Abstract
The fibroblast growth factor receptor (FGFR) cascade plays crucial roles in tumor cell proliferation, angiogenesis, migration and survival. Accumulating evidence suggests that in some tumor types, FGFRs are bona fide oncogenes to which cancer cells are addicted. Because FGFR inhibition can reduce proliferation and induce cell death in a variety of in vitro and in vivo tumor models harboring FGFR aberrations, a growing number of research groups have selected FGFRs as targets for anticancer drug development. Multikinase FGFR/vascular endothelial growth factor receptor (VEGFR) inhibitors have shown promising activity in breast cancer patients with FGFR1 and/or FGF3 amplification. Early clinical trials with selective FGFR inhibitors, which may overcome the toxicity constraints raised by multitarget kinase inhibition, are recruiting patients with known FGFR(1–4) status based on genomic screens. Preliminary signs of antitumor activity have been demonstrated in some tumor types, including squamous cell lung carcinomas. Rational combination of targeted therapies is expected to further increase the efficacy of selective FGFR inhibitors. Herein, we discuss unsolved questions in the clinical development of these agents and suggest guidelines for management of hyperphosphatemia, a class-specific mechanism-based toxicity. In addition, we propose standardized definitions for FGFR1 and FGFR2 gene amplification based on in situ hybridization methods. Extended access to next-generation sequencing platforms will facilitate the identification of diseases in which somatic FGFR(1–4) mutations, amplifications and fusions are potentially driving cancer cell viability, further strengthening the role of FGFR signaling in cancer biology and providing more possibilities for the therapeutic application of FGFR inhibitors.
Keywords: fibroblast growth factor receptor FGFR, amplification, cancer, hyperphosphatemia, oncogene, targeted therapy
introduction
Fibroblast growth factor receptor (FGFR) signaling plays crucial roles in cancer cell proliferation, migration, angiogenesis, and survival. The FGFR pathway was primarily studied in cancer as a direct promoter of endothelial cell proliferation and tumor neoangiogenesis, having complementary and synergistic effects with the vascular endothelial growth factor (VEGF) signaling [1, 2]. Recent studies have uncovered increasing evidence that in addition to its role as an escape mechanism of anti-VEGF therapies, deregulated FGFRs can function as driving oncogenes in certain tumor types, acting in a cell autonomous fashion to maintain the malignant properties of cancer cells [2, 3]. When FGFRs are mutated or amplified, aberrant activation of downstream pathways results in mitogenic, mesenchymal, and antiapoptotic responses in cells. The combination of knockdown studies and selective pharmacological inhibition in preclinical models confirms that FGFRs are attractive targets for therapeutic intervention in cancer [2]. In this article, we will focus on the main FGFR genomic alterations found in human cancer to date, how they may contribute to specific tumor types, describe the range of treatment strategies currently employed or in development to inhibit deregulated FGFRs and discuss unsolved questions in the clinical development of these agents.
FGFR pathway
The FGFR family includes four receptor tyrosine kinases FGFR(1–4) comprised of an extracellular domain, a transmembrane domain, and a cytoplasmic domain. The extracellular portion contains three immunoglobulin-like (Ig) folds (IgI, IgII, and IgIII) with a stretch of eight consecutive acidic residues between IgI and IgII (the acidic box). While the IgII and IgIII domains are necessary and sufficient for ligand binding, the amino-terminal portion of the receptor containing IgI and the acidic box has an auto-inhibitory function. Alternative splicing of the IgIII extracellular fragment of FGFR1, 2, or 3 may generate isoforms that differ in terms of ligand-binding specificity, with IgIIIb and IgIIIc specifically expressed in the epithelium and mesenchyme, respectively. The intracellular region of FGFRs contains a juxta-membrane domain, a split kinase domain with the classical tyrosine kinase motifs, and a carboxy-terminal tail [4].
Fibroblast growth factors (FGFs) are secreted glycoproteins that are readily sequestered by the extracellular matrix and the cell surface by heparan sulfate proteoglycans (HPSGs). Cell-surface HPSGs stabilize the FGF ligand–receptor interaction by protecting FGFs from protease-mediated degradation [2]. In the case of hormone-like FGFs (FGF19, 21, and 23), the FGF–FGFR interaction requires a cell surface co-receptor, klotho or β-klotho, for high-affinity binding and signaling. Upon ligand binding, FGFR substrate 2 (FRS2) functions as a key adaptor protein that associates with the receptor and initiates downstream signaling with activation of mitogen activated protein kinase (MAPK) and the phosphoinositide-3-kinase (PI3K)/AKT pathways. FGFR signaling also couples to phospholipase C-gamma (PLC-γ) in an FRS2-independent manner and stimulates protein kinase C (PKC), which partly reinforces the MAPK pathway activation by phosphorylating RAF. Depending on the cellular context, several other pathways are also activated by FGFRs including the p38 MAPK and Jun N-terminal kinase pathways, signal transducer and activator of transcription signaling and ribosomal protein S6 kinase 2 (RSK2) [2, 4, 5].
The mechanisms of attenuation and negative feedback control of FGFR signaling are poorly understood and are likely to vary depending on the cell type. Downstream signaling can be attenuated through the induction of MAPK phosphatases (MAPK3), Sprouty (SPRY) proteins, and SEF family members that modulate receptor signaling at several points in the signal transduction cascade. In addition, following activation, FGFRs are internalized and then degraded or recycled according to the level of ubiquitination [2, 4, 5].
In cancer, different FGFR pathway aberrations have been identified and include: (i) gene amplification or post-transcriptional regulation giving rise to receptor overexpression; (ii) FGFR mutations producing receptors that are either constitutively active or exhibit a reduced dependence on ligand binding for activation; (iii) translocations resulting in expression of FGFR-fusion proteins with constitutive FGFR kinase activity; (iv) alternative splicing of FGFR and isoform switching, which substantially alters ligand specificity increasing the range of FGFs that can stimulate tumor cells; and (v) upregulation of FGF expression in cancer or stromal cells and the enhanced release of FGFs from the extracellular matrix, resulting in paracrine/autocrine activation of the pathway. In humans, several gain-of-function germline mutations in the FGFR genes result in skeletal dysplasias, with FGFR2 mutations a common cause of craniosynostosis and FGFR3 mutations frequent in chondrodysplasia syndromes. Mutations in cancer resemble those seen in hereditary disorders and interestingly, they are not limited to the kinase domain but are spread over the complete length of the gene. Notably, FGFR signaling in cancer exhibits clear context-dependence, with aberrations differing according to tumor type [4–8]. Table 1 summarizes the most frequent FGFR genomic deregulations in solid tumors and the details are discussed subsequently.
Table 1.
Common FGFR genomic deregulations in solid tumors
| Aberration |
Tumor | Prevalence (%) | |
|---|---|---|---|
| FGFR1 | Amplification | Breast (hormone receptor positive) | 10 |
| Lung (squamous cell carcinoma) | 10–20 | ||
| Lung (small cell) | 6 | ||
| Head and neck (squamous cell carcinoma) | 10–17 | ||
| Esophageal (squamous cell carcinoma) | 9 | ||
| Ovarian | 5 | ||
| Osteosarcoma | 5 | ||
| FGFR2 | Amplification | Breast (triple-negative) | 4 |
| Gastric | 5–10 | ||
| Mutation | Endometrial | 12 | |
| FGFR3 | Mutation | Bladder (nonmuscle invasive) | 50–60 |
| Bladder (muscle-invasive) | 10–15 | ||
| Translocation | Bladder (muscle-invasive) | 6 | |
| Glioblastoma | 3–7 | ||
| FGFR4 | Amplification | Colorectal | 5 |
| Mutation | Rhabdomyosarcoma | 8 | |
FGFRs as oncogenic drivers in cancer
breast cancer
FGFR family members are infrequently mutated but frequently overexpressed in breast cancer, and this is often accompanied by increased, or altered, expression of FGF ligands [9]. The 8p11-12 amplicon, which contains FGFR1, is observed in about 10% of breast cancer patients, predominantly hormone receptor positive (HR+) disease [9–12]. Importantly, genes other than FGFR1 in the 8p11-12 amplicon are also likely to contribute to carcinogenesis [13–15]. In addition, it is noteworthy to mention that FGFR1 is simultaneously amplified with an amplicon containing CCND1, FGF3, FGF4, and FGF19 on chromosome 11q12-14 in one-third of the samples, and in vitro studies suggests substantial functional interaction between the genes on 8p11-12 and 11q [16]. The 11q 12-14 amplicon is seen in ∼15%–20% of human breast tumors [17, 18], and was shown to correlate with increased invasiveness in node-negative breast carcinoma [17]. FGFR1-overexpressed cancers are more likely to be progesterone receptor negative and present high proliferation, characteristics of the luminal-B subtype [19]. Large series have shown that FGFR1 amplification is an independent predictor of poor outcome [19, 20] and drives resistance to endocrine therapy [19]. No significant heterogeneity in FGFR1 amplification status has been observed after matching primary and metastatic carcinoma samples, although this observation is based on a small sample size [12].
