Abstract
The ability to study nonhematologic cancers through noninvasive sampling of blood is one of the most exciting and rapidly advancing fields in cancer diagnostics. This has been driven both by major technologic advances, including the isolation of intact cancer cells and the analysis of cancer cell–derived DNA from blood samples, and by the increasing application of molecularly driven therapeutics, which rely on such accurate and timely measurements of critical biomarkers. Moreover, the dramatic efficacy of these potent cancer therapies drives the selection for additional genetic changes as tumors acquire drug resistance, necessitating repeated sampling of cancer cells to adjust therapy in response to tumor evolution. Together, these advanced noninvasive diagnostic capabilities and their applications in guiding precision cancer therapies are poised to change the ways in which we select and monitor cancer treatments.
Significance
Recent advances in technologies to analyze circulating tumor cells and circulating tumor DNA are setting the stage for real-time, noninvasive monitoring of cancer and providing novel insights into cancer evolution, invasion, and metastasis.
INTRODUCTION
Blood contains two types of cancer-derived materials that are susceptible to detailed molecular analysis: intact circulating tumor cells (CTC) and cell-free circulating tumor DNA (ctDNA). The former are shed from primary or metastatic tumor deposits, and although they are rare, they are thought to be enriched for metastatic precursors. Initially detected in an 1869 autopsy within the blood of a patient with widespread breast cancer (1), CTCs are now isolated with increasingly sophisticated technologies (2–4). However, the advantage of applying multiple DNA, RNA, and protein-based assays to study whole tumor cells in the circulation (so-called liquid biopsies) is currently restricted by the need for complex cellular isolation platforms.
Cancer-derived molecules in the blood include well-established protein markers, such as carcinoembryonic antigen (CEA) or prostate-specific antigen (PSA), as well as circulating cell fragments such as exosomes. However, among cell-free biomarkers, it is ctDNA that offers the greatest opportunity for the application of detailed molecular techniques. Although cell-free DNA (cfDNA) in the circulation was first described in 1948 (5), abnormalities in patients with cancer were observed only decades later (6, 7). ctDNA is thought to be derived from tumor deposits and lysed CTCs. As such, although its isolation is far simpler than CTCs, it is the variable contribution of tumor-derived ctDNA versus the typically much larger amount of cfDNA shed from normal cells that has limited analyses to date. The application of next-generation sequencing (NGS) together with advanced computational methods has recently allowed ctDNA-based tumor genotyping.
As both CTC and ctDNA technologies evolve, they will likely have similar as well as distinct clinical applications, reflecting their relative biologic and technologic strengths and weaknesses (Fig. 1; see also ref. 8). However, they are both integral to the emerging view of cancer as comprising a heterogeneous and dynamic molecular landscape; ultimate therapeutic success will require a high level of integration between real-time diagnostic measurements and targeted interventions. In this regard, we first address the various clinical indications in which blood-based molecular diagnostics may play a significant role.
Figure 1.
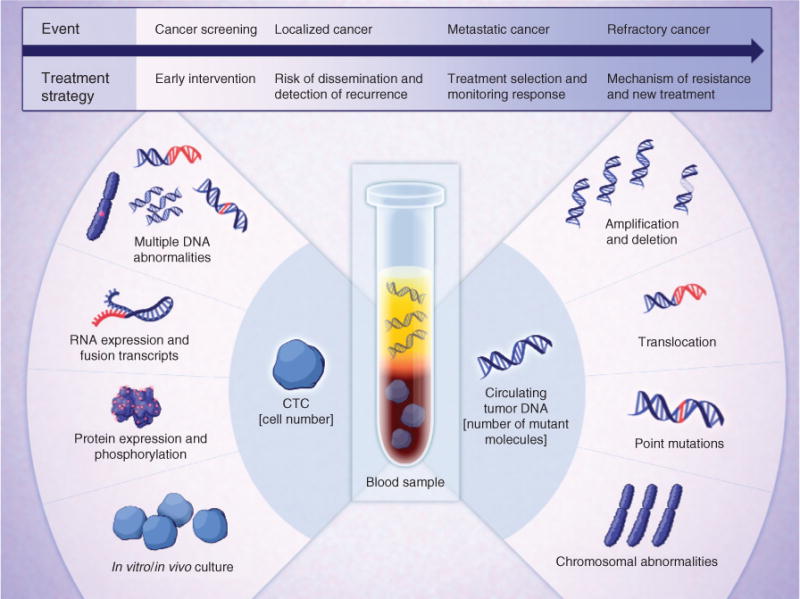
Clinical applications of CTC and ctDNA analyses in cancer care. The molecular analyses that are enabled by the isolation of CTCs and ctDNA from blood specimens are illustrated. These may be applied to guide different treatment strategies at different events in the initial diagnosis and treatment of patients with cancer.
BLOOD-BASED MEASUREMENTS IN THE DIAGNOSIS AND TREATMENT OF CANCER
The application of blood-based protein markers in quantifying tumor response to therapy is well established in clinical practice, especially in settings in which the cancer itself is not readily measurable. For instance, bone metastases in prostate cancer do not show rapid radiographic changes following hormonal therapy, and hence serum PSA levels are routinely used as a surrogate marker of drug response (9). In the selected cases studied to date, both CTCs and ctDNA measurements show rapid responses following administration of effective therapy (10, 11). Such blood-based markers may prove particularly useful as the choice of potentially effective therapies increases with novel targeted drug regimens. Indeed, we anticipate a time when brief therapeutic trials of different regimens followed by blood-based measurements of tumor burden, or even cell-based signaling studies, may allow rapid selection of effective therapies without waiting for radiographic evidence of response or nonresponse.
The choice of therapeutic agent itself may be based on blood-based diagnostics. Early studies of CTCs identified their presence as conferring a negative prognostic significance in patients with metastatic cancers of the breast, colon, and prostate (12–14). The therapeutic implications of such information, however, were indirect, without compelling data that more-aggressive chemotherapeutic regimens are more effective in patients whose metastatic cancer is associated with high levels of CTCs. More-recent studies have focused on the presence of genetic mutations, identified in CTCs or in ctDNA, whose presence is predictive of response to targeted inhibitors (10, 15). Blood-based molecular genotyping in non–small cell lung cancer, melanoma, and breast cancer, for instance, may guide the administration of drugs targeting mutant EGFR, BRAF, and PIK3CA, or the EML4–ALK translocation. In cases in which the primary tumor cannot be readily biopsied, CTC- or ctDNA-based genotyping may provide a rapid and noninvasive strategy to obtain clinically relevant genotypes needed for treatment selection. Tumors acquire resistance to targeted drugs, through either mutations that reduce drug binding, activation of alternative signaling pathways, increased expression of antiapoptotic genes, or cellular transformations to mesenchymal and even distinct histologic phenotypes (16, 17). Appropriate selection of second-line therapies is key to achieving effective response (18, 19), and serial blood-based monitoring for emerging mechanisms of drug response may prove to be one of the most compelling applications of these diagnostic strategies.
