Abstract
Development of complex multicellular organisms requires careful regulation at both transcriptional and post-transcriptional levels. Post-transcriptional gene regulation is in part mediated by a class of non-coding RNAs of 21–25 nucleotides in length known as microRNAs (miRNAs). β-catenin, regulated by the canonical Wnt signaling pathway, has a highly evolutionarily conserved function in patterning early metazoan embryos, in forming the Anterior-Posterior axis, and in establishing the endomesoderm. Using reporter constructs and site-directed mutagenesis, we identified at least three miRNA binding sites within the 3’ untranslated region (3’UTR) of the sea urchin β-catenin. Further, blocking these three miRNA binding sites within the β-catenin 3’UTR to prevent regulation of endogenous β-catenin by miRNAs resulted in a minor increase in β-catenin protein accumulation that is sufficient to induce aberrant gut morphology and circumesophageal musculature. These phenotypes are likely the result of increased transcript levels of Wnt responsive endomesodermal regulatory genes. This study demonstrates the importance of miRNA regulation of β-catenin in early development.
Keywords: endoderm, mesoderm, larval gut, circumesophageal muscles, PMCs, sea urchin, microRNA target protectors, post-transcriptional regulation
Introduction
β-catenin, the key effector molecule of the canonical Wnt pathway, has a highly conserved function in regulating cell proliferation, differentiation and fate decisions (Komiya and Habas, 2008; Moon, 2005). Aberrant Wnt signaling has been associated with many human diseases, including cancer, skeletal disorders, neuronal diseases, and cardiovascular diseases (Anastas and Moon, 2013; Clevers, 2006; Clevers and Nusse, 2012; Kim et al., 2013; MacDonald et al., 2009; Moon et al., 2004). In the absence of a Wnt ligand, β-catenin is phosphorylated at several N-terminal serine and threonine residues by casein kinase I and Glycogen Synthase Kinase 3β (GSK3β). Phosphorylation of β-catenin occurs in a multiprotein complex composed of GSK3β, a scaffolding protein Axin, and the tumor suppressor gene product Adenomatous Polyposis Coli protein (APC). Phosphorylation by GSK3β leads to the ubiquitination and proteasome-mediated degradation of β-catenin. Upon binding of the Wnt ligand to the Frizzled and LRP5/6 receptors, activated Disheveled (Dvl) recruits Axin to cell membrane leading to the disassembly of the destruction complex and inhibition of the degradation of β-catenin (Logan and Nusse, 2004). Increased cytoplasmic levels of β-catenin leads to its accumulation in the nucleus, where β-catenin interacts with TCF/LEF transcription factors and activates the transcription of target genes that mediate body plan determination, tissue patterning, and endoderm specification (Komiya and Habas, 2008; Moon, 2005; Peter and Davidson, 2010).
In addition to its essential role as a transcription coactivator, β-catenin is also a central structural component of the Cadherin/Catenin adhesion complex (Aberle et al., 1996; Nelson and Nusse, 2004). The Cadherin/Catenin-based adhesion system is the major mechanism by which cells adhere to one another. The abundance of β-catenin in the adhesion complex at the plasma membrane affects its accumulation and function with the signaling complex in the nucleus. This is demonstrated with experiments in which perturbation of cadherin complexes has an effect on Wnt/β-catenin regulated processes. For example, overexpression of cadherins in Xenopus embryos inhibited dorsal axis formation which is known to be dependent on canonical Wnt signaling (Heasman et al., 1994). E-cadherin knockout embryonic stem cells showed accumulation of β-catenin/Lef1 in the nucleus and activation of a Wnt reporter, which could be reversed by expression of E-cadherin (Orsulic et al., 1999).
The initial regionalization of β-catenin in the early embryo contributes to polarity establishment, patterning, and germ layer specification (Logan et al., 1999; Petersen and Reddien, 2009). In numerous deuterostome embryos, including amphibians, fish, chicks, ascidians and sea urchins, β-catenin becomes localized in the nuclei of blastomeres at one pole of the cleavage stage embryo (Imai et al., 2000; Larabell et al., 1997; Logan et al., 1999; Roeser et al., 1999; Rowning et al., 1997; Schneider et al., 1996). In general, the pole of the embryo in which β-catenin is detected in the nucleus gives rise to endodermal and mesodermal tissues. Similar to many deuterostomes, the sea urchin β-catenin is required for the specification of the endoderm and mesoderm. (Logan et al., 1999; Wikramanayake et al., 1998). Overexpression of proteins that interfere with nuclear localization and/or function of β-catenin such as cadherins, GSK3β, and dominant forms of TCF/LEF, lead to embryos with excess ectodermal tissues and a lack of mesenchyme cells and gut (Emily-Fenouil et al., 1998; Logan et al., 1999; Vonica et al., 2000; Wikramanayake et al., 1998). Conversely, overexpression of β-catenin leads to embryos deprived of ectodermal tissue, consisting of mainly endodermal and mesodermal derivatives (Wikramanayake et al., 1998).
While the Wnt signaling pathway has been examined in the sea urchin (Emily-Fenouil et al., 1998; Logan et al., 1999; Vonica et al., 2000; Wikramanayake et al., 1998), the regulatory roles of microRNAs (miRNAs) in this developmental pathway have not been examined. miRNAs are a relatively novel class of 22-bp non-coding RNA molecules that fine tune gene expression by pairing to the 3’ untranslated region (3’UTR) of protein coding mRNAs to repress their translation and/or induce mRNA degradation (Bartel, 2004; Rajewsky, 2006). They are pivotal regulators of nearly all biological processes, including cell fate specification and differentiation (Bartel, 2004; Mukherji et al., 2011).
The vast majority of miRNAs are transcribed by RNA polymerase II and initially processed by the enzyme Drosha and its cofactor DGCR8 into stem-loop structures which get transported out from the animal nucleus to the cytoplasm (Lee et al., 2003). This stem-loop precursor is further processed into mature miRNAs by the riboendonuclease Dicer. The mature miRNA is loaded onto the RNA Induced Silencing Complex (RISC) and used as a guide to direct the binding of miRNA 5’ seed (nucleotides 2–8) and anchor nucleotides to the 3’UTR of target mRNAs to mediate translational silencing and promote targeted mRNA degradation (Baek et al., 2008; Bartel, 2009; Ghildiyal and Zamore, 2009; Guo et al., 2010; Hendrickson et al., 2009; Liu, 2008; Selbach et al., 2008). The regulatory role of miRNAs in early development was demonstrated by deleting or knocking down Dicer, an essential enzyme in miRNA processing, which causes either developmental defects or embryonic lethality in many animal systems (Bernstein et al., 2003; Giraldez et al., 2005; Saurat et al., 2013; Song et al., 2012).
Our laboratory has previously demonstrated that knockdowns of key enzymes in the miRNA biogenesis pathway in the sea urchin embryos lead to gastrulation failure and embryonic lethality (Song et al., 2012). Dicer knockdown embryos at the gastrula stage express significantly reduced endodermal and mesodermal antigens, suggesting a failure to properly specify these cell types. This current study tests the hypothesis that the highly conserved canonical Wnt/β-catenin pathway which is required for endomesodermal specification is regulated by miRNAs and may in part contribute to the previous Dicer knockdown phenotype. The strength of our study is to test miRNA regulation of β-catenin in the context of a developing embryo at the systems level. Our results indicate that β-catenin is post-transcriptionally regulated by at least two miRNAs at three binding sites. Removal of miRNA regulation using miRNA target protector morpholinos (miRNA TPs) specific to the β-catenin 3’UTR resulted in a minor increase of the β-catenin protein level that led to aberrant gut morphology, endodermal differentiation, and less developed circumpharyngeal musculature. Further, at the molecular level, a greater than 2-fold increase of Wnt responsive gene transcript levels, including endodermal regulatory genes Krl (Howard et al., 2001), FoxA (de-Leon and Davidson, 2010; Oliveri et al., 2006), Eve (Peter and Davidson, 2010, 2011), Wnt8 (Wikramanayake et al., 2004), Bra (Gross and McClay, 2001), and ectodermal regulatory gene Nodal (Duboc et al., 2010; Yaguchi et al., 2008) was observed in β-catenin miRNA TP-treated embryos. These results demonstrate the importance of miRNA regulation on β-catenin in early development.
Materials and Methods
Animals
Adult Strongylocentrotus purpuratus were obtained from California (Point Loma Marine Company) and cultured in an aquarium (Marineland, Moonpark, CA) with artificial sea water (Instant Ocean). Gametes were obtained by intracoelomic injection of 0.5 M KCl and embryos were cultured at 15°C in filtered natural sea water collected from the Indian River Inlet; University of Delaware Lewes campus.
Microinjections
Control morpholino antisense oligonucleotides (MASO) 5’ CCTCTTACCTCAGTTACAATTTATA 3’, dicer MASO 5’ GGACTCGATGGTGGCTCATCCATTC 3’ (Song et al., 2012), and miRNA target protector MASOs (miRNA TPs) were ordered from GeneTools (Philomath, OR). 1.5 mM of Dicer MASO stock injection solution was used. Three miRNA TPs were designed to protect one miRDeep2-30364-35240 binding site and two miR-2007 binding sites from respective miRNA binding to the β-catenin 3’UTR. The morpholino that blocks miR-2007 at position +922 bp is 5’ CTATTTCAGATATAATCTTGACGAG 3’; the morpholino that blocks miR-2007 at position +2647 bp is 5’ TTATTTCAGACATGAAAAATGGAAG 3’ and the morpholino that blocks miRDeep2-30364-35240 is 5’ TTATTGCACCTTTTTAAGAGGCATC 3’. These miRNA target protectors were diluted to various stock injection solutions (3 nM to 3 mM). The estimated injected miRNA TPs were the following: from the 30 µM stock of injection solution, approximately 35 attomoles (35 × 10−18) was injected. From the 300 µM stock of injection solution, approximately 350 attomoles (350 × 10−18) was injected. The injected negative control morpholino contained the same molar concentration as the corresponding sum of injected miRNA TPs. 300 µM stock of each of the β-catenin miRNA TPs were used in all real time, quantitative PCR, dual luciferase, Western blotting and phenotyping experiments.