Importantly, in vitro studies reinforce the potential oncogenic nature of FGFR1 amplification. Inhibition of FGFR1 kinase activity causes death of breast cancer-derived cell lines that overexpress FGFR1, indicating that these cells are addicted to the pathway for viability [11]. In vivo models of FGFR1-amplified breast cancer are challenging and still missing.
Additionally, FGFR2 has also been implicated in some cases of breast cancer, with gene amplification in 4% of triple-negative tumors [21]. FGFR2 amplification appears to promote breast tumorigenicity through maintenance of breast tumor-initiating cells [22]. In vitro studies showed that FGFR2-amplified cell lines have constitutive activation of the receptor and are highly sensitive to FGFR inhibition, with induction of apoptosis and decrease in the tumor-initiating cells population [21, 22].
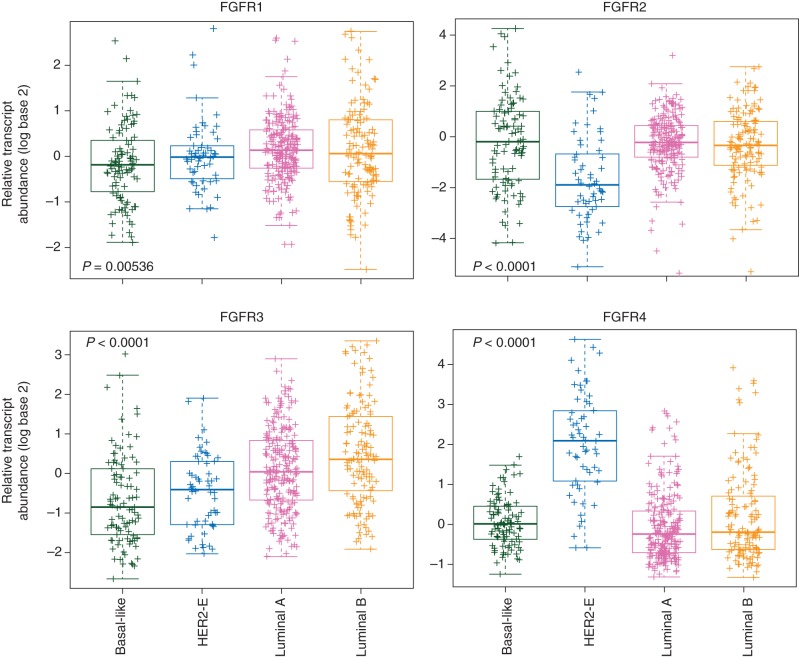
Ligand-dependent signaling is also likely to play a key role in breast cancer. Basal-like breast cancer cell lines with epithelial–mesenchymal transition features and triple-negative breast cancers frequently express FGF2. RNA interference targeting of FGF2 in basal-like cell lines significantly reduced growth in vitro. Notably, small-molecule tyrosine kinase inhibitors (TKIs) of FGFR induce tumor shrinkage in xenogafts models with an active autocrine FGF2 signaling loop [23]. These experiments highlight the potential driving role of the FGFR pathway in different subtypes of breast cancer. Figure 1 portrays the relative expression of the FGFR(1-4) genes according to The Cancer Genome Atlas (TCGA) breast cancer microarray-based dataset, emphasizing the differences across the intrinsic subtypes of breast tumors [9]. The significance of FGFR4 activation in HER2-positive tumors still needs further investigation but it is noteworthy to mention that it is one of the HER2-enriched specific genes included in the 50-gene intrinsic subtype predictor (PAM50) [24].
Figure 1.
Relative expression of the FGFR(1–4) genes across the intrinsic subtypes of breast cancer. Gene expression data and subtype calls have been obtained from The Cancer Genome Atlas (TCGA) breast cancer microarray-based dataset (http://cancergenome.nih.gov/). P-values have been obtained by comparing the mean expression across the groups (ANOVA test).
lung cancer
FGFR1 is amplified in 10%–20% of squamous non-small-cell lung cancer (NSCLC) [25–28]. In early-stage disease, it appears to correlate with increased cigarette smoking and poor survival [29]. Interestingly, recurrent FGFR1 amplification is also seen in 10%–17% of head and neck [30] and 9% of esophageal squamous cell carcinomas [31].
Preclinical studies clearly demonstrate that FGFR1 amplification confers dependence upon FGFR signaling. Treatment of FGFR1-amplified lung cancer cell lines with selective FGFR TKIs resulted in growth inhibition and apoptosis. Moreover, in vivo xenograft models derived from both cell lines and patient tumors have also shown increased sensitivity with stasis or regressive effects [25, 32, 33]. Ligand-dependent epithelial–mesenchymal transition in NSCLC cell lines has been shown in vitro, and inhibition of FGFR signaling through antisense RNA or anti-FGF2 monoclonal antibodies (mAbs) led to inhibition of cellular proliferation and tumor growth [34].
In small-cell lung cancer (SCLC), exome sequencing identified FGFR1 locus amplification in 6% of the tumors [35]. There is a clear correlation of high copy number gains in cytogenetic bands encoding FGFR1 and in vitro/in vivo sensitivity of SCLC models to FGFR inhibition [36, 37]. Paracrine production of FGFs has also been reported and high levels of serum FGF2 in SCLC are associated with a poor prognosis [38].
gastric cancer
FGFR2 amplification is found in 5%–10% of gastric cancers, mainly in the aggressive diffuse subtype [39, 40]. Aberrations are mutually exclusive with other receptor tyrosine kinase amplifications (ERBB2, MET) [40]. In vitro, FGFR2-amplified gastric cancer cell lines are selectively sensitive to the growth inhibitory effects of FGFR TKIs [41]. FGFR2 downregulation led to significant inhibition of cell growth and survival that further translated into tumor growth regression in vivo [42–44]. Additional evidence of target dependence comes from xenograft models treated with anti-FGFR2 mAbs, which exhibit potent antitumor activity against gastric cancer driven by activated FGFR2 signaling [45, 46].
endometrial cancer
FGFR2 mutations occur in 12% of endometrioid endometrial carcinomas [47–49]. The S252W and P253R substitutions in the extracellular domain, in particular, confer a gain in ligand-binding promiscuity by isoform-switching, thereby establishing an autocrine loop of pathway activation [50]. The N549K and K659E mutations in the kinase domain lead to ligand-independent, constitutively active FGFR2 [48]. Interestingly, FGFR2 mutations in endometrial cancer show a mutually exclusive pattern with mutations in KRAS [51]. In early-stage disease, they are associated with shorter disease-free and overall survival [51]. FGFR2-mutant endometrial cancer cell lines and in vivo tumor xenograft models are highly sensitive to FGFR TKIs, which reflects oncogenic addiction to the aberrant receptor [43, 44, 47].
urothelial cancer
Up to 80% of low-grade and low-stage tumors express mutant FGFR3, but this abnormality is found in less than 20% of invasive high-grade bladder cancer [52–57]. The most common FGFR3 mutations in urothelial cell carcinoma occur in the extracellular and transmembrane domains (R248C, S249C, G370C, and Y373C) and lead to ligand-independent dimerization and constitutive activation [58]. Several mutations in the kinase domain of FGFR3 (K650E, K650M), leading to enhanced kinase activity, have also been described [58]. FGFR3 mutations in bladder cancer are mutually exclusive with mutations in HRAS [59]. Multiple preclinical studies including bladder cell culture experiments and mouse models treated with small-molecule inhibitors and anti-FGFR3 mAbs reported decrease in cell proliferation and reduction in tumor growth [43, 60–62]. Activity of an FGFR3-specific antagonistic antibody likely to disrupt dimerization, and hence activation, of the FGFR3 R248C and S249C mutants, was also extended to wild-type tumors that overexpress the receptor [60].
