Abstract
We propose biofunctionalized nanofluidic slits (nanoslits) as an effective platform for real-time fluorescence-based biosensing in a reaction-limited regime with optimized target capture efficiency. This is achieved by the drastic reduction of the diffusion length, thereby a boosted collision frequency between the target analytes and the sensor, and the size reduction of the sensing element down to the channel height comparable to the depletion layer caused by the reaction. Hybridization experiments conducted in DNA-functionalized nanoslits demonstrate the analyte depletion and the wash-free detection ∼10 times faster compared to the best microfluidic sensing platforms. The signal to background fluorescence ratio is drastically increased at lower target concentrations, in favor of low-copy number analyte analysis. Experimental and simulation results further show that biofunctionalized nanoslits provide a simple means to study reaction kinetics at the single-pixel level using conventional fluorescence microscopy with reduced optical depth.
INTRODUCTION
Most conventional open-top fluorescence-based biosensor platforms, such as DNA or protein microarrays, provide only end-point detection where the information of binding reaction kinetics is often missing due to the need of a washing step after reaction.1 On the other hand, microfluidic devices offer tremendous potentials for biological and chemical applications due to the reduction of dimensions and the possibility to integrate different functionalities on a single chip.2 The use of microfluidics for DNA or protein arrays and biosensing enhances the device performances mainly in terms of reduced sample consumption and assay time (i.e., the time to reach target-probe reaction equilibrium).3 This is typically achieved by overcoming the diffusion-limited nature of typical biological reactions using approaches such as forced convection,4 microfluidic mixing,5 sample oscillation,6 or electrophoretic transport.7 However, the simplest and most efficient way to decrease the reaction time is to further shrink the channel dimensions in order to bring target molecules closer to the surface immobilized probes, providing that molecular transport through the channel is assisted by convection.8 Lately, we have witnessed the rapid development of nanofluidics, allowing biomolecular manipulation and observation down to the single molecule level.9–19 While most applications of nanochannels have taken advantage of either the confined fluidic environment for biophysics studies and single molecule separation/analysis or the influence of the electrical double layer to create ion depletion zones for molecular sieving or concentration, there have been surprisingly few studies using nanochannels as biosensors.20 This is partly due to the technical difficulty to fabricate nanoscale devices coupled to the challenge to locally immobilize probe molecules in nanofluidic channels for sensing purposes.
Recent attempts of using biofunctionalized nanochannels demonstrated the detection of streptavidin molecules by impedance spectroscopy.21,22 As expected, the reaction rate in 50 nm deep nanochannels may be increased by a factor of 50 compared with diffusion-limited reactions.21 In both studies, though the measurement of the channel conductance enables a label-free detection method, the sensor response is in fact integrated over the whole channel length without taking full advantage of a locally reduced detection time because of the limited number of target molecules brought by the one-dimensional flux along the sensor. Yet another approach employed a porous block copolymer membrane, with ∼15 nm diameter pores functionalized with single-stranded DNA, and fluorescence microscopy to detect single nucleotide polymorphism (SNP) using a flow-through competitive assay.23 While being effective, as it benefits from the enhanced reaction efficiency caused by molecular confinement, this approach does not rely on planar fabrication technology and thus is not easy to integrate into a lab-on-a-chip.
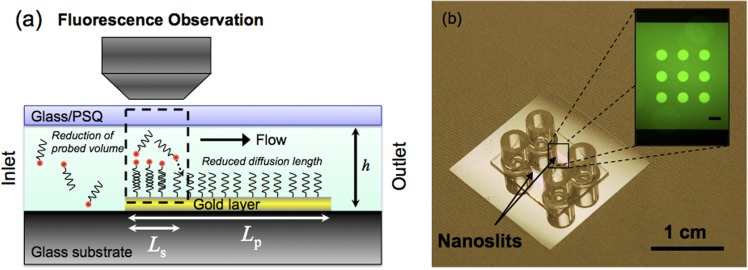
In this work, we present both hybridization experiments and finite element simulations to demonstrate that biofunctionalized uniaxial-confined planar nanochannels, or nanoslits, combined with highly localized sensing elements (where the sensor critical dimension, Ls for single pixels, is comparable to the channel depth h, Figure 1(a)) permit to operate in a sweet spot where diffusion/convection/reaction rates are all comparable and with an optimized target capture efficiency for biodetection or molecular filtering based on rapid biorecognition through highly enhanced collision frequency between the target analyte and the sensing element. In addition, when coupled to fluorescence microscopy, it enables the detection of fluorescently labeled target molecules in real-time with spatially resolved (0.42 μm2 for single camera pixels) reaction kinetics. Figure 1(a) also illustrates that the sampling volume reduction during observation may translate into a decreased background and allow us to directly correlate the fluorescence observed in the nanoslit to the number of hybridized molecules without the need of reagent wash. Since this condition is satisfied along with real-time measurement, it is then possible to use nanofluidics to probe the sensing surface and have access to kinetic data with conventional fluorescence microscopes, i.e., without the need of using complicated and expensive setups, such as surface plasmon resonance (SPR) spectroscopy,24 total internal reflection fluorescence microscopy (TIRFM),25 or quartz crystal microbalance (QCM).26 The consistency between our simulation results and the experimental data confirms that the hybridization sigmoid can be used to extract reaction kinetic parameters. This proposed biofunctionalized nanoslit approach combined with classical fluorescence microscopy thus offers a novel, but simple, solution perfectly suited for the detection of ultra-low copy number analytes and the analysis of ultra-low volumes (e.g., for single cell or purified compounds studies).
