Abstract
Neural crest cells (NCC) comprise a multipotent, migratory stem cell and progenitor population that gives rise to numerous cell and tissue types within a developing embryo, including craniofacial bone and cartilage, neurons and glia of the peripheral nervous system, and melanocytes within the skin. Here we describe two novel stable transgenic mouse lines suitable for lineage tracing and analysis of gene function in NCC. Firstly, using the F10N enhancer of the Mef2c gene (Mef2c-F10N) linked to LacZ, we generated transgenic mice (Mef2c-F10N-LacZ) that express LacZ in the majority, if not all migrating NCC that delaminate from the neural tube. Mef2c-F10N-LacZ then continues to be expressed primarily in neurogenic, gliogenic and melanocytic NCC and their derivatives, but not in ectomesenchymal derivatives. Secondly, we used the same Mef2c-F10N enhancer together with Cre recombinase to generate transgenic mice (Mef2c-F10N-Cre) that can be used to indelibly label, or alter gene function in, migrating NCC and their derivatives. At early stages of development, Mef2c-F10N-LacZ and Mef2c-F10N-Cre label NCC in a pattern similar to Wnt1-Cre mice, with the exception that Mef2c-F10N-LacZ and Mef2c-F10N-Cre specifically label NCC that have delaminated from the neural plate, while premigratory NCC are not labeled. Thus, our Mef2c-F10N-LacZ and Mef2c-F10N-Cre transgenic mice provide new resources for tracing migratory NCC and analyzing gene function in migrating and differentiating NCC independently of NCC formation.
Keywords: Neural crest cell, Mef2C enhancer (Mef2c-F10N), β-galactosidase reporter (LacZ), Cre recombinase
Introduction
The discovery of neural crest cells (NCC) is credited to Wilhelm His (His, 1868) and for the past 150 years, NCC have fascinated scientists as a model system for studying cell specification, cell delamination, epithelia to mesenchymal transformation, cell migration and cell differentiation. Derived from the neuroectoderm, NCC are a transient multipotent stem and progenitor cell population that gives rise to a diverse array of tissues and cell lineages. Within the head NCC give rise to osteocytes, osteoblasts, chondrocytes and chondroblasts of craniofacial bone and cartilage (Le Lievre and Le Douarin, 1975; Noden, 1982) odontoblasts of the teeth (Achilleos and Trainor, 2012; Chai et al., 2000), neurons and glia of the peripheral nervous system (PNS) (D'Amico-Martel and Noden, 1983), the meninges covering the surface of cerebral cortex (Jiang et al., 2002; Yoshida et al., 2008), smooth muscle cells of the craniofacial vasculature (Etchevers et al., 2001), and myoblasts of the extrinsic ocular muscles (Noden, 1983). Other NCC derivatives include chromaffin cells of the adrenal medulla (Jiang et al., 2000) and melanocytes in the skin (Jiang et al., 2000).
In mouse, analyses of migrating NCC traditionally relied upon classic histological stains and dye labeling of cultured embryos (Nichols, 1986; Osumi-Yamashita et al., 1994, 1996; Tam and Trainor, 1994; Trainor and Tam, 1995). More recently genetic approaches using transgenic or homologous recombinant reporters have come to the fore. These modern molecular approaches rely on promoters and enhancers that drive gene expression in NCC to indelibly label them or to alter gene function therein. Some of the lines used for these purposes include a knock-in into the 3’ untranslated region of the Activator protein 2 alpha transcription factor locus; AP2α-IRES-Cre (Macatee et al., 2003), human tissue plasminogen activator promoter; Ht-PA-Cre (Pietri et al., 2003), myelin protein zero promoter; P0-Cre (Yamauchi et al., 1999), knock-in at Paired box family 3 locus; Pax3-Cre (Li et al., 2000); PAC transgene insertion of Sox (SRY-related HMG-box) family transcription factor 10 locus; Sox10-Cre (Matsuoka et al., 2005), Wnt1 promoter and enhancer; Wnt1-Cre (Chai et al., 2000) and Tyrosinase enhancer and promoter; Tyrosinase-Cre (Tonks et al., 2003).
Of the Cre drivers available for analyses of NCC, the Wnt1-Cre transgenic mouse line exhibits the most robust and consistent activity within NCC and their derivatives and thus has become the standard for lineage tracing and investigation of gene function in NCC (Brault et al., 2001; Chai et al., 2000; Danielian et al., 1998; Jiang et al., 2000). However, Wnt1-Cre is expressed in the dorsal neuroepithelium which encompasses the territory in which NCC are induced to form. Therefore, Wnt1-Cre can’t distinguish effects in premigratory NCC from delaminated migratory NCC and does indeed elicit effects in premigratory and migratory NCC as well as in dorsal brain tissue (Brault et al., 2001; Chai et al., 2000; Jiang et al., 2000; Lewis et al., 2013). Many of the other Cre transgenic mice that have been used for analyses of NCC have confounding properties of expression in non-NCC-derived tissue and/or lack of expression in some NCC lineages (Hari et al., 2002; Lee et al., 2013; Li et al., 2000; Macatee et al., 2003; Pietri et al., 2003). Thus, in order to specifically investigate migratory NCC and their derivatives independently of premigratory NCC within the neural plate and to investigate gene function during NCC migration independently of NCC formation, new mouse lines are needed.
The Myocyte-specific enhancer factor 2C (Mef2c) gene is expressed broadly in the developing mouse embryo, including in NCC (Edmondson et al., 1994; Potthoff and Olson, 2007; Verzi et al., 2007). Furthermore, tissue specific conditional inactivation of Mef2c in NCC via Wnt1-Cre, illustrates that Mef2c is important for NCC-dependent craniofacial and melanocyte development (Agarwal et al., 2011; Verzi et al., 2007). Analyses of cis-regulatory sequences in the Mef2c locus in transient transgenic embryos identified an enhancer element called F10N (Mef2c-F10N) that marked migrating NCC in embryonic day (E) 9.5 embryos (De Val et al., 2008).
In this study, we used the Mef2c-F10N enhancer to establish new tools and resources for studying NCC development and function. Firstly, we made a stable version of the transgenic lacZ reporter line (Mef2c-F10N-LacZ) and secondly, a transgenic Cre recombinase line (Mef2c-F10N-Cre). We observed the Mef2c-F10N-LacZ reporter to initially mark all or nearly all NCC that delaminate from the neuroepithelium. Mef2c-F10N-LacZ expression then subsequently becomes restricted primarily to neurogenic, gliogenic and melanocytic lineages of the NCC but not ectomesenchymal derivatives. The Mef2c-F10N-Cre line induces recombination in a similar pattern when mated with Rosa26 LacZ reporter mice (R26R). Mef2c-F10N-Cre indelibly labels migrating NCC and their derivatives in the head (bone, cartilage, cranial nerve and connective tissue), heart, trunk, (neurons and glia, thymus and adrenal gland cells) and skin (melanocytes) during embryonic development. Thus, our Mef2c-F10N transgenic mice provide useful new tools and resources for tracing and analyzing gene function during NCC migration and differentiation, independently of NCC formation.
Results
Mef2c-F10N-LacZ specifically marks migrating neural crest cells in early E8–9 embryos, becoming limited to neuronal, glial and melanocyte lineages after E11
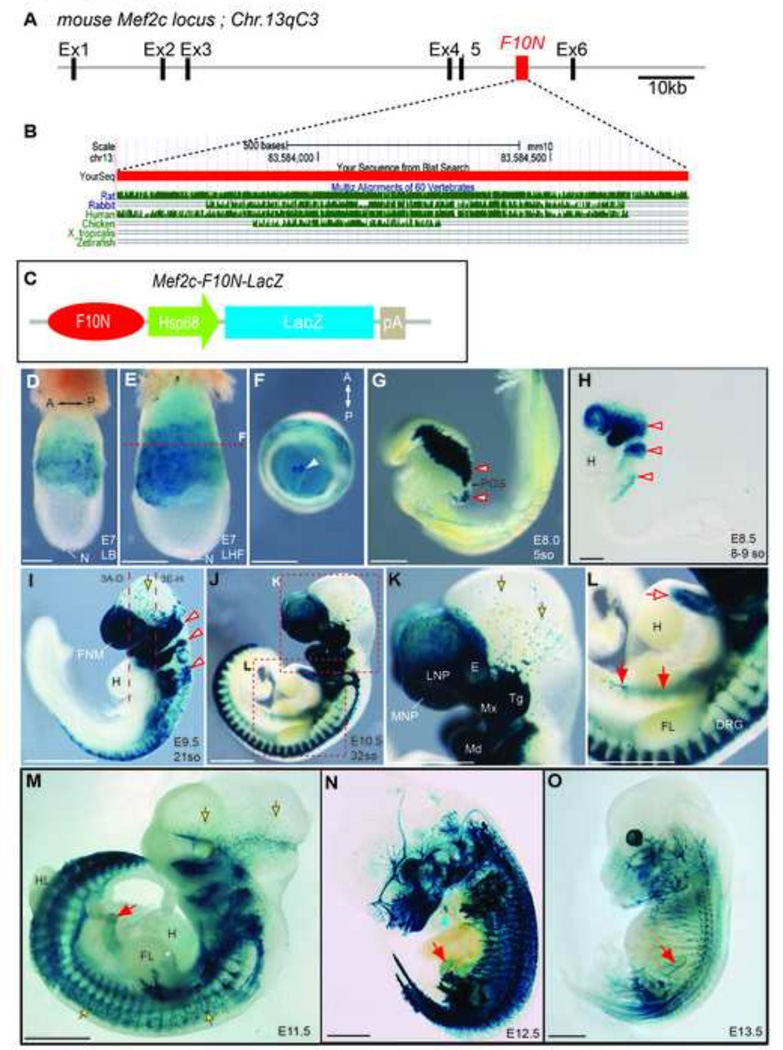
The F10N enhancer was originally identified as a 1,227 base pair fragment (chromosome 13qC3; 83,583,444–83,584,687) in intron 5 of the mouse Mef2c, (also known as MADS box transcription enhancer factor 2 polypeptide C) locus (Fig.1 A). In E9.5 transient transgenic mouse embryos, this enhancer appeared to mark migrating NCC (De Val et al., 2008). Comparative sequence analysis indicates that the Mef2c-F10N enhancer is highly conserved in human, mouse, rat, rabbit and chicken, but not in aquatic species (Fig.1 B). In order to establish new genetic tools for investigating fate and function of NCC, we sought to establish stable transgenic lines with a Mef2c-F10N NCC specific pattern. Therefore a Mef2c-F10N-LacZ construct (Fig.1 C) was injected into pronuclei of one cell stage CBA; B10 F1 hybrid embryos and three stable mouse lines were established. Presence of the Mef2c-F10N-LacZ transgene was confirmed by PCR genotyping. Expression of Mef2c-F10N-LacZ in the newly established lines was examined via whole-mount X-gal staining of embryos at various developmental stages. In E7.0–7.5 gastrulation stage embryos Mef2c-F10N-LacZ is detected in extraembryonic tissues surrounding the exocoelomic cavity including the allantois, but β-galactosidase activity was not present in the embryonic epiblast that gives rise to the embryo (Fig.1D–F). Beginning around E8.0–8.5 (5 somite stage), Mef2c-F10N-LacZ expression appeared in embryonic tissues resembling cranial NCC as they delaminate from the dorsal neuroectoderm (Fig.1G, H, red outline arrowheads). Mef2c-F10N-LacZ continued to be expressed in a pattern consistent with the stereotypical streams of cranial NCC that colonize the pharyngeal arches (Fig.1H, red arrows). In E9.5 embryos, Mef2c-F10N-LacZ continued to label NCC that colonized the craniofacial primordia, including the frontonasal mass (FNM) and pharyngeal arches (Fig.1I, arrowheads). Some scattered Mef2c-F10N-LacZ labeled cells however remained in association with the dorsal midbrain (Fig.1I, K yellow arrows). Furthermore, Mef2c-F10N-LacZ expression in the cardiac, vagal and trunk NCC populations was also clearly evident at this stage. These results suggest that Mef2c-F10N-LacZ marks migrating neural crest cells but does not label premigratory NCC within the neuroepithelium.