Although the application of blood-based molecular diagnostics in patients with known metastatic cancer constitutes the most immediate application of these technologies, it is probably the early diagnosis of cancer, at a stage in which it may be curable, where they may achieve their greatest impact. For instance, the detection of minimal residual disease, after potentially curative therapy, may lead to second-line salvage therapies in sarcomas, prostate cancer, and colorectal cancer. In patients with localized cancers, evidence of either circulating cancer cells or abundant ctDNA might identify subsets at increased risk of recurrence, in whom adjuvant therapy might be considered. As the sensitivity of the detection assays improves, it is conceivable that blood-based assays may be useful in early cancer screening, particularly in high-risk individuals who may be repeatedly monitored. Finally, as our understanding evolves about fundamental mechanisms that drive cancer cell invasion into the bloodstream, novel drug targets may be identified, ultimately leading to therapies aimed at preventing metastasis. Thus, from immediate application to more futuristic goals, the introduction of blood-based molecular diagnostics into the clinic is likely to fundamentally alter the way in which we treat patients with cancer.
CIRCULATING TUMOR CELLS
CTCs are shed into the vasculature from primary and/or metastatic tumor deposits (2, 20). The process underlying the intravasation of tumor cells is not well understood and may involve both active invasion of cells with increased migratory potential [resulting from epithelial-to-mesenchymal transition (EMT); ref. 21] and passive shedding of individual cells or tumor cell clusters resulting from compromised tumor vasculature (22, 23). Once in the circulation, CTCs seem to persist for a short time; in patients with localized cancer who have detectable CTCs, most no longer have evidence of such cells at 24 hours following surgical resection (24). On the basis of their morphology, CTCs are highly heterogeneous. Many seem apoptotic or damaged, even following the most gentle isolation techniques, whereas others seem similar in appearance to cells from matched tumor biopsies. Some cancer cells in the circulation travel in clusters, ranging from two CTCs caught in mitosis to large microemboli with >50 cells detectable in the peripheral vasculature (22–26). The proliferative index of CTCs, defined by Ki67 staining, is highly variable among different patients (24), whereas single-cell analyses have revealed heterogeneity in signaling pathways among CTCs from individual patients (27, 28–30). Most importantly, identifying the subset of CTCs capable of initiating a metastatic lesion, and weighing the relative contributions of “seed versus soil,” remain major challenges. Culturing CTCs in vitro (31, 32) and testing their tumorigenic properties, as well as their susceptibilities to various clinically relevant drug regimens, are exciting future applications of the technology. Taken together, the biology of CTCs provides two fundamental avenues for research: first, understanding and ultimately targeting the process of blood-borne metastasis and, second, using CTC analyses as a readout of tumor status for therapeutic and early-detection applications.
CTC Isolation Technologies and Platforms
Technologies for isolating intact CTCs from the circulation are faced with the challenge of finding extremely rare cells among abundant normal blood cells in a specimen drawn from a patient with cancer. Although some rare outliers may have hundreds or even thousands of CTCs/mL of blood, most patients with metastatic cancer have fewer than 10 cells/mL [1 mL of blood contains 1 million white blood cells (WBC) and 1 billion red blood cells]. Many different CTC isolation technologies have emerged over the past few years, but all share the fundamental challenge of sorting through massive numbers of blood cells without losing or damaging the few CTCs present, purifying these efficiently while limiting the number of contaminating leukocytes, and finally correctly identifying CTCs based on unique immunophenotypes, cytopathologic features, or molecular genetic features.
CTC isolation strategies fall broadly within different classes, depending on whether they rely on physical properties of tumor cells, their expression of unique cell surface markers, or, more recently, the effective depletion of normal leukocytes to reveal untagged CTCs. There are innumerable technologic approaches that fall within these broad categories, all at different stages of development, from “proof of concept” using cancer cell lines spiked into blood, to more advanced testing with blood specimens from patients with different cancers. Representative technologies are listed in Table 1.
Table 1.
Technologies for isolation of CTCs
| Underlying technology | Representative | Rationale platforms | Selected references |
|---|---|---|---|
| Antibody capture | Selection for EpCAM on tumor cells | Veridex/CellSearch | 54–56 |
| Magsweeper | 57 | ||
| Microfluidic CTC-Chip | 59, 60, 110, 111 | ||
|
| |||
| High-throughput imaging | Scanning of cells on slide | Epic | 26, 38–41 |
|
| |||
| Physical properties | Differential size, density, others | Physical filter | 33–37 |
| Density gradient | 42 | ||
| Dielectric | 43, 44 | ||
| Photoacoustic | 45, 46 | ||
| Microfluidic | 47 | ||
|
| |||
| Functional characteristics | Protein secretion, migratory properties | EPISPOT secretion assay | 48–50 |
| Invasion assay | 51–53 | ||
|
| |||
| Leukocyte depletion | Negative depletion of leukocytes | Batch cell lysis | 112–114 |
| Microfluidic CTC-iChip | 28 | ||
Size-based filtering approaches take advantage of the fact that many epithelial cancer cells are larger (median diameter 15 μm) than leukocytes (10 μm; refs. 33–37). These platforms have the advantage of ease of use, although processing large volumes of cells through a static filter poses significant hemodynamic stress on cells, which may reduce their integrity. Moreover, measurements of CTCs isolated using other parameters reveal considerable variation in the size of CTCs, even those derived from a single patient. In some cases, CTCs may be similar in size to or even smaller than leukocytes, while large circulating cells in patients undergoing chemotherapy may include bone marrow–derived megakaryocytes (28). Nonetheless, filtering technologies are continuing to improve and may provide a relatively simple way to assess CTC burden, particularly in cancer types associated with larger tumor cell diameters.
High-throughput microscopic scanning of blood specimens depleted of red blood cells and plated onto a large adherent surface has been tested to screen for CTCs (26, 38–41). This strategy is unbiased by cell size in initial selection of CTCs and it relies on staining for epithelial or tumor markers to identify CTCs, although molecular characterization of cancer cells within such unpurified blood populations presents significant challenges. Other isolation technologies that are based on physical properties of cancer cells make use of their differential density (42), electrical charge (43, 44), photoacoustic resonance (45, 46), or size-based flow kinetics (47). Secretion of marker proteins by CTCs (48–50) or their invasion through collagen-coated surfaces (51–53) have also been tested as detection strategies.
The most popular CTC isolation technologies have involved antibody-mediated capture of cancer cells. The commercial technology (CellSearch) makes use of magnetically tagged antibodies against the common epithelial cell surface marker EpCAM (54–56). In this approach, blood cells are first fixed, exposed to antibody, and then separated in a batch process by application of a magnetic field. Although effective and highly reproducible, the relatively low yield of CTCs recovered using this technology may reflect the loss of rare cells through a multistep batch purification and the inefficient magnetic separation of labeled cells traveling across a dense population of unlabeled cells. EpCAM-positive cells have also been captured by incubation with an antibody-coated magnetic stir-bar, followed by cell release (57, 58). Microfluidic technologies are particularly well suited to the field of rare cell purification, as they can be applied directly to unprocessed whole blood, making use of optimized cell–antibody contact under precisely controlled low shear stress flow conditions that can be multiplexed to readily increase throughput. These so-called “CTC-Chip” platforms include flowing blood through 80,000 anti-EpCAM antibody-coated microposts or through a mixing chamber whose walls are coated with antibody (59, 60). Although these highly sensitive CTC capture platforms have enabled detailed molecular characterization of CTCs (10, 24, 27, 61, 62), the capture of cells within three-dimensional chambers poses limitations to both high-throughput imaging and single-cell molecular analyses.