FIG. 1.
(a) Schematic of a biofunctionalized nanoslit (channel height h), with embedded probe-immobilized Au patterns, used as a real-time biosensor, where Lp is the sensor patch length and Ls is the single-pixel resolved sensor length. The drastic reduction of the target diffusion time (t) and the enhanced target-probe collision frequency, fm = 1/(2t), using a nanoslit sensor may be apparent with the estimated t from top to bottom of the slit, using a diffusion constant D = 150 μm2/s for 25 mer DNA and a simple one-dimensional diffusion model, which is 33 s and 33 μs for h = 100 μm and 100 nm, respectively, with the corresponding fm = 0.015 Hz and 15 kHz. (b) Photograph of a fabricated nanoslit sensor chip. Inset: fluorescence image of Cy3-labeled DNA solution flowing in the channel, on top of a 3 × 3 array of functionalized gold patterns (scale bar: 100 μm).
MATERIALS AND METHODS
Lately, we have proposed a unique fabrication strategy enabling the realization of nanoslit biosensors where the grafting of probe molecules is carried out before sealing the nanofluidic network.27 Based on the use of polysilsesquioxane (PSQ) as a bonding gasket layer and asymmetric oxygen plasma activation, our approach offers many advantages, when compared to post-sealing functionalization techniques commonly practiced, such as the possibility to immobilize several probes in a single channel for multiplexed sensing. This fabrication technique was used to obtain biosensors embedded in nanoslits for hybridization experiments. Figure 1(b) displays images of a fabricated chip that bears two independent and parallel straight channels; the first one containing activated sensors and the second one being used as a negative control. Each nanoslit is 1 cm long, 1 mm wide, and 600 nm deep, and the gold patterns are 100 μm in diameter prefunctionalized with the thiolated-single-stranded oligodeoxynucleotide (ODN) probes designed for demonstrating the detection of the breast cancer BRCA1 gene.
Nanoslit sensor device fabrication
The 600 nm deep nanoslits were first etched in a fused silica wafer then, to selectively functionalize specific locations of the chips, gold dot arrays were patterned at the surface of the etched channels. The chips were diced and access holes drilled by means of a sandblasting machine. An immobilization protocol provided in the literature28 was used to graft probe molecules by creating a mixed monolayer surface containing thiolated-single-stranded ODN and mercaptohexanol (MCH) self-assembled monolayer. The chips were finally sealed using a modified protocol of the PSQ-bonding technique,29 where PSQ was spun-coated onto a cover slip, cured and activated by oxygen plasma. Bringing the PSQ layer into contact with the glass chip led to a permanent bond through a silanol group condensation process and thus to the encapsulation of the nanoslits with embedded sensors.27 Finally, for packaging purposes, quartz reservoirs were epoxy-glued onto the glass surface (Fig. 1(b)). The chips were loaded with TE buffer (10 mM Tris-HCl, 1 mM EDTA) by capillary forces, and the outlet reservoirs were connected to a vacuum pump to induce liquid flow in the slit channels.
Sample solution and fluorescence imaging
The ODN probe was designed for demonstrating the detection of the breast cancer BRCA1 gene, so the target ODN (complementary DNA or cDNA) included a 25 mer sequence of the gene while the non-complementary ODN sequence (ncDNA) was randomly selected from the human genome (see details of the sequences in Ref. 27). The complementary and non-complementary ODN were labeled with Cy5 and Cy3 dyes, respectively, for fluorescence imaging and were diluted in TE buffer (with 500 mM NaCl) at final concentrations ranging from 1 nM to 1 μM. The hybridization experiments were conducted at room temperature. The chips were imaged using an inverted microscope (Leica DMI 6000B) and an EMCCD (iXonEM + 888, Andor). An acquisition time of 1 s was chosen for all experiments with frame rates varied from 1/30 to 1/1.16 fps. Fluorescence images were obtained with a 10× or a 20× objective and two Cy3-Cy5 dedicated filter sets (41007a-Cy3 and 41008-Cy5, Chroma). The fluorescence of the gold patterns was recorded in real-time after injecting solutions of fluorescently labeled ODN in the nanoslits. After the hybridization and saturation of the sensors, the chip was regenerated by flowing in 1M NaOH solution to the nanoslits.
