Abstract
Drug resistance remains a major clinical challenge for cancer treatment. Multiple myeloma (MM) is an incurable plasma cell cancer selectively localized in the bone marrow (BM). The main cause of resistance in myeloma is the minimal residual disease (MRD) cells that are resistant to the original therapy including bortezomib treatment and high dose melphalan in stem cell transplant. In this study, we demonstrate that altered tumor cell metabolism is essential for the regulation of drug resistance in MM cells. We show the unprecedented role of the metabolic phenotype in inducing drug resistance through LDHA and HIF1A in MM; and that specific inhibition of LDHA and HIF1A can restore sensitivity to therapeutic agents such as bortezomib and can also inhibit tumor growth induced by altered metabolism. Knockdown of LDHA can restore sensitivity of bortezomib resistance cell lines while gain of function studies using LDHA or HIF1A induced resistance in bortezomib sensitive cell lines. Taken together, these data suggest that HIF1A and LDHA are important targets for hypoxia-driven drug resistance. Novel drugs that regulate metabolic pathways in MM, specifically targeting LDHA, can be beneficial to inhibit tumor growth and overcome drug resistance.
Keywords: metabolites, hypoxia, multiple myeloma, drug resistance
INTRODUCTION
Cellular metabolic versatility is essential for the maintenance of energy production throughout a range of oxygen concentrations (1). Metabolic changes occurring in cancer cells are considered to be fundamental for the transformation of normal cells into cancer cells. A common property of invasive cancers is an altered glucose metabolism or glycolysis (1). Glycolysis converts glucose into pyruvate and in normal cells; this process is inhibited by the presence of oxygen, which allows mitochondria to oxidize pyruvate to CO2 and H2O. This inhibition is termed as the “Pasteur effect” (2). Conversion of glucose to lactic acid in the presence of oxygen is known as aerobic glycolysis or the “Warburg effect” and the increase of aerobic glycolysis is often observed in tumor cells (3–5).
One of the most recognized reasons for altered tumor metabolism is hypoxia in the tumor microenvironment (6,7). Cells respond to the hypoxic microenvironment with the activation of hypoxia-inducible factor 1 (HIF1) transcription factor. The net result of hypoxic HIF1 activation is to shift energy production by increasing glycolysis and decreasing mitochondrial function (6). The largest functional group of genes consistently regulated by HIF1 is associated with glucose metabolism. HIF1 increases the expression of the glucose transporters, enzymatic breakdown of glucose into pyruvate and enzymes involved in the pyruvate metabolism as well as lactate production (8,9). Although HIF1 was initially identified because of its response to low O2 concentrations, it is now apparent that HIF1 can be regulated by other factors such as oncogene activation (RAS, MYC and PI3K) or loss of tumor suppressors including VHL (von Hippel-Lindau) or PTEN leading to increased glycolysis, angiogenesis and drug resistance (10–12).
Multiple myeloma (MM) is an incurable plasma cell cancer selectively localized in the bone marrow (BM). The introduction of novel agents, including bortezomib in combination with autologous stem cell transplantation, has led to a significant advancement in the treatment of patients leading to complete response in many patients. Unfortunately, most patients ultimately relapse due to the presence of surviving tumor cells at the minimal residual disease (MRD) state, suggesting the presence of drug resistance within specific niches in the BM. The BM has heterogeneous areas of hypoxia and these specific niches are altered during chemotherapy and radiation therapy (1). We hypothesize that hypoxia in specific bone marrow niches regulates the maintenance of minimal residual disease cells which are resistant to treatment and have the capability to induce relapse. We sought to investigate the mechanism underlying this drug resistance through the cellular metabolic profile of MM cells in normoxic and hypoxic conditions. Our results reveal unprecedented features of MM cells metabolism and further demonstrate that LDHA and HIF1A are valid targets to prevent MM drug resistance and progression in vivo.
MATERIALS AND METHODS
Cells
MM cell lines MM1S, H929 and RPMI8226 (were kindly provided by Prof. Jesús F. San Miguel, Salamanca, Spain). Cell lines were cultured in RPMI 1640 medium with L-glutamine and supplemented with antibiotics (penicillin at 100 U/ml, streptomycin at 100 μg/ml) and 10% fetal bovine serum (FBS). The luciferase (luc)–expressing MM1S-GFP/luc cell lines were generated by retroviral transduction with the pGC-gfp/luc vector (kind gift of Dr. A. Kung, Dana-Farber Cancer Institute, Boston, MA).
Growth inhibition assay
The inhibitory effect of different drugs was assessed by measuring 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Chemicon International, Temecula, CA) dye absorbance, as previously described (13,14).
Cell cycle and apoptosis assays
Cell-cycle analysis was profiled by flow cytometry using propidium iodide (PI) staining (5μg/mL, Sigma Chemical) after 24 hours culture in normoxic or hypoxic conditions. Apoptosis was measured using Annexin-V-FITC staining and flow cytometric analysis according to manufacturer’s protocol.
Gene expression profile
Gene expression profile has been performed on MM1S using Affymetrix Human Genome U133 Plus 2.0 Array, (GEO accession number: GSE52315). Comparison between Normoxia (n=3), and Hypoxia (n=3) was performed by using dChip (2 fold change; P <0.05). Differentially expressed genes were classified using dChip software. Gene set enrichment analysis (GSEA) was performed using the publicly available desktop application from the Broad Institute (http://www.broad.mit.edu/gsea/software/software_index.html). The gene sets database used was that of functional sets, s2.symbols.gmt. p values were calculated by permuting the genes 1000 times. The classic enrichment statistic was selected. The gene expression datasets from Schaefer CF et al. provided in the Pathway Interaction Database (National Cancer Institute and Nature Publishing Group) were used for HIF1A and HIF2A analysis in GSEA (15). The gene expression datasets REACTOME_ GLUCOSE_ METABOLISM and KEGG_ PYRUVATE_ METABOLISM were used respectively for Glucose metabolism and TCA cycle analysis in GSEA. Gene Pattern (http://www.broadinstitute.org/cancer/software/genepattern.html) analysis was performed using the “Comparative Marker Selection” tool to find the genes that are most closely correlated with the two phenotypes normoxia and hypoxia.
To determine the Gene enrichments sets of hypoxia-related pathways, glucose metabolism and TCA cycle in plasma cells isolated from normal subjects or from newly diagnosed MM patients, as well as from responder and relapsed patients to bortezomib, we used published datasets from the Gene Expression Omnibus by Chng W.J. et al and Mulligan G. et al (series numbers GSE6477 and GSE9782, respectively)) (16,17).
Metabolite profiling
Metabolites were extracted in ice-cold methanol and endogenous metabolite profiles were obtained using two liquid chromatography-tandem mass spectrometry (LC-MS) methods as described (18). Data were acquired using a 5500 QTRAP triple quadrupole mass spectrometer (AB/Sciex) coupled to a Prominence UFLC system (Shimadzu) via selected reaction monitoring (SRM) of a total of 289 endogenous water soluble metabolites for steady-state analyses of samples.
