Abstract
Current therapy for controlling HIV-1 infection and preventing AIDS progression has profoundly decreased viral replication in cells susceptible to HIV-1 infection, but it does not eliminate the low level of viral replication in latently infected cells which contain integrated copies of HIV-1 proviral DNA. There is an urgent need for the development of HIV-1 genome eradication strategies that will lead to a permanent or “sterile” cure of HIV-1/AIDS. In the past few years, novel nuclease-initiated genome editing tools have been developing rapidly, including ZFNs, TALENs, and the CRISPR/Cas9 system. These surgical knives, which can excise any genome, provide a great opportunity to eradicate the HIV-1 genome by targeting highly conserved regions of the HIV-1 long terminal repeats or essential viral genes. Given the time consuming and costly engineering of target-specific ZFNs and TALENs, the RNA-guided endonuclease Cas9 technology has emerged as a simpler and more versatile technology to allow permanent removal of integrated HIV-1 proviral DNA in eukaryotic cells, and hopefully animal models or human patients. The major unmet challenges of this approach at present include inefficient nuclease gene delivery, potential off-target cleavage, and cell-specific genome targeting. Nanoparticle or lentivirus-mediated delivery of next generation Cas9 technologies including nickase or RNA-guided FokI nuclease (RFN) will further improve the potential for genome editing to become a promising approach for curing HIV-1/AIDS.
Keywords: Genome editing, CRISPR/Cas9, HIV-1 integration, latent reservoir, cure, animal models
Viral latency as a complex barrier to an HIV-1 cure
An effective cure for HIV-1 infection can only be achieved with the complete purging of the viral reservoirs within an infected individual in order to ultimately prevent the spread of the virus to healthy cells (Stone et al, 2013). Viral latency is a phenomenon common to several viruses including cytomegalovirus (CMV), herpes simplex virus (HSV), human T-lymphotropic virus 1 (HTLV-1), and human immunodeficiency virus (HIV-1). Latency has been defined as a state of non-productive infection (Siliciano and Greene, 2011) and it is thought to have evolved as a mechanism to evade triggering of an immune system response, therefore allowing viral persistence. In the context of HIV-1, latency is established within specific cell populations (latent reservoir) (Battistini and Sgarbanti, 2014; Kumar et al, 2014; Van Lint et al, 2013).
Several approaches have been proposed to eliminate the latent HIV-1 viral infection. One of the most studied consists of a “purging strategy” to induce reactivation of the virus in latently infected T-cells in order to make the latently infected cells susceptible to treatment without inducing T-cell activation (Shan and Siliciano, 2013). Indeed, small molecules that can target the DNA and impact viral gene expression offer alternatives that avoid activation of cell surface receptors (Matalon et al, 2011). Histone deacetylase inhibitors (HDACi), like valproic acid or voninostat (SAHA), are well suited to the purpose, with the ability to increase histone acetylation of the integrated viral promoter and therefore trigger reactivation of the latent reservoir. This strategy was tried in patients but unfortunately was unsuccessful in eliminating all latent viruses, likely due to only partial activation of the latent reservoir (Archin et al, 2014; Manson McManamy et al, 2014; Rasmussen et al, 2013; Siliciano and Siliciano, 2014).
Two different strategies have been described as potential “cures” for HIV-1 infection: a functional cure and a sterilizing cure (Van Lint et al, 2013). A functional cure would be obtained when the latent reservoir remains but there is permanent control of viral replication without the need for continuous anti-retroviral therapy. Indeed, CCR5 impairment is the most advanced approach for a functional cure for HIV-1 to date (Allers et al, 2011; Tebas et al, 2014; Ye et al, 2014). On the other hand, a sterilizing cure would only be obtained upon complete eradication of all replication-competent forms of HIV-1 by eliminating the latent reservoir and removing all traces of the virus from infected patients. At present, a path leading to a sterilizing cure has not been achieved, however, such a cure could be achieved by excision of the integrated viral genome from the host DNA utilizing one of several genome editing strategies which are still in the early stages of development (Hu et al, 2014; Manjunath et al, 2013; Stone et al, 2013).
Genome editing technologies and their application to HIV-1 infection
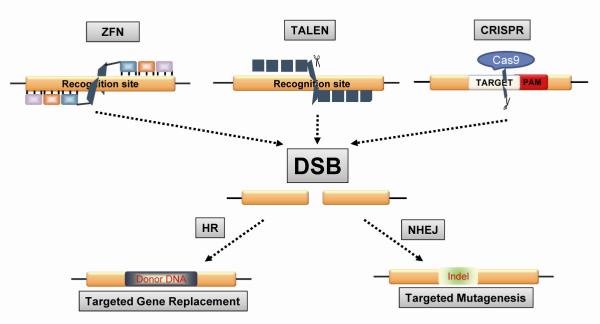
Genome engineering strategies have been largely explored in the past decade and a better understanding of their functions and mechanisms has been achieved. Traditional gene targeting, such as replacement of genomic DNA with exogenous DNA (donor DNA) by homologous recombination, has been extensively employed to manipulate the eukaryotic genome. In particular, gene targeting in mouse embryonic stem cells has generated a large number of transgenic knockin/knockout mice. However, several challenges for traditional gene targeting exist, including extremely low target efficiency, limited gene target size, species limitations, time-consuming and labor-intensive selection/screening, and uncertain germline transmission. These challenges have been improved by inducing site-specific DNA double-strand breaks (DSBs), which dramatically increase the recombination efficiency for gene targeting by utilizing the intrinsic cellular DNA repair machinery. DSBs stimulate restoration of the intact genome either by homologous recombination (HR) in the presence of DNA donor or, more commonly, via error-prone non-homologous end joining (NHEJ), which has a high probability of gene alteration due to insertion and deletion mutations, known as indels. Several types of nucleases have been identified that induce DSBs (Fig. 1) and implicated in targeting HIV-1 virus and host cellular genes (Table 1).
Figure 1. Nuclease-induced double strand breaks (DSBs) initiate the cellular homologous recombination (HR) and non-homologous end joining (NHEJ) DNA repair pathways exploited by the ZFN, TALEN, and CRISPR genome editing technologies.
Yellow squares indicate target genome. PAM, protospacer adjacent motif. Indel, insertion or deletion mutants.
Table 1.