RESULTS AND DISCUSSION
Feature of reaction/depletion zone
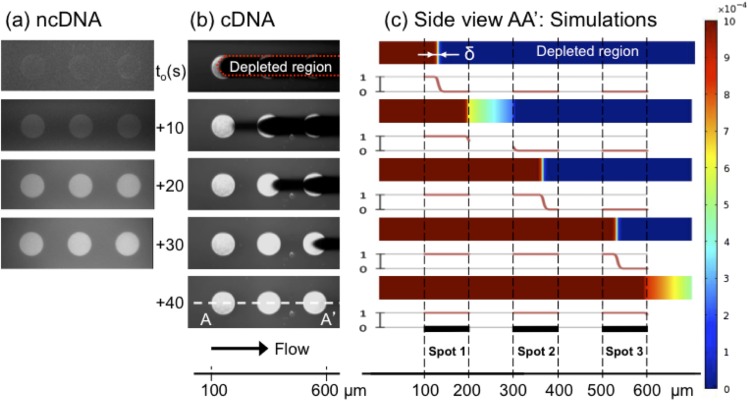
Figure 2 presents experimental observations of the fluorescence images recorded in the channel after flowing in 1 μM ncDNA (Fig. 2(a), Multimedia view) and cDNA (Fig. 2(b), Multimedia view) solutions at different times, where sequential images show three subsequent functionalized gold sensors in the direction of the fluid flow. The injected dye-labeled ncDNA targets do not bind to the probe DNA but slightly increase the background observed in the channel in a uniform way: the two-fold increase of the background on top of the gold sensors is due to the reflectivity of the gold layer that reaches almost 100% at the operating wavelength30 so that the fluorescence emitted by the target molecules flowing on top of the sensor is collected twice by the camera. However, shortly after loading the cDNA solution, concurrent to the fluorescence background increase in the channel, a strong signal is observed on the confronting edge of the first gold pattern. Then, the high fluorescence signal front moves along the pattern (at a much slower pace than the fluid flow itself) until reaching saturation of the sensor. This fluorescent front is followed by a signal loss that corresponds to the dark tail indicated in the top of Fig. 2(b), noted as a target depletion zone. Ultimately, the fluorescence reaches a steady-state level on the whole first sensor pattern and the depletion zone disappears.
FIG. 2.
Simulation results by finite-element-method (FEM, COMSOL Multiphysics 4.0a) in Fig. 2(c) (Multimedia view) describe accurately the experimental observations and show that indeed a diffusing layer, i.e., a depleted layer, forms at the reaction site, which is theoretically predicted for low-aspect ratio fluidic channels, exhibiting low enough Péclet numbers (the ratio of convection rate over diffusion), Pe, combined with a high enough ratio of sensors length, L, with respect to the channel height (h) defined as λ = L/h,31 where L = Lp if the whole sensor patch is used for signal integration and L = Ls if the sensor signal is from a sub-set of the sensor patch, for example, a single pixel of the patch. Analysis of the key figure numbers, Pe along with the Damköhler number (Da), provides the full picture of our sensor's regime of operation. These two dimensionless numbers, which, respectively, relate the rate of reaction and convection to that of diffusion, can in our case be expressed as and , where kon is the forward kinetic constant (4 × 106 M−1 s−1, see Eq. (1) and below), P0 is the surface density of immobilized probe molecules (104 molecules/μm2),28 h is the channel height (600 nm), Um is the mean flow velocity (300 μm/s), and D is the diffusion coefficient (150 μm2/s).32 The calculated value for the Damköhler number being smaller than one (∼0.27) indicates that hybridization in the nanoslit operates under a slight reaction-limited regime. This is not surprising given the fact that it takes only few milliseconds for the target molecules to travel across the nanoslit in the vertical direction. Besides, the flow rate used in our experiments (∼9 nl/min) results in Pe ≈ 1, indicating that neither diffusion nor convective mass transport is limiting. The reduced time, , i.e., the time it takes for the target molecules to travel along the bioactive area (l = 100 μm) related to the time to diffuse across the channel vertically is thus much larger than 2, indicating the reaction efficiency approaches 100%,33 hence the full target collection on the sensor and the formation of the observed depletion zone (δ ∼ h).31
The full target collection feature supports the idea that biofunctionalized nanoslits constitute a means to restrain specific molecules from a solution via immobilized probe molecules. In other words, it can act as a molecular filter based on molecular affinity.34 Considering the total length of the biofunctionalized area, where λ = Lp/h ∼ 160 (which is not the case at the camera pixel size, since the apparent sensor length is much smaller, i.e., λ = Ls/h = 1), with a hybridization efficiency of ∼100%, and neglecting the lateral diffusion (i.e., taking only those pixels located at the center of the sensor), we can roughly estimate the time required to saturate the entire sensor pattern. For a 1 μM target solution, it takes about 11 s to bring enough target molecules to equal the theoretical number of probes grafted along the 100 μm pattern, which is consistent with the timescale observed in our experiments, i.e., ∼10 s (cf. Multimedia view of Fig. 2(b)). Hence, the filtering capacity of the nanoslit, in terms of volume consumed at a given concentration, can be easily evaluated using an analogous simple calculation.
