Abstract
Effective systemic treatment of cancer relies on the delivery of agents with optimal therapeutic potential. The molecular age of medicine has provided genomic tools that can identify a large number of potential therapeutic targets in individual patients, heralding the promise of personalized treatment. However, determining which potential targets actually drive tumor growth and should be prioritized for therapy is challenging. Indeed, reliable molecular matches of target and therapeutic agent have been stringently validated in the clinic for only a small number of targets. Patient-derived xenografts (PDX) are tumor models developed in immunocompromised mice using tumor procured directly from the patient. As patient surrogates, PDX models represent a powerful tool for addressing individualized therapy. Challenges include humanizing the immune system of PDX models and ensuring high quality molecular annotation, in order to maximise insights for the clinic. Importantly, PDX can be sampled repeatedly and in parallel, to reveal clonal evolution, which may predict mechanisms of drug resistance and inform therapeutic strategy design.
Keywords: personalized medicine, patient-derived xenografts, genomics, targeted therapy, therapeutic targets
1. Introduction: Identification of therapeutic targets in the clinical setting
Through our improved understanding of cancer biology, identification of molecular drivers of cancer growth, and the development of targeted therapeutics, we have an increased ability to deliver treatment matched to a patient’s cancer. The reality, however, is that for the majority of patients, this approach is still beyond their reach. The process of personalized medicine focuses on treating a patient as an individual, rather than as a representative member of a group of patients with similar histological designation, as has been the historical mechanism for assigning treatment1. However, this approach entails significant challenges in terms of logistics and identification of the best model systems in which to validate the utility of personalized therapies. The use of patient-derived xenografts (PDX), or human tissue transplanted into immune-deficient mice without any intervening in vitro culture step, provides powerful models in which to determine the efficacy of therapies targeted to specific molecular aberrations2.
In the past five decades of cancer therapeutic discoveries, the way in which a cancer case has been described and matched to treatment has focused on the organ in which the cancer was thought to have arisen1, the histopathologic appearance of the cancer tissue and draining lymph nodes and the staining of between one and ten protein markers present on or in the cancer cell. Indeed, apart from a number of molecular tests involving the analysis of one or two genes, such as the routine use of in situ hybridization analysis to determine amplification of the HER2 gene in breast cancer or DNA sequencing to determine mutations in KRAS in lung cancer or colorectal cancer4; or in melanoma or colorectal cancer, histopathology and immunohistochemstry underpins the majority of treatment decisions for many for many patients today.
We are currently in the middle of the most extraordinary technological revolution6, which has led us from the mammoth task of proposing to sequence the first human genome, predicted to take 15 years and cost three billion USD, to the current availability of whole genome sequencing (WGS), of an entire genome (or a cancer genome) in only a few days, for the cost of around one thousand USD. Indeed, genomic technologies, such as high-throughput sequencing of DNA, RNA (RNASeq), microRNA and the epigenome, now provide the first systematic approaches to discover the genes and cellular pathways underlying disease6. Although these technologies provide a tremendous opportunity, being able to read individual base pairs and compare them with a reference sequence does not tell us what we urgently need to know: who will get cancer, what type and when and how should that cancer best be treated? There is hope, however, that companion technologies that allow us to determine gene expression and epigenetic marks, or silencing or accessibility of the genome, will enhance our ability to interpret gene sequence variations.
Thanks to exponential improvements in the speed and depth of DNA sequencing, next-generation sequencing (NGS) can analyse entire human genomes in days, at a reassuring read depth7. Sequencing a cancer genome is more complex than a germline genome, due to the variety of complex aberrations found in cancer, including multiple gene copies, structural changes, epigenomic changes and intra-tumoral genetic heterogeneity7. This complexity necessitates greater read depth, or coverage (how many times a specific region has been sequenced by unique reads with a different start/end site/read length), with a median coverage of 50× (excluding duplicate reads), rather than the 30× generally accepted for standard germline genomes. Criteria for ensuring quality NGS data and interpretations are being addressed by the Next-generation Sequencing Standardization of Clinical Testing (Nex-StoCT) workgroup8 and the College of American Pathologists9.
Many diagnostic cancer samples are preserved in formalin fixed paraffin embedded (FFPE) tissue blocks, containing fragmented or cross-linked DNA, with few whole genomes reported from FFPE samples to date. It has been suggested that if cancer tissue is not preserved appropriately (for example, snap frozen in addition to formalin fixed) that this could constitute willful destruction of evidence, necessitating that clear practice guidelines are generated to describe acceptable standard of care around tissue preservation for treatment-focused testing1. In response to this practical problem, new approaches are being developed to ensure optimal use of FFPE sections, such that sufficient information may be obtainable10.
The availability of NGS has resulted in datasets ripe for interrogation and new insights. Companies are racing to provide panel tests, which interrogate hundreds of cancer genes, each gene included because it has been proven or hypothesised to be a cancer-causing or cancer-driving gene. This includes panels such as the Foundation Medicine T5a test11. If a potentially actionable aberration is detected by sequencing, for example, a mutation which is known or predicted to cause a non-functional (tumor suppressor) or activated (onco)-gene, then a recommendation may be made regarding the utility of a targeted therapy which may impact on that gene, or its associated pathway. The level of evidence underlying such a recommendation is variable12. Access to the right drug may be problematic and the chance or durability of response in that tumor type usually unknown. How should we validate potential actionable aberrations to aid in clinical trial design and choice of treatments for patients?
2. What constitutes an actionable aberration?
2.1 Human tumor cohort association studies
The molecular analysis of human tumors has the potential to unlock a series of molecular alerts or flags that may be predictive of drug response or resistance. In this setting, an actionable aberration is a molecular flag, which is underpinned by variable levels of evidence to suggest that a therapy targeting this aberration could be effective12. To date, molecular interrogation of cancer specimens has varied from analysis of expression of single genes or proteins by in situ hydridization or immunohistochemistry, or DNA sequencing of single genes (eg KRAS in lung cancer or colorectal cancer4 or BRAF in melanoma or colorectal cancer through to DNA sequencing of up to several hundred genes (such as the Foundation One T5a test11). Analysis of the whole exome or whole genome is available but the interpretation is problematic and these are not approved to guide treatment, outside research studies.
In the research setting, massive parallel sequencing of multiple tumor genomes has produced a plethora of data, with some emergent themes. The Cancer Genome Atlas (TCGA) is on its way to providing comprehensive characterization of cancer genomes from 24 of the most common as well as nine of the most rare tumor types. Much of this work has already been published, with insights stretching from the analysis of cell of origin across cancer types13 to detailed subsetting within cancer types eg for ovarian cancer14, glioblastoma15 and most recently, gastric cancer16. These data are valuable in helping us to understand the diverse biology of different tumors, for example, that genomic stability is a major determinant of subtype13,16. Similarly, the identification of a constellation of aberrations within a particular gene or pathway can indicate a therapeutic approach for a subset of cancers. For example, PIK3CA mutations were found in one of four subsets of gastric cancer16. This subset was defined by enrichment of high Epstein-Barr virus (EBV) burden, extensive DNA promoter methylation and 80% mutation rate in PIK3CA, suggesting that PI(3)-kinase inhibition should be examined in this patient group. Of interest, the PIK3CA mutations were more dispersed in EBV-positive gastric cancers, whereas in EBV-negative gastric cancers, PIK3CA mutations were localized in the kinase domain (exon 20) and present at a lower rate (3–42%) in the other three subtypes. The therapeutic implications of these findings remain to be determined.
The TCGA studies to date suggest that multiple novel cancer-associated genes remain to be discovered, particularly as new tumor types are analysed in depth17. Most cancer genes identified to date, appear to be altered in only 2–20% of cancers and will need to better understood, in order to be efficiently targeted in the clinic. Importantly, these studies of cancer genome associations are largely correlative and do not provide proof that a specific gene or pathway is indeed actionable. Pre-clinical models allowing functional interrogation can provide additional evidence that a target may be worth addressing in the clinic, as will be discussed in Sections 4 and 5.
3. Does the context of a molecular aberration matter?
An increasing number of distinct tumor types are being recognized in the clinic, as a result of newly defined molecular subsets. In order to design therapy targeted to specific tumor subsets, as well as to rare cancer types, many of which have limited treatment options18, it is important to know whether a therapeutic outcome is informative from one tumor context to another. This requires consideration of histologic subtype, the gene or drug target involved, the type of molecular aberration (eg amplification versus activating mutation) and the molecular context, as will be discussed in Sections 3.1–3.3. Understanding this complexity, which cannot be encompassed in the clinical trial setting, lends weight to the need for pre-clinical models for proof of principle analysis, in order to underpin clinical trial directions.
3.1 Histologic tumor type
During the early stages of the National Cancer Institute’s (NCI) Developmental Therapeutics Program (DTP) in the 1950’s, initially only three transplanted rodent models were used (sarcoma, leukemia and carcinoma), hoping that they would be informative for a wide range of tumor types19. As that proved increasingly unlikely, other approaches were adopted including human xenografts and the NCI panel of 61 human tumor cell lines. Studies were also performed comparing whether cell line xenograft responses from one tumor type would be predictive for clinical outcomes of other tumor types in the clinic. These approaches yielded some clues but were largely disappointing20.