Microinjections were performed as previously described with modifications (Cheers and Ettensohn, 2004; Song et al., 2012; Stepicheva and Song, 2014). MASO oligonucleotides were resuspended in sterile water and heated for 10 minutes at 60 °C prior to use. Injection solutions contained 20% sterile glycerol, 2 mg/ml 10,000 MW Texas Red lysine charged dextran (Molecular Probes, Carlsbad, CA) and various concentrations of MASO. Approximately 1–2 pl were injected into each newly fertilized egg. Eggs from S. purpuratus were collected and dejellied in acidic sea water (pH 5.15) for 10 minutes on ice, followed by two sea water washes. Dejellied eggs were rowed onto 60 × 15 mm petri dishes that were previously coated with protamine sulfate (1% w/v). Eggs were fertilized with sperm in the presence of 1 mM 3-amino-1,2,4 triazole (Sigma, St. Louis, MO). Injections were performed using the Femto Jet injection system (Eppendorf; Hamberg, Germany) (Stepicheva and Song, 2014). A vertical needle puller PL-10 (Narishige, Tokyo, Japan) was used to pull the injection needles (1×90 mm glass capillaries with filaments) (Narashige; Tokyo, Japan).
Preparation of anti-β-catenin polyclonal antibodies
To investigate the level of the β-catenin protein, two affinity-purified anti-β-catenin polyclonal rabbit antibodies, β-catenin1 and β-catenin55, were made against two short sequences (NH2- CYKKRISVELGNSLFRGDS-COOH) and (NH2-CHSLHSNHGEYRQPPPT-COOH) of the S. purpuratus β-catenin protein. Antibodies were generated by Bethyl Labs (Montgomery, TX). To test the specificity of the antibodies, a preabsorption assay was performed by incubating the antibody with the peptide (provided by Bethyl labs) at a 10 fold molar excess for one hour at room temperature prior to incubating with the samples for Western blot analysis (Fig. S2). 30 µg of 32-cell embryonic extracts were loaded on to the SDS-PAGE gel. The S. purpuratus β-catenin protein is predicted to be 825 amino acids in length, and the molecular size is estimated to be approximately 90 kDa.
Real time, quantitative PCR (qPCR)
For Dicer morpholino, control morpholino, or β-catenin miRNA TP experiments (stock of injection solution at 300 µM for each miRNA TP), 80–100 injected embryos were used. 200 uninjected embryos were collected at various time points to determine transcript levels of endonenous β-catenin throughout development. Total RNA was extracted using Qiagen microRNeasy kit according to manufacturer’s instructions (Qiagen Inc., Valencia, CA). cDNA was synthesized using TaqMan Reverse Transcription Reagents kit (Invitrogen, Carlsbald, CA). qPCR was performed using 2.5 embryo equivalent for each reaction with the Applied Biosystem power SYBER Green PCR Master Mix (Invitrogen) in the 7300 Real-Time PCR cycler system. Results were normalized to the mRNA expression of the housekeeping gene ubiquitin and shown as fold changes compared to control embryos that were injected with the control morpholino using the ΔΔCt method. The estimated numbers of β-catenin transcripts are calculated based on the level of ubiquitin in various developmental stages described previously (Materna and Davidson, 2012; Materna and Oliveri, 2008). Three to five biological replicates were conducted. Primers used for qPCR analysis are listed in Table S1.
Western blotting
200 embryos injected with control MASO and miRNA TP were pelleted by spinning them down at 15,000 RPM for 30 seconds and resuspended in 15 µl of 4X sample buffer (1 mM Tris HCl, 0.1 mM sucrose, 4% SDS and 10 mM EDTA) prior to heating at 100°C for 10 minutes. Samples were stored at −80°C until later use. 1 mM DTT was added to the protein sample and heated at 100°C for 10 minutes before loading samples onto Tris-Glycine 4–20% gradient gel (Bio-Rad, Hercules, CA). The proteins were electrophoresed at 120 V for 1 hour in Tris-glycine electrophoresis buffer (25 mM Tris, 250 mM glycine at pH 8.3, 0.1% SDS). Proteins were transferred onto nitrocellulose membranes (Pall Corporation, Pensacola, FL) using 250 AMP for 2 hours at 4°C. Membranes were blocked with Blotto (0.05 mM Tris at pH 7.5, 0.18 M NaCl, 3% dry milk and 0.05% Tween 20) for 2 hours at RT or overnight at 4°C and then incubated with β-catenin55 antibody at 1:250 in Blotto overnight at 4°C, followed by three TBST (0.05 M Tris, 0.18 M NaCl, 0.05% Tween) washes. The blot was incubated with secondary antibody goat anti-rabbit HRP (Jackson Immuno Research, West Grove, PA) diluted to 1:3000 in Blotto for 45 minutes at RT. The blots were washed 3 times with TBST and signals were detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific, Rockford, IL) according to the manufacturer’s instructions. Blots were treated with Restore™ Western blot Stripping Buffer (Thermo Scientific, Rockford, IL) according to manufacturer’s instructions and reprobed with the rabbit polyclonal actin antibody (Sigma-Aldrich, St. Louis, MO). Images were acquired with Fluorchem (TM) Q imaging system (Proteinsimple, Santa Clara, CA). Quantification of bands was conducted using the Alpha view-FluorChem QSA, version 3.2.2.0.
Cloning of the reporter constructs
In order to test post-transcriptional regulation of β-catenin by miRNAs and miRNA TPs, its 3’UTR was cloned downstream of GFP coding sequence (in pGEMT vector background) and Renilla luciferase from psiCHECK vector (Promega, Madison, WI). The mCherry coding region was cloned into the pSP6T4 vector flanked with Xenopus β-globin UTRs (Gustafson and Wessel, 2010). Primers were designed using the Primer 3 program (Rozen and Skaletsky, 2000). Primers used for β-catenin 3’UTR cloning are: Forward, 5’ AACGCTCGAGATCCAGATGTACCAAGCCAA 3’ and Reverse 5’ GGCACTAGTAGGAACTACAAGAAAGTCTC 3’. Restriction sites used for subcloning are underlined. PCR products were purified with Promega Wizard SVgel and PCR cleanup kit (Promega, Madison, WI) prior to ligation. The ligated products were cloned into PCRII cloning kit dual promoter (Invitrogen, Carlsbad, CA) and transformed into DH5-α cells (Invitrogen, Carlsbald, CA). β-catenin 3’UTR positive clones (in pCRII) were sequenced (Genewiz, Inc., South Plainfield, NJ) and subcloned downstream of eGFP coding region into a pGEMT vector backbone. Renilla luciferase was subcloned into the pGEMT vector to replace the eGFP coding sequence. Firefly luciferase from psiCHECK vector was PCR amplified with primers Forward 5’ GCATAGATCTATGGCCGATGCTAAGAACATT 3’ and Reverse 5’ ATAGCTCGAGATACCACATTTGTAGAGGTTTTA 3’ with restriction enzyme sites underlined, and cloned into the pSP6T4 flanked by the Xenopus β-globin 3’UTRs. Positive clones were sequenced to confirm cloning of the β-catenin 3’UTR downstream of the eGFP or Renilla luciferase reporters and the Firefly luciferase construct in pSP6T4 (Genewiz, Inc., South Plainfield, NJ).
Site-directed mutagenesis
The miR-2007 seed sequence was identified to have two binding sites within the β-catenin 3’UTR. The miRDeep2-30364-35240 seed sequence was identified to have one binding site within the β-catenin 3’UTR. All seed sequences were mutated at positions 3 and 5 within the miRNA seed sequence (Gregory et al., 2008). miR-2007 seed sequence 5’ CTGAAAT 3’ was modified to 5’ CTCACAT 3’, and miRDeep2-30364-35240 was modified from 5’ GTGCAAT 3’ to 5’ GTCCTAT 3’. Modified bases are underlined.
Mutageneic primers were designed according to the primer design program at www.agilent.com/genomics/qcpd. All mutations were generated using the QuikChange Lightning kit according to manufacturer’s instructions (Agilent Technologies, Santa Clara, CA). Primer sequences used for site-directed mutagenesis for miR-2007 at position +922 are 5’ CAGGCCTCGTCAAGATTATATCTCACATAGATATCTCATGATTGGCTAC 3’ and 5’ GTAGCCAATCATGAGATATCTATGTGAGATATAATCTTGACGAGGCCTG 3’. The second set of primers for miR-2007 at position +2647 (Fig. 2A) are 5’ AATCTAAGCTACTTCCATTTTTCATGTCTTAGATAAACTGAATACATTCTTTTAAGAT TG 3’ and 5’ GCAATCTTAAAAGAATGTATTCAGTTTATCTAAGACATGAAAAATGGAAGTAGCTTA GAT 3’. The primers used for miRDeep2-30364-35240 are 5’ GCATATTGATGCCTCTTAAAAAGGTCCTATAAAGAGTACAATATGCAACAATC 3’ and 5’ GATTGTTGCATATTGTACTCTTTATAGGACCTTTTTAAGAGGCATCAATATGC 3’. Altered seed sequences are underlined. Positive clones were sequenced to validate the fidelity of the mutagenesis (Genewiz, South Plainfield, NJ).
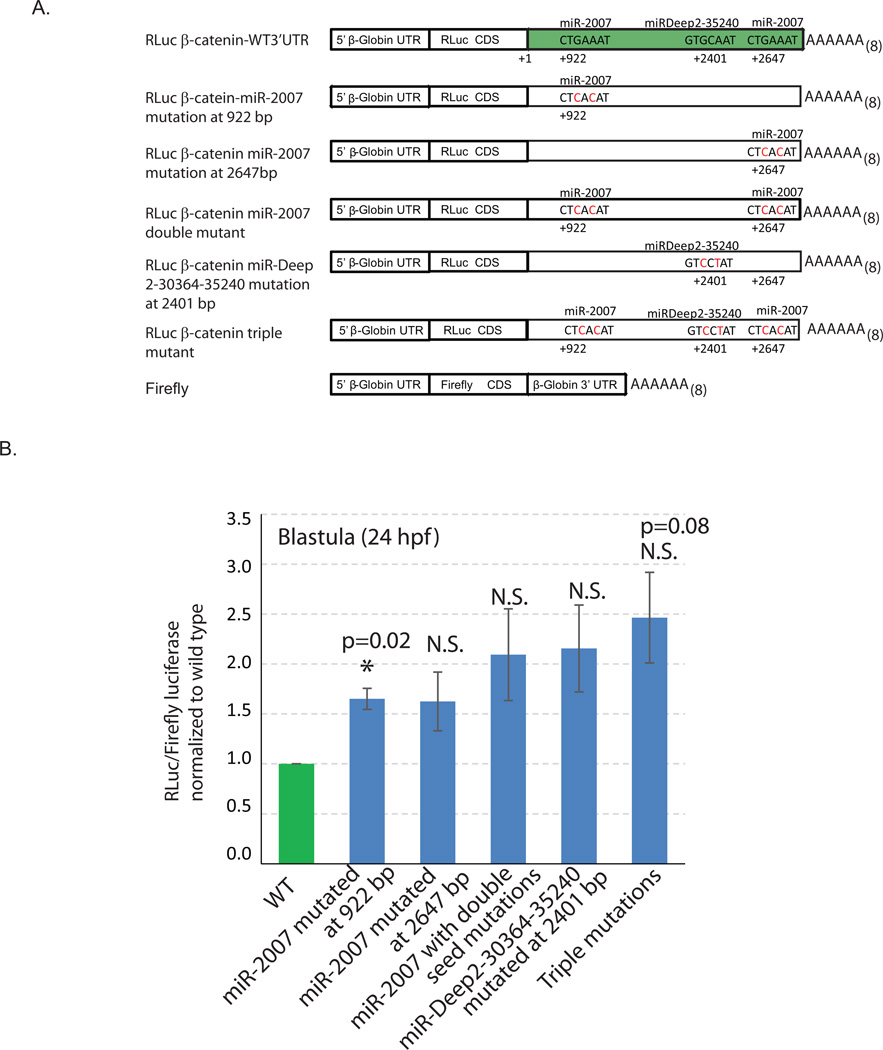
Fig. 2. Mutagenesis analysis of potential miRNA binding site within the β-catenin 3’UTR.
