Abstract
Epidermal growth factor receptor (EGFR) is highly amplified, mutated and overexpressed in human malignant gliomas. Despite its prevalence and growth-promoting functions, therapeutic strategies to inhibit EGFR kinase activity have not been translated into profound beneficial effects in glioma clinical trials. To determine the roles of oncogenic EGFR signaling in gliomagenesis and tumor maintenance, we generated a novel glioma mouse model driven by inducible expression of a mutant EGFR (EGFR*). Using combined genetic and pharmacological interventions, we revealed that EGFR*-driven gliomas were insensitive to EGFR tyrosine kinase inhibitors although they could efficiently inhibit EGFR* auto-phosphorylation in vitro and in vivo. This is in contrast to genetic suppression of EGFR* induction which led to significant tumor regression and prolonged animal survival. But in spite of their initial response to genetic EGFR* extinction, all tumors would relapse and propagate independent of EGFR*. We further showed that EGFR*-independent tumor cells existed prior to treatment and were responsible for relapse following genetic EGFR* suppression. And addition of PI3K/mTOR inhibitor could significantly delay relapse and prolong animal survival. Our Findings shed mechanistic insight into EGFR drug resistance in glioma and provide a platform to test therapies targeting aberrant EGFR signaling in this setting.
Keywords: malignant glioma, EGFR, therapeutic resistance, molecular heterogeneity
Introduction
Malignant glioma is the most common and lethal type of brain tumor (1, 2). The current standard care for malignant glioma includes maximal surgical resection, followed by radiation with adjuvant chemotherapy for the residual infiltrative tumor component. Despite such aggressive treatment efforts, the disease invariably returns. In its most aggressive form, glioblastoma (GBM) has a median survival of only 12–15 months after initial diagnosis (2, 3). Unfortunately, refinements of available therapeutic modalities including microneurosurgery, radiation and chemotherapy in the last several decades have not substantially improved patient survival.
While clinical progress has been slow, the past decade has witnessed major advances in our understanding of malignant glioma molecular pathogenesis. Detailed genetic and genomic characterization of malignant gliomas has yielded a comprehensive atlas of genomic rearrangements, genetic mutations and epigenetic alterations that drive disease pathogenesis and biology (4–8). These analyses also defined clinically relevant malignant glioma molecular subtypes (8). Specifically, genetic alterations of IDH1/PDGFRA and NF1 define proneural and mesenchymal subtypes, whereas the classical subtype is highly enriched for EGFR amplification/mutation and loss of PTEN and CDKN2A (4). These revelations underscore the selective cooperativity among different genetic and genomic alterations during malignant glioma formation.
EGFR gene amplification occurs in ~30–50% of malignant gliomas and is often associated with gene rearrangements (9). Among them, a constitutively activated mutant form of EGFR with in-frame deletion of 2–7 exons (referred hereafter as EGFR*) is the most common, and presents in ~40–50% of EGFR amplified malignant gliomas (4, 10–12). The high-level EGFR amplifications often comprise extrachromosomal double minutes and are heterogeneously distributed within the tumors (13–16). To date, abundant experimental evidences including mouse models have firmly established the causal role of aberrant EGFR amplification/mutation and its overexpression on glioma pathogenesis (10, 17–20), making it a compelling candidate for targeted therapies. But despite the remarkable therapeutic benefits of EGFR tyrosine kinase inhibitors (TKIs) on non-small cell lung cancer (NSCLC) patients carrying EGFR mutations (21–23), these agents have showed meager efficacy in malignant glioma clinical trials (24–26). The clinical observations have raised questions about whether EGFR signaling is a viable therapeutic target for malignant glioma treatment. In this study, we present a novel inducible glioma mouse model to interrogate the role of oncogenic EGFR signaling on glioma maintenance.
Materials and Methods
Mice
All mouse manipulations were approved and performed under the guidelines of the Institutional Animal Care and Use Committee of the Cold Spring Harbor Laboratory. The conditional Ink4a/Arf (27), Pten (28), transgenic tetO-EGFR* (29), Nestin-CreERT2 (30), and hGFAP-tTA mice (31) (from Jackson Laboratory) have been described previously. All combinations of compound mice were generated by interbred and maintained on FvB/C57BL/6 hybrid background in specifically pathogen-free conditions at Cold Spring Harbor Laboratory. The breeding pairs and neonatal pups until 4-week-old age were kept continuously on doxycycline (Dox) containing drinking water (2 g/L) unless otherwise indicated. Genotypes were confirmed using PCR. To induce glioma formation, 4-week-old compound mice containing Nestin-CreERT2 transgene were injected intraperitoneally with tamoxifen (124 mg/kg body weight) dissolved in sunflower oil daily for 5 consecutive days. Mice were monitored daily for signs of ill-health, and euthanized and necropsied when moribund following NIH guidelines.
Reagents
Erlotinib, gefitinib, crizotinib, and Bez-235 were purchased from LC Laboratories. Doxycycline was ordered from Research Products International. Tamoxifen was purchased from Sigma. D-Luciferin was ordered from Goldbio Technology. The antibodies used in this study are described in Supplemental Experimental Procedures.
Histology and Immunohistochemistry
At time of sacrifice, mice were perfused with 4% paraformaldehyde (PFA), and brains were dissected, followed by overnight post-fixation in 4% PFA at 4°C. Tissues were processed and embedded in paraffin by CSHL Research Pathology Core. Serial sections were prepared at 5 μm for paraffin sections with every tenth slide stained by hematoxylin and eosin. All slides were examined by S.K., tumor grading was determined by H.Z. assisted by P.C. based upon the WHO grading system for malignant astrocytoma (1). Immunohistochemical (IHC) and immunofluorescence (IF) analyses were performed as previously described (28). Images were captured using an Olympus BX53 or a Zeiss 710 LSM confocal microscope.
Cell culture
Primary tumor cells were isolated from tumor regions of affected mice using a stereo-dissection microscope (Zeiss). Single-cell suspensions made from enzymatically dissociated tissues were cultured in neurobasal media supplemented with EGF (20 ng/mL) and bFGF (10 ng/mL) as previously described (28). Murine astrocytes were prepared as previously described (32) and maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). For EGFR TKI treatment, control or EGFR mutant transduced astrocytes were seeded in equal cell numbers and serum starved for 24 hours before treatment. EGFR TKIs erlotinib (250 nM) and gefitinib (50 nM) in dimethyl sulfoxide (DMSO) were added to the cells for 4 hours before collection.