Of the new microfluidic CTC isolation strategies, the most promising involves depletion of leukocytes from a blood sample, leaving untagged CTCs for analysis (28). There is a powerful rationale for this strategy: Leukocyte cell surface markers are well characterized and invariant, whereas cancer cells may express multiple different epitopes, even within a single patient. Furthermore, nonepithelial cancers, such as melanoma, do not express EpCAM, while others undergo EMT, losing their expression of EpCAM and other epithelial cell surface markers. However, the massive depletion of leukocytes required to achieve a highly pure CTC population requires sophisticated microfluidic technologies. In the so-called “CTC-iChip,” an integrated microfluidic device first achieves size-based separation of all nucleated cells (i.e., WBCs and CTCs) from red blood cells, platelets, and plasma. The nucleated cells are then arrayed within a single file as they travel through specially configured curved channels, taking advantage of a physical phenomenon termed “inertial focusing” (63). Magnetic deflection of inertially focused tagged leukocytes as they travel through a microfluidic channel requires minimal force and is highly efficient, allowing 104 depletion of WBCs at a flow rate of 10 mL/h. Untagged and unmanipulated CTCs are delivered at an average cell purity of 1% in solution, where they can be stained for enumeration, lysed for molecular characterization, or picked individually for single-cell analyses (28). Automation followed by broad dissemination of such powerful CTC isolation platforms to the cancer research community will allow widespread investigation of CTCs as molecular markers and their application to large-scale clinical trials.
Molecular Characterization of CTCs
Once captured, CTCs may be stained and enumerated. Traditional criteria have defined these cells as positive for pan-cytokeratin and negative for the common leukocyte antigen CD45 (55). The use of fluorescent-conjugated antibodies requires care in setting appropriate signal thresholds; cellular fragments are excluded using nuclear dyes, as are “double-positive” cells staining for both cytokeratin and CD45. Most recently, microfluidic isolation technologies have enabled high-resolution light microscopy, using clinical laboratory cytopathology protocols, including standardized immunohistochemistry (28). Such integration between a research platform and clinically accepted diagnostic standards is particularly important to the acceptance of blood-based diagnostics as a clinical tool in the management of patients with cancer.
As noted above, baseline enumeration of CTCs has been demonstrated to have prognostic value in patients with known metastatic cancers of the breast, colon, and prostate (12–14). However, it is the change in CTCs within a given patient following therapeutic intervention that is most likely to have significant clinical benefit. Across most platforms that yield a dynamic range in CTC numbers, these counts drop rapidly and significantly in patients who have bona fide responses to effective therapies (10, 24, 64). Interestingly, whereas CTC counts within individual patients are correlated with clinical response, across different patients, CTC counts are not correlated with tumor burden as measured radiographically or using serum protein markers. Baseline CTC numbers in each patient may therefore reflect additional parameters, possibly including tumor invasiveness, vascularity, or other factors. In genetically uniform mouse tumor models, CTC numbers are relatively well correlated with tumor burden (61).
Characterization of CTCs for expression of protein markers is readily achieved using fluorescence microscopy, although not all platforms have sufficient channels to allow staining of cells for multiple markers, in addition to those required to identify cytokeratin-positive/CD45-negative cells. Examples of promising protein-based analyses include dual Ki67/PSA staining in prostate cancer CTCs, demonstrating an increasing proliferation index as patients progress from responsive castration-sensitive disease to the more refractory castration-resistant form (24). Dual staining for androgen-induced PSA and androgen-suppressed prostate-specific membrane antigen (PSMA) markers also allows quantification of heterogeneity in androgen signaling status within prostate CTCs, before and after hormonal therapy (27). However, the application of increasingly multiplexed protein markers to the analysis of these rare cells also requires careful calibration of each antibody with respect to signal intensity, background levels in rare hematopoietic subpopulations, and cross-reactivity with other antibody stains. Together, cancer type–specific panels of antibody stains may ultimately provide valuable information to monitor the status of a tumor and guide therapeutic choices.
RNA-based expression monitoring in CTCs is most successful using isolation techniques that do not involve formaldehyde fixation. Early studies demonstrated reverse transcription PCR (RT-PCR) amplification of lineage-specific transcripts in CTC-enriched cell populations (65, 66). Tumor-specific trans-locations (e.g., EML4–ALK in non–small cell lung cancer and TMPRSS2–ERG in prostate cancer) are also readily detectable in such populations (24, 28, 56). More recently, whole-genome expression profiling using NGS technologies has been achieved (58, 61, 62). When using partially pure CTC populations, digital subtraction of background leukocyte reads is essential to deriving CTC-based expression signatures, and NGS technologies that do not require cDNA amplification (i.e., single-molecule sequencing) have an important advantage in detecting the low fraction of CTC-derived templates (61, 62). Most recently, isolation of single CTCs and derivation of single-CTC transcription profiles offers great promise for a more comprehensive transcriptome coverage, as well as shedding light on the heterogeneity of CTCs. Finally, the development of highly sensitive and robust RNA-in situ hybridization (ISH) techniques has allowed their application to CTCs. Early applications of CTC-based RNA-ISH has included detection of CTC-specific transcripts, as well as scoring for the relative abundance of epithelial versus mesenchymal transcripts within individual CTCs (61, 62).
From a clinical standpoint, genotyping of CTCs is likely to be one of the most immediate applications of the technology. Allele-specific PCR-based assays of CTC-enriched cell populations have been demonstrated for EGFR-mutant non–small cell lung cancer, with a high concordance between tumor biopsies at presentation and CTC-derived genotypes (10, 28, 30, 56). The treatment-associated emergence of drug resistance mutations can also be documented using allele-specific PCR or targeted NGS analysis. However, whole-exome sequencing is complicated by both the very low levels of tumor-specific templates and contamination by abundant leukocyte-derived sequences. Advances in NGS strategies and computational analyses may be successful in resolving this challenge; however, the most promising results may emerge from single-CTC sequencing strategies, which would provide direct insight into CTC heterogeneity and the emergence of distinct subsets of tumor cells during the course of therapy.
CIRCULATING TUMOR DNA
The presence of cfDNA in the circulation is a well-established phenomenon. Fragments of DNA are shed into the bloodstream from dying cells during cellular turnover or other forms of cell death (67). Normally, apoptotic or necrotic cells are cleared, and the levels of cfDNA are relatively low. Several thousand genome equivalents of DNA are typically present in 1 mL of circulating plasma, with more than 90% of healthy individuals having less than 25 ng cfDNA per mL (7, 68). In certain conditions, including inflammation, exercise, or tissue injury, cfDNA levels can be substantially higher. Recent analyses have shown that levels may increase by more than an order of magnitude during surgery (69). cfDNA levels in patients with cancer are typically several-fold higher than those in healthy individuals, but the levels can vary widely (7, 68). cfDNA in the circulation is typically fragmented to 160 to 180 bp in length, corresponding to nucleosome-protected DNA observed in apoptotic cells (70).
In patients with cancer, a fraction of cfDNA is tumor derived and is termed ctDNA (71). Conceptually, ctDNA may be derived from primary tumors, metastatic lesions, or CTCs. The fraction of cfDNA that is tumor derived in patients with cancer has a variable contribution ranging from <0.1% to >10% of the DNA molecules (69). The variability in levels of ctDNA is not well understood and is thought to be affected by tumor burden, stage, cellular turnover, accessibility to the circulation, and factors affecting blood volume. Although patients with similar tumor types may have varying absolute levels of ctDNA at the time of diagnosis, the relative levels of ctDNA within an individual have been shown to correlate with tumor burden and response to therapy (69).
