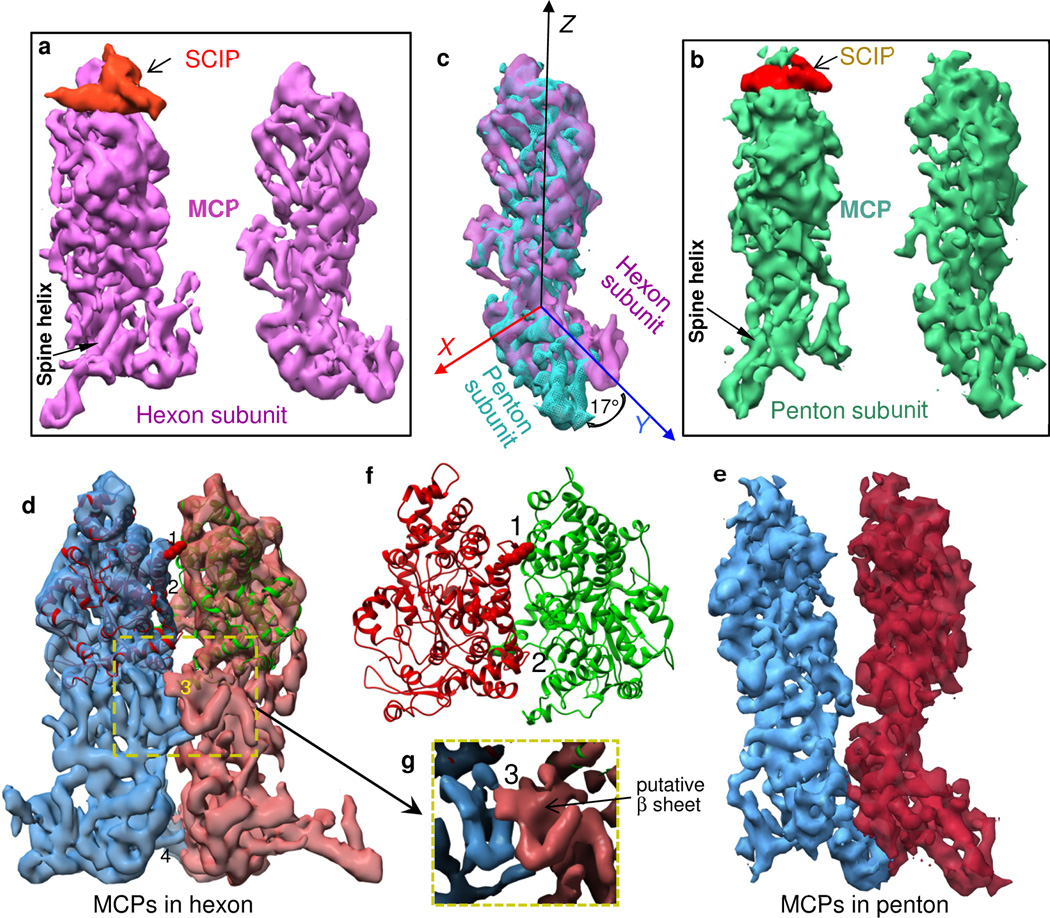
Figure 4. MCP interactions within pentons and hexons (organizational level 1).
(a, b) Two side views of a hexon subunit (a), including SCIP on the left panel, and the corresponding views of a penton subunit (b). (c) Superposition of the hexon and penton MCP monomers from the right panels of (a) and (b), showing conformational differences between penton MCP and hexon MCP. (d, e) Two interacting MCP subunits extracted from a hexon (d) and from a penton (e), viewed from inside the capsomer channel. The inter-MCP interaction sites in the upper domain (#1 and #2) and in the channel domain (#3) are present between hexon MCPs that are vertically aligned (in a ‘closed’ conformation, d) but absent between penton MCPs that tilt away from each other (in an ‘open conformation, e). By contrast, the interaction site in the floor region (#4) is present between MCPs of both hexon and penton. (f) Zoom-in view of the interaction sites #1 and #2 in a ribbon model of the MCPud. (g) Surface view of the yellow dotted boxed region of (d), showing interaction site #3, involving putative β sheets, located in the channel domain. Interaction sites #1 and #3 create constriction rings inside the hexon channel. See also Movie S5.
First level of hierarchical organization: intra-capsomer MCP-MCP interactions
Both penton and hexon have an axial channel running from the upper region through the floor region of the MCP, which constitutes the floor of the capsid shell (Fig. 1b–d). Side views of a pair of neighboring MCPs in a hexon (Fig. 4d) and in a penton (Fig. 4e) show the interior walls of their axial channels. The most striking differences between the penton and hexon channels are the sites of interaction or lack thereof. Specifically, adjacent MCPs in a hexon have four interaction sites (Fig. 4d and Movie S5), two between their upper domains (#1, #2), one between the small, putative β sheet of their channel domains (#3), and one between their Johnson-fold domains in the floor (#4). Only #4 is present between subunits in pentons (Fig. 4d vs. Fig. 4e). Even though the limited resolution of our cryoEM map has precluded modeling of side chains of amino acid residues, we are confident about the assignment of the amino-acid segments contributing to these interactions. We will suggest in Discussion that these interaction sites offer insight into capsid maturation.
The interaction site #1 is located between helices in the interfacial region of neighboring MCPud, near the interior of the channel (Fig. 4f). The densities arising from six interaction sites #1 inside each hexon form a bonded ring near the top of the channel. A fit of our pseudo-atomic model of MCPud into hexon cryoEM density reveals the details of these interactions, specifically a region containing polar amino-acid residues from two neighboring MCPud (Fig. 4f and Movie S5). On one subunit, the most likely contributors to this interaction are Ser670, Lys671 and Asp672, which are located at the end of a helix (a.a. 670 to 694). On the other subunit, the most likely contributors are Asn643 and Asn644, located in a kink between two helices (a.a. 624 to 645 and 654 to 665) (Figs. 4f, S2).