3.1.1 TP53 mutation, ubiquitous in some and variable in other tumor types
Mutations in the TP53 tumor suppressor gene have been correlated with poor prognosis and poor treatment response in many cancers. The incidence of mutations in TP53 varies by cancer type, ranging from near 100% in high-grade serous ovarian cancer (HGSC)21, with much lower rates of mutation seen in some hematological malignancies, including follicular lymphoma or diffuse large B cell lymphoma. In HGSC, mutation of TP53 is an early event in tumorogenesis, which together with its ubiquitous nature in HGSC, may mean it is less likely to represent an actionable target. In contrast, this mutation in lymphoma is associated with relative drug resistance and may be used in a predictive fashion, to alter therapeutic decisions22.
3.1.2 BRAF mutation in melanoma compared with colorectal cancer
The context in which a specific molecular vulnerability occurs (such as the tumor of origin) may influence its biologic behavior. Activating mutations of BRAF (V600E) occur in both melanoma and colorectal cancer. In BRAF mutant melanoma, the BRAF inhibitor, vemurafenib has shown impressive clinical benefit23. However, relative resistance is seen BRAF (V600E) mutant colorectal cancers treated with BRAF inhibitors. In contrast with melanoma, BRAF (V600E) inhibition in colorectal cancer is mediated by alternative signaling pathways including EGFR24,25 or PI3K/AKT pathway activation5, resulting in malignant growth. Potent synergy is seen with relevant inhibitor combinations in in vitro and in vivo in colon cancer models,24,25 suggesting the synthetic lethality of combination therapies as a viable clinical strategy. Thus, the environment in which a mutation occurs, which may be specific to the original tumor type, may heavily influence the effect of a given molecular mutation26.
3.1.3 ERBB2 amplification or over-expression in breast cancer compared with other cancer types
Molecular therapies are transforming the practice of oncology. In breast cancer, as a result of large scale, well organized clinical trials, treatment with trastuzumab, pertuzumab, lapatinib, TDM-1 and other therapies targeting ERBB2 (also known as human epidermal growth factor receptor 2 (HER2)), have established roles in adjuvant and metastatic disease, with clear survival advantages in ERBB2-amplified disease 3,27,28,29,30. In gastric cancer, the ToGA trial has conclusively shown that ERBB2 amplification is a predictive marker for response to trastuzumab, with significant improvements in median overall and progression free survival with the addition of trastuzumab to chemotherapy31. ERBB2 gene amplification or protein overexpression has also been reported in 11% of esophageal squamous cell cancer32 and in 18 – 35% of mucinous epithelial ovarian cancer, providing a potential actionable target for the management of these cancers33.
3.2 The type of aberration in a validated target
3.2.1 ERBB2 activating mutation, rather than amplification
Since 2004, activating mutations have been identified in ERBB2, across multiple tumor types, including non-small cell lung cancer and mucinous ovarian cancer34. As these aberrations do not involve over-expression of the receptor, small molecule tyrosine kinase inhibitors acting on the intra-cellular domain would be most likely to be of use, rather than antibodies directed against the extra-cellular domain34. Thus, the choice of therapeutic strategy would be dictated by the aberration and although the mutation rate in each particular tumor type is low, clinical trials are needed to address whether the pipeline of ERBB2-directed TKIs could be of clinical utility for these patients.
3.2.2 ALK amplification, rather than ALK translocation
Molecular inhibitors of specific aberrancies may have broader efficacy in anomalies of the putative signaling pathway. The identification of Anaplastic lymphoma kinase (ALK) translocations as oncogenic drivers in non-small cell lung cancer, coupled with the rapid development of crizotinib, has dramatically altered the therapeutic landscape in this disease. Preclinical responses to crizotinib have been demonstrated in inflammatory breast cancer showing ALK amplifications (increased ALK copy number, as distinct from gene translocations) stimulating clinical trials evaluating ALK targeted therapeutics in patients with broader ALK abnormalities35. ALK amplifications are common events in esophageal cancers (where ALK translocations are not present), providing potential targets for selective inhibition as an attractive clinical strategy.
3.3 Molecular context
3.3.1 PARP inhibitor response in DNA-repair defective HGSC
PARP inhibitor efficacy is known to be dependent on the presence of a DNA repair defect in the homologous recombination pathway, such as seen with HGSC mutated for either BRCA1 or BRCA236,37,38. It was initially proposed that PARP inhibitor efficacy was mediated by effects on the Base Excision Repair pathway, however, Patel and colleagues showed impaired efficacy of PARP inhibition in HGSC cell lines in which non-homologous end-joining (NHEJ) had been disabled39. This suggested that NHEJ hyperactivation may be responsible for PARP inhibitor induced lethality in HR-incompetent cells. Thus, in the expected context, that of BRCA1/2 mutant HR defective HGSC, PARP inhibitor action may be prevented by the lack of an essential collaborating pathway, that of intact, indeed, hyperactivated NHEJ.
In addition to better understanding the complexity of genetic alterations in multiple tumor types, we need to predict the implications of these alterations, in order to improve design of combination therapy studies. Indeed, we need a coordinated effort to map genetic dependences, understand feedback loops and recognize crosstalk circuits which might indicate successful combination therapeutic approaches, as simply trying two drugs which might be predicted to work together is proving to be inefficient26. Equally, the concept of embracing complexity and adopting a combination of systems biology methods and integrated analyses may be necessary40. This complexity requires rigorous analysis in improved pre-clinical models.
4. Tractable pre-clinical models for prioritizing targets
4.1 Tractable models of human cancer
In order to provide accurate prediction as to whether a particular molecular aberration is indeed actionable, models systems are required which accurately represent human disease and are reasonably stable from passage to passage such that repeat analysis over time yields the same result41. These models need to generate material suitable for a full range of molecular analyses, including selection experiments to determine the impact of genome-wide screens or drug library screens. As cancer cells evolve to evade whatever selective pressure they are placed under, for example, drug pressure, the ability to model response after multiple lines of therapy should be an essential component of a translational model, recapitulating the patient journey and allowing analysis of clonal evolution.
4.2 Long-established cancer cell lines
The pre-clinical use of carefully curated, genomically-annotated cancer cell lines can provide biologic insights which help to validate putative targets, including those identified by molecular analysis42. Cell culture systems however, have inherent insufficiencies undermining their validity as comprehensive models and have failed to reliably predict clinical responses, as demonstrated by the National Cancer Institute drug development program over the last several decades43. Selection during tissue culture over time, may artificially eradicate features of the host tumor, pertinent for faithful replication of drug response and may activate spurious cell signaling pathways44,45. Domcke and colleagues have recently showed that many HGSC cell lines fail to recapitulate the genomic features of this disease46. None of the five popular cell lines accounting for 90% of the relevant literature were considered good quality lines, with the two cell lines, SK-OV-3 and A2780, which account for 60% of the literature, considered poorly suited as models of HGSC.
Cancer cell lines show low fidelity compared with complex genetic and epigenetic abnormalities existing in human tumors, and lack the stromal and immune influence of the human tumor microenvironment. As demonstrated by drug testing, xenografts generated from long-established human cancer cell lines may show inconsistent therapeutic responses47 and poor correlation with clinical outcomes, in comparison with human primary orthotopic tumor xenografts48,20. Whilst the judicious use of molecularly curated cancer cell lines may inform mechanistic evaluation, cell culture systems lack the capacity to comprehensively model malignant processes, underscoring the urgent requirement for improved model systems.
4.3 Patient-derived xenografts (PDX)
An increasingly accepted preclinical model producing translational advances, is that of the patient-derived xenograft (PDX)2. The transplantation of tumor obtained at the time of surgery or biopsy, unmanipulated, into recipient mice, generates in vivo models which are tractable, renewable and have massive potential for parallel, sequential and long-term therapy experiments (Figure 1). The ability to drive drug resistance, as happens in a patient, allows the comparison of tumor and circulating tumor DNA from plasma, in a way that is not possible in the clinic49.
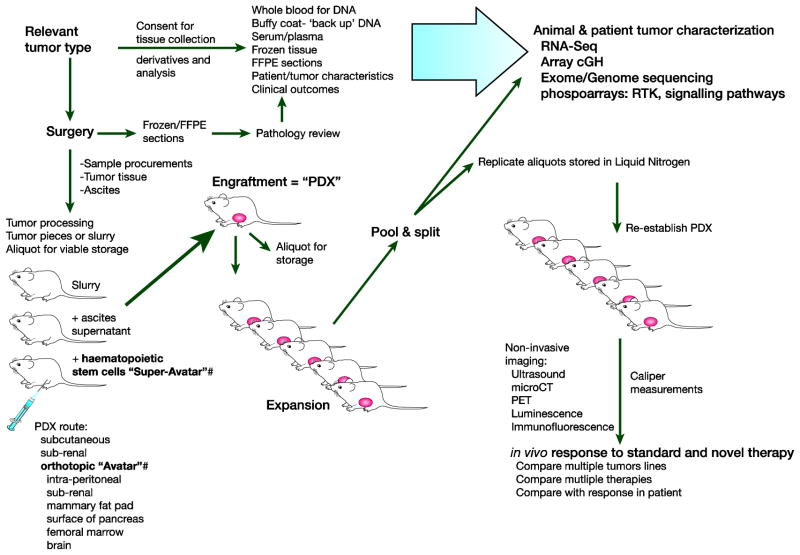
Figure 1. Flexibility of patient-derived xenograft models.