HIV genomic and cellular targets by genome editing technology
| Viral targets | Nuclease | Sequence/Strategy | Cells/Organism | Gene delivery | Efficiency | Reference |
|---|---|---|---|---|---|---|
| LTR | Cas9 | Single, U3 region Duplex, U3 region |
TZMbI, U1, J- Lat |
Transient or Stable transfection |
30-90% | (Hu et al, 2014) |
| Cas9 | Single: TAR and U3 |
HEK293T cell, Hela cells, Jurkat cell line |
Repeated transient transfection |
30-96% | (Ebina et al, 2013) | |
| ZFN | Single, R and U3 region |
Primary T cells Jurkat T cell line |
Transient transfection |
46% | (Qu et al, 2013) (Qu et al, 2014) |
|
| Tre | 34 bp sequence (loxLTR) |
T cells, HSC, Humanized mice |
Lentivirus Cell transplantation |
(Hauber et al, 2013; Mariyanna et al, 2012) | ||
| Gag/Pol/Env | ZFN | Disruption | T cells, TZMbI | Lentivirus | 80% | (Wayengera, 2011) |
| Vpr | Meganuclease | Disruption | CHO | Lentivirus | (Izmiryan et al, 2011) | |
|
Cellular
targets |
Nuclease | Sequence/Strategy | Cells/Organism | Gene delivery | Efficiency | Reference |
| CCR5 (Too many reports, only representatives are cited) |
Cas9 | Disruption | HEK293T | Transfection | 18-50% | (Cradick et al, 2013) (Fu et al, 2014) |
| Disruption | iPSC | Stable transfection |
33- 100% | (Ye et al, 2014) | ||
| Disruption | K562 | Plasmid transfection |
11% | (Cho et al, 2013) | ||
| ZFN | Delta32 | T cells | Transfection Adenovirus Lentivirus NILV |
10-54% | (Mani et al, 2005) (Lee et al, 2010) (Perez et al, 2008) (Maier et al, 2013) (Lombardo et al, 2011) |
|
| Disruption | ESC | Transfection | (Yao et al, 2012) | |||
| Disruption | HSC | Transfection Adenovirus Transplantation |
10-50% | (Lombardo et al, 2007) (Holt et al, 2010) (Li et al, 2013) |
||
| Disruption | iPSC | Transfection | (Lombardo et al, 2011) (Yao et al, 2012) (Ye et al, 2014) |
|||
| Disruption | Neural stem cells |
Transfection | 5% | (Lombardo et al, 2011) | ||
| Delta32 | Humanized mice |
Adenovirus NILV Cell transplantation |
(Lee et al, 2010) (Perez et al, 2008) (Yi et al, 2014) |
|||
| Delta32 | Patients | Adenovirus Cell transplantation |
(Maier et al, 2013) (Tebas et al, 2014) |
|||
| TALEN | CCR5delta32 | HEK293T | Transfection | (Nerys-Junior et al, 2014) (Liu et al, 2014) |
||
| CCR5 knockin | Rabbit animal model |
Oocyte injection |
(Tang et al, 2014) | |||
| CCR5 | iPSC | Transfection | (Ramalingam et al, 2014) | |||
| Disruption | HEK293T NuFF cells |
Plasmid | 15-45% | (Mussolino et al, 2011) (Mussolino et al, 2014) |
||
| CXCR4 | ZFN | Disruption | T cells Humanized mice |
Ad5/F35 Cell transplantation |
30-34% | (Wilen et al, 2011) |
| CCR5 and CXCR4 |
ZFN | Disruption | T cells, Humanized Mice |
Plasmid, Adenovirus, Cell transplantation |
(Yuan et al, 2012) | |
| Disruption | T cells Humanized mice |
Ad5/F35 Cell transplantation |
(Didigu et al, 2014) | |||
| PSIP1 | Talen | whole-gene deletion and exon deletion |
HEK293T Jurkat |
Transfection | (Fadel et al, 2014a) | |
| Restriction factors |
ZFN | TRIM5α and APOBEC3G |
T cells | Transfection | (Voit et al, 2013) |
Note: HSC, hematopoietic stem/progenitor cells; ESC, embryonic stem cells; iPSC, induced pluripotent cells; Tre, Tailored site-specific recombinase. NILV, non-integrated lentivirus.
Homing endonucleases
More than a decade ago, meganuclease families, also called homing endonucleases, were described which are sequence-specific endonucleases with recognition sites larger than 12 bp (usually 16-30 bp) (Karvelis et al, 2013; Pingoud and Silva, 2007). Homing endonucleases have been used for site-directed genome modification and for several years were considered to be a very promising tool for genome editing. As proof of concept, homing endonucleases were tested for their ability to disrupt integrated HIV-1 provirus DNA using an integrated lentiviral reporter assay. The main limitation of the technology is their size and their potential disruption of endogenous genes or transcriptional activation of neighboring genes (Arnould et al, 2011). An added challenge to the engineering of meganucleases is the fact that the DNA recognition and cleavage functions of these enzymes are combined within a single domain.
Zinc finger nucleases (ZFNs)
The zinc finger nucleases (ZFNs), however, are much more compact in size. ZFNs are nucleases engineered to contain a specific class of transcription factor that targets the DNA via zinc finger motifs and a non-specific cleavage domain from the Type IIS restriction endonuclease, FokI. FokI dimerization through the binding of paired zinc finger proteins activates the ZFN, leading to DSBs within the target region. Since their discovery in 1996 (Kim et al, 1996), ZFNs have been extensively used to manipulate the genomes of plants, animals, and humans. The ability of ZFNs to disrupt CCR5 and CXCR4 genes has largely been tested in HIV-1 infected CD4+ T cells (Mani et al, 2005; Perez et al, 2008; Yuan et al, 2012). Due to the reduced number of CCR5Δ32 donors, strategies using ZFNs have been employed to obtain engineered CCR5 knockout cells in HIV-1 patients (Holt et al, 2010) and a clinical trial has demonstrated that the ZFN technique appears to be safe and effective in humans (Tebas et al, 2014). For the first time, Tebas and colleagues used ZFNs to target and disrupt the CCR5 gene in CD4+ T-cells, inducing a “beneficial” mutation in the CCR5 gene within those cells. CCR5-modified autologous CD4+ T-cells were then infused into the patients to repopulate the immune system with CCR5-deficient central memory T cells. This approach increased resistance to HIV-1 virus in 12 patients with chronic aviremic HIV infection under cART (Tebas et al, 2014). Indeed, 6 of the 12 participants suspended cART, and their HIV levels rebounded more slowly than normal, suggesting that the attempt was not yet a “permanent cure” but a delay in progression.