3’UTR of β-catenin was cloned downstream of the Renilla luciferase (RLuc) construct for testing miRNA regulation. (A) Schematic of various reporter constructs with wild type (WT) or mutated miRNA binding sites within the β-catenin 3’UTR is depicted. The Firefly luciferase reporter construct is flanked with Xenopus β-globin UTRs and used as a microinjection control for normalization. Newly fertilized eggs were coinjected with mRNAs of either RLuc CDS fused with the wild type β-catenin 3’UTR or fused with the β-catenin with miRNA seed mutations at miR-2007 and/or miRDeep2-30364-35240 sites and Firefly luciferase. (B) The embryos were collected at mesenchyme blastula stage (24 hpf). The RLuc readings were normalized to the Firefly luciferase readings. These RLuc/Firefly luciferase ratios of the mutated constructs were normalized to the control. miR-2007 at 922 bp and miRDeep2-30364-35240 contain functional miRNA binding sites (3 biological replicates with 50 embryos each). *Student T-test is used to analyze the significance between the constructs with wild type β-catenin 3’UTR and β-catenin 3’UTR with mutated miRNA seeds. N.S.=not significant.
In vitro transcription
All reporter constructs were linearized prior to in vitro transcription. mCherry was linearized with Sal1 enzyme; β-catenin eGFP, Renilla luciferase and Firefly luciferase reporter constructs were linearized with Spe1 enzyme (Promega, Madison, WI). Linearized reporter constructs were in vitro transcribed using mMessage machine kit (Invitrogen, Carlsbad, CA). The T7 RNA polymerase was used for generating the Renilla luciferase β-catenin mRNAs, and the Sp6 RNA polymerase was used for generating the Firefly luciferase and mCherry mRNAs according to the manufacturer’s instructions. mRNAs were purified by using Qiagen microRNeasy kit according to manufacturer’s instructions (Qiagen Inc., Valencia, CA) and purified mRNAs were loaded onto the Millipore spin columns for further clean up (Millipore, Billerica, MA). For luciferase assays, the 2.5 µl of injection solution contained 100 to 500 ng of Renilla luciferase and 150 ng of Firefly luciferase mRNA.
Quantitative microscopy analysis of GFP and mCherry signals
To test if miRNA TPs bind to the three functional miRNA sites within the β-catenin 3’UTR, we tested the level of translated eGFP in the presence of all three β-catenin miRNA TPs. Injected embryos were first treated with 2X sea water for 2 minutes to induce a temporary shedding of the cilia to immobilize the swimming embryos (Ettensohn et al., 2004). They were washed with 1X sea water twice and imaged with Zeiss Observer Z1 microscope (Carl Zeiss Incorporation, Thorwood, NY) as living embryos at low magnification (total magnification at 100x) to maximally capture fluorescent pixels. The imaging settings were determined with all sets of embryos prior to image acquisition to ensure that the setting captures the GFP and the mCherry fluorescence within the linear range. All embryos within an experiment were imaged under the exact same conditions. The GFP and mCherry fluorescence signals were quantified using Zeiss software (Axio vision SE 64 Rel.4.8). The GFP signal in each embryo was normalized to the mCherry signal to account for injection volume differences. Unpaired Student T-test was used to determine statistical significance (Fig. S1).
Dual luciferase quantitation
Dual luciferase quantitation was performed using the Promega™ Dual-Luciferase™ Reporter (DLR™) Assay Systems with the Promega™ GloMax™ 20/20 Luminometry System (Promega, Madision, WI). 50 embryos at the mesenchyme blastula stage (24 hours post fertilization; hpf) were collected in 22 µl of 1X Promega passive lysis buffer and vortexed for at least 5 minutes. Embryonic lysates were either stored at −80°C or processed immediately. Prior to luciferase readings, 100 µl of the Luciferase Activating Reagent II (LAR-II) was added to the 96-well plate (BD Falcon, Franklin Lakes, NJ). 20 µl of the embryonic lysates was then added to the corresponding wells and luciferase reading for the Firefly was obtained. 100 µl of the Stop and Glow solution was added into the 96-well plate to quench Firefly luciferase while the Renilla luciferase readings were obtained.
Immunofluorescence
For Endo1 and 1D5 immunolabeling, embryos were fixed in 4% paraformaldehyde (16% stock; EMS, Hatfield, PA) in artificial sea water for 10 minutes at room temperature. Four 10 minute PBST (Phosphate Buffered Saline-Tween) washes were performed. For myosin immunolabeling, embryos were fixed in 100% methanol at −20°C for 30 minutes, followed by PBST washes. Embryos were blocked with 4% sheep serum (Sigma Aldrich, St. Louis, MO) for 1 hour at room temperature. Primary antibody incubation was performed with Endo1 antibody (Wessel and McClay, 1985) at 1:200, 1D5 antibody at 1:50 and myosin heavy chain antibody (Wessel et al., 1990) at 1:100 overnight at 4°C. Embryos were washed three times 15 minutes with PBST followed by goat anti mouse Alexa 488 or goat anti rabbit Alexa 488 conjugated secondary antibody at 1:300 (Invitrogen, Carlsbad, CA). Immunolabeled embryos were imaged using a Zeiss LSM 780 scanning confocal microscope (Carl Zeiss Incorporation, Thorwood, NY) and data were obtained and analyzed using the Zen software (Carl Zeiss Incorporation, Thorwood, NY).
Detection of endogenous alkaline phosphatase
The endogenous alkaline phosphatase was used to assay for differentiated endoderm of the larvae (Annunziata et al., 2013; Drawbridge, 2003; Hinman and Degnan, 1998; Kumano and Nishida, 1998; Nishida and Kumano, 1997; Whittaker, 1990). Three day old larvae were fixed in MOPS-paraformaldehyde based fixative (4% paraformaldehyde, 100 mM MOPS pH 7.0, 2 mM MgSO4, 1 mM EGTA, and 0.8 M NaCl) for 10 min at room temperature. Embryos were washed with alkaline phosphatase buffer three times (100 mM Tris pH 9.5, 100 mM NaCl, 50 mM MgCl2, 0.1% Tween-20), followed by staining until desired color development with the staining solution (0.1 M Tris pH 9.5, 50 mM MgCl2, 0.1 M NaCl, 1 mM Levamisole, 10% Dimethylformamide, 45 µl of 75 mg/mL NBT and 35 µl of 50 mg/mL BCIP per 10 ml of solution). The staining step was stopped with washes with MOPS buffer (0.1 M MOPS pH 7.0, 0.5 M NaCl, and 0.1% Tween-20). Images were acquired with Nikon D90 digital camera connected to a Zeiss Observer Z1 microscope.
Whole mount in situ hybridization
β-catenin, Krl, Blimp1b and FoxA were PCR-amplified from sea urchin embryonic cDNA. PCR primers used to clone the RNA probes are listed in Table S1. Krl probe was made directly from the PCR product; Blimp1b, FoxA and β-catenin were cloned into Blunt-TOPO, PGEMT and PCRII vectors, respectively. The plasmids containing β-catenin, Blimp1b, FoxA were linearized with Not1, EcoR1 and Pst1, respectively (Promega, Madison, WI). Antisense probes were labeled with digoxigenin with the DIG RNA Labeling Kit (Roche Diagnostics Corporation, Indianapolis, IN, USA). 0.1 µg probe/mL was used to detect native transcript in embryos according to previously published protocols (Minokawa et al., 2004).
Results
β-catenin transcripts are destabilized by miRNAs
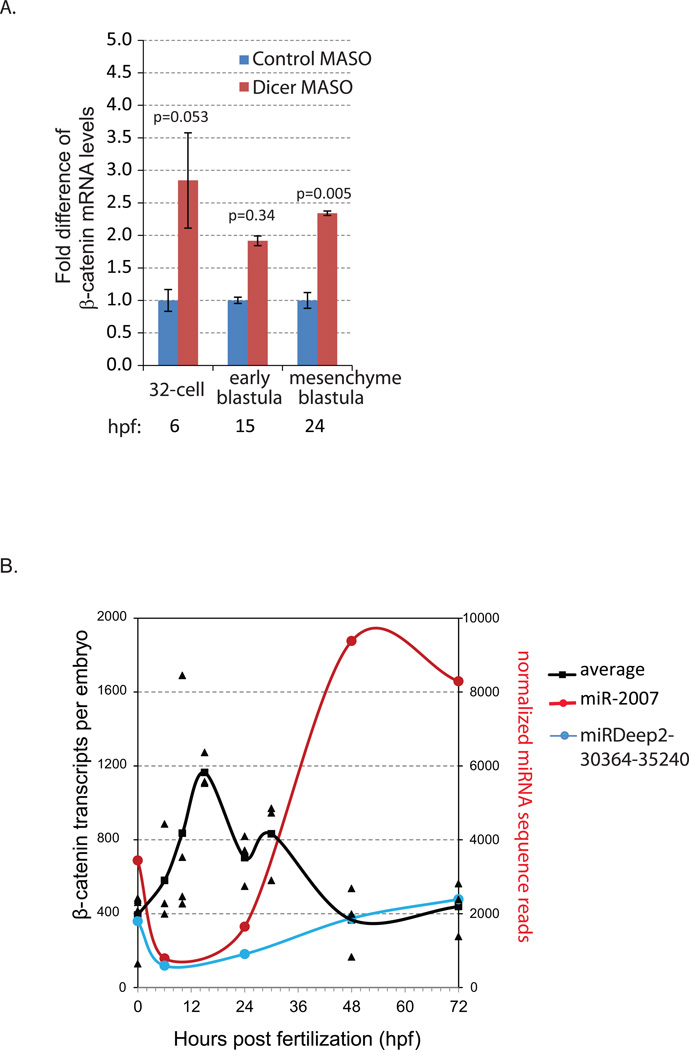
To determine if β-catenin was potentially regulated by miRNAs, we examined its transcript levels in control-MASO and Dicer MASO-injected embryos with real time, quantitative PCR (qPCR). Newly fertilized eggs were microinjected with Dicer morpholino (MASO) to perturb the miRNA biogenesis pathway, leading to global miRNA depletion (Song et al., 2012). Embryos were collected at the 32-cell stage (5–6 hpf), early blastula (15 hpf), and mesenchyme blastula (24 hpf) stages. Transcript levels of β-catenin at all stages were increased in Dicer knockdown embryos (Fig.1A). At the mesenchyme blastula stage, β-catenin transcript level was significantly increased (Fig. 1 A), suggesting that miRNAs regulate β-catenin mRNA stability at 24 hpf.