By transplanting fresh, unmanipulated tumor at the time of biopsy or surgery into mice, stable patient-derived xenografts (PDX) can be generated with considerable similarity to the primary human cancer. Detailed functional and molecular analysis can be performed of the PDX can be compared with the primary human sample and with patient outcome. In vivo response to conventional and novel therapy can be performed in parallel for multiple drugs or sequentially, in order to drive drug resistance, as occurs in the clinic. A renewable resource can be generated by using viable freezings of minced tissue slurry. Orthotopic chemonaiive PDX can be referred to as “Avatars”. Mice in which humanized immune reconstitution has also been performed can be termed “Super-Avatars”. Receptor Tyrosine Kinase = RTK. Photoemission Tomography = PET.
4.3.1 Terminology: PDX, Avatars and Super-Avatars
The development of genetically modified immunodeficient mice has allowed for the generation of tumor xenograft models in which patient tumor obtained at the time of surgery or biopsy can be transplanted directly into mice without any in vitro manipulation. This type of model is known as a PDX the methodology of which has recently been extensively reviewed2. Tumors that successfully engraft can then be serially transplanted into subsequent generations of mice, generating a renewable resource, which can be annotated in detail and used to study novel therapies. Tumors engraft typically 2–4 months after transplantation2; however many factors can influence successful engraftment including method of processing and mode of transplantation, the recipient murine strain used and the tumor type being transplanted50. Within the field of cancer research, PDX models have taken various forms and as many of these adaptations have not previously been described and defined, the authors here attempt for the first time to define subsets of PDX models. “Avatars” represent a subset of PDX models that have the distinction of being transplanted in an orthotopic location and have not been previously subjected to cancer therapeutics50 (Figure 1). In contrast, we define “super-avatars” as models generated by co-transplantation of hematopoietic stem cells (HPSC) and patient tumor, via an orthotopic route, which offer the potential of studying novel immunotherapies, described in more detail below (Figure 1).
4.3.2 The generation of PDX models
Orthotopic models are some of the most clinically translatable models in oncology research, as they recapitulate aspects of the clinical disease that are not shared by cell lines. It is important to note that different types of tumors may have different requirements for optimal transplantation and some may be easier (HGSC) and some more difficult (ER-positive breast cancer) to transplant2,49,51,52. Many PDX models are established by transplanting tumor from the patient into immunodeficient mice via the subcutaneous route or other locations for improved engraftment and easy of injection. Using this approach many aspects of the tumor microenvironment may be lost with successive rounds of transplantation, including stromal cells (although the human stroma from the initial graft tends to be replaced with murine stroma which often takes on a very similar morphologic appearance to the initial human graft) and various aspects of the innate and adaptive immune system53. Some subcutaneous xenografts fail to progress or to metastasize with the same pattern as human disease and therefore do not reflect all patterns of tumor progression seen in patients54. Certain tumor types when injected orthotopically recapitulate the tumor microenvironment and better track patient disease progress. These tumor types include ovarian51,55,56, breast57–61, pancreatic62–67, renal68–72, non-small cell lung73–76 and melanoma77–79. The use of orthotopic tumor transplantation has been shown by many groups to accurately recapitulate patient tumor in an in vivo setting, DeRose and colleagues demonstrated that breast cancer tumor grafts developed metastases with frequencies from 38% to 100% in sites corresponding to patient metastatic sites, recapitulating the original patient metastatic cascade51,57,80. For all PDX, it is important to note whether the starting material was derived from a chemotherapy-naive tumor (upon first diagnosis, prior to any treatment) or after specific lines of treatment. Paired PDX from the same patient, before and after systemic therapy, have been reported using tumor from breast cancer patients52. The generation of similar models in other cancer types could be very valuable for exploring clonal evolution in response to therapy, including with analysis of circulating tumor DNA (ctDNA) (Figure 2).
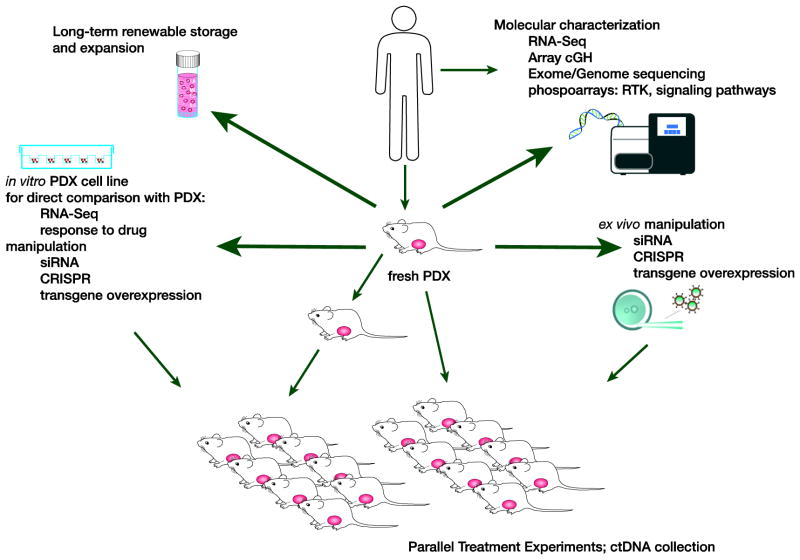
Figure 2. Derivatives generated from patient-derived xenografts.
Once patient-derived xenografts (PDX) have been generated and annotated, additional derivatives can be generated, which further increase the utility of the original model, adding to its functionality. These include the generation of a cell line, which once compared molecularly with the baseline tumor and PDX, can be manipulated using techniques such as siRNA, CRISPR or transgene over-expression. Such cell lines can then be sub-cloned in vivo for further therapeutic analysis. Similar techniques can be applied directly to fresh PDX material using a short ex vivo process (24–48 hours in vitro culture). Parallel treatment experiments can be accompanied by surveillance for markers of drug resistance using ctDNA analysis. Receptor Tyrosine Kinase = RTK. Circulating tumor DNA = ctDNA.
A range of PDX models derived from unmanipulated primary human solid tumors without intervening in vitro culture, have been successfully used to demonstrate utility in predicting efficacy of a range of therapeutic approaches (Table 1). In these studies, PDX were annotated for specific molecular biomarkers of relevance for the drug in question. Response to the novel targeted therapeutic was documented in the light of the molecular context and could be utilized to underpin clinical trial design.
Table 1. Targeted therapeutic studies performed in PDX derived from primary patient samples with biomarker analysis.
Examples of the use of PDX derived directly from primary solid human tumor with no intervening in vitro culture step, to study the efficacy of a targeted therapeutic in the setting of an exploratory or known biomarker.