Fusions of the FGFR3 gene with transforming acidic coiled-coil (TACC3) were recently identified in a subset of invasive bladder tumors (2 of 32 samples) [63]. The FGFR3 component lacks the final exon that includes PLCγ binding site, leading to loss of negative regulation and higher expression of an active receptor. Notably, cell lines were extremely sensitive to selective FGFR inhibition [63].
brain tumors
A small subset of glioblastomas (3%–7%) also harbors oncogenic chromosomal translocations that fuse in-frame the tyrosine kinase coding domains of FGFR1 or FGFR3 to TACC genes [64, 65]. This aberration is mutually exclusive with epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), or MET amplifications, known drivers in brain tumors [65]. The fusion, caused by tandem duplication on 4p16.3, leads to loss of the 3′-UTR of FGFR3, blocking negative gene regulation and enhancing its expression [65]. In agreement with in vitro data showing constitutive kinase activity, in vivo models show that administration of an FGFR inhibitor prolongs survival of mice harboring FGFR3-TACC3-initiated gliomas [64].
sarcomas
Recurring somatic FGFR4 kinase domain mutations (K535 and E550) have been described in 8% of rhabdomyosarcoma patients. FGFR4 knockdown in a human rhabdomyosarcoma cell line transplanted into mice reduced tumor growth and metastasis [66]. FGFR1 amplification was detected in a small subset of osteosarcomas (1 of 17 samples) and predicted sensitivity to a selective FGFR inhibitor in vitro [44]. Activation of FGFR signaling pathway as a result of amplification of the FRS2 adaptor was recently identified in dedifferentiated liposarcomas. Selective FGFR inhibitor was able to inhibit growth of cell lines with FRS2 and FGFR overexpression [67].
colorectal cancer
Colon cancer has been shown to overexpress FGF18 and FGF19, acting through the FGFR3-IIIc and FGFR4 receptors on the tumor cells [5]. An anti-FGF19 mAb that selectively blocks the interaction of FGF19 with FGFR4 inhibited growth of colon tumor xenografts in vivo, supporting a role for paracrine/autocrine FGFR activation in cancer maintenance [68].
hepatocellular carcinoma
Ligand-dependent signaling is also likely to play a role in hepatocellular carcinomas (HCCs). FGF2 and FGFs 8, 17, 18, and 19 are upregulated and have been shown to initiate autocrine growth stimulation, cell survival, and neoangiogenesis in liver cancer [5]. Genetic knockdown of FGF19 inhibits the growth of HCC cell lines carrying the amplicon [69]. Targeting FGF19 with a mAb effectively prevented HCCs in FGF19 transgenic mice [68] and a humanized anti-FGFR4 mAb was reported to inhibit tumor growth in HCC xenograft models [70]. Interestingly, FGF19 amplification predicts sensitivity to selective FGFR inhibitors in vitro in HCC cell lines that express the coreceptor β-klotho, which is essential for high-affinity interactions of FGF19 with FGFR4 [44].
prostate cancer
Extensive correlative studies in human prostate cancer as well as preclinical models indicate that FGFR signaling plays an important role in prostate cancer progression, epithelial–mesenchymal transition and angiogenesis. FGFR1 and FGFR4 are frequently overexpressed, several FGFs are upregulated, while expression of SEF, a negative regulator of the FGFR pathway, is reduced in aggressive cancers [5, 71]. Pathway addiction and a potential role for FGFR targeting in prostate cancer have also been documented. Neutralizing antibody against FGF8 displayed potent anti-tumor activity in mouse models [72] and FGFR TKI treatment was able to completely inhibit tumor growth and angiogenesis in vivo [73].
FGFRs as angiogenic drivers in cancer
FGFs directly promote endothelial cell proliferation and indirectly synergize with the VEGF and PDGF pathways, promoting tumor neoangiogenesis through complementary and overlapping functions. FGFR pathway activation has been shown to mediate resistance to anti-VEGF therapy [1, 3]. In the preclinical setting, tumors progressing to anti-VEGF treatment showed a higher expression of FGF2 and combined VEGFR and FGFR blockade led to increased antitumor activity in the setting of adaptive/evasive resistance [74]. Interestingly, FGF2 is higher in patients with colorectal cancer after the failure of bevacizumab-containing regimens [75] and in glioblastoma patients after treatment with a VEGFR TKI [76].
FGFRs as suppressors in cancer
It is well recognized that context-dependent differences in signaling can lead to either tumor promotion or senescence in response to activated FGFR pathway. FGFR2 signaling is clearly oncogenic in many tumor models. However, several studies support a tumor protective effect of FGFR2 signaling, with FGFR2IIIb expression blocking proliferation in vitro and being downregulated on progression of multiple tumor types [2]. Loss-of-function FGFR2 mutations have been identified in 10% of melanoma tumors and cell lines and are the clearest indication of opposing roles of FGFRs in cancer [77]. These mutations result in receptor loss-of-function through several distinct mechanisms, including loss of ligand binding affinity (R251Q), impaired receptor dimerization (E219K), destabilization of the extracellular domains (G271E), and reduced kinase activity (D530N, I642V, and A648T) [77]. Nevertheless, the mechanisms underlying the tumor suppressive effects of FGFR2 are unknown and oncogene-induced senescence is a possible explanation for the distinct gene functions according to molecular context and tumor type.
resistance to FGFR targeting
Limited preclinical data are available regarding resistance to FGFR inhibitors. Using functional RNA interference screens, loss of PTEN was identified as a potential mechanism of resistance that was specific to FGFR2-amplified cell lines [78]. Remarkably, ponatinib and the mTOR inhibitor ridaforolimus had a synergistic effect on the in vitro growth of endometrial cell lines bearing an activating FGFR2 mutation, irrespective of PTEN status [79]. In contrast, EGFR activation was identified as a resistance mechanism in FGFR3-mutant bladder cancer cell lines. Combinations of FGFR and EGFR inhibitors had increased efficacy both in vitro and in vivo [78]. The concept that activation of the HER family members could compensate for inhibition of FGFRs was also shown in other tumor types, including triple-negative breast cancer [80, 81]. In addition, in xenograft models of FGFR1 and MET co-activation, the combination of selective FGFR and MET inhibitors led to increased antitumor efficacy [80]. It should be noted that few data have been published on the molecular mechanism of cell death induced by FGFR inhibition in different cellular contexts [7]. In order to exploit the oncogene dependency for therapeutic gain, we need to understand the exact mechanism of oncogene addiction that exists in different cell types.
Furthermore, FGFR activation could potentially serve as a mechanism of acquired resistance to targeted therapies possibly related to the extensive cross-talk between FGF and oncogenic pathways. Pathway activation as a result of FGFs released in the tumor microenvironment has been implicated in resistance to gefitinib and erlotinib in NSCLC, cetuximab in KRAS wild-type squamous carcinoma cells and to vemurafenib in BRAF V600E melanoma cells [82–87]. These data will allow rationale clinical testing of combination of FGFR inhibitors with other targeted agents.
translation to the clinic
The strong evidence of increased sensitivity of multiple FGFR-aberrant cell lines and tumor models to FGFR inhibitors reveals a substantial therapeutic opportunity for selective intervention, validates oncogene addiction for proliferation and/or viability, and provides a rationale for the use of FGFR inhibitors in such instances.
Most FGFR inhibitors currently in development are small molecules kinase inhibitors of the ATP-binding domain. As the catalytic domains of various kinases exhibit significant structural homology, TKIs generally demonstrate activity against more than one kinase. This is a common feature of the first-generation of FGFR inhibitors, which generally have activity against VEGFR and/or PDGFR, two structurally related receptor tyrosine kinases. Multikinase inhibition may increase effectiveness in the treatment of a particular tumor type by disrupting redundant pathways that drive resistance. This might be valid when these agents are used as antiangiogenic therapies. Nevertheless, toxicity is expected to increase significantly and off-targets effects may be detrimental when FGFR is targeted in the context of the oncogene-addicted tumors discussed above, as side-effects may limit the ability to achieve doses required for effective FGFR inhibition. We are now in a unique position to validate clinically the many hypotheses that have been generated preclinically. In addition to selective FGFR TKIs, mAbs that bind to the extracellular domain of different FGFRs and compete with endogenous ligands (FGFs), thereby blocking FGFR dimerization and downstream activation, are also undergoing clinical testing.
toxicology and pharmacodynamics markers
FGFRs are widely expressed in normal tissues and have a key role in development and physiology, notably the phosphate and vitamin D homeostasis. Preclinical models with highly potent selective FGFR TKIs have caused hyperphosphatemia-mediated tissue calcification owing to blockade of FGF23 release from bone and its signal in the kidney [88]. FGF23 binds FGFR4 and the IIIc isoforms of FGFR1 and FGFR3, but uncertainty remains about the relative contribution of individual FGFR subtypes to hyperphosphatemia [3, 8]. Therefore, increase in serum FGF23, phosphate, and vitamin D levels are potential biomarkers for effective FGFR inhibition. In early clinical trials with FGFR inhibitors, development of hyperphosphatemia as a class-specific toxicity may help in the definition of an optimal biological dose [89].
Additional mechanism-based toxicities observed in toxicology models include skin and other cutaneous events, as well as dose-dependent keratopathy and retinal pigment epithelial detachment. In contrast to multikinase VEGFR/FGFR inhibitors, efficacious doses of potent selective FGFR inhibitors do not induce elevations in blood pressure or proteinuria [90, 91].
multikinase (nonselective) FGFR inhibitors
The most clinically advanced FGFR TKIs have dominant pharmacological activity in other kinases, such as VEGFR, PDGFR, FLT3, RET, KIT, and BCR-ABL. These include brivanib, cediranib, nintedanib, lenvatinib, sulfatinib, dovitinib, ponatinib, and lucitanib. Although in vitro kinase and cellular activity of these compounds against FGFR1-3 is variable (IC50 ranging from 2 to >500 nM), multiple preclinical models showing increased activity in FGFR-deregulated tumors have been published. FGFR1 amplification in breast cancer cell lines predicts sensitivity to brivanib, dovitinib, ponatinib, and lucitanib [43, 92–94]. In vitro and in vivo models of FGFR1-amplified lung cancer are highly sensitive to ponatinib and lucitanib as single agents [43, 94]. Cediranib, dovitinib, and ponatinib are very active in FGFR2-amplified gastric cancer models [40, 42, 43]. In addition, dovitinib and ponatinib also show significant antitumor activity in FGFR2-mutant endometrial and FGFR3-mutant bladder cancer xenografts [43, 62, 95].