Cancers contain tumor-specific (somatic) genetic alterations that are present in most, if not all, cancer cells in an individual patient (72). By virtue of the clonal nature of tumor cells, somatic changes are present in many copies that are continuously released and can be detected in the circulation. Several studies have shown that mutations in ctDNA exactly correspond to mutations from the primary tumor, including both point mutations and structural alterations such as copy-number changes and rearrangements. These analyses demonstrate that somatic alterations detected in ctDNA are directly derived from an individual tumor. Somatic DNA alterations therefore can be thought to define the presence and level of ctDNA. Importantly, ctDNA mutations can be used to identify potentially actionable changes affecting driver genes, such as EGFR, KRAS, BRAF, and PIK3CA, as well as providing personalized biomarkers that can be used to detect residual disease or monitor tumor levels during therapy.
Technologies for Analysis of ctDNA
Because tumor-specific alterations in ctDNA are not present in normal cells, they offer an exquisitely sensitive and specific approach for cancer detection. From a clinical perspective, the preparation of cfDNA for analyses of DNA alterations is simple to implement. Isolation of cfDNA typically requires 5 to 10 mL of blood, collected in tubes treated with an anticoagulant such as EDTA. Cells are separated by centrifugation, and the plasma supernatant is removed. Circulating DNA is extracted from plasma using commercially available kits. Serum can also be used, but is less preferable due to the possibility of lysed cellular DNA that may affect the relative levels of ctDNA. Technical aspects of plasma collection may affect ctDNA levels. ctDNA has limited stability in the blood because of the presence of DNase activity, and as such cfDNA preparation should not exceed several hours after blood draw. Once cfDNA is isolated, the challenge is to detect genetic alterations even when ctDNA is present in a small fraction of the total DNA in the circulation. With the advent of new technologies, ctDNA can now be analyzed not only for specific mutations but also for larger alterations in the genome. Representative approaches for analyzing ctDNA are summarized in Table 2.
Table 2.
Technologies for detection and characterization of ctDNA
| Underlying technology | Mutation detection approach | Type of alteration | Example alterations | Selected references |
|---|---|---|---|---|
| Real-time or end- point PCR | ARMS-Scorpion PCR | Known point mutations | KRAS, EGFR hotspot changes | 74 |
| PCR-SSCP | 73 | |||
| Mutant allele–specific PCR | 75 | |||
| Mass spectrometry | 68 | |||
| Bi-PAP amplification | 76 | |||
|
| ||||
| Digital PCR | BEAMing | Known point mutations | KRAS, EGFR hotspot changes | 78 |
| Droplet-based digital PCR | 80 | |||
| Digital droplet PCR | 79 | |||
|
| ||||
| Gene sequencing | SafeSeqs | Point mutations in gene regions | PIK3CA, EGFR, TP53 coding mutations | 81 |
| OnTarget | 83 | |||
| TamSeq | 82 | |||
|
| ||||
| Whole-genome sequencing | Digital karyotyping | Genome-wide copy-number changes | Personalized amplifications | 87, 88, 108, 115 |
|
| ||||
| Whole-genome sequencing | PARE | Genome-wide rearrangements | Personalized rearrangements | 85, 86, 88 |
|
| ||||
| Targeted sequencing | Digital karyotyping/PARE | Structural alterations in gene regions | MET, ERBB2 amplification | 88, 98, 108 |
Abbreviations: SSCP, single-strand conformational polymorphism; BEAM, Beads, Emulsions, Amplification, and Magnetics; PARE, Personalized Analysis of Rearranged Ends.
Analysis of Point Mutations
For detection of somatic point mutations as biomarkers, the earliest analyses involved mutation-specific real-time or endpoint PCR approaches (68, 73–75). A particularly sensitive and specific method used the combination of pyrophosphorolysis-activated polymerization and allele-specific amplification during PCR (Bi-PAP-A; ref. 76).
More recently, a variety of digital genomic methods have been developed to improve identification of genetic alterations in ctDNA. These approaches are based on the concept that the most effective method to detect and quantify mutations is to analyze individual template molecules (77). This can be achieved by performing thousands of PCR reactions on template diluted to the point at which one or less template molecule is present in each reaction. Simply by counting the number of reactions containing wild-type or mutant PCR product, a sensitive and accurate quantification can be obtained. For individual mutations, this digital PCR approach can be used directly. However, such analyses are labor intensive and expensive when applied to more than a few hundred templates. To overcome these limitations, an approach called BEAMing was developed (78). This PCR-based method allows single-molecule PCR reactions to be performed on magnetic beads in water-in-oil emulsions. To distinguish mutant from wild-type coated beads, allele-specific fluorescent probes complementary to the known wild-type or mutant sequences are added to the beads for hybridization. Because each bead contains thousands of molecules of the identical sequence, the signal-to-noise ratio obtained by hybridization or enzymatic assays is high, and millions of beads can be analyzed rapidly using flow cytometry. BEAMing is sensitive and cost-effective when a limited number of potentially mutated positions are evaluated.
Other digital PCR approaches have been developed that can be applied for analyses of ctDNA in a similar manner. These include droplet digital PCR (Bio-Rad; ref. 79), picoliter droplet-based digital PCR (RainDance; ref. 80), and micro-fluidic systems for parallel PCR reactions (Fluidigm; ref. 78). When combined with PCR-based mutation detection strategies, these approaches can be used for sensitive detection of individual mutations at specific positions within the analyzed sequences. More recently, digital amplification and sequencing approaches using NGS methods have been developed (81, 82). These include PCR or capture of specific genomic loci and massively parallel sequencing to identify sequence alterations in the analyzed regions. In these approaches, unique identifiers are applied to the template molecules to help distinguish bona fide alterations from artifacts of PCR or sequencing. Hybridization methods have been developed to enrich for mutant alleles in the sample population (83). Overall, these analyses have included multiple exons of key genes and have been extended to allow for whole-exome analyses (84). The digital sequencing–based approaches have the advantage of allowing a larger number of loci to be evaluated simultaneously for potential alterations throughout the sequence of the template molecule rather than only at specific locations.
Whole-Genome Analyses
In addition to using somatic point mutations as markers for the detection of tumor-derived DNA, other strategies for the detection of ctDNA have been developed, including genome-wide detection of rearrangements and chromosomal copy-number changes. Two genome-wide methods to identify alterations that can be applied to detection of tumor DNA in the circulation include Personalized Analysis of Rearranged Ends (PARE) and related approaches (85, 86) and digital karyotyping (87). PARE is a method for identifying genome rearrangements in human tumors and using these alterations for development and detection of tumor biomarkers in the circulation (85, 88). Digital karyotyping is a genome-wide method for detection of copy-number alterations and novel sequences that has been applied to detect previously uncharacterized chromosomal changes and exogenous sequences in human cancer (87, 89, 90).
Chromosomal rearrangements, defined as the joining of DNA sequences that are normally not adjacent in the human genome, have the potential to serve as highly sensitive biomarkers for tumor detection. Virtually all tumors of clinical consequence are thought to have rearranged DNA sequences, and these sequences are not present in normal human plasma or nontumor tissues. Gains and losses of chromosomal regions are similarly widespread in human cancer. Using PARE, rearrangements detected in tumor DNA, including those resulting from copy-number changes, have been used to develop PCR-based biomarker tests to quantitatively measure the level of ctDNA in patient blood specimens. Initial analyses demonstrated that the sensitivity of this approach (i.e., the ability to detect tumor DNA in a mixture of tumor and normal DNA) is lower than 0.001% (85). This approach provides an exquisitely sensitive and broadly applicable approach for the development of personalized biomarkers to enhance the clinical management of patients with cancer.