Metaboanalyst software was used for analysis. Metabolite levels were normalized to the total of all metabolites detected on a triplicate set of cells treated identically to the experimental cells.
Hexokinase activity and lactate measurement
Hexokinase activity was measured with the Hexokinase Assay Kit (Abcam) and cellular lactate levels were measured using the Lactate Colorimetric Assay Kit (Biovision) according to the manufacturer’s instructions.
RNA purification, reverse transcription and quantitative RT-PCR (qRT-PCR)
Total RNA was prepared with QIAzol reagent (Invitrogen) according to the manufacturer’s instructions. 1 μg of total RNA were reverse-transcribed using SuperScript III First-Strand Synthesis (Invitrogen). Diluted cDNAs were analyzed by real-time PCR using Sybr Green I Mastermix on Lightcycler 480 (Roche)/on an ABI Prism 7900 Fast instrument. The level of gene expression was normalized to 18S. The primers sequences are provided below:
| Gene | Primers |
|---|---|
| human HIF1A | F: TTGGACACTGGTGGCTCATTAC R: TGAGCTGTCTGTGATCCAGCAT |
| human HIF2A | F: GTGTTGTGGACACTGCAGACTTGT R: ATGACTCCACTGCTCGGATTGTCA |
| human HK2 | F: AGCCCTTTCTCCATCTCCTT R: AACCATGACCAAGTGCAGAA |
| human LDHA | F: GGAGATCCATCATCTCTCC R: GGCCTGTGCCATCAGTATCT |
| human 18S | F: TCAACTTTCGATGGTAGTCGCCGT R: TCCTTGGATGTGGTAGCCGTTTCT |
In vivo studies
Six weeks old female SCID-beige mice from Charles River Labs (Wilmington, MA) were intravenously (IV) implanted with 100 μl of 5 × 106 MM1S-GFP/luc-scramble, MM1S-GFP/luc-shHIF1A or MM1S-GFP/luc-shLDHA. Mice were treated with bortezomib, 0.75 mg/Kg in PBS once weekly by intraperitoneal (IP) injection beginning 10 days after tumor implantation until moribund. Mice with different stages of tumor development based on tumor size detected by bioluminescence (BLI) were treated with the hypoxia marker pimonidazole hydrochloride (PIMO; 100 mg/kg by IP injection; Hypoxyprobe Store). After 4 hours, BM was isolated from one femur by flushing with cold PBS and prepared for RNA isolation as described; the other femur was used for immunohistochemistry (IHC).
In vivo tumor growth has been assessed by using in vivo bioluminescence imaging. Mice were injected with 75 mg/kg of Luciferin (Xenogen, Hopkington, MA), and tumor growth was detected by bioluminescence 3 min after the injection, using Xenogen In Vivo Imaging System (Caliper Life Sciences, Hopkinton, MA). Mice were monitored and sacrificed when they developed side effects of tumor burden in accordance with approved protocol of the Dana-Farber Cancer Institute (DFCI) Animal Care and Use Committee.
Immunohistochemistry
In the in vivo model, femurs were fixed in 4% paraformaldehyde and embedded in paraffin. Sections were stained with hematoxylin and eosin (H&E) in accordance with standard procedures. Immunohistochemistry was performed using antibodies against Mab1 (Hypoxyprobe Store), HK2 (Cell signaling) and LDHA (Cell signaling) according to manufacturer’s instructions. All MM biopsies were evaluated at DFCI, and the histological diagnosis was based on H&E. Histologic IHC images were obtained with the Olimpus AH2 Microscope Camera from Center Valley, PA. Image acquisition and processing software were performed using an Olimpus DP12 camera and software.
Knockdown constructs
Stable knockdowns of HIF1A, HIF2A, HK2 and LDHA were generated by lentiviral transduction of MM1S and MM1S-GFP/luc cells with five independent shRNA hairpin sequences targeting human HIF1A, HIF2A, HK2 and LDHA respectively. A scrambled shRNA sequence was used as control (TRCN0000072212).
Lentiviral shRNAs were obtained from The RNAi Consortium (TCR) collection of the Broad Institute. The TCR numbers for the shRNA used are:
| Gene | Clones |
|---|---|
| human HIF1A | TRCN0000003809 TRCN0000003810 TRCN0000010819 TRCN0000318675 TRCN0000349634 |
| human HIF2A | TRCN0000003803 TRCN0000003804 TRCN0000003805 TRCN0000352630 TRCN0000342501 |
| human HK2 | TRCN0000037670 TRCN0000195171 TRCN0000196260 TRCN0000232927 TRCN0000232928 |
| human LDHA | TRCN0000026538 TRCN0000026536 TRCN0000158762 TRCN0000164922 TRCN0000166246 |
Statistics
P values described in the in vitro assays are based on T-tests (two-tailed; α 0.05). P values are provided for each figure.
RESULTS
Hypoxia promotes drug resistance in multiple myeloma
We first examined hypoxia regulated pathways in primary MM patients to test the enrichment of HIF1A and HIF2A (15) pathways in plasma cells isolated from BM of normal donors and MM patients; we analyzed the published gene-expression datasets (series numbers, GSE6477 and GSE9782, respectively) (16,17). Newly diagnosed MM patients showed an enrichment of HIF1A and HIF2A pathways compared with normal donors (Supplemental Figure 1A), we also found an enrichment of both pathways in patients with relapsed and bortezomib-refractory myeloma compared with patients responding to bortezomib (Supplemental Figure 1B).
We next investigated whether hypoxia plays a role in drug resistance in MM. Given the importance of hypoxia in facilitating tumor progression and resistance to chemotherapy in solid tumors, we evaluated the effect of several conventional agents used in the treatment of MM such as dexamethasone, melphalan and bortezomib, either under normoxia (20% O2) or hypoxia (1% O2) in MM cells. Our findings indicate that hypoxia inhibited the effect of bortezomib and melphalan in MM1S (Supplemental Figure 1C), RPMI8226 (Supplemental Figure 1D) and H929 (Supplemental Figure 1E) cells. As hypoxia has been shown to cause cell cycle arrest and this could potentially impact therapeutic responses, we analyzed the effect of hypoxia on cell cycle regulation and apoptosis by flow cytometry. After 24 hours, hypoxia induced G1 arrest (Supplemental Figure 1F) but not apoptosis (Supplemental Figure 1G) in MM cells. However, induction of G1 arrest through serum starvation (Supplemental Figure 1H) did not impact the response to bortezomib in MM1S, RPMI8226 and H929 (Supplemental Figure 1I). Carfilzomib showed similar activity to bortezomib on MM1S, RPMI8226 and H929 cells (Supplemental Figure 1J). Therefore, the drug resistance effect observed with hypoxia was not due to the G1 arrest observed in hypoxic conditions. To further confirm the effect of hypoxia, we pretreated the cells for 12 hours under normoxic or hypoxic conditions and then added the therapeutic agent for an additional 24 hours under both normoxia and hypoxia. We found MM cells responded to bortezomib treatment under normoxic conditions but not under hypoxia even with previous culture in normal oxygen levels (Supplemental Figure 1K).