| Author | Tumor type | PDX type, site, mouse strain | Actionable target | n = independent PDX tested | Targeted therapy | Outcome |
|---|---|---|---|---|---|---|
| Fichtner et. al. Clin Cancer Res, 2008 112 | NSCLC | subcutaneous NOD/SCID mice; nu/nu mice for drug testing | EGFR mutation, KRAS mutation | 25 PDX | Cetuximab (EGFR mAb) and erlotinib (EGFR inhibitor) | >50% tumor growth inhibition: seen with cetuximab in 12 of 25 NSCLC xenografts, seen with erlotinib in 6 of 25. No correlation with EGFR mutations, K-ras mutations correlated with erlotinib resistance. |
| Zhang et. al. Clin Cancer Res, 2012 113 | NSCLC | subcutaneous nu/nu mice | FGFR1 amplification | 4 PDX | AZD4547 (selective small molecule FGFR inhibitor) | Potent tumor stasis/regression in 4 of 5 FGFR1-amplified models |
| Zhang et. al. J Transl Med, 2013 114 | NSCLC | subcutaneous SCID and nu/nu mice | EGFR activating mutation; KRAS mutation; cMET gene amplification; FGFR1 amplification | 10 PDX tested for EGFR, KRAS, cMET and FGFR1 | Gefitinib (EGFR inhibitor) | EGFR activating mutation was most sensitive, FGFR1 gene amplification were insensitive, KRAS mutation +/− cMET amplification was resistant (no model with cMET mutation alone) |
| Kortmann et. al. Clin Cancer Res, 2011 115 | Ovarian (not noted as HGSC) | renal capsule, NOD-SCID mice | BRCA2 germline mutation | 2 PDX 1 sBRCA1/2 WT 1 BRCA2m |
Olaparib (PARP inhibitor) | BRCA2 mutant PDX was sensitive to Olaparib |
| Das Thakur et. al. Nature, 2013 116 | Melanoma | subcutaneous nu/nu mice | BRAF mutation (V600E) | 2 PDX | Vemurafenib (BRAF inhibitor) | Demonstrated that resistant tumors show dependency on BRAF due to elevated BRAF(V600E) expression, and become drug-dependent for their continued proliferation; cessation of drug led to tumor regression |
| Marangoni et. al. Clin Cancer Res, 2007 117 | Breast cancer | interscapular fat pad, Swiss nu/nu mice | HER2 amplification | 2 PDX HER2 amplified |
Trastuzumab (HER2 mAb) | One of two HER2-amplified xenografts responded to trastuzumab |
| Ma et. al. JCI, 2012 118 | Breast cancer | mammary fat pad*, NOD-SCID mice | TNBC, p53 mutation | 3 PDX 1 p53 WT 2 p53 mutant |
Chk1 inhibitor +/− irinotecan | Combination therapy induced checkpoint bypass and apoptosis in p53 mutant tumors; inhibited tumor growth and prolonged survival of p53 mutant TNBC. (PDX cell line retrovirally infected with shp53) |
| Dave et. al. PLoS One, 2012 119 | Breast cancer | mammary fat pad*, SCID Beige mice | TNBC, pSTAT3 expression | 2 PDX | STAT3 inhibitor +/− docetaxel | Tumor growth and recurrence inhibited by combination in one chemoresistant PDX (high pSTAT3 expression) and not in one chemosensitive PDX; pSTAT3 expression reduced post STAT3 inhibitor |
| Oakes et. al. PNAS, 2012 120 | Breast cancer | mammary fat pad*, NOD-SCID-IL2Rγc−/− mice | BCL-2 expression | 5 PDX variable expression of BCL-2 | BH3 mimetic +/− docetaxel, | Improved tumor response and overall survival with combination for PDX with elevated levels of BCL-2 |
| Vaillant et. al. Cancer Cell, 2013 121 | Breast cancer | mammary fat pad*, NOD-SCID-IL2Rγc−/ − mice | ER positive, BCL-2 expression, pAKT expression | 4 PDX variable expression of BCL-2 | BH3 mimetic/tamoxifen +/− PI3K/mTOR inhibitor | BH3 mimetic improved tumor response to tamoxifen. Median survival was significantly prolonged with triple therapy compared to tamoxifen/ABT-737 in the PDX with highest expression of BCL-2 |
| Julien et. al. Clin Cancer Res, 2012 122 | CRC | subcutaneous Swiss nu/nu mice or rats | KRAS | 52 PDX | Cetuximab (EGFR mAb) | 25 PDX models responded to cetuximab. Improved survival for wild-type KRAS models (grouped) versus mutated KRAS models (grouped); 12/28 KRAS WT models did not respond to cetuximab. |
| Pawaskar et. al. Cancer Chemother Pharmacol, 2013 123 | Pancreas cancer | subcutaneous CB-17 SCID mice | KRAS/Raf pathway, PI3 k-Akt-mTOR pathway | 2 PDX | Sorafenib, (Raf, VEGFR and PDGF-b inhibitor) + everolimus (mTOR inhibitor) | Higher doses of single agent sorafenib and everolimus inhibited tumor growth; complete inhibition of tumor growth in combination |
| Rubio-Viqueira et. al. Clin Cancer Res, 2006 124 | Pancreas cancer | subcutaneous nu/nu mice | EGFR, mTOR and Erk activation | 14 PDX | Erlotinib (EGFR inhibitor), temsirolimus (mTOR inhibitor), and CI-1040 (Erk inhibitor) | Inhibition by CI-1040 seen with Erk activation (14%), 7% response rate to Temsirolimus, no Erlotinib responses |
| Cao et. al. Br J Cancer, 2009 125 | Pancreas cancerContact | subcutaneous SCID mice; surface of pancreas* | PI3K/Akt/mTOR deregulation | 5 PDX | NVP-BEZ235 (dual PI3K/mTOR inhibitor) | Suppression of phosphorylation of PKB/Akt. Chronic dosing produced modest tumor growth inhibition in three of five PDX models |
| Al-Ejeh et. al. Clin Cancer Res, 2014 126 | Pancreas cancer | subcutaneous NOD-SCID-IL2Rγc −/− and nu/nu mice | CHK1, EGFR | 1 PDX | PF-477736 (CHK1 inhibitor), radioimmuno therapy (Lutetium-labeled anti-EGFR) and gemcitabine | Complete regression with triple combination therapy |
| Sivanand et. al. Sci Transl Med, 2012 127 | Renal cancer | renal capsule* NOD/SCID mice (similar results for subcutaneous vs subrenal) | mTORC pathway | 9 PDX | Dovitinib, sunitinib (multikinase inhibitors), sirolimus (mTORinhibi tor), erlotinib (EGFR inhibitor) | PDX retained sensitivities to sunitinib and (tem)sirolimus observed in the clinic and failed to respond to the control drug, erlotinib. Dovitinib more potently inhibited PDX than did sunitinib or sirolimus (tested in 1 PDX) |
| Su et. al. Molecular Cancer 2014 128 | Renal cancer | subcutaneous NOD/SCID mice | microRNA (let-7d) | 1 PDX | intratumoral injection of cholesterol-conjugated let-7d mimics | Tumor growth suppressed, tumor weight decreased. Number of metastatic colonies and the quantification of human-specific Alu-sequence in mouse lung were also reduced |
| Stewart E, et al. Cell Reports, 2014 129 | Ewings Sarcoma | Femoral marrow* CD1 nu/nu mice | PARP-1 | 1 PDX | Veliparib, Olaparib, BMN-673 +/− irinotecan, or temozolomide | Complete remission for PARPi + chemotherapy in > 80% of mice |
orthotopic site
4.3.3 Therapy response in PDX or Avatars reflects that seen in patients
Chemotherapies are used to treat a wide variety of tumor types81. Platinum agents are capable of directly binding to DNA, causing adducts that lead to the formation of single and double strand DNA breaks during replication and translation82. Coordinated platinum salts, such as cisplatin and carboplatin, are the cornerstone, for example, of epithelial ovarian cancer treatment and are the components of first-line treatment in this tumor type. Sensitivity to platinum-based therapy in ovarian cancer is disease-defining and prognostic, as platinum resistance correlates with poor outcomes. As such, being able to predict sensitivity, and potentially identify alternative therapies early, can have a clinical impact. Two recent reports demonstrated that HGSC PDX faithfully recapitulated response to platinum when compared with the outcome of treatment of the patient: Topp and colleagues defined platinum response for subcutaneous HGSC PDX, with three of four platinum sensitive HGSC PDX containing DNA repair gene mutations, and the fourth being methylated for BRCA1, whereas in contrast, all three platinum refractory PDX overexpressed dominant oncogenes (such as CCNE1, LIN28B and/or BCL2)49. In keeping with this, Weroha and colleagues used nine HGSC intra-peritoneal Avatar models, treated with four rounds of carboplatin/paclitaxel. When compared to patient response, nine out of nine tumorgrafts demonstrated in vivo platinum response reflective of the patient’s clinical response51. Patient Avatar models can be valuable tools in predicting which patients might benefit from the use of platinum-based therapies and more importantly, highlighting those for whom platinum might have limited potential, with the requirement of other therapies in the short-term.
In a breast cancer model, Zhang and colleagues established PDX models representing a variety of breast cancer subtypes, which were treated with single agent docetaxel, doxorubicin, or combined trastuzumab and lapatinib, depending on the treatment received by the patient from which the PDX was derived. In this report a significant association between the PDX and patient treatment response was observed, with 12 of the 13 PDX responses matching the patient’s clinical response52. Garralda and colleagues, used Avatar models along with whole-exome sequencing analysis in order to inform the treatment of patients with advanced stage solid tumors, including colorectal cancer, glioblastoma, non-small cell lung cancer, melanoma and pancreatic cancer83. A total of 13 treatments were directed by genomic and/or PDX model data, with 11 of the 13 models response mimicking the patient response.
Finally, PDX/Avatars can be generated from tumor samples obtained from warm autopsy. The aim of this approach is to obtain multiple biopsies from different metastatic sites from the one patient at the time of treatment failure and then to directly compare similarities and differences between samples. As the immediate engraftment of all sites of procurement may be cost prohibitive, it is feasible to viably preserve the material, molecularly screen it and then determine which metastatic sites to engraft and compare functionally. This would address the issue of heterogeneity at the end of the patient journey, in direct comparison with PDX/Avatars generated from chemo-naive patients at the time of diagnostic surgery, prior to any cancer treatment, when tumor heterogeneity may be less of an issue. The fact that many studies report comparable outcomes from a PDX derived from a single tumor site, when compared with the patient response to treatment, suggests that tumor heterogeneity may not be such an issue at first diagnosis, at least for cancers such as HGSC49,51.
5. Next-generation Avatars
5.1 Super-Avatars: co-engrafted in vivo models; human tumor cells and hematopoietic stem cells
PDX/Avatar approaches are gaining in applicability and becoming powerful tools for studying tumor biology and assessing novel therapeutic approaches in the preclinical setting. While these systems recapitulate many aspects of the tumor microenvironment, they preclude studying interactions between immune and cancer cells84. The knowledge that tumor cells have evolved complex mechanisms to evade immunological response has led to the development of many immuno-oncology targeted treatments85. In colorectal cancer, Old and colleagues showed that patients whose tumors were infiltrated by lymphocytes had a better chance of survival86. Subsequent work in other tumor types has sought to further understand and leverage the immune system’s ability to recognize tumor cells, a concept known as immunosurveillance, and the immune system’s ability to protect against tumor growth and metastasis, a process known as immunoediting87. There is an urgent need to develop models that allow us to characterize the interactions between immune and cancer cells in the tumor microenvironment.