Recent implementation of NGS with the above approaches has allowed direct sequence-based detection of chromosomal alterations in patient plasma (88, 91, 92). A challenge in adapting these methods for detection of rearrangements directly from plasma DNA is distinguishing the relatively few somatic structural alterations present in ctDNA from the much larger number of structural variants resulting from copy-number variations in the germline of all individuals. Bioinformatic filters have been developed that enrich for high-confidence somatic structural alterations while removing germline and artifactual changes (88). For rearrangements, such filters include sequencing template molecules in the plasma from both ends and selecting paired-end sequences that map to different chromosomes or to the same chromosome but at large distances apart, span rearrangement junctions, or contain sequenced rearrangement breakpoints, and map to genomic regions that do not contain known germline copy-number variants or repeated sequences.
As a proof-of-principle of this approach, a recent analysis examined paired-end NGS data from plasma DNA of 10 patients with cancer and 10 normal controls (88). Application of the above criteria identified candidate rearrangements or chromosomal alterations that could be detected in all colorectal and breast cancer plasma samples analyzed but not in the plasma samples from healthy individuals (nor in a large number of additional normal genomes). The rearranged sequences were evaluated by PCR amplifications across the rearrangement junctions in plasma, tumor, and normal lymphocyte DNA from the same individuals, and all were confirmed to be present in the plasma and tumor samples, but not in the matched normal DNA. Several of the identified structural alterations included changes that contained actionable genes, including amplification of ERBB2 and amplification of CDK6, showing that ctDNA genotyping can be performed through a combination of whole-genome plasma sequencing and the approaches described above. Furthermore, the approach showed an association between the level of rearranged tumor markers and tumor burden during therapy.
Implementation of whole-genome NGS with approaches using the principles of digital karyotyping has similarly been used to identify copy-number alterations in maternal plasma DNA for detection of fetal aneuploidy (93, 94). These analyses highlight the utility of identifying copy-number alterations in cfDNA for prenatal diagnosis. In a complementary approach, light-coverage whole-genome analyses have also been used to analyze alterations in repetitive sequences in cfDNA in the circulation of patients with breast cancer (95).
Promise and Challenges of ctDNA
Analysis of ctDNA provides opportunities for noninvasive detection of human cancers. Detection of somatic genetic alterations in the circulation has been challenging, but new approaches for such analyses have facilitated sensitive and specific detection at low levels. As discussed above, such approaches were initially focused on known alterations in commonly altered genes, allowing only a limited number of mutations to be analyzed at one time. These approaches have now been extended to de novo mutations through unbiased analyses in a larger number of gene exons or through genome-wide approaches. Sensitivity of detection by focused approaches ranges from approximately 1:500,000 for mutant:wild-type DNA sequences when analyzing rearrangements (85) to approximately 1:20,000 for point mutations (69). The practical sensitivity of NGS approaches for detecting such alterations may be as low as one alteration in several thousand wild-type molecules using a single lane of an NGS instrument (81, 82), and is expected to continue improving with the decreasing cost of sequencing and through new error-reducing approaches. This holds the promise of extending ctDNA from applications in late-stage tumors for genotyping and monitoring, to detection of residual disease after surgery and to early detection.
Despite progress in the analysis of ctDNA, many challenges remain. The most immediate applications focus on detection of hotspot alterations in commonly altered oncogenes. Although such analyses have important clinical uses, they miss the vast majority of somatic alterations in cancer that would require discovery of mutations rather than simply recognizing existing alterations. Likewise, although the technical sensitivity of the various approaches for mutation detection is known, the biologic level of ctDNA in early-stage patients, among different tumor types, or in various clinical scenarios has not been characterized. Some of this information has recently become available and suggests that there is a wide range of levels of ctDNA among individual patients (15). For early-stage disease, the sensitivity of ctDNA approaches has been shown to be ≥50% in patients with localized colo rectal, breast, esophageal, and pancreatic tumors (15, 96, 97), suggesting that this approach may be feasible for early detection in these and other tumor types. The amount of blood collected may be a limitation in some settings and may need to be increased to elevate the sensitivity of the approach. As with other diagnostic approaches, the use of ctDNA analyses in some clinical settings may result in detection of nonprogressing benign lesions that would not benefit from early intervention. In addition, the contribution of multiple heterogeneous tumor lesions to ctDNA will need to be evaluated, as clonal alterations common to all lesions in an individual will be present at a higher level than those that are heterogeneous or that may be present in only a single metastatic site. Approaches focusing on DNA changes may miss other molecular alterations that occur in patients with cancer, including increased levels of transcripts or protein biomarkers, although such changes lack the specificity of DNA-based somatic alterations.
CLINICAL APPLICATIONS OF CTCs AND ctDNA
We have presented the technologic considerations in both CTC and ctDNA analyses, highlighting the promise as well as challenges facing both of these strategies. Both platforms are evolving rapidly, with considerable improvements expected over the coming years. Hence, we can provide only general guidelines about their comparative utility in addressing current and future needs in clinical oncology. Overall, the analysis of CTCs brings extraordinary depth by allowing analysis of the whole cell, with RNA- and protein-based diagnostic tests, as well as DNA-based genotyping. Most significantly, as single-cell technologies evolve, CTC analyses will allow precise measurements of cancer heterogeneity and subclonal populations. Ultimately, real-time studies of CTCs cultured ex vivo could allow drug-sensitivity testing and transform individualized therapeutics. However, CTC studies will become widespread only when the most promising technologies currently under development are commercialized and broadly available to the cancer research and clinical community.
In contrast, ctDNA analysis has the great attribute of ease of collection and high-throughput analysis. As such, ctDNA genotyping may be rapid, economical, and reliable for clinical applications. Furthermore, there are indications that the levels of ctDNA may be higher than CTCs in certain tumor types, facilitating direct analyses (15, 98). The limitations to ctDNA analyses are its restriction to measurable DNA mutations, gene copy abnormalities, and potential DNA methylation abnormalities. Although the purity and level of tumor-derived sequences within total free plasma DNA are variable (15), further improvements in DNA-sequencing technologies are likely to allow whole-genome analyses and associated gene discoveries. Together, CTC and ctDNA technologies are likely to be synergistic, rather than strictly competitive, in their applications to clinical oncology. In fact, the driving rationale for both technologies stems from the concept that tumors evolve, especially in response to powerful and effective therapies, and hence repeated sampling is essential for optimal patient management. As clinical decisions become increasingly dependent on real-time monitoring of tumor status, both CTC and ctDNA analyses, each with its own particular capabilities and applications, are likely to become essential components of cancer management.
Tumor Genotyping
The most immediate applications for both CTC and ctDNA analyses are likely to be the genotyping of cancers for which mutation-targeted therapies are effective. Currently, these involve predominantly the approved indications for non–small cell lung cancer (EGFR and EML4–ALK mutations) and melanoma (BRAF), as well as upcoming applications for BRAF + EGFR–directed therapies in colorectal cancer and PIK3CA-targeted treatments in breast cancer and other cancers (99, 100). These applications are likely to increase as additional genotype-driven therapies are developed, and though they constitute a small subset of all cancers, broad testing even in cases at relatively low risk is important, given their significant impact on therapeutic choices. The availability of real-time, noninvasive, and inexpensive blood-based tumor genotyping, through either CTC or ctDNA analyses, is likely to greatly increase its application in clinical oncology practice.