Quantitative Real Time PCR
Total RNAs were extracted from tissues using RNeasy (Qiagen) and first-strand cDNAs were prepared with SuperScript VILO cDNA Synthesis Kit (Applied Biosystems, ABI). Quantitative real-time PCR (qPCR) was performed using QuantiTect SYBR Green PCR kit (Qiagen) on Applied Biosystems StepOne. The primer sequences used in this study are described in Supplementary Experimental Procedures.
Grafting Experiments and In Vivo Inhibitor Treatments
For orthotopic grafting, 10,000 primary mouse glioma cells transduced with either luciferase or GFP expressing vector were injected into front-lobe caudate nucleus of 4–6 week-old Nu/Nu mice (Charles River) using a sterotaxic frame as previously described (32, 33). For subcutaneous grafting, 200,000 cells were injected into flanks of 4–6 week-old Nu/Nu mice. Mice were monitored daily and subjected to weekly bioluminescent imaging for tumor development. In vivo treatments were initiated at the indicated time (for orthotopic transplants) or tumor volume (~200mm3). The tumor-bearing mice were randomized into different groups and treated with Dox (2 g/L in drinking water), or daily oral administration of control vehicle, erlotinib (50 mg/kg/d), gefitinib (150 mg/kg/d), crizotinib (25 mg/kg/d), Bez-235 (45 mg/kg/d), alone or in combination as indicated. Mice were monitored daily, and sacrificed at onset of symptoms. At the time of sacrifice, tumor tissues were processed for further experiments.
In Vivo Bioluminescent Imaging
Mice were given intraperitoneal injections of D-Luciferin (150 mg/kg) and imaged 5–8 min post injection using the Caliper Xenogen IVIS Spectrum imaging system following the manufacturer’s instructions.
Expression Analysis
Expression Profiling was performed at the Cold Spring Harbor Laboratory Microarray Core facility using the Mouse Genome 430 2.0 Array (Affymetrix). Student’s t test was used to identify differentially expressed genes according to expression profiles. The ingenuity pathway analysis application was performed according to the manufacturer’s instruction (Ingenuity Systems). In brief, differentially expressed genes with threshold p ≤ 0.05 were mapped into Ingenuity Pathways Analysis database as input. Complete profiles are deposited on the GEO website under accession GSE64751.
Statistical Analysis
Animal survivals were analyzed using Graphpad Prism5. Statistical analyses were performed using Log-rank (Mantel-Cox) Test. Student’s t test was used for comparisons of experiments from two groups. For all experiments with error bars, standard deviation was calculated to indicate the variation within each experiment and data, and values represent mean ± SD.
Results
EGFR TKIs effectively block EGFR* auto-phosphorylation in murine astrocytes
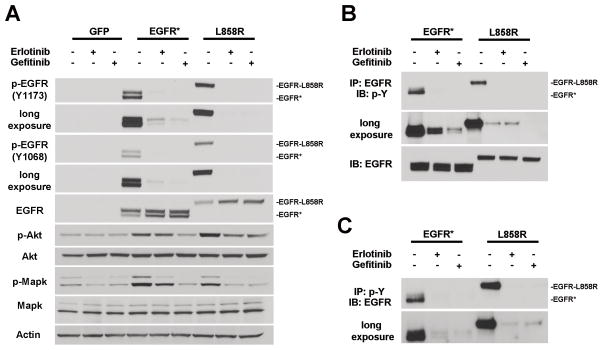
The poor response of glioma patients carrying EGFR amplification/mutation to EGFR TKIs is in stark contrast to their remarkable therapeutic benefits for lung cancer patients harboring activated EGFR mutations. Intriguingly, although the glioma and lung cancer EGFR mutants both exhibit ligand-independent auto-phosphorylation and activation, the locations of the mutations are conspicuously different - lung cancer mutations generally reside in the EGFR intracellular kinase domain, while glioma mutations mainly cluster in the extracellular domain (such as EGFR*) (34, 35). To investigate whether the extracellular domain mutants respond to EGFR TKIs differently than the lung cancer mutants, EGFR* and a TKI-sensitive lung cancer mutant EGFR-L858R were transduced individually into murine astrocytes. To analyze their auto-phosphorylation, control and mutant EGFR transduced astrocytes were serum starved before being subjected to treatment with two clinically approved small molecule EGFR TKIs - erlotinib or gefitinib. Immunoblot analysis of phosphorylation of tyrosine residues Y1068 and Y1173 indicated that the inhibitors could effectively block auto-phosphorylation in both EGFR-L858R and EGFR* (Fig. 1A). Similar results were observed when measuring total phosphotyrosine content of the immunoprecipitated EGFR receptors or total EGFR in the immunoprecipitated phosphotyrosine protein (Fig. 1B and C). These observations indicated that the EGFR* auto-phosphorylation could be inhibited comparably as the TKI-sensitive lung cancer mutant EGFR-L858R. TKI treatment also resulted in visible inhibition of EGFR downstream signaling pathways as evidenced by decreased phospho-Akt and phospho-Mapk levels, although to a lesser extent in erlotinib treated EGFR* transduced samples (Fig. 1A). Similar results were also obtained for two other glioma specific EGFR extracellular domain mutants – A289V and G598V (Supplementary Fig. S1). Together, these data indicate that auto-phosphorylation of the brain cancer EGFR mutants can be efficiently suppressed by EGFR TKIs.
Figure 1.
Auto-phosphorylation of EGFR* and EGFR-L858R is efficiently inhibited by EGFR TKIs. A, murine Ink4a/Arf−/− Pten −/− astrocytes transduced with control or indicated EGFR mutants were serum starved for 24 hours followed by 4-hour treatment of vehicle (DMSO), 250 nM erlotinib or 50 nM gefitinib. Cell lysates were prepared and subjected to immunoblot analysis. Note, the molecular weight of EGFR (wt) and EGFR-L858R is ~170KD and EGFR* ~140 KD. B and C, cell lysates from the indicated treatments as in (A) were immunoprecipitated with anti-EGFR (B) or anti-phospho-tyrosine antibody (PT-66) (C), and immunoblot analysis was performed using anti-phospho-tyrosine (4G10) (B) or anti-EGFR (C) antibody, respectively.