The target depletion zone in the flow direction is a distinct feature in our nanoslit sensor that common microfluidic sensors do not exhibit (since the depletion zone forms above the sensor and in parallel to its surface). Despite the high probability of finding such depletion zone in other nanochannel sensing platforms (where at least λ = Lp/h ≫ 1), observation of the detection front has never been reported so far since the measurement is always integrated over the whole channel length. In this work, by locally probing the concentration of fluorescent targets through the use of a microscope, we are able to capture this depletion front. From Fig. 2, we clearly see that the depletion front exhibits a characteristic length along which the concentration drops from maximum to zero. This situation occurs because transverse diffusion time, longitudinal convection time, and surface absorption times are all comparable. Indeed, in the case of too fast convection time, molecules flowing on top of the sensors are not trapped. If diffusion is too fast, then not enough molecules are fed from convection in order to sustain the front. Finally, the thickness of the depletion layer directly comes from the fact that the reaction is not fast enough to consume all injected molecules. Thus, the reason for observing such a depletion layer with non-negligible thickness is because we operate in the sweet spot of where Pe and Da are both close to unity. In fact, in the present case where no hydrodynamic dispersion occurs,35 the depletion zone characteristics could probably be used to estimate the target concentration.36 However, here we have rather observed and measured the local concentration of target molecules at a fixed position as a function of time and as the depletion front moves along the sensor patch.
Performance of nanoslit sensors
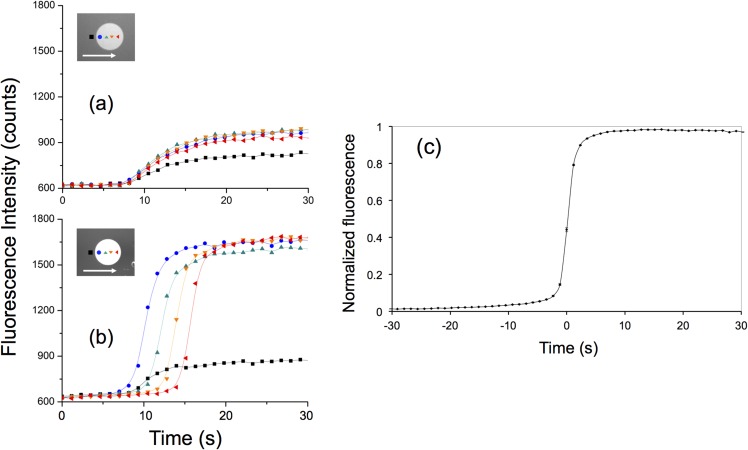
We studied the response of our nanoslit biosensor with high spatial resolution, by plotting in time the fluorescence recorded from single pixels of the CCD camera with sizes ranging from 0.65 × 0.65 to 1.3 × 1.3 μm2 (using a 20× or 10× objective, respectively). Figure 3 presents the variation of fluorescence for 4 pixels of the same sensor pattern when injecting 1 μM of ncDNA (Fig. 3(a)) and cDNA (Fig. 3(b)) solutions where ncDNA can be discriminated from the one with matching sequence. When matching targets are injected in the nanoslit, we observed a strong correlation between the signals from the subsequent sensing pixels indicating the high degree of reproducibility of the results down to the single-pixel level (Fig. 3(c)), which highlights another feature of our device, i.e., each pixel represents an independent measurement of the reaction kinetics. Hence, large sampling area over a number of pixels ensures much reduced statistical errors (higher precision) than non-spatially resolved sensing platforms.
FIG. 3.
Fluorescence measurements from single CCD image pixels (size: 0.65 × 0.65 μm) when flowing (a) 1 μM ncDNA solution and (b) 1 μM cDNA solution. The location of the probed pixels along the functionalized sensing pattern and the direction of the fluid flow are shown in the insets. The black curve on each plot corresponds to the fluorescence background measured in the channel. (c) Averaged plot of 70 response curves obtained by measuring the fluorescence onto 0.42 μm2 pixels located within different sensing patterns from single nanoslit sensor chip. The average values were obtained after superimposing pixels with normalized fluorescence signal and shifted time origin. Measurements were carried out in a single experiment where 1 μM cDNA was injected in the nanoslit. The small standard errors illustrate the excellent reproducibility of the measurements.