Transcription activator-like effector nucleases (TALENs)
Similar to ZFNs, transcription activator-like effector (TALE) nucleases (TALENs) have been developed and tested for controlling HIV-1 infection. TALEs are natural proteins secreted by the bacteria Xanthomonas spp. to regulate gene transcription in host plant cells. TALEN is a synthesized nuclease composed of a nonspecific FokI nuclease domain in fusion with a customizable TALE-derived DNA-binding domain with highly conserved 33-35 amino acid repeats. Each TALE repeat binds to a single nucleotide of DNA, which is dependent on the two hypervariable residues which are usually found at positions 12 and 13 of the repeat. The domain containing Asn/Ile for adenine (A), Asn/Gly for thymine (T), Asn/Asn for guanine (G) and His/Asp for cytosine (C) has been identified and is currently used. The number of repeats in the array corresponds to the length of the target site. The dimerization of the fused FokI brought by the opposite pair of TALE repeats allows the cleavage of the target DNA at the spacer region (10-15 bp). In the past 3-4 years, TALEN has been extensively used in a large number of applications due to its easy design, high cleavage efficiency and seemingly unlimited targeting range (Guilinger et al, 2014a; Joung and Sander, 2013). For HIV-1 infection, both TALENs and ZFNs induced ~45% efficiency in disrupting the CCR5 gene but TALEN induced much lower cytotoxicity and off-target effects (Mussolino et al, 2014; Mussolino et al, 2011). In inducible pluripotent stem cells, TALEN is capable of inducing up to 100% targeting of CCR5 32 on one allele with ~14% biallelic targeting (Ye et al, 2014). TALEN knockout of the PSIP1 gene in HEK293T cells and Jurkat cells rendered them resistant to HIV-1 infection in a cell culture model (Fadel et al, 2014b). In contrast to ZFNs that recognize three nucleotides per finger, TALENs recognize only one nucleotide per domain. Therefore, the ability to construct functional TALEN pairs with large numbers of repeats may increase their potential specificity when compared to ZFNs (Holkers et al, 2013). However, the sequence homology between each of the repeat variable diresidues (RVDs) makes it very difficult to build constructs containing multiple TALENs. Furthermore, a crucial limitation to their potential therapeutic application is their delivery into host cells, due to TALEN’s susceptibility to induce rearrangements resulting in genomic instability. Indeed, previous studies have shown that HIV-1-based lentiviral vector genomes containing TALEN sequences are prone to rearrangements in the target cells and new optimization strategies will likely be required if they are to be used in future clinical applications (Hockemeyer et al, 2011).
RNA-guided CRISPR/Cas9 technology
In the past several years, ZFNs and TALENs have attracted extensive attention in the field of genome editing because of their advantages over traditional gene targeting technologies, including unlimited species application, higher target efficiency, rapid animal engineering, and faithful germline transmission. One major challenge for ZFNs and TALENs is the costly and time-consuming engineering of the target gene-specific fusion proteins. Thus, a new easy and versatile genome editing tool has recently entered the spotlight: the CRISPR-associated (Cas) RNA-guided endonuclease Cas9 technology (Hsu et al, 2014).
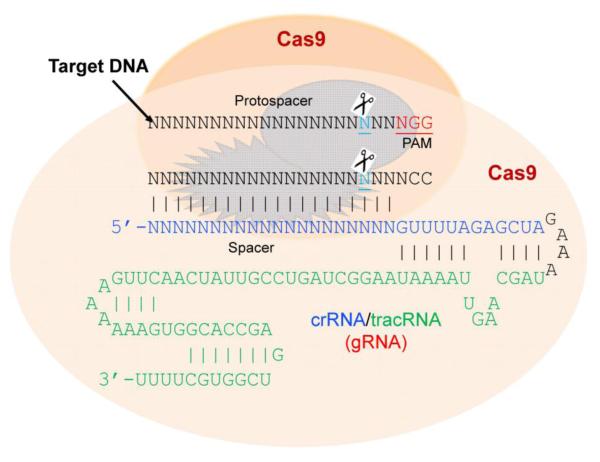
The clustered regularly interspaced short palindromic repeats (CRISPR) system has been detected in the majority of bacteria and archaea where it acts as a natural defense mechanism against bacteriophages. Due to the ubiquity and abundance of bacteriophages, prokaryotes have developed and refined this notable defense mechanism to neutralize their infectivity (Barrangou et al, 2007). The CRISPR machinery works as a kind of primitive yet highly selective and efficient immune system to preserve genetic integrity after exogenous DNA exposure and uptake. CRISPR consists of an array of small conserved repeat sequences interspaced by small DNA sequences called “spacers”, which derive from the phage DNA. CRISPR loci, together with Cas genes, form the CRISPR-Cas adaptive immune system. The type II system has been engineered to incorporate nucleic acids of the invading host into CRISPR loci and uses the corresponding CRISPR RNA (crRNA) or “guide” RNA (gRNA) to direct the degradation of homologous target sequences (protospacer).
Engineered gRNAs contain a 20 bp guide sequence (spacer, seed) that directs Cas9 to the genomic target using canonical base pairing which can be easily programmed (Ran et al, 2013). Hence, the multifunctional Cas9 protein and its appropriate gRNA are fundamental components for the formation of the functional Cas9/gRNA complex (Mali et al, 2013b). In order to recognize a target sequence (protospacer) and perform precise cleavage of the target DNA, the system requires complementarity between the spacer and the target sequence (protospacer) together with the presence of the protospacer-adjacent motif (PAM) sequence at 3′ end of the target sequence (Fig. 2). The PAM allows strict selectivity to the target array. The type II engineered system, mimicking the strategy of Streptococcus pyogenes (SpCas9), requires either NGG or NAG as the PAM sequence, in which N is any nucleotide. In just over a year from its first application in mammalian cells (Cong et al, 2013; Mali et al, 2013c), the engineered CRISPR/Cas9 system has been employed by thousands of laboratories for genome editing in a large number of cells and species including human cell lines, bacteria, zebrafish, yeast, mouse, fruit flies, roundworms, rats, common crops, pigs, and monkeys (Hsu et al, 2014). Since Cas9 works as a universal nuclease, researchers in any lab need only a tiny custom RNA molecule which, like small interfering RNA (siRNA), can be chemically synthesized or in vitro transcribed for direct RNA transfection or expressed from an RNA expression vector containing a U6 or H1 promoter. Thus, simple oligonucleotide synthesis and a single cloning step are all that is needed to utilize this versatile genome-editing tool. The simplicity of Cas9 targeting lends itself to the design of large gRNA libraries, which can be adapted to include multiple gRNAs covering the entire genome of a host organism, and thus can be employed for forward genetic screening and selection approaches. In addition gRNA libraries with the Cas9 nuclease have been used to induce knockout mutations in a variety of cells, in contrast with short hairpin RNA libraries which only induce gene knockdown (Sander and Joung, 2014; Zhou et al, 2014).