The interaction site #2, also located in the interfacial region between two neighboring MCPud but close to the outer surface of the hexon, rather than near the interior of the channel (Movie S5), likely involves charged residues, Arg973 of one MCPud and Glu707 of its neighboring MCPud (Figs. 4f, S2).
The interaction site #3, which lies in the channel domain of each MCP, is mediated through the small, putative β sheets on neighboring MCP monomers near the interior of the channel (Fig. 4g), thus six interaction sites #3 comprise a second, lower constricting ring inside each hexon channel.
While MCP monomers in both hexons and pentons show the interaction site #4 in the floor region, the absence of the interaction sites #1 – #3 among pentons, including the absence of the two constricting rings, is accompanied by a splaying outward of the middle and upper regions of MCP monomers in the penton (compare Fig. 4e with Fig. 4d). Because the pentameric MCP subunits are spaced apart from each other, the structures corresponding to the helix and helix-loop-helix interactions conspicuous in interaction site #1 of the hexon are unable to support interactions between adjacent MCP subunits in the penton and are partially disordered.
The difference in binding between MCP subunits in hexons and that in pentons is accompanied by a corresponding difference in the orientation of the floor domains – the Johnson-fold domain and the dimerization domain – with respect to the middle and upper domains of the hexon and penton MCPs. Although it remains unclear whether this change in orientation is the cause or the result of the MCP binding differences, the structural variation reflects a context-dependent conformational switch of the MCP subunits.
To illustrate the difference in orientation, we superimposed hexon and penton MCP subunits with the ‘ fit in map’ tool in Chimera. The upper and middle regions of the hexon and penton MCP subunits (right panel of Fig. 4a, right panel of Fig. 4b) match (Fig. 4c). In addition to the upper and middle regions of the MCP subunits, the channel-proximal ends of the long spine helices in the Johnson-fold domain in the floor regions of the hexon and penton MCPs also coincide (Fig. 4c). We describe these coincident ends as a pivot point and identify that point as the origin of the X-Y-Z coordinate system depicted in Figure 4c. We find that the rest of the spine helix in the penton MCP is oriented 17° down with respect to the spine helix in the hexon MCP (Fig. 4c).
Second level of hierarchical organization: inter-capsomer linkage by MCP dimerization domains and stapling by triplex heterotrimers
Linking capsomers, two at a time via MCP dimerization domains
Across the interface between neighboring capsomers at the local two-fold axis, the dimerization domains (Fig. 3h) of neighboring MCPs link neighboring capsomers laterally (boxes and inset in Fig. 2b).
Triplex heterotrimer
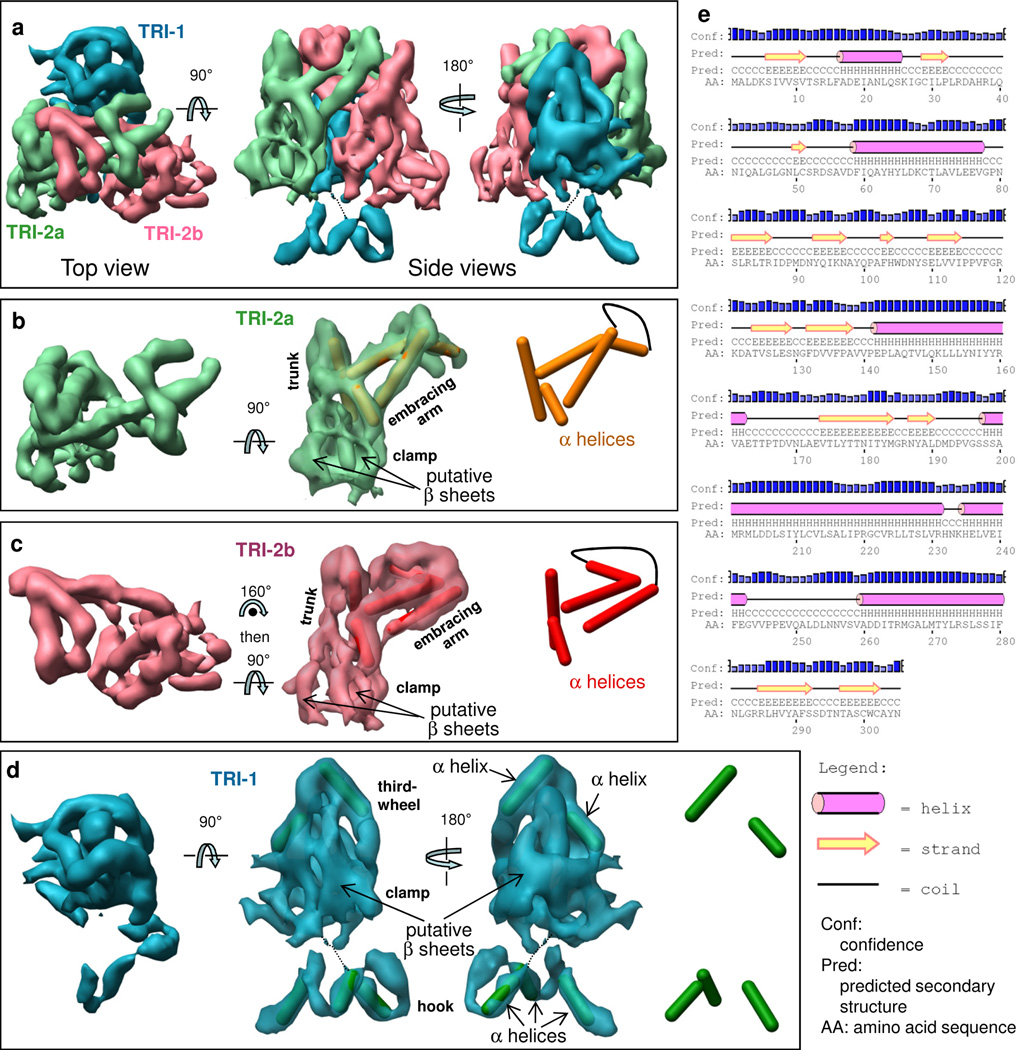
Each of the 320 triplex heterotrimers sits at a local three-fold symmetric position, where the floor domains of three neighboring capsomers (either three hexons or two hexons and a penton) meet (Fig. 1b,d). Each triplex heterotrimer is composed of one molecule of TRI-1 and two molecules of TRI-2 (O'Connor et al., 2003; Yu et al., 2003), which are now resolved from one another (Fig. 5a–d). Our density map shows that TRI-2 has 5 long helices (rods in Fig. 5b,c), whereas TRI-1 has 6 short helices (arrows in Fig. 5d). The observed secondary structures