Fig. 1. Real time, quantitative PCR (qPCR) of β-catenin transcript levels.
(A) The transcript levels of β-catenin were measured in control MASO and Dicer MASO-injected (1.5 mM stock injection solution) embryos. Results were normalized to the mRNA expression of the housekeeping gene ubiquitin and shown as fold changes compared to control embryos that were injected with the control MASO using the ΔΔCt method. The transcript level of β-catenin was significantly more abundant in Dicer MASO than in the control MASO-injected embryos at the mesenchyme blastula stage. Standard error bars are graphed. At least three independent biological replicates with 80–100 embryos per replicate were tested. hpf= hours post fertilization. *Student T-test, p=0.005.
(B) Time course expression profile of β-catenin mRNA. The transcript levels of β-catenin were measured in uninjected embryos collected at various developmental stages. Results were normalized to the mRNA expression of the housekeeping gene ubiquitin. The estimated numbers of β-catenin transcripts are calculated based on the level of ubiquitin in various developmental stages described previously (Materna and Davidson, 2012; Materna and Oliveri, 2008). At least three independent biological replicates with 200 embryos per replicate were tested. 2.5 embryo equivalents were used. Individual data points are shown and the average is graphed as a line. β-catenin transcripts are most abundant between the 32-cell and early blastula stages. Its level decreases and stabilizes from the mesenchyme blastula stage to the larval stage. Normalized sequence reads of miR-2007 and miRDeep2-30364-35240 from our previous study are plotted (Song et al., 2012). β-catenin mRNAs and these miRNAs have inverse expression patterns.
We tested the possibility that miRNAs may play a role in the post-transcriptional regulation of β-catenin by analyzing the 3’UTR of β-catenin for potential miRNA regulatory sites. The miRNA prediction tools that perform best according to high-throughput validation all scan 3'UTRs for short motifs complementary to the miRNA seed sequences (Baek et al., 2008; Selbach et al., 2008). However, current miRNA prediction programs such as PicTar and TargetScan rely heavily on conservation information, which is not yet available for the S. purpuratus genome. Therefore, the best tool we have now is to search for 100% miRNA seed matches within 3’UTRs. We searched for 100% miRNA seed matches within the 3’UTR of β-catenin and found that of the 16 bioinformatically identified miRNA predicted sites, only spu-miRDeep2-30364-35240 and spu-miR-2007 have greater than 1,000 normalized deep sequence reads during early developmental stages of the sea urchin (Table S2) (Song et al., 2012).
Next, we examined the mRNA expression profile of endogenous β-catenin and their potential miRNA regulators, spu-miRDeep2-30364-35240 and spu-miR-2007, at various developmental stages. Results indicate that the levels of β-catenin mRNA changes over time throughout development (Fig. 1B). The level of β-catenin mRNA peaks at early blastula stage (15 hpf), followed by a decrease at the mesenchyme blastula stage (24 hpf) and stabilization from the gastrula to larval stages. Coinciding with this decrease and stabilization of β-catenin mRNA from 24 hpf to larval stages, spu-miRDeep2-30364-35240 and spu-miR-2007 have increased sequence reads from 24 hpf to the larval stage (Fig. 1B) (Song et al., 2012). This inverse expression pattern of β-catenin and these two miRNAs is consistent with the possibility that β-catenin mRNA may be regulated by these miRNAs (Baek et al., 2008; Bartel, 2009; Brodersen and Voinnet, 2009; Hendrickson et al., 2009; Liu, 2008; Selbach et al., 2008).
β-catenin is directly regulated by miRNAs
To test if β-catenin is directly regulated by miRNAs, we cloned the β-catenin 3’UTR downstream of Renilla luciferase (Rluc) to analyze post-transcriptional regulation of β-catenin by spu-miRDeep2-30364-35240 and spu-miR-2007 (Fig. 2A). The 3’UTR of β-catenin was amplified with PCR based on 3’UTR sequences identified from previous analyses (Samanta et al., 2006; Sodergren et al., 2006). miR-2007 has two binding sites (at positions +922 bp and +2647 bp downstream of the stop codon at +1 bp) and miRDeep2-30364-35240 has one binding site (at position +2401 bp downstream of the stop codon) within the β-catenin 3’UTR. We used site-directed mutagenesis to alter the third and fifth base pairs of the seed sequences to disrupt the miRNA to target recognition to test its direct regulation by miRNAs (Gregory et al., 2008). Firefly luciferase reporter construct was fused to the Xenopus β-globin 3’UTR and used as a control for Rluc luciferase normalization.
Newly fertilized eggs were co-injected with in vitro transcribed mRNAs of Renilla reporter constructs fused to the β-catenin 3’UTR and the Firefly reporter construct fused to the Xenopus β-globin 3’UTR. We collected mesenchyme blastula stage embryos, when β-catenin mRNA is significantly increased in Dicer knockdown embryos, to measure miRNA regulation on β-catenin using the dual luciferase assay. Results indicate that miR-2007 regulates both binding sites of β-catenin, although only the miR-2007 mutated at 922 bp reached statistical significance (Fig. 2B). miR-2007 regulation at these two sites are likely to be redundant, since the normalized luciferase reading of miR-2007 double mutant was not significantly different than the normalized luciferase reading of miR-2007 single mutants (Student T-test, p=0.6). The Renilla luciferase reporter construct with triple mutants abolishing binding and regulation by both miR-2007 and miR-Deep2–30364–35240 resulted in additional increase in luciferase protein levels, indicating additive regulation by miRNAs. The relatively small change in the luciferase expression caused by the removal of miRNA regulation is consistent with the previous studies (Nicolas, 2011; Pelosi et al., 2013; Selbach et al., 2008; Wang et al., 2014). Overall, these results indicate that β-catenin 3’UTR contains at least three functional miRNA regulatory sites.
β-catenin miRNA target protector morpholinos (miRNA TP) modulate β-catenin mRNA and protein levels
Once we identified the three functional miRNA sites within the β-catenin 3’UTR, we designed miRNA target protector morpholinos (miRNA TP) against these three sites to compete with the endogenous miRNAs for β-catenin regulation (Staton and Giraldez, 2011). These miRNA TP sequences are unique to the β-catenin 3’UTR, as BLASTN searches in the sea urchin genome only identified these sequences to be located within the β-catenin 3’UTR. To test if miRNA TPs bind to the three functional miRNA binding sites within the β-catenin 3’UTR, we cloned β-catenin 3’UTR downstream of eGFP and tested the level of translated eGFP in the presence of miRNA TP morpholino treatments at the early blastula stage (Fig. S1A). We determined that blocking functional spu-miRDeep2-30364-35240 and spu-miR-2007 miRNA sites with miRNA TPs resulted in significant increase in translated eGFP protein, indicating that miRNA TPs are able to compete with endogenous miRNAs for binding and block endogenous miRNA-mediated translational repression (Fig. S1A). The amount of miRNA TP injected was designed to be a 1000x molar excess of the amount of TP calculated to block the injected amount of eGFP reporter construct and did not seem to be toxic to the blastula embryos at the morphological level (Fig. S1B).
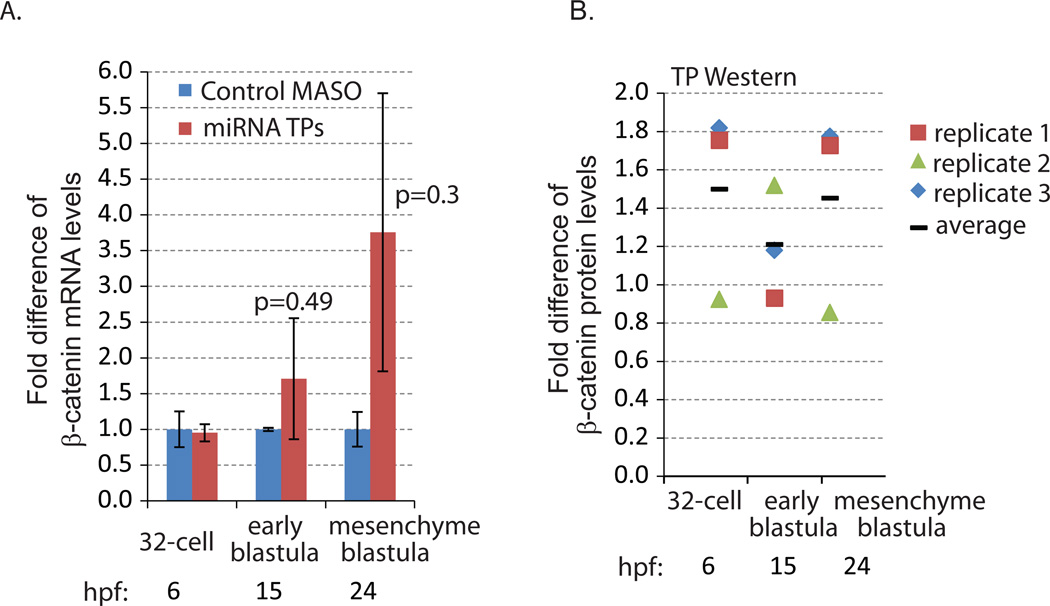
We tested the effect of miRNA TPs on β-catenin mRNA level (Fig. 3A). Introducing β-catenin miRNA TPs caused an increase in endogenous β-catenin mRNA levels at 15 and 24 hpf; however, this difference was not statistically significant (Fig. 3A). Further, to examine if the removal of miRNA regulation on β-catenin would lead to a change in its protein levels, we used S. purpuratus specific β-catenin antibodies for detection. Preabsorption studies indicated the specificity of the β-catenin antibodies (Fig. S2). We determined that the microinjection of 300 µM of each miRNA TP (injection stock concentration) was able to elicit on average a 1.5 fold increase in β-catenin protein compared to the MASO control at 32-cell and mesenchyme blastula stages, indicating that blocking miRNA mediated regulation in vivo is sufficient to cause a minor increase in β-catenin protein level (Fig. 3B). Because this β-catenin antibody does not work in whole mount immunolabeling, we are not able to address the spatial distribution of β-catenin or if this increase in β-catenin is of the cytoplasmic or nuclear β-catenin populations.
