Abstract
Muscle-invasive urothelial carcinomas of the bladder (MIUCB) exhibit frequent receptor tyrosine kinase alterations but the precise nature of their contributions to tumor pathophysiology is unclear. Using mutant HRAS (HRAS*) as an oncogenic prototype, we obtained evidence in transgenic mice that RTK/RAS pathway activation in urothelial cells causes hyperplasia that neither progresses to frank carcinoma nor regresses to normal urothelium through a period of one year. This persistent hyperplastic state appeared to result from an equilibrium between pro-mitogenic factors and compensatory tumor barriers in the p19-MDM2-p53-p21 axis and a prolonged G2 arrest. Conditional inactivation of p53 in urothelial cells of transgenic mice expressing HRAS* resulted in carcinoma-in-situ and basal-subtype MIUCB with focal squamous differentiation resembling the human counterpart. The transcriptome of microdissected MIUCB was enriched in genes that drive epithelial-mesenchyme transition, the upregulation of which is associated with urothelial cells expressing multiple progenitor/stem cell markers. Taken together, our results provide evidence for RTK/RAS pathway activation and p53 deficiency as a combinatorial theranostic biomarker which may inform the progression and treatment of urothelial carcinoma.
Keywords: oncogenic HRAS, p53, epithelial-mesenchymal transition, progenitor cells, invasive urothelial carcinoma subtype
Introduction
Muscle-invasive urothelial carcinoma of the bladder (MIUCB) is amongst the most aggressive and deadliest cancers (1). Due to its high risk of progression to metastatic stages, MIUCB often calls for multi-agent neoadjuvant chemotherapy followed by radical cystectomy or adjuvant chemotherapy after the surgery or radiotherapy concomitant with systemic chemotherapy (2,3). Despite such debilitating therapies, over 50% of MIUCB advance to local and distant metastasis at which point the 5-year survival rates are only about 30% and 5%, respectively (1).
A significant recent development is the recognition that MIUCB is not a single disease entity but comprises distinct subtypes distinguishable by combinatorial molecular signatures and divergent clinical outcomes (4-11). While the exact number, interrelationship and spectra of the molecular signatures between different subtypes from different studies remain to be delineated, a consensus is emerging pointing to at least two major subtypes: luminal and basal. The luminal subtype bears features of the luminal umbrella cells of normal urothelium, e.g., high-levels of uroplakins, cytokeratin 20 and E-cadherin (4-6,9-11). Mutations of fibroblast growth factor 3 (FGFR3) and tuberous sclerosis 1 (TSC1) are prevalent along with alterations involving many other genes. The basal subtype, on the other hand, expresses abundant proteins associated with the basal cells of normal urothelium, such as cytokeratins 14, 5 and 6B as well as markers signifying increased stemness and epithelial-mesenchymal (EMT) transition a (e.g., high CD44, TWIST1/2, SNAI2, ZEB2, VIM and N-cadherin and low E-cadherin and claudin) (4-6,9,10). Focal squamous differentiation is common in this subtype and, as suspected, the basal subtype is much more aggressive and correlates with more advanced stage and poorer prognosis than is the luminal subtype (4,8,10). Notably, the frequency of p53 mutations that characterize MIUCB in general does not differ significantly between the two major subtypes, although one study found RB1 pathway alterations to be more prevalent in the basal subtype than in the luminal subtype (5).
Notwithstanding the recent progress in subtyping MIUCB, several critical issues remain. First and foremost, are different subtypes of MIUCB caused by distinct genetic drivers? Thus far, most sub-classification studies are based on expression signatures including those of uroplakins, cytokeratins and cadherins (9-11), which are not genetic tumor drivers but phenotypic consequences of urothelial differentiation vis-à-vis de-differentiation. Those making use of gene mutations for sub-classification often involve multiple alterations (8) whose relationship with a particular subtype remains correlative. A definitive cause-consequence effect between a minimum essential set of genetic drivers and a given subtype requires experimental verification using biologically relevant systems. Such biological studies are important because defining the genetic driver(s) could not only simplify the subtyping of MIUCB and reduce the number of prognosticators, but also narrow down druggable targets for precise therapeutic intervention (12). Second, do different subtypes of MIUCB progress via divergent phenotypic pathways? Clinicopathological studies have long held that MIUCB can (i) arise de novo (i.e., without a defined precursor), (ii) progress from flat, carcinoma-in-situ (CIS) precursor lesions, or (iii) progress from high-grade, non-invasive papillary urothelial carcinomas (13-16). It is crucially important to determine whether some of the MIUCB subtypes are actually a result of tumor progression from a particular premalignant lesion, so that specific strategies can be devised to predict and prevent progression. Third, do different MIUCB subtypes originate from different normal urothelial cell types? Normal urothelium can be divided into at least three different compartments: basal, intermediate and luminal (17). While all urothelial carcinomas were previously thought to derive from the normal urothelial stem cells residing in the basal zone, recent studies suggest otherwise (16,18). In particular, chemical carcinogenesis employing a bladder-specific carcinogen, N-butyl-N-(4-hydroxybutyl)nitrosamine (BBN), coupled with lineage tracing, suggests that low-grade non-invasive and high-grade MIUCB originate from intermediate and basal compartments, respectively (19,20). It remains an open question, however, as to whether the different subtypes within MIUCB can also originate from different normal urothelial subtypes. Finally, are different MIUCB subtypes molecularly and phenotypically static or are they quite dynamic and interchangeable reflecting different stages of de-dedifferentiation and tumor progression? In other words, could the luminal subtype de-differentiate and transition into the basal subtype during the course of tumor progression? Conversely, could the basal subtype re-gain the ability to differentiate into the luminal subtype thus becoming less aggressive subsequent to radio- and/or chemo-therapy?
To begin to tackle some of these questions, we took an in-depth look of the effects of HRAS activation and p53 deficiency using a blend of in vitro and in vivo approaches. Activation of the RTK/RAS pathway and inactivation of the p53 pathway, events that were previously thought to define low-grade non-invasive and high-grade MIUCB, respectively (13,21,22), were recently found in whole-genome analyses to be equally prevalent in high-grade MIUCB (72% with RTK/RAS activation and 76% with p53 pathway activation) (23). This suggests that alterations affecting both signaling pathways could overlap, simply by chance, in at least 50% of the MIUCB. One scenario is that this overlap is merely due to genetic drifting of two common events that do not necessarily cross-talk and are of no consequence to tumorigenesis. Another scenario is that these two events functionally converge as a result of selective pressure in tumor cells and that they collaborate or even synergize to exert a tumor-driving role leading to the formation of MIUCB. In this study, we examine these two competing hypotheses and our results have important implications on the molecular pathogenesis of MIUCB and shed light on how some of the MIUCB subtypes can be better managed clinically.
Materials and Methods
Transgenic, Knockout and Compound Mice
The transgenic mouse line, Upk2-HRAS*, harbored a single-copy transgene comprising a 3.6-kB murine uroplakin II promoter (UPII) and a constitutively active HRAS gene (24). The urothelial expression level of the HRAS* in this low-copy Upk2-HRAS* line is equivalent to that of the endogenous wild-type Ras, as evidenced by Real-time PCR and Western blotting (24). The second transgenic line, Upk2-cre harbored a transgene comprising the UPII and a 1.4-kB cre recombinase gene (25). The third transgenic line harbored a “floxed” p53 allele (e.g., p53LOX) where loxP sites were inserted in introns 4 and 6, allowing deletion of exons 5 and 6 upon cre expression (26). The identity of Upk2-HRAS* and Upk2-cre were verified by Southern blotting and that of p53LOX by genomic PCR. Intercrosses were carried out among these three lines with additional crosses among their offspring, yielding a number of genotypes, from which four major genotypes were chosen for phenotypic characterization: (i) Upk2-cre (as negative control), (ii) Upk2-HRAS*/WT, (iii) Upk2-cre/p53LOX/LOX and (iv) Upk2-HRAS*/WT/Upk2-cre/p53LOX/LOX. All animal experiments were approved by Institutional Animal Care and Use Committee.
Laser-capture Microdissection (LCM) and Expression Arrays
Since urinary bladders of Upk2-cre mice exhibited normal urothelia and those of Upk2-HRAS*/WT/Upk2-cre/p53LOX/LOX compound mice exhibited CIS and muscle-invasive lesions, these bladders were used for cross-sectioning and LCM. Briefly, 30 μm-thick frozen sections were lightly stained with hemotoxylin and the aforementioned lesions were dissected out using Leica LMD6000 Laser Micro-Dissection System. Total RNAs were extracted using RNeasy Micro Kit (Qiagen, Valencia, CA) and the RNA quality was verified by HPLC. Microarray was carried out with Affymetrix 3′ IVT mouse expression arrays at our in-house facility (GEO accession number: GSE64756). Primary data were analyzed at the Center for Applied Genomics in University of Medicine and Dentistry of New Jersey and pathway and bioprocess analyses were performed online using Ingenuity® iReport™.