EGFR* induction alone is insufficient to promote gliomagenesis in adult mice
Since EGFR TKIs could effectively inhibit auto-phosphorylation of the glioma-specific EGFR mutants, we reasoned that the poor response to EGFR TKIs observed in glioma patients might be due to inefficient drug penetration through the blood-brain barrier, or the possibility that aberrant EGFR signaling might not be required for glioma maintenance. To address these questions, we generated compound transgenic mice that harbored a tetracycline-inducible EGFR* (tetO-EGFR*) allele driven by hGFAP-tTA, which targets tTA expression to neural progenitor cells and cortical astrocytes (31) (Fig. 2A). Since early induction of EGFR* gene expression caused profound brain developmental abnormalities that led to embryonic or early postnatal death in bitransgenic hGFAP-tTA tetO-EGFR* mice, the breeding pairs and neonatal pups were maintained on Dox-containing drinking water to repress transgene expression.
Figure 2.
EGFR* induction cooperates with conditional Ink4a/Arf and Pten deletions to induce malignant glioma formation. A, schematic of inducible EGFR* allele under regulation of human GFAP promoter element. B, EGFR* induction was efficiently repressed by Dox administration. Brain tissue lysates were prepared from littermate single- or bi-transgenic mice maintained on- (+) or off- (−) Dox and subjected to immunoblot analysis for EGFR and Actin. C, Kaplan-Meier brain tumor-free survival analysis of tamoxifen-treated mouse cohorts consisting of tetO-EGFR* cInkL cPtenL (iEIP) maintained off-Dox (n = 25), tetO-EGFR* cInkL cPtenL (iEIP) on-Dox (n = 5), tetO-EGFR* cInkL (iEI) off-Dox (n = 6), tetO-EGFR* cPtenL (iEP) off-Dox (n = 5), tetO-EGFR* (iEGFR) off-Dox (n = 7), and cInkL cPtenL (cIP) off-Dox (n = 8). D, iEIP gliomas displayed high mitotic indices and low levels of apoptosis. H&E and IHC staining against Ki67 and activated Caspase-3 (Act-Cas3) were performed using sections from normal mouse brains or iEIP malignant gliomas. E, sections of normal mouse brain tissue or iEIP tumors were stained with antibodies against Pten and EGFR. The arrow heads point to embedded Pten-positive endothelial cells in the tumors. F, shown are representative images of IHC staining against phosphorylated Akt (p-Akt), Mapk (p-Mapk) and Stat3 (p-Stat3) performed on sections from normal mouse brains and iEIP malignant gliomas. Scale bars represent 50 μm.
To evaluate EGFR* transgene inducibility, we first compared EGFR* mRNA and protein expression of 12-week-old off-Dox bitransgenic mice (n = 2) with their on-Dox littermate controls (n = 3). qPCR and immunoblot analysis demonstrated strong EGFR* induction in the off-Dox animal brains (Fig. 2B and Supplementary Fig. S2A). IF analysis confirmed that EGFR* expression in the off-Dox brains was confined to Gfap-positive cells across all CNS regions (Supplementary Fig. S2B). Besides neurogenic subventricular zone (SVZ)/subgranular (SGL) regions, EGFR* expression was induced in a subset of astrocytes characterized by stellate morphology and Gfap protein expression. As control, bitransgenic mice kept on Dox showed no discernible EGFR* protein expression, indicating that Dox could penetrate the blood-brain barrier in adult mice and enable efficient repression of EGFR* transgene induction.
Despite abundant EGFR* induction, gross examination of whole brains from 12-week-old mice that were switched to off-dox at 4 weeks of age revealed no major CNS developmental abnormalities. One exception was noticed in the neurogenic SVZ region, where we found off-Dox mice harbored a modestly expanded Gfap-positive neural precursor cell (NPC) population with strong EGFR* induction. Occasional EGFR-positive cells were seen migrating out of SVZ into the adjacent sub-striatum of cortical white matter (Supplementary Fig. S2C). But despite the elevated NPC proliferation, none of the mice (n = 18) kept off-Dox from 4 weeks of age developed brain tumors up to 18 months of age, indicating that EGFR* induction alone is not sufficient to induce glioma formation in adult mice.
EGFR* induction cooperates with Ink4a/Arf and Pten inactivation to induce malignant glioma formation
Malignant glioma pathogenesis is driven by the accumulation of genetic and epigenetic alterations (5, 7). In human gliomas, EGFR amplification/mutation is frequently associated with deletions of PTEN and CDKN2A (encoding for both p16INK4A and p14ARF) tumor suppressor genes (8). To model these cooperative genetic events, we crossed the hGFAP-tTA/tet-EGFR* mice to conditional knockout alleles of Ink4a/Arf (cInk4a/ArfLox) and Pten (cPtenLox), together with a tamoxifen (TMX)-activated Nestin-CreERT2 allele that allowed TMX activated Cre activation in NPCs at various developmental stages (30). Nestin and Gfap proteins were co-expressed in a subset of SVZ/SGL NPCs but not in differentiated astrocytes (Supplementary Fig. S3). As the result, combined hGFAP-tTA and Nestin-CreERT2 alleles enabled EGFR* induction and inactivation of Ink4a/Arf and Pten in the same NPCs. The compound experimental mice (Nestin-CreERT2 cInk4a/ArfLox/Lox cPtenLox/Lox hGFAP-tTA tetO-EGFR*, hereafter termed iEIP; Nestin-CreERT2 cInk4a/ArfLox/Lox cPtenLox/Lox, termed cIP; Nestin-CreERT2 cInk4a/ArfLox/Lox hGFAP-tTA tetO-EGFR*, termed iEI; Nestin-CreERT2 cPtenLox/Lox hGFAP-tTA tetO-EGFR*, termed iEP; cInk4a/ArfLox/Lox cPtenLox/Lox hGFAP-tTA tetO-EGFR*, termed iEGFR) were generated in the expected Mendelian ratios without visible developmental defects.
To induce glioma formation, compound mice were administered with TMX to transiently activate the Cre recombinase at 4 weeks of age and subsequently kept off-Dox to initiate EGFR* induction. Between 16 to 45 weeks of age, 23/25 (92%) of TMX-treated iEIP mice developed brain tumors with acute-onset neurological symptoms including seizure, ataxia, and/or paralysis (Fig. 2C). In comparison, 1/6 (17%) of TMX-treated iEI mice and none from other control groups (cIP, n = 8; iEP, n = 5; iEGFR off-Dox, n = 7; iEIP on-Dox, n = 5) developed brain tumors through 65 weeks of age, indicating combinatorial EGFR* induction and inactivation of both Ink4a/Arf and Pten tumor suppressors are required for tumor formation in this model.