Fig. 3. β-catenin miRNA TP induced increased β-catenin protein and mRNA levels.
(A) qPCR was used to measure the transcript levels of β-catenin in the embryos injected with control and miRNA TP MASOs against the three functional miRNA binding sites. 100 embryos were collected at the 32-cell (6 hpf), early blastula (15 hpf) and mesenchyme blastula (24 hpf) stages. The mRNA level of β-catenin was increased at early blastula and mesenchyme blastula stages when miRNA regulation of β-catenin was abolished. However, these increases were not significant in β-catenin miRNA TP and control MASO-treated embryos (Student T-test). Hpf = hours post fertilization. (B) Western blot of 200 embryos injected with control or miRNA TPs. The embryos were collected at the 32-cell (6 hpf), early blastula (15 hpf) and mesenchyme blastula (24 hpf) stages. Compared to control embryos, embryos injected with miRNA TPs accumulated on average 1.5 times more β-catenin protein at the 32-cell and mesenchyme blastula stages. β-catenin protein levels are normalized to actin (3 biological replicates). Individual data points were indicated by the different icons. The average is graphed as a black bar.
Removal of miRNA regulation of β-catenin does not affect spicule formation in the sea urchin embryos
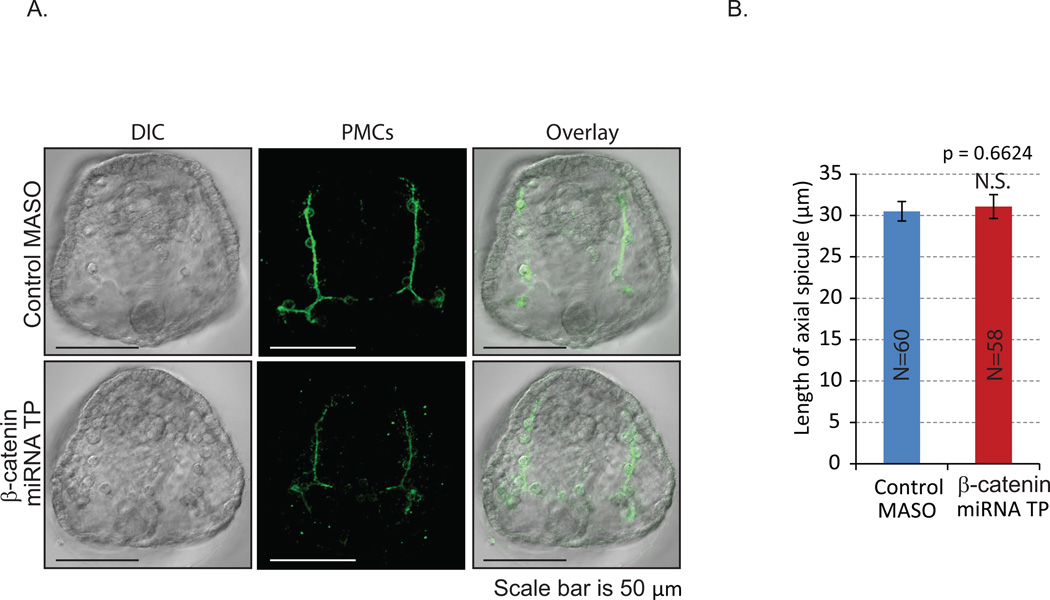
Since the specification of mesodermally-derived PMCs is downstream of the Wnt/β-catenin signaling pathway (Oliveri et al., 2003), we examined PMCs and skeleton formation in β-catenin miRNA TPs-treated embryos. Results indicate that of removal of miRNA regulation β-catenin resulted in reduced PMC (1D5) staining but did not affect PMC patterning or the spicule lengths (Fig. 4). Thus, the change in β-catenin protein levels induced by miRNA TPs is not sufficient to perturb PMC specification, patterning, or skeleton formation.
Fig. 4. Removal of miRNA regulation of β-catenin does not affect skeletogenesis.
Gastrula stage embryos (48 hpf) were immunolabeled with the 1D5 antibody against primary mesenchyme cell (PMC)-specific membrane protein (McClay et al., 1983) and imaged with an LSM 780 scanning confocal microscope. Z-stacks were reconstructed into a single projected image. (A) Removal of miRNA regulation of β-catenin did not affect PMC patterning. (B) The length of axial skeletogenic spicules was not significantly changed in the miRNA TP injected gastrula embryos compared to control. Student T-test, p=0.662. N is the total number of axial spicules measured. N.S.=not significant.
Treatment of β-catenin miRNA TPs resulted in aberrant gut morphology
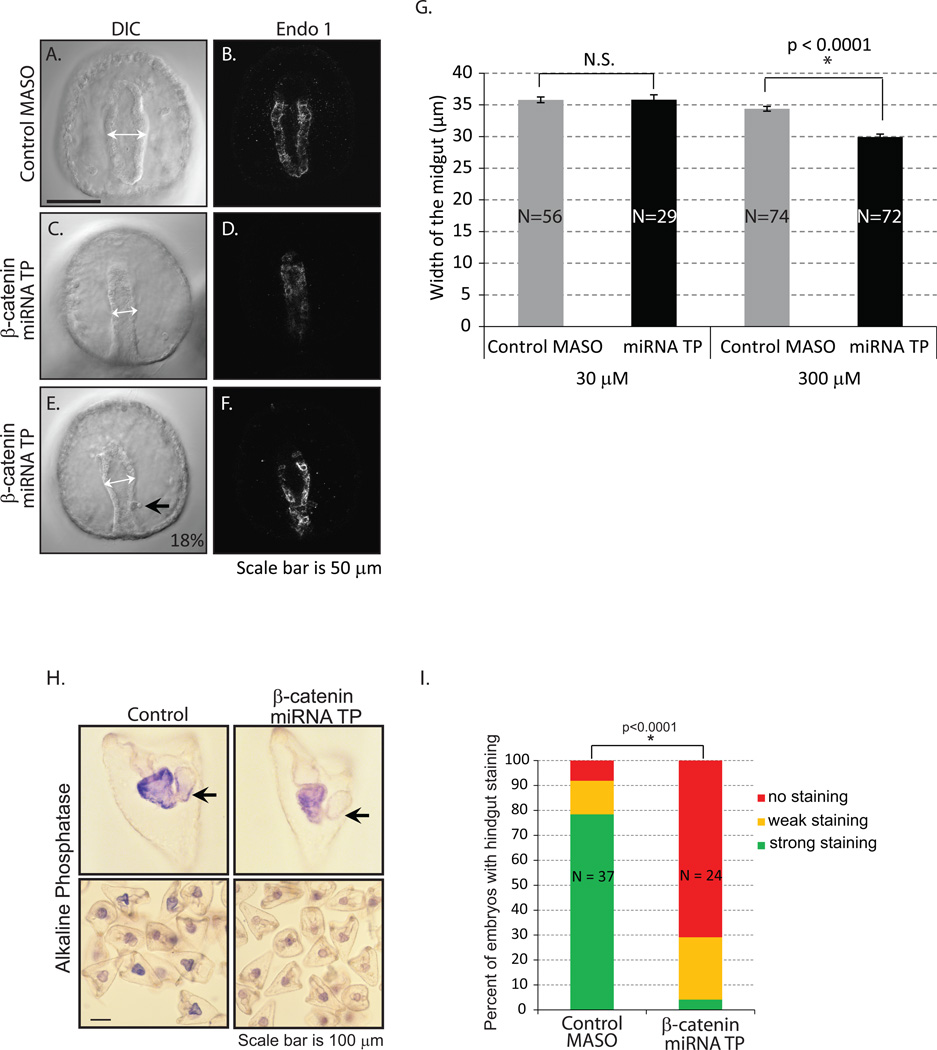
Next, we examined the effect of this minor increase in β-catenin protein accumulation induced by the removal of miRNA regulation of β-catenin on gut development. We injected β-catenin miRNA TPs and examined the morphology of the gut. The larval digestive system has a tripartite structure, consisting of a muscular esophagus (foregut), a large stomach (midgut), and a short tubular intestine (hindgut) (Burke, 1981; Burke and Alvarez, 1988). A gastrula embryo has a straight tubular gut that is lined with single epithelial cells (Burke, 1981). The β-catenin miRNA TP-treated embryos had a significantly narrower gut at the gastrula stage than the control embryos (Fig. 5A,B vs. 5C,D). We tested a variable amount of β-catenin miRNA TPs, ranging from stocks of injection solutions from 3 nM to 3 mM (Fig. 5G and data not shown), and only 300 µM and 3 mM β-catenin miRNA TP-treated embryos had statistically significantly smaller midgut widths compared to the control (Fig. 5G). However, at the 3 mM stock concentration, the control embryos had less than 60% healthy embryos, indicating non-specific toxic response to the morpholino, so these data were not included in our analysis. In addition, 18% of the β-catenin miRNA TP-treated embryos (in 2 replicates out of 4) have aberrant intestine (hindgut) morphology (Fig. 5E,F). In β-catenin miRNA TP-treated embryos, the shape of their intestine (hindgut) is trapezoidally shaped instead of a cylindrical structure (Fig. 5A,B vs. E,F).
Fig. 5. Removal of miRNA regulation on β-catenin results in aberrant gut morphology.