Cell Culture, Transfection and Establishment of Stable Lines
Human bladder urothelial carcinoma cell line, RT4, originally isolated from a low-grade, non-invasive urothelial carcinoma (27), was purchased from American Type Cell Culture (ATCC), maintained in McCoy’s 5A medium containing 10% fetal bovine serum and used within 6 months of receipt. Authentication of RT4 at ATCC employed short tandem repeat profile and isoenzyme analysis. An sh-RNA of mouse p53 (5′-gactccagtggtaatctact-3′) was sub-cloned into retroviral vector, pMKO.1-puro (Addgene, Cambridge, MA) and the resultant pMKO.1-puro/sh-p53 was co-transfected with pCL-10A1 packaging vector (Novus Biologicals) into cultured Phoenix cells. The packaged virus in the supernatant was collected and used to infect RT4 cells. Following a 10-day selection in culture medium containing 100 μg/ml puromycin, survived single clones were verified for p53 knockdown. HRASWT and HRASV12 were sub-cloned separately into retroviral vector, pBABE-hygro (Addgene), and co-transfected with the pCL-10A1 packaging vector into the Phoenix cells. The packaged retroviruses were isolated and infected into RT4 cells stably expressing the sh-RNA-p53. Stable clones was selected in culture medium containing 200 μg/ml hygromycin for 10 days and the resultant stable clones were verified for desired gene expression.
Cell Migration and Invasion Assays
Cell migration of stable cell lines was first compared by wound healing assay. When cultured cells reached 80% confluence, wounds were introduced under an inverted microscope using a sterile pipette tip. Wounded cells were cultured in fresh medium for 3 days before phase-contrast images were recorded. For invasion assay, BioCoat Matrigel Invasion Chamber (BD Biosciences, San Jose, CA) was employed. Briefly, stable clones (2.5 × 104 cells) were seeded in 24-well chambers (in triplicate) containing 20 ng/ml 12-O-tetradecanoylphorbol-13-acetate. After incubation for 72 h, the non-invading cells atop the membrane were removed by scrapping and, the invading cells underneath the membrane were visualized using Diff-Quik stain and counted in five high-power (200 ×) microscopic fields (one-center and four-peripheral).
Cell Proliferation Assay
Stably transfected cells (2×103/well) were cultured for 48 h and quantified by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide) method (Bio-Rad, Hercules, CA).
Cell Cycle Analysis
Urinary bladders were inverted to expose the mucosa. After incubation in a solution containing 1 mg/ml dispase at 4°C overnight, the urothelial cells were gently scraped off and digested with a solution containing 0.25% Trypsin-EDTA at 37°C for 30 min. The cells were washed in PBS by centrifugation at 800 g for 5 min and filtered through a 100 μm pore-size filter, fixed with pre-cooled 70% ethanol at 4°C and stained with 40 μg/ml propidium iodide containing 100 μg/ml RNase. Cell sorting was carried out using Facscan (Beckman) and the data were analyzed using ModiFit 3.2 (Verity Software House).
Quantitative Real-time PCR
Total RNA was isolated from bladder urothelia using RNeasy Mini Kit (Qiagen) and 2 μg of it was used for cDNA synthesis using High Capacity cDNA Reverse Transcription Kit (Applied Biosystem, Grand Island, NY). Real-time PCR was carried out with 7500 System (Applied Biosystems) under 95°C for 15′ for the first cycle; 95°C for 15″, 58°C for 20″ and 72°C for 30″ for 50 cycles, and 72°C for 5′ for the last cycle. PCR products were quantified by direct SYBR Green incorporation, with the relative abundance expressed as ratios to β-actin. The primers were: p19ARF-forward: gtcgcaggttcttggtcact, p19ARF-reverse: cgaatctgcaccgtagttga; p53-forward: agagaccgccgtacagaaga, p53-reverse: ctgtagcatgggcatccttt; p21-forward: cggtggaactttgacttcgt, p21-reverse: cagggcagaggaagtactgg.
Western Blotting, Histological, Immunohistochemical and Immunofluorescent Staining
Total proteins from mouse urothelia or cultured RT4 cells were dissolved in a lysis buffer (10% SDS, 20 mM Tris/HCl (pH7.5), 50 mM NaCl, 5 mM β-mercaptoethanol and a mixture of protease inhibitors). After SDS-PAGE, the proteins were transferred onto PVDF membrane and reacted consecutively with primary (Supplemental Table S1) and peroxidase-conjugated secondary antibodies.
Freshly dissected urinary bladders were fixed in PBS-buffered 10% formalin and embedded routinely in paraffin. Four μm thick sections were stained with H&E for histological examination. For immunohistochemistry, de-paraffinized sections were microwaved in a citrate buffer (pH 6.0) for 20 min to unmask the antigens and then incubated with primary (Supplemental Table S1) and secondary antibodies conjugated with horseradish peroxidase.
Statistical Analysis
Student’s t test (two-tailed) was used to evaluate the statistical significance between Upk2-HRAS* mice and wild-type (Upk2-cre) mice in urothelial expression of p53 pathway components and between different groups of stably transfected cultured cells in their proliferation and invasion rates, with P value <0.05 considered statistically significant.
Results
Oncogenic HRAS*-induced Persistent Urothelial Hyperplasia Results from an Equilibrium between Mitogenic Signals and Anti-tumor Defenses
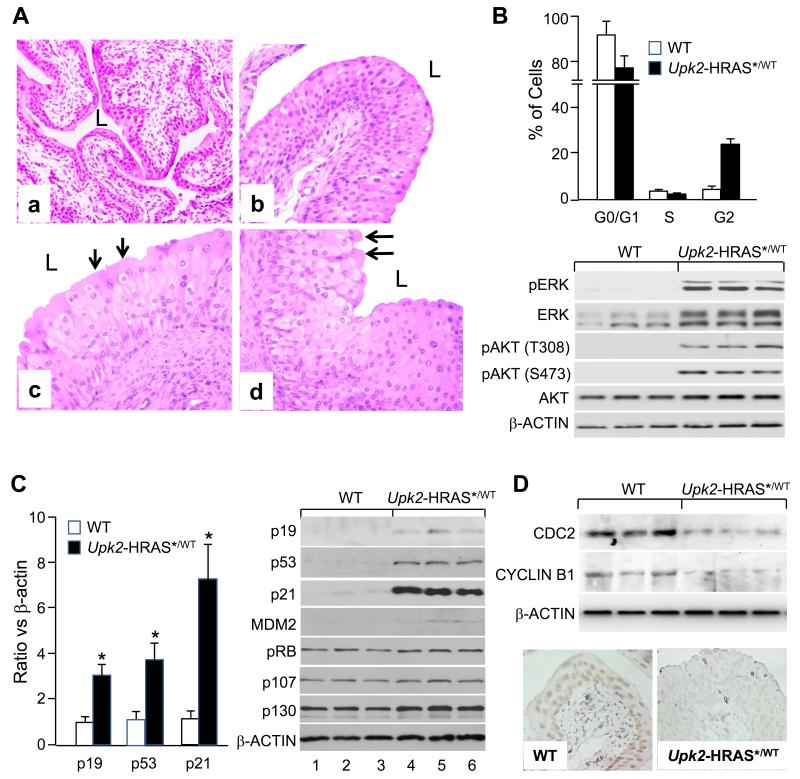
A highly reproducible phenotype in transgenic mice bearing a single copy of oncogenic HRAS* under the control of the UPII promoter (Upk2-HRAS*) was the persistent urothelial hyperplasia (24). Compared to normal urothelium from the wild-type littermates (Fig. 1A/a), the hyperplastic lesions of the Upk2-HRAS* mice appeared as highly thickened, nonetheless well-differentiated urothelia with excellent polarity (Fig. 1A/b-d) and retention of superficial umbrella cells (arrows in Fig. 1A/c and 1A/d); and they started around 2 months of age and persisted through 12 months, without progressing, in grade or stage, to full-fledged urothelial tumor or reverting to normal urothelium. To understand the molecular underpinning of this phenomenon, we examined the cell-cycle status and found that at the steady state there was a significant reduction of G0/G1 urothelial cells and increase of G2 cells in Upk2-HRAS* transgenic mice (12 months of age), as compared with the wild-type controls (Fig. 1B, upper panel). This corresponded well with elevated mitogenic signals including phosphorylated ERK and AKT (both T308 and S473) in the transgenic mice (Fig. 1B lower panel; Supplemental Fig. S1). However, S-phase cells were not significantly higher (Fig. 1B), suggesting that the DNA synthesis was held in check and that a prolonged G2 arrest existed, possibly due to concurrent induction of growth inhibitors/tumor suppressors (28). Of the tumor-suppressive pathways surveyed, that of p53, including p19, p53 and p21, exhibited marked upregulation on mRNA (Fig. 1C, left panel) and protein (Fig 1C, right panel) levels. Such overt upregulation was not observed in pRB family proteins (e.g., pRB, p107 and p130). Interestingly, factors key to promoting G2/M transition such as CDC2 and CYCLIN B1 were significantly down-regulated in the hyperplastic lesions of the Upk2-HRAS* mice (Fig. 1D), a phenomenon observed in non-urothelial cells with an upregulated p53 pathway (28). Our results suggest that oncogenic HRAS*-triggered proliferative forces are counter-balanced by anti-proliferative forces, especially by the p53 signaling axis, thus reaching an equilibrium and resulting in a non-progressive and non-regressive state of persistent urothelial hyperplasia, that is quite different from oncogenic RAS-induced premature senescence and apoptosis in primary-cultured cells (29).