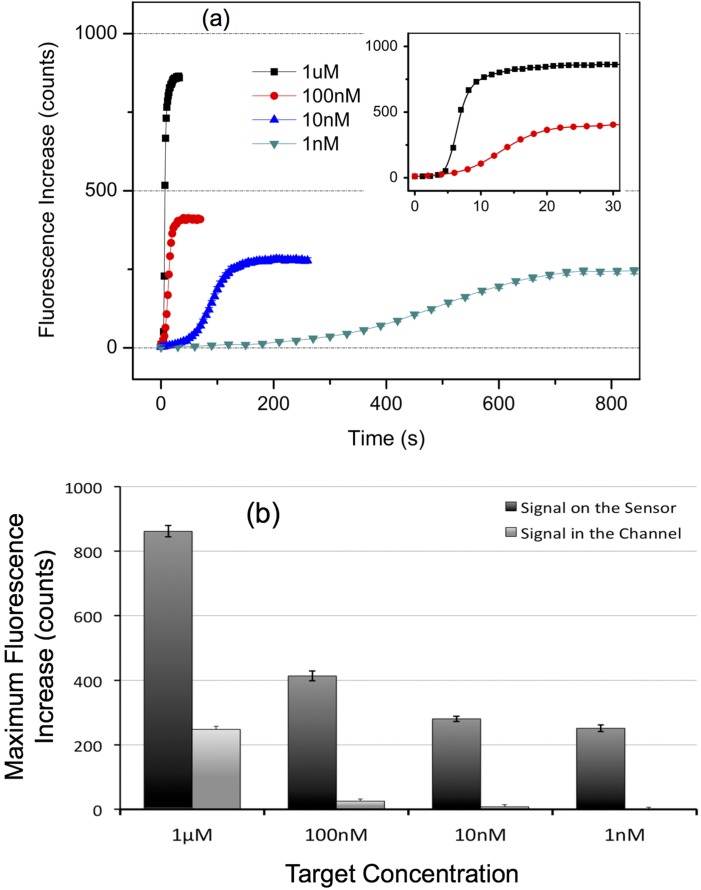
Finally, the response of our nanoslit sensor was studied for various target molecule concentrations, ranging from 1 nM to 1 μM. Figure 4(a) displays the time variation of the mean fluorescence level (obtained using 20 pixels selected from 2 sensor patterns in the channel) as a function of target concentration. We observed that, as expected, the time to reach signal saturation increases with decreasing target concentration since it takes more time to bring the equivalent number of target molecules to reach the steady-state saturation of the sensor. Also, as expected, the final fluorescence level decreases with target concentration because the background due to the fluorescent molecules flowing on top of the sensing region is reduced (Figure 4(b)). After taking into account the reflectivity of the gold layer, i.e., where the final fluorescence levels are corrected by subtracting twice the fluorescence background, the steady-state fluorescence related to the hybridized targets is actually comparable for all the working concentrations: this is a result commonly observed in other sensing platforms for concentrated solutions when the response is characterized by a Langmuir adsorption isotherm.37 Further, Figure 4 demonstrates that the reaction equilibrium is fast-reached and that, to our knowledge, the response time of the nanoslit sensor is at least 10 times faster than the best published results.38
FIG. 4.
(a) Kinetic curves from single CCD image pixels (size: 1.3 × 1.3 μm) as a function of cDNA target concentration. Each plot was obtained by averaging the fluorescence intensity measured onto 20 different pixels (10 flow direction-aligned pixels on 2 different patterns) during a single experiment (the error bars: standard errors). The time origin was arbitrarily chosen. The inset is a closed-in plot, with the same coordinate units, for 1 μM and 100 nM target concentration. Note that the data presented here include the background due to the fluorescence emitted by the target molecules flowing in the channel and the one reflected from the gold sensor surface. (b) Saturated fluorescence intensity measured in the channel and on the sensor with various flow-in target concentrations. The fluorescence background due to the flowing molecules constitutes 28% of the sensor signal at 1 μM. However, this level falls down to 6%, 2.9%, and 0.05% for target concentrations of 100, 10, and 1 nM, respectively, indicating an increase of S/B ratio to 560 fold when the target concentration is reduced from 1 μM to 1 nM (the error bars: standard errors).