Metabolic reprogramming delineates a role for drug resistance in myeloma cells
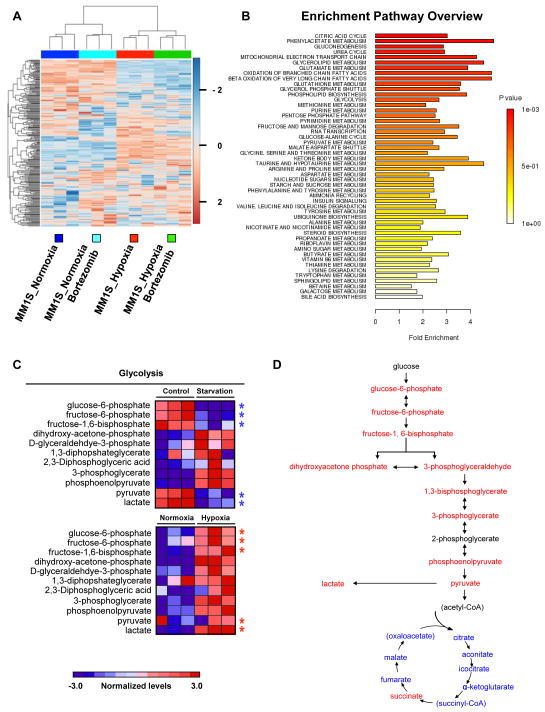
Because one of largest functional groups regulated by HIF1 is associated with glucose metabolism, we hypothesized that changes in cell metabolism could serve as the main factor of drug resistance in MM cells. To examine mechanisms underlying metabolism-associated drug resistance, we performed targeted metabolomic profiling in MM1S cells before and after bortezomib treatment under both normoxic and hypoxic conditions. The metabolic profile of hypoxic cells demonstrated a clear shift when compared with normoxic cells; however cells treated with bortezomib were similar to their non-treated counterparts in both conditions (Figure 1A). In hypoxic cells, intermediates of TCA cycle and mitochondrial electron transport chain were reduced whereas intermediates of glycolysis were elevated (Figure 1B).
Figure 1. Metabolic profiles of MM cells reflect an increase in glycolysis in response to hypoxia.
(A) Unsupervised hierarchical clustering was performed using the 289 metabolites whose intensity varied across the 12 samples. Samples were clustered using Metaboanalyst software. For each metabolite, data were median-centered (white), with the lowest and highest intensity values in blue and red, respectively. (B) Metabolite enrichment pathway overview. (C) Heat map comparing relative levels of metabolites on MM cells in response to serum starvation for 12 hours and to hypoxia for 24 hours. Heat maps show the relative levels of starved cells (Figure 1C, top panel) and hypoxic cells (Figure 1C, bottom panel) in comparison with MM1S-Control-Normoxic cells. (D) Schema illustrating the specific metabolites that are increased (red) or decreased (blue) in MM cells in response to hypoxia after 24 hours.
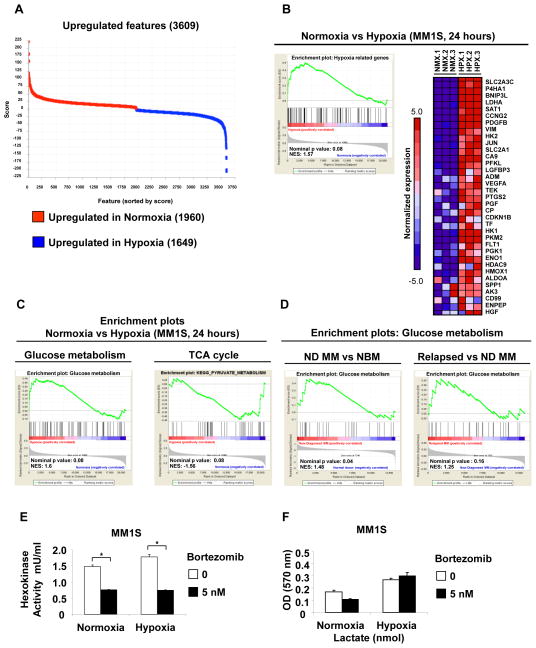
To determine specific metabolic changes are due to hypoxia and not to the G1 cell cycle arrest observed in hypoxic cells, we starved MM1S cells for 12 hours and the metabolic profile of MM1S cells arrested at G1 phase showed a decrease on glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, pyruvate and lactate metabolites (Figure 1C). In contrast to G1 arrested cells, hypoxic cells showed an increase in glycolysis metabolites including pyruvate and lactate (Figures 1C and 1D). To further examine metabolic alterations in hypoxic conditions, we performed GEP on MM1S under normoxic or hypoxic conditions and performed GSEA using the ranked gene list from Dchip (changes in gene expression greater than two-fold with p value lower than 0.05 were considered significant). In order to identify the most significant pathways altered by hypoxia, we compared normoxic samples (n=3) vs hypoxic samples (n=3) and found that after 24 hours of hypoxia 3609 genes were deregulated (1649 genes were upregulated in hypoxia and 1960 genes were upregulated in normoxia) (Figure 2A). The main pathways altered by hypoxic culture were those involved in glucose metabolism (including glycolytic enzymes) (Figure 2B and 2C) and TCA cycle (Figure 2C). We also validated our data on additional cell lines; and showed that similar changes on key metabolic genes (HK2, PFKFB3, PFKFB4 and LDHA) were observed among all the cell lines tested MM1S; H929 and RPMI8226 (Supplemental Figures 2A–C).
Figure 2. Gene expression profiles evidence an increase in glycolytic and lactate enzymes in response to hypoxia.
(A) Gene pattern comparative marker showing upregulated genes in normoxic and hypoxic MM cells, n=3 samples in each condition. (B) Microarray data were analyzed using GSEA software to identify functionally related groups of genes (gene sets) with statistically significant enrichment. The figure shows the enrichment plot and the top 35 enriched genes for Hypoxia related set. (C) GSEA enrichment plots for Glucose metabolism and TCA cycle in Normoxia vs Hypoxia conditions in MM cells. (D) GSEA enrichment plots for Glucose metabolism and TCA cycle in newly diagnosed (ND) patients vs normal bone marrows (NBM) (GSE6477) and in relapsed vs new diagnosed (ND) myeloma patients (GSE9782) (E) Hexokinase activity in MM1S cells treated with bortezomib (5 nM) under normoxic and hypoxic conditions for 24 hours. (F) Lactate levels in MM1S cells treated with bortezomib (5 nM) under normoxic and hypoxic conditions for 24 hours. *p<0.05, ** p<0.005, and *** p<0.0005.