Figure 1.
Simultaneous induction of pro- and anti-mitogenic signals in transgenic mice expressing oncogenic HRAS* in urothelia, leading to persistent non-progressive hyperplasia. Urothelia from Upk2-HRAS*/WT transgenic mice and wild-type littermates were subject to (A) histopathology, (B, upper panel) flow cytometry, (B, lower panel) Western blotting of key RAS-downstream effectors, (C, left panel) Real-time PCR of p53 pathway components, (C, right panel) Western blotting of key tumor suppressors, and (D, upper panel) Western blotting and (D, lower panel) immunohistochemical staining of promoters of G2/M transition CDC2 and CYCLIN B1. Note that, the WT mouse (12-months old (A-a)) exhibited normal urothelium, whereas the Upk2-HRAS*/WT transgenic mice (A-b, A-c, and A-d, 4-, 8- and 12-months, respectively) exhibited urothelial hyperplasia. Note the reduced G0/G1 and increased G2 phase (B, upper panel) and elevated pERK and pAKT (B, lower panel) in the transgenic mouse urothelia as compared with the WT controls. Note the overexpression of p19, p53 and p21 on both mRNA (C, left panel) and protein level (C, right panel) and largely unchanged pRB family proteins in the transgenic mice. Also note the reduced levels of CDC2 and CYCLIN B1 in the Upk2-HRAS*/WT transgenic mice (D). Magnification: 200 × in (A) and (D).
Removal of p53 Confers Invasive Property to Cultured Non-invasive Urothelial Tumor Cells Expressing Oncogenic HRAS*
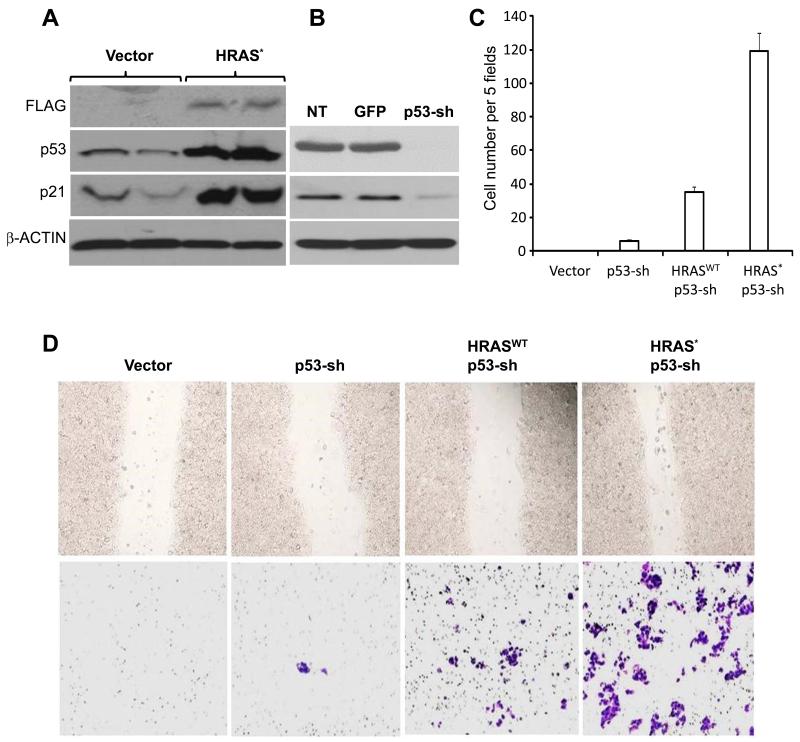
To determine whether the tumor-barrier effects of p53 upregulation by oncogenic HRAS* in urothelial cells were coincidental or causative, we introduced oncogenic HRAS* along with sh-RNA of p53 into cultured RT4 cells, which were originally derived from a human low-grade, superficial papillary bladder tumor (27) and lacked RAS or p53 mutation/deletion (30). Enforced expression of oncogenic HRAS* in RT4 elicited a marked upregulation of p53 and p21 (Fig. 2A). Knockdown of p53 or that along with the expression of a wild-type HRAS enhanced cell proliferation (Supplemental Fig. S2), but only slightly increased cell migration and invasion (Fig. 2C). In contrast, knocking down p53 and expressing an oncogenic HRAS* resulted in a dramatic increase of cell migration and invasion of RT4 cells (Fig. 2C and 2D). Thus, p53 deficiency and RAS activation appear to be synergistic in conferring the invasive property to human urothelial tumor cells and triggering the conversion of non-invasive human urothelial tumor cells into invasive ones.
Figure 2.
Combined effects of oncogenic HRAS* and p53 deficiency on cultured urothelial cells. (A, left panel) Western blotting showing that cultured RT4 cells stably transfected with FLAG-tagged HRAS* overexpress p53 and p21, compared with vector-only transfected cells. (B) Western blotting showing that RT4 cells stably transfected with sh-RNA of p53 had marked decrease of p53 itself and p21. RT4 cells stably transfected with Sh-RNA of green fluorescent protein (GFP) served as a negative control; NT stands for no transfection. (C) Cell invasion assay using Matrigel (invasive cells counted per 5 high-powered (200 ×) fields microscopically), showing a very small increase of invasive cells in p53-knockdown cells, a moderate increase in p53-knockdown/WT-HRAS co-expressing cells and a marked increase in p53-knockdown/HRAS* co-expressing cells. (D) Microscopic images of wound healing (upper panel) and Matrigel-invasion (lower panel) experiments showing marked increase of both cell migration and invasion in cells co-expressing HRAS* and p53-shRNA.