Histopathological examination revealed that all neurologically symptomatic iEIP mice harbored malignant gliomas that, by World Health Organization (WHO) classification criteria (1), ranging from anaplastic astrocytomas (WHO Grade III, n = 13, 56.5%) to glioblastoma (WHO Grade IV, n = 10, 43.5%). All the tumors displayed marked pleomorphism, high mitotic indices with minimal apoptosis (Fig. 2D). The tumors classified as Grade IV also exhibited necrosis with pseudopalisading and to a lesser extent, necrosis combined with microvascular proliferation. Similar to their human counterparts, these murine tumor cells featured diffuse infiltrative spread at single cell-manner and frequently formed secondary structures of Scherer, including perineuronal, perivascular satellitosis and subpial collections in the cerebral cortex (Supplementary Fig. S4). On the molecular level, the iEIP gliomas registered complete loss of Pten protein expression in the tumor cells along with robust EGFR* induction (Fig. 2E). Consequentially, the tumor cells exhibited strong activation of downstream signaling pathway components including Akt, Mapk and Stat3 as evidenced by their enhanced phosphorylation (Fig. 2F).
Heterogeneous features of the tumor
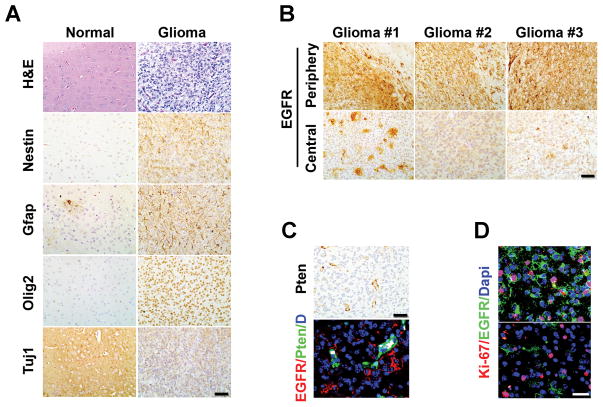
A classical feature of human malignant glioma is its high degree of inter- and intra-tumoral histological heterogeneity, hence the moniker of glioblastoma “multiforme”. Similar phenotypic and molecular heterogeneity were mirrored in the iEIP tumors. As illustrated in Fig. 3A, the iEIP glioma cells expressed an assortment of stem or lineage progenitor markers commonly observed in human gliomas, including the stem/progenitor marker Nestin (N), astrocytic lineage marker Gfap (G), neuronal lineage marker Tuj1 (Tuj), oligodendroglial progenitor marker Olig2 (O), but not mature neuronal and oligodendrocyte markers, such as NeuN and MBP (Supplementary Fig. S5). Importantly, a heterogeneous pattern of intra-tumoral EGFR expression was also observed in the tumors despite EGFR* induction was required for iEIP glioma initiation. Specifically, the tumor periphery and infiltrating borders tended to have high EGFR protein expression, whereas cancer cells in the tumor centers generally showed lower levels of EGFR expression (Fig. 3B), reminiscent of the heterogeneous EGFR gene amplification/expression pattern observed in human malignant glioma samples (13–16). In some regions, glioma cells, distinguished by their negative Pten staining, displayed mosaic EGFR expression pattern with interspersed EGFR-high and -low tumor cells (Fig. 3C). Co-IF staining further revealed that the EGFR-low tumor cell population also contained Ki67-positive proliferative cells (Fig. 3D), suggesting that not all tumor cells depended on high EGFR* levels for survival and propagation.
Figure 3.
The iEIP malignant gliomas are heterogeneous. A, iEIP glioma cells display multi-lineage differentiation. Normal mouse brain and iEIP glioma sections were stained with H&E or antibodies against indicated lineage markers. B, IHC staining against EGFR was performed on sections from different regions of three independent iEIP gliomas. Stronger EGFR expression was found at invasive edges of tumor periphery compared to relatively solid tumor centers. Scale bars represent 50 μm. C, shown are representative IF images of iEIP gliomas with interspersed EGFR-high and -low tumor cells. Tumor cells were distinguished by their negative Pten expression compared to embedded Pten-positive normal cells (green). D, co-staining of EGFR and Ki67 antibodies revealed that EGFR-high and EGFR-low tumor cells both retained proliferation capacity. Scale bars represent 100 μm.
iEIP gliomas are sensitive to EGFR* transgene ablation but resist to EGFR TKI treatment
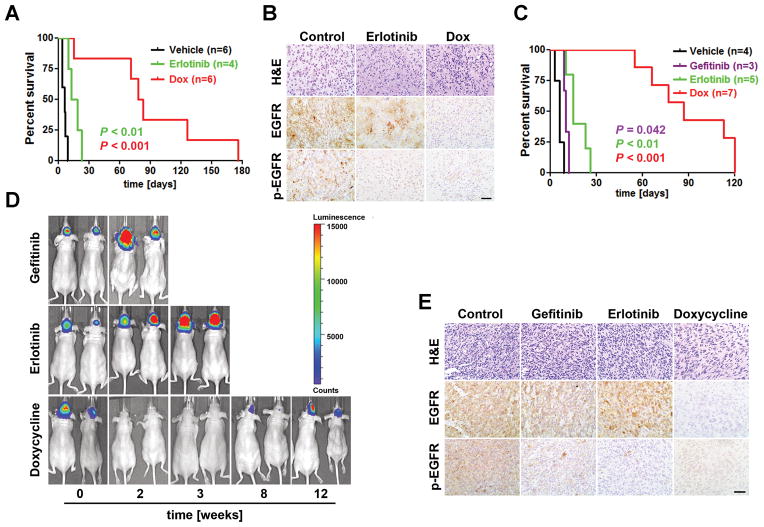
Tumor cells often retain their dependence on an initiating oncogene even after serial passages (36). To determine whether EGFR* induction and its protein phosphorylation were required for tumor maintenance, a cohort of glioma-bearing iEIP animals were randomly assigned to receive control vehicle (n = 6), EGFR TKI erlotinib (n = 4), or Dox (n = 6). Consistent with previous clinical observations (24), erlotinib treatment only marginally prolonged median survival of the affected mice compared to the vehicle treated group despite markedly attenuated EGFR phosphorylation in the tumors (Fig. 4A and B). This minor effect of erlotinib was in contrast to Dox treatment, which elicited a universal response within 3–5 days. And the affected mice exhibited generally alleviated symptoms thereafter, indicating EGFR* expression was required for tumor maintenance. However, this regression was not durable and all the mice under continuous Dox treatment succumbed to relapse within 5–18 weeks. IHC staining confirmed the absence of EGFR* induction in all Dox-treated relapsed tumors (n = 6, Fig. 4B), indicating relapse was not caused by escape from Dox-mediated EGFR* repression or from compensatory mechanism(s) that upregulated endogenous EGFR gene.