Glucose metabolism gene set was also enriched in the datasets comparing plasma cells from newly diagnosed MM patients with normal donors and in plasma cells from relapsed myeloma patients vs newly diagnosed (ND) MM (Figure 2D).
Importantly, hypoxia regulates expression of HIF1A target genes that are critical for increased glucose uptake and catabolism such us hexokinase II (HK2) and lactate dehydrogenase A (LDHA) (19–21). Both genes were significantly upregulated in our expression datasets (FC=2.1 and FC=5.2, respectively). To test whether bortezomib affects the enzymatic activities of HK2 and LDHA, we measured the hexokinase and lactate activities in cells treated with bortezomib under normoxic and hypoxic conditions. Bortezomib was able to decrease the hexokinase activity even under hypoxic conditions (Figure 2E) but not the lactate activity that was significantly increased under hypoxic conditions (Figure 2F). We found that bortezomib inhibited HK2 activity of MM cells under hypoxic conditions but not lactate activity, suggesting that LDHA may play a role in modulating drug resistance of MM cells in hypoxia.
HIF1A knockdown decreases lactate levels and partially restores the effect of bortezomib under hypoxic conditions
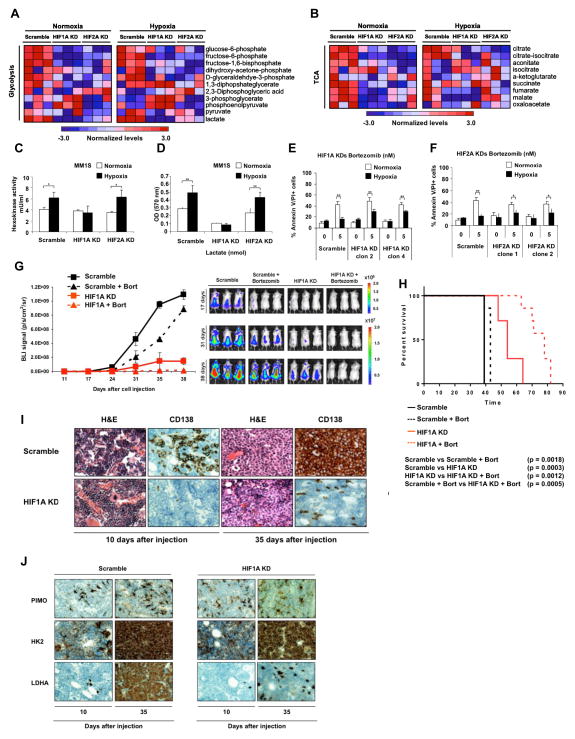
HIF1A is the main pathway to induce glycolysis and lactate production during hypoxia (20,22,23). Under low oxygen conditions, HIF1A is stabilized and promotes transcription of several genes critical for the cellular response to hypoxia (10). We hypothesized that cells lacking HIF1A will fail to upregulate glycolytic enzymes and lactate production in response to hypoxia and will render them more sensitive to chemotherapy. To test this hypothesis, we performed HIF1A and HIF2A-loss of function studies in MM cells. HIF1A and HIF2A knockdown efficiencies were evaluated using qRT-PCR and western blot (Supplemental Figures 3A–C). The metabolic profile of MM1S-HIF1A knockdown cells demonstrated a clear shift away from glycolytic metabolism when compared with MM1S-scramble under normoxic conditions (Figure 3A), and a similar shift occurred under hypoxic conditions (Figure 3A). In MM1S- HIF2A knockdown, intermediates of glycolysis were slightly reduced, but pyruvate and lactate levels increased after hypoxia exposure (Figure 3A). The levels of TCA cycle intermediates decreased in HIF1A and HIF2A knockdowns compared with MM1S-scramble after either normoxia or hypoxia exposures (Figure 3B).
Figure 3. Stable knockdown of HIF1 decreases lactate levels and partially restore the effect of bortezomib under hypoxic conditions.
(A and B) Heat map comparing metabolite patterns of HIF1A and HIF2A knockdowns (KDs) in normoxia and hypoxia. (A) Glycolysis and (B) TCA cycle. Red and blue indicate up- or downregulation, respectively. Scramble, HIF1A and HIF2A knockdowns (n=12) were cultured in 20% O2 (normoxia, NMX) or 1% O2 (hypoxia, HPX) for 24 hours, and metabolites were analyzed by LC-MS. (C) Hexokinase activity of HIF1A and HIF2A knockdowns under normoxic and hypoxic conditions, normalized to scramble in normoxia. (D) Lactate levels in HIF1A and HIF2A knockdowns under normoxic and hypoxic conditions, normalized to scramble in normoxia. (E and F) MM1S scramble, (E) HIF1A and (F) HIF2A knockdowns were incubated with bortezomib (5 and 10 nM) under normoxic or hypoxic conditions for 24 hours and apoptosis was evaluated by Annexin/PI staining and flow cytometric analysis. Graphs and bars represent the mean ± s.d. for cell apoptosis in three independent experiments performed in four replicates. Two clones of each knockdown were tested for apoptosis assays. (G) Mice were intravenously injected with 5 × 106 of MM1S-GFP/luc scramble or MM1S-GFP/luc-shHIF1A. Vehicle or bortezomib were administered IP, weekly, at 0.75mg/kg, starting 10 days after injection and xenograft growth was tested by BLI. (H) Kaplan-Meier curve comparing survival of different groups. (I) Histopathology was performed on different groups 10 and 35 days after injection, H&E and CD138 staining were detected by IHC (J). Histopathology was performed on different groups 10 and 35 days after injection, PIMO, HK2 and LDHA staining were detected by IHC.
To confirm that the HIF1A and HIF2A knockdown metabolites patterns reflected a shift in glycolysis and mainly in lactate production, we measure hexokinase activity and lactate production in both cells under normoxic and hypoxic conditions. As expected, HIF1A knockdown cells showed a reduction in hexokinase activity under hypoxic conditions similar to normoxic levels (Figure 3C) and a significant decrease in lactate production in both normoxic and hypoxic levels compared with the scramble (Figure 3D). Hexokinase and lactate activities did not change in HIF2A knockdown cells (Figures 3C and 3D).
Consistent with the metabolite profile pattern and the reduction of hexokinase and lactate levels, the HIF1A knockdown partially overcomes drug resistance to bortezomib (Figure 3E) and melphalan (Supplemental Figure 3D) after hypoxia exposure. However, HIF2A knockdown did not produce similar results indicating that HIF2A is not essential for the induced drug resistance (Figure 3F).
To determine the in vivo effect of HIF1A loss, we knockdown HIF1A expression in MM1S-GFP/luc. HIF1A knockdown efficiency was evaluated by qRT-PCR (Supplemental Figure 3E). MM1S-GFP/luc-scramble (n=14) and MM1S-GFP/luc-shHIF1A (HIF1A knockdown) (clone 2) (n=14) were injected intravenously into NOD-SCID mice. After 10 days we started the treatment of each group (n=7 per group), 28 mice were treated with vehicle (n=14) or bortezomib (0.75 mg/Kg, weekly, intraperitoneal) (n=14). Tumor growth was measured using luciferin and bioluminescence (BLI).