Conditional Compound Mice Expressing Oncogenic HRAS* and Lacking p53 Develop High-grade, Muscle-invasive Urothelial Carcinoma
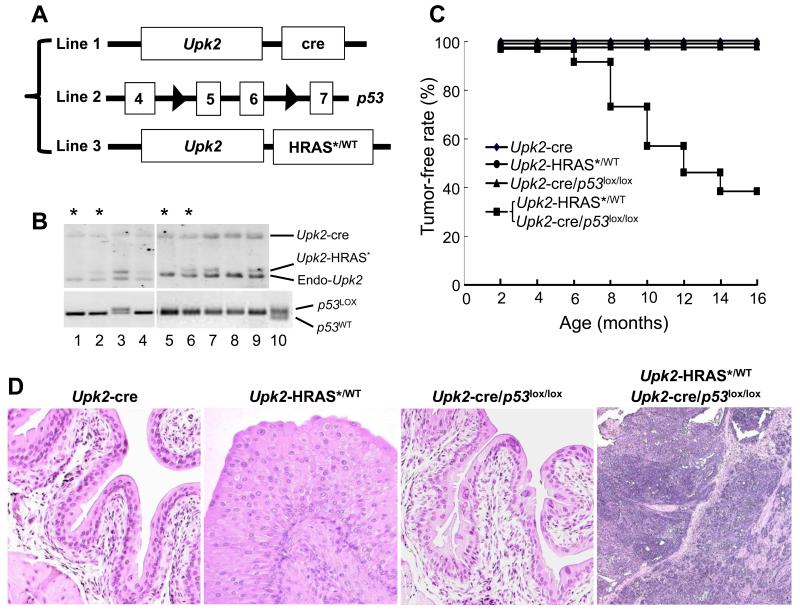
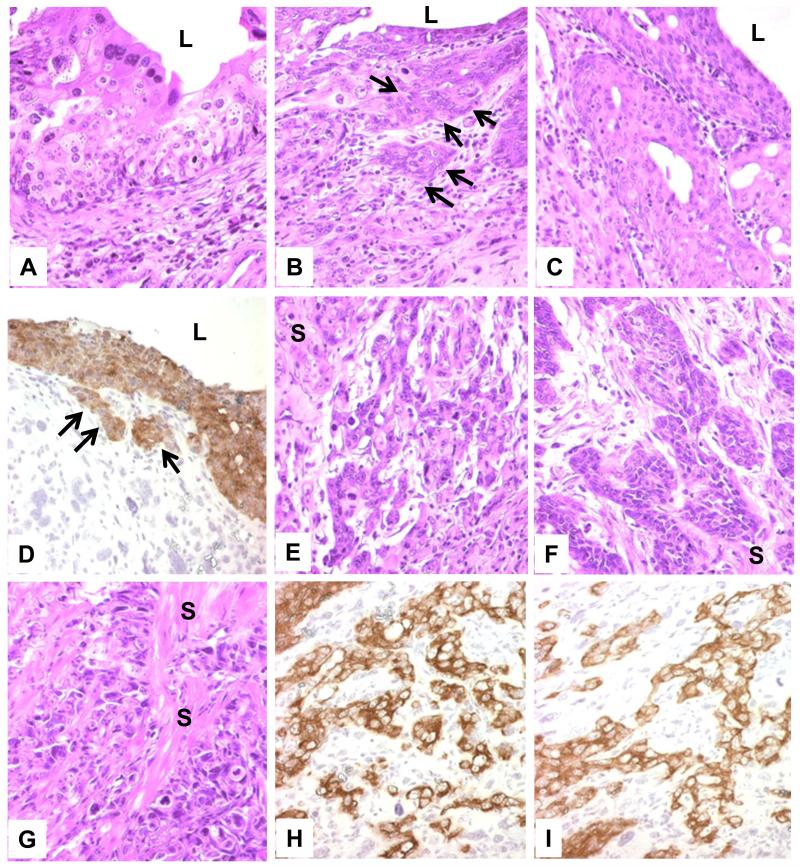
To further define the interactive effects between oncogenic HRAS* and p53 deficiency in vivo, we developed compound mice by ablating p53 from urothelial cells expressing oncogenic HRAS*. To do so, we cross-bred three independent mouse lines: Upk2-HRAS* (24), Upk2-cre (in which the UPII drives the expression of a cre recombinase in urothelium) (25) and floxed p53 (in which the exons of 5 and 6 were flanked two loxP sites) (Fig. 3A) (26). We chose four resultant genotypes for phenotypic analyses: (i) Upk2-cre (as negative controls), (ii) Upk2-HRAS*/WT, (iii) Upk2-cre/p53LOX/LOX and (iv) Upk2-HRAS*/WT/Upk2-cre/p53LOX/LOX (Fig. 3B and 3C). These four groups were followed for 16-months and, upon histopathological examination, the Upk2-cre and Upk2-cre/p53LOX/LOX lines exhibited normal urothelia, and the Upk2-HRAS*/WT line exhibited urothelial hyperplasia, as expected, throughout the 16-month observation (Fig. 3D). In stark contrast, the compound line expressing oncogenic HRAS* and lacking p53 developed exclusively high-grade bladder tumors in the form of carcinoma-in-situ (CIS) and muscle-invasive tumors (Fig. 3C and 3D and Fig. 4A-C, 4E-G). The invasive tumors arose as early as 6 months of age and, by 16 months, a majority (60%) of the mice harbored muscle-invasive bladder tumors (Fig. 3C). The CIS lesions were relatively flat with microinvasive lesions in adjacent lamina propria (Fig. 4B and 4C). The microinvasive and muscle-invasive lesions were confirmed by cytokeratin 5 staining (Fig. 4D, 4H and 4I). These lesions bear strong resemblance to those found in human patients with muscle-invasive urothelial carcinoma, and lend strong support to the sequence of urothelial tumor progression from CIS to invasive tumors (13,14,31,32). Finally, focal squamous differentiation within the muscle-invasive lesions was common as evidenced by H&E staining and by immunohistochemical staining using antibodies against keratin 1 and TRIM29 (Supplemental Fig. S3), markers for identifying squamous components in human muscle-invasive urothelial carcinomas (33).
Figure 3.
Inactivation of p53 in urothelial cells of transgenic mice expressing oncogenic HRAS*. (A) Three transgenic lines for intercrossing contained transgene of uroplakin II promoter (Upk2) driving cre recombinase (Line 1); floxed p53 where exons 5 and 6 were flanked by loxP sites (Line 2); and Upk2 driving oncogenic HRAS*. (B) Offspring of two representative crosses were genotyped by Southern blotting of restriction-digested tail genomic DNA (upper panels) using a mouse Upk2 probe, revealing the 5.4-kB Upk2–cre transgene fragment, the 1.4-kB Upk2-HRAS* fragment and the 1.1-kB endogenous Upk2 fragment (endo-Upk2). The same DNA samples were subject to PCR using primers specifically detecting the first loxP site of the p53 mutant allele. Asterisks denote the four major genotypes chosen for additional analyses (also see C). (C) The rate of the four major genotypes free of high-grade muscle-invasive urothelial carcinoma (tumor-free rate). Note that only mice expressing oncogenic HRAS* as well as lacking p53 in urothelia developed invasive urothelial carcinoma. (D) Representative H&E images of the four genotypes (all 8 months old) (see text). Magnification: 200 ×.
Figure 4.
Morphological features of urothelial lesions in compound transgenic mice expressing oncogenic HRAS* and lacking p53. Urinary bladders of transgenic mice expressing oncogenic HRAS* and lacking p53 (8-12 months old) were stained by H&E (A-C; E-G) or anti-keratin 5 (D, H and I). Note the high-grade lesions resembling carcinoma-in-situ (A-C) with lamina propria invasion (B and D, arrows) and muscle-invasive lesions (E-G) that were strongly labeled (brown) by anti-keratin 5 (D, H and I). L denotes lumen. S in (E-F) denotes smooth muscle. Magnification: 200 × for all panels.
Epithelial-mesenchymal Transition Signifies CIS-invasive Tumor Conversion
That compound mice urothelially expressing oncogenic HRAS* and lacking p53 developed CIS and then invasive lesions also provided a unique opportunity for us to utilize these well-defined lesions to identify the molecular events that underlie this poorly defined progression step. Toward this end, we performed laser-capture microdissection of normal urothelia from the Upk2-cre mice, and CIS and muscle-invasive lesions from Upk2-HRAS*/WT/Upk2-cre/p53LOX/LOX compound mice. After high-quality mRNAs were isolated from freshly dissected lesions, the cDNAs were hybridized to oligonucleotide arrays representing all mouse genes (Affymatrix). Of differentially upregulated genes, those functioning in the epithelial-mesenchymal transition (EMT) dominate the muscle-invasive lesions when CIS lesions were used as a reference (Table 1). Three groups of genes were particularly worth noting: (i) transcription factors that drive EMT (e.g., twist homolog 1 (TWIST), zinc finger E-box binding homeobox 2 (ZEB2) and ZEB1); (ii) matrix-degrading enzymes (e.g., matrix metallopeptidases 13, 3, 2 and 9); and (iii) extracellular matrix components (e.g., collagen (type I, III and IV), versican, vimentin, and fibronectin) (Table 1). Not surprisingly, those upregulated in the muscle-invasive tumor/CIS comparison were also upregulated in the muscle-invasive tumor/normal urothelium comparison. However, few of those upregulated in muscle-invasive tumors were also upregulated in the CIS lesions, indicating that the EMT genes are primarily switched on during muscle invasion. The only genes that showed more than 2-fold increase in CIS over normal urothelium were MMP13 and platelet-derived growth factor receptor, suggesting their potential role(s) in CIS formation. Antibody staining confirmed that MMP2, 3, 9 and 13 were all over-expressed almost exclusively in the muscle-invasive lesions, with MMPs 2 and 9 primarily associated with tumor cells and MMP3 and 13 in both tumor cells and matrix (Fig. 5).
Table 1.