The fluorescence background due to the molecules flowing in the nanoslit is compared to the saturated fluorescence signal measured on the sensor in Figure 4(b). This background constitutes 28% of the sensor signal at 1 μM, meaning that this signal does not directly relate the amount of captured targets. However, this level falls down to 6%, 2.9%, and 0.05% for target concentrations of 100, 10, and 1 nM, respectively. It indicates that the signal to background (S/B) ratio may be increased 560 fold when the target concentration is reduced from 1 μM to 1 nM, a feature enabling us to perform a virtually background-free sensing with no washing step, in contrary to most microfluidics-based microarray systems with optical detection,1 thus making it possible to directly relate the fluorescence signal to the amount of hybridized targets, Ph, at the sensor surface at particularly low target concentrations. As a result, the fluorescence signals displayed in Figure 4(a) constitute kinetic curves that, in addition to providing means for real-time detection where no washing step is necessary, can be used to derive the reaction kinetic parameters, which is usually achieved by using more complex setups, e.g., SPR spectroscopy or QCM, where high spatial resolution is not readily achievable.
Extraction of kinetic constant
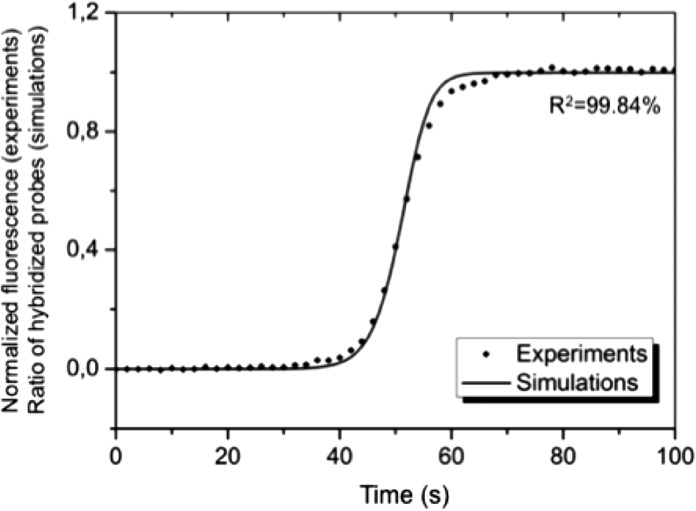
As a demonstration, we have used the finite-element-method (FEM, COMSOL Multiphysics 4.0a) to model the response of our nanoslit biosensor. Since the width of the slit is much larger than the channel depth, the structure was assumed to be only two dimensional, thus the influence of lateral diffusion of the target molecules was neglected. The nanoslit model was built via the Chemical Reaction Engineering Module, where the fluid flow was computed using the Transport of Diluted Species Interface (that supposes a chemical species transport through diffusion and convection with the mass balance equation implemented). Then, assuming first-order Langmuir kinetics, the number of molecules captured on the sensing surfaces was estimated from31
| (1) |
where C is the concentration of the flowing analytes, Ph is the surface density of the hybridized probes, and koff is the corresponding reverse kinetic constant. The concentration of target molecules flowing in the channel and that at the sensor surface and the hybridization rates were then calculated as a function of time using experimental (e.g., Um = 300 μm/s) and literature values (koff = 10−5 s−1).39 For simplicity, only the forward kinetic constant was used to adjust the simulation results, since the in-depth parametric fit requires access to the dissociation phase and is not the main focus of this work. When the signal obtained from flowing 100 nM cDNA solution compared with computed values of captured target molecules (Figure 5), we found an optimal fitting value of kon = 4 × 106 M−1 s−1 that is in agreement with previous report.39 The fairly good fit between the sigmoid provided by the simulated results and the experimental data demonstrates that a similar model can indeed be used to extract kinetic constants from the output of our nanoslit biosensor since the fraction of hybridized probes (Ph) can be derived from the total fluorescence intensity recorded at the pixel level, I, providing the channel height is small enough
| (2) |
Hence, our platform not only provides reaction kinetics with better spatial resolution (single-pixel) and precision (if averaged over a number of pixels), and optimized reaction conditions (full target collection and reaction-limited-regime) over the state-of-the-art microfluidic array platforms40 but also offers a low-cost alternative to SPR or QCM technologies.
FIG. 5.
Experimental and simulated response of the nanoslit biosensor at the single-pixel level (size: 1.3 × 1.3 μm). The normalized measured fluorescence is compared to the fraction of hybridized probe molecules (target concentrations: 100 nM), with an optimal fitting value of kon = 4 × 106 M−1s−1, is in agreement with a previous report39 (the error bars: standard errors).