Several of these molecules are being developed as antiangiogenic agents and trials are underway in a variety of tumor types irrespective of FGFR aberrations, such as nintedanib and brivanib in endometrial cancer (clinical trials.gov NCT01225887 and NCT00888173), as well as nintedanib and levantinib in lung cancer (NCT01441297, NCT01529112). Should there be any partial or complete responses in these trials it will be interesting to correlate with the amplification/mutation status of FGFR1/FGFR2. Levantinib has shown promising efficacy in HCC, but correlation with FGF upregulation when its indication is mainly based on the potent antiangiogenic activity is more challenging [96]. As seen in Table 2, clinical trials with multikinase FGFR inhibitors in patients selected based on FGFR aberrations are also in progress.
Table 2.
Genomically driven clinical trials of FGFR inhibitors
| Agent | Phase | Clinical trials.gov | Description |
|---|---|---|---|
| Multikinase inhibitors | |||
| Dovitinib | Phase II | NCT01379534 | FGFR2-mutant or wild-type endometrial cancer |
| Phase II | NCT01732107 | FGFR3-mutant or overexpressed BCG refractory urothelial carcinoma | |
| Phase II | NCT01719549 | FGFR2-amplified gastric cancer | |
| Lucitanib | Phase I/II | NCT01283945 | Expansion cohort in FGFR1-amplified tumors |
| Ponatinib | Phase II/III | NCT01761747 | Advanced squamous cell lung cancers with FGFR kinase alterations |
| Selective FGFR inhibitors | |||
| AZD4547 | Phase I | NCT00979134 | Expansion cohort in FGFR1- or FGFR2 amplified tumors |
| Phase II | NCT01457846 | Gastric or lower-esophageal cancer, FGFR2-amplified or not, randomized to AZD4547 or paclitaxel | |
| Phase I/II | NCT01202591 | Estrogen receptor + and FGFR1-amplified breast cancer, randomized to AZD4547 plus fulvestrant or fulvestrant alone | |
| BGJ398 | Phase I | NCT01004224 | FGFR1- or FGFR2-amplified, FGFR3-mutant advanced cancer |
| LY2874455 | Phase I | NCT01212107 | Advanced cancer with FGFR aberrations during dose expansion |
| JNJ-42756493 | Phase I | NCT01703481 | Expansion cohort in FGFR1-, FGFR2-, or FGFR4-amplified tumors |
The first reported trial, which evaluated the VEGFR/PDGFR/FGFR inhibitor dovitinib in FGFR1-amplified and nonamplified metastatic breast cancer, failed to reach its primary end point of improved overall response rate in the genomically selected arm [92]. However, in the subgroup with FGFR1 amplification, 13% had unconfirmed partial responses. Activity was observed primarily in the subgroup of patients with co-amplification of FGF3 as measured by quantitative real-time polymerase chain reaction (RT-PCR). Pharmacodynamic analysis indicates FGFR and VEGFR inhibition at tolerable doses, with consistent and maintained increase in serum FGF23, VEGF, and PDGF [92, 97].
More recently, the therapeutic potential of FGFR inhibition in breast cancer was uncovered with preliminary results of phase I trial with lucitanib, a potent VEGFR/FGFR inhibitor [98]. Enrollment in the expansion cohort at recommended doses was limited to patients whose tumors harbored FGFR1 amplification by fluorescent in situ hybridization (FISH) or 11q amplification (locus of FGF3) as measured by comparative genomic hybridization (CGH) array. Seven of 10 assessable patients had a partial response, 4 with FGFR1-amplified breast tumors, and 3 with 11q amplification (one of them without concomitant FGFR1 amplification). Pharmacodynamic data confirming pathway inhibition is still pending [98]. Interestingly, hyperphosphatemia, an adverse event specific of potent FGFR inhibitors, was not consistently observed with lucitanib or dovitinib [92, 98]. The toxicity profile of these agents is mainly related to VEGFR inhibition, including hypertension, proteinuria, and hypothyroidism, but off-target effects such as gastrointestinal toxicity and asthenia were also dose-limiting [92, 98]. It remains to be proven whether the impressive efficacy of lucitanib is related to FGFR inhibition, VEGFR inhibition or the combination of targets, although prior clinical trials with other multikinase VEGFR inhibitors as single agents in unselected breast cancer patients showed disappointing results [99, 100]. The results of clinical trials with more selective FGFR inhibitors in FGFR-deregulated breast cancer will shed light on this issue. A confirmatory phase II trial with lucitanib will start recruitment soon.
selective FGFR inhibitors
Many selective FGFR inhibitors have recently started clinical development. Phase I trials with BGJ398, AZD4547, LY2874455, and JNJ-42756493 are currently recruiting patients. The in vitro kinase activity of these compounds against FGFR1, FGFR2, and FGFR3 is very high (IC50 < 10 nM), with variable anti-FGFR4 activity. A patient-derived xenograft model of FGFR1-amplified lung cancer was highly sensitive to AZD4547 as single agent [32], and BGJ398 also demonstrated potent in vivo activity in FGFR2-amplified gastric and FGFR2-mutant endometrial cancer [44, 95]. Following stringent preclinical data suggesting oncogene addiction, clinical development is centered in FGFR-aberrant tumors, as shown in Table 2.
Early results of the ongoing phase I trials with BGJ398 and AZD4547 have been presented. In the BGJ398 study, patients with advanced solid tumors showing FGFR1 or FGFR2 amplification or FGFR3 mutation are still been recruited. Results of the first 29 patients (18 with FGFR1-amplified tumors) are available [101]. The most frequently observed adverse events were diarrhea, fatigue, nausea and hyperphosphatemia (about one-third of the patients) and dose-limiting toxicities included grade 3 elevations in transaminase levels and grade 2 corneal events. The incidence of hyperphosphatemia increased at higher doses of BGJ398 but could be managed with phosphate binders and diuretics. One lung cancer patient with FGFR1 amplification by FISH analysis had a confirmed partial response. Preliminary efficacy data in the breast cancer population with FGFR aberrations was disappointing, with no partial responses in the first 13 patients recruited [101]. With regards to AZD4547, dose-limiting toxicities included renal failure (pyelonephritis, dehydration), mucositis, increase in transaminase levels and hyperphosphatemia [102]. In addition to dose-proportional increase in phosphate and vitamin D levels, other potential mechanism-based toxicities included alopecia, nail disorders, dry skin, and asymptomatic retinal pigment epithelial detachment. Investigators did not report soft-tissue calcification. Human exposure at recommended phase II doses was consistent with exposures in preclinical models that induced tumor regressions. Of 20 patients with FGFR pathway aberrations, 5 had clinical benefit as assessed by investigators, 3 of them with significant tumor shrinkage: 1 patient with FGFR1-amplified squamous NSCLC had a confirmed partial response; 1 with FGFR1-amplified breast cancer patient presented 25% decrease in the size of target lesions; and 1 with FGFR3-mutant bladder cancer had 23% reduction in tumor size for more than 6 months. Clinical benefit appeared to be higher in those with high-level FGFR1 amplification (FGFR1/CEP8 ratio >2.8 by FISH) [102]. A phase II study evaluating AZD4547 in combination with endocrine therapy in breast cancer has recently started accrual (NCT01202591).
monoclonal antibodies targeting the FGFR pathway
Therapeutic mAbs can be highly specific for a particular FGF ligand or FGFR isoform, hence displaying a more narrow range of toxicity when compared with pan-FGFR inhibitors. Specific inhibition of a particular oncogenic FGFR molecule, including splice variants that are selectively upregulated in tumor cells, would avoid targeting different FGFR isoforms that have opposing effects in cancer cells. In addition, by recruiting the immune system via antibody-dependent cellular cytotoxicity or complement-dependent cytotoxicity, antitumor activity might be increased. Several antibodies targeting the FGFR pathway have been assessed in preclinical studies in a variety of solid tumors, including gastric, bladder, prostate cancer, and HCC, as described above.
The first agent to move into the clinic was FP-1039, a soluble fusion protein consisting of the extracellular domain of human FGFR1 linked to the Fc portion of human IgG1. It was engineered to spare the metabolic hormone FGFs (including FGF23 involved in phosphate and vitamin D metabolism) and to bind tightly to all of the mitogenic FGF ligands [103]. In vivo models with genetic aberrations in the FGFR pathway, including FGFR1-amplified lung cancer and FGFR2-mutated endometrial cancer, were particularly sensitive to FP-1039-mediated tumor inhibition. In addition, it was shown to block FGF- and VEGF-induced angiogenesis in vivo [103]. The final report of the phase I trial with FP-1039 is still pending but it does not appear to significantly increase serum calcium and phosphate levels or induce hypertension or proteinuria [103]. Unfortunately, a phase II study testing FP-1039 in FGFR2 S252W or P253R mutant endometrial cancer patients was not feasible. The original assumption was that at least 5% of patients screened would qualify, but after screening 70 patients, none qualified (NCT01244438).