Differential expression of genes important for epithelial-mesenchymal transition (EMT) between muscle-invasive urothelial carcinoma (MIUCB), carcinoma-in-situ (CIS) and normal urothelium. Laser-capture, microdissected tissues were subject to mRNA extraction/cDNA synthesis and expression array analysis (see Methods; GEO accession number: GSE64756). Fold changes in three pair-wise comparisons are shown, with ranking from the highest to the lowest (>2 fold) expression in consecutive order for the MIUCB/CIS comparison chosen for practical purposes (see text).
| Gene Name | CIS vs. Normal Urothelium |
MIUCB vs. Normal Urothelium |
MIUCB vs. CIS |
|---|---|---|---|
| Matrix metallopeptidase 3 | −1.26 | 339.46 | 427.06 |
| Matrix metallopeptidase 13 | 4.04 | 732.76 | 181.25 |
| Collagen, type I, alpha 2 | 1.39 | 84.52 | 60.83 |
| Secreted frizzled-related protein 2 | −1.56 | 34.86 | 54.23 |
| Versican | −1.63 | 24.55 | 39.99 |
| Insulin-like growth factor binding protein 4 | 1.97 | 77.03 | 39.14 |
| Collagen, type III, alpha 1 | −1.89 | 19.24 | 36.32 |
| Matrix metallopeptidase 2 | 1.09 | 38.11 | 34.92 |
| Collagen, type V, alpha 2 | −1.05 | 29.23 | 30.65 |
| Lysyl oxidase | −1.16 | 26.38 | 30.64 |
| Secreted frizzled-related protein 1 | −1.10 | 20.77 | 22.88 |
| Fibroblast growth factor receptor 1 | −4.50 | 3.24 | 14.59 |
| Vimentin | 1.46 | 20.30 | 13.91 |
| Platelet derived growth factor receptor, beta polypeptide | 4.23 | 54.52 | 12.88 |
| Transforming growth factor, beta 2 | −2.37 | 4.88 | 11.56 |
| Twist homolog 1 / Twist-Related Protein 1 | −1.09 | 9.84 | 10.77 |
| Zinc finger E-box binding homeobox 2 | 1.50 | 14.60 | 9.73 |
| Fibronectin 1 | 1.32 | 11.13 | 8.44 |
| Calcium/calmodulin-dependent protein kinase II inhibitor 1 | 1.56 | 12.23 | 7.84 |
| Zinc finger E-box binding homeobox 1 | 1.35 | 9.38 | 6.97 |
| Wingless-type MMTV integration site 9A | −1.57 | 4.21 | 6.60 |
| Wingless-related MMTV integration site 7A | 1.23 | 7.53 | 6.14 |
| Transforming growth factor, beta 3 | 1.20 | 6.87 | 5.73 |
| Wingless-related MMTV integration site 2 | −1.93 | 2.32 | 4.47 |
| Matrix metallopeptidase 9 | 1.67 | 7.29 | 4.36 |
| Fibroblast growth factor 13 | −1.05 | 3.83 | 4.04 |
| Wingless related MMTV integration site 10a | 1.23 | 4.76 | 3.86 |
| Fibroblast growth factor 7 | −1.23 | 2.96 | 3.64 |
| Transcription factor 4 | 1.33 | 4.62 | 3.47 |
| Thymoma viral proto-oncogene 3 | 1.89 | 5.80 | 3.07 |
| Recombination signal binding protein for immunoglobulin kappa J region | 1.14 | 3.35 | 2.93 |
| Wingless-related MMTV integration site 5B | −1.53 | 1.64 | 2.51 |
| Phosphoinositide-3-kinase, regulatory subunit 5 | −1.16 | 2.17 | 2.51 |
Figure. 5.
Upregulation of matrix metalloproteinases in muscle-invasive urothelial carcinoma cells. Immunochemical staining using anti-MMP antibodies followed by hematoxylin counter staining was performed on bladder tissues from age-matched (8-months) Upk2-cre, Upk2-HRAS*, Upk2-cre/p53lox/lox and Upk2-HRAS*/Upk2-cre/p53lox/lox mice. Note the significant upregulation of MMP2, MMP3, MMP9 and MMP13 in the muscle-invasive urothelial carcinoma cells of transgenic mice expressing the oncogenic HRAS* and deficient for p53. MMP3 and MMP13 were also detected strongly in some matrix cells. Magnification: 200× for all panels.
EMT Occurs in Urothelial Carcinoma Progenitor/Stem Cells
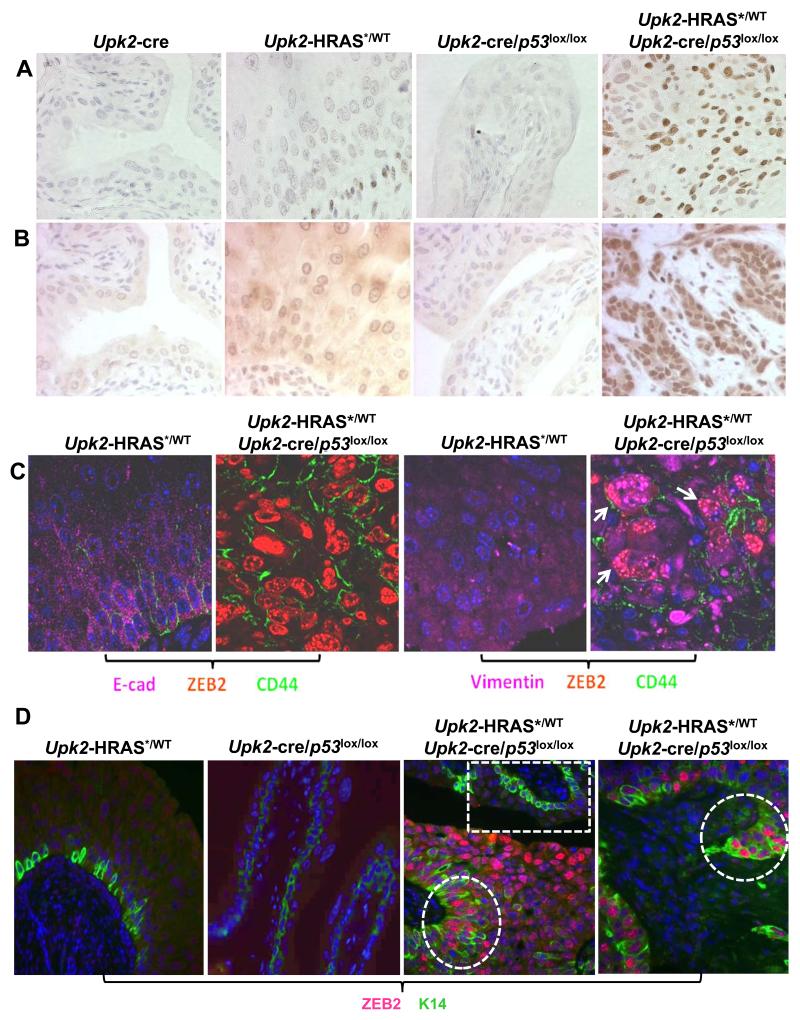
To explore whether overexpression of EMT drivers occurred in more differentiated urothelial cells or in progenitor cells thus potentially playing a role in invasive tumor initiation, we performed immunohistochemical and double- and triple-immunofluorescent localization. Both ZEB1 and ZEB2 were strongly labeled in the nuclei of the invasive tumors cells of Upk2-HRAS*/WT/Upk2-cre/p53LOX/LOX compound mice, but were barely detectable in normal urothelia of Upk2-cre or Upk2-cre/p53LOX/LOX mice and were only weakly labeled in Upk2-HRAS* mice (Fig. 6A and 6B). Upon triple fluorescent staining, ZEB2 was found to be associated with cells specifically expressing CD44, a urothelial/carcinoma progenitor cell marker (Fig. 6C) (18,19,34). Interestingly, these ZEB2- and CD44-positive cells had a marked decrease of E-cadherin, an epithelial marker (35), and a marked increase of vimentin, a mesenchymal cell marker (Fig. 6C) (36). Double staining of ZEB2 with keratin 14, another urothelial progenitor cell marker (34), again showed excellent co-localization (Fig. 6D). Whereas normal-appearing urothelial regions showed K14-positive cells which lacked ZEB2 labeling, areas with tumor morphology showed strong co-expression of ZEB2 and K14 (Fig. 6D). Furthermore, areas with leading edge of invasion showed strong ZEB2 and K14 co-expression (Fig. D). Interestingly, invasive tumors cells of the Upk2-HRAS*/WT/Upk2-cre/p53LOX/LOX compound mice, but not the non-invasive cells of the single transgenic mice, strongly co-expressed in the nuclei activated AKT, activated β–catenin and ZEB2 (Supplemental Fig. S4), suggesting this signaling pathway as the underlying cause of EMT activation. These results establish that urothelial tumor progenitor cells in our compound transgenic mice expressing oncogenic HRAS* and lacking p53 strongly express EMT drivers and their expression may play a central role in initiating muscle-invasive urothelial carcinoma. Finally, in contrast to the expansion of K14-positive cells in the muscle-invasive lesions, cells positive for keratin 20, a marker expressed in urothelial superficial umbrella cells and used for terminal differentiation of normal urothelium (37), were completely absent from the muscle-invasive lesions (Supplemental Fig. S5). These results, together with our observation of focal squamous differentiation of the muscle-invasive lesions, strongly indicate that the muscle-invasive urothelial carcinoma of the bladder that we observed in our Upk2-HRAS*/Upk2-cre/p53LOX/LOX mice belong to the “basal-subtype” recently classified in patients (4-6,8-10).