CONCLUSIONS
In summary, the reduction of the characteristic length in fluidic systems is a key to avoid the diffusion-limiting nature of target-probe reactions commonly observed in bioarrays. The typical low Reynolds number characterizing microfluidic channels is not an obstacle to fast kinetics anymore because the molecules convectively carried on top of the sensing area instantaneously react with the probe molecules, through the much shortened diffusion length and the enhanced collision frequency. In this work, we have demonstrated that the reduction of channel height down to the nanoscale permits not only to operate in an optimized target capture and a reaction-limited regime but also to detect fluorescently labeled biomolecules in real-time with the possibility to extract kinetic parameters. We realized the following features in our nanoslit sensors. First, the adsorption efficiency is expected to reach unity in a flow-through configuration, hence biofunctionalized nanoslits can act as molecular filters based on biorecognition and specificity, as indicated by the unique target depletion zone observed. Second, the time to reach reaction equilibrium is optimized with a limited amount of target molecules that are all detected, thus providing means for ultrafast real-time biosensing of ultra-low copies of molecules in ultra-small volumes. Most importantly, the results presented here indicate that the reduction of target concentration screened during observation enables wash-free sensing with a direct correlation of the signal from the fluorescent targets to the number of hybridized molecules, thanks to the dramatic increase of the S/B ratio. When combined with the predictive power of FEM simulation, our fluorescence-based nanoslit biosensor platform enables a simple and low-cost way to study spatiotemporally resolved reaction kinetics.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Chau-Hwang Lee for fruitful discussions and technical assistance from the Academia Sinica (AS) Nano Core Facilities. This work was gratefully supported by the AS Postdoctoral Fellowship (Nos. 097-2811-M-001-136 and 098-2811-M-001-162) to T.L., and AS Integrated Thematic Project (AS-103-TP-A01), Ministry of Science and Technology (MOST), Taiwan (Nos. 102-2112-M-001-005-MY3 and 103-2923-M-001-007-MY3), and Asian Office for Aerospace Research and Development (#FA2386-12-1-4002) to C.F.C., and the Orchid Program from MOST and the Bureau Français de Taipei.
References
- 1.Sassolas A., Leca-Bouvier B. D., and Blum L. J., Chem. Rev. 108, 109 (2008). 10.1021/cr0684467 [DOI] [PubMed] [Google Scholar]
- 2.Yager P., Edwards T., Fu E., Helton K., Nelson K., Tam M. R., and Weigl B. H., Nature 442, 412 (2006). 10.1038/nature05064 [DOI] [PubMed] [Google Scholar]
- 3.Situma C., Hashimoto M., and Soper S. A., Biomol. Eng. 23, 213 (2006). 10.1016/j.bioeng.2006.03.002 [DOI] [PubMed] [Google Scholar]
- 4.Pappaert K., Vanderhoeven J., Van Hummelen P., Dutta B., Clicq D., Baron G. V., and Desmet G., J. Chromatogr., A 1014, 1 (2003). 10.1016/S0021-9673(03)00715-5 [DOI] [PubMed] [Google Scholar]
- 5.Toegl A., Kirchner R., Gauer C., and Wixforth A., J. Biomol. Tech. 14(3), 197; see http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2279950/. [PMC free article] [PubMed] [Google Scholar]
- 6.Lenigk R., Liu R. H., Athavale M., Chen Z. J., Ganser D., Yang J. N., Rauch C., Liu Y. J., Chan B., Yu H. N., Ray M., Marrero R., and Grodzinski P., Anal. Biochem. 311, 40 (2002). 10.1016/S0003-2697(02)00391-3 [DOI] [PubMed] [Google Scholar]
- 7.Heller M. J., Forster A. H., and Tu E., Electrophoresis 21, 157 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Kim J. H. S., Marafie A., Jia X. Y., Zoval J. V., and Madou M. J., Sens. Actuators, B 113, 281 (2006). 10.1016/j.snb.2005.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tegenfeldt J. O., Prinz C., Cao H., Huang R. L., Austin R. H., Chou S. Y., Cox E. C., and Sturm J. C., Anal. Bioanal. Chem. 378, 1678 (2004). 10.1007/s00216-004-2526-0 [DOI] [PubMed] [Google Scholar]
- 10.Duan C. H., Wang W., and Xie Q., Biomicrofluidics 7, 026501 (2013). 10.1063/1.4794973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakajin O. B., Duke T. A. J., Chou C. F., Chan S. S., Austin R. H., and Cox E. C., Phys. Rev. Lett. 80, 2737 (1998). 10.1103/PhysRevLett.80.2737 [DOI] [PubMed] [Google Scholar]