Another antibody targeting the FGFR pathway tested in the clinic is the human anti-FGFR3 agent MFGR1877S [104]. Based on preclinical data showing activity of the compound in bladder cancer with FGFR3 overexpression or mutations, the phase I trial was enriched with patients diagnosed with advanced urothelial carcinomas. Five of the 10 bladder cancer patients had stable disease as their best response (4 remained on study for more than three cycles). Dose-limiting toxicity was thrombocytopenia in one patient, and the drug was escalated until effective doses based on preclinical models [104]. Future development of this compound and other mAbs targeting the FGFR pathway is unknown at this time.
challenges in the clinical development of FGFR inhibitors
Despite promising preliminary results with FGFR inhibition in genetically selected tumors, many logistical challenges related to prescreening strategies and biomarker platform selection will need to be overcome for successful clinical development of this class of agents. We need to define precisely the alterations associated with pathway addiction taking into consideration context-dependency of the FGFR signaling. In parallel, optimal molecular diagnostic procedures for FGFR aberrations need to be developed, as companion diagnostics will be required to enroll patients in clinical trials. As an example, the definition of FGFR1/2 amplification based on in situ hybridization techniques varied significantly in the published literature, as shown in Table 3.
Table 3.
Definition of FGFR1/2 amplification based on in situ hybridization techniques in retrospective studies and or clinical trials
| Aberration | Method | Tumor | Threshold | References |
|---|---|---|---|---|
| FGFR1 amplification | FISH | Squamous lung cancer | Average copy number/nucleus ≥6, FGFR1/CEN8 ratio ≥2, or ≥10% tumor cells with ≥15 FGFR1 signals or large copy number clusters | [27] |
| Average copy number/nucleus ≥9 | [25, 29] | |||
| FGFR1/CEN8 ratio ≥2.2 | [26] | |||
| Breast cancer | Average copy number/nucleus ≥6 | [92] | ||
| Average copy number/nucleus ≥6 or FGFR1/CEN8 ≥2.2 | [98] | |||
| Any tumor | FGFR1/CEN8 ratio >2 in ≥10% tumor cells | [102] | ||
| CISH | Breast cancer | Average copy number/nucleus ≥5 or large copy number clusters | [19, 20] | |
| Average copy number/nucleus >6 or large copy number clusters | [12] | |||
| FGFR2 amplification | FISH | Gastric cancer | FGFR2/CEN10 ratio >2 | [39] |
| Any tumor | FGFR2/CEN10 ratio >2 in ≥10% tumor cells | [102] |
CEN, centromere; CISH, chromogenic in situ hybridization; FISH, fluorescence in situ hybridization.
Many limitations for a consensus definition of FGFR pathway activation became clear during the last years. First, there is a marked genomic heterogeneity in the FGFR1 amplicon structure in breast (broad) and squamous NSCLC (focal) and the influence of these differences on the degree of FGFR addiction is unknown. Second, in situ hybridization analyses showed that tumors might exhibit a focal and heterogenous pattern of amplification with frequent polysomy of CEN8; this could lead to an FGFR1/CEN8 ratio below 2.0 despite an increase in absolute numbers of FGFR1 signals compared with normal tissue. Empiric data will be required to define the exact amplification cut-off that predicts anti-FGFR therapy response. In the meantime, our molecular pathology laboratories have been applying the HER2 FISH CAP/ASCO (College of American Pathologists/American Society of Clinical Oncology) guidelines to score FGFR (amplified is either FGFR/CEP ratio >2.2 or average FGFR gene copy number >6 signals/nucleus). Assays for detecting FGFR fusions will be challenging, as at least some of the variants such as FGFR3-TACC3 are not amenable to traditional FISH analysis as the genes map too close to each other.
Standardized definition of pathway activation becomes an even more difficult problem for identifying tumors in which paracrine/autocrine signaling is potentially driving tumor cell proliferation. Ligand amplification can be detected by many different techniques, such as RT-PCR, array CGHs, microarrays, and RNA sequencing. Thus far, the inconsistent definitions of FGFs amplifications make their use investigational in nature, although early trials have clearly shown that ligand amplification might help define the population of patients with higher chances of response. Importantly, molecular testing is also evolving, moving from ‘one test-one drug’ paradigm to multiplex approaches looking for mutations, amplifications, and gene fusions [105, 106]. More robust and reproducible genomic platforms that can screen alterations of multiple FGF-pathway components in a single assay are being gradually incorporated into the prescreening process of FGFR inhibitors trials. With massively parallel sequencing, the number of clinically significant and potentially predictive oncogenic aberrations in the FGFR pathway is expected to increase. This is illustrated by the recent finding of intragenic duplications of the portion of FGFR1 encoding the tyrosine kinase domain in grade II diffuse childhood gliomas [107], recurrent FGFR1 hotspot mutations in pediatric pilocytic astrocytomas [108], as well as TKI-sensitive FGFR2 and FGFR3 mutations in squamous NSCLC [109]. Newly rare but recurrent FGFR2 and FGFR3 fusions were also described across multiple solid tumors, including cholangiocarcinomas and squamous NSCLC [110, 111]. These rearrangements apparently share the mechanism of pathway activation, with different fusion partners mediating oligomerization, which triggers overexpression of the respective FGFR kinase. Patients whose tumors harbor these FGFR fusions are being referred to clinical trials with FGFR inhibitors and the preliminary results are eagerly anticipated.
Finally, investigators should keep an open mind during biomarker-driven clinical development of targeted therapies. First, selective FGFR inhibitors might have a more favorable safety profile when compared with multikinase inhibitors and hence could be combined with other targeted agents. Taking into consideration preliminary data showing that the clinical responses of FGFR-amplified tumors to selective FGFR inhibitors have not been impressively high, we envision that rational combination strategies with other targeted agents should be further explored, promising partners being endocrine therapies, anti-EGFR and downstream PI3K pathway or RAF/MEK inhibitors. On the other hand, the clinical activity of selective FGFR inhibitors as single agents in highly addicted FGFR(1–4) mutated and FGFR(2–3) rearranged tumors is still unknown. Nevertheless, mechanism-based toxicities of FGFR inhibitors may be particularly difficult to control and their long-term consequences are unknown. Table 4 summarizes a specific toxicity management protocol for hyperphosphatemia based on our experience during early clinical development of these agents. Second, multikinase nonselective FGFR inhibitors may show significant antitumor activity in both FGFR wild-type and mutant tumors as well as superior efficacy in FGFR-aberrant tumors by co-targeting parallel pathways, therefore allowing greater flexibility in patient selection. Correctly designing clinical trials to address these different hypotheses is crucial [112]. Third, antiangiogenic activity of potent VEGFR/FGFR TKIs makes this class of agents suitable in the setting of resistance to prior anti-VEGF therapies, directing clinical investigation of these compounds toward tumor types without FGFR genomic aberrations.
Table 4.
Guidelines for management of hyperphoshatemia related to FGFR inhibitors
| Serum phosphate ≥5.5–7 mg/dl |
| Continue FGFR inhibitor |
| Dietary phosphate intake restriction |
| Phosphate binder |
| Sevelamer 1 tablet (800 mg) per meal; i.e. every 8 h |
| Increase the dose of sevelamer up to 1200 mg every 8 h if needed (phosphate levels still increasing after 1 week) |
| Serum phosphate >7–9 mg/dl |
| Continue FGFR inhibitor |
| If serum phosphate levels continue >7–9 mg/dl despite phosphorus lowering therapy for 2 weeks, the dose of the FGFR inhibitor should be reduced. If serum phosphate levels >7–9 mg/dl despite dose reduction and optimal phosphate lowering therapy for 2 weeks, temporarily discontinue FGFR inhibitor. Restart at reduced dose level when serum phosphate <7 mg/dl. |
| Dietary phosphate intake restriction |
| Phosphate binder |
| Sevelamer 2 tablets (1600 mg) per meal; i.e. every 8 h |
| Phosphaturic agents |
| Acetazolamide 1 tablet (250 mg) 2–3 × per day |
| Serum phosphate >9 mg/dl |
| Discontinue FGFR inhibitor |
| Restart at reduced dose level when serum phosphate <7 mg/dl. |
| Dietary phosphate intake restriction |
| Phosphate binder |
| Sevelamer 2 tablets (1600 mg) per meal; i.e. every 8 h |
| Phosphaturic agents |
| Acetazolamide 1 tablet (250 mg) 2–3 × per day |
| Repeated episodes of serum phosphate >9 mg/dl or simultaneous alteration in renal function |
| Discontinue FGFR inhibitor permanently |
conclusion
Targeting FGFRs is a promising therapeutic strategy in a variety of cancers. Early clinical data confirm preclinical studies suggesting that in some tumor types, FGFRs may act as oncogenes to which cancer cells are addicted. Nevertheless, in the clinical scenario, questions such as what class of agents is the most promising (nonselective versus selective FGFR inhibitors) and whether combination therapies are needed in order to obtain meaningful clinical benefit are still unsolved. Successful development will ultimately depend on the selection of tumors in which FGF signaling is driving proliferation and survival. Widespread use of in situ hybridization techniques and massively parallel sequencing tests will facilitate the identification of additional diseases in which the therapeutic use of FGFR inhibitors is worth testing. In this context, histology-independent trials (‘basket’ design) enrolling patients with different FGFR genomic aberrations could be very informative with regard to future directions for clinical investigation of these compounds. Notably, not all activating mutations in FGFR(1–4) can be effectively targeted by current selective FGFR inhibitors. Therefore, functional experiments designed to determine which aberrations play causative roles in tumorigenesis are required in order to fully understand the clinical implications of genomic data. Eventually, continued translational research in the field will further strengthen the role of FGF signaling in cancer biology and allow personalized use of FGFR inhibitors at the clinic.