Figure 6.
Detection of transcriptional factors driving EMT in urothelial progenitor cells. (A and B) Urinary bladders from age-matched (8-months) Upk2-cre, Upk2-HRAS*/WT, Upk2-cre/p53lox/lox and Upk2-HRAS*/WT/Upk2-cre/p53lox/lox mice were immunohistochemically stained with anti-ZEB1 (A) and anti-ZEB2 (B) and counter-stained by hemotoxylin. Note the marked upregulation of both proteins almost exclusively in the muscle-invasive lesions of the Upk2-HRAS*/WT/Upk2-cre/p53lox/lox mice. (C) Urinary bladders from Upk2-HRAS*/WT and Upk2-HRAS*/WT/Upk2-cre/p53lox/lox mice were triple-stained using immuofluorescent method with anti-E-cadherin (E-cad), -ZEB2 and -CD44 (left two panels) or with anti-vimentin, -ZEB2 and -CD44 (right two panels). DAPI was used to visualize the nuclei. Note the marked down-regulation of E-cadherin and dramatic up-regulation of ZEB2 in CD44-positive cells in the muscle-invasive lesions of the Upk2-HRAS*/WT/Upk2-cre/p53lox/lox mice. Also note the co-localization of vimentin, ZEB2 and CD44 in the invasive tumor cells (far-right panel, arrows). (D) Urinary bladders from Upk2-HRAS*/WT, Upk2-cre/p53lox/lox and Upk2-HRAS*/WT/Upk2-cre/p53lox/lox mice were subject to immunofluorescent staining with anti-ZEB2 and -keratin 14 antibodies, with DAPI as counter-staining to visualize the nuclei. Note the lack of ZEB2 staining in K14-positive cells in Upk2-HRAS*/WT and Upk2-cre/p53lox/lox mice and the strong staining of ZEB2 in K14-positive cells in Upk2-HRAS*/WT/Upk2-cre/p53lox/lox mice (middle panel, dashed circle). Dashed box illustrates an area of normal-appearing urothelium showing the lack of ZEB2 staining. Also note that the leading edge of an early invasive lesion in Upk2-HRAS*/WT/Upk2-cre/p53lox/lox mice had marked upregulation of ZEB2 in K14-positive cells (right panel). Magnification: 200× for all panels.
Discussion
The recent expansion of whole-genome and whole-exome sequencing into a broad range of human cancers has yielded unprecedented details about somatic gene mutations, making it possible to classify cancers in genomic terms and to devise target-specific, precision therapies (38). Urothelial carcinoma of the bladder (UCB) is no exception. In a landmark paper (23), The Cancer Genome Atlas (TCGA) Research Network reported a comprehensive, multiplatform analysis of 131 high-grade, muscle-invasive urothelial carcinomas of the bladder (MIUCB) on their somatic mutation, DNA copy number, messenger and micro-RNA expression, protein and phosphorylated protein expression and DNA methylation. Of the several surprises from that report, one relates to the high frequency of alterations in the RTK/RAS/PIK3K signaling axis. Up to 72% of the high-grade MIUCB harbored activation mutations in the FGFR3, EGFR, ERBB2, ERBB3, HRAS/NRAS and PIK3CA or inactivating mutations in NF1, PTEN, INPP4B, STK11, TSC1 and TSC2 (23). This is surprising because alterations in this pathway were previously assigned primarily to low-grade, non-invasive UCB and to predict low risk of progression and favorable clinical outcome (13,21,22) – a concept supported by independent studies using genetically engineered mice. For instance, urothelial expression of an FGFR3 mutant (K644E) that constitutively activates the tyrosine kinase of FGFR3, either alone or in combination with KRAS and β-catenin mutations or with PTEN deletion, in transgenic mice failed to elicit any urothelial carcinoma (39). Similarly, urothelial overexpression of an epidermal growth factor receptor (EGFR) in our transgenic mice induced proliferation but not tumor formation even after an exhaustively long (28-month) follow-up (40). Furthermore, urothelium-specific expression in our transgenic mice of oncogenic HRAS* at a level comparable to the endogenous RAS elicited urothelial hyperplasia that only occasionally progressed to low-grade, papillary non-invasive UCB in aged mice (>12 months) (24). High-grade MIUCB was never observed in any of these RTK/RAS-pathway-activated mouse models (24,39,40). The fact that gene mutations that activate the RTK/RAS pathway are highly prevalent in human high-grade MIUCB from the TCGA study (23) raises an important question as to whether these mutations are tumor “drivers” or “passengers” and whether the mutations require additional genetic alterations to be tumorigenic.
Our present study provides experimental evidence establishing that RAS activation per se is non-tumorigenic in urothelial cells in vivo due, in large part, to a compensatory tumor barrier that RAS elicits in the p53 tumor suppressor pathway (Figs. 1, 2 and Supplemental Fig. S1). Although p53 deficiency by itself is also non-tumorigenic, it is highly synergistic with RAS activation, and these two alterations together are necessary and sufficient to initiate high-grade, carcinoma-in-situ (CIS) and MIUCB (Figs. 3 and 4). Of note, the MIUCB we observed in our double transgenic mice expressing oncogenic Ha-RAS and lacking p53 bear strong resemblance to the basal-subtype of MIUCB recently classified in patients (4-11) in their (i) high expression of basal cell markers such as K5, K14 and CD44 (Figs. 4 and 6 and Supplemental Fig. S5), (ii) low or lack of expression of luminal cell markers such as E-cadherin and K20 (Fig. 6 and Supplemental Fig. S5), (iii) focal squamous differentiation (Supplemental Fig. S3), and (iv) high expression of EMT transcription factors (Twist, ZEB1 and ZEB2) (Table 1; Fig. 6), EMT markers (vimentin, MMPs 2, 3, 9 and 13) (Table 1; Figs. 5 and 6) and extracellular matrix components (collagen, versican and fibronectin) (Table 1). Our study therefore functionally defines RAS pathway activation and p53 deficiency as the highly synergistic co-drivers for the basal-subtype MIUCB, and it has several significant implications. First, as has been demonstrated in other cancer types, tumor drivers (as opposed to the passengers) are more reliable biomarkers for cancer sub-classification and prediction of chemotherapeutic response and clinical outcome (12). RAS pathway activation together with p53 deficiency could potentially serve as a new biomarker set for the genetic identification of the basal-subtype of MIUCB that may be associated with an unfavorable prognosis, hence requiring aggressive therapeutic modalities. Second, our study reveals a previously unrecognized molecular cross-talk between RAS and p53 pathways in converting low-grade non-invasive urothelial lesions (e.g., hyperplasia and low-grade papillary) into becoming high-grade non-invasive (e.g., high-grade papillary and CIS) and invasive ones (e.g., MIUCB). Not only did we demonstrate such a relationship in transgenic mice (Figs. 3-6; Supplemental Figs. S3-S5), we also showed that introducing oncongenic HRAS* and knocking down p53 in cultured RT4 cells confer invasive properties to these otherwise non-invasive human UCB cells (Fig. 2). It has been suggested, based on clinical longitudinal studies, that approximately 25% of the low-grade, non-invasive UCB can eventually progress in grade and/or stage to muscle invasion (41,42). This occurs in an unpredictable manner that necessitates lifelong, vigilant follow-up by repeated cystoscopy and biopsy - a main cause for morbidity, time lost from work and high medical expenses. Thus far, no biomarker exists that can reliably predict the risk of progression of non-invasive UCB to the invasive stage (43,44). Perhaps it is not surprising that p53 alterations are not very predictive of UCB progression (45), based on data from genetically engineered mice indicating the lack of tumorigenicity by p53 deficiency alone (Fig. 3; (26,46,47)). It is possible, however, that a combination of RAS pathway activation and p53 deficiency, as we demonstrated here, are better biomarkers for UCB surveillance and prediction of tumor progression. It is worth noting that ablation of both PTEN and p53 in mouse urothelia also led to MIUCB (48), consistent with the fact that PTEN acts in the RAS pathway and PTEN inactivation is functionally akin to RAS activation. Third, because RAS activation is a co-driver of the basal-type MIUCB, inhibition of this pathway might be of significant value in treating and/or preventing the progression of this MIUCB subtype. The fact that the basal-type MIUCB in humans is often resistant to the existing chemotherapeutics (4) makes RAS pathway inhibition a particularly attractive avenue to explore. Since suppressing activated RAS per se remains challenging (49), it is likely that effectors of RAS will have to be targeted and that inhibition of more than one signaling branch (e.g., PI3K-AKT as well as MAPK) is required to achieve satisfactory results (50). Finally, the development of a new transgenic mouse model that consistently develops the basal-type MIUCB provides a novel in vivo platform for dissecting the evolutionary steps and the potential cross-talks among the different MIUCB subtypes and for testing subtype-specific diagnostic, preventive and therapeutic strategies. Clearly, many of these ideas require clinical validation studies before they can be translated to the bedside.