The co-engraftment of hematopoietic stem cells (HPSC) and patient tumor presents new challenges, as engraftment of HPSC and patient tumor may occur at different rates (Figure 1). Successful co-engraftment depends on two main factors: isolation of the HPSC and the murine strain used. HPSC are adult stem cells capable of repopulating all the hematopoietic lineages in vivo and sustaining production of these cells for the life span of an individual88. Transplantation and xenograft repopulation assays are routinely achieved by isolation and re-injection of CD34+ cells, a cell-cell adhesion factor that also mediate the attachment of stem cells to bone marrow89,90. The strain of mouse used can also influence engraftment success; the development of three different murine strains with IL-2 receptor mutations, has increased rates of engraftment: NOD.Cg-PrkdcscidIl2rgtm1Wjl (NSG mice), NODShi.Cg-PrkdcscidIl2rgtm1Sug (NOG mice) and C;129S4-Rag2tm1FlvIl2rgtm1Flv (BRG mice)91,92. Of these models, NSG lack the IL-2 receptor, whilst NOG and BRG mice express a truncated IL-2 receptor, resulting in all these models lacking cytokine responses and expressing defective NK cells92. Subsequent mouse strains, denoted MITRG and MISTRG have been generated to increase engraftment success. Human versions of genes encoding human M-CSF (csf1), human interleukin 3 (IL-3) and GM-CSF, and human thrombopoietin were generated as a transgenic model in respective mouse loci in Rag2−/− Il2rg−/− mice (MITRG mice). The resulting human cytokines support the development and function of monocytes, macrophages and NK cells derived from human fetal liver or adult CD34(+) progenitor cells co-injected into the mice93. MISTRG mice also bear a bacterial artificial chromosome (BAC) transgene encoding human SIRPα, which binds CD47 and the resulting signal suppresses phagocytosis of CD47-expressing cells, enabling mouse phagocytes to “tolerate” and not clear engrafted human cells. These models may prove very useful for studies of therapies targeting both the tumor and the host immune system.
Various groups have laid the ground-work for such studies through the generation of cancer models designed to examine the interaction between immune and tumor cells54,94. Bankert and colleagues investigated tumor-associated T cells present in the donor graft (patient-derived) and found that T-cells remained active following transplantation for seven days54. This model can be utilized to provide valuable information of the events leading to inactivation of T-cells in human ovarian tumors54. In a breast cancer model, Lehmann and colleagues co-engrafted mice with human umbilical cord hematopoietic stem cells and breast cancer cell lines95. This model offers a novel approach to generate completely humanized monoclonal antibodies for all tumor types, particularly those cancer subtypes with no currently available antibody therapy95. A more complete understanding of the complex interaction between the interaction of the cancer cells and the human immune is required in order to design novel immunotherapies that allow for strategies to leverage the immune systems ability to help target and attack primary and dissemination tumor cells54.
5.2 Avoiding host-derived lymphoma in PDX/Avatar models
PDX represent a powerful experimental tool, however, several groups have observed the unanticipated formation of lymphomas96–98,49,51. Although mechanisms leading to the not infrequent development of lymphoma are poorly understood and few studies have properly characterized the presence of lymphoma, T-cell activation is implicated98. Work by Ghanekar and colleagues demonstrated that the development of these lymphoma were Epstein-Barr Virus (EBV)-associated B-cell lymphomas, likely to result from reactivation of latent EBV introduced following transplantation into immunodeficient mice96. The SCID mouse strains commonly used for PDX development lack B and T lymphocytes, which allows for the unwanted, unregulated growth of lymphoma97,99,100. SCID mice with the addition of a Beige mutation, known as SCID/Beige, result in mice without B- and T-lymphocytes and defective natural killer (NK) cells 101. NSG mice, which are lacking mature NK cell and mature B- and T-lymphoctes, have been used for PDX models and to study EBV92,102. Given the high prevalence of latent EBV infection in adults and the universal presence of B lymphocytes in solid tumors, this potentially cofounding process represents a potential pitfall of solid tumor xenografting96. The presence of B-cell lymphocytes can be addressed by having multiple (eg three) recipient mice implanted with each human tumor, as it is uncommon for more than one mouse per implanted tumor to develop lymphoma (Topp et al). Immunohistochemical straining of the tumor arising in each first passage recipient mouse for expression of pan-cytokeratin to confirm the epithelial origin of each tumorgraft, provides a simple way of differentiating engrafted tumors from solid lymphomas49,51 Other approaches include the exclusion of leukocytes from source tissue or targeted therapies. Single-cell sorting by flow cytometry prior to transplantation would be the most stringent technique to remove any lymphocytes, as even the presence of a few B-cells can be sufficient to establish a lymphoma96,103. However, that requires solid tumor digestion, which may contribute to PDX drift. An alternative approach is the use of targeted treatment of recipient mice to prevent the development of lymphoma. Rituximab is a monoclonal antibody targeting the B-cell specific antigen CD20, which upon antibody binding signals for its destruction by NK cells104. The success of rituximab has spurred the development of other CD20 targeted agents such as ocerizumab, ofatumumab, and obinutuzumab105. Given that these mice are immunocompromised, the use of rituximab or any of these other targeted therapies would be helpful to reduce the occurrence of lymphoma.
5.3 Avatar models and their use in clinical trials
PDX provide a useful pre-clinical model to bridge the gap between in vitro studies and patient trials (Figure 3). These models remain histologically and genetically similar to their donor. They have also been shown to be predictive of clinical outcome and are being used for drug evaluation, biomarker identification, biological studies and personalized medicine studies2. Recent work has shown that response rates in PDX and Avatar models correlate with those observed in clinic, both for targeted agents and for classic cytotoxic drugs49,51,52. Thus, the potential for using these models for translational biomarkers development and directing individualize therapy in patients is increasingly recognized2. The generation of PDX can be established in co-clinical trials, which refers to trials conducted simultaneously in human patients and in PDX, in which both the patient and the PDX receive the same treatment50. This strategy is helpful as it permits the simultaneous assessment of drug response in both patient and mouse for correlative studies, identification of biomarkers of susceptibility and resistance, and investigation of novel therapies to address the emergences of resistance2. This may also aid in the investigation of exceptional responders exploring the molecular basis of why some tumors are very sensitive to certain treatments106. Such studies can be conducted in the PDX, even when little or no tissue remains in the source patient who experiences a dramatic response to therapy. In order for correlative models to aid the study of therapies in co-clinical trials, PDX can provide additional important in vivo functional information in parallel with in vitro assays such as the more commonly used spheroid and colony formation assays.
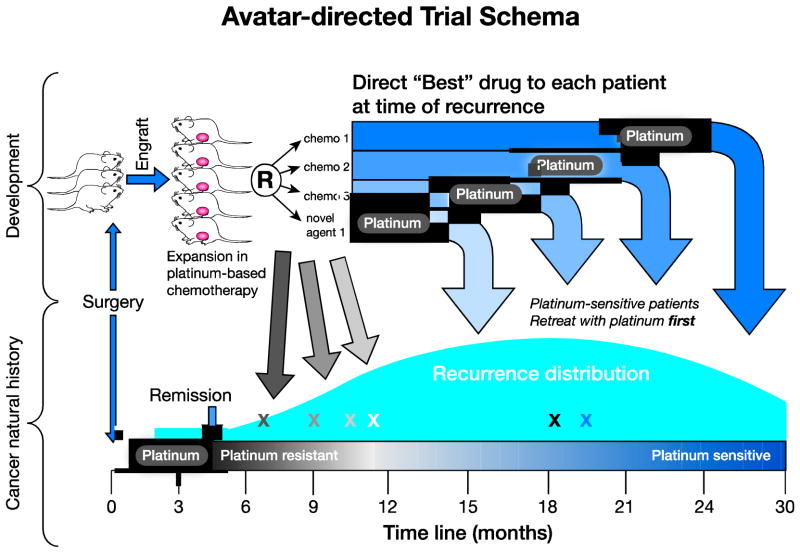
Figure 3. Avatar-directed trial schema.
At the time of initial diagnosis, a chemonaiive patient-derived xenograft (PDX) can be generated and treated with the standard therapy for that tumor type (for example, a tumor-type for which platinum is a very important treatment, such as high-grade serous ovarian cancer). As clones emerge during platinum-based therapy, additional chemotherapeutics or novel drugs can be tested, in order to determine the best treatment for that patient upon relapse after standard therapy. chemotherapy = chemo.
Co-clinical trials are often hampered by the fact that PDX and Avatar models are a costly and time consuming addition to any clinical trial, which are substantial barriers50. PDX can also be utilized to determine patient-specific targeted therapies, although outside clinical trials, we do not have sufficient evidence to suggest that study of a single PDX should guide patient treatment. This concept of personalized treatment presents with it challenges that must also be addressed at a conceptual level, as often only a small biopsy representing one portion of large heterogeneous tumor is transplanted. However, this may be mitigated by data that has demonstrated that PDX outcomes are broadly representative of patient outcomes51,49. Additionally, patients and mice may demonstrate different levels of drug toxicity, which may in part be due dosing schedules not always being fully translatable. Importantly, patients are often on additional medications and diets, which are not factored into PDX treatments2,50. When considering PDX with mutations in a particular gene of interest, it would be inappropriate for the study of only a few PDX representing a small spectrum of possible mutations, to be considered informative for all mutations in that gene.