Gastrulae were immunolabeled with Endo1, an antigen expressed in the midgut and hindgut of the embryo (Wessel et al., 1990), and imaged with an LSM 780 scanning confocal microscope. (A–B) Gastrulae injected with control MASO had a straight tubular gut that is lined with a single layer of epithelial cells. (C–F) β-catenin miRNA TP treatment resulted in embryos with narrower gut structure. (E–F) 18% of the 97 embryos miRNA TP injected embryos had an aberrant hindgut structure (black arrow) (2 out of 4 biological replicates). (G) The width of the midgut was measured with embryos injected with either 30 µM or 300 µM stock of β-catenin miRNA TP (white arrows). Only the 300 µM miRNA TP treated embryos had a significantly narrower gut than the control. *Student T-test, p<0.0001. N is the total number of embryos imaged and phenotyped. N.S. = not significant. (H) The alkaline phosphatase staining was used to assess endodermal differentiation of the larvae. Control MASO-injected embryos have more intense alkaline phosphate staining than the β-catenin miRNA TP treated embryos. Many of the embryos exposed to the β-catenin miRNA TP lack alkaline phosphatase staining in the intestine (hindgut) (arrows). (I) The percentage of embryos with hindgut staining in the miRNA TP treatment group is significantly lower than in the control embryos. *Fisher’s Exact Test (no hindgut staining vs. hindgut staining, p<0.0001). N is the total number of embryos examined.
Since we observed gut morphological difference in β-catenin miRNA TP-treated embryos, we tested if endodermal differentiation was affected. Alkaline phosphatase is a digestive enzyme expressed in the guts of most animals. In sea urchin pluteus larvae, alkaline phosphatase is expressed only in the gut epithelium and serves as a marker for differentiated endoderm (Drawbridge, 2003; Kumano and Nishida, 1998; Nishida and Kumano, 1997; Whittaker, 1990). Although the β-catenin miRNA TP treated embryos expressed alkaline phosphatase in the gut, it seems to be at a reduced level as compared to the control (Fig. 5H,I). Further, a significant number of β-catenin miRNA TP-treated embryos have little or no alkaline phosphatase present in the intestine (hindgut) as compared to the control (Fig. 5I). Thus, β-catenin miRNA TP-treated embryos may have defects in endodermal differentiation.
β-catenin miRNA TP-treated embryos have normal sphincters but less well developed circumesophageal musculature
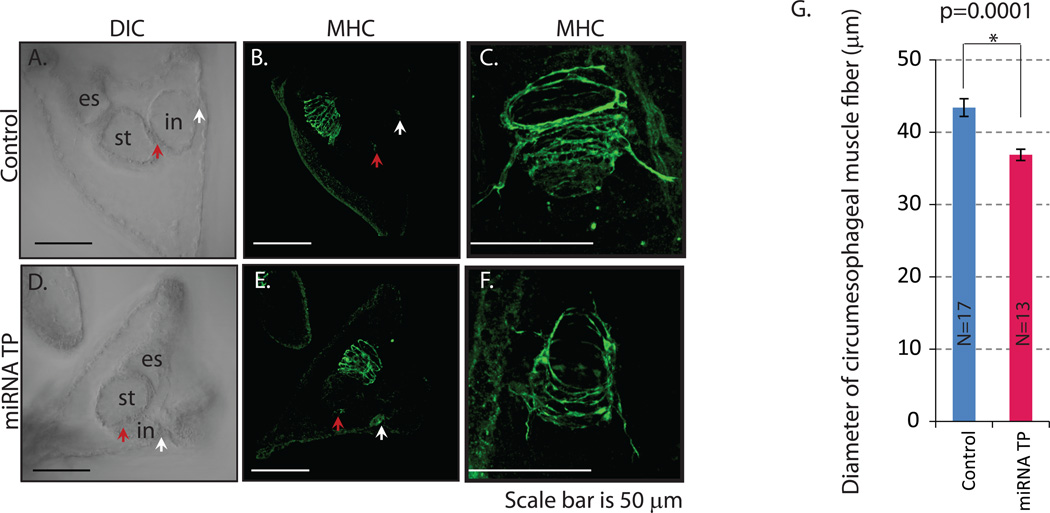
We further examined the circumesophageal muscles of mesodermal origin and the sphincters of endodermal origin with the antibody against the sea urchin myosin heavy chain (MHC) (Wessel et al., 1990). In sea urchin embryos, a set of muscle cells partition the various compartments of the digestive system. The cardiac sphincter, a constriction made from a simple striated myoephithelium with myofibrils, separates the esophagus (foregut) and the stomach (midgut) (Burke, 1981; Wessel et al., 1990). Separating the stomach (or midgut) and the intestine (hindgut) is the pyloric sphincter. The digestive tract ends with an anal sphincter. All these sphincters are endodermally derived and contain myosin heavy chain (Annunziata et al., 2013; Burke, 1981; Wessel et al., 1990). These constrictions give the overall gut structure an hourglass-like shape in the larva. The β-catenin miRNA TP treated embryos have normal pyloric and anal sphincters (Fig. 6A,B vs. D,E). However, in general, embryos treated with the β-catenin miRNA TPs had a less well developed circumesophageal musculature as compared to the control (Fig. 6C vs. F). Further, the diameter of the muscle fibers is significantly larger in control than in the β-catenin miRNA TP-treated embryos (Fig. 6G). These results indicate that blocking miRNA regulation of β-catenin impacted mesodermally-derived circumesophageal musculature.
Fig. 6. β-catenin miRNA TP-treated embryos have normal sphincters but less well developed circumesophageal musculature.
Three day old larvae were immunolabeled with the myosin heavy chain antibody (MHC) (Wessel et al., 1990) to detect circumpharyngeal muscle fibers and sphincters (red arrow-pyloric sphincter; white arrow-anal sphincter) that compartmentalize the larval gut. (A) DIC image of the control larva. (B) Control larva immunolabeled with MHC. (C) Zoomed in view of a normal larval circumpharyngeal muscles. (D) DIC of the β-catenin miRNA TP-treated embryo. (E) β-catenin miRNA TP treated embryo immunolabeled with MHC. (F) Zoomed in view of the larval circumpharyngeal muscles from β-catenin miRNA TP injected larvae. es=esophagus/foregut; st=stomach/midgut; in=intestine/hindgut. (G) The diameter of the circumesophageal muscle fibers is significantly smaller in the β-catenin miRNA TP treated embryos compared to the control embryos.
Removal of miRNA regulation of β-catenin resulted in increased transcripts of endodermal regulatory genes
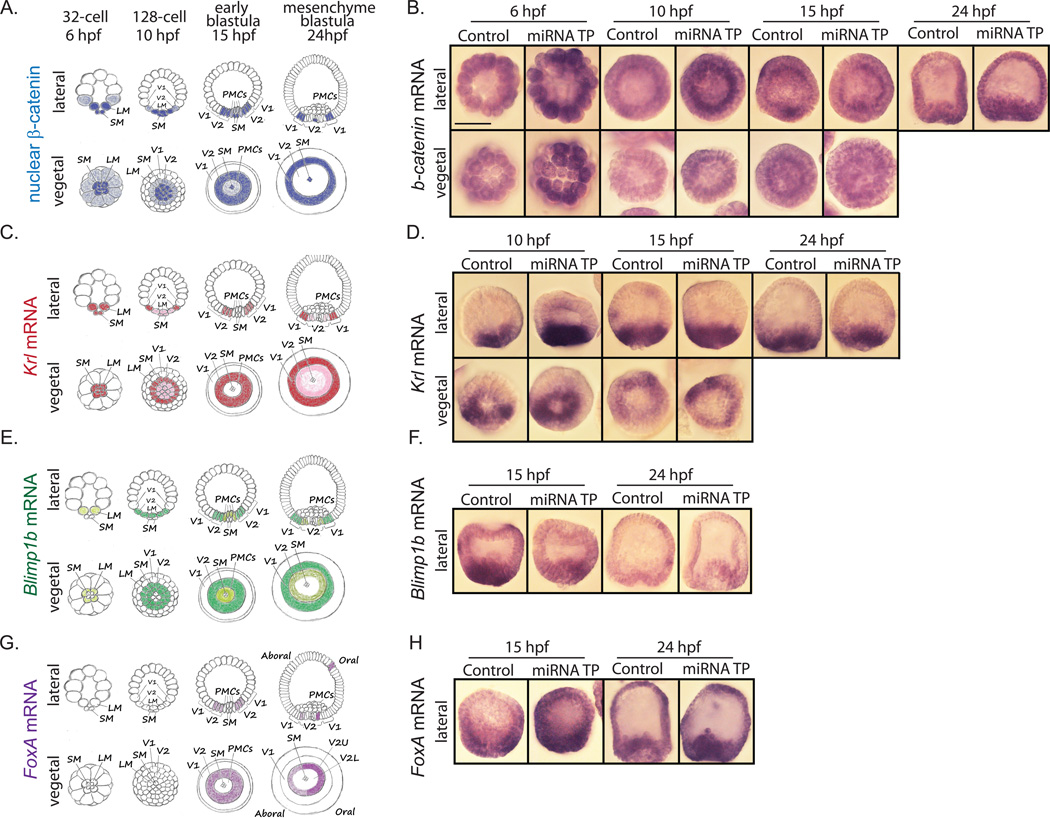
The sea urchin endoderm is derived from two layers of cells that are specified early in development: Veg2 and Veg1 cells (Logan and McClay, 1997; Ransick and Davidson, 1998). The Veg2 cells give rise to the non-skeletogenic mesodermal cells, the foregut and ventral/oral midgut, while the Veg1 cells give rise to the ectoderm, the dorsal/aboral midgut and hindgut (Logan and McClay, 1997; Ransick and Davidson, 1998). Nuclearization of β-catenin was shown to play a critical role in differentiation of both Veg2 and Veg1 cells. Previous studies demonstrated that β-catenin protein localizes into the nucleus of specific cells according to the developmental stage (Fig. 7A): at the 32-cell stage (6 hpf) it is mostly restricted to the micromeres, by 128-cell stage (10 hpf) it extends to the Veg2 cells, and by early blastula stage (15 hpf) its expression begins to fade from the PMCs but remains in the Veg2 cells. By mesenchyme blastula stage (24 hpf) nuclear β-catenin clears from the Veg2 region and shifts to the Veg1 region. Interestingly, small micromeres always contain high amounts of nuclear β-catenin (Logan et al., 1999). Wnt/β-catenin signaling along with Otx induce Veg2 cells to become endoderm by activating endodermal regulatory genes such as Blimp1b, Krl, Eve, and Hox11/13b (Arenas-Mena et al., 2006; Howard et al., 2001; Peter and Davidson, 2010, 2011; Smith et al., 2008). Activation of these genes in turn activates more endodermal regulators, including Brachyury, FoxA, and GataE, thus promoting endoderm differentiation (Fig. 7A) (Peter and Davidson, 2011; Smith et al., 2008).