From a mechanistic standpoint, RAS activation and p53 deficiency could synergize on several fronts to affect cellular processes that govern urothelial tumorigenesis and progression. As shown recently, RAS activation increases the replicative pressure on urothelial cells, causing them to undergo DNA damage (51). Under normal circumstances, i.e., when p53 pathway is intact, urothelial cells can sense DNA damage and upregulate p19Arf which in turn upregulates p53 and downstream effectors such as p21 (Fig. 1). This helps restrain G1/S and G2/M transition and allow time for DNA damage repair to take place. When p53 pathway is defective, however, cell cycle progression proceeds with amplification of the damaged DNA, setting a stage for malignant transformation. Another level of interaction is the collaborative nature of RAS activation and p53 deficiency on cell motility. Activated RAS is a strong enhancer of cell motility (52), whereas a functional p53 is a potent cell motility inhibitor (53). As we showed in our in vitro assay, activated RAS or p53 knock down alone only had a marginal increase on cell motility, but combining these two events resulted in a marked increase of cell motility and triggered invasion (Fig. 2). Finally, as with other epithelial cells, RAS activation and p53 deficiency are both strong promoters of EMT (54,55). MAPK and AKT pathways, both shown to be prominently activated in our transgenic mice (Fig. 1; Supplemental Fig. S1), can activate factors such as β-catenin that drive EMT (Table 1; Supplemental Fig. S4). While normal p53 negatively regulates this process, p53 deficiency fuels EMT and sets the tumor cell invasion in motion (Figs. 2-6; Supplemental Fig. S3-S5). There is mounting evidence suggesting that EMT can lead to drug resistance (35). Since EMT enhances the stemness and the plasticity of urothelial cells, it may also fuel the trans-differentiation of some of the urothelial progenitor cells toward the squamous lineage and squamous differentiation, another potential cause of drug resistance. In this regard, inhibiting RAS effectors that drive EMT and/or inhibiting EMT effectors such as MMPs may play a critical role in reducing chemoresistance that has been observed in the basal-type MIUCB (4). Because EMT is highly activated in progenitor/stem cells that give rise to the basal-type MIUCB (Fig. 6 and Supplemental Fig. S5), its suppression may present a unique opportunity for controlling the root cause of tumor cell expansion and invasion.
In summary, the data presented in this paper provide the first experimental evidence demonstrating that the loss of p53 is critical in allowing hyperplastic urothelial cells in vivo to bypass G2 arrest induced by activated HRAS and proceed to tumor formation; that RAS pathway activation and p53 pathway inactivation together confer invasive properties to non-invasive urothelial tumor cells and these two synergistic events are necessary and sufficient to convert carcinoma in situ to basal-subtype, muscle-invasive urothelial carcinoma of the bladder; and that activation of EMT and increased stemness in urothelial progenitor cells are crucial epigenetic events for invasive tumorigenesis. Our data also strongly suggest that increased urothelial plasticity due to EMT may underlie urothelial transdifferentiation to the squamous lineage, leading to focal squamous differentiation in urothelial carcinomas. From a clinical standpoint, combined RAS pathway activation and p53 pathway inactivation, events highly prevalent in human urothelial carcinomas as evidenced by whole-genome analyses, may serve as a new biomarker set to predict urothelial carcinoma progression, and inhibition of receptor tyrosine kinase/RAS pathway components may be used as therapeutic targets for basal-subtype, muscle-invasive urothelial carcinomas that are resistant to conventional chemotherapeutics.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the United States National Institutes of Health (P01 CA165980) and Veterans Affairs Office of Research and Development (Biomedical Laboratory Research and Development Service; 1I01BX002049) and a grant-in-aid from the Goldstein Fund for Urological Research of the New York University School of Medicine.
Footnotes
The authors declare that there are no conflicts of interest in the work reported in this manuscript.
References
- 1.Lotan Y, Kamat AM, Porter MP, Robinson VL, Shore N, Jewett M, et al. Key concerns about the current state of bladder cancer: a position paper from the Bladder Cancer Think Tank, the Bladder Cancer Advocacy Network, and the Society of Urologic Oncology. Cancer. 2009;115(18):4096–103. doi: 10.1002/cncr.24463. [DOI] [PubMed] [Google Scholar]
- 2.Dancik GM, Theodorescu D. Pharmacogenomics in bladder cancer. Urologic oncology. 2014;32(1):16–22. doi: 10.1016/j.urolonc.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meeks JJ, Bellmunt J, Bochner BH, Clarke NW, Daneshmand S, Galsky MD, et al. A systematic review of neoadjuvant and adjuvant chemotherapy for muscle-invasive bladder cancer. European urology. 2012;62(3):523–33. doi: 10.1016/j.eururo.2012.05.048. [DOI] [PubMed] [Google Scholar]
- 4.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer cell. 2014;25(2):152–65. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(8):3110–5. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurst CD, Knowles MA. Molecular subtyping of invasive bladder cancer: time to divide and rule? Cancer cell. 2014;25(2):135–6. doi: 10.1016/j.ccr.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Riester M, Taylor JM, Feifer A, Koppie T, Rosenberg JE, Downey RJ, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18(5):1323–33. doi: 10.1158/1078-0432.CCR-11-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volkmer JP, Sahoo D, Chin RK, Ho PL, Tang C, Kurtova AV, et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(6):2078–83. doi: 10.1073/pnas.1120605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sjodahl G, Lovgren K, Lauss M, Patschan O, Gudjonsson S, Chebil G, et al. Toward a molecular pathologic classification of urothelial carcinoma. The American journal of pathology. 2013;183(3):681–91. doi: 10.1016/j.ajpath.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Huang HY, Shariat SF, Sun TT, Lepor H, Shapiro E, Hsieh JT, et al. Persistent uroplakin expression in advanced urothelial carcinomas: implications in urothelial tumor progression and clinical outcome. Human pathology. 2007 doi: 10.1016/j.humpath.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livshits G, Lowe SW. Accelerating cancer modeling with RNAi and nongermline genetically engineered mouse models. Cold Spring Harbor protocols. 2013;2013(11) doi: 10.1101/pdb.top069856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5(9):713–25. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 14.Dinney CP, McConkey DJ, Millikan RE, Wu X, Bar-Eli M, Adam L, et al. Focus on bladder cancer. Cancer cell. 2004;6(2):111–6. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Grossman HB. Superficial bladder cancer: decreasing the risk of recurrence. Oncology (Huntingt) 1996;10(11):1617–24. discussion 24, 27-8. [PubMed] [Google Scholar]
- 16.Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, Matos T, Cordon-Cardo C. Molecular pathways of urothelial development and bladder tumorigenesis. Urologic oncology. 2010;28(4):401–8. doi: 10.1016/j.urolonc.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Wu XR, Kong XP, Pellicer A, Kreibich G, Sun TT. Uroplakins in urothelial biology, function, and disease. Kidney Int. 2009;75:1153–65. doi: 10.1038/ki.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandhi D, Molotkov A, Batourina E, Schneider K, Dan H, Reiley M, et al. Retinoid signaling in progenitors controls specification and regeneration of the urothelium. Developmental cell. 2013;26(5):469–82. doi: 10.1016/j.devcel.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Batavia J, Yamany T, Molotkov A, Dan H, Mansukhani M, Batourina E, et al. Bladder cancers arise from distinct urothelial sub-populations. Nature cell biology. 2014 doi: 10.1038/ncb3038. [DOI] [PubMed] [Google Scholar]