Despite these potential short-comings, Avatar mouse models have been used in pancreatic and non-small cell lung cancer to direct personalized cancer treatment107,108. In non-small cell lung cancer, Avatar models were used to test the efficacy of three common first-line chemotherapeutics, revealing that patients fall into different subgroups, with some showing sensitivity to various treatments and resistance to others107. Garralda and colleagues performed whole-exome sequencing on patients with advanced stage solid tumors and used Avatar models to test targeted therapies in the setting of patient-specific mutations. Using this strategy, six out of thirteen patients achieved durable remission83. Zhang and colleagues established 32 breast cancer PDX models representing a variety of breast cancer sub-types, which were all shown to be genomically consistent with the patient sample and demonstrated comparable treatment responses in both PDX and patient52. These early clinical results warrant further investigation. In conclusion, the use of PDX is proving to be a powerful tool to assess novel therapies in the co-clinical setting, however, in order to utilize the full potential, we must develop rigorous clinical trial platforms to allow Avatars to inform patient treatment. It is likely that Avatar models will contribute to development of personalized medicine as technology and our ability to reliably engraft tumors makes this approach more cost effective.
5.4 Generation of Avatar derivatives
PDX/Avatar cohorts, which have been molecular annotated and functionally analysed for response to standard and relevant novel therapies, become a valuable resource, for interrogation of specific hypotheses relevant for that histologic and molecular subtype. In addition to driving drug resistance and studying clonal evolution in tumor and ctDNA (matched tumor and plasma samples), PDX can be studied ex vivo, following a brief cellular digestion and fluorescent activated cell sorting (FACS) to generate a single cell suspension for analysis in drug screens, including drug library screens over 1–5 days (Figure 2). Cell lines can also be generated, although with in vitro culture the risk of generating culture artifact is ever present, requiring monitoring of cell line drift. Such PDX-derived cell lines could be used to support parallel ex vivo and in vivo PDX studies and are valuable additions to PDX models because of the depth of analysis performed on the PDX in vivo. Derivatives of PDX or PDX-derived cell lines (of limited passage number eg passage 10–50) can be generated and studied in vitro or in vivo, with the incorporation of additional modifications by siRNA, TALENS or CRISPR to enable specific gene editing, gene activation or reversible gene knockdown42. These robust technologies assist in the identification of critical genetic regulators of cellular processes. The incorporation of high-throughput library screens may facilitate powerful exploration of fundamental drivers of cellular machinery or drug effect109,110. By harnessing these techniques, well-annotated PDX and derivatives of specific molecular subtype may provide exquisite opportunities for studying relevant drug response and resistance mechanisms (Figure 2).
6. Conclusions
In conclusion, PDX provide a proof of principle opportunity, if carefully curated, to inform clinical trial design and improve outcomes for patients participating in clinical trials. While it is not practical for a PDX or Avatar to be developed in real time to direct most patient’s front-line therapy, it may be possible for later lines of treatment. However, there is no doubt that much can be learned from the study of PDX. By employing these models, drugs brought to the clinic in clinical trials are more likely to be successful for that tumor type, due to improved treatment choice, based on better informed treatment-indication and knowledge of the drug resistance pattern likely to emerge. During treatment with one carefully chosen inhibitor, break-out resistance could be predicted, with the next inhibitor chosen as a preventive maintenance therapy to pre-empt that mode of resistance or as the next line therapy once relapse has emerged111. This approach can be trialed in PDX/Avatar models prior to being tested in the clinic. Indeed, in this way, more toxic combination therapies can be compared with less toxic sequential approaches, in the future with liquid biopsy of ctDNA surveillance for known drug resistance mechanisms.
Used in this way, PDX could allow prediction of likely mechanisms of drug resistance and in doing so, inform design of appropriate therapeutic strategies. By defining the rules of engagement in the fight against cancer evolution under treatment pressure, we should be better placed to prioritize treatment for our patients, who unlike PDX models, have far fewer chances to try multiple therapeutic options.
Supplementary Material
Highlights.
Prioritizing potential therapeutic targets identified by genomics is challenging
Well-annotated patient-derived xenografts (PDX) can be powerful patient surrogates
PDX can be sampled serially and in parallel to reveal clonal evolution on treatment
PDX could predict likely mechanisms of drug resistance to inform therapeutic strategy
Challenges include high quality annotation and humanizing the murine immune system
Acknowledgments
This work was supported by fellowships and grants from the National Health & Medical Research Council of Australia grant 1062702 (CLS) and scholarship 1076048 (AMH); the Cancer Council Victoria Sir Edward Dunlop Fellowship in Cancer Research (CLS); the Victorian Cancer Agency Clinical Fellowship (CLS); United States National Institute of Health Grants: R01 CA184502 (PH) and Mayo Clinic SPORE in Ovarian Cancer CA136393 (PH), Minnesota Partnership for Biotechnology and Medical Genomics (PH, KL) and Ovarian Cancer Research Fund OCRF258797 (PH, KL); and Ginkgo, LLC (PH). This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS and the Australian Cancer Research Foundation.
Abbreviations
- WT
wildtype
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tursz T, Bernards R. Hurdles on the road to personalized medicine. Mol Oncol. 2014 doi: 10.1016/j.molonc.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hidalgo M, Amant F, Biankin AV, et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014;4(9):998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 4.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao M, Tian F, Mariadason JM, et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin Cancer Res. 2013;19(3):657–667. doi: 10.1158/1078-0432.CCR-11-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 7.Ulahannan D, Kovac MB, Mulholland PJ, Cazier JB, Tomlinson I. Technical and implementation issues in using next-generation sequencing of cancers in clinical practice. Br J Cancer. 2013;109(4):827–835. doi: 10.1038/bjc.2013.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gargis AS, Kalman L, Berry MW, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30(11):1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sah S, Chen L, Houghton J, et al. Functional DNA quantification guides accurate next-generation sequencing mutation detection in formalin-fixed, paraffin-embedded tumor biopsies. Genome Med. 2013;5(8):77. doi: 10.1186/gm481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meric-Bernstam F, Farhangfar C, Mendelsohn J, Mills GB. Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol. 2013;31(15):1849–1857. doi: 10.1200/JCO.2012.45.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robson M. Multigene panel testing: planning the next generation of research studies in clinical cancer genetics. J Clin Oncol. 2014;32(19):1987–1989. doi: 10.1200/JCO.2014.56.0474. [DOI] [PubMed] [Google Scholar]
- 13.Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatta G, van der Zwan JM, Casali PG, et al. Rare cancers are not so rare: the rare cancer burden in Europe. Eur J Cancer. 2011;47(17):2493–2511. doi: 10.1016/j.ejca.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Venditti JM. Preclinical drug development: rationale and methods. Semin Oncol. 1981;8(4):349–361. [PubMed] [Google Scholar]
- 20.Johnson JI, Decker S, Zaharevitz D, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84(10):1424–1431. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221(1):49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knoops L, de Jong D. The role of the p53 pathway in the treatment of follicular lymphoma. Cell Cycle. 2008;7(4):436–439. doi: 10.4161/cc.7.4.5441. [DOI] [PubMed] [Google Scholar]
- 23.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2(3):227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 26.Bernards R. A missing link in genotype-directed cancer therapy. Cell. 2012;151(3):465–468. doi: 10.1016/j.cell.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 28.Perez EA, Romond EH, Suman VJ, et al. Four-year follow-up of trastuzumab plus adjuvant chemotherapy for operable human epidermal growth factor receptor 2-positive breast cancer: joint analysis of data from NCCTG N9831 and NSABP B-31. J Clin Oncol. 2011;29(25):3366–3373. doi: 10.1200/JCO.2011.35.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 30.Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17(9):2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 31.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 32.Kato H, Arao T, Matsumoto K, et al. Gene amplification of EGFR, HER2, FGFR2 and MET in esophageal squamous cell carcinoma. Int J Oncol. 2013;42(4):1151–1158. doi: 10.3892/ijo.2013.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anglesio MS, Kommoss S, Tolcher MC, et al. Molecular characterization of mucinous ovarian tumours supports a stratified treatment approach with HER2 targeting in 19% of carcinomas. J Pathol. 2013;229(1):111–120. doi: 10.1002/path.4088. [DOI] [PubMed] [Google Scholar]