Figure 4.
The iEIP gliomas are sensitive to genetic suppression of EGFR* induction but are refractory to EGFR TKI treatment. A, Kaplan-Meier survival analysis of cohorts of tumor bearing iEIP transgenic mice treated with vehicle (n = 6), erlotinib (n = 4), or Dox (n = 6). Day 0 represents the day when treatment was initiated. B, EGFR* phosphorylation but not EGFR* protein was downregulated in erlotinib treated tumors. Shown are representative images of H&E and IHC staining against EGFR and phospho-EGFR (p-EGFR) on tumor sections from (A). C, Kaplan-Meier survival analysis of mouse cohorts that were orthotopically transplanted with iEIP glioma cells and treated with vehicle (n = 4), gefitinib (n = 3), erlotinib (n = 5), or Dox (n = 7) after tumors were established. Day 0 represents the day when treatment was initiated. D, shown are representative bioluminescence images of animals subjected to the indicated treatment from (C). E, EGFR* phosphorylation but not EGFR* protein levels were diminished in tumors treated with gefitinib or erlotinib. Representative tumor sections from (C) were stained with H&E, anti-EGFR, or anti-p-EGFR. Scale bars represent 50 μm.
To further analyze the development of resistance, we employed an orthotopic transplantation model by using freshly isolated primary iEIP glioma cells. Compared to the primary tumors, the orthotopic model provided a flexible and more controlled system to investigate therapeutic response and resistance development in a large group of animals. To facilitate in vivo imaging, isolated glioma cells were transduced with luciferase expressing vector. In general, animals carrying grafted iEIP glioma cells started to develop measurable bioluminescent imaging (BLI) signals by 4–6 weeks. At week eight, the mice were randomly assigned to four groups to receive vehicle control (n = 4), erlotinib (n = 5), gefitinib (n = 3), or Dox (n = 7), respectively. Similar to observation in the transgenic iEIP cohorts, neither erlotinib nor gefitinib treatment significantly slowed down tumor growth despite their efficient inhibition of intra-tumoral EGFR phosphorylation (Fig. 4C–E). In contrast, the Dox-treated mice displayed strong initial response as evidenced by their substantially attenuated BLI output, although tumor relapse would later resume and eventually proved fatal. The response to Dox nevertheless translated into significant overall survival benefit compared to erlotinib or gefitinib treatment.
To fully exclude possibility that the poor response to EGFR TKIs might be caused by inefficient drug penetration through blood-brain barrier, the treatment was repeated in a cohort of immunocompromised Nu/Nu mice subcutaneously transplanted with iEIP glioma cells. Again, erlotinib only transiently slowed down tumor progression compared to Dox treatment, which caused robust initial tumor regression followed by an extended period of stasis before the eventual relapse (Supplementary Fig. S6A). IHC staining and immunoblot analysis confirmed the inhibition of EGFR* phosphorylation (Supplementary Fig. S6B and C). Collectively, our data indicate that EGFR* protein expression is important for the iEIP tumor maintenance, but its phosphorylation level might not be an accurate indicator of its tumor maintenance functions.
EGFR*-independent glioma cells are pre-existed
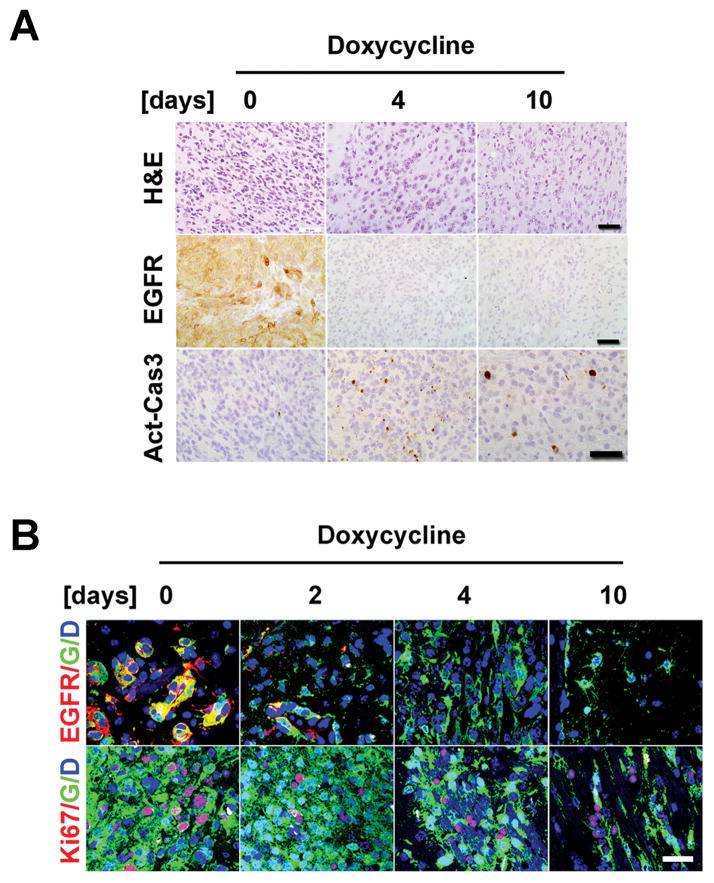
Although Dox treatment elicited robust initial response and glioma regression, tumors invariably relapsed under continued suppression of EGFR* induction. To examine whether the resistant tumor cells were present before treatment or acquired resistance during treatment, we conducted a serial time-course analysis to define the acute response following genetic EGFR* suppression. To facilitate tumor cell tracking, the freshly isolated iEIP glioma cells were transduced with a GFP-expressing vector before being orthotopically transplanted into recipient mice. At 8 weeks post transplantation, the tumor-bearing mice were switched to Dox drinking water and sacrificed at 0, 2, 4, 6, 8, 10 days, respectively. As illustrated in Fig. 5A, Dox treatment caused rapid EGFR* protein downregulation and completely silenced EGFR* transgene induction by 4 days post-treatment. The suppression of EGFR* induction was accompanied by markedly enhanced apoptosis (activated caspase3) and progressively decreased tumor cellularity and phospho-Akt staining (Fig. 5A and Supplementary Fig. S7). In spite of pronounced cell death, however, Dox treatment did not completely eradicate the tumor cell population. IF staining revealed the retention of residual GFP-positive but EGFR-negative tumor cells following 4- and 10-day treatment (Fig. 5B). The presence of mitotically active tumor cells suggests that these EGFR*-independent tumor cells existed prior to treatment and likely fueled the later relapse.