After 35 days, the scramble group demonstrated significant tumor progression; whereas minimal tumor growth was detected in mice injected with HIF1A knockdown (Figure 3G). Mice groups injected with scramble cells and treated with bortezomib showed a delay in tumor growth, but after 35 days the tumor growth increased reaching similar BLI signal to the control group (Figure 3G). Bortezomib showed a significant effect in the HIF1A knockdown group (Figure 3G), increasing the survival significantly compared with the non-treated group and compared with the control group treated with bortezomib (Figure 3H). Furthermore, mice femurs were collected at day 10 and at day 35; and CD138 expression was analyzed by IHC. In concordance with our BLI data, we observed an increase of CD138+ population with a strong staining in the scramble biopsies whereas the HIF1A knockdown group remained very low even at day 35 (Figure 3I). The hypoxic state of MM cells in the BM was also examined by IP injection of pimonidazole (PIMO) prior to BM isolation and IHC was performed to detect HK2 and LDHA protein levels in tumor cells. Our results demonstrate that the increase of the hypoxic microenvironment induces the overexpression of HK2 and LDHA in plasma cells (Figure 3J).
Loss of LDHA sensitizes MM cells to drug effect
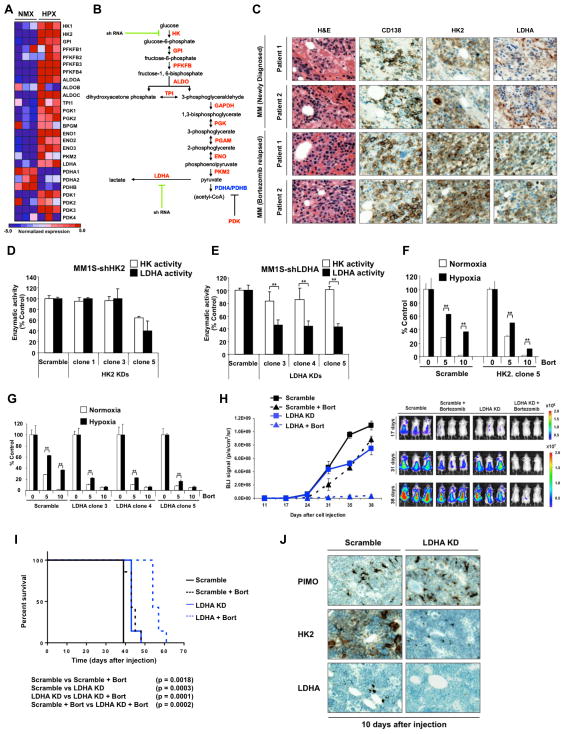
To better define the functional role of glycolysis pathways in drug resistance on MM cells, we explored the expression of the glycolytic enzymes deregulated in our genes expression profile (GEP) (Figure 4A) and performed HK2 and LDHA-loss of function studies on MM1S cells (Figure 4B). HK2 and LDHA knockdown efficiency was evaluated by qRT-PCR (Supplemental Figure 4A–B).
Figure 4. Stable loss of LDHA sensitizes MM cells to bortezomib effect.
(A) Heat map comparing glycolytic enzymes expression in MM1S cells under normoxia or hypoxia. (B) Schematic overview of glycolytic enzymes illustrating the targeted genes. (C) Bone marrow biopsies from MM patients relapsed to bortezomib treatment (N=20) were fixed in Zanker’s formalin, embedded in paraffin blocks, and sectioned. Sections were stained for HK2 and LDHA. (D) Hexokinase activity and lactate production of HK2 knockdowns normalized to the scramble cells. (E) Hexokinase activity and lactate production of LDHA knockdowns normalized to the scramble cells. (F) MM1S scramble and HK2 knockdown were incubated with bortezomib (5 and 10 nM) under normoxic or hypoxic conditions for 24 hours and apoptosis was evaluated by MTT. Graphs and bars represent the mean ± s.d. for cell viability in three independent experiments performed in four replicates. (G) MM1S scramble and LDHA knockdown were incubated with bortezomib (5 and 10 nM) under normoxic or hypoxic conditions for 24 hours and apoptosis was evaluated by MTT. Graphs and bars represent the mean ± s.d. for cell apoptosis in three independent experiments performed in four replicates. Three clones of LDHA knockdown were tested for apoptosis assays. (H) Mice were intravenously injected with 5 × 106 of MM1S-GFP/luc scramble or MM1S-GFP/luc-shLDHA. Vehicle or bortezomib were administered IP, weekly, at 0.75mg/kg, starting 10 days after injection and xenograft growth was tested by BLI. (J) Kaplan-Meier curve comparing survival of different groups. *p<0.05, ** p<0.005, and *** p<0.0005. (I) Histopathology was performed on different groups 10 days after injection. PIMO, HK2 and LDHA staining was detected by IHC.
To confirm that HK2 and LDHA expression are increased in multiple myeloma, we analyzed HK2 and LDHA protein levels by IHC in plasma cells of newly diagnosed and refractory myeloma patients. Out of 20 patient samples, all samples showed high levels of HK2 and LDHA staining in tumor CD138+ plasma cells (Figure 4C). Similarly, GEP of an independent set of CD138+ cells (16,17), revealed that the expression of HK2 and LDHA increased with disease progression (Supplemental Figure 4C).
We evaluated the effect of LDHA and HK2 knockdown on hexokinase activity and intracellular levels of lactate and found that knockdown of HK2 leads to inhibition of both hexokinase activity and lactate (Figure 4D), however LDHA knockdown showed a reduction in lactate but did not affect the cellular hexokinase activity compared with scramble control cells (Figure 4E).
We next examined the effect of bortezomib in HK2 and LDHA knockdowns. MM1S-shHK2 (HK2 knockdown) cells were sensitive to the effect of bortezomib after exposure to hypoxia (Figure 4F) whereas the effect of bortezomib was markedly higher in MM1S-shLDHA (LDHA knockdown) under both normoxic and hypoxic conditions (Figure 4G). We also examined the effect in response to melphalan treatment, and LDHA KD restored the effect of melphalan under hypoxic conditions (Supplemental Figure 4D). Therefore, this effect shows that metabolism-induced drug resistance represents a common mechanism of resistance to drug therapy and not just specific to bortezomib or proteasome inhibition.
LDHA expression as well as pyruvate dehydrogenase kinase 1(PDK1) were found to be upregulated in the GEP data analysis. Both of these enzymes can limit the use of pyruvate as a carbon source for the TCA cycle (24,25). Indeed, our metabolomics data shows an overall decrease in TCA pool sizes under hypoxic conditions. To determine if pyruvate utilization plays a role in drug resistance, we performed gain of function experiments with PDK1 (Supplemental Figure 4E) and examined the effect of bortezomib in PDK1 expressing cells under normoxic conditions. We observed that MM1S-PDK1 expressing cells were resistant to the effect of bortezomib (Supplemental Figure 4F), this effect was similar to that observed in MM1S wild type under hypoxic conditions where PDK1 was upregulated by microarray analysis.