in TRI-2 are consistent with sequence-based predictions (Fig. 5e). Each TRI-2 monomer consists of three domains: a trunk domain, an embracing arm domain and a clamp domain. Since TRI-1 accompanies but does not embrace the TRI-2 monomers, we describe the upper portion of TRI-1 as the “third-wheel” domain, reflecting its supporting role to the embracing couple of TRI-2 dimer. Each TRI-1 monomer does contain a clamp domain. Additionally, it contains a hook domain with three helices (Fig. 5d and Fig. S3). This connection between the hook domain and the clamp domain is weak, as it can only be seen when displayed at lower density threshold.
Figure 5. Structure of the triplex heterotrimer.
(a) Triplex structure obtained by averaging triplexes Tb, Tc, Td, Te but not Ta (around the penton) and not Tf (at the three-fold axis) (Fig. 1d), composed of one TRI-1 (cyan) and two TRI-2 conformers [TRI-2a (green) and TRI-2b (pink)]. (b) TRI-2a, top view (left), side view of the surface superimposed with α helices (cylinders) (middle), and same side view of the secondary structure model by itself (right). (c) Same as (b) but for TRI-2b. (d) TRI-1, top surface view (left) and side surface views superimposed with helices (middle panels), and helices alone (right panel). (e) Secondary structure prediction of TRI-2. α helices, β strands and loops are shown as pink cylinders, yellow arrows and black lines, respectively. See also Fig. S3 and Movie S6.
TRI-1 and TRI-2 bind to each other to form the triplex heterotrimer. Reaching out from the trunk domain, the embracing arm domain in each TRI-2 contains a series of three long helices (Fig. 5b,c). The embracing arm of each monomer reaches around the embracing arm of the other (pink and green in Fig. 5a, Movie S6), an interlocking association that could bring together the triplex monomers and consequently the subjacent MCPs of the associated capsomers. The TRI-2 embracing arm also interacts with the trunk domain of the adjacent TRI-2 monomer and the third-wheel domain of the TRI-1 monomer (cyan in Fig. 5a,d). The TRI-2 and TRI-1 clamp domains are rich in putative β sheets (middle panels in Fig. 5b–d) that resemble those in gpD in phage lambda (Yang et al., 2000). Although TRI-1 has a clamp domain like TRI-2, TRI-1 has no sequence similarity with TRI-2, and the arrangement of predicted secondary structures of TRI-1 based on sequence bears no resemblance to that of TRI-2 (Compare Fig. 5e with Fig. S3). We will discuss the three helices in the hook domain of TRI-1 below.
Stapling capsomers in groups of three via triplex-MCP interactions
With respect to the triplex heterotrimer, the blue, orange and red MCP monomers may be regarded as an ‘inner’ group of MCPs, with the light blue, yellow and magenta as an ‘outer’ group (Figs. 2d, 6a–g, and S4). Using its putative β sheets, the TRI-2b (pink) triplex monomer mediates interactions that link the Johnson-fold domains of three MCPs, inner blue, outer yellow, and inner orange at the sites marked by dotted circles in side views (Fig. 6d,e). Specifically, it has one contact on the putative β sheet of the P subdomain (Fig. 3i) of the Johnson-fold domain of the inner blue MCP subunit (the dotted circle in Fig. 6d). In addition, it has one contact on the E loop (Fig. 3i) of the outer yellow MCP (the middle dotted circle in Fig. 6e). It also has two contacts on the putative β sheet of the P subdomain of the Johnson-fold domain of the inner orange MCP subunit (the left and right dotted circles in Fig. 6e).