Figure 2. Precise cleavage of the third nucleotide from protospacer adjacent motif (PAM) by RNA-guided Cas9 dual nucleases induces double strand breaks in the target DNA.
The guide RNA (gRNA) is a chimeric stem-loop structure consisting of CRISPR RNA (crRNA) and trans-activating crRNA (tracRNA) with a GAAA tetraloop.
Similar to ZFNs and TALENS, Cas9 has been extensively tested for its ability to disrupt CCR5 due to its critical importance as a receptor for HIV-1 infection (Cho et al, 2013; Cradick et al, 2013; Niu et al, 2014; Yang et al, 2013). For biallelic targeting of the CCR5 32 transgene in induced pluripotent stem cells, Cas9 induced a better targeting efficiency (33%) than TALENs (14%) (Ye et al, 2014). Thus, extensive application of Cas9 to edit the HIV-1 genome and host cellular genes important in each step of the viral life cycle is now possible.
Genome editing to eradicate the whole HIV-1 genome
As described above, all three well-established genome editing tools (ZFN, TALEN and Cas9) have been tested for their efficiency in disrupting CCR5 because of the very promising cure of HIV-1 in the “Berlin” patient with a CCR5 32 mutation (Burke et al, 2014). However, CCR5 is not uniquely a receptor for HIV-1 infection as it exhibits many other cellular functions as well. Targeting the CCR5 gene will not offer protection against viruses using alternate co-receptors, such as CXCR4. Also, cells already infected with the virus still persist within the host organism after CCR5 or CXCR4 disruption which affects only the entry of HIV-1 into the host cells. Therefore, manipulation of the integrated HIV-1 genome in latently infected cells is critical to achieve a permanent or sterilizing cure for HIV-1/AIDS. Such proof of principal is also applicable to other latently-integrated chronic viral infections such as hepatitis B virus, and herpes simplex virus (Bi et al, 2014; Kennedy et al, 2014; Schiffer et al, 2012; Suenaga et al, 2014; Wang and Quake, 2014; Zhen et al, 2014).
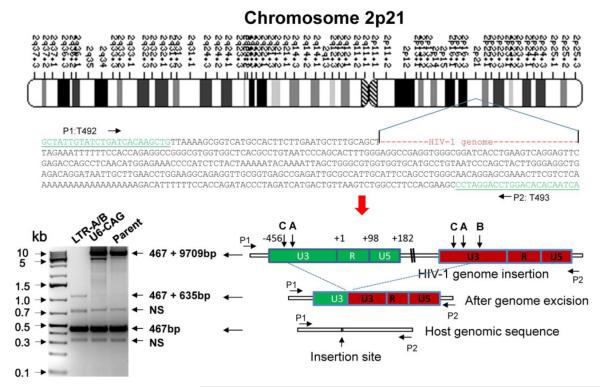
Targeting genes encoding HIV-1 proteins within the proviral DNA has been tested using homing endonuclease (Aubert et al, 2011) and ZFN (Wayengera, 2011), and is anticipated using TALEN and Cas9. However, such a strategy has limitations in that it will not eliminate the entire genome, and the remnant long terminal repeat (LTR) sequences may continue promoting transcription of potential toxic proteins after cleavage. Therefore, targeting the highly conserved LTRs of the HIV-1 proviral DNA is an ideal approach to eradicate the entire viral genome between the 5′ and 3′ LTRs while simultaneously disrupting the LTR sequences. We have tested this proof of concept using the novel Cas9/gRNA technology in several types of latently infected cells (Hu et al, 2014). During the course of our investigation, two labs reported the successful editing of the HIV-1 LTR by ZFN and Cas9 targeting a single conserved transcription factor binding site (Ebina et al, 2013) or the HIV-1 trans-activation response (TAR) element (Qu et al, 2013), which may potentially affect the host genome. In both studies, single cleavage of the targeted 5′ and 3′ LTRs induced excision of the entire integrated HIV-1 proviral genome in latently infected T cells as determined by PCR genotyping, Sanger sequencing, and functional assays (Ebina et al, 2013; Qu et al, 2013). In our studies, we demonstrated higher efficiency of HIV-1 genome eradication by single and multiplex Cas9/LTR-gRNAs not only in latently infected T cells but also in myeloid lineage cells (microglia and monocyte/macrophage) which are major cell types that can serve as HIV-1 reservoirs. Most interestingly, the HIV-1 Cas9/gRNA system is capable of eliminating more than one copy of the viral genome in an infected cell, even when the virus is integrated within different regions of the host genome. For example, we demonstrated the ability of the HIV-1 Cas9/gRNA system to delete both copies of the HIV-1 proviral DNA contained within the monocytic U1 cell line, one located on chromosome X (Hu et al, 2014) and the other on chromosome 2 (Fig. 3), suggesting that this novel genome editing system can alter the DNA sequences of HIV-1 in latently infected cells containing multiple copies of the proviral DNA. Despite the high promise of engineered nuclease-induced genome editing, a key barrier to its clinical and basic research applications is potential off-target effects due to permanent host genome modifications. The specificities of ZFN, TALEN and Cas9 have been extensively investigated using a combination of in vitro and in vivo experimental assays, computational characterization, and next generation sequencing. Although unbiased assays for off-target effects and continuous improvement of genome editing tools are still required, apparent genotoxicity, cytotoxicity, and off-target effects were not observed in most cases, particularly for Cas9-induced genome editing in both cells and animals (Cho et al, 2014; Fu et al, 2013; Gabriel et al, 2011; Hsu et al, 2014; Hsu et al, 2013; Pattanayak et al, 2013; Wang et al, 2013; Wu et al, 2013). In addition, long-term stable expression of Cas9 and/or gRNAs in cells (Heckl et al, 2014; Koike-Yusa et al, 2014; Zhou et al, 2014) and the viability of transgenic mice which constitutively express Cas9 show no signs of toxicity (Platt et al, 2014 and Hu et al., unpublished observations). However, reliable whole genome sequencing for careful assessment of genome stability and potential genotoxicity is warranted. In terms of foreign viral DNA editing, the potential off-target effects of Cas9/gRNAs on the host genome may not be a big concern because of the fairly low homology between the exogenous viral genome and endogenous human genomes including human endogenous retrovirus (van der Kuyl, 2012), although the transcription factor binding sites within the HIV-1 LTR are highly homologous to sites within the host cellular genome and can be avoided by selective gRNA design. In addition, careful bioinformatics screening of HIV-1 gRNA target sites against the host genome could eventually improve the genome editing specificity of Cas9/HIV-1 gRNAs. Nevertheless, whole genome sequencing of homogeneous individual cells with deep sequencing coverage and higher calling confidence will be urgently needed to evaluate the potential risks of off-target effects in clinical applications. Recent whole genome sequencing in human iPS cells revealed high specificity and very rare off-target effects of Cas9/gRNA technology (Smith et al, 2014; Veres et al, 2014).