Fig.1. Mef2c-F10N-LacZ expression corresponds to delaminating and migrating NCC in whole mount embryos.
(A) Schematic representation of the F10N enhancer element in mouse Mef2c locus in intron 5 between Exon 5 (Ex5) and Exon 6 (Ex6) on chromosome 13 (Chr13). (B) Analysis of sequence conservation of the Mef2c-F10N enhancer among rat, rabbit, human, chicken, Xenopus tropicalis and zebrafish. (C) Schematic of Mef2c-F10N-LacZ transgene construct composed of the F10N enhancer, heat shock protein 68 (Hsp68) minimal promoter, a cDNA encoding β-galactosidase (LacZ) and SV40 polyadenylation signal sequences (pA). (D–O) Mef2c-F10N-LacZ embryos whole mount stained for β-galactosidase activity at progressive stages of development reveal the expression pattern of Mef2c-F10N-LacZ. At early gastrulation stages, expression is observed in extraembryonic tissues. At the onset of NCC migration Mef2c-F10N-LacZ is expressed in NCC newly delaminated form the neural ectoderm. Later, expression persists in NCC and neurogenic, gliogenic and melanocyte derivatives. (D) Lateral view of E7 late bud stage (LB). (E) Lateral view of E7 late head fold stage (LHF) embryo and (F) axial view of exocoelomic cavity opened at red dashed line in (E), allantois (white arrowhead). (G) Lateral view of E8.0, 5 somite stage embryo, in which β-galactosidase activity corresponds to NCC newly delaminated from the neural ectoderm. (H) E8.5, 8–9 somite stage embryos with expression apparent in pre-otic, post-otic and vagal NCC streams. (I) E9.5, 21-somite stage embryo with transgene expression in NCC of the fronotnasal mass, pharyngeal arches, and anterior trunk. (J) E10.5, 32-somite stage embryo with expression in developing facial tissues and trunk PNS. (K–L) Enlarged images of regions outlined in (J). (K) Transgene expression in NCC-filled prominences of the developing face, and in trigeminal ganglion and melanocytes. (L) Transgene expression is apparent in the cardiac outflow tract, ENS and DRG. (M–O) At E11.5 and later stages of development expression is observed in PNS, ENS and melanocytes. (M) E11.5 (N), E12.5 and (O) E13.5. White-red arrowheads indicate pre-otic, postotic and vagal NCC streams. Red solid tailed arrows indicate migrating ENS NCC. Red open tailed arrow, cardiac outflow tract. Yellow-black outlined arrows, presumptive melanocyte progenitors or melanocytes. Blue arrowhead indicates location of thymus. Abbreviations: A, anterior; DRG, dorsal root ganglia; E, eye, FL, fore limb; FNM, frontonasal mass; H, heart; HL, hind limb; LNP, lateral nasal process; MNP, medial nasal process; Md, mandibular process; Mx, maxillary process; N, node; P, posterior; POS, post otic sulcus; so, somites Tg, trigeminal ganglion. Scale bars: 0.2 mm in D–H, 1mm in I–O.
In E10.5 embryos (Fig.1J–L), Mef2c-F10N-LacZ was strongly expressed in NCC-derived facial mesenchyme including the medial and lateral nasal processes and pharyngeal arches. Mef2c-F10N-LacZ expression was also apparent within the cardiac outflow tract (Fig.1L, open red arrow), in trunk DRG (Fig 1J, L) and in the vagal region where NCC enter the gastrointestinal tract (Fig.1L, closed red arrows). At no stage is expression observed within the neural tube. These results suggest that Mef2c-F10N-LacZ marks migrating NCC but does not label premigratory NCC within the neuroepithelium.
Whereas Mef2c-F10N-LacZ expression corresponds to migrating NCC during early stages of embryonic development, from E11.5 onward transgene expression becames more refined. β-galactosidase activity was no longer observed in ectomesenchymal or cardiac NCC. Instead, Mef2c-F10N-LacZ expression appeared to be limited to neuronal and glial NCC derivatives in cranial nerves and the trunk PNS (Fig.1M–O, Fig.2, Supplemental Fig.1). Enhancer activity corresponded to the location of known NCC derivatives in the enteric ganglia of the gut (Fig.1M–O, Fig.2D, red arrow). At E12.5 transgene expression was detected in the region of the thymus (Fig.1N, Fig.2D, blue arrowheads). Expression was also observed in a pattern consistent with that of putative melanocyte progenitors, which migrate into the cranial and vagal region as early as E10.0 (Adameyko et al., 2012) (Fig.1K–O, Fig.2E–H, Fig.3B, F, Supplemental Fig.1G, H, yellow arrows). The distribution of Mef2c-F10N-LacZ expressing presumptive melanocytes at E10.5 and E11.5 suggests that these cells are derivatives of the scattered cells detected within the midbrain of E9.5 embryos (Fig. 1I, yellow arrows). The arrangement and location of X-gal stained cells in the facial and trunk epidermis of E11.5–12.5 embryos (Fig.2 E–H, yellow arrows) is consistent with the known distribution of melanocytes at these stages (Mayer, 1973). Together, these results suggest that between E11.5–13.5, Mef2c-F10N-LacZ expression becomes restricted to the NCC derivatives of the neural and glial lineages within the PNS and to the melanocyte lineage (Fig. 1 M–O, Fig.2 A–D).
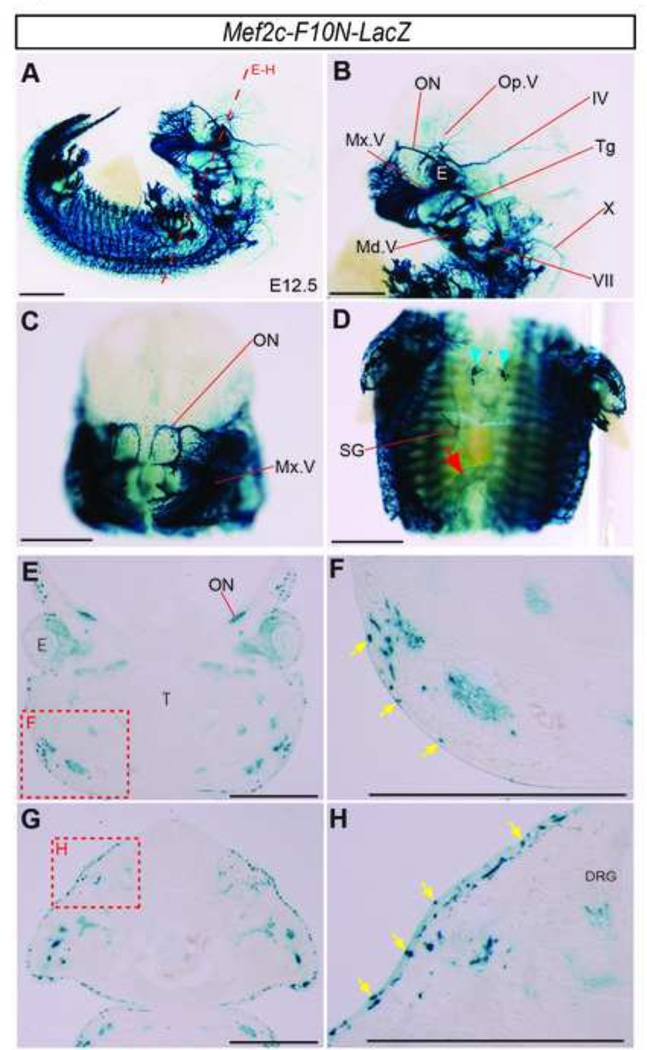
Fig.2. Mef2c-F10N-LacZ expression pattern corresponds to PNS and melanocyte NCC lineages.
(A–D) Whole-mount E12.5 X-gal stained embryos. Lateral (A, B), and frontal (C–D) images of whole embryo (A), head (B–C), and trunk (D). Red arrow indicates ENS NCC staining. Blue arrowheads indicate the location of the thymus. Mef2c-F10N-LacZ expression is apparent in the cranial PNS, trunk PNS, the thymus, and the ENS of the gut. (E–H) Section images of the face (E) and trunk (G) at the level of long red dashed line in (A). (F, H) High magnification images of red dashed insets in (E) and (G), respectively. Transgene expression is detected in epidermal cells in a distribution corresponding to melanocytes, yellow arrows. Abbreviations: DRG, dorsal root ganglion; IV, trochlear nerve; E, eye; Md.V, mandibular brunch of trigeminal nerve; Mx,V, maxillary brunch of trigeminal nerve; ON, olfactory nerve; Op.V, ophthalmic brunch of trigeminal nerve; SG, sympathetic ganglion; T, tongues; Tg, trigeminal ganglion; VII facial nerve; X, vagus nerve. Scale bars: 1 mm in A–H.