Figure 5.
EGFR*-independent glioma cells exist prior to treatment. A, tumor bearing animals grafted with GFP-expressing iEIP glioma cells were switched to Dox and sacrificed at indicated time-points (n = 2 for each). H&E and IHC staining against EGFR or activated caspase3 (Act-Cas3) revealed complete suppression of EGFR* induction at 4-day post treatment and enhanced apoptosis in Dox treated tumors. Scale bars represent 50 μm. B, IF staining revealed a subpopulation of GFP-labeled Ki67-positive proliferative tumor cells persisted through genetic suppression of EGFR* induction. G - GFP; D - DAPI. Scale bars represent 100 μm.
Hgf/Met signaling is activated in relapsed tumors refractory to EGFR* ablation
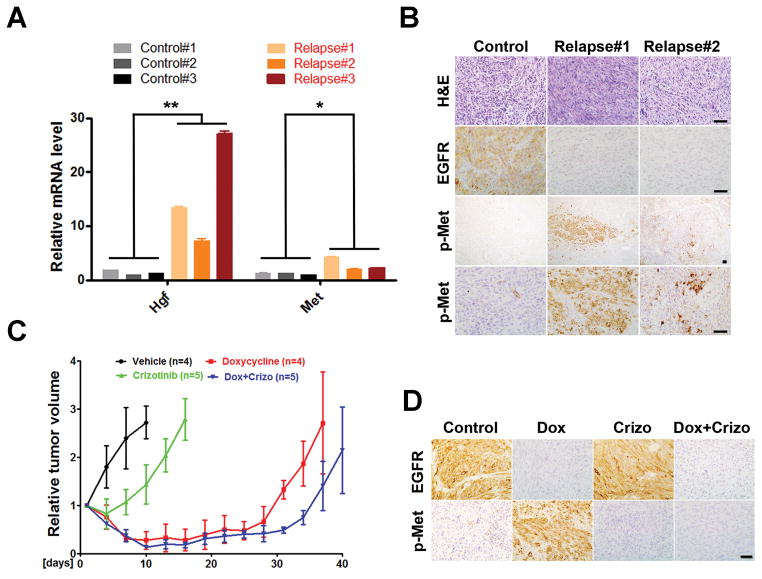
In an effort to identify the molecular mechanism underlying the tumor relapse, we performed a gene expression analysis comparing Dox-treated relapsed tumors with untreated controls. Ingenuity pathway analysis of the two groups identified “hepatic fibrosis/hepatic stellate cell activation” as the most enriched canonical pathway (Supplementary Fig. S8). qPCR revealed that hepatocyte growth factor (Hgf), and to a lesser extent its receptor Met, was significantly upregulated in the Dox-treated relapsed tumors (Fig. 6A). IHC further confirmed markedly enhanced regional Met activation in the relapsed tumors compared to their untreated controls (Fig. 6B). Notably, the Met-activated cells were not distributed evenly within the relapsed tumors, but instead, were focally patched and generally comprised less than 10% of total tumor cell population.
Figure 6.
Hgf/Met signaling is activated in EGFR*-independent relapsed tumors. A, Total RNAs were prepared from untreated control and Dox-treated relapsed tumors and subjected to qPCR analysis for Hgf, Met and β-Actin. Results were normalized with β-Actin expression and shown as mean ± SD. Student’s t test was used for the comparison between untreated control and Dox-treated relapsed group (**, p = 0.034; *, p = 0.046). Data were from two independent experiments with triplicates. B, Met activated tumor cells were focally distributed in Dox-treated relapsed tumors. Shown are representative images of untreated control and relapsed tumor sections stained for H&E, EGFR and phospho-Met (p-Met). C and D, Met inhibitor had limited effect on iEIP tumor growth and relapse prevention. Mice with subcutaneously grafted iEIP glioma cells were treated with vehicle (n = 4), crizotinib (n = 5), Dox (n = 4), or Dox + crizotinib (n = 5). Day 0 represents the day when treatment was initiated. Tumor growth was measured at indicated time and calculated relative to initial tumor volume. The data are presented as mean ± SD.
MET activation either by amplification or overexpression of its ligand HGF, has been shown to cause de novo resistance to EGFR TKIs in human NSCLC (37, 38). To determine its impact on tumor relapse in our model, we next examined the function of Met inhibitor, alone or in combination with Dox, on in vivo iEIP glioma growth. Freshly isolated iEIP glioma cells were subcutaneously transplanted into the flanks of recipient Nu/Nu mice. As tumor volumes reached a palpable size (~200mm3), mice were randomly assigned to four groups to receive vehicle (n = 5), Met inhibitor crizotinib (n = 5), Dox (n = 5), or a combination of Dox + crizotinib (n = 5). Consistent with the minor presence of Met-activated tumor cells in the iEIP gliomas, crizotinib treatment alone only modestly slowed tumor growth compared to the vehicle control (Fig. 6C and D). The combination of Dox + crizotinib treatment elicited a slightly deeper initial tumor regression response but still failed to significantly delay tumor relapse when compared to Dox treatment alone. These data suggest that besides the Met/Hgf signaling axis, other survival signaling pathways might have evolved to sustain EGFR*-independent relapsed tumors.