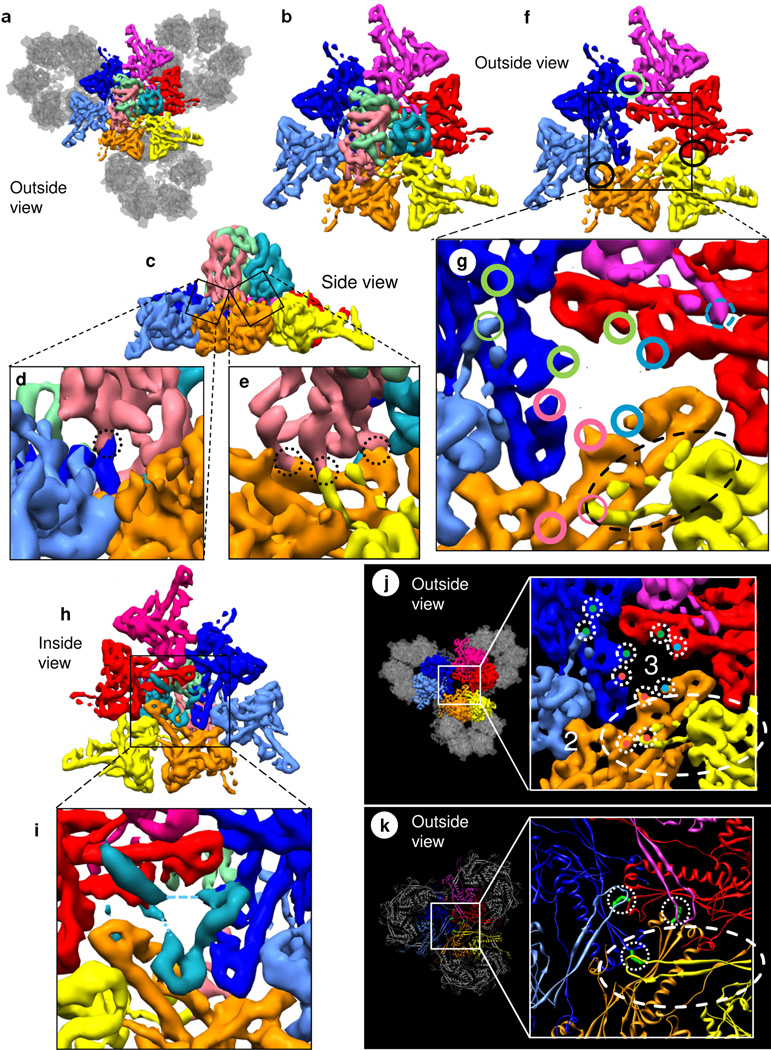
Figure 6. Interactions among triplex and capsomers.
(a) Top view showing inter-capsomer interactions among three neighboring hexons (colored as in Fig. 2d: yellow/orange/four gray; red/magenta/four gray; and dark blue/light blue/four gray) mediated by the Tc triplex heterotrimer (colored as in Fig. 5a). Only the floor regions of MCPs are shown. (b) Magnified view of the colored MCPs in (a). (c) Side view of (b). (d, e) Close-up and slightly rotated view of the boxes in (c) showing interactions (dotted circles) among the pink triplex monomer and the Johnson-fold domains of MCPs. (f) View in (b) after removing the triplex with circles marking inter-MCP interactions at dimerization domains. (g) Close-up of (f) showing triplex contacts on the P subdomain of the Johnson fold (thick circles) and triplex interactions with the E loop of the Johnson fold [two thin circles and the dashed (weak) circle]. The colors of the circles - green, pink and cyan - identify contacts made by the green, pink and cyan triplex monomers with red and dark blue MCPs, dark blue and orange MCPs, and orange and red MCPs, respectively. Dashed ellipse marks the E loop overlying the P subdomain of the orange Johnson fold. See also Fig. S4. (h) Bottom view of (b) and its close-up (i) showing the three helix densities of the hook domain of TRI-1 (cyan). The dashed line indicates weak densities only visible at low threshold. (j, k) Different mechanisms are employed for clamping adjacent capsomers in RRV and bacteriophage HK97. In RRV (j), the β sheets of the Johnson-fold domains are clamped together by interactions above (markings taken from g) and below (shown in i). (k) By contrast, HK97 uses a covalent chainmail mechanism (Wikoff et al., 2000), where the E loop forms an isopeptide bond (green) with the β-sheet from the adjacent hexon. See also Movie S7.
Computational removal of the triplex (Fig. 6f) clarifies the topographic organization of these linking sites. Specifically, these TRI-2b linking interactions are now marked by pink circles in top view (Fig. 6g). As depicted in Figures 2d and S4, the clamp domain of the (pink) TRI-2b triplex monomer links the end of the P subdomain of the Johnson-fold domain of the inner blue MCP (at the upper end of the pink curly brace in Figs. 2d and S4) to the end of the E loop of the Johnson-fold domain of the outer yellow MCP (at the lower end of the pink curly brace in Fig. 2d). This link is supported by the middle region of the P subdomain of the Johnson-fold domain of the inner orange MCP (underneath the pink curly brace in Fig. 2d).
Similarly, from the P subdomain of one Johnson-fold domain to the E loop of another Johnson-fold domain, the (green) TRI-2a triplex monomer links the inner red MCP to the outer light blue MCP, supported by the blue MCP. Likewise, supported by the red MCP, the (cyan) TRI-1 monomer probably links the inner orange MCP to the outer magenta MCP (dashed circle in Fig. 6g). Together, the three clamp domains of each triplex trimer link the Johnson-fold domains of three pairs of MCP subunits (e.g., blue to yellow + red to light blue + orange to magenta in Figs. 2d, 6g and S4) that are members of three hexon capsomers (orange/yellow/four gray, blue/light blue/four gray, and red/magenta/four gray in Fig. 6a). Thus, each triplex heterotrimer interacts with elements of the Johnson-fold domains of the five (and probably the six) surrounding MCP subunits belonging to three capsomers (Fig. 6a).
Conversely, each (blue, orange or red) subunit in the inner group of MCP subunits is linked to two triplex monomers (green and pink, or pink and cyan, or cyan and green). Each (light blue, yellow or magenta) subunit in the outer group of MCP subunits is linked to one triplex monomer (green, or pink, or perhaps cyan).
Unlike the TRI-2 triplex monomers, the TRI-1 triplex monomer (cyan) has a hook domain with three helices (Fig. 5a,d) that reaches through a hole underneath the triplex heterotrimer at the local 3-fold symmetric position (Fig. 6f,g) in the floor to interact with three grooves of the three Johnson-fold domains on the inner surfaces of the inner three (blue, red and orange) MCP subunits (Fig. 6h,i).