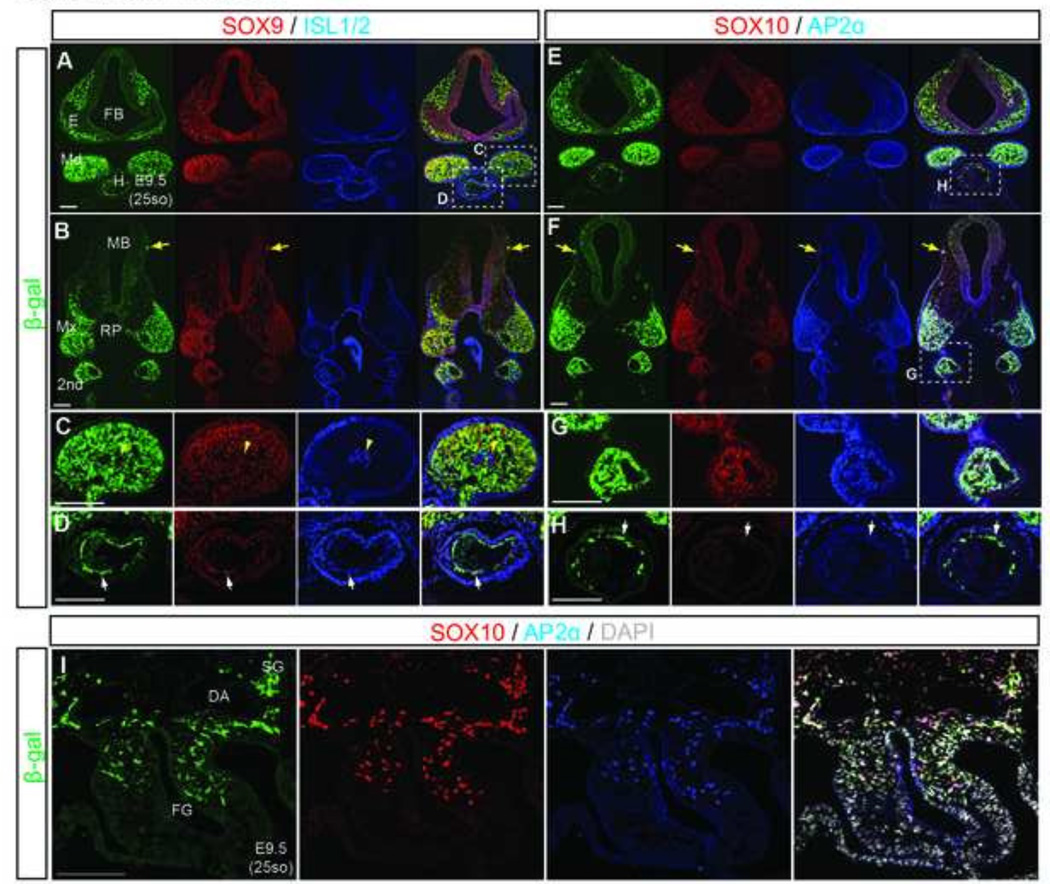
Fig.3. Mef2c-F10N-LacZ labels facial mesenchyme, cardiac and gut NCC.
(A–H) Frontal sections of E9.5 Mef2c-F10N-LacZ embryos at the level of the eye primordium and Rathke’s pouch (A, E) as indicated in Fig.1I. Triple fluorescence staining with NCC markers, SOX9, ISL1/2, SOX10 and AP2α. (C, D, G, H) Enlarged images of areas identified by white dashed insets in A, E, F. Yellow tailed arrows indicate isolated melanocytes under head epithelium. Yellow arrowheads indicate ISL1/2 positive mesenchymal core. White arrows indicate cardiac NCC. (I) Triple staining of frontal foregut sections. DAPI is used to stain nuclei. Abbreviations: 2nd, second pharyngeal arch; DA, dorsal aorta; E, Eys; FB, forebrain; FG, foregut; H, heart; MB, midbrain; Md, mandibular process; Mx, maxillary process; Rp, Rathke’s pouch; SG, sympathetic ganglion. Scale bars: 0.2 mm.
While staining of whole embryos suggested that Mef2c-F10N-LacZ was expressed in cranial, cardiac and gut NCC, in order to properly validate the identity of the cells expressing Mef2c-F10N-LacZ, we performed section immunohistochemistry to visualize LacZ expression with an anti-β-galactosidase antibody in conjunction with standard markers of NCC. To detect NCC we used antibodies against, SOX9, SOX10, and AP2α (Fig.3). In order to identify mesodermal cells within the core of developing pharyngeal arches, we stained for the LIM homeodomain transcriptional factors, Islet1/2 (ISL1/2). Double immunostaining indicates that Mef2c-F10N-LacZ was expressed in NCC derivatives within the mesenchyme of facial primordial (Fig.3A–C, E–G). Importantly there was a clear segregation of transgene expression from the ISL1/2 expressing mesodermal core of the mandibular process (Fig.3C, yellow arrowhead). Mef2c-F10N-LacZ was co-expressed with SOX9/10 and AP2α markers in migrating cardiac NCC (Fig.3D, H, white arrow). Expression of β-galactosidase overlapped with that of SOX10 and AP2α at the entrance to the foregut (Fig.3G, I), indicating that the Mef2c-F10N-LacZ transgene is expressed in vagal NCC progenitors within the enteric nervous system (ENS) as well. Thus, taken together, these data reveal that Mef2c-F10N-LacZ expressing cells which appear in a stereotypical NCC distribution as early as E8.0 are cranial, cardiac and vagal NCC.
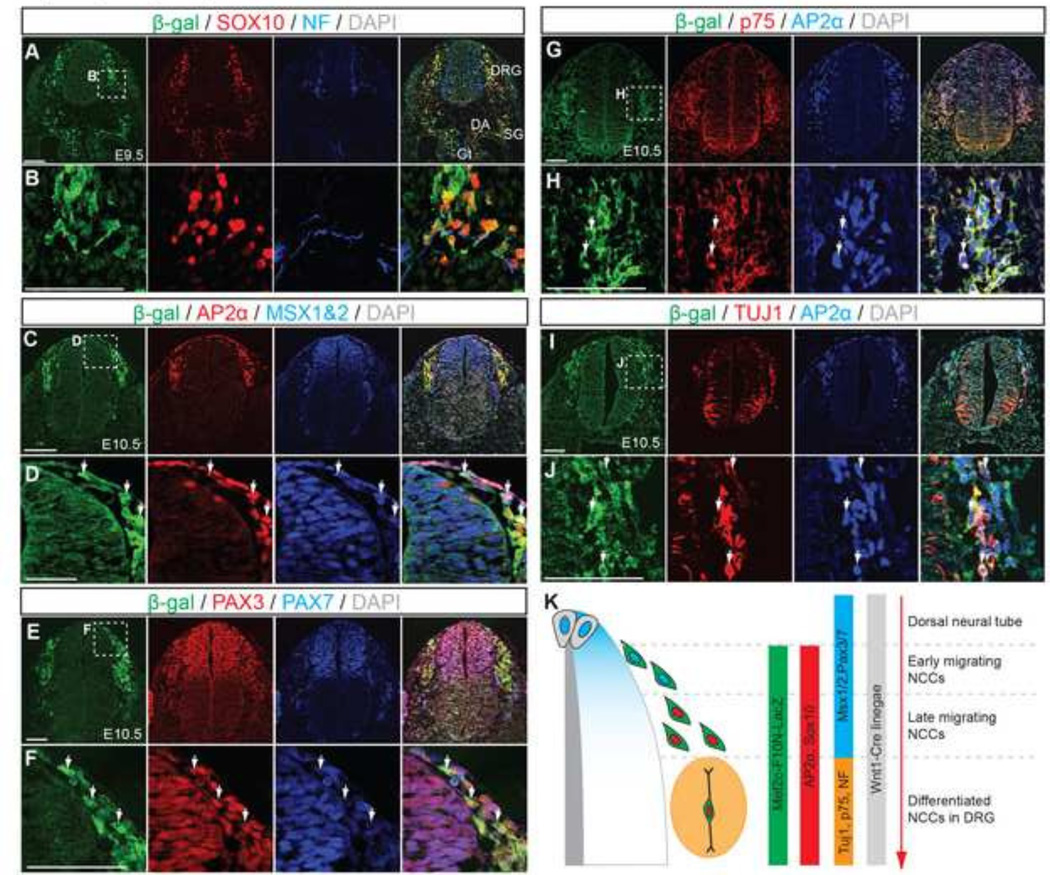
Within the trunk region of the embryo, NCC are known to contribute to neuronal and glial cells of the PNS, including the ENS of the gut, sympathetic ganglia (SG), dorsal root ganglia (DRG), and they also give rise to melanocytes (Jiang et al., 2000) (Fig.4). To examine whether Mef2c-F10N-LacZ demarcates NCC within the trunk region as it does in the head, and to determine if it labels neural and/or glial fated NCC, we performed immunohistochemistry on transverse sections from the trunk level of E9.5–10.5 embryos (Fig.4A–J). Mef2c-F10N-LacZ-positive cells co-stained with SOX10 and AP2α, indicating that the transgene is expressed in NCC in the trunk as it is in the head (Fig.4A–D, G–J). Msh homeobox 1/2 (Msx1/2) and Paired box protein 3 and 7 (PAX3 and PAX7) are expressed in the dorsal neuroepithlium where they label premigratory NCC, and they are also expressed in migrating NCC after delamination (Murdoch et al., 2012). Mef2c-F10N-LacZ labeling overlapped with MSX1/2 and PAX3/7, but only in migrating NCC. (Fig.4E, F). Although we cannot absolutely rule out the occasional Mef2C-F10N-LacZ positive premigratory or delaminating neural crest cell, the fact that MSX1/2 and PAX3/7-positive cells within the neuroepithelium, which include premigratory NCC were not stained for β-galactosidase, indicates that in the trunk, as in the head, Mef2c-F10N-LacZ specifically labels migratory NCC that have delaminated from the neuroepithlium.