Combined PI3K/mTOR and EGFR* repression delay tumor relapse and prolong survival
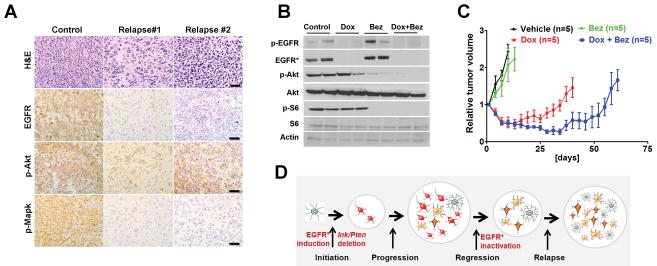
The focal presence and clonal nature of Met-activated cells within the relapsed tumors suggested that multiple independent survival pathways might be activated to compensate EGFR* repression. We therefore reasoned that inhibition of single or even multiple RTK signaling might not elicit a durable cure. And a combinatorial blockade of EGFR* and major downstream signaling outputs might represent a better option to counter relapse. By examining common downstream pathways, we found that while the phospho-Mapk was significantly diminished, the relapsed tumors still maintained activated PI3K/mTOR signaling as evidenced by their strong phospho-Akt staining (Fig. 7A). These findings suggested that activated PI3K/mTOR signaling but not Mapk pathways, might play an important role in relapse development. To determine whether the addition of dual PI3K/mTOR inhibitor (Bez-235) could prevent relapse, we evaluated the effect of a regime with either single or combined Dox and Bez-235 on subcutaneous iEIP tumor growth. Despite it significantly inhibited PI3K/mTOR signaling as evidenced by markedly diminished Akt and S6 phosphorylation (Fig. 7B and Supplementary Fig. S9), Bez-235 treatment alone had limited effect on iEIP tumor growth (Fig. 7C). In contrast, combined Dox + Bez-235 significantly delayed relapse and prolonged animal survival compared to Dox treatment alone. Notably, focally patched p-Met cells again were found in both Dox and Dox + Bez treated relapsed tumors, confirming that Met activation likely acted as one source of resistance against EGFR* suppression. Nevertheless, the synergistic therapeutic effect of combined EGFR* and PI3K/mTOR inhibition suggests that the combination might be useful for treatment of aberrant EGFR signaling-driven malignant gliomas in clinic.
Figure 7.
Combining PI3K/mTOR inhibition with genetic EGFR* suppression prolongs animal survival by delaying tumor relapse. A, Akt but not Mapk phosphorylation was maintained in Dox-treated relapsed tumors. Shown are representative images of untreated control and relapsed tumor sections stained with antibodies against phospho-Akt (p-Akt) and phospho-Mapk (p-Mapk). Scale bars represent 50 μm. B and C, mice with subcutaneously grafted iEIP glioma cells were treated with vehicle (n = 5), Bez-235 (n = 5), Dox (n = 4), or Dox + Bez-235 (n = 5). Day 0 represents the day when treatment was initiated. Immunoblot analysis (B) was performed using tumor lysates prepared from the indicated treatments. Tumor growth (C) was measured at indicated time and calculated relative to initial tumor volume. The data are presented as mean ± SD. D, model of EGFR*-induced glioma initiation/progression, and its relation to EGFR-targeted therapeutic resistance development.
Discussion
In the current study, we have described a novel malignant glioma mouse model driven by inducible EGFR* expression and provided genetic evidence that oncogenic EGFR* serves a tumor maintenance role in fully established EGFR*-driven malignant gliomas. Genetic suppression of EGFR* induction in the affected animals led to significant tumor regression. This was in contrast to treatment using two different EGFR TKIs despite they could efficiently inhibit EGFR* auto-phosphorylation both in vitro and in vivo. Our findings therefore suggested that poor response to EGFR TKIs in glioma clinical trials might not be due to their inability to inhibit EGFR phosphorylation but rather glioma’s less dependence on EGFR kinase activity relative to lung cancer. In addition to the EGFR-dependent resistance mechanism, our studies also uncovered an EGFR-independent mechanism in which preexisting resistant tumor cells persist through EGFR* ablation and lead to tumor relapse. Addition of PI3K/mTOR inhibitor together with genetic EGFR* suppression could significantly delay relapse and prolong animal survival. These observations have significant implications for our understanding of glioma biology as well as future therapeutic development.
EGFR TKIs have emerged as effective therapeutic entities for lung cancer patients carrying EGFR kinase domain mutations (21–23). But perplexingly, the same TKIs were ineffective in glioma patients with EGFR amplification/mutation (24–26, 39). This clinical observation is also confirmed in our current studies using iEIP animal model. But despite their marginal effect on glioma progression, we and others showed that EGFR TKIs could efficiently inhibit EGFR* auto-phosphorylation both in vitro and in vivo (40–42). In addition, treatment of subcutaneously grafted iEIP tumors with EGFR TKIs in our studies also excluded the possibility of inefficient drug penetration through blood-brain barrier as the main cause of the poor response, suggesting that glioma maintenance, unlike lung cancer, might not require high level EGFR kinase activity. Intriguingly, glioma-specific EGFR mutants have a very unique mutation pattern compared to other cancer types. For example, the EGFR intracellular kinase domain mutations frequently seen in lung cancers are conspicuously absent in gliomas (40, 42, 43). Instead, the glioma-specific mutations mainly cluster in EGFR extracellular domain (such as EGFR*). Whether this tissue specific mutational pattern reflect glioma’s less dependence on EGFR kinase activity remains to be determined.
Two recent reports proposed that distinct conformational requirements and/or kinase-site occupancy rates may contribute to the different sensitivities to EGFR TKIs observed in brain versus lung cancer EGFR mutants (40, 42). It is plausible that low levels of EGFR kinase activity might be sufficient for glioma maintenance. As such, more potent or selective EGFR TKIs could theoretically overcome the resistance. However, since a substantially higher dose of EGFR TKIs is generally required to achieve comparable cell death in cultured mutant expressing-glioma versus -lung cancer cells (40, 42, 43), deeper target inhibition might not be feasible owing to potential toxicity to normal tissues. In addition, besides the much studied kinase-dependent features, EGFR kinase-independent function was also shown to endow tumor cells with increased survival capacity against EGFR TKIs by maintaining the basal intracellular glucose level (44). Therefore, in order to achieve optimal efficacy against EGFR-driven gliomas, the next generation of targeted therapeutics might need to consider other EGFR functional domains in addition to its kinase activity.