Figure 3. Eradication of the entire HIV-1 genome spanning between the 5′- and 3′-LTRs by Cas9/LTR-A/B gRNAs in the human U1 monocytic cell line.
The integration site of the HIV-1 genome within chromosome 2 of the U1 cell line is shown. Cells were transfected with plasmids encoding Cas9 and gRNAs targeting two sequences within the 5′ and 3′ HIV-1 LTRs, called LTR-A and LTR-B. Sanger sequencing of a 1.1 kb fragment from long-range PCR using a primer pair (T492/T493) targeting the flanking sequences of the HIV-1 integration site (467 bp) validated the elimination of the entire HIV-1 genome (9,709 bp). NS, non-specific band. Similar results were seen for the second copy of the HIV-1 genome integrated into chromosome X of the U1 cell line. (See Hu et al., 2014 for a detailed description of the LTR-A and -B sequences and the results for chromosome X).
Pre-existing genome editing tool to vaccinate cells against HIV-1 infection
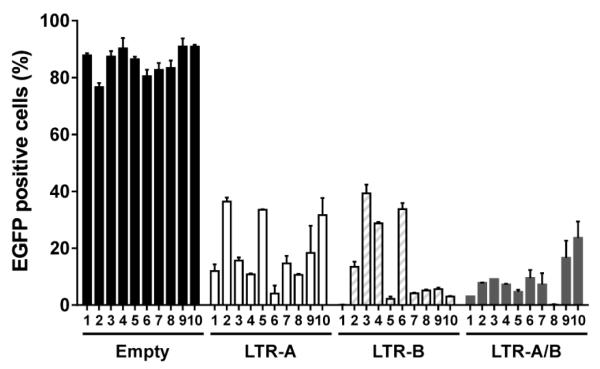
Given that the Cas9 genome editing system originated in bacteria as an immune defense system against invading bacteriophages, we hypothesized that pre-existing genome editing in cells may immunize them against new HIV-1 infection. To test this hypothesis, we established Cas9/gRNA-expressing stable cell lines from TZM-bI (Hu et al, 2014) and CHME5 cells (Fig. 4) by puromycin selection and limiting dilution. The target genome editing efficiency for the subclones depends upon the simultaneous expression of both Cas9 and the gRNA, which varied with different subclones (Fig. 4) and cell types (Hu et al, 2014). Using these effective subclones, we demonstrated that Cas9/gRNA genome editing effectively immunizes cells against new HIV-1 infection. Most interestingly, the preexistence of the Cas9/gRNA system in cells leads to a rapid elimination of the new HIV-1 before it integrates into the host genome (Hu et al, 2014). Similarly, the vaccinating system would be functional in eradicating newly packaged proviruses from the post-integrated HIV-1 genome in cells already infected with the virus. Further investigation of such HIV-1 vaccination in various latent reservoir cells and transgenic animals with stable or inducible expression of Cas9/LTR-gRNAs (Platt et al, 2014) is urgently needed. In clinical practice, the delivery of Cas9 protein plus LTR gRNAs (in the form of nanoparticles, for example) in healthy subjects (including vaginal, rectal, or systemic deliveries) would be a promising approach to prevent HIV-1 infection, because efficient in vitro genome editing using a mixture of Cas9, gRNA, and target genome has been well established (Chen et al, 2014; Jinek et al, 2013; Karvelis et al, 2013; Kim et al, 2014). Like any gene delivery, it is possible that the promoters for stably-expressing Cas9 and/or gRNA could be silenced in some cells, particularly in memory T cells and macrophages. The likelihood of promoter silencing, although very low, may dampen the therapeutic application of Cas9/gRNA approach.
Figure 4. Stable expression of Cas9 plus LTR-A/B efficiently eradicates the HIV-1 genome in latently infected CHME5 microglial cells.
Cells were transfected with plasmids expressing Cas9 and LTR-A, -B, or -A/B. After a 2-week selection with puromycin (1 g/ml), subclones were cultured by limiting dilution and single cell clones were treated with trichostatin A (TSA, 250 nM) for 2 d to induce viral reactivation before EGFP flow cytometry. A dramatic reduction in TSA-induced reactivation of latent pNL4-3- Gag-d2EGFP reporter virus was detected in most of the subclones with single or duplex LTR-gRNAs as compared with the empty pX260 control vector. (See Hu et al., 2014 for further details.)