Fig.4. Mef2c-F10N-LacZ marks migrating neural crest cells in trunk region.
(A–J) Transverse sections of Mef2c-F10N-LacZ embryos immunostained for β-galactosidase (β-gal) and NCC markers, SOX10, AP2α, MSX1/2, PAX3, PAX7 and differentiated neuronal markers, neurofilament (NF), p75-NGF receptor (p75) at E9.5 and E10.5. (B, D, F, H, J) Enlarged images of insets identified by white dashed squares in (A, C, E, G, I). DAPI was used to stain nuclei. White arrows indicate overlapping expression. (K), Summary of Mef2c-F10N-LacZ expression relative to sequential expression of markers of migrating NCC after delaminating from dorsal neural tube. Abbreviations: DA, dorsal aorta; DRG, dorsal root ganglion; Gt, gut; H, heart; SG, sympathetic ganglion. Scale bars: 0.1 mm.
Within the PNS of the trunk, NCC contribute to both neuronal and glial cell lineages. Neurons of the DRG, derived from NCC can be identified by expression of Neurofilament (NF), Nerve growth factor receptor, (NGFR, known as p75) and neuron-specific class III β-tubulin (Tuj1) (Fig.4A, B, G–J). Immunostaining for each of these molecules reveals the presence of cells in developing DRG co-labeled with β-galactosidase and each of the neuronal markers, demonstrating that Mef2c-F10N-LacZ labels NCC derived neurons in the PNS (Fig.4A, B, G–J). Similarly at later embryonic stages, Mef2c-F10N-LacZ labeled cells co-stained with Tuj1 in cranial and trunk peripheral nerves (Fig.2, Supplemental Fig. 1). These results indicate that the Mef2c-F10N enhancer labels migrating NCC after their delamination from the neuroepithelium and continues to label NCC fated to become neurons within the PNS (Fig.4K).
It is important to note that we have not observed β-galactosidase expression in skeletal derivatives even as early as E12.0 (Figure 2) nor in skeletal tissues in older E15.5 and E18.5 embryos. Thus Mef2c-F10N-LacZ must be switched off in putative ectomesenchyme prior to any mesenchymal condensation. These observations are consistent with Mef2c-F10N-LacZ labeling migratory neural crest cells and their neurogenic, but not mesenchymal derivatives.
Comparison of Mef2c-F10N-LacZ transgenk embryos and Wnt1-Cre; R26R and Rosa-YFP embryos
To further validate the utility of the Mef2c-F10N-LacZ line in labeling NCC and their derivatives, we compared its pattern of expression to that generated when Wnt1-Cre is crossed to Rosa26R Cre-dependent reporter lines, which has been widely used for genetic tracing of NCC lineages, cell fate specification and gene function. E8.5–9.5 Mef2c-F10N-LacZ embryos exhibit a remarkably similar staining patterning to Wnt1-Cre;R26R embryos, with the exception that Mef2c-F10N-LacZ labels only migrating NCC (Fig.5A, C, I), whereas Wnt1-Cre;R26R marks the dorsal neural plate and premigratory NCC in addition to migratory NCC (Fig.5 B, D, F, J red line and bracket). In E8.5 embryos both transgenes mark NCC that were delaminating from the forebrain and midbrain neuroepithelium (Fig. 5A–D), but Wnt1-Cre;R26R additionally labels the dorsal neuroepithelium (Fig.5B red line; Fig.5D red bracket).
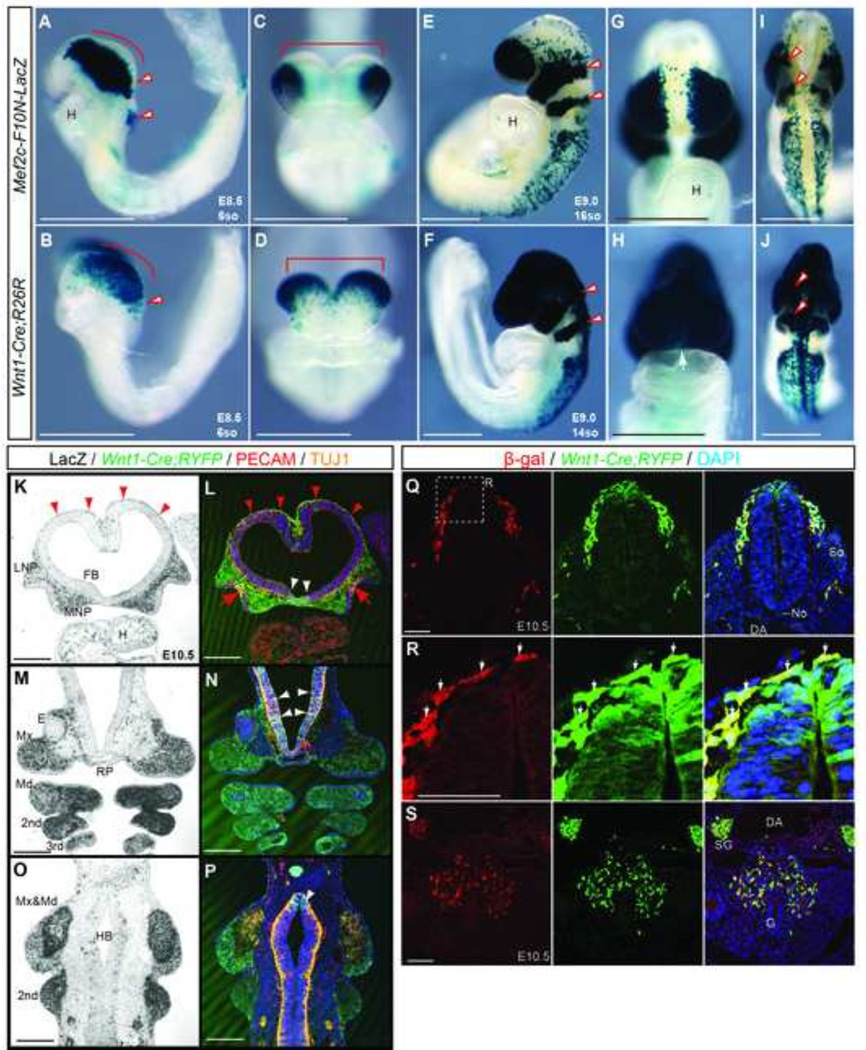
Fig.5. Mef2c-F10N-LacZ marks cells in a pattern very similar to Wnt1-Cre but is specific for migratory NCC.
(A–J) Comparison between Mef2c-F10N-LacZ (A, C, E, G, I) expression and Wnt1-Cre;R26R lineage marking (B, D, F, H, J) in whole embryos. Distribution of β-galactosidase activity in lateral (A, B, E, F), frontal (C, D, G, H) and dorsal (I, J) views of E8.5, 6-somite stage (A–D) and E9.0, 16-somite stage (E–J) embryos. Red outline arrowheads indicate pre-otic NCC streams. (K–P) Frontal sections of E10.5 Mef2c-F10N-LacZ and Wnt1-Cre;Rosa-YFP compound embryos. Mef2c-F10N-LacZ expression is revealed by β-galactosidase activity (K, M, O), and Wnt1-Cre;YFP lineage marking is revealed by immunostaining for YFP in conjunction with the endothelial marker CD31, and the neural marker Tuj1. Red arrowheads indicate NCC of dorsal forebrain that may contribute to the meninges. Red tailed arrows indicate delaminating olfactory nerve from the olfactory epithelium. White arrows indicate the overlap with migrating NCC. (Q–S) Double staining of β-galactosidase expression from the Mef2c-F10N-LacZ transgene and YFP expression from Wnt1-Cre;Rosa26YFPi+ in neural tube (Q, R) and gut (S) in E10.5 embryos. (R) High magnification images of white dashed inset in (Q). White arrows indicate the overlap with migrating NCC. DAPI is used to stain nuclei. Abbreviations: 2nd second pharyngeal arch; 3rd pharyngeal arch; DA, dorsal aorta; E, eye; FB, forebrain ventricle; G, gut; H, heart; HB, hindbrain; LNP, lateral nasal process; Md, mandibular process; Mx, maxillary process; MNP, medial nasal process No, notochord; RP, Ratke’s pouch; So, somite; SG, sympathetic ganglion; 3rd, third pharyngeal arch; TG, trigeminal ganglion. Scale bars: 0.5 mm in A–P, 0.1 mm in Q–S.
In developing craniofacial tissues of Mef2c-F10N-LacZ embryos, labeled cells primarily populated the frontonasal process and pharyngeal arches (Fig.5E red arrowheads), while in contrast, labeled cells in Wnt1-Cre;R26R embryos populated the entire head, including the frontonasal process and pharyngeal arches (Fig.5F red arrowheads). In E9.0 embryos, NCC migrating into the trunk region were labeled to a similar extent by both transgenes (Fig.5E, F). However, the midline region of the frontonasal process, brain and spinal cord did not stain in Mef2c-F10N-LacZ embryos, but does stain in Wnt1-Cre;R26R embryos (Fig.5 G–J). These data are consistent with the observation that Mef2c-F10N-LacZ specifically marks migrating NCC after delamination, in contrast to Wnt1-Cre;R26R which labels the dorsal neuroepithelium and consequently premigratory NCC, as well as migratory NCC
To more precisely determine the extent of overlapping expression between Mef2c-F10N-LacZ and Wnt1-Cre-labeled cells, we assessed the expression of Mef2c-F10N-LacZ within the background of Wnt1-Cre;RYFP (ROSA-yellow fluorescent protein) embryos. In cranial tissues of E10.5 embryos, Mef2c-F10N-LacZ was expressed in NCC derived facial mesenchyme and cranial nerves (Fig.5 K, M, O) in a pattern very similar to that of Wnt1-Cre;RYFP (Fig.5L, N, P). In the trunk, Mef2c-F10N-LacZ was expressed in migrating NCC contributing to the DRG and the gastrointestinal tract in a pattern similar to Wnt1-Cre;RYFP (Fig.5Q–S). Consistent with previous observations, Wnt1-Cre;RYFP embryos also exhibited expression in the dorsal regions of the brain and spinal cord (Fig.5L, N, P, Q, R), whereas Mef2c-F10N-LacZ did not (Fig.5K–R). Thus, Mef2c-F10N-LacZ is expressed in NCC in a manner highly overlapping with that the Wnt1-Cre lineage map, but is specific for migratory NCC that have delaminated form the neuroepithelium.