disclosure
The authors have declared no conflicts of interest related to this review article. R. Dienstmann is a recipient of “La Caixa International Program for Cancer Research & Education”.
acknowledgement
We thank all physicians from the Vall d'Hebron University Hospital referring patients with FGFR aberrations for treatment with matched targeted agents at the Molecular Therapeutics Research Unit, as well as the whole team involved in the prescreening process.
references
- 1.Lieu C, Heymach J, Overman M, et al. Beyond VEGF: inhibition of the fibroblast growth factor pathway and antiangiogenesis. Clin Cancer Res. 2011;17:6130–6139. doi: 10.1158/1078-0432.CCR-11-0659. doi:10.1158/1078-0432.CCR-11-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner N, Grose R. Fibroblast growth factor signaling: from development to cancer. Nature Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. doi:10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 3.Crawford Y, Ferrara N. Tumor and stromal pathways mediating refractoriness/resistance to anti-angiogenic therapies. Trends Pharmacol Sci. 2009;30:624–630. doi: 10.1016/j.tips.2009.09.004. doi:10.1016/j.tips.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Knights V, Cook SJ. De-regulated FGF receptors as therapeutic targets in cancer. Pharmacol Ther. 2010;125:105–117. doi: 10.1016/j.pharmthera.2009.10.001. doi:10.1016/j.pharmthera.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199–213. doi: 10.1042/BJ20101603. doi:10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 6.Heinzle C, Sutterlüty H, Grusch M, et al. Targeting fibroblast-growth factor receptor-dependent signaling for cancer therapy. Expert Opin Ther Targets. 2011;5:829–846. doi: 10.1517/14728222.2011.566217. doi:10.1517/14728222.2011.566217. [DOI] [PubMed] [Google Scholar]
- 7.Greulich H, Pollock PM. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol Med. 2011;17:283–292. doi: 10.1016/j.molmed.2011.01.012. doi:10.1016/j.molmed.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. doi:10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Network. Comprehensive molecular characterization of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. doi:10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courjal F, Cuny M, Simony-Lafontaine J, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57:4360–4367. [PubMed] [Google Scholar]
- 11.Reis-Filho JS, Simpson PT, Turner NC, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. doi:10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- 12.Brunello E, Brunelli M, Bogina G, et al. FGFR-1 amplification in metastatic lymph-nodal and haematogenous lobular breast carcinoma. J Exp Clin Cancer Res. 2012;31:103. doi: 10.1186/1756-9966-31-103. doi:10.1186/1756-9966-31-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray ME, Yang ZQ, Albertson D, et al. Genomic and expression an analysis of the 8p11–12 amplicon in human breast cancer cell lines. Cancer Res. 2004;64:40–47. doi: 10.1158/0008-5472.can-03-1022. doi:10.1158/0008-5472.CAN-03-1022. [DOI] [PubMed] [Google Scholar]
- 14.Garcia MJ, Pole JC, Chin SF, et al. A 1Mbminimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–5245. doi: 10.1038/sj.onc.1208741. doi:10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- 15.Haverty PM, Fridlyand J, Li L, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–542. doi: 10.1002/gcc.20558. doi:10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 16.Kwek SS, Roy R, Zhou H, et al. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene. 2009;28:1892–1903. doi: 10.1038/onc.2009.34. doi:10.1038/onc.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fioravanti L, Cappelletti V, Coradini D, et al. Int-2 oncogene amplification and prognosis in node-negative breast carcinoma. Int J Cancer. 1997;74:620–624. doi: 10.1002/(sici)1097-0215(19971219)74:6<620::aid-ijc11>3.0.co;2-9. doi:10.1002/(SICI)1097-0215(19971219)74:6<620::AID-IJC11>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Gruel N, Lucchesi C, Raynal V, et al. Lobular invasive carcinoma of the breast is a molecular entity distinct from luminal invasive ductal carcinoma. Eur J Cancer. 2010;46:2399–2407. doi: 10.1016/j.ejca.2010.05.013. doi:10.1016/j.ejca.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. doi:10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbauomy Elsheikh S, Green AR, Lambros MB, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. doi:10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner N, Lambros MB, Horlings HM, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. doi:10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S, Dubrovska A, Salamone RJ, et al. FGFR2 promotes breast tumorigenicity through maintenance of breast tumor-initiating cells. PLoS One. 2013;8:e51671. doi: 10.1371/journal.pone.0051671. doi:10.1371/journal.pone.0051671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharpe R, Pearson A, Herrera-Abreu MT, et al. FGFR signaling promotes the growth of triple-negative and basal-like breast cancer cell lines both in vitro and in vivo. Clin Cancer Res. 2011;17:5275–5286. doi: 10.1158/1078-0432.CCR-10-2727. doi:10.1158/1078-0432.CCR-10-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. doi:10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. doi:10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heist RS, Mino-Kenudson M, Sequist LV, et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7:1775–1780. doi: 10.1097/JTO.0b013e31826aed28. doi:10.1097/JTO.0b013e31826aed28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol. 2012;25:1473–1480. doi: 10.1038/modpathol.2012.102. doi:10.1038/modpathol.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. doi:10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013;31:731–737. doi: 10.1200/JCO.2012.43.8622. doi:10.1200/JCO.2012.43.8622. [DOI] [PubMed] [Google Scholar]
- 30.Freier K, Schwaenen C, Sticht C, et al. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC) Oral Oncol. 2007;43:60–66. doi: 10.1016/j.oraloncology.2006.01.005. doi:10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 31.von Loga K, Kohlhaussen J, Marx AH, et al. FGFR1 amplification is linked to the squamous cell carcinoma subtype in esophageal carcinoma. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research (AACR); 6–10 April; Washington DC. 2013. Abstr 3024. [Google Scholar]
- 32.Zhang J, Zhang L, Su X, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18:6658–6667. doi: 10.1158/1078-0432.CCR-12-2694. doi:10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 33.Semrad TJ, Mack PC. Fibroblast growth factor signaling in non-small-cell lung cancer. Clin Lung Cancer. 2012;13:90–95. doi: 10.1016/j.cllc.2011.08.001. doi:10.1016/j.cllc.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Marek L, Ware KE, Fritzsche A, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. doi:10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. doi:10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pardo OE, Latigo J, Jeffery RE, et al. The fibroblast growth factor receptor inhibitor PD173074 blocks small cell lung cancer growth in vitro and in vivo. Cancer Res. 2009;69:8645–8651. doi: 10.1158/0008-5472.CAN-09-1576. doi:10.1158/0008-5472.CAN-09-1576. [DOI] [PubMed] [Google Scholar]
- 37.Thomas A, Lee J, Wang Y, et al. The role of fibroblast growth factor receptor 1 in small cell lung cancer. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research (AACR); 6–10 April; Washington DC. 2013. Abstr 5466. [Google Scholar]
- 38.Ruotsalainen T, Joensuu H, Mattson K, et al. High pretreatment serum concentration of basic fibroblast growth factor is a predictor of poor prognosis in small cell lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:1492–1495. [PubMed] [Google Scholar]
- 39.Matsumoto K, Arao T, Hamaguchi T, et al. FGFR2 gene amplification and clinicopathological features in gastric cancer. Br J Cancer. 2012;106:727–732. doi: 10.1038/bjc.2011.603. doi:10.1038/bjc.2011.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng N, Goh LK, Wang H, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–684. doi: 10.1136/gutjnl-2011-301839. doi:10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kunii K, Davis L, Gorenstein J, et al. FGFR2- amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008;68:2340–2348. doi: 10.1158/0008-5472.CAN-07-5229. doi:10.1158/0008-5472.CAN-07-5229. [DOI] [PubMed] [Google Scholar]