Paired Nickase or Cas9-FokI strategies for genome editing to reduce off-target effects
CRISPR-dependent targeting of crucial genetic loci is a highly selective method to degrade foreign DNA with great versatility due to its ability to target multiple gene locations. In order to obtain Cas9 specificity for editing of a target gene, Zhang and colleagues have described four golden rules to minimize potential ‘off-target’ effects within the host genome (Hsu et al, 2013). A powerful computational tool (http://crispr.mit.edu/) has been designed to aid in the selection of gRNAs and to predict off-target loci for specificity analyses (Hsu et al, 2013). Also, several other programs are available for designing specific gRNAs with various levels of scores, such as the DNA2.0 CRISPR gRNA design tool (https://www.dna20.com/eCommerce/cas9/input) and the E-CRISP (http://www.e-crisp.org/E-CRISP/designcrispr.html), sgRNAcas9 software package (www.biootools.com) (Xie et al, 2014), Cas-OFFinder (Bae et al, 2014) and CasOT (Xiao et al, 2014). However, the specificity of the gRNAs in host cells remain to be explored and, so far, three human cell types have acquired bona fide off-target mutations induced by Cas9 (Fu et al, 2013; Kuscu et al, 2014). A better understanding of gRNA-mediated off-target effects in eukaryotic cells is essential for this approach to find potential therapeutic applications in the clinic. As mentioned above, the wild-type Cas9 nucleases are able to induce site-specific DSBs through the activity of their RuvC and HNH nuclease domains. Catalytically inactivating the RuvC or HNH nuclease domains via point mutations can convert Cas9 into a DNA “nickase” (Gasiunas et al, 2012; Jinek et al, 2012; Sapranauskas et al, 2011). Nickase activity typically induces repair either seamlessly or through high-fidelity homology directed repair (Cong et al, 2013; Gasiunas et al, 2012), therefore its action causes less off-target effects. Recently, a ‘paired nickase’ strategy has been developed in which adjacent off-set nicks are generated at the target site using two offset gRNAs and Cas9 nickases (Cho et al, 2014; Mali et al, 2013a; Ran et al, 2013). This strategy is similar to the dimeric ZFNs or TALENs, leading to the doubling of the target sequence (from 23 bp to 46 bp) and thus significantly increasing the editing specificity (by up to 1500-fold) (Ran et al, 2013; Shen et al, 2014). The cooperative nicks with tail-to-tail offset (−10 to +30 bp) mimic DSBs and mediate efficient indel formation. The nickase strategy has been shown to be effective for gene knockout in mouse zygotes without reducing on-target cleavage efficiency (Mali et al, 2013a; Ran et al, 2013; Shen et al, 2014).
However, the paired nickase approach requires two single monomeric nickases that may induce potential off-target indel mutations through an undefined mechanism (Mali et al, 2013a; Ran et al, 2013; Tsai et al, 2014). Recently, a dimerization-dependent RNA-guided FokI-fused catalytically-dead Cas9 (dCas9) nuclease (RFN) has been established (Guilinger et al, 2014b; Tsai et al, 2014). Similar to ZFN and TALEN, this novel RFN takes advantage of the non-specific cleavage function of the well-characterized, dimerization-dependent FokI nuclease domains that are brought together by two RNA-guided dCas9 monomers that simultaneously bind opposite target sites separated by approximately 14-17 bp or 25 bp (Guilinger et al, 2014b; Tsai et al, 2014). In contrast to the co-localized, paired Cas9 nickases, the cleavage activity of RFN is largely dimerization-dependent, requiring simultaneous binding of two gRNAs to the target DNA with stringent spacing and orientation, and substantially increasing the specificity of genome editing. RFNs guided by a single gRNA generally induce very little or no off-target effects (Guilinger et al, 2014b; Tsai et al, 2014). However, the very stringent requirements of the off-set size (only 14-17 bp or 25 bp) between two gRNA target sites dramatically reduces the number of candidate gRNAs, limiting the opportunity to identify effective paired gRNAs. Therefore, new approaches to improve the specificity remain to be developed.
Conclusions and future directions
In conclusion, DSB-mediated nuclease-initiated DNA editing tools that can be used as novel genome surgical knives have extensive applications in biomedical science. The facile and versatile Cas9/gRNA technology platform (Ran et al, 2013) is growing rapidly, and shows promise for successful development into novel therapeutic platforms for treating human genetic diseases, infectious diseases, and cancer (Cho et al, 2013; Manjunath et al, 2013; Zhang et al, 2014). By targeting both 5′ and 3′ HIV-1 LTRs with single or multiplex gRNAs, this genome editing tool is capable of excising the entire genome spanning between the LTRs from the host genome, leading to disruption of latent provirus and protecting cells against new HIV-1 infection (Fig. 4). There are, however, several important issues that need to be considered. First, whether or not the Cas9/gRNA system will have any effect on episomal DNA. In this respect, one may consider several recent studies illustrating the capability of this novel genome editing system in cleaving several other viral genes including adenovirus, herpes simplex virus, Epstein-Barr virus and human papillomavirus (Bi et al, 2014; Kennedy et al, 2014; Suenaga et al, 2014; Wang and Quake, 2014; Zhen et al, 2014), leading to the belief that this strategy can also be effective in targeting episomal HIV-1 genomes. This is supported by our observation that expression of Cas9/LTR-gRNA in cells significantly prevents new HIV-1 infection before integration (Hu et al, 2014). The second issue concerns the ability of the Cas9/gRNA to effectively target every single cell which harbors a complete HIV-1 genome. While at present time, we can offer no conclusive evidence in the absence of in vivo data, one may hope that the elimination of a large fraction of activatable HIV-1 genomes from infected cells by Cas9/gRNA may provide a strong head start for the host immune system to overcome any remaining cells harboring HIV-1. Thus, a genetic approach for eliminating HIV-1 in some population of latently infected cells may rejuvenate immune cells to combat viral infection as a whole. Furthermore, rapid developing technologies such as nanoparticle and lentivirus delivery systems hold promise for highly efficient HIV-1 eradication in latently infected cells. Another important issue that needs attention relates to the robust expression of therapeutic genes, i.e. Cas9 and gRNAs in cells which are not actively replicating virus. In this respect, one may consider to employ an inducible promoter to actively control the expression levels of these genes in the target cells. Two other major barriers need to be overcome to bring this powerful genome editing tool into the clinic, including the potential off-target effects as well as the design of a safe and efficient delivery system for cell-specific distribution and genome editing within recipient organisms. Several promising strategies are under development to surmount these hurdles. In terms of HIV-1 genetic variation and quasispecies in individual patients, one may develop personalized treatment modalities based on the data from deep sequencing of the patient-derived viral genomes prior to engineering therapeutic gRNA molecules, which can be generated quickly and easily.