Fig. 7. Removal of miRNA regulation of β-catenin does not affect the spatial localization of Wnt responsive genes.
(A) Schematic depicts the localization of nuclear β-catenin (Logan et al., 1999) (C) Schematic depicts the localization of Krl mRNA (Howard et al., 2001; Minokawa et al., 2004; Peter and Davidson, 2010) (E) Schematic depicts the localization of Blimp1b mRNA (Livi and Davidson, 2006; Peter and Davidson, 2010, 2011; Smith et al., 2007) (G) Schematic depicts the localization of FoxA mRNA (Oliveri et al., 2006; Peter and Davidson, 2010). SM = small micromeres, LM = large micromeres, V1 = Veg1 tier, V2 = Veg2 tier, V2U = Veg2 upper region, V2L = Veg2 lower region, PMCs = primary mesenchyme cells. Whole mount in situ hybridization was performed to address the spatial localization of (B) β-catenin and Wnt responsive genes, including (D) Krl, (F) Blimp1b and (H) FoxA as a result of β-catenin miRNA TP treatment. Embryos were collected at various developmental stages. Spatial localizations of Krl, Blimp1b and FoxA transcripts were not affected by removal of miRNA regulation of β-catenin.
To dissect the molecular mechanism of the observed endomesodermal morphological defects, we examined if increase in β-catenin protein induced by removal of its miRNA regulation has an impact on the spatial distribution of endodermal genes. First we performed whole mount in situ hybridization of β-catenin itself to test if the expression domains of its mRNA would be affected upon removal of miRNA regulation (Fig. 7B). Results indicate that the ubiquitous spatial distribution of β-catenin mRNA was not altered upon β-catenin miRNA TP treatment. However, a noticeable increase in the mRNA levels of β-catenin in all blastomeres of miRNA TP-treated embryos at 6 hpf and 10 hpf was observed. This observed increased level of β-catenin mRNA may be translated to the protein that would become either cytoplasmic protein where it associates with the adherens junctions or acting as a nuclear transcriptional activator (Logan, et al., 1999). Because the β-catenin antibody does not work in whole mount immunolabeling, we cannot address the potential domain expansion of β-catenin protein in the cytoplasmic or the nuclear β-catenin population.
We also examined the spatial localization of known Wnt/β-catenin downstream targets, including Krl, Blimp1b, and FoxA (Fig. 7). Krl is a transcriptional repressor containing a Zn-finger DNA-binding domain that was shown to be important for β-catenin-mediated endoderm specification (Howard et al., 2001; Peter and Davidson, 2010, 2011) . In S. purpuratus, knockdown of Krl resulted in the lack of endoderm (Howard et al., 2001). The expression of Krl mRNA (Fig. 7C,D) is similar to the expression of nuclear β-catenin (Fig. 7A) (Logan et al., 1999). It is initially restricted to the micromere descendants at the 32-cell stage, followed by shifting to the Veg2 region upon hatching (Howard et al., 2001; Peter and Davidson, 2011). However, unlike the nuclear β-catenin, Krl clears from the small micromeres by early blastula stage (Livi and Davidson, 2006). We did not observe noticeable difference in spatial localization of Krl.
Blimp1b is a member of the SET domain family of proteins (which are directly involved in histone modifications) that was shown to promote specification of endodermal cells from the Veg1 tier (Livi and Davidson, 2006). Blimp1b MASO resulted in blastula embryos lacking Veg1 descendants and gastrula embryos with short truncated archenteron or gut invagination defects (Livi and Davidson, 2006). Similar to the nuclear β-catenin and Krl, the expression of Blimp1b mRNA begins in large micromere descendants at 10 hpf, then shifts to Veg2 tier of cells by 18 hpf (Fig.7E) (Livi and Davidson, 2006). By 24 hpf, Krl is expressed in the Veg2 endoderm (Peter and Davidson, 2010, 2011), as well as in the Veg1 tier (Livi and Davidson, 2006). However, unlike the nuclear β-catenin and Krl, Blimp1b mRNA was never observed to be expressed in small micromeres (Livi and Davidson, 2006). We did not detect noticeable difference in the expression domain of Blimp1b mRNA upon miRNA TP treatment (Fig. 7F).
FoxA is an evolutionary conserved forkhead transcription factor ortholog that specifies endoderm in many bilaterians and cnidarians (Boyle and Seaver, 2008; Burtscher and Lickert, 2009; Friedman and Kaestner, 2006; Hiruta et al., 2005; Oliveri et al., 2006). Knockdown of FoxA resulted in severe defects in archenteron formation with truncated gut, missing foregut or entire gut, and a lack of stomadaeum (Oliveri et al., 2006). Even though FoxA is a direct downstream target of β-catenin, its expression pattern does not completely mimic the pattern of β-catenin nuclearization (Fig.7A,G). At 18 hpf FoxA mRNA was reported to be restricted to Veg2 endomesoderm followed by its expression in Veg2-derived endodermal ring by 21 hpf. The oral side of the endodermal ring was shown to express more FoxA compared to the aboral side (Oliveri et al., 2006). FoxA is initially expressed in Veg2 tier of cells at 15 hpf and is restricted to Veg2U that gives rise exclusively to the endoderm (Oliveri et al., 2006). We did not observe changes in FoxA mRNA spatial distribution in β-catenin miRNA TP-treated embryos compared to the control (Fig. 7H).
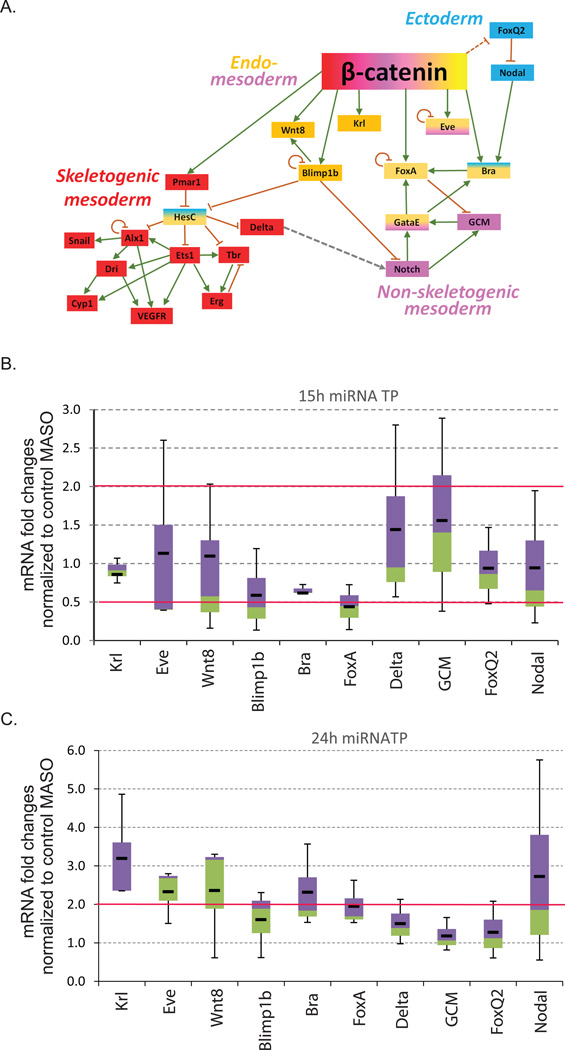
Using qPCR, we assayed for transcript level changes in Wnt targeted transcription factors and signaling molecules involved in cell fate specification (Fig. 8). Transcript levels in the miRNA TP-treated embryos were normalized to the control MASO-treated embryos. Results indicated that transcript levels of endodermal regulators, Krl, Eve, Wnt8, Bra and, FoxA and ectodermal regulator, Nodal, were increased on average of at least 2-fold in embryos treated with β-catenin miRNA TPs as compared to embryos treated with the control MASO in mesenchyme blastulae, suggesting that post-transcriptional regulation by miRNAs on β-catenin may contribute to endodermal cell specification and differentiation (Fig. 8C). Consistent with the in situ data, Krl mRNA had the highest transcript increase upon β-catenin miRNA TP treatment (Figs. 7D and 8C).
Fig. 8. Removal of miRNA regulation of β-catenin results in the transcript changes of Wnt responsive genes.
(A) Simplified gene regulatory network activated by Wnt/β-catenin signaling pathway. Skeletogenesis (red) is indirectly regulated by β-catenin through a single transcriptional repressor Pmar1 (Oliveri et al., 2003). Endoderm (orange) and mesoderm (purple) formation is regulated by multiple direct targets of β-catenin such as Krl (Howard et al., 2001; Minokawa et al., 2004; Peter and Davidson, 2011), FoxA (Oliveri et al., 2006), Blimp1b (Livi and Davidson, 2006; Smith et al., 2007), Eve (Peter and Davidson, 2010, 2011), Wnt8 (Wikramanayake et al., 2004) and Bra (Gross and McClay, 2001). In addition, β-catenin indirectly regulates ectoderm differentiation through indirect regulation of FoxQ2 (Angerer et al., 2011; Range et al., 2013; Yaguchi et al., 2008).
(B) qPCR was used to measure the transcript levels of genes involved in the specification of endoderm, mesoderm, and ectoderm in control and β-catenin miRNA TP-injected embryos at the early blastula stage (15 hpf). (C) qPCR was used to measure the transcript levels of genes involved in the specification of endoderm, mesoderm, and ectoderm in control and β-catenin miRNA TP-injected embryos at the mesenchyme blastula stage (24 hpf). Most of the endomesodermal regulatory genes have ≥2-fold increase in transcript levels in miRNA TP treated embryos in comparison to the control embryos at mesenchyme blastula stage but not at early blastula stage (3–5 biological replicates). The box plot represents the third quartile (purple) and the first quartile (green). Median is the junction between the first and the third quartile. The whiskers represent the maximum and the minimum of the data. The average of the data is shown by the black line. Red line indicates the 2-fold changes.
Discussion
Our results for the first time demonstrated that miRNAs modulate β-catenin in the canonical Wnt signaling pathway in the early sea urchin embryo. We identified at least three miRNA regulatory sites in the sea urchin β-catenin 3’UTR, as supported by the luciferase reporter data (Fig. 2) and by our observation that blocking miRNA regulation of β-catenin resulted in an increase of the β-catenin mRNA and protein levels (Fig. 3). Importantly, this miRNA TP-induced increase in β-catenin protein is sufficient to cause transcriptional changes in downstream Wnt responsive endodermal and mesodermal regulatory genes (Fig. 8), potentially impacting endodermal specification/differentiation, gut development, and circumesophageal muscle structure in vivo (Figs. 5,6).