Understanding, circumventing, and ultimately treating acquired resistance to targeted therapies will require monitoring for multiple molecular mechanisms; acquired drug resistance-associated mutations (such as the T790M-EGFR mutation in lung cancer or emergence of KRAS mutations in colorectal cancer; refs. 101–104) will be measured by either ctDNA or CTC analyses, whereas more complex mechanisms (including EMT or transversions from non–small cell to small cell histologies; ref. 17) will require whole-cell analyses. Most importantly, monitoring evolving mechanisms of resistance before they overtake the tumor mass will involve measuring subclonal populations and assessing heterogeneity for molecular markers. Such analyses are possible using ctDNA through measurements of read numbers, comparing the frequency of different mutated alleles with each other. In CTC-based analysis, it will require analysis of single CTCs, a capability that is under active development but not yet routinely available.
Surrogates of Drug Response
There is currently a strong interest in the oncology community in considering whether CTC numbers or tumor-specific ctDNA levels can be used as a surrogate of treatment response. In some cancers, such as castration-resistant prostate cancer, neither serum PSA levels nor bone scan changes are predictive of long-term patient outcome following treatment with second-line hormonal therapies, and surrogate markers are essential to facilitate the selection of experimental agents (105, 106). Both CTC numbers and ctDNA have the potential to serve as markers of tumor burden within a given patient followed longitudinally (10, 11, 65, 86). However, long-term studies will be required to see if these measurements are correlated with disease-free survival (DFS) in specific clinical settings, and whether they outperform standard radiologic measurements of disease. For ctDNA, the timing of such measurements in relation to therapy may be important, as dying tumor cells may actually lead to increased DNA release into the circulation during treatment. In addition to such long-term predictive values, short-term readouts to guide choices among multiple therapies may one day become feasible. For instance, monitoring CTCs shortly after drug administration could measure rapid shifts in intracellular phospho-signaling, apoptosis, or proliferative indices. Alternatively using ctDNA, rapid shifts in allele fractions for specific mutations could identify responsiveness of subclonal tumor populations.
Detecting Early Relapse
The ability to detect molecular evidence of cancer recurrence after initial surgical or radiation treatments has been controversial. Early studies using RT-PCR analyses to detect known translocations were confounded by the absence of relevant therapeutic options (107). However, with the advent of increasingly effective therapies, diagnosing relapse early may allow more effective treatment while the tumor burden is still low. In this context, ctDNA analyses are particularly sensitive in that the primary tumor can be sequenced for tumor-specific “driver” or “passenger” translocations that provide an exquisitely sensitive way to monitor for early tumor recurrence (85, 88, 98).
High-Risk Localized Cancers
Most curative cancer therapies are administered in the adjuvant setting, where the low tumor burden is likely to result in eradication of tumor cells following appropriately administered therapy. Distinguishing individuals with localized tumors at high risk for recurrence who would benefit from such therapies, versus those who can be safely monitored without adjuvant treatment, has traditionally relied on histopathologic criteria within the primary tumor (e.g., grade, size, and vascular invasion), presence of tumor cells within draining “sentinel” lymph nodes, or the use of molecular markers, such as Oncotype Dx, which may help predict tumor aggressiveness in some contexts. However, these criteria have imperfect predictive value, and it is possible that levels of ctDNA or presence of tumor cells within the vasculature may provide additional information with respect to risk of relapse. For both ctDNA and CTC analyses, additional assay sensitivity will be critical to enable reliable analysis of small localized cancers, and rigorous clinical trials will be essential to test whether the amount of ctDNA or CTCs will correlate with high-risk localized cancers, or whether particular molecular or cellular subsets harbor such information.
Novel Therapeutic Targets
Long-term goals of both ctDNA and CTC analyses involve testing drug sensitivity regimens ex vivo in cells derived from an individual tumor. That goal, an ultimate achievement for personalized cancer therapy, would require the robust culture of viable CTCs, a promising area of investigation. It is also possible that CTC analyses will identify particular pathways that support the viability of tumor cells during their transit in the circulation; although such drug targets may not be evident from analyses of primary tumors, their identification in CTCs may enable therapeutic strategies to suppress blood-borne metastasis. Similarly, repeated deep sequencing of ctDNA during the course of treatment may identify key targets that have become dominant in a tumor in real time and help focus therapeutics on such targets. As an example of this approach, whole-genome analyses of plasma ctDNA during patient treatment have recently identified MET amplification as a mechanism of resistance to EGFR blockade with cetuximab (108, 109). Such analyses can be used for the discovery of molecular resistance as well as identification of new therapeutic targets.
Early Detection of Cancer
Finally, it is likely that blood-based diagnostics will have their greatest impact in early detection of cancer. Even localized cancers may shed some DNA into the circulation, and CTCs have been detected in some patients with localized cancer. Thus, the presence of these biomarkers in the blood does not by itself indicate advanced or incurable cancer, and both ctDNA and CTC analyses may prove suitable for early diagnosis of cancer. Much optimization remains to be done, both in terms of increasing the sensitivity of both assays and guarding against false positives, which may doom any population-based screening. Nonetheless, it is possible to imagine a time when individuals at high risk of developing cancer due to either genetic or environmental risk factors (e.g., women with inherited BRCA gene mutations at risk for breast cancer, or heavy smokers at risk for lung cancer) could be serially monitored using either ctDNA or CTC analyses. The choice of technology in such cases would be driven by cost, sensitivity, specificity, and robustness of the assays, but such approaches may change forever the approach to screening for cancers that are currently incurable unless diagnosed at an early stage.
CONCLUDING REMARKS
We see very rapid progress in technologic developments toward blood-based diagnostics in clinical oncology, with multiple applications throughout different stages and disease types. Much remains to be done to optimize the diverse technologies and their applications, standardize these across different platforms, and enable their broad dissemination throughout the cancer research and clinical oncology communities. Nonetheless, these technologies are poised to radically change our approaches to the treatment of cancer. We see ctDNA and CTC analyses as complementary in the types of information that they will provide in different clinical settings. Where they are competitive, primarily in DNA genotyping analyses, it is likely that cost and reliability will dictate the most relevant technology, although even there, it is likely that different assays will be required in diverse clinical contexts (e.g., measurements of point mutations in DNA vs. chimeric translocated RNA transcripts, dominant mutations vs. rare subclones, known recurrent mutations in primary tumors vs. novel drug resistance associated variants). Together, ctDNA and CTC analyses are ushering in a new era in oncology, where “real-time” monitoring of tumor status is paramount for effective therapy.
The revolution in targeted cancer treatments, now accompanied by rapid changes in the ability to genotype cancers and measure their evolving functional properties through noninvasive blood monitoring, links cancer therapeutics and diagnostics as never before. These two previously disparate fields are now rapidly co-evolving, with the success of each depending on the capabilities of the other. This realization should lead to more integration of research in both academic and pharmaceutical efforts, as well as supporting research and ultimately clinical applications at the federal regulatory level. As these technologies mature over the next few years and are disseminated to the cancer community, we anticipate that they will prove enabling for major new directions in the diagnosis and treatment of diverse cancers.
Acknowledgments
The authors thank their respective laboratory members and collaborators for critical review of this article and E. Cook for assistance with artwork. The authors apologize that space constraints prevent them from citing all relevant publications.