Similarly to our previous in vivo studies, we knocked down LDHA expression in MM1S-GFP/luc. LDHA knockdown efficiency was evaluated by qRT-PCR (Supplemental Figure 4G). MM1S-GFP/luc-shLDHA (LDHA knockdown) (clone 4) (n=14) were injected intravenously into NOD-SCID mice and treatment was initiated after 10 days. Mice were treated with vehicle (n=7) or bortezomib (0.75 mg/kg, weekly, intraperitoneal) (n=7).
After 35 days, the tumor progression was lower in the LDHA knockdown mice compared with their controls and the LDHA knockdown group treated with bortezomib showed a significant delay in tumor growth (Figure 4H). Survival curves showed a significant difference between the scramble and LDHA knockdown groups and bortezomib highly increased the survival percentage of the LDHA knockdown mice compared with the non-treated group and with the scramble treated group (Figure 4I). The observed changes may be due to an impact of HIF1A- and LDHA-silencing on both bone marrow homing/engraftment of MM cells; together with a reduced MM cell growth. These data are indeed supported by the IHC studies where a reduced human-CD138 cell infiltration was documented in those mice that were injected with HIF1A- and LDHA-silenced MM1S cells compared to the related scramble control.
Mice femurs were collected at day 10 and IHC was performed to detect PIMO, HK2 and LDHA levels. In concordance with our in vitro data, even under hypoxic conditions as demonstrated by PIMO staining, LDHA knockdown group showed a lower HK2 expression in plasma cells (Figure 4J).
HIF1A and LDHA expression are associated with drug resistance
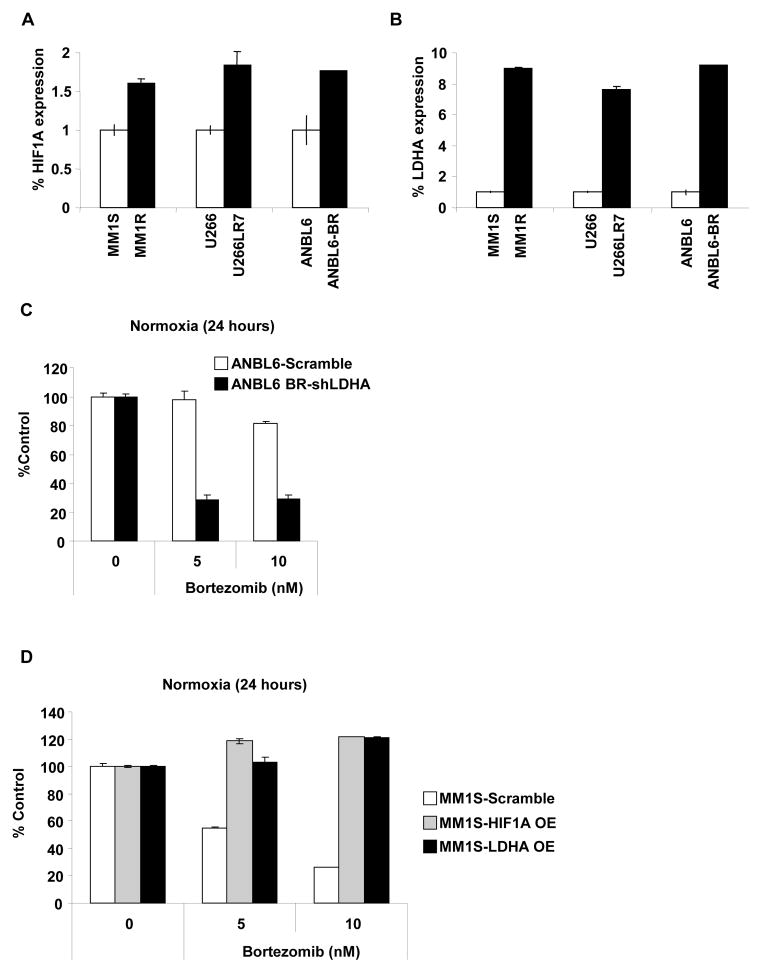
To further confirm the dependence between HIF1A, LDHA and drug resistance, we examined the relative levels of HIF1A and LDHA in a panel of six MM representative cell lines: MM1S and MM1R (sensitive and resistant to dexamethasone), U266 and U266LR7 (sensitive and resistant to melphalan) and ANBL6 and ANBL6-bortezomib-resistant (BR) (sensitive and resistant to bortezomib, respectively).
We found that HIF1A (Figure 5A) and particularly LDHA (Figure 5B) levels are upregulated in all the cell lines resistant to different drugs as compared with their drug sensitive counterparts. To better characterize the role of LDHA in drug resistant to bortezomib, we knocked down LDHA expression in the bortezomib resistance cell line ANBL6-BR (Supplemental Figure 5A) and overexpressed HIF1A and LDHA in MM1S cells (sensitive to bortezomib) (Supplemental Figure 5B–D). In ANBL6-BR, LDHA knockdown restored the sensitivity to bortezomib (Figure 5C) and in MM1S the overexpression (OE) of HIF1A and LDHA induced resistant to bortezomib (Figure 5D).
Figure 5. HIF1A and LDHA expression are associated with drug resistance.
(A–B) Relative mRNA levels of (A) HIF1A and (B) LDHA in MM1S and MM1R, U266 and U266LR7 and ANBL6 and ANBL6-BR cells measured by qRT-PCR. (C) Stable knockdown of LDHA restores the effect of bortezomib in ANBL6-bortezomib resistant. ANBL6 scramble and ANBL6-LDHA knockdown were incubated with bortezomib (5 and 10 nM) for 24 hours and cell viability was evaluated by MTT. Graphs and bars represent the mean ± s.d. for cell viability in three independent experiments performed in four replicates. (D) Stable overexpression of LDHA induces resistance to bortezomib in MM1S cells. MM1S Scramble, HIF1A overexpressed cells and LDHA overexpressed cells were incubated with bortezomib (5 and 10 nM) for 24 hours and cell viability was evaluated by MTT. Graphs and bars represent the mean ± s.d. for cell viability in three independent experiments performed in four replicates.
Taken together, these data suggest that HIF1A and LDHA are important targets for hypoxia-associated drug resistance and the reduction of LDHA increase the effect of bortezomib not only under hypoxic conditions but also in normoxic cells.
DISCUSSION
Targeting cancer energy metabolism has been partly elusive because of the poor understanding of metabolic phenotypes of different cancers. One of the reasons for altered tumor metabolism is the physiological stress that exists within the tumor. The tumor microenvironment suffers from hypoxia and the net result of hypoxia-inducible transcription factors activation is to shift energy production by increasing glycolysis and decreasing mitochondrial function (6,7).