Mef2c-F10N-Cre transgene permanently marks delaminated migrating neural crest cells
To expand the utility of the Mef2c-F10N enhancer and to allow for indelible labeling or genetic modification of all migrating and differentiating NCC independently of premigratory NCC within the dorsal neuroepithelium, we constructed a Cre transgene using the Mef2c-F10N enhancer (Fig.6A). Mef2c-F10N-Cre transgenic mice were generated via pronuclear injection of one cell CBA; B10 F1 hybrid embryos. Six Cre positive founder lines were produced. To check the activity of the Mef2c-F10N-Cre lines, we mated each founder line with R26R reporter mice and analyzed for spatiotemporal activity at multiple developmental stages (Fig.6A). Two founder lines (#2 and #5) indelibly labeled migrating NCC and their derivatives (Fig.6B, C and Supplemental Fig. 2). The frequency of inherited transgene expression in Line 2 and Line 55 was 46.9% (15/32) and 6.61% (8/121), respectively. Line 2 exhibited some non-NCC associated labeling in the heart and forelimb (Supplemental Fig.2). Therefore, we used Mef2c-F10N-Cre line 5 (Fig.6) for the subsequent detailed analysis of NCC and their derivatives (Fig.7–10).
Fig.6. Mef2c-F10N-Cre Line 5.
(A, B) E9.5 and (C) E10.5 Mef2c-F10N-Cre;R26R embryos stained for β-galactosidase activity illustrating the pattern of migrating NCC contribution to the head, heart and trunk. Black arrow indicates labeled cells in the heart (C). Mef2c-F10N-Cre labeled NCC also contribute to the aortic arch and ductus arteriosus (arrows in D, E), and valve leaflets (arrow in F) in E14.5 Mef2c-F10N-Cre;R26R embryos. Scale bars: 1 mm.
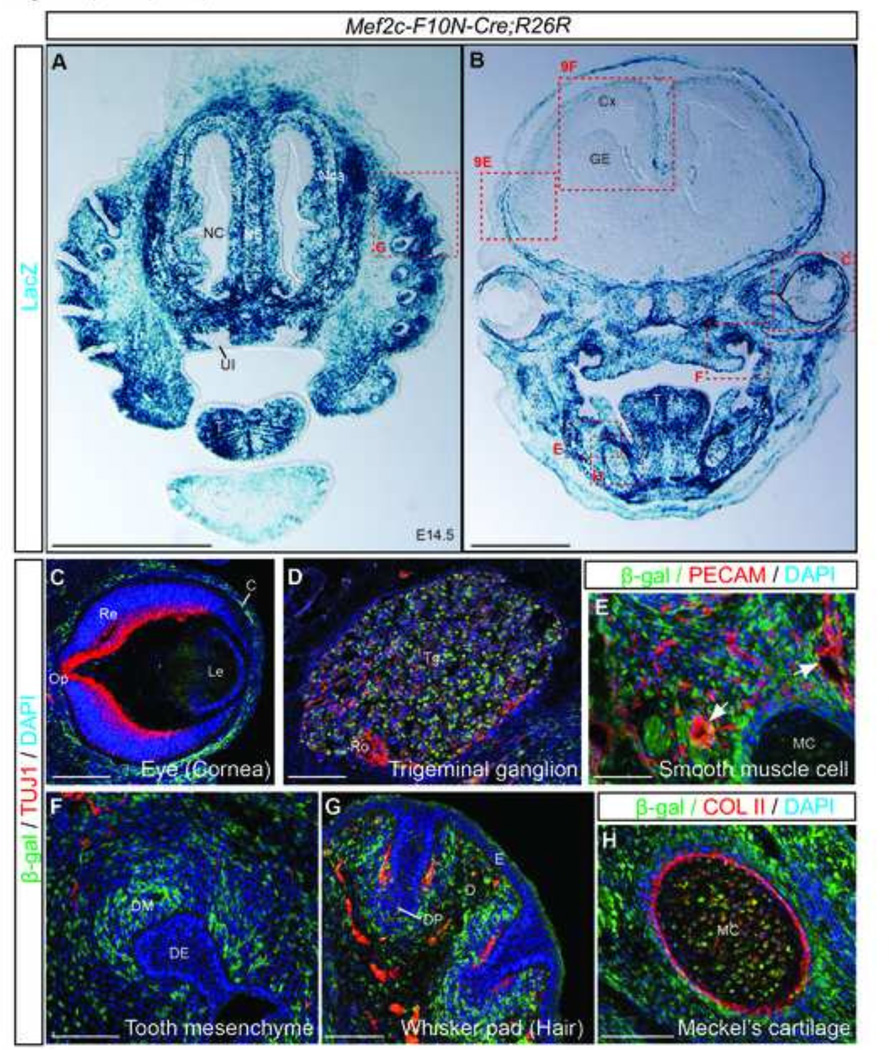
Fig.7. The Mef2c-F10N-Cre lineage includes cranial NCC derivatives.
(A, B) Frontal sections of E14.5 Mef2c-F10N-Cre;R26R embryos at the level of the nasal region (A), and eye (B) stained for β-galactosidase activity. (C–F) Triple fluorescence staining of cells marked by Cre excision in Mef2c-F10N-Cre embryos (β- β-galactosidase), mature neurons (Tuj1), endothelial cells (CD31), or collagen II (Col II) to identify cartilage. Mef2c-F10N-Cre lineage labeled cells are present in the cornea of the eye (C), trigeminal ganglion (D), vascular smooth muscle (E), tooth mesenchyme (F), whisker pad (G) and Meckel’s cartilage (H). Abbreviations: C, cornea; Cx, cortex; D, dermis; DE, dental epithelium; DM, dental mesenchyme; DP, dermal papilla; E, epidermis; GE, ganglionic eminence; Le, lens; MC, Meckel cartilage; NC, nasal cavity; Nca, nasal capsule; NS, nasal septum; Op, optic nerve; Re, retina; Ro, route of trigeminal nerve; T, tongue; Tg, trigeminal ganglion; UI, upper incisor. Scale bars: 1 mm in A, B, 0.1 mm in C–H.
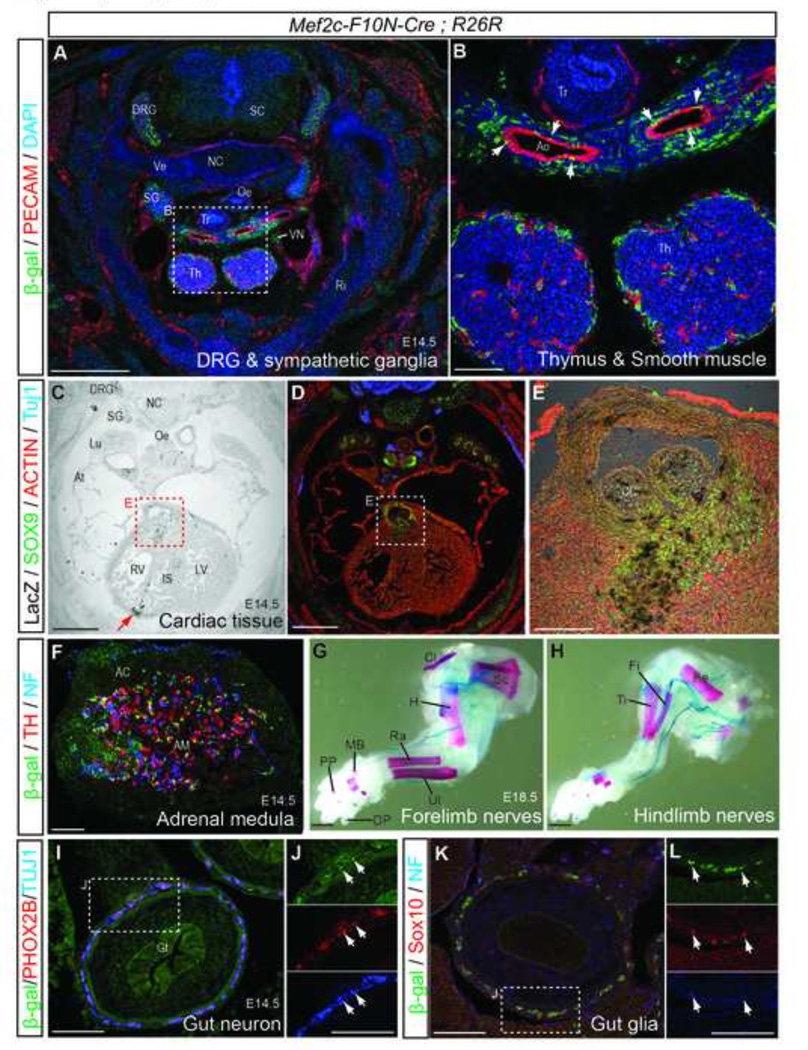
Fig.10. Mef2c-F10N-Cre lineage labels trunk NCC derivatives.
(A, B) Transverse sections of an E14.5 Mef2c-F10N-Cre;R26R embryo at upper chest level imunostained to detect cell linages labeled by Cre excision (β-gal) and endothelial cells (CD31). (B) High magnification images in A. White arrows indicate NCC in smooth muscle of the aorta. (C–E) Transverse section at cardiac level stained for β-galactosidase activity (X-gal) and immunostained to identify NCC (SOX9), mature neurons (Tuj1), and muscle (Actin) (D, E). Red arrow indicates expression in non-NCC cardiac tissue. (E) High magnification of inset in (C, D). (F) Section through adrenal gland of E14.5 embryo immunostained for β-galactosidase, tyrosine hydroxylase (TH) and Tuj1. (G, H) LacZ and Alizarin red staining of E18.5 fore limb (G) and hind limb (H). (I–L) Section of gut stained for Cre excision (β-gal) and for mature neurons (PHOX2B/Tuj1) (I, J), or for NCC and glia SOX10/neurofilament (NF) (K, L). J, L, High magnification images of insets in I, J. White arrows indicate triple overlapping expression. Abbreviations: AC, adrenal cortex; AM, adrenal medulla; Ao, aorta arch; At, atrium; CC, conotruncal seminlunar valve leaflets; Cl, clavicle; DRG, dorsal root ganglion; DP, distal phalange; Fe, femure; Fi, fibula, Gt, gut; H, humerus; IS, interventricular septum; LV, left ventricle;, Lu, lung; N, nasal bone; NC, notochord; Oe, oesophagus; PP, proximal phalange; Ra, radius bone; Ri, rib; RV, right ventricle; Th, thymus; Ti, tibia; Tr, trachea; Ul, ulna bone; Sc, scapula bone; SC, spinal cord; SG, sympathetic ganglion; Ve, vertebral body; VN trigeminal nerve. Scale bars: 0.5 mm in A, C, D, 0.1 mm in B, E, F, I–L, 1 mm in G, H.