Grant Support: D.A. Haber is supported by the Howard Hughes Medical Institute, NIH (CA-129933, NIBIB-EB008047), a Stand Up To Cancer Dream Team Translational Cancer Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0309), the Breast Cancer Research Foundation, the National Foundation for Cancer Research, and the Johnson & Johnson Center for Excellence in CTCs at Massachusetts General Hospital. V.E. Velculescu is supported by NIH (CA-121113), The European Community’s Seventh Framework Programme, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the John G. Ballenger Trust, a Stand Up To Cancer Dream Team Translational Cancer Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0509), and the Commonwealth Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest
D.A. Haber has received commercial research support from Johnson & Johnson and is a consultant/advisory board member of Life Technologies. V.E. Velculescu is member of the Board of Directors of Personal Genome Diagnostics and has ownership interest (including patents) in the same.
References
- 1.Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust Med J. 1869;14:146–9. [Google Scholar]
- 2.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells:approaches to isolation and characterization. J Cell Biol. 2011;192:373–82. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients:challenges and perspectives. Trends Mol Med. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Maheswaran S, Haber DA. Circulating tumor cells:a window into cancer biology and metastasis. Curr Opin Genet Dev. 2010;20:96–9. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil. 1948;142:241–3. [PubMed] [Google Scholar]
- 6.Stroun M, Anker P, Lyautey J, Lederrey C, Maurice PA. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol. 1987;23:707–12. doi: 10.1016/0277-5379(87)90266-5. [DOI] [PubMed] [Google Scholar]
- 7.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–50. [PubMed] [Google Scholar]
- 8.Alix-Panabieres C, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- 9.Scher HI, Morris MJ, Larson S, Heller G. Validation and clinical utility of prostate cancer biomarkers. Nat Rev Clin Oncol. 2013;10:225–34. doi: 10.1038/nrclinonc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson SJ, Rosenfeld N, Caldas C. Circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;369:93–4. doi: 10.1056/NEJMc1306040. [DOI] [PubMed] [Google Scholar]
- 12.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 13.Cohen SJ, Alpaugh RK, Gross S, O’Hara SM, Smirnov DA, Terstappen LW, et al. Isolation and characterization of circulating tumor cells in patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2006;6:125–32. doi: 10.3816/CCC.2006.n.029. [DOI] [PubMed] [Google Scholar]
- 14.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–8. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 15.Bettegowda C, Sausen M, Leary RJ, Kinde I, Want Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–9. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 19.Xu L, Kikuchi E, Xu C, Ebi H, Ercan D, Cheng KA, et al. Combined EGFR/MET or EGFR/HSP90 inhibition is effective in the treatment of lung cancers codriven by mutant EGFR containing T790M and MET. Cancer Res. 2012;72:3302–11. doi: 10.1158/0008-5472.CAN-11-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alix-Panabieres C, Pantel K. Circulating tumor cells:liquid biopsy of cancer. Clin Chem. 2013;59:110–8. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 21.Kalluri R, Weinberg RA. The basics of epithelial–mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fidler IJ. The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis. Eur J Cancer. 1973;9:223–7. doi: 10.1016/s0014-2964(73)80022-2. [DOI] [PubMed] [Google Scholar]
- 23.Liotta LA, Saidel MG, Kleinerman J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res. 1976;36:889–94. [PubMed] [Google Scholar]
- 24.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, et al. Isolation and characterization of circulating tumor cells from localized and metastatic prostate cancer patients. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duda DG, Duyverman AM, Kohno M, Snuderl M, Steller EJ, Fukumura D, et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proc Natl Acad Sci U S A. 2010;107:21677–82. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol. 2012;9:016001. doi: 10.1088/1478-3975/9/1/016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, et al. Single cell profiling of circulating tumor cells:transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–75. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539–44. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 33.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–8. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–53. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohamed H, Murray M, Turner JN, Caggana M. Isolation of tumor cells using size and deformation. J Chromatogr A. 2009;1216:8289–95. doi: 10.1016/j.chroma.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 36.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, et al. Isolation by size of epithelial tumor cells:a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microdevices. 2009;11:883–92. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 38.Lazar DC, Cho EH, Luttgen MS, Metzner TJ, Uson ML, Torrey M, et al. Cytometric comparisons between circulating tumor cells from prostate cancer patients and the prostate-tumor-derived LNCaP cell line. Phys Biol. 2012;9:016002. doi: 10.1088/1478-3975/9/1/016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marrinucci D, Bethel K, Bruce RH, Curry DN, Hsieh B, Humphrey M, et al. Case study of the morphologic variation of circulating tumor cells. Hum Pathol. 2007;38:514–9. doi: 10.1016/j.humpath.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 40.Pachmann K, Clement JH, Schneider CP, Willen B, Camara O, Pachmann U, et al. Standardized quantification of circulating peripheral tumor cells from lung and breast cancer. Clin Chem Lab Med. 2005;43:617–27. doi: 10.1515/CCLM.2005.107. [DOI] [PubMed] [Google Scholar]
- 41.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, et al. A rare-cell detector for cancer. Proc Natl Acad Sci U S A. 2004;101:10501–4. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gertler R, Rosenberg R, Fuehrer K, Dahm M, Nekarda H, Siewert JR. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 2003;162:149–55. doi: 10.1007/978-3-642-59349-9_13. [DOI] [PubMed] [Google Scholar]
- 43.Gascoyne PR, Noshari J, Anderson TJ, Becker FF. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis. 2009;30:1388–98. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, et al. Membrane micro-filter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162:154–61. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 45.Weight RM, Dale PS, Viator JA. Detection of circulating melanoma cells in human blood using photoacoustic flowmetry. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:106–9. doi: 10.1109/IEMBS.2009.5335145. [DOI] [PubMed] [Google Scholar]
- 46.Galanzha EI, Shashkov EV, Kelly T, Kim JW, Yang L, Zharov VP. In vivo magnetic enrichment and multiplex photoacoustic detection of circulating tumour cells. Nat Nanotechnol. 2009;4:855–60. doi: 10.1038/nnano.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casavant BP, Mosher R, Warrick JW, Maccoux LJ, Berry SM, Becker JT, et al. A negative selection methodology using a microfluidic platform for the isolation and enumeration of circulating tumor cells. Methods. 2013;64:137–43. doi: 10.1016/j.ymeth.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alix-Panabieres C, Vendrell JP, Pelle O, Rebillard X, Riethdorf S, Muller V, et al. Detection and characterization of putative metastatic precursor cells in cancer patients. Clin Chem. 2007;53:537–9. doi: 10.1373/clinchem.2006.079509. [DOI] [PubMed] [Google Scholar]
- 49.Alix-Panabieres C, Rebillard X, Brouillet JP, Barbotte E, Iborra F, Segui B, et al. Detection of circulating prostate-specific antigen-secreting cells in prostate cancer patients. Clin Chem. 2005;51:1538–41. doi: 10.1373/clinchem.2005.049445. [DOI] [PubMed] [Google Scholar]
- 50.Ramirez JM, Fehm T, Orsini M, Cayrefourcq L, Maudelonde T, Pantel K, et al. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin Chem. 2014;60:214–21. doi: 10.1373/clinchem.2013.215079. [DOI] [PubMed] [Google Scholar]
- 51.Friedlander TW, Ngo VT, Dong H, Premasekharan G, Weinberg V, Doty S, et al. Detection and characterization of invasive circulating tumor cells (ictcs) derived from men with metastatic castration resistant prostate cancer (mCRPC) Int J Cancer. 2014;134:2284–93. doi: 10.1002/ijc.28561. [DOI] [PubMed] [Google Scholar]