ACKNOWLEDGMENTS
The authors thank past and present members of the Department of Neuroscience and the Center for NeuroVirology. We also thank C. Papaleo for editorial assistance. This work was supported by R01NS087971 (W.H., K.K.) and P30MH092177 (K.K.).
Footnotes
CONFLICT OF INTERESTS The authors declare that they have no conflict of interests.
REFERENCES
- Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, Schneider T. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood. 2011;117:2791–9. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang KH, Dahl NP, Kearney MF, Anderson EM, Coffin JM, Strain MC, Richman DD, Robertson KR, Kashuba AD, Bosch RJ, Hazuda DJ, Kuruc JD, Eron JJ, Margolis DM. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis. 2014;210:728–35. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould S, Delenda C, Grizot S, Desseaux C, Paques F, Silva GH, Smith J. The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng Des Sel. 2011;24:27–31. doi: 10.1093/protein/gzq083. [DOI] [PubMed] [Google Scholar]
- Aubert M, Ryu BY, Banks L, Rawlings DJ, Scharenberg AM, Jerome KR. Successful targeting and disruption of an integrated reporter lentivirus using the engineered homing endonuclease Y2 I-AniI. PLoS One. 2011;6:e16825. doi: 10.1371/journal.pone.0016825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–5. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Battistini A, Sgarbanti M. HIV-1 latency: an update of molecular mechanisms and therapeutic strategies. Viruses. 2014;6:1715–58. doi: 10.3390/v6041715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Sun L, Gao D, Ding C, Li Z, Li Y, Cun W, Li Q. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog. 2014;10:e1004090. doi: 10.1371/journal.ppat.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke BP, Boyd MP, Impey H, Breton LR, Bartlett JS, Symonds GP, Hutter G. CCR5 as a Natural and Modulated Target for Inhibition of HIV. Viruses. 2014;6:54–68. doi: 10.3390/v6010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Choi J, Bailey S. Cut Site Selection by the Two Nuclease Domains of the Cas9 RNA-guided Endonuclease. J Biol Chem. 2014;289:13284–94. doi: 10.1074/jbc.M113.539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–41. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–92. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didigu CA, Wilen CB, Wang J, Duong J, Secreto AJ, Danet-Desnoyers GA, Riley JL, Gregory PD, June CH, Holmes MC, Doms RW. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123:61–9. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel HJ, Morrison JH, Saenz DT, Fuchs JR, Kvaratskhelia M, Ekker SC, Poeschla EM. TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol. 2014a;88:9704–17. doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel HJ, Morrison JH, Saenz DT, Fuchs JR, Kvaratskhelia M, Ekker SC, Poeschla EM. TALEN Knockout of the PSIP1 Gene in Human cells: Analyses of HIV-1 Replication and Allosteric Integrase Inhibitor Mechanism. J Virol. 2014b doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, Holmes MC, Gregory PD, Glimm H, Schmidt M, Naldini L, von Kalle C. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–23. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Pattanayak V, Reyon D, Tsai SQ, Sander JD, Joung JK, Liu DR. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat Methods. 2014a;11:429–35. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014b;32:577–82. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber I, Hofmann-Sieber H, Chemnitz J, Dubrau D, Chusainow J, Stucka R, Hartjen P, Schambach A, Ziegler P, Hackmann K, Schrock E, Schumacher U, Lindner C, Grundhoff A, Baum C, Manz MG, Buchholz F, Hauber J. Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice. PLoS Pathog. 2013;9:e1003587. doi: 10.1371/journal.ppat.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014 doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, Recchia A, Cathomen T, Goncalves MA. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–47. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, Mo X, Khalili K. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A. 2014;111:11461–6. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izmiryan A, Basmaciogullari S, Henry A, Paques F, Danos O. Efficient gene targeting mediated by a lentiviral vector-associated meganuclease. Nucleic Acids Res. 2011;39:7610–9. doi: 10.1093/nar/gkr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Gasiunas G, Siksnys V. Programmable DNA cleavage in vitro by Cas9. Biochem Soc Trans. 2013;41:1401–6. doi: 10.1042/BST20130164. [DOI] [PubMed] [Google Scholar]
- Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–72. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kim D, Kim S, Kim JS. Genotyping with CRISPR-Cas-derived RNA-guided endonucleases. Nat Commun. 2014;5:3157. doi: 10.1038/ncomms4157. [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–60. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Kumar A, Abbas W, Herbein G. HIV-1 latency in monocytes/macrophages. Viruses. 2014;6:1837–60. doi: 10.3390/v6041837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014 doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–9. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao LF, Tran CA, Torres-Coronado M, Gardner A, Gonzalez N, Kim K, Liu PQ, Hofer U, Lopez E, Gregory PD, Liu Q, Holmes MC, Cannon PM, Zaia JA, DiGiusto DL. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21:1259–69. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gaj T, Patterson JT, Sirk SJ, Barbas CF., 3rd Cell-penetrating peptide-mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS One. 2014;9:e85755. doi: 10.1371/journal.pone.0085755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo Riso P, Damo M, Pello OM, Holmes MC, Gregory PD, Gritti A, Broccoli V, Bonini C, Naldini L. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8:861–9. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Maier DA, Brennan AL, Jiang S, Binder-Scholl GK, Lee G, Plesa G, Zheng Z, Cotte J, Carpenito C, Wood T, Spratt SK, Ando D, Gregory P, Holmes MC, Perez EE, Riley JL, Carroll RG, June CH, Levine BL. Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther. 2013;24:245–58. doi: 10.1089/hum.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013b;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013c;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S. Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun. 2005;335:447–57. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- Manjunath N, Yi G, Dang Y, Shankar P. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5:2748–66. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson McManamy ME, Hakre S, Verdin EM, Margolis DM. Therapy for latent HIV-1 infection: the role of histone deacetylase inhibitors. Antivir Chem Chemother. 