- 34.Herter-Sprie GS, Greulich H, Wong KK. Activating Mutations in ERBB2 and Their Impact on Diagnostics and Treatment. Front Oncol. 2013;3:86. doi: 10.3389/fonc.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoppmann SF, Streubel B, Birner P. Amplification but not translocation of anaplastic lymphoma kinase is a frequent event in oesophageal cancer. Eur J Cancer. 2013;49(8):1876–1881. doi: 10.1016/j.ejca.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Audeh MW, Carmichael J, Penson RT, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376(9737):245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 37.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366(15):1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 38.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15(8):852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 39.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108(8):3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du W, Elemento O. Cancer systems biology: embracing complexity to develop better anticancer therapeutic strategies. Oncogene. 2014 doi: 10.1038/onc.2014.291. [DOI] [PubMed] [Google Scholar]
- 41.Tentler JJ, Tan AC, Weekes CD, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ocana A, Pandiella A, Siu LL, Tannock IF. Preclinical development of molecular-targeted agents for cancer. Nat Rev Clin Oncol. 2011;8(4):200–209. doi: 10.1038/nrclinonc.2010.194. [DOI] [PubMed] [Google Scholar]
- 43.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2(2):247–250. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- 44.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26(1):54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang HL, Hsing HW, Lai TC, et al. Trypsin-induced proteome alteration during cell subculture in mammalian cells. J Biomed Sci. 2010;17:36. doi: 10.1186/1423-0127-17-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buick RN, Pullano R, Trent JM. Comparative properties of five human ovarian adenocarcinoma cell lines. Cancer Res. 1985;45(8):3668–3676. [PubMed] [Google Scholar]
- 48.Kerbel RS. Human tumor xenografts as predictive preclinical models for anticancer drug activity in humans: better than commonly perceived-but they can be improved. Cancer Biol Ther. 2003;2(4 Suppl 1):S134–139. [PubMed] [Google Scholar]
- 49.Topp MD, Hartley L, Cook M, et al. Molecular correlates of platinum response in human high-grade serous ovarian cancer patient-derived xenografts. Mol Oncol. 2014;8(3):656–668. doi: 10.1016/j.molonc.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malaney P, Nicosia SV, Dave V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett. 2014;344(1):1–12. doi: 10.1016/j.canlet.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weroha SJ, Becker MA, Enderica-Gonzalez S, et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin Cancer Res. 2014;20(5):1288–1297. doi: 10.1158/1078-0432.CCR-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Claerhout S, Prat A, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73(15):4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson-Abelson MR, Sonnenberg GF, Takita H, et al. Long-term engraftment and expansion of tumor-derived memory T cells following the implantation of non-disrupted pieces of human lung tumor into NOD-scid IL2Rgamma(null) mice. J Immunol. 2008;180(10):7009–7018. doi: 10.4049/jimmunol.180.10.7009. [DOI] [PubMed] [Google Scholar]
- 54.Yokota SJ, Facciponte JG, Kelleher RJ, Jr, et al. Changes in ovarian tumor cell number, tumor vasculature, and T cell function monitored in vivo using a novel xenograft model. Cancer Immun. 2013;13:11. [PMC free article] [PubMed] [Google Scholar]
- 55.Dolmans MM, Marinescu C, Saussoy P, Van Langendonckt A, Amorim C, Donnez J. Reimplantation of cryopreserved ovarian tissue from patients with acute lymphoblastic leukemia is potentially unsafe. Blood. 2010;116(16):2908–2914. doi: 10.1182/blood-2010-01-265751. [DOI] [PubMed] [Google Scholar]
- 56.Bankert RB, Balu-Iyer SV, Odunsi K, et al. Humanized mouse model of ovarian cancer recapitulates patient solid tumor progression, ascites formation, and metastasis. PLoS One. 2011;6(9):e24420. doi: 10.1371/journal.pone.0024420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeRose YS, Wang G, Lin YC, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visonneau S, Cesano A, Torosian MH, Miller EJ, Santoli D. Growth characteristics and metastatic properties of human breast cancer xenografts in immunodeficient mice. Am J Pathol. 1998;152(5):1299–1311. [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beckhove P, Schutz F, Diel IJ, et al. Efficient engraftment of human primary breast cancer transplants in nonconditioned NOD/Scid mice. Int J Cancer. 2003;105(4):444–453. doi: 10.1002/ijc.11125. [DOI] [PubMed] [Google Scholar]
- 61.Liu H, Patel MR, Prescher JA, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107(42):18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walters DM, Stokes JB, Adair SJ, et al. Clinical, molecular and genetic validation of a murine orthotopic xenograft model of pancreatic adenocarcinoma using fresh human specimens. PLoS One. 2013;8(10):e77065. doi: 10.1371/journal.pone.0077065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Metildi CA, Kaushal S, Hoffman RM, Bouvet M. In vivo serial selection of human pancreatic cancer cells in orthotopic mouse models produces high metastatic variants irrespective of Kras status. J Surg Res. 2013;184(1):290–298. doi: 10.1016/j.jss.2013.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez-Torras S, Vidal-Pla A, Miquel R, et al. Characterization of human pancreatic orthotopic tumor xenografts suitable for drug screening. Cell Oncol (Dordr) 2011;34(6):511–521. doi: 10.1007/s13402-011-0049-1. [DOI] [PubMed] [Google Scholar]
- 65.Qiu W, Su GH. Development of orthotopic pancreatic tumor mouse models. Methods Mol Biol. 2013;980:215–223. doi: 10.1007/978-1-62703-287-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chai MG, Kim-Fuchs C, Angst E, Sloan EK. Bioluminescent orthotopic model of pancreatic cancer progression. J Vis Exp. 2013;(76) doi: 10.3791/50395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat Protoc. 2009;4(11):1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kudo-Saito C, Wansley EK, Gruys ME, Wiltrout R, Schlom J, Hodge JW. Combination therapy of an orthotopic renal cell carcinoma model using intratumoral vector-mediated costimulation and systemic interleukin-2. Clin Cancer Res. 2007;13(6):1936–1946. doi: 10.1158/1078-0432.CCR-06-2398. [DOI] [PubMed] [Google Scholar]
- 69.Norian LA, Kresowik TP, Rosevear HM, et al. Eradication of metastatic renal cell carcinoma after adenovirus-encoded TNF-related apoptosis-inducing ligand (TRAIL)/CpG immunotherapy. PLoS One. 2012;7(2):e31085. doi: 10.1371/journal.pone.0031085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sicklick JK, Leonard SY, Babicky ML, et al. Generation of orthotopic patient-derived xenografts from gastrointestinal stromal tumor. J Transl Med. 2014;12:41. doi: 10.1186/1479-5876-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tracz A, Mastri M, Lee CR, Pili R, Ebos JM. Modeling spontaneous metastatic renal cell carcinoma (mRCC) in mice following nephrectomy. J Vis Exp. 2014;(86) doi: 10.3791/51485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hillman GG, Wang Y, Che M, et al. Progression of renal cell carcinoma is inhibited by genistein and radiation in an orthotopic model. BMC Cancer. 2007;7:4. doi: 10.1186/1471-2407-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mathieu A, Remmelink M, D’Haene N, et al. Development of a chemoresistant orthotopic human nonsmall cell lung carcinoma model in nude mice: analyses of tumor heterogenity in relation to the immunohistochemical levels of expression of cyclooxygenase-2, ornithine decarboxylase, lung-related resistance protein, prostaglandin E synthetase, and glutathione-S-transferase-alpha (GST)-alpha, GST-mu, and GST-pi. Cancer. 2004;101(8):1908–1918. doi: 10.1002/cncr.20571. [DOI] [PubMed] [Google Scholar]
- 74.Mordant P, Loriot Y, Lahon B, et al. Bioluminescent orthotopic mouse models of human localized non-small cell lung cancer: feasibility and identification of circulating tumour cells. PLoS One. 2011;6(10):e26073. doi: 10.1371/journal.pone.0026073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Justilien V, Fields AP. Utility and applications of orthotopic models of human non-small cell lung cancer (NSCLC) for the evaluation of novel and emerging cancer therapeutics. Curr Protoc Pharmacol. 2013;62(Unit 14):27. doi: 10.1002/0471141755.ph1427s62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Onn A, Isobe T, Itasaka S, et al. Development of an orthotopic model to study the biology and therapy of primary human lung cancer in nude mice. Clin Cancer Res. 2003;9(15):5532–5539. [PubMed] [Google Scholar]
- 77.Rozenberg GI, Monahan KB, Torrice C, Bear JE, Sharpless NE. Metastasis in an orthotopic murine model of melanoma is independent of RAS/RAF mutation. Melanoma Res. 2010;20(5):361–371. doi: 10.1097/CMR.0b013e328336ee17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mueller AJ, Maniotis AJ, Freeman WR, et al. An orthotopic model for human uveal melanoma in SCID mice. Microvasc Res. 2002;64(2):207–213. doi: 10.1006/mvre.2002.2398. [DOI] [PubMed] [Google Scholar]
- 79.Bobek V, Kolostova K, Pinterova D, et al. A clinically relevant, syngeneic model of spontaneous, highly metastatic B16 mouse melanoma. Anticancer Res. 2010;30(12):4799–4803. [PubMed] [Google Scholar]
- 80.Landis MD, Lehmann BD, Pietenpol JA, Chang JC. Patient-derived breast tumor xenografts facilitating personalized cancer therapy. Breast Cancer Res. 2013;15(1):201. doi: 10.1186/bcr3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7(8):573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 82.Telli M. Optimizing chemotherapy in triple-negative breast cancer: the role of platinum. Am Soc Clin Oncol Educ Book. 2014:e37–42. doi: 10.14694/EdBook_AM.2014.34.e37. [DOI] [PubMed] [Google Scholar]