Third level of hierarchical organization: belts created by rings of alternating Johnson folds and triplex heterotrimers
As described above, the Johnson-fold domains are linked by the interactions with triplex heterotrimers. Indeed, the ring of yellow Johnson folds in Figure 2c and triplex heterotrimers constitutes one belt that encircles a whole capsomer (magenta/red/four transparent blue MCPs). Likewise, the ring of red Johnson folds and triplex heterotrimers constitutes a second belt that encircles a whole capsomer (magenta/yellow/four transparent green MCPs), and the ring of magenta Johnson folds and triplex heterotrimers constitutes a third belt about a whole capsomer (red/yellow/four transparent orange MCPs). These belts involve alternations of triplex heterotrimer and Johnson folds as follows: The Johnson fold at one end – the P subdomain – of one MCP is linked via a triplex heterotrimer to the Johnson fold at the other end – the E loop – of another MCP (Fig. 6g, j). In bacteriophage HK97, the linkage is achieved by an isopeptide bond within the 280 a.a. gp5 protein (Fig. 6k), without the intervention of additional proteins. In RRV, the linkage is achieved by the Johnson-fold domain of similar size (Fig. S1d–f) and non-covalent interactions with the triplex heterotrimer. The effect of the belt is to draw together the six capsomers surrounding each hexon or the five capsomers surrounding each penton. Moreover, in addition to being encircled by one belt, every hexon/penton itself is also part of six/five belts (Fig. 2c).
Fourth level of hierarchical organization: concatenated belts or chainmail
All three belts of MCP subunits (yellow, red and magenta) in Figure 2c loop around the triplex heterotrimer at the local three-fold symmetric axis, as illustrated in the inset of Figure 2e. Topologically, one belt (e.g., red) crosses over a second belt (magenta), over a third (yellow), under the second (purple) and under the third (yellow) (Fig. 2e). In more detail, the E loop of the #1 red Johnson fold in Figure 2c crosses over the P subdomain of a magenta Johnson fold, and the E loop of the #2 red Johnson fold in Figure 2c crosses over the P subdomain of a yellow Johnson fold. The P subdomain of the #3 red Johnson fold in Figure 2c crosses under the E loop of the magenta Johnson fold, and the P subdomain of the #4 red Johnson fold in Figure 2c crosses under the E loop of the yellow Johnson fold. Due to symmetry, the magenta belt and the yellow belt cross the other belts in the same fashion as the red belt just described. The three belts are thus concatenated (left in Fig. 2e), and a break in one of the concatenated belts in RRV does not cause the others to unlink. For Borromean rings, the situation would be the same at each local three-fold axis (inset of Fig. 2e), but the order of crossing of the rings/belts is over-under-over-under (right in Fig. 2e). Consequently, breaking one belt of linked Borromean rings would cause the other two belts to unlink as well. Therefore, the linked belts in RRV constitute a single chainmail for the entire capsid, a mail that would hold together in the face of the random making and breaking of the non-covalent bonds between interacting subunits, as would be expected in biological assemblies.
DISCUSSION
Comparison with capsid shell structures of other herpesviruses
The main functions of the capsid shells in all herpesviruses are to encapsidate the viral genomic DNA and to eject it into the nucleus of a host cell for replication. As such, capsid shells of different herpesviruses would be expected to have highly conserved structures. For the MCP, the Johnson-fold domain and most of the structural features described here, particularly the spine helix, are conserved across different subfamilies of herpesviruses (Baker et al., 2005; Homa et al., 2013; Zhou et al., 2000). Other MCP structural features in the HSV-1 capsid, not explicitly described due to limited resolution (8.5 Å), correspond to the newly described dimerization domain, helix-rich domain and the channel domain in RRV (Zhou et al., 2000). Among triplex proteins, the helical structures in the trunk and embracing arm domains of RRV TRI-2 protein are similar to those described previously for HSV-1 VP23 (Zhou et al., 2000).
Among the most notable differences are regions related to the attachment of tegument proteins. For example, the SCIP homolog VP26 in HSV-1 does not bind the upper domain of penton MCP (Trus et al., 1995; Zhou et al., 2002; Zhou et al., 1995), whereas it does bind both penton and hexon MCP in the capsid of the gammaherpesvirus here and the murine cytomegalovirus, a betaherpesvirus (Dai et al., 2013). Functional studies further indicate that the SCIP homolog in the betaherpesvirus stabilizes DNA-encapsidation by recruiting the unique tegument protein pp150 into a net that encloses the capsid (Dai et al., 2013). Indeed, this position on the upper domain of penton MCP in alphaherpesvirus capsids is occupied by the CVSC proteins (Conway et al., 2010; Homa et al., 2013; Trus et al., 2007; Zhou et al., 1999), suggesting that CVSC proteins play a role similar to that of betaherpesvirus pp150 during virion assembly.
Among triplex proteins, VP19c of HSV-1 differs from its RRV TRI-1 homolog in having an additional region on top of the third-wheel domain (Zhou et al., 2000), consistent with its 40% longer sequence. Additionally, at the bottom of RRV TRI-1, the structure reveals a prominent hook domain containing three helices (Fig. 5d). We would not be surprised if a similar hook domain is identified in HSV-1 VP19c when the resolution of its structure improves.