2014;23:145–9. doi: 10.3851/IMP2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariyanna L, Priyadarshini P, Hofmann-Sieber H, Krepstakies M, Walz N, Grundhoff A, Buchholz F, Hildt E, Hauber J. Excision of HIV-1 proviral DNA by recombinant cell permeable tre-recombinase. PLoS One. 2012;7:e31576. doi: 10.1371/journal.pone.0031576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matalon S, Rasmussen TA, Dinarello CA. Histone deacetylase inhibitors for purging HIV-1 from the latent reservoir. Mol Med. 2011;17:466–72. doi: 10.2119/molmed.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Alzubi J, Fine EJ, Morbitzer R, Cradick TJ, Lahaye T, Bao G, Cathomen T. TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 2014;42:6762–73. doi: 10.1093/nar/gku305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–93. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerys-Junior A, Costa LC, Braga-Dias LP, Oliveira M, Rossi AD, da Cunha RD, Goncalves GS, Tanuri A. Use of the heteroduplex mobility assay and cell sorting to select genome sequences of the CCR5 gene in HEK 293T cells edited by transcription activator-like effector nucleases. Genet Mol Biol. 2014;37:120–6. doi: 10.1590/s1415-47572014000100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Zhang B, Chen H. Applications of TALENs and CRISPR/Cas9 in Human Cells and Their Potentials for Gene Therapy. Mol Biotechnol. 2014 doi: 10.1007/s12033-014-9771-z. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–43. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–16. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A, Silva GH. Precision genome surgery. Nat Biotechnol. 2007;25:743–4. doi: 10.1038/nbt0707-743. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell. 2014;159:440–55. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Wang P, Ding D, Li L, Wang H, Ma L, Zhou X, Liu S, Lin S, Wang X, Zhang G, Liu S, Liu L, Wang J, Zhang F, Lu D, Zhu H. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013;41:7771–82. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Wang P, Ding D, Wang X, Zhang G, Zhou X, Liu L, Zhu X, Zeng H, Zhu H. Zinc finger nuclease: a new approach for excising HIV-1 proviral DNA from infected human T cells. Mol Biol Rep. 2014;41:5819–27. doi: 10.1007/s11033-014-3456-3. [DOI] [PubMed] [Google Scholar]
- Ramalingam S, Annaluru N, Kandavelou K, Chandrasegaran S. TALEN-Mediated Generation and Genetic Correction of Disease-specific Human Induced Pluripotent Stem Cells. Curr Gene Ther. 2014 doi: 10.2174/1566523214666140918101725. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TA, Schmeltz Sogaard O, Brinkmann C, Wightman F, Lewin SR, Melchjorsen J, Dinarello C, Ostergaard L, Tolstrup M. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum Vaccin Immunother. 2013;9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–82. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer JT, Aubert M, Weber ND, Mintzer E, Stone D, Jerome KR. Targeted DNA mutagenesis for the cure of chronic viral infections. J Virol. 2012;86:8920–36. doi: 10.1128/JVI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Siliciano RF. From reactivation of latent HIV-1 to elimination of the latent reservoir: the presence of multiple barriers to viral eradication. Bioessays. 2013;35:544–52. doi: 10.1002/bies.201200170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Siliciano RF. Recent developments in the search for a cure for HIV-1 infection: targeting the latent reservoir for HIV-1. J Allergy Clin Immunol. 2014;134:12–9. doi: 10.1016/j.jaci.2014.05.026. [DOI] [PubMed] [Google Scholar]
- Siliciano RF, Greene WC. HIV latency. Cold Spring Harb Perspect Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–3. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Curr Opin HIV AIDS. 2013;8:217–23. doi: 10.1097/COH.0b013e32835f736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga T, Kohyama M, Hirayasu K, Arase H. Engineering large viral DNA genomes using the CRISPR-Cas9 system. Microbiol Immunol. 2014;58:513–22. doi: 10.1111/1348-0421.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Zhang Q, Li X, Fan N, Yang Y, Quan L, Lai L. Targeted modification of CCR5 gene in rabbits by TALEN] Yi Chuan. 2014;36:360–8. [PubMed] [Google Scholar]
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–10. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–76. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kuyl AC. HIV infection and HERV expression: a review. Retrovirology. 2012;9:6. doi: 10.1186/1742-4690-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C, Bouchat S, Marcello A. HIV-1 transcription and latency: an update. Retrovirology. 2013;10:67. doi: 10.1186/1742-4690-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit RA, McMahon MA, Sawyer SL, Porteus MH. Generation of an HIV resistant T-cell line by targeted “stacking” of restriction factors. Mol Ther. 2013;21:786–95. doi: 10.1038/mt.2012.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci U S A. 2014;111:13157–62. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayengera M. Proviral HIV-genome-wide and pol-gene specific zinc finger nucleases: usability for targeted HIV gene therapy. Theor Biol Med Model. 2011;8:26. doi: 10.1186/1742-4682-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen CB, Wang J, Tilton JC, Miller JC, Kim KA, Rebar EJ, Sherrill-Mix SA, Patro SC, Secreto AJ, Jordan AP, Lee G, Kahn J, Aye PP, Bunnell BA, Lackner AA, Hoxie JA, Danet-Desnoyers GA, Bushman FD, Riley JL, Gregory PD, June CH, Holmes MC, Doms RW. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 2011;7:e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. Correction of a Genetic Disease in Mouse via Use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–62. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, Zhang B. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]
- Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014;9:e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, Huang PY, Daley G, Church G. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–61. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Nashun B, Zhou T, Qin L, Qin L, Zhao S, Xu J, Esteban MA, Chen X. Generation of CD34+ cells from CCR5-disrupted human embryonic and induced pluripotent stem cells. Hum Gene Ther. 2012;23:238–42. doi: 10.1089/hum.2011.126. [DOI] [PubMed] [Google Scholar]
- Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, Chang JC, Bao G, Muench MO, Yu J, Levy JA, Kan YW. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc Natl Acad Sci U S A. 2014;111:9591–6. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G, Choi JG, Bharaj P, Abraham S, Dang Y, Kafri T, Alozie O, Manjunath MN, Shankar P. CCR5 Gene Editing of Resting CD4(+) T Cells by Transient ZFN Expression From HIV Envelope Pseudotyped Nonintegrating Lentivirus Confers HIV-1 Resistance in Humanized Mice. Mol Ther Nucleic Acids. 2014;3:e198. doi: 10.1038/mtna.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Wang J, Crain K, Fearns C, Kim KA, Hua KL, Gregory PD, Holmes MC, Torbett BE. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol Ther. 2012;20:849–59. doi: 10.1038/mt.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun. 2014;450:1422–6. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–91. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]