Studies in multiple genetically engineered mouse models indicate that tumor maintenance is often dependent on the driver oncogene that initiates tumor development (36). Indeed, genetic suppression of EGFR* induction in our study elicited rapid cell death and tumor regression. However, the gliomas did not regress fully, and a small population of residual tumor cells persisted through EGFR* suppression and likely formed the basis for later relapse. The incomplete remission was consistent with the heterogeneous EGFR* expression pattern observed in fully established iEIP gliomas, as one would expect that tumor cells with diverse EGFR* expression would have varied degrees of dependence on oncogenic EGFR signaling. Considering that EGFR* induction is required for iEIP glioma initiation, our findings support a model directly linking glioma plasticity with therapeutic resistance (Fig. 7D). In this model, EGFR* induction cooperates with Ink4a/Arf and Pten inactivation to initiate tumor formation. But later on, a glioma progression process amplifies diverse lineages of tumor cells that vary in their molecular characteristics and dependence on oncogenic EGFR signaling. Genetic EGFR* suppression generates initial tumor regression by eradicating the bulk of EGFR*-dependent glioma cells before the relapse, during which the previous minor or even dormant EGFR*-independent tumor cells take over. Due to the high tumor plasticity and clonal nature of relapsed tumors, it is quite possible that different resistant tumor cell clones carrying varied EGFR-independent survival signaling pathways can evolve in parallel within the same lesion. If so, one would expect that therapies targeted single or even combinations of RTKs will not be able to suppress all types of relapsed tumor cells. Indeed, our current study identified Met activation as one source of resistance against EGFR* ablation in relapsed gliomas. But addition of a Met inhibitor to genetic EGFR* suppression induced only slightly deeper initial tumor regression without significantly delaying tumor relapse. In contrast, combined EGFR* ablation with administration of a PI3K/mTOR inhibitor significantly delayed tumor relapse, suggesting that combinatorial blockade of key downstream outputs together with EGFR suppression might represent a better therapeutic approach to overcome innate resistance.
Supplementary Material
Acknowledgments
Grant Support
This work is supported by a V Foundation Scholar Grant and a Sontag Foundation Distinguished Scientist Grant (to H.Z.). Additional funding was provided by the Bradley Zankel Foundation. H.Z. is also supported by Sidney Kimmel Foundation Scholars Award and National Cancer Institute Cancer Center Support Grant Development Funds (Grant CA45508).
We thank P. Tarr for carefully reading the manuscript, E. Earl at CSHL animal facility for excellent mouse husbandry and care, Y. Jin and M. Hammell from CSHL Bioinformatic Core for assistance with Ingenuity Pathways Analysis, S. Hearn from CSHL Microscopy facility for help on imaging, A. Nourjanova and R. Puzis from CSHL Histology Core for help on histology.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 7.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–7. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 10.Fan QW, Cheng CK, Gustafson WC, Charron E, Zipper P, Wong RA, et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24:438–49. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87:8602–6. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zadeh G, Bhat KP, Aldape K. EGFR and EGFRvIII in glioblastoma: partners in crime. Cancer Cell. 2013;24:403–4. doi: 10.1016/j.ccr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 13.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada Y, Hurwitz EE, Esposito JM, Brower MA, Nutt CL, Louis DN. Selection pressures of TP53 mutation and microenvironmental location influence epidermal growth factor receptor gene amplification in human glioblastomas. Cancer Res. 2003;63:413–6. [PubMed] [Google Scholar]
- 15.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–7. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–6. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–85. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–45. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukasa A, Wykosky J, Ligon KL, Chin L, Cavenee WK, Furnari F. Mutant EGFR is required for maintenance of glioma growth in vivo, and its ablation leads to escape from receptor dependence. Proc Natl Acad Sci U S A. 2010;107:2616–21. doi: 10.1073/pnas.0914356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci U S A. 2009;106:2712–6. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 22.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 23.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandes AA, Franceschi E, Tosoni A, Hegi ME, Stupp R. Epidermal growth factor receptor inhibitors in neuro-oncology: hopes and disappointments. Clin Cancer Res. 2008;14:957–60. doi: 10.1158/1078-0432.CCR-07-1810. [DOI] [PubMed] [Google Scholar]
- 25.Wen PY, Chang SM, Lamborn KR, Kuhn JG, Norden AD, Cloughesy TF, et al. Phase I/II study of erlotinib and temsirolimus for patients with recurrent malignant gliomas: North American Brain Tumor Consortium trial 04–02. Neuro Oncol. 2014;16:567–78. doi: 10.1093/neuonc/not247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yung WK, Vredenburgh JJ, Cloughesy TF, Nghiemphu P, Klencke B, Gilbert MR, et al. Safety and efficacy of erlotinib in first-relapse glioblastoma: a phase II open-label study. Neuro Oncol. 2010;12:1061–70. doi: 10.1093/neuonc/noq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–33. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006;103:7817–22. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imayoshi I, Sakamoto M, Ohtsuka T, Takao K, Miyakawa T, Yamaguchi M, et al. Roles of continuous neurogenesis in the structural and functional integrity of the adult forebrain. Nat Neurosci. 2008;11:1153–61. doi: 10.1038/nn.2185. [DOI] [PubMed] [Google Scholar]
- 31.Lin W, Kemper A, McCarthy KD, Pytel P, Wang JP, Campbell IL, et al. Interferon-gamma induced medulloblastoma in the developing cerebellum. J Neurosci. 2004;24:10074–83. doi: 10.1523/JNEUROSCI.2604-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, et al. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb Symp Quant Biol. 2008;73:427–37. doi: 10.1101/sqb.2008.73.047. [DOI] [PubMed] [Google Scholar]
- 34.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 35.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 36.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–4. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 37.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 38.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–84. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barkovich KJ, Hariono S, Garske AL, Zhang J, Blair JA, Fan QW, et al. Kinetics of inhibitor cycling underlie therapeutic disparities between EGFR-driven lung and brain cancers. Cancer Discov. 2012;2:450–7. doi: 10.1158/2159-8290.CD-11-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hegi ME, Diserens AC, Bady P, Kamoshima Y, Kouwenhoven MC, Delorenzi M, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial. Mol Cancer Ther. 2011;10:1102–12. doi: 10.1158/1535-7163.MCT-11-0048. [DOI] [PubMed] [Google Scholar]
- 42.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–71. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–93. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.