Herpesvirus capsid maturation
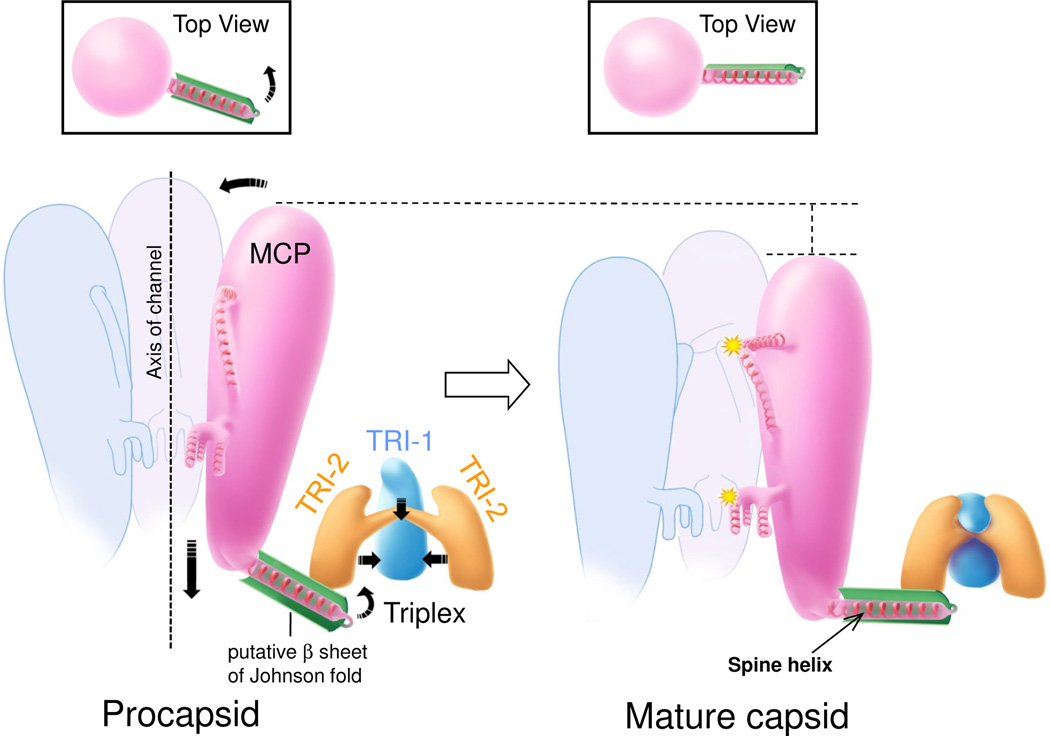
Capsid maturation in HSV-1 proceeds from spherical procapsid to angular mature capsid (Heymann et al., 2003), similar to what occurs in dsDNA bacteriophages (Conway et al., 2001; Steven et al., 2005). In the (immature) procapsid of HSV-1, both hexon and penton capsomers have an ‘open’ configuration, evident even at ~20 Å resolution (Heymann et al., 2003). This open configuration is similar to what we see in the penton in the mature RRV capsid, with upper and middle regions tilted away from the axis of the channel running up the center of the penton (Fig. 4e), in contrast to the closed configuration of the hexon (Fig. 4d). If RRV matures similarly from a spherical procapsid to an angular capsid, accompanied by a change in hexon configuration from open to closed, then we can propose a testable model of capsid maturation in RRV and perhaps in herpesviruses in general (Fig. 7 and Movie S8). The test would begin with the isolation and structural characterization of the hypothesized RRV procapsid. In this model, it is the conformational switches that bring specific domains of neighboring MCP upper and middle regions together that would accomplish large scale structural change during capsid maturation. We would then infer that it was the local conformational changes initiated by the action of the triplex heterotrimers, each associated with three hexons, that would be responsible for the global transformation from the spherical procapsid to the angular, mature capsid. This model has the additional virtue that it draws together the intricacies of the RRV structure reported here.
Figure 7. Model of conformational changes during capsid maturation.
Arrows indicate directions of movement. In the open configuration, MCP subunits in the procapsid (left) lack significant inter-subunit interactions, specifically interaction sites #1 to #3 along their upper and middle regions along the hexon channels, that will engage after rotation into the closed configuration in the mature capsid (right). Starting from the procapsid, TRI-1 and TRI-2 in a triplex embrace more tightly and engage their three clamp domains with the three Johnson-fold β sheets (green) of three MCP subunits. This process pulls the three long Johnson-fold helices toward a point centered beneath the triplex heterotrimer, away from the channel axis. Insets: the pull would also rotate upward the peripheral subdomain of the Johnson fold to allow the spine helix to reach out to its neighboring MCP floor region and establish inter-capsomeric contacts. As a result, in the mature capsid (right), the MCPs in hexons tilt into alignment, facilitating interactions between neighboring MCPs at interaction sites #1 to #3 (yellow stars), and enter the closed configuration. In addition, the tilt enables the three helices of the TRI-1 hook domain to pull in concert the β sheets connected to the spine helix, thus adding to the pull of the floor of the MCP away from the channel axis. See also Movie S8.
Pressure, chainmail and DNA release
The task of any viral capsid is to protect its genome in the extracellular environment and then to release its genome into its host cell or host nucleus in a controlled manner. One complicating factor for dsDNA viruses is the high stiffness and long persistence-length of dsDNA. Indeed, the genome in some of these viruses are so tightly packed – in liquid crystal form (Booy et al., 1991) – that the spacing between adjacent DNA duplex strands can be only ~25Å (Earnshaw and Harrison, 1977; Earnshaw et al., 1978; Harrison, 1980), just slightly larger than the diameter of the DNA double helix. Herpesviruses resemble HK97 in this regard, with a spacing in RRV of 25Å (Yu et al., 2003). This tight packing produces internal pressures as high as 50 atmospheres (Gelbart and Knobler, 2009). Here for RRV capsid, we describe four levels of organization that cooperate to prepare the virus to withstand high internal pressure. The bacterial viruses also employ some of these levels, including concatenated belts, thus chainmail – covalent chainmail in the case of HK97 and non-covalent chainmail in the cases of bacteriophages BPP-1 (Zhang et al., 2013), Epsilon 15 (Baker et al., 2013), lambda (Lander et al., 2008) and perhaps T4 (Fokine et al., 2005). These viruses, having stabilized their capsids so firmly, face the problem of how to release and inject DNA into the host. They solve this problem by the use of a portal complex located at one of the twelve pentons of the capsid (Cardone et al., 2007; Chang et al., 2007; Deng et al., 2007).