It is well established that hypoxia and consequently HIF1A activation is associated with metastasis in solid tumors and with poor patients outcome (26–28), but the metabolic-induced phenotype driven to drug resistance has been poorly explored mainly because the lack of understanding of cellular responses to inhibition of specific enzymes involved in energy metabolism. In this study we demonstrate that altered metabolism has important implications for tumor cell growth and drug resistance.
We find that increased HIF1A-metabolic related targets such as HK2 and LDHA are present in plasma cells from newly diagnosed MM patients and even more upregulated in relapsed MM patients, highlighting the importance of elevated glucose metabolism in relapsed patients compared with newly diagnosed myeloma cells. This fact is further validated by the finding of a similar response and gene expression pattern in MM cell lines under hypoxic conditions that persist even after bortezomib treatment and by the fact that both HIF1A and LDHA are overexpressed in MM cell lines resistant to different treatments such as dexamethasone, melphalan and bortezomib. Based on this, we hypothesized that metabolic alterations play a crucial role in drug resistance in MM. This is most critical in the minimal residual disease state when cells can survive in a hypoxic bone marrow niche after treatment with bortezomib and high dose melphalan in stem cell transplantation leading to ultimate recurrence of the disease.
In this study, we demonstrate that the increase of glycolytic metabolism results in increased lactate levels in MM cells. Importantly, we show that increased glycolysis is not a consequence of the G1 arrest that cells undergo after hypoxia conditions. Rather, hypoxia actively regulates cellular glucose metabolism by activation of glycolytic enzymes transcription as we demonstrate by GEP analysis and at the functional level by enzymatic assays. Thus, taken together, our study shows that increased glycolysis leads to chemotherapy resistance in MM cells.
The role of dysregulated metabolism in therapeutic resistance has not been examined previously (12,29–34). The ability to reduce chemoresistance through the inhibition of metabolic pathways would be an important research area to improve patient response to therapy. As a central energetic resource for the cell, glucose metabolism is quite complex. Many enzymes contribute to the glycolytic breakdown of glucose. In the glycolytic pathway, the first rate-limiting step is the transport of glucose across the plasma membrane through glucose transporters (GLUT family). GLUT family of proteins are often found upregulated in malignant cells (35). GLUT1 inhibitors, such us WZB117 and phloretin, decrease glucose uptake and display synergistic anticancer effects in lung, colon and breast cancer as well as in leukemia in vitro (36,37). Under hypoxia, the GLUT1 inhibitor, phloretin, significantly enhances daunorubicin effect and overcomes hypoxia-conferred drug resistance (38). Another key rate-limiting enzyme in glycolysis is HK, which has important roles in both glycolysis and apoptosis, and inhibitors of HK, such us 2-deoxyglucose (2-DG), 3-bromopyruvate (3-BrPA) and lonidamine (LND) are in pre-clinical and early phase clinical trials (31). Although there are several reports showing that HK inhibitors enhance the drug response in vitro under normoxic conditions (39–41), there are no ongoing trials with HK inhibitors as a single agent due to the lack of response in vivo (42). Consistent with these data, we did not observe any effect in MM cells treated with 2-DG, 3-BrPA or LND under hypoxic conditions or in our in vivo model (data not shown). These findings together with the fact that bortezomib was able to decrease HK2 activity under hypoxic conditions and that our HK2 knockdown revealed a decrease in LDHA activity led us to hypothesize that the main enzyme to target in our hypoxia-drug resistant model was LDHA. This hypothesis is further supported by the fact that both HIF1A and LDHA overexpression lead to drug resistance in MM1S while LDHA knockdown sensitizes ANBL6 bortezomib-resistant cells to the effect of bortezomib. LDHA catalyzes the final step in the glycolytic pathway and has a critical role in tumor maintenance. LDHA contributes to paclitaxel/trastuzumab resistance in breast cancer (33,34) and knockdown of LDHA increased mitochondrial respiration, decreased cellular ability to proliferate under hypoxic conditions, and suppressed tumorigenicity in tumor cells (43). In line with this data, our LDHA knockdown improves the effect of bortezomib in both normoxic and hypoxic conditions and showed a significant effect in our in vivo model.
Our in vitro experiments demonstrate that stable inhibition of HIF1A in MM cells decreased proliferation and sensitized cells to the therapeutic effect of bortezomib. Stable depletion of HIF1A inhibited glycolysis and decreased lactate levels and this reduction of lactate remained at low levels even after the exposure to hypoxia. Reduction of HK2 and LDHA expression also increased the effect the bortezomib under hypoxic conditions, however due to the LDHA reduction observed in our stable HK2 knockdown and due to the fact that hexokinase activity was also decreased in cells treated with bortezomib under hypoxic conditions; we proposed that LDHA could be one of the main targets to overcome drug resistance induced through hypoxia. Depletion of HIF1A and LDHA in MM cells also restored drug sensitivity to therapeutic agents such as bortezomib in vivo. MM tumors with HIF1A knockdown clearly demonstrated a proliferative disadvantage compared with the scramble-control tumors and a significant increase of survival in the bortezomib treated group. LDHA knockdown significantly improved the response to bortezomib treatment; however the overall survival in the non-treated group was similar to the scramble non-treated group. A possible explanation of the decreased proliferation in MM tumor with HIF1A knockdown may relate to the downregulation of the entire HIF1A pathway that involves not only the effect on therapeutic response but also a decrease of proliferation and angiogenesis (28,44,45).
In summary, our studies reveal that regulation of tumor cell metabolism in MM cells is essential to target drug resistance in MM. We show that the metabolic phenotype in MRD cells induced by hypoxia leads to drug resistance through HIF1A and that specific regulation of HIF1A or LDHA can restore sensitivity to therapeutic agents such as bortezomib and melphalan and can also inhibit tumor growth induced by altered metabolism.
Supplementary Material
Acknowledgments
Supported in part by EHA (European Hematology Association), SPORE P50 CA100707, NIH R01CA154648 and Leukemia and Lymphoma Society funding.
Footnotes
Conflicts of Interest
Irene M. Ghobrial is on the advisory board of Millennium, Celgene, and Onyx.
Author Contributions
P.M. and I.M.G. conceived of and designed the experiments, analyzed data, and wrote the manuscript. P.M., D.H. and M.M. designed and performed the in vitro and in vivo functional experiments. A.S., Y.A. and Y.M. performed in vitro assays. J.M.A. performed metabolite profiling. A.M.R. and A.C.K. revised the paper.