- 52.Paris PL, Kobayashi Y, Zhao Q, Zeng W, Sridharan S, Fan T, et al. Functional phenotyping and genotyping of circulating tumor cells from patients with castration resistant prostate cancer. Cancer Lett. 2009;277:164–73. doi: 10.1016/j.canlet.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 53.Lu J, Fan T, Zhao Q, Zeng W, Zaslavsky E, Chen JJ, et al. Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients. Int J Cancer. 2010;126:669–83. doi: 10.1002/ijc.24814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer:a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–8. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 55.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 56.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A’Hern R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–8. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 57.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A. 2009;106:3970–5. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cann GM, Gulzar ZG, Cooper S, Li R, Luo S, Tat M, et al. mRNA-Seq of single prostate cancer circulating tumor cells reveals recapitulation of gene expression and pathways found in prostate cancer. PLoS ONE. 2012;7:e49144. doi: 10.1371/journal.pone.0049144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–7. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–3. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Carlo D, Irimia D, Tompkins RG, Toner M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc Natl Acad Sci U S A. 2007;104:18892–7. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 65.Seiden MV, Kantoff PW, Krithivas K, Propert K, Bryant M, Haltom E, et al. Detection of circulating tumor cells in men with localized prostate cancer. J Clin Oncol. 1994;12:2634–9. doi: 10.1200/JCO.1994.12.12.2634. [DOI] [PubMed] [Google Scholar]
- 66.Xi L, Nicastri DG, El-Hefnawy T, Hughes SJ, Luketich JD, Godfrey TE. Optimal markers for real-time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers. Clin Chem. 2007;53:1206–15. doi: 10.1373/clinchem.2006.081828. [DOI] [PubMed] [Google Scholar]
- 67.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–42. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 68.Perkins G, Yap TA, Pope L, Cassidy AM, Dukes JP, Riisnaes R, et al. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS ONE. 2012;7:e47020. doi: 10.1371/journal.pone.0047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem. 2010;56:1279–86. doi: 10.1373/clinchem.2010.144188. [DOI] [PubMed] [Google Scholar]
- 71.Jen J, Wu L, Sidransky D. An overview on the isolation and analysis of circulating tumor DNA in plasma and serum. Ann N Y Acad Sci. 2000;906:8–12. doi: 10.1111/j.1749-6632.2000.tb06581.x. [DOI] [PubMed] [Google Scholar]
- 72.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang JY, Hsieh JS, Chang MY, Huang TJ, Chen FM, Cheng TL, et al. Molecular detection of APC, K-ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J Surg. 2004;28:721–6. doi: 10.1007/s00268-004-7366-8. [DOI] [PubMed] [Google Scholar]
- 74.Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120:461–7. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 75.Yamada T, Nakamori S, Ohzato H, Oshima S, Aoki T, Higaki N, et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma:correlation with clinicopathological features. Clin Cancer Res. 1998;4:1527–32. [PubMed] [Google Scholar]
- 76.Shi J, Liu Q, Sommer SS. Detection of ultrarare somatic mutation in the human TP53 gene by bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification. Hum Mutat. 2007;28:131–6. doi: 10.1002/humu.20423. [DOI] [PubMed] [Google Scholar]
- 77.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A. 2003;100:8817–22. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pekin D, Skhiri Y, Baret JC, Le Corre D, Mazutis L, Salem CB, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11:2156–66. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- 81.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–5. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 83.Thompson JD, Shibahara G, Rajan S, Pel J, Marziali A. Winnowing DNA for rare sequences:highly specific sequence and methylation based enrichment. PLoS ONE. 2012;7:e31597. doi: 10.1371/journal.pone.0031597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 85.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer. 2010;49:1062–9. doi: 10.1002/gcc.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang TL, Maierhofer C, Speicher MR, Lengauer C, Vogelstein B, Kinzler KW, et al. Digital karyotyping. Proc Natl Acad Sci U S A. 2002;99:16156–61. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra54. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang TL, Diaz LA, Jr, Romans K, Bardelli A, Saha S, Galizia G, et al. Digital karyotyping identifies thymidylate synthase amplification as a mechanism of resistance to 5-fluorouracil in metastatic colorectal cancer patients. Proc Natl Acad Sci U S A. 2004;101:3089–94. doi: 10.1073/pnas.0308716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duncan CG, Leary RJ, Lin JC, Cummins J, Di C, Schaefer CF, et al. Identification of microbial DNA in human cancer. BMC Med Genomics. 2009;2:22. doi: 10.1186/1755-8794-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, et al. Cancer genome scanning in plasma:detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2012;59:211–24. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 92.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–71. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:20458–63. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beck J, Urnovitz HB, Mitchell WM, Schutz E. Next generation sequencing of serum circulating nucleic acids from patients with invasive ductal breast cancer reveals differences to healthy and nonmalignant controls. Mol Cancer Res. 2010;8:335–42. doi: 10.1158/1541-7786.MCR-09-0314. [DOI] [PubMed] [Google Scholar]
- 96.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–73. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beaver JA, Jelovac D, Balukrishna S, Cochran R, Croessmann S, Zabransky D, et al. Detection of cancer DNA in plasma of early stage breast cancer patients. Clin Cancer Res. 2014 Feb 8; doi: 10.1158/1078-0432.CCR-13-2933. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–7. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 100.McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N Engl J Med. 2011;364:340–50. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 101.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 103.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer:a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–9. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sklar J. Polymerase chain reaction:the molecular microscope of residual disease. J Clin Oncol. 1991;9:1521–4. doi: 10.1200/JCO.1991.9.9.1521. [DOI] [PubMed] [Google Scholar]
- 108.Diaz LA, Jr, Sausen M, Fisher GA, Velculescu VE. Insights into therapeutic resistance from whole-genome analyses of circulating tumor DNA. Oncotarget. 2013;4:1856–7. doi: 10.18632/oncotarget.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3:658–73. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kirby BJ, Jodari M, Loftus MS, Gakhar G, Pratt ED, Chanel-Vos C, et al. Functional characterization of circulating tumor cells with a prostate-cancer-specific microfluidic device. PLoS ONE. 2012;7:e35976. doi: 10.1371/journal.pone.0035976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu YT, Zhao L, Shen Q, Garcia MA, Wu D, Hou S, et al. NanoVelcro Chip for CTC enumeration in prostate cancer patients. Methods. 2013;64:144–52. doi: 10.1016/j.ymeth.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tkaczuk KH, Goloubeva O, Tait NS, Feldman F, Tan M, Lum ZP, et al. The significance of circulating epithelial cells in breast cancer patients by a novel negative selection method. Breast Cancer Res Treat. 2008;111:355–64. doi: 10.1007/s10549-007-9771-9. [DOI] [PubMed] [Google Scholar]
- 113.Balasubramanian P, Yang L, Lang JC, Jatana KR, Schuller D, Agrawal A, et al. Confocal images of circulating tumor cells obtained using a methodology and technology that removes normal cells. Mol Pharm. 2009;6:1402–8. doi: 10.1021/mp9000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, et al. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng. 2009;102:521–34. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, et al. Cancer genome scanning in plasma:detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–24. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]