Mef2c-F10N-Cre labeled NCC contribute to numerous organs and tissues
NCC contribute to many tissues and organs throughout the body. To validate Mef2c-F10N-Cre as a useful line for indelibly labeling migrating and differentiating NCC, we characterized the distribution of labeled cells in Mef2c-F10N-Cre;R26R embryos throughout gestation. In frontal sections of E14.5 Mef2c-F10N-Cre;R26R embryos (Fig.7A, B), many cell types were stained with X-gal. Mef2c-F10N-Cre;R26R marked the cornea of the eye, trigeminal ganglion, tooth mesenchyme, whisker pad and Meckel’s cartilage (Fig.7C–H). The mesenchymal cell labeling in these facial tissues was consistent with NCC lineage tracing in Wnt1-Cre and other NCC-specific reporter mice.
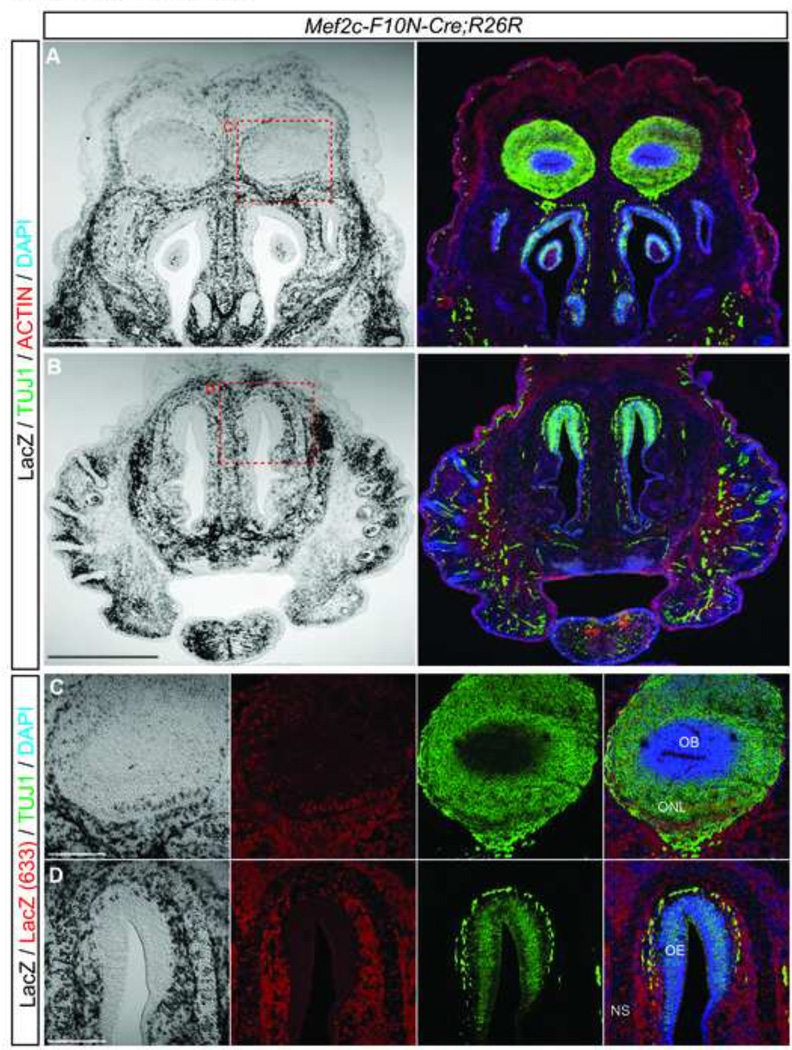
A number of reports have indicated that NCC invade the olfactory epithelium and contribute to olfactory neurons and olfactory ensheathing glial cells that support olfactory axons projecting to the olfactory bulb (Barraud et al., 2010; Katoh et al., 2011; Murdoch et al., 2012). E14.5 Mef2c-F10N-Cre;R26R embryos exhibit β-galactosidase expression within the olfactory bulb, and the mesenchyme surrounding the olfactory epithelium, but not the olfactory epithelium itself (Fig.8A–D). Of the β-galactosidase expressing cells in the olfactory bulb most were negative for Tuj1 (Fig.8C). The lack of overlapping β-galactosidase expression and neuronal maker expression in the olfactory bulb suggests that NCC do not invade the olfactory epithelium to become neurons, but instead contribute primarily to the olfactory ensheathing glial cell lineage in the peripheral and central olfactory region (Fig.8D).
Fig.8. Mef2c-F10N-Cre lineage labeling in the olfactory bulb.
(A–D) Frontal sections through developing olfactory structures of an E14.5 Mef2c-F10N-Cre;R26R embryo. (A–B) X-gal staining and immunostaining for markers of mature neurons (Tuj1) and muscle (Actin). (C, D) High magnification images of olfactory bulb and nasal epithelium as outlined in red dashed insets in (A, B) with X-gal staining directly detected by 633nm fluorescence emission in combination with antibody staining of mature neurons (Tuj1). Abbreviations: NS, nasal septum; OB, olfactory bulb; OE, olfactory epithelium; ONL, olfactory nerve layer. Scale bars: 0.5 mm in A, 0.2 in B–D.
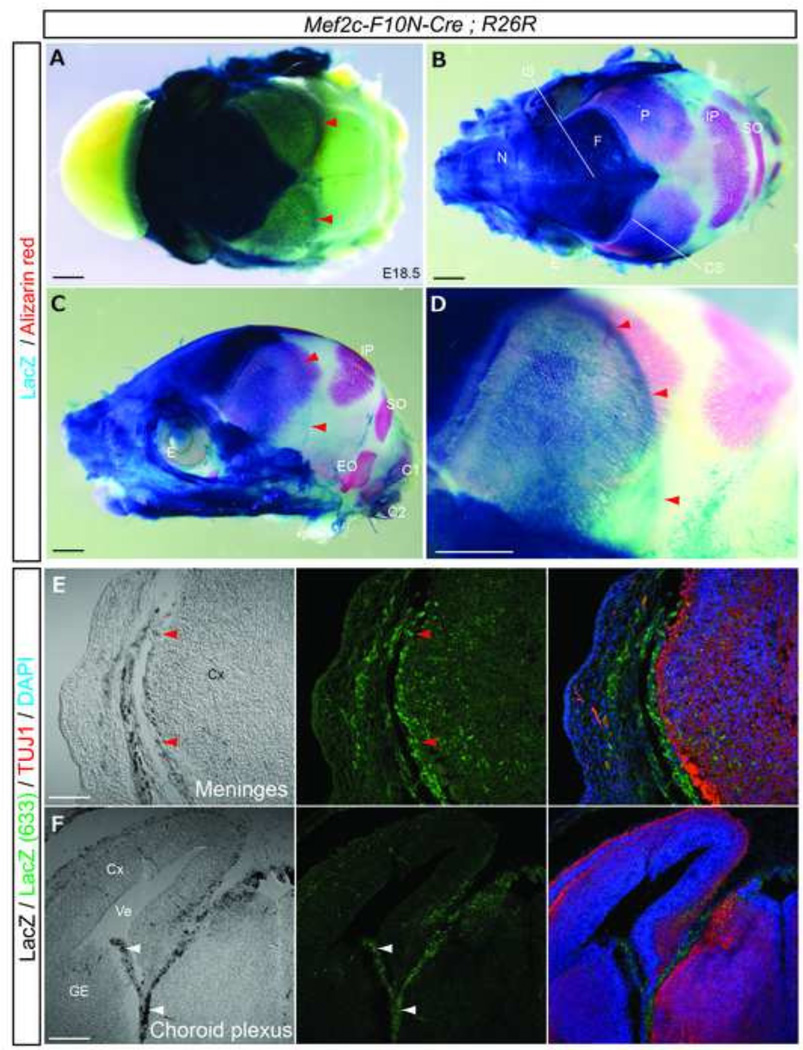
NCC contribution to the cranial vault and brain as assessed using Wnt1-Cre;R26R mice or chicken-quail chimeras indicates that the frontal bone and a portion of the interparietal bone of the calvaria, and the meninges (dura, arachnoid and pia mater) of the forebrain are NCC-derived (Etchevers et al., 2001; Jiang et al., 2002; Siegenthaler and Pleasure, 2011). To check whether Mef2c-F10N-Cre marks a similiar set of NCC derivatives, we performed whole-mount X-gal staining in conjunction with Alizarin red staining to identify bone. Staining of E18.5 Mef2c-F10N-Cre;R26R embryos revealed the frontal bone was strongly β-galactosidase positive, and labeled cells were evident in the coronal suture (Fig.9A–D). However, it is evident histologically that neither the parietal nor interparietal bones were labeled Sections of embryos immunostained for Tuj1/DAPI revealed that NCC contribute to the meninges surrounding the forebrain derived cortex (Fig.9A, C–E, red arrowheads) and choroid plexus (Fig.9F, white arrows). These results are consistent with other analyses of neural crest cells (Chai et al., 2000; Kague et al., 2012; Matsuoka et al., 2005; Noden, 1978; Sailer et al., 2005; Sternberg et al., 2014). Thus, cranial NCC labeled by Mef2c-F10N-Cre-labeled contribute to the frontal bone and meninges of cortex and diencephalon, but not the parietal and interparietal bones, nor to the meninges of the midbrain.
Fig.9. Mef2c-F10N-Cre lineage labels calvarial bones of the skull and meninges and choroid plexus of the brain.