- 12.Guo L. J., Cheng X., and Chou C. F., Nano Lett. 4, 69 (2004). 10.1021/nl034877i [DOI] [Google Scholar]
- 13.Reisner W., Larsen N. B., Flyvbjerg H., Tegenfeldt J. O., and Kristensen A., Proc. Natl. Acad. Sci. U.S.A. 106, 79 (2009). 10.1073/pnas.0811468106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh J. W., Taoni A., Chen Y. L., and Chou C. F., Nano Lett. 12, 1597 (2012). 10.1021/nl2045292 [DOI] [PubMed] [Google Scholar]
- 15.Matsuoka T., Kim B. C., Moraes C., Han M., and Takayama S., Biomicrofluidics 7, 041301 (2013). 10.1063/1.4816835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S. N. and Lee L. J., Biomicrofluidics 7, 011301 (2013). 10.1063/1.4774071 [DOI] [Google Scholar]
- 17.Sriram K. K., Yeh J. W., Lin Y. L., Chang Y. R., and Chou C. F., Nucleic Acids Res. 42, e85 (2014). 10.1093/nar/gku254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lesser-Rojas L., Sriram K. K., Liao K. T., Lai S. C., Kuo P. C., Chu M. L., and Chou C. F., Biomicrofluidics 8, 016501 (2014). 10.1063/1.4861435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sriram K. K., Chang C.-L., Rajesh Kumar U., and Chou C.-F., Biomicrofluidics 8, 052102 (2014). 10.1063/1.4892515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han J., in Nanofluidics Nanoscience and Nanotechnology, edited by Edel J. B. and deMello A. J. (RCS Publishing, 2009), p. 31. [Google Scholar]
- 21.Schoch R. B., Cheow L. F., and Han J., Nano Lett. 7, 3895 (2007). 10.1021/nl0724788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karnik R., Castelino K., Fan R., Yang P., and Majumdar A., Nano Lett. 5, 1638 (2005). 10.1021/nl050966e [DOI] [PubMed] [Google Scholar]
- 23.Yang S. Y., Son S., Jang S., Kim H., Jeon G., Kim W. J., and Kim J. K., Nano Lett. 11, 1032 (2011). 10.1021/nl200357y [DOI] [PubMed] [Google Scholar]
- 24.Ouellet E., Lausted C., Lin T., Yang C. W. T., Hood L., and Lagally E. T., Lab Chip 10, 581 (2010). 10.1039/b920589f [DOI] [PubMed] [Google Scholar]
- 25.Peter C., Meusel M., Grawe F., Katerkamp A., Cammann K., and Borchers T., Fresenius J. Anal. Chem. 371, 120 (2001). 10.1007/s002160101006 [DOI] [PubMed] [Google Scholar]
- 26.Okahata Y., Kawase M., Niikura K., Ohtake F., Furusawa H., and Ebara Y., Anal. Chem. 70, 1288 (1998). 10.1021/ac970584w [DOI] [PubMed] [Google Scholar]
- 27.Leïchlé T., Lin Y.-L., Chiang P.-C., Hu S.-M., Liao K.-T., and Chou C.-F., Sens. Actuators, B 161, 805 (2012). 10.1016/j.snb.2011.11.036 [DOI] [Google Scholar]
- 28.Herne T. M. and Tarlov M. J., J. Am. Chem. Soc. 119, 8916 (1997). 10.1021/ja9719586 [DOI] [Google Scholar]
- 29.Gu J., Gupta R., Chou C.-F., Wei Q., and Zenhausern F., Lab Chip 7, 1198 (2007). 10.1039/b704851c [DOI] [PubMed] [Google Scholar]
- 30.Handbook of Optics, 2nd ed., Sponsored by the Optical Society of America, edited by Bass M. ( McGraw-Hill, New York, 1995). [Google Scholar]
- 31.Squires T. M., Messinger R. J., and Manalis S. R., Nat. Biotechnol. 26, 417 (2008). 10.1038/nbt1388 [DOI] [PubMed] [Google Scholar]
- 32.Stellwagen E. and Stellwagen N. C., Electrophoresis 23, 2794 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Sjolander S. and Urbaniczky C., Anal. Chem. 63, 2338 (1991). 10.1021/ac00020a025 [DOI] [PubMed] [Google Scholar]
- 34.Wang Y. Y., Cheng P., and Chan D. W., Proteomics 3, 243 (2003). 10.1002/pmic.200390036 [DOI] [PubMed] [Google Scholar]
- 35.Balakotaiah V. and Chang H. C., Philos. Trans. R. Soc. A 351, 39 (1995). 10.1098/rsta.1995.0025 [DOI] [Google Scholar]
- 36.Balakotaiah V. and Chang H. C., Siam J. Appl. Math. 63, 1231 (2003). 10.1137/S0036139901368863 [DOI] [Google Scholar]
- 37.Su X. D., Wu Y. J., and Knoll W., Biosens. Bioelectron. 21, 719 (2005). 10.1016/j.bios.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 38.Wei C. W., Cheng J. Y., Huang C. T., Yen M. H., and Young T. H., Nucleic Acids Res. 33, e78 (2005). 10.1093/nar/gni078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Hammer D. A., and Graves D. J., Biophys. J. 89, 2950 (2005). 10.1529/biophysj.104.058552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noerholm M., Bruus H., Jakobsen M. H., Telleman P., and Ramsing N. B., Lab Chip 4, 28 (2004). 10.1039/b311991b [DOI] [PubMed] [Google Scholar]