- 83.Garralda E, Paz K, Lopez-Casas PP, et al. Integrated next-generation sequencing and avatar mouse models for personalized cancer treatment. Clin Cancer Res. 2014;20(9):2476–2484. doi: 10.1158/1078-0432.CCR-13-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ishikawa F. Modeling normal and malignant human hematopoiesis in vivo through newborn NSG xenotransplantation. Int J Hematol. 2013;98(6):634–640. doi: 10.1007/s12185-013-1467-9. [DOI] [PubMed] [Google Scholar]
- 85.Eggermont AM, Robert C. Melanoma: smart therapeutic strategies in immuno-oncology. Nat Rev Clin Oncol. 2014;11(4):181–182. doi: 10.1038/nrclinonc.2014.36. [DOI] [PubMed] [Google Scholar]
- 86.Odunsi K, Old LJ. Tumor infiltrating lymphocytes: indicators of tumor-related immune responses. Cancer Immun. 2007;7:3. [PMC free article] [PubMed] [Google Scholar]
- 87.Finn OJ. Immuno-oncology: understanding the function and dysfunction of the immune system in cancer. Ann Oncol. 2012;23(Suppl 8):viii6–9. doi: 10.1093/annonc/mds256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wognum AW, Eaves AC, Thomas TE. Identification and isolation of hematopoietic stem cells. Arch Med Res. 2003;34(6):461–475. doi: 10.1016/j.arcmed.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 89.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333(6039):218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 90.Furness SG, McNagny K. Beyond mere markers: functions for CD34 family of sialomucins in hematopoiesis. Immunol Res. 2006;34(1):13–32. doi: 10.1385/IR:34:1:13. [DOI] [PubMed] [Google Scholar]
- 91.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol. 2012;12(11):786–798. doi: 10.1038/nri3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rongvaux A, Willinger T, Martinek J, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32(4):364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Werner-Klein M, Proske J, Werno C, et al. Immune humanization of immunodeficient mice using diagnostic bone marrow aspirates from carcinoma patients. PLoS One. 2014;9(5):e97860. doi: 10.1371/journal.pone.0097860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wege AK, Schmidt M, Ueberham E, et al. Co-transplantation of human hematopoietic stem cells and human breast cancer cells in NSG mice: a novel approach to generate tumor cell specific human antibodies. MAbs. 2014;6(4):968–977. doi: 10.4161/mabs.29111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen K, Ahmed S, Adeyi O, Dick JE, Ghanekar A. Human solid tumor xenografts in immunodeficient mice are vulnerable to lymphomagenesis associated with Epstein-Barr virus. PLoS One. 2012;7(6):e39294. doi: 10.1371/journal.pone.0039294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.John T, Yanagawa N, Kohler D, et al. Characterization of lymphomas developing in immunodeficient mice implanted with primary human non-small cell lung cancer. J Thorac Oncol. 2012;7(7):1101–1108. doi: 10.1097/JTO.0b013e3182519d4d. [DOI] [PubMed] [Google Scholar]
- 98.Zenger E, Abbey NW, Weinstein MD, et al. Injection of human primary effusion lymphoma cells or associated macrophages into severe combined immunodeficient mice causes murine lymphomas. Cancer Res. 2002;62(19):5536–5542. [PubMed] [Google Scholar]
- 99.Shibata S, Asano T, Ogura A, et al. SCID-bg mice as xenograft recipients. Lab Anim. 1997;31(2):163–168. doi: 10.1258/002367797780600107. [DOI] [PubMed] [Google Scholar]
- 100.Bosma MJ, Carroll AM. The SCID mouse mutant: definition, characterization, and potential uses. Annu Rev Immunol. 1991;9:323–350. doi: 10.1146/annurev.iy.09.040191.001543. [DOI] [PubMed] [Google Scholar]
- 101.Green M, Rogers D, Russell P, et al. The SCID/Beige mouse as a model to investigate protection against Yersinia pestis. FEMS Immunol Med Microbiol. 1999;23(2):107–113. doi: 10.1111/j.1574-695X.1999.tb01229.x. [DOI] [PubMed] [Google Scholar]
- 102.Shultz LD, Lyons BL, Burzenski LM, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174(10):6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 103.Dainiak MB, Kumar A, Galaev IY, Mattiasson B. Methods in cell separations. Adv Biochem Eng Biotechnol. 2007;106:1–18. doi: 10.1007/10_2007_069. [DOI] [PubMed] [Google Scholar]
- 104.Rudnicka D, Oszmiana A, Finch DK, et al. Rituximab causes a polarization of B cells that augments its therapeutic function in NK-cell-mediated antibody-dependent cellular cytotoxicity. Blood. 2013;121(23):4694–4702. doi: 10.1182/blood-2013-02-482570. [DOI] [PubMed] [Google Scholar]
- 105.Cang S, Mukhi N, Wang K, Liu D. Novel CD20 monoclonal antibodies for lymphoma therapy. J Hematol Oncol. 2012;5:64. doi: 10.1186/1756-8722-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dienstmann R, Rodon J, Tabernero J. Optimal design of trials to demonstrate the utility of genomically-guided therapy: Putting Precision Cancer Medicine to the test. Mol Oncol. 2014 doi: 10.1016/j.molonc.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dong X, Guan J, English JC, et al. Patient-derived first generation xenografts of non-small cell lung cancers: promising tools for predicting drug responses for personalized chemotherapy. Clin Cancer Res. 2010;16(5):1442–1451. doi: 10.1158/1078-0432.CCR-09-2878. [DOI] [PubMed] [Google Scholar]
- 108.Garber K. Personal mouse colonies give hope for pancreatic cancer patients. J Natl Cancer Inst. 2007;99(2):105–107. doi: 10.1093/jnci/djk046. [DOI] [PubMed] [Google Scholar]
- 109.Zhou Y, Zhu S, Cai C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 110.Wei Y, Xu F, Li P. Technology-driven and evidence-based genomic analysis for integrated pediatric and prenatal genetics evaluation. J Genet Genomics. 2013;40(1):1–14. doi: 10.1016/j.jgg.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 111.Giles FJ, DeAngelo DJ, Baccarani M, et al. Optimizing outcomes for patients with advanced disease in chronic myelogenous leukemia. Semin Oncol. 2008;35(1 Suppl 1):S1–17. doi: 10.1053/j.seminoncol.2007.12.002. quiz S18–20. [DOI] [PubMed] [Google Scholar]
- 112.Fichtner I, Rolff J, Soong R, et al. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin Cancer Res. 2008;14(20):6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- 113.Zhang J, Zhang L, Su X, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18(24):6658–6667. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 114.Zhang XC, Zhang J, Li M, et al. Establishment of patient-derived non-small cell lung cancer xenograft models with genetic aberrations within EGFR, KRAS and FGFR1: useful tools for preclinical studies of targeted therapies. J Transl Med. 2013;11:168. doi: 10.1186/1479-5876-11-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kortmann U, McAlpine JN, Xue H, et al. Tumor growth inhibition by olaparib in BRCA2 germline-mutated patient-derived ovarian cancer tissue xenografts. Clin Cancer Res. 2011;17(4):783–791. doi: 10.1158/1078-0432.CCR-10-1382. [DOI] [PubMed] [Google Scholar]
- 116.Das Thakur M, Salangsang F, Landman AS, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494(7436):251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marangoni E, Vincent-Salomon A, Auger N, et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin Cancer Res. 2007;13(13):3989–3998. doi: 10.1158/1078-0432.CCR-07-0078. [DOI] [PubMed] [Google Scholar]
- 118.Ma CX, Cai S, Li S, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest. 2012;122(4):1541–1552. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dave B, Landis MD, Tweardy DJ, et al. Selective small molecule Stat3 inhibitor reduces breast cancer tumor-initiating cells and improves recurrence free survival in a human-xenograft model. PLoS One. 2012;7(8):e30207. doi: 10.1371/journal.pone.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oakes SR, Vaillant F, Lim E, et al. Sensitization of BCL-2-expressing breast tumors to chemotherapy by the BH3 mimetic ABT-737. Proc Natl Acad Sci U S A. 2012;109(8):2766–2771. doi: 10.1073/pnas.1104778108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vaillant F, Merino D, Lee L, et al. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell. 2013;24(1):120–129. doi: 10.1016/j.ccr.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 122.Julien S, Merino-Trigo A, Lacroix L, et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res. 2012;18(19):5314–5328. doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- 123.Pawaskar DK, Straubinger RM, Fetterly GJ, et al. Synergistic interactions between sorafenib and everolimus in pancreatic cancer xenografts in mice. Cancer Chemother Pharmacol. 2013;71(5):1231–1240. doi: 10.1007/s00280-013-2117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rubio-Viqueira B, Jimeno A, Cusatis G, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12(15):4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 125.Cao P, Maira SM, Garcia-Echeverria C, Hedley DW. Activity of a novel, dual PI3-kinase/mTor inhibitor NVP-BEZ235 against primary human pancreatic cancers grown as orthotopic xenografts. Br J Cancer. 2009;100(8):1267–1276. doi: 10.1038/sj.bjc.6604995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Al-Ejeh F, Pajic M, Shi W, et al. Gemcitabine and CHK1 inhibition potentiate EGFR-directed radioimmunotherapy against pancreatic ductal adenocarcinoma. Clin Cancer Res. 2014;20(12):3187–3197. doi: 10.1158/1078-0432.CCR-14-0048. [DOI] [PubMed] [Google Scholar]
- 127.Sivanand S, Pena-Llopis S, Zhao H, et al. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci Transl Med. 2012;4(137):137ra175. doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Su B, Zhao W, Shi B, et al. Letf7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 2014;13:206. doi: 10.1186/1476-4598-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stewart EGR, Bradley C, Griffiths LM, Benavente C, Twarog NR, Miller GM, Caufield W, Freeman BB, Bahrami A, Pappo A, Wu J, Loh S, Karlstrom A, Calabrese C, Gordon B, Tsurkan L, Hatfield MJ, Potter PM, Snyder SM, Thiagarajan S, Shirinifard A, Sablauer A, Shelat A, Dyer MA. Targeting the DNA Repair Pathway in Ewing Sarcoma. Cell Reports. 2014 doi: 10.1016/j.celrep.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.