(A), Dorsal view of the skull vault of an E18.5 Mef2c-F10N-Cre;R26R embryo stained with X-gal. (B–D) Skull double stained for X-gal and Alizarin red in dorsal (B) and lateral (C, D) views. (D), Higher magnification image of specimen in (C). (E, F) Frontal sections through the forebrain region of an E14.5 embryo corresponding to inset in Fig.7B. Distribution of β-galactosidase activity revealed by bright field microscopy and also by direct detection of far-red (633nm) X-gal fluorescence in conjunction with immunostaining for neural maturation, (Tuj1) and nuclei, (DAPI). Red arrowheads in (A, C–E) indicate β-galactosidase activity of meninges covered cortex. White arrowheads in F indicate choroid plexus. Abbreviations: C1, C1 vertebra; C2, C2 vertebra; CS, coronal suture; Cx, cortex; E, eye; EO, exoccipital bone; F, frontal bone; GE, ganglionic eminence; IP, interparietal bone; IS, interfrontal suture; N, nasal bone; P, parietal bone; SO, supraoccipital bone; Ve, ventricle. Scale bars: 1 mm in A–D, 0.2 in E, F.
We also explored the distribution of Mef2c-F10N-Cre-labeled NCC in the trunk of E14.5 embryos. Here we observed β-galactosidase positive cells in dorsal root ganglia (DRG), sympathetic ganglia, in the thymus, and in smooth muscle cells of the aortic arches (Fig.10A, B). In the heart, Mef2c-F10N-Cre labeled NCC were observed in the mesenchyme at the base of the conotruncus and in the forming semilunar valve leaflets (Fig.6F and Fig.10C–E). Mef2c-F10N-Cre labeled NCC also contributed to the aortic arch and ductus arteriosus (Fig. 6D, E). Earlier in E10.5 embryos, β-galactosidase positive cells are clearly evident migrating through the outflow tract of the heart (Fig. 6C). NCC labeled by Mef2c-F10N-Cre also populated the medulla of the adrenal gland, where they co-stained with the chromaffin cell marker, tyrosine hydroxylase (TH) and the neural intermediate filament marker, neurofilament (NF) (Fig.10F). Furthermore, Mef2c-F10N-Cre-labeled NCC also contribute to the nerves of the forelimbs and hindlimbs (Fig.10G, H) and to the ENS of the gut, where they co-stained with the autonomic neuron marker, paired-homeodomain transcriptional factor (PHOX26) (Nagashimada et al., 2012) the differentiated neuron marker Tuj1 (Fig.10I, J), the glial marker SOX10, and neurofilament protein (NF) (Fig.10K, L). Thus, the Mef2c-F10N-Cre lineage includes migrating trunk NCC and their stereotypical derivatives.
Discussion
Several transgenic lines have been generated for lineage tracing and analyses of gene function in NCC. However, some of these lines have non-NCC specific expression and gene activity or other unwanted characteristics. For example, the widely used Wnt1-Cre mice exhibit dorsal cranial and trunk neuroepithelial expression (Fig.5L, N, P, Q, R). Furthermore, although a new version of Wnt1-Cre has been generated termed Wnt1-Cre2 (Lewis et al., 2013), the widely used original version of the Wnt1-Cre transgenic line exhibits elevated Wnt signaling mediated by Wnt1 coding sequence in the transgenic construct (Lewis et al., 2013). Other Cre lines that label migrating NCC additionally express in cells other than NCC. For example, Sox10-Cre labels non-NCC derivatives in the cochlea owing to its expression in the otic vesicle (Jacques-Fricke et al., 2012) and P0-Cre labels non-NCC derivatives in the brain. The activity of these Cre lines makes them less than ideal for lineage tracing of NCC and for analyses of gene function in migratory and differentiating NCC independently of NCC formation. Thus new tools that enable labeling or genetic modification specifically within NCC are needed.
Here we describe two novel transgenic lines Mef2c-F10N-LacZ and Mef2c-F10N-Cre, which mark migrating NCC after delamination from the neural tube. Aside from the distinction between pre-migratory and migratory NCC, the major difference between the Mef2c-F10N-Cre and Wnt1-Cre lines appears to lie with respect to lineage tracing of derivatives of migrating NCC in the calvarial bones (Fig.9). Our data, together with published reports of Wnt1-Cre;R26R embryos, indicate that in addition to the frontal bone, the Wnt1-Cre driver also labels the medial part of the interparietal bone (Jiang et al., 2002; Yoshida et al., 2008). In contrast, the Mef2c-F10N-Cre lineage tracing does not appear to include cells in the interparietal bone (Fig.9B–D). Other than that, the similarities between the Mef2c-F10N-Cre and Wnt1-Cre lines are strong. The close concordance lends confirmation that the Mef2c-F10N-Cre transgene may serve as an independent tool to genetically label or modify NCC and their derivatives.
The precise contribution of NCC to olfactory tissues, particularly the olfactory epithelium and neurons remains unresolved. Recently published P0-Cre, Pax7-Cre, Wnt1-Cre and Sox10-Venus lineage tracing experiments have documented labeling of olfactory neurons and olfactory ensheathing glia. However, a contribution of NCC to the olfactory epithelium remains controversial (Barraud et al., 2010; Katoh et al., 2011; Murdoch et al., 2010). GFP transgenic chick grafting experiments suggest that NCC do not contribute to the olfactory epithelium (Barraud et al., 2010), but more recent EGFP electroporation experiments in chick embryos suggest that a few NCC do invade the olfactory epithelium (Katoh et al., 2011). Interestingly, in Mef2c-F10N-Cre;R26R embryos, olfactory ensheathing glia surrounding the olfactory epithelium and within the olfactory bulb were labeled (Fig.8A, B), but neurons of the olfactory epithelium were not (Fig.2C, Fig.5K, Fig.7A, Fig.8B). Thus, our Mef2c-F10N-LacZ and Mef2c-F10N-Cre embryo labeling data strongly supports the view that NCC contribute to olfactory ensheathing glia but not to neurons of the olfactory epithelium.
In conclusion, the Mef2c-F10N-LacZ transgenic line labels migrating NCC that have delaminated from the neuroepithelium and continues to predominantly mark neuronal and glial NCC derivatives in the PNS and melanocytes in the skin throughout embryonic development. The Mef2c-F10N-Cre similarly induces Cre recombination in NCC after their delamination from the neuroectoderm. The final distribution pattern of indelibly labeled NCC is very similar to that of Wnt1-Cre-labeled NCC. Therefore, our Mef2c-F10N transgenic lines provide useful new tools and resources for lineage tracing and analyses of gene function in migrating NCC independently of NCC formation.
Materials and methods
Generation of Mef2c-F10N-LacZ and Mef2c-F10N-Cre transgenic mice
Mef2c-F10N was cloned into the transgenic reporter plasmid hsp68-LacZ (Kothary et al., 1989a; Kothary et al., 1989b) to generate Mef2c-F10N-LacZ (De Val et al., 2008). Mef2c-F10N-Cre was made by replacing the LacZ gene of Mef2c-F10N-LacZ with the Cre cDNA from p8S185-CMV-Cre (Sauer and Henderson, 1990). To generate transgenic mice, a 5.5-kb Sall fragment of Mef2c-F10N-LacZ and a 3.8-kb Not1/Sall fragment of Mef2c-F10N-Cre were gel purified, ethanol precipitated and used for pronuclear injection into CBA; B10 fertilized eggs.
Genotyping
Genotyping by PCR with LacZ and Cre specific probes was performed by Transnetxy Inc.
β-galactosidase detection and bone staining
Mouse embryos were dissected and fixed in 1% formalin/0.1%PBS or 4% Paraformaldehyde (PFA) 0.1% PBS or 0.2% glutalaldehyde/2% formalin/0.1% PBS for 15 min (E7–8), 30 min (E9–10), 1hr (E11–12), 1.5hr (E13 and older) at 4°C. The strength of LacZ staining varied depending on fixation with glutaraldehyde/formalin producing the strongest signal followed by PFA and then formalin. Fixing with glutaraldehyde/formalin was the most suitable for visualizing melanocytes, but was not used for co-immunofluorescent staining because glutaraldehyde fixation can result in tissue autofluorescence (Collins and Goldsmith, 1981). For whole-mount X-gal staining, embryos were washed 3 × 15 minutes in cold PBS prior to β-galactosidase detection. For cryosection staining, embryos were washed with cold PBS, then processed through a graded series of sucrose solutions 5%/10%/15%/20% in PBS, before being embedded in 2:1 mixture of 20% sucrose/OCT compound (Tissue-Tek) and stored at −80°C until sectioned. β-galactosidase activity was detected with X-gal solution (40mg/ml X-gal, 5mM potassium ferricyanide, 5mM potassium ferrocyanide, 2mM MgCl, 0.01% sodium deoxycholate, 0.02% Nonidet P-40). For combined X-galand Alizarin red staining, E18.5 embryos were first stained with X-galand then stained with 0.005% Alizarin red S in 3% KOH (A5533, Sigma-Aldrich) to visualize bone. For combined X-gal staining with immunostaining, slides were first stained with X-gal, then washed with PBS and post-fixed with 10% formalin/PBS for 10min, prior to antibody immunostaining. β-galactosidase signal was detected under bright field microscopy and also fluorescently via confocal microscopy as an emission signal in the 650–770 nm range (far red) after excitation at 633 nm (Levitsky et al., 2013).
Immunofluorescence
Dissected mouse embryos were fixed in 4% PFA/PBS for 2 hours or 1% formalin overnight at 4°C and then embedded in OCT as described above. Frozen sections (10–14 µm) were cut, then dried with a hair dryer for 30–60 sec. Sections were washed 3 × 5 minutes in PBS, incubated in a blocking solution of 3% bovine serum albumin (BSA) / 0.01% Tween 20 in PBS (PBT) for 1hr at room temperature, and then incubated with primary antibodies overnight at 4°C Antibodies used include mouse anti-Actin HHF35 (1/100, M0635, DAKO), mouse anti-AP2α (1/50, 3B5, Iowa hybridoma), rabbit anti-β-galactosidase (1/2,000–1/5,000, #559761, Capple), goat anti-β-galactosidase (1/2,000–1/5,000, #4600–1409, Biogenesis), mouse anti-collagen 11 (1/50, II-II6B3, Iowa hybridoma), chick anti-GFP (1/1,000, GFP-1020, Aves), rabbit anti-GFP (1/1,000, A6455, Invitrogen), mouse anti-Islet1/2 (1/10, 4D5, Iowa hybridoma), mouse anti-MSX1/2 (1/50, 4G1, Iowa hybridoma), mouse anti-neurofilament (1/100, 2H3, Iowa hybridoma), rabbit anti-p75 NGFR (1/500, G3231, Promega), mouse anti-PAX3 (1/50, Iowa hybridoma), mouse anti-PAX7 (1/100, Iowa hybridoma), rat anti-CD31 (1/100, #553370, BD pharmingen), goat anti-PHOX2b (kindly provide form Dr. Enomoto), mouse anti-SOX2 (1/100, MAB2018, R&D system), rabbit anti-SOX9 (1/500, AB5535, Millipore), goat anti-SOX10 (1/100, AF2864, R&D system), rabbit anti-Tuj1 (1/2,000, PRB-435P, Covance) and rabbit anti-tyrosine hydroxylase (1/100, #2792, Cell signaling). Following incubation with primary antibodies, slides were washed 3×5 minutes in PBS, and then incubated with secondary antibodies for one hour at room temperature. Secondary antibodies included donkey anti-goat, -mouse or -rabbit fluorescent Alexa Fluor 488, 546, 647 conjugated antibodies (1/200, Invitrogen), DAPI was used to stain the nuclei (1/1,000, Sigma). Fluorescent confocal images were acquired using an LSM-510-VIS, Upright Pascal or LSM-700 (Carl Zeiss).