- 20.Shin K, Lim A, Odegaard JI, Honeycutt JD, Kawano S, Hsieh MH, et al. Cellular origin of bladder neoplasia and tissue dynamics of its progression to invasive carcinoma. Nature cell biology. 2014;16(5):469–78. doi: 10.1038/ncb2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff EM, Liang G, Jones PA. Mechanisms of Disease: genetic and epigenetic alterations that drive bladder cancer. Nat Clin Pract Urol. 2005;2(10):502–10. doi: 10.1038/ncpuro0318. [DOI] [PubMed] [Google Scholar]
- 22.van Rhijn BW, van der Kwast TH, Vis AN, Kirkels WJ, Boeve ER, Jobsis AC, et al. FGFR3 and P53 characterize alternative genetic pathways in the pathogenesis of urothelial cell carcinoma. Cancer research. 2004;64(6):1911–4. doi: 10.1158/0008-5472.can-03-2421. [DOI] [PubMed] [Google Scholar]
- 23.Network TCGAR Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mo L, Zheng X, Huang HY, Shapiro E, Lepor H, Cordon-Cardo C, et al. Hyperactivation of Ha-ras oncogene, but not Ink4a/Arf deficiency, triggers bladder tumorigenesis. J Clin Invest. 2007;117(2):314–25. doi: 10.1172/JCI30062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo L, Cheng J, Lee EY, Sun TT, Wu XR. Gene deletion in urothelium by specific expression of Cre recombinase. Am J Physiol Renal Physiol. 2005;289(3):F562–8. doi: 10.1152/ajprenal.00368.2004. [DOI] [PubMed] [Google Scholar]
- 26.He F, Mo L, Zheng XY, Hu C, Lepor H, Lee EY, et al. Deficiency of pRb family proteins and p53 in invasive urothelial tumorigenesis. Cancer research. 2009;69(24):9413–21. doi: 10.1158/0008-5472.CAN-09-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rigby CC, Franks LM. A human tissue culture cell line from a transitional cell tumour of the urinary bladder: growth, chromosone pattern and ultrastructure. British journal of cancer. 1970;24(4):746–54. doi: 10.1038/bjc.1970.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20(15):1803–15. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 29.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12(19):3008–19. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeGraff DJ, Robinson VL, Shah JB, Brandt WD, Sonpavde G, Kang Y, et al. Current preclinical models for the advancement of translational bladder cancer research. Molecular cancer therapeutics. 2013;12(2):121–30. doi: 10.1158/1535-7163.MCT-12-0508. [DOI] [PubMed] [Google Scholar]
- 31.Cohen SM, Ohnishi T, Clark NM, He J, Arnold LL. Investigations of rodent urinary bladder carcinogens: collection, processing, and evaluation of urine and bladders. Toxicol Pathol. 2007;35(3):337–47. doi: 10.1080/01926230701197115. [DOI] [PubMed] [Google Scholar]
- 32.Dalbagni G, Presti J, Reuter V, Fair WR, Cordon-Cardo C. Genetic alterations in bladder cancer. Lancet. 1993;342(8869):469–71. doi: 10.1016/0140-6736(93)91595-d. [DOI] [PubMed] [Google Scholar]
- 33.Huang W, Williamson SR, Rao Q, Lopez-Beltran A, Montironi R, Eble JN, et al. Novel markers of squamous differentiation in the urinary bladder. Human pathology. 2013;44(10):1989–97. doi: 10.1016/j.humpath.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):14016–21. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McConkey DJ, Choi W, Marquis L, Martin F, Williams MB, Shah J, et al. Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer. Cancer metastasis reviews. 2009;28(3-4):335–44. doi: 10.1007/s10555-009-9194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kokkinos MI, Wafai R, Wong MK, Newgreen DF, Thompson EW, Waltham M. Vimentin and epithelial-mesenchymal transition in human breast cancer--observations in vitro and in vivo. Cells, tissues, organs. 2007;185(1-3):191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Beltran A, Montironi R, Vidal A, Scarpelli M, Cheng L. Urothelial dysplasia of the bladder: diagnostic features and clinical significance. Analytical and quantitative cytopathology and histopathology. 2013;35(3):121–9. [PubMed] [Google Scholar]
- 38.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmad I, Singh LB, Foth M, Morris CA, Taketo MM, Wu XR, et al. K-Ras and beta-catenin mutations cooperate with Fgfr3 mutations in mice to promote tumorigenesis in the skin and lung, but not in the bladder. Disease models & mechanisms. 2011;4(4):548–55. doi: 10.1242/dmm.006874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng J, Huang H, Zhang ZT, Shapiro E, Pellicer A, Sun TT, et al. Overexpression of epidermal growth factor receptor in urothelium elicits urothelial hyperplasia and promotes bladder tumor growth. Cancer research. 2002;62(14):4157–63. [PubMed] [Google Scholar]
- 41.Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27(3):361–73. doi: 10.1093/carcin/bgi310. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka M, Grossman HB. Tumor suppressor genes of bladder cancer and potential for gene therapy. Adv Exp Med Biol. 2003;539(Pt A):185–91. doi: 10.1007/978-1-4419-8889-8_14. [DOI] [PubMed] [Google Scholar]
- 43.Xylinas E, Kluth LA, Lotan Y, Daneshmand S, Rieken M, Karakiewicz PI, et al. Blood- and tissue-based biomarkers for prediction of outcomes in urothelial carcinoma of the bladder. Urologic oncology. 2014;32(3):230–42. doi: 10.1016/j.urolonc.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Shirodkar SP, Lokeshwar VB. Potential new urinary markers in the early detection of bladder cancer. Current opinion in urology. 2009;19(5):488–93. doi: 10.1097/MOU.0b013e32832eb3a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stadler WM, Lerner SP, Groshen S, Stein JP, Shi SR, Raghavan D, et al. Phase III study of molecularly targeted adjuvant therapy in locally advanced urothelial cancer of the bladder based on p53 status. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(25):3443–9. doi: 10.1200/JCO.2010.34.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng J, Huang H, Pak J, Shapiro E, Sun TT, Cordon-Cardo C, et al. Allelic loss of p53 gene is associated with genesis and maintenance, but not invasion, of mouse carcinoma in situ of the bladder. Cancer research. 2003;63(1):179–85. [PubMed] [Google Scholar]
- 47.Gao J, Huang HY, Pak J, Cheng J, Zhang ZT, Shapiro E, et al. p53 deficiency provokes urothelial proliferation and synergizes with activated Ha-ras in promoting urothelial tumorigenesis. Oncogene. 2004;23(3):687–96. doi: 10.1038/sj.onc.1207169. [DOI] [PubMed] [Google Scholar]
- 48.Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23(6):675–80. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer cell. 2014;25(3):272–81. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 50.Zhou H, Huang HY, Shapiro E, Lepor H, Huang WC, Mohammadi M, et al. Urothelial tumor initiation requires deregulation of multiple signaling pathways: implications in target-based therapies. Carcinogenesis. 2012;33(4):770–80. doi: 10.1093/carcin/bgs025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evangelou K, Bartkova J, Kotsinas A, Pateras IS, Liontos M, Velimezi G, et al. The DNA damage checkpoint precedes activation of ARF in response to escalating oncogenic stress during tumorigenesis. Cell death and differentiation. 2013;20(11):1485–97. doi: 10.1038/cdd.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker NM, Yee Chow H, Chernoff J, Der CJ. Molecular Pathways: Targeting RAC-p21-Activated Serine-Threonine Kinase Signaling in RAS-Driven Cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(18):4740–6. doi: 10.1158/1078-0432.CCR-13-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. The Journal of cell biology. 2011;192(2):209–18. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of disease: epithelial-mesenchymal transition--does cellular plasticity fuel neoplastic progression? Nature clinical practice Oncology. 2008;5(5):280–90. doi: 10.1038/ncponc1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ansieau S, Courtois-Cox S, Morel AP, Puisieux A. Failsafe program escape and EMT: a deleterious partnership. Seminars in cancer biology. 2011;21(6):392–6. doi: 10.1016/j.semcancer.2011.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.