References
- 1.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature reviews Cancer. 2004;4(11):891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 2.Racker E. History of the Pasteur effect and its pathobiology. Mol Cell Biochem. 1974;5(1–2):17–23. doi: 10.1007/BF01874168. [DOI] [PubMed] [Google Scholar]
- 3.Semenza GL, Artemov D, Bedi A, Bhujwalla Z, Chiles K, Feldser D, et al. ‘The metabolism of tumours’: 70 years later. Novartis Found Symp. 2001;240:251–60. discussion 60–4. [PubMed] [Google Scholar]
- 4.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–70. [PubMed] [Google Scholar]
- 5.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8(6):519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nature reviews Cancer. 2008;8(9):705–13. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 7.Milosevic M, Fyles A, Hedley D, Hill R. The human tumor microenvironment: invasive (needle) measurement of oxygen and interstitial fluid pressure. Seminars in radiation oncology. 2004;14(3):249–58. doi: 10.1016/j.semradonc.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3(3):e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vengellur A, Woods BG, Ryan HE, Johnson RS, LaPres JJ. Gene expression profiling of the hypoxia signaling pathway in hypoxia-inducible factor 1alpha null mouse embryonic fibroblasts. Gene Expr. 2003;11(3–4):181–97. doi: 10.3727/000000003108749062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Molecular cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–6. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nature reviews Cancer. 2010;10(4):267–77. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 13.Maiso P, Carvajal-Vergara X, Ocio EM, Lopez-Perez R, Mateo G, Gutierrez N, et al. The histone deacetylase inhibitor LBH589 is a potent antimyeloma agent that overcomes drug resistance. Cancer research. 2006;66(11):5781–9. doi: 10.1158/0008-5472.CAN-05-4186. [DOI] [PubMed] [Google Scholar]
- 14.Maiso P, Liu Y, Morgan B, Azab AK, Ren P, Martin MB, et al. Defining the role of TORC1/2 in multiple myeloma. Blood. 2011;118(26):6860–70. doi: 10.1182/blood-2011-03-342394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: the Pathway Interaction Database. Nucleic acids research. 2009;37(Database issue):D674–9. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer research. 2007;67(7):2982–9. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 17.Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109(8):3177–88. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 18.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. Journal of chromatography A. 2007;1147(2):153–64. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 19.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer cell. 2011;19(3):416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer research. 2011;71(22):6921–5. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- 21.Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer cell. 2013;24(2):213–28. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheaton WW, Chandel NS. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol. 2011;300(3):C385–93. doi: 10.1152/ajpcell.00485.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nature reviews Cancer. 2011;11(6):393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 24.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 25.Ader I, Brizuela L, Bouquerel P, Malavaud B, Cuvillier O. Sphingosine kinase 1: a new modulator of hypoxia inducible factor 1alpha during hypoxia in human cancer cells. Cancer research. 2008;68(20):8635–42. doi: 10.1158/0008-5472.CAN-08-0917. [DOI] [PubMed] [Google Scholar]
- 26.Shin DH, Chun YS, Lee DS, Huang LE, Park JW. Bortezomib inhibits tumor adaptation to hypoxia by stimulating the FIH-mediated repression of hypoxia-inducible factor-1. Blood. 2008;111(6):3131–6. doi: 10.1182/blood-2007-11-120576. [DOI] [PubMed] [Google Scholar]
- 27.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440(7088):1222–6. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 28.Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer cell. 2009;15(1):35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birsoy K, Sabatini DM, Possemato R. Untuning the tumor metabolic machine: Targeting cancer metabolism: a bedside lesson. Nat Med. 2012;18(7):1022–3. doi: 10.1038/nm.2870. [DOI] [PubMed] [Google Scholar]
- 30.Dang CV, Hamaker M, Sun P, Le A, Gao P. Therapeutic targeting of cancer cell metabolism. Journal of molecular medicine. 2011;89(3):205–12. doi: 10.1007/s00109-011-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Mjiyad N, Caro-Maldonado A, Ramirez-Peinado S, Munoz-Pinedo C. Sugar-free approaches to cancer cell killing. Oncogene. 2011;30(3):253–64. doi: 10.1038/onc.2010.466. [DOI] [PubMed] [Google Scholar]
- 32.Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med. 2012;209(2):211–5. doi: 10.1084/jem.20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP, Wilson GL, et al. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer research. 2011;71(13):4585–97. doi: 10.1158/0008-5472.CAN-11-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou M, Zhao Y, Ding Y, Liu H, Liu Z, Fodstad O, et al. Warburg effect in chemosensitivity: targeting lactate dehydrogenase-A re-sensitizes taxol-resistant cancer cells to taxol. Mol Cancer. 2010;9:33. doi: 10.1186/1476-4598-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. Journal of cellular physiology. 2005;202(3):654–62. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H, et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Molecular cancer therapeutics. 2012;11(8):1672–82. doi: 10.1158/1535-7163.MCT-12-0131. [DOI] [PubMed] [Google Scholar]
- 37.Monti E, Gariboldi MB. HIF-1 as a target for cancer chemotherapy, chemosensitization and chemoprevention. Curr Mol Pharmacol. 2011;4(1):62–77. doi: 10.2174/1874467211104010062. [DOI] [PubMed] [Google Scholar]
- 38.Cao X, Fang L, Gibbs S, Huang Y, Dai Z, Wen P, et al. Glucose uptake inhibitor sensitizes cancer cells to daunorubicin and overcomes drug resistance in hypoxia. Cancer chemotherapy and pharmacology. 2007;59(4):495–505. doi: 10.1007/s00280-006-0291-9. [DOI] [PubMed] [Google Scholar]
- 39.Kurtoglu M, Gao N, Shang J, Maher JC, Lehrman MA, Wangpaichitr M, et al. Under normoxia, 2-deoxy-D-glucose elicits cell death in select tumor types not by inhibition of glycolysis but by interfering with N-linked glycosylation. Molecular cancer therapeutics. 2007;6(11):3049–58. doi: 10.1158/1535-7163.MCT-07-0310. [DOI] [PubMed] [Google Scholar]
- 40.Maher JC, Krishan A, Lampidis TJ. Greater cell cycle inhibition and cytotoxicity induced by 2-deoxy-D-glucose in tumor cells treated under hypoxic vs aerobic conditions. Cancer chemotherapy and pharmacology. 2004;53(2):116–22. doi: 10.1007/s00280-003-0724-7. [DOI] [PubMed] [Google Scholar]
- 41.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25(34):4633–46. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 42.Maschek G, Savaraj N, Priebe W, Braunschweiger P, Hamilton K, Tidmarsh GF, et al. 2-deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo. Cancer research. 2004;64(1):31–4. doi: 10.1158/0008-5472.can-03-3294. [DOI] [PubMed] [Google Scholar]
- 43.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer cell. 2006;9(6):425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 44.Martin SK, Diamond P, Gronthos S, Peet DJ, Zannettino AC. The emerging role of hypoxia, HIF-1 and HIF-2 in multiple myeloma. Leukemia. 2011;25(10):1533–42. doi: 10.1038/leu.2011.122. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Li M, Yao Q, Chen C. Recent advances in tumor hypoxia: tumor progression, molecular mechanisms, and therapeutic implications. Medical science monitor : international medical journal of experimental and clinical research. 2007;13(10):RA175–80. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.