EXPERIMENTAL PROCEDURES
Capsid purification
Cell culture, RRV infection and capsid purification were performed as described previously (O'Connor et al., 2003; Yu et al., 2003). Briefly, capsids were isolated from the media of RRV-infected rhesus fibroblasts through a 600 µl 20–50% sucrose-MTNE gradient by centrifugation at 75,000 g for 40 minutes. The gradient fraction containing capsids was diluted in MTNE buffer (20mM Tris HCl at pH 8.0, 250 mM NaCl and 1 mM EDTA), pelleted by centrifugation at 42,300 g for 30 minutes at 4°C, and resuspended in 20 µl MTNE buffer for cryoEM sample preparation.
CryoEM
An aliquot (3 µl) of RRV A-capsids was applied to a Quantifoil R2/1 grid, blotted, and plunge-frozen. Focal-pair images were recorded on Kodak SO163 films at 300kV with a magnification of 33,000 and an electron dose of ~10 electrons/Å2/micrograph. Films were digitized in a Zeiss SCAI densitometer using a step size equivalent to 2.1 Å/pixel on the specimen.
Data processing was carried outwith IMIRS (Liang et al., 2002) with modifications (Liu et al., 2008). We boxed out 22,889 particle images from 320 micrographs and included 14,374 particles in the final reconstruction. The effective resolution, 7.2Å, is based on the Fourier shell correlation (FSC) criterion of 0.143 (about 8.5 Å at 0.5 FSC) (Rosenthal and Henderson, 2003) between two maps from half datasets independently processed and reconstructed (Fig. S1a).
Visualization, segmentation and comparative modeling
3D visualization was carried out using Chimera (Pettersen et al., 2004). Much of the molecular boundaries were resolved by varying the density threshold, and individual molecules were segmented interactively. Segmentation of regions involving direct molecular interactions, such as that between the triplex and the floor domains of a hexon/penton MCP, was more challenging. In such cases, segmentation was facilitated through comparison of local (i.e. quasi-) symmetry-related subunit arrangements, such as those observed between two interacting hexon subunits as depicted in the schematic representation of the floor interaction (#4 in Fig. 4d). Segmentation of triplex subunits was aided similarly by considering dimerization of TRI-2; boundary densities not distributed in two copies were assigned to the TRI-1 monomer (see Fig. 5). Inasmuch as interactions are strong, segmentation of different subunits become more difficult and thus less certain (e.g., between SCIP and MCP subunits as in Fig. 3a). In these rare cases, we color subunits differently for purpose of illustration but refrain from basing interpretations on precise boundaries or details of interactions.
To obtain a pseudo-atomic model of the upper domain of RRV MCP using our cryoEM density map, we used the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2) (Kelley and Sternberg, 2009), which generated a homology model using the atomic structure of HSV-1 MCPud (Bowman et al., 2003) as template. The homology model was superimposed with the RRV density map of a hexon subunit to assess secondary structure conformity with Chimera (Pettersen et al., 2004). Refinement of this preliminary model was performed by adjusting the orientation and position of the secondary structure elements using Coot (Guex and Peitsch, 1997) such that the modified model fit within the spatial constraints provided by the cryoEM map. The new model was further refined through several rounds of bond angle and energy optimization via Swiss-PdbViewer (Emsley et al., 2010) and finally validated by matching with the cryoEM map (e.g., Fig. 3b).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Xuekui Yu and Ivo Atanasov technical assistance in sample preparation, and Xiaorui Zhang for preparing Fig. 7 and Movie S8. This research is supported in part by grants from National Institute of Health (AI046420 and GM071940 to Z.H.Z. and CA088768 to D.H.K.). We thank one of the anonymous reviewers for advising caution in identifying β sheets.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The EMDB access number for the cryoEM density map of the RRV capsid is EMD-2550.
SUPPLEMENTAL INFORMATION
Supplemental Information includes results, three figures and eight movies and can be found with this article online at XXXX.
Author Contribution
Z.H.Z. and D.H.K designed the project; C.M.O. and B.H.K. prepared the sample; Z.H.Z. and M.B.S. recorded experimental data. W.H.H., S. Shah and Z.H.Z. processed the data; J.J., Z.H.Z. and W.H.H. did structural prediction and built the pseudo-atomic model; Z.H.Z. and S. Schein interpreted the structure and wrote the manuscript. All authors reviewed the paper.