- 42.Takeda M, Arao T, Yokote H, et al. AZD2171 shows potent antitumor activity against gastric cancer over-expressing fibroblast growth factor receptor 2/keratinocyte growth factor receptor. Clin Cancer Res. 2007;13:3051–3057. doi: 10.1158/1078-0432.CCR-06-2743. doi:10.1158/1078-0432.CCR-06-2743. [DOI] [PubMed] [Google Scholar]
- 43.Gozgit JM, Wong MJ, Moran L, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–699. doi: 10.1158/1535-7163.MCT-11-0450. doi:10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 44.Guagnano V, Kauffmann A, Wöhrle S, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2:1118–1133. doi: 10.1158/2159-8290.CD-12-0210. doi:10.1158/2159-8290.CD-12-0210. [DOI] [PubMed] [Google Scholar]
- 45.Zhao WM, Wang L, Park H, et al. Monoclonal antibodies to fibroblast growth factor receptor 2 effectively inhibit growth of gastric tumor xenografts. Clin Cancer Res. 2010;16:5750–5758. doi: 10.1158/1078-0432.CCR-10-0531. doi:10.1158/1078-0432.CCR-10-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai A, Meetze K, Vo NY, et al. GP369, an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer Res. 2010;70:7630–7639. doi: 10.1158/0008-5472.CAN-10-1489. doi:10.1158/0008-5472.CAN-10-1489. [DOI] [PubMed] [Google Scholar]
- 47.Dutt A, Salvesen HB, Chen TH, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci USA. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. doi:10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Hara AJ, Bell DW. The genomics and genetics of endometrial cancer. Adv Genomics Genet. 2012;2012:33–47. doi: 10.2147/AGG.S28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kandoth C, Schultz N, Cherniack AD, et al. Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. doi:10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu K, Herr AB, Waksman G, et al. Loss of fibroblast growth factor receptor 2 ligand binding specificity in Apert syndrome. Proc Natl Acad Sci USA. 2000;97:14536–14541. doi: 10.1073/pnas.97.26.14536. doi:10.1073/pnas.97.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byron SA, Gartside M, Powell MA, et al. FGFR2 point mutations in 466 endometrioid endometrial tumors: relationship with MSI, KRAS, PIK3CA, CTNNB1 mutations and clinicopathological features. PLoS One. 2012;7:e30801. doi: 10.1371/journal.pone.0030801. doi:10.1371/journal.pone.0030801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cappellen D, De Oliveira C, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 53.Kimura T, Suzuki H, Ohashi T, et al. The incidence of thanatophoric dysplasia mutations in FGFR3 gene is higher in low-grade or superficial bladder carcinomas. Cancer. 2001;92:2555–2561. doi: 10.1002/1097-0142(20011115)92:10<2555::aid-cncr1607>3.0.co;2-m. doi:10.1002/1097-0142(20011115)92:10<2555::AID-CNCR1607>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 54.Billerey C, Chopin D, Aubriot-Lorton MH, et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am J Pathol. 2001;158:1955–1959. doi: 10.1016/S0002-9440(10)64665-2. doi:10.1016/S0002-9440(10)64665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomlinson DC, Baldo O, Harnden P, et al. FGFR3 protein expression and its relationship tomutation status and prognostic variables in bladder cancer. J Pathol. 2007;213:91–98. doi: 10.1002/path.2207. doi:10.1002/path.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Ahmadie HA, Iyer G, Janakiraman M, et al. Somatic mutation of fibroblast growth factor receptor-3 (FGFR3) defines a distinct morphological subtype of high-grade urothelial carcinoma. J Pathol. 2011;224:270–279. doi: 10.1002/path.2892. doi:10.1002/path.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Rhijn BW, van der Kwast TH, Liu L, et al. The FGFR3 mutation is related to favorable pT1 bladder cancer. J Urol. 2012;187:310–314. doi: 10.1016/j.juro.2011.09.008. doi:10.1016/j.juro.2011.10.172. [DOI] [PubMed] [Google Scholar]
- 58.Bernard-Pierrot I, Brams A, Dunois-Lardé C, et al. Oncogenic properties of the mutated forms of fibroblast growth factor receptor 3b. Carcinogenesis. 2006;27:740–747. doi: 10.1093/carcin/bgi290. doi:10.1093/carcin/bgi290. [DOI] [PubMed] [Google Scholar]
- 59.Jebar AH, Hurst CD, Tomlinson DC, et al. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218–5225. doi: 10.1038/sj.onc.1208705. doi:10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 60.Qing J, Du X, Chen Y, et al. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009;119:1216–1229. doi: 10.1172/JCI38017. doi:10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyake M, Ishii M, Koyama N, et al. PD173074, a selective tyrosine kinase inhibitor of FGFR3, inhibits cell proliferation of bladder cancer carrying the FGFR3 gene mutation along with up-regulation of p27/Kip1 and G1/G0 arrest. J Pharmacol Exp Ther. 2010;332:795–802. doi: 10.1124/jpet.109.162768. doi:10.1124/jpet.109.162768. [DOI] [PubMed] [Google Scholar]
- 62.Lamont FR, Tomlinson DC, Cooper PA, et al. Small molecule FGF receptor inhibitors block FGFR-dependent urothelial carcinoma growth in vitro and in vivo. Br J Cancer. 2011;104:75–82. doi: 10.1038/sj.bjc.6606016. doi:10.1038/sj.bjc.6606016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. doi:10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. doi:10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parker BC, Annala MJ, Cogdell DE, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123:855–865. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor JG, Cheuk AT, Tsang PS, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–3407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang K, Chu K, Wu X, et al. Amplification of FRS2 and activation of FGFR/FRS2 signaling pathway in high-grade liposarcoma. Cancer Res. 2013;73:1298–1307. doi: 10.1158/0008-5472.CAN-12-2086. doi:10.1158/0008-5472.CAN-12-2086. [DOI] [PubMed] [Google Scholar]
- 68.Desnoyers LR, Pai R, Ferrando RE, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. doi: 10.1038/sj.onc.1210623. doi:10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- 69.Sawey ET, Chanrion M, Cai C, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. doi:10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bumbaca D, Wong A, Drake E, et al. Highly specific off-target binding identified and eliminated during the humanization of an antibody against FGF receptor 4. MAbs. 2011;3:376–386. doi: 10.4161/mabs.3.4.15786. doi:10.4161/mabs.3.4.15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murphy T, Darby S, Mathers ME, et al. Evidence for distinct alterations in the FGF axis in prostate cancer progression to an aggressive clinical phenotype. J Pathol. 2010;220:452–460. doi: 10.1002/path.2657. [DOI] [PubMed] [Google Scholar]
- 72.Maruyama-Takahashi K, Shimada N, Imada T, et al. A neutralizing anti-fibroblast growth factor (FGF) 8 monoclonal antibody shows anti-tumor activity against FGF8b-expressing LNCaP xenografts in androgen-dependent and -independent conditions. Prostate. 2008;68:640–650. doi: 10.1002/pros.20728. doi:10.1002/pros.20728. [DOI] [PubMed] [Google Scholar]
- 73.Feng S, Shao L, Yu W, et al. Targeting fibroblast growth factor receptor signaling inhibits prostate cancer progression. Clin Cancer Res. 2012;18:3880–3888. doi: 10.1158/1078-0432.CCR-11-3214. doi:10.1158/1078-0432.CCR-11-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Allen E, Walters I, Hanahan D. Brivanib, a dual FGF/VEGF inhibitor, is active both first and second line against mouse pancreatic neuroendocrine tumors developing adaptive/evasive resistance to VEGF inhibition. Clin Cancer Res. 2011;17:5299–5310. doi: 10.1158/1078-0432.CCR-10-2847. doi:10.1158/1078-0432.CCR-10-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kopetz S, Hoff PM, Morris JS, et al. Phase II trial of infusional fluorouracil, irinotecan, and bevacizumab for metastatic colorectal cancer: efficacy and circulating angiogenic biomarkers associated with therapeutic resistance. J Clin Oncol. 2010;28:453–459. doi: 10.1200/JCO.2009.24.8252. doi:10.1200/JCO.2009.24.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Batchelor TT, Sorensen AG, di Tomaso E, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11:83–95. doi: 10.1016/j.ccr.2006.11.021. doi:10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gartside MG, Chen H, Ibrahimi OA, et al. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Mol Cancer Res. 2009;7:41–54. doi: 10.1158/1541-7786.MCR-08-0021. doi:10.1158/1541-7786.MCR-08-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Herrera Abreu M, Garcia-Murilla I, Pearson A, et al. functional screens to identify mechanisms of resistance to FGFR inhibitors in FGFR amplified and mutated cell lines. Eur J Cancer. 2012;48(suppl 6) Abstr 163. [Google Scholar]