Genome comparison
The UCSC genome browser (http://genome.ucsc.edu/) was used to compare the Mef2c-F10N enhancer from rat, rabbit, human, chicken, Xenopus tropicalis and zebrafish.
Supplementary Material
Acknowledgements
We wish to thank Dr. Robb Krumlauf for providing the Cre recombinase expression vector and Dr. Hideki Enomoto for the PHOX2B antibody. The AP2a, collagen II, Islet1/2, MSX1/2, 2H3, PAX3, and PAX7 antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was supported by the Stowers Institute for Medical Research and the National Institute of Dental and Craniofacial Research (DE 016082).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achilleos A, Trainor PA. Neural crest stem cells: discovery, properties and potential for therapy. Cell Res. 2012;22:288–304. doi: 10.1038/cr.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adameyko I, Lallemend F, Furlan A, Zinin N, Aranda S, Kitambi SS, Blanchart A, Favaro R, Nicolis S, Lubke M, Muller T, Birchmeier C, Suter U, Zaitoun I, Takahashi Y, Ernfors P. Sox2 and Mitf cross-regulatory interactions consolidate progenitor and melanocyte lineages in the cranial neural crest. Development. 2012;139:397–410. doi: 10.1242/dev.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P, Verzi MP, Nguyen T, Hu J, Ehlers ML, McCulley DJ, Xu SM, Dodou E, Anderson JP, Wei ML, Black BL. The MADS box transcription factor MEF2C regulates melanocyte development and is a direct transcriptional target and partner of SOX10. Development. 2011;138:2555–2565. doi: 10.1242/dev.056804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Seferiadis AA, Tyson LD, Zwart MF, Szabo-Rogers HL, Ruhrberg C, Liu KJ, Baker CV. Neural crest origin of olfactory ensheathing glia. Proc Natl Acad Sci U S A. 2010;107:21040–21045. doi: 10.1073/pnas.1012248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Collins JS, Goldsmith TH. Spectral properties of fluorescence induced by glutaraldehyde fixation. J Histochem Cytochem. 1981;29:411–414. doi: 10.1177/29.3.6787116. [DOI] [PubMed] [Google Scholar]
- D'Amico-Martel A, Noden DM. Contributions of placodal and neural crest cells to avian cranial peripheral ganglia. Am J Anat. 1983;166:445–468. doi: 10.1002/aja.1001660406. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- Etchevers HC, Vincent C, Le Douarin NM, Couly GF. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- Hari L, Brault V, Kleber M, Lee HY, Ille F, Leimeroth R, Paratore C, Suter U, Kemler R, Sommer L. Lineage-specific requirements of beta-catenin in neural crest development. J Cell Biol. 2002;159:867–880. doi: 10.1083/jcb.200209039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- His W. Die erste Entwicklung des Hunchens im Ei. Leipzig: F.C.W. Vogel; 1868. Untersuchungen uber die erste Analge des Wirbeltierleibes. [Google Scholar]
- Jacques-Fricke BT, Roffers-Agarwal J, Gammill LS. DNA methyltransferase 3b is dispensable for mouse neural crest development. PloS one. 2012;7:e47794. doi: 10.1371/journal.pone.0047794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kague E, Gallagher M, Burke S, Parsons M, Franz-Odendaal T, Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PloS one. 2012;7:e47394. doi: 10.1371/journal.pone.0047394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Shibata S, Fukuda K, Sato M, Satoh E, Nagoshi N, Minematsu T, Matsuzaki Y, Akazawa C, Toyama Y, Nakamura M, Okano H. The dual origin of the peripheral olfactory system: placode and neural crest. Molecular brain. 2011;4:34. doi: 10.1186/1756-6606-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989a;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Kothary RK, Allen ND, Surani MA. Transgenes as molecular probes of mammalian developmental genetics. Oxf Surv Eukaryot Genes. 1989b;6:145–178. [PubMed] [Google Scholar]
- Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J Embryol Exp Morphol. 1975;34:125–154. [PubMed] [Google Scholar]
- Lee RT, Nagai H, Nakaya Y, Sheng G, Trainor PA, Weston JA, Thiery JP. Cell delamination in the mesencephalic neural fold and its implication for the origin of ectomesenchyme. Development. 2013;140:4890–4902. doi: 10.1242/dev.094680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitsky KL, Toledo-Aral JJ, Lopez-Barneo J, Villadiego J. Direct confocal acquisition of fluorescence from X-gal staining on thick tissue sections. Scientific reports. 2013;3:2937. doi: 10.1038/srep02937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O'Neill AK, Soriano P, Bush JO. The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Biol. 2013;379:229–234. doi: 10.1016/j.ydbio.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen F, Epstein JA. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–164. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, Richardson WD, McMahon AP, Koentges G. Neural crest origins of the neck and shoulder. Nature. 2005;436:347–355. doi: 10.1038/nature03837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TC. The migratory pathway of neural crest cells into the skin of mouse embryos. Dev Biol. 1973;34:39–46. doi: 10.1016/0012-1606(73)90337-0. [DOI] [PubMed] [Google Scholar]
- Murdoch B, DelConte C, Garcia-Castro MI. Embryonic Pax7-expressing progenitors contribute multiple cell types to the postnatal olfactory epithelium. J Neurosci. 2010;30:9523–9532. doi: 10.1523/JNEUROSCI.0867-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, DelConte C, Garcia-Castro MI. Pax7 lineage contributions to the mammalian neural crest. PloS one. 2012;7:e41089. doi: 10.1371/journal.pone.0041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashimada M, Ohta H, Li C, Nakao K, Uesaka T, Brunet JF, Amiel J, Trochet D, Wakayama T, Enomoto H. Autonomic neurocristopathy-associated mutations in PHOX2B dysregulate Sox10 expression. J Clin Invest. 2012;122:3145–3158. doi: 10.1172/JCI63401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DH. Formation and distribution of neural crest mesenchyme to the first pharyngeal arch region of the mouse embryo. Am J Anat. 1986;176:221–231. doi: 10.1002/aja.1001760210. [DOI] [PubMed] [Google Scholar]
- Noden D. The control of avian cephalic neural crest cytodifferentiation I. skeletal and connective tissues. Devl Biol. 1978;67:296–312. doi: 10.1016/0012-1606(78)90201-4. [DOI] [PubMed] [Google Scholar]
- Noden DM. Patterns and organization of craniofacial skeletogenic and myogenic mesenchyme: a perspective. Prog Clin Biol Res. 1982;101:167–203. [PubMed] [Google Scholar]
- Noden DM. The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am J Anat. 1983;168:257–276. doi: 10.1002/aja.1001680302. [DOI] [PubMed] [Google Scholar]
- Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K. The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of mouse embryos. Dev Biol. 1994;164:409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K. Rhombomere formation and hind-brain crest cell migration from prorhombomeric origins in mouse embryos. Develop Growth Differ. 1996;38:107–118. doi: 10.1046/j.1440-169X.1996.00013.x. [DOI] [PubMed] [Google Scholar]
- Pietri T, Eder O, Blanche M, Thiery JP, Dufour S. The human tissue plasminogen activator-Cre mouse: a new tool for targeting specifically neural crest cells and their derivatives in vivo. Dev Biol. 2003;259:176–187. doi: 10.1016/s0012-1606(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Sailer MH, Hazel TG, Panchision DM, Hoeppner DJ, Schwab ME, McKay RD. BMP2 and FGF2 cooperate to induce neural-crest-like fates from fetal and adult CNS stem cells. J Cell Sci. 2005;118:5849–5860. doi: 10.1242/jcs.02708. [DOI] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. The New biologist. 1990;2:441–449. [PubMed] [Google Scholar]
- Siegenthaler JA, Pleasure SJ. We have got you 'covered': how the meninges control brain development. Curr Opin Genet Dev. 2011;21:249–255. doi: 10.1016/j.gde.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg H, Jiang J, Sim P, Kidd J, Janus J, Rinon A, Edgar R, Shitrit A, Larocca D, Chapman KB, Binette F, West MD. Human embryonic stem cell-derived neural crest cells capable of expressing markers of osteochondral or meningeal-choroid plexus differentiation. Regenerative medicine. 2014;9:53–66. doi: 10.2217/rme.13.86. [DOI] [PubMed] [Google Scholar]
- Tam PP, Trainor PA. Specification and segmentation of the paraxial mesoderm. Anat Embryol (Berl) 1994;189:275–305. doi: 10.1007/BF00190586. [DOI] [PubMed] [Google Scholar]
- Tonks ID, Nurcombe V, Paterson C, Zournazi A, Prather C, Mould AW, Kay GF. Tyrosinase-Cre mice for tissue-specific gene ablation in neural crest and neuroepithelial-derived tissues. Genesis. 2003;37:131–138. doi: 10.1002/gene.10242. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Tam PP. Cranial paraxial mesoderm and neural crest cells of the mouse embryo: co-distribution in the craniofacial mesenchyme but distinct segregation in branchial arches. Development. 1995;121:2569–2582. doi: 10.1242/dev.121.8.2569. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Agarwal P, Brown C, McCulley DJ, Schwarz JJ, Black BL. The transcription factor MEF2C is required for craniofacial development. Dev Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, Kuratani S, Yamamura K. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.