REFERENCES
- Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, Chiu W. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12301–12306. doi: 10.1073/pnas.1309947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. Structure of the herpesvirus major capsid protein. EMBO J. 2003;22:757–765. doi: 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 2007;81:2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 2010;397:575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- Dai W, Hodes A, Hui WH, Gingery M, Miller JF, Zhou ZH. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4347–4352. doi: 10.1073/pnas.0915008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Yu X, Gong H, Jiang X, Abenes G, Liu H, Shivakoti S, Britt W, Zhu H, Liu F, Zhou Z. The smallest capsid protein mediates binding of the essential tegument protein pp150 to stabilize DNA-containing capsids in human cytomegalovirus. PLoS Pathog. 2013 doi: 10.1371/journal.ppat.1003525. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B, O’Connor CM, Kedes DH, Zhou ZH. Direct visualization of the putative portal in the Kaposi's sarcoma-associated herpesvirus capsid by cryoelectron tomography. J. Virol. 2007;81:3640–3644. doi: 10.1128/JVI.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J. Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Harrison SC. DNA arrangement in isometric phage heads. Nature. 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, King J, Harrison SC, Eiserling FA. The structural organization of DNA packaged within the heads of T4 wild-type, isometric and giant bacteriophages. Cell. 1978;14:559–568. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D. In: Kaposi’s sarcoma-associated herpesvirus In Fields Virology. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Philadelphia: Lippincott-Williams & Wilkins; 2007. pp. 2847–2888. [Google Scholar]
- Gelbart WM, Knobler CM. Virology. Pressurized viruses. Science. 2009;323:1682–1683. doi: 10.1126/science.1170645. [DOI] [PubMed] [Google Scholar]
- Germi R, Effantin G, Grossi L, Ruigrok RW, Morand P, Schoehn G. Three-dimensional structure of the Epstein-Barr virus capsid. J. Gen. Virol. 2012;93:1769–1773. doi: 10.1099/vir.0.043265-0. [DOI] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Harrison SC. Molecular packing of nucleic acids in spherical viruses. Prog. Clin. Biol. Res. 1980;40:301–310. [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Homa F, Huffman J, Toropova K, Lopez H, Makhov A, Conway J. Structure of the Pseudorabies Virus Capsid: Comparison with Herpes Simplex Virus Type 1 and Differential Binding of Essential Minor Proteins. J. Mol. Biol. 2013 doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong PA, Yu X, Zhou ZH, Jensen GJ. Correcting for the ewald sphere in high-resolution single-particle reconstructions. Methods Enzymol. 2010;482:369–380. doi: 10.1016/S0076-6879(10)82015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ke EY, Zhou ZH. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J. Struct. Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- Liu H, Cheng L, Zeng S, Cai C, Zhou ZH, Yang Q. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J. Struct. Biol. 2008;161:64–73. doi: 10.1016/j.jsb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Shenk T, Pass RF. In: Cytomegaloviruses In Fields Virology. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Philadelphia: Lippincott-Williams & Wilkins; 2007. pp. 2702–2772. [Google Scholar]
- Nakagawa A, Miyazaki N, Taka J, Naitow H, Ogawa A, Fujimoto Z, Mizuno H, Higashi T, Watanabe Y, Omura T, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure (Camb) 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Trus BL, Cheng N, Steven AC, Sheaffer AK, Tenney DJ, Weller SK, Brown JC. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 2000;74:1663–1673. doi: 10.1128/jvi.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Damania B, Kedes DH. De novo infection with rhesus monkey rhadinovirus leads to the accumulation of multiple intranuclear capsid species during lytic replication but favors the release of genome-containing virions. J. Virol. 2003;77:13439–13447. doi: 10.1128/JVI.77.24.13439-13447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Kedes DH. Rhesus monkey rhadinovirus: a model for the study of KSHV. Curr. Top. Microbiol. Immunol. 2007;312:43–69. doi: 10.1007/978-3-540-34344-8_2. [DOI] [PubMed] [Google Scholar]
- Orzechowska BU, Powers MF, Sprague J, Li H, Yen B, Searles RP, Axthelm MK, Wong SW. Rhesus macaque rhadinovirus-associated non-Hodgkin lymphoma: animal model for KSHV-associated malignancies. Blood. 2008;112:4227–4234. doi: 10.1182/blood-2008-04-151498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rickinson AB, Kieff E. Epstein-Barr Virus. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. Philadelphia: Lippincott-Williams & Wilkins; 2007. pp. 2656–2700. [Google Scholar]
- Rixon FJ. Structure and assembly of herpesviruses. Seminars in Virology. 1993;4:135–144. [Google Scholar]
- Roizman B, Knipe DM, Whitley RJ. In: Herpes simplex viruses In Fields Virology. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Philadelphia: Lippincott-Williams & Wilkins; 2007. pp. 2502–2601. [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 2005;15:227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Homa FL, Booy FP, Newcomb WW, Thomsen DR, Cheng N, Brown JC, Steven AC. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol. Cell. 2007;26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D, Ng TS, Sendamarai AK, Eilers BJ, Lawrence CM, Lok SM, Young MJ, Johnson JE, Fu CY. Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5504–5509. doi: 10.1073/pnas.1300601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D, Quispe J, Grigorieff N, Potter CS, Carragher B, Johnson JE. Maturation in action: CryoEM study of a viral capsid caught during expansion. Structure. 2012;20:1384–1390. doi: 10.1016/j.str.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wu L, Lo P, Yu X, Stoops JK, Forghani B, Zhou ZH. Three-dimensional structure of the human herpesvirus 8 capsid. J. Virol. 2000;74:9646–9654. doi: 10.1128/jvi.74.20.9646-9654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Pluckthun A, Wlodawer A. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat. Struct. Biol. 2000;7:230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XK, O’Connor CM, Atanasov I, Damania B, Kedes DH, Zhou ZH. Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging. J. Virol. 2003;77:13182–13193. doi: 10.1128/JVI.77.24.13182-13193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo H, Jin L, Czornyj E, Hodes A, Hui WH, Nieh AW, Miller JF, Zhou ZH. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution. Elife. 2013;2:e01299. doi: 10.7554/eLife.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat. Struct. Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou ZH. Limiting factors in atomic resolution cryo electron microscopy: no simple tricks. J. Struct. Biol. 2011;175:253–263. doi: 10.1016/j.jsb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 2002;76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Baker ML, Jiang W, Dougherty M, Jakana J, Dong G, Lu G, Chiu W. Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat. Struct. Biol. 2001;8:868–873. doi: 10.1038/nsb1001-868. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Chiu W. Determination of icosahedral virus structures by electron cryomicroscopy at subnanometer resolution. Adv. Protein Chem. 2003;64:93–124. doi: 10.1016/s0065-3233(03)01003-9. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, He J, Jakana J, Tatman JD, Rixon FJ, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.