- 79.Gozgit JM, Squillace RM, Wongchenko MJ, et al. Combined targeting of FGFR2 and mTOR by ponatinib and ridaforolimus results in synergistic antitumor activity in FGFR2 mutant endometrial cancer models. Cancer Chemother. 2013;71:1315–1323. doi: 10.1007/s00280-013-2131-z. doi:10.1007/s00280-013-2131-z. [DOI] [PubMed] [Google Scholar]
- 80.Harbinski F, Craig VJ, Sanghavi S, et al. Rescue screens with secreted proteins reveal compensatory potential of receptor tyrosine kinases in driving cancer growth. Cancer Discov. 2012;2:948–959. doi: 10.1158/2159-8290.CD-12-0237. doi:10.1158/2159-8290.CD-12-0237. [DOI] [PubMed] [Google Scholar]
- 81.Issa A, Gill JW, Heideman MR, et al. Combinatorial targeting of FGF and ErbB receptors blocks growth and metastatic spread of breast cancer models. Breast Cancer Res. 2013;15:R8. doi: 10.1186/bcr3379. doi:10.1186/bcr3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ware KE, Marshall ME, Heasley LR, et al. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One. 2010;5:e14117. doi: 10.1371/journal.pone.0014117. doi:10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ye X, Yue P, Patel R, et al. Activation of FGFR signaling as a mechanism of acquired resistance to erlotinib in EGFR-mutant lung cancer associated with an EMT. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research (AACR); 6–10 April; Washington DC. 2013. Abstr 4463. [Google Scholar]
- 84.Ware KE, Hinz TK, Kleczko E, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. doi:10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Terai H, Soejima KN, Yasuda H, et al. Activation of the FGF2-FGFR1 autocrine pathway: a novel mechanism of acquired resistance to gefitinib in NSCLC cells. Mol Cancer Res. 2013;11:759–767. doi: 10.1158/1541-7786.MCR-12-0652. doi:10.1158/1541-7786.MCR-12-0652. [DOI] [PubMed] [Google Scholar]
- 86.Oliveras-Ferraros C, Cufí S, Queralt B, et al. Cross-suppression of EGFR ligands amphiregulin and epiregulin and de-repression of FGFR3 signalling contribute to cetuximab resistance in wild-type KRAS tumour cells. Br J Cancer. 2012;106:1406–1414. doi: 10.1038/bjc.2012.103. doi:10.1038/bjc.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yadav V, Zhang X, Liu J, et al. Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem. 2012;287:28087–28098. doi: 10.1074/jbc.M112.377218. doi:10.1074/jbc.M112.377218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown AP, Courtney CL, King LM, et al. Cartilage dysplasia and tissue mineralization in the rat following administration of a FGF receptor tyrosine kinase inhibitor. Toxicol Pathol. 2005;33:449–455. doi: 10.1080/01926230590961845. doi:10.1080/01926230590961845. [DOI] [PubMed] [Google Scholar]
- 89.Dienstmann R, Braña I, Rodon J, et al. Toxicity as a biomarker of efficacy of molecular targeted therapies: focus on EGFR and VEGF inhibiting anticancer drugs. Oncologist. 2011;16:1729–1740. doi: 10.1634/theoncologist.2011-0163. doi:10.1634/theoncologist.2011-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gavine PR, Mooney L, Kilgour E, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. doi:10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 91.Zhao G, Li WY, Chen D, et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther. 2011;10:2200–2210. doi: 10.1158/1535-7163.MCT-11-0306. doi:10.1158/1535-7163.MCT-11-0306. [DOI] [PubMed] [Google Scholar]
- 92.Andre F, Bachelot TD, Campone M, et al. A multicenter, open-label phase II trial of dovitinib, an FGFR1 inhibitor, in FGFR1 amplified and non-amplified metastatic breast cancer. J Clin Oncol. 2011;29(Suppl) Abstr 508. [Google Scholar]
- 93.Shiang CY, Qi Y, Wang B, et al. Amplification of fibroblast growth factor receptor-1 in breast cancer and the effects of brivanib alaninate. Breast Cancer Res Treat. 2010;123:747–755. doi: 10.1007/s10549-009-0677-6. doi:10.1007/s10549-009-0677-6. [DOI] [PubMed] [Google Scholar]
- 94.Colella G, Damia G, D'Incalci M, et al. E-3810 antitumor activity in human xenografts expressing different levels of FGF receptor 1. Cancer Res. 2011;71(suppl 1) Abstr 595. [Google Scholar]
- 95.Konecny GE, Kolarova T, O'Brien NA, et al. Activity of the fibroblast growth factor receptor inhibitors dovitinib (TKI258) and NVP-BGJ398 in human endometrial cancer cells. Mol Cancer Ther. 2013;12:632–642. doi: 10.1158/1535-7163.MCT-12-0999. doi:10.1158/1535-7163.MCT-12-0999. [DOI] [PubMed] [Google Scholar]
- 96.Okita K, Kumada H, Ikeda K, et al. Phase I/II study of E7080 (lenvatinib), a multitargeted tyrosine kinase inhibitor, in patients with advanced hepatocellular carcinoma: initial assessment of response rate. J Clin Oncol. 2012;30(suppl) Abstr 320. [Google Scholar]
- 97.Kim KB, Chesney J, Robinson D, et al. Phase I/II and pharmacodynamic study of dovitinib (TKI258), an inhibitor of fibroblast growth factor receptors and VEGF receptors, in patients with advanced melanoma. Clin Cancer Res. 2011;17:7451–7461. doi: 10.1158/1078-0432.CCR-11-1747. doi:10.1158/1078-0432.CCR-11-1747. [DOI] [PubMed] [Google Scholar]
- 98.Dienstmann R, André F, Soria JC, et al. Significant antitumor activity of E-3810, a novel FGFR and VEGFR inhibitor, in patients with FGFR1amplified breast cancer. Ann Oncol. 2012;23(suppl 9) Abstr 3190. [Google Scholar]
- 99.Barrios CH, Liu MC, Lee SC, et al. Phase III randomized trial of sunitinib versus capecitabine in patients with previously treated HER2-negative advanced breast cancer. Breast Cancer Res Treat. 2010;121:121–131. doi: 10.1007/s10549-010-0788-0. doi:10.1007/s10549-010-0788-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moreno-Aspitia A, Morton RF, Hillman DW, et al. Phase II trial of sorafenib in patients with metastatic breast cancer previously exposed to anthracyclines or taxanes: North Central Cancer Treatment Group and Mayo Clinic Trial N0336. J Clin Oncol. 2009;27:11–15. doi: 10.1200/JCO.2007.15.5242. doi:10.1200/JCO.2007.15.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wolf J, LoRusso PM, Camidge RD, et al. A phase I dose escalation study of NVP-BGJ398, a selective pan FGFR inhibitor in genetically preselected advanced solid tumors. Proceedings of the103rd Annual Meeting of the American Association for Cancer Research (AACR); 31 March–4 April; Chicago, IL. 2012. Abstr LB-122. [Google Scholar]
- 102.Andre F, Ranson M, Dean E, et al. Results of a phase I study of AZD4547, an inhibitor of fibroblast growth factor receptor, in patients with advanced solid tumors. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research (AACR); 6–10 April; Washington DC. 2013. Abstr LB-145. [Google Scholar]
- 103.Harding TC, Long L, Palencia S, et al. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Sci Transl Med. 2013;5:178ra39. doi: 10.1126/scitranslmed.3005414. doi:10.1126/scitranslmed.3005414. [DOI] [PubMed] [Google Scholar]
- 104.O'Donnell P, Goldman JW, Gordon MS, et al. A phase I dose-escalation study of MFGR1877S, a human monoclonal anti-fibroblast growth factor receptor 3 (FGFR3) antibody, in patients with advanced solid tumors. Eur J Cancer. 2012;48:191–192. Abstr 621 doi:10.1016/S0959-8049(12)72418-8. [Google Scholar]
- 105.Rodón J, Saura C, Dienstmann R, et al. Molecular prescreening to select patient population in early clinical trials. Nat Rev Clin Oncol. 2012;9:359–366. doi: 10.1038/nrclinonc.2012.48. doi:10.1038/nrclinonc.2012.48. [DOI] [PubMed] [Google Scholar]
- 106.Dienstmann R, Rodon J, Tabernero J. Biomarker-driven patient selection for early clinical trials. Curr Opin Oncol. 2013;25:305–312. doi: 10.1097/CCO.0b013e32835ff3cb. [DOI] [PubMed] [Google Scholar]
- 107.Zhang J, Wu G, Miller CP, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. doi:10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jones DTW, Hutter B, Jäger N, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. doi:10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liao RG, Jung J, Tchaicha J, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. doi:10.1158/0008–5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. doi:10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Majewski IJ, Mittempergher L, Davidson NM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230:270–276. doi: 10.1002/path.4209. doi:10.1002/path.4209. [DOI] [PubMed] [Google Scholar]
- 112.Dieci MV, Arnedos M, Andre F, et al. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 2013;3:264–279. doi: 10.1158/2159-8290.CD-12-0362. doi:10.1158/2159-8290.CD-12-0362. [DOI] [PubMed] [Google Scholar]