Across species from fruit fly to zebrafish to human, miRNAs have been found to regulate various components of the Wnt/β-catenin pathway (Hashimi et al., 2009; Kennell et al., 2008; Saydam et al., 2009; Silver et al., 2007; Thatcher et al., 2008; Wang and Xu, 2010; Xia et al., 2010). Recently 38 miRNAs were identified to regulate the canonical Wnt pathway in human HEK293 cells, including miR-25 which inhibits human β-catenin by targeting its coding sequence (Anton et al., 2011). Interestingly, miRDeep2-30364-35240 that we found to regulate sea urchin β-catenin has the same seed sequence as miR-25, suggesting that miRDeep2-30364-35240 may play a potential conserved role as the mammalian miR-25 in regulating β-catenin. We also found β-catenin to be post-transcriptionally regulated by miR-2007 in the early embryo. miR-2007 is conserved within Eleutherozoa that is shared by Stronylocentrotus (Echinoidea) and Henricia (Asteroidea) (Wheeler et al., 2009).
Of note is that the mammalian β-catenin gene is regulated by miR-200a (Saydam et al., 2009; Xia et al., 2010). Even though a single miR-200 exists in the sea urchin, we did not bioinformatically identify any miR-200 seed sequences within the sea urchin β-catenin 3’UTR. Interestingly, the miR-200* does have one predicted binding site within the β-catenin 3’UTR (Table S2). It is likely that using seed sequence as the only criterion for bioinformatic identification of miRNA binding sites is insufficient in predicting all the functional miRNA regulatory sites. Additional miRNAs may also regulate the β-catenin 3’UTR, even though they are not highly expressed in the embryo (Table S2) (Song et al., 2012). miRNAs with conserved seed sequences may share conserved gene targets, as in the case of sea urchin miRDeep2-30364-35240 and he mammalian miR-25 in mediating post-transcriptional regulation of the β-catenin gene.
Our results indicate that the removal of miRNA regulation of β-catenin by using miRNA TP MASO is able to induce an increase in β-catenin protein level by the 32-cell stage (6 hpf) and in both mRNA and protein levels by mesenchyme blastula stage (Fig. 3). The change in β-catenin protein level upon miRNA TP treatment at the mesenchyme blastula stage (24 hpf) coincides with increased levels of miRDeep2-30364-35240 and miR-2007 sequence reads reported from our previous study (Song et al., 2012)(Fig. 1B). Removing miRNA regulation of β-catenin at the early blastula stage (15 hpf) did not result in changes in the transcript levels of endomesodermal regulators (Fig. 8B), consistent with the data that these miRNAs have relatively low sequence reads prior to the blastula stage (Fig 1B). miRNA regulation of β-catenin is also supported by the Dicer knockdown experiments where the mRNA level of β-catenin was significantly increased at the mesenchyme blastula stage (24 hpf) but not by early blastula stage (15 hpf) (Fig. 1A). One caveat is that in the complex background of Dicer knockdown, the change in β-catenin mRNA would be a result of global depletion of all miRNAs and the effect of their target gene expression changes.
β-catenin is highly regulated at the transcriptional, post-transcriptional, translational, and post-translational levels (Huang et al., 2010; Zhao et al., 2011). We propose that the type of regulatory mechanism that controls β-catenin is dependent on the developmental stage of the embryo. The most well understood regulatory mechanism of β-catenin is mediated by Wnt ligand binding that leads to the disassembly of the destruction complex and inhibition of the degradation of β-catenin (Logan and Nusse, 2004). The binding interactions of β-catenin is also regulated by the transforming growth factor β (TGFβ), epidermal growth factor receptor (EGFR), hepatocyte growth factor, erythroblastic leukemia viral oncogene-2 and Janus Kinase 2 (JAK2)-mediated signaling pathways (Blume-Jensen et al., 1998; Gonzalez and Medici, 2014; Incassati et al., 2010; Liu et al., 2010; Paul et al., 2013; Zeng et al., 2006). Early in development, β-catenin may be predominantly regulated by activated Disheveled that disrupts the targeted degradation of β-catenin in the vegetal blastomeres (Peng and Wikramanayake, 2013), while miRNAs may facilitate the clearing of β-catenin later at and after the blastula stage.
β-catenin may be crucial for activating signals that pattern ectoderm (Wikramanayake et al., 1998). Previous studies demonstrate that an overexpression of β-catenin in the animal halves of the sea urchin embryos (through microinjection of 0.01–0.02 pg of β-catenin mRNA into the newly fertilized eggs followed by isolation of animal halves) could induce ectoderm patterning along the dorsal/ventral (oral-aboral) axis, as supported by morphological and molecular alterations (Wikramanayake et al., 1998). Ectoderm patterning is mediated by FoxQ2 and Nodal which are important for the formation of the anterior neuroectoderm territory and the primary (anterior-posterior) and secondary (dorsal-ventral) axes (Angerer et al., 2011; Yaguchi et al., 2008). Recently it was shown that the nuclear β-catenin dependent signal indirectly restricts FoxQ2 expression to the animal plate, allowing Nodal signaling to increase through autoregulation to initiate dorsal-ventral patterning (Angerer et al., 2011; Range et al., 2013; Yaguchi et al., 2008). To examine if ectodermal specification regulators were affected by β-catenin miRNA TP treatment, we tested the transcript levels of FoxQ2 and Nodal. The transcript of FoxQ2 was not altered but the Nodal transcript is increased in the presence of increased β-catenin by mesenchyme blastula stage (Fig. 8C). Thus, Nodal is likely to be regulated by the Wnt/β-catenin pathway, independent from FoxQ2. The exact molecular mechanism of how this regulation occurs still needs to be explored.
The Wnt/β-catenin signaling is important for the skeletogenesis of the sea urchin embryos (Fig. 8A). PMCs arise from the large micromere lineage in response to the canonical Wnt signaling that leads to the activation of the transcriptional repressor Pmar1 (paired class homeodomain repressor) in the large micromeres (Oliveri et al., 2003). Activation of Pmar1 results in transcriptional inhibition of an ubiquitous transcriptional repressor HesC (Revilla-I-Domingo et al., 2007), leading to the activation of skeletogenic transcription factors which regulate various differentiation effector genes that are essential for PMC patterning and spicule formation (Damle and Davidson, 2011; Koga et al., 2010; Kurokawa et al., 1999; Sharma and Ettensohn, 2010, 2011; Wahl et al., 2009). Our data indicate that removal of miRNA regulation of β-catenin did not affect skeletogenesis of the sea urchin embryos (Fig. 4). In contrast to the specification of endomesoderm, which is regulated by multiple genes that are directly activated by the Wnt/β-catenin pathway, PMCs are specified indirectly through a single β-catenin-responsive gene Pmar1 (Fig.8A). Thus, a relatively small accumulation of β-catenin protein observed in the miRNA TP-treated embryos is not sufficient to alter PMC development.
We then examined whether endodermal structures were affected by the removal of β-catenin regulation by miRNAs. The Wnt/β-catenin signaling pathway plays a central role in endomesoderm formation and it was shown that overexpression of β-catenin (0.05 to 0.1 pg RNA) in the sea urchin embryos led to strong vegetalization of an embryo, including formation of excessive endoderm and mesoderm (Wikramanayake et al., 1998). However, the effect of low level of β-catenin overexpression on gut development in whole embryos is not known. Our data indicate that β-catenin miRNA TP treatment led to a significantly narrower gut (Fig. 5A,B vs. 5C,D), reduced alkaline phosphatase staining (Fig. 5H,I), and at least two fold increase in the levels of endodermal regulators Krl, Eve, Wnt8 and Bra mRNAs in the vegetal pole (Fig.7 and Fig.8C). A relatively minor increase in β-catenin protein level resulted in 2-fold increase in the expression of endodermal regulators (Figs. 3 and 8). This result can potentially be explained by restricted spatial expression of β-catenin regulatory miRNAs in a fraction of cells in the embryo. Since we measured the total levels of β-catenin protein, whereas only changes in the nuclear β-catenin would affect the expression of its downstream targets, the actual effect on β-catenin in endodermal cells may be larger than the average effect measured for the entire embryo. Although we do not understand how increased levels of these essential endodermal transcription factors may result in gut developmental defects, we speculate that a combination of downstream targets of these transcription factors that are critical for proper endodermal specification/differentiation and gut development result in the β-catenin miRNA TP-induced phenotypes.
We also tested the expression of mesodermal regulatory genes, Delta and GCM, in β-catenin miRNA TP-treated embryos (Fig. 8). The Delta/Notch signaling pathway is known to be important for mesodermal specification which targets GCM at the top of the early mesoderm gene regulatory hierarchy (Croce and McClay, 2010; Ransick and Davidson, 2006; Sherwood and McClay, 1999; Sweet et al., 2002; Sweet et al., 1999). Our data indicate that both Delta and Gcm mRNAs are not altered by β-catenin miRNA TP treatment. However, we observed a noticeable and significant difference in the structure of the circumesophageal musculature (Fig. 4). Recently a set of genes were identified in the sea urchin myoblasts that appear in the late gastrula stage, including MyoD2, FoxF, FoxC, FoxL1, Myocardin, Twist, and Tbx6 (Andrikou et al., 2013). It is not clear if any of these molecules are directly regulated by the canonical Wnt signaling pathway. In vertebrates, the Wnt pathway regulates myogenic regulatory factors such as MyoD and Myf5 (Borello et al., 2006; Tajbakhsh et al., 1998; von Maltzahn et al., 2012). Thus, the canonical Wnt pathway may regulate the structure of mesodermally-derived circumesophageal musculature (Figs. 5 and 8C).
Conclusions
In summary, our results identified specific miRNAs that regulate β-catenin and demonstrated that blocking endogenous miRNA regulation of β-catenin resulted in increased transcript levels of Wnt responsive endodermal regulatory genes in the sea urchin, revealing potential molecular mechanism to β-catenin miRNA TP induced phenotypes. This study demonstrates that miRNA regulation of β-catenin at the post-transcriptional level in the developing embryo has an important impact on the development of the larval gut and the structure of circumesophageal musculature.
Supplementary Material
Reporter constructs were used to test the binding of miRNA TPs to β-catenin 3’UTR. eGFP reporter constructs were fused to the β-catenin 3’UTR containing wild type miRNA binding sites and mCherry constructs were fused to the Xenopus β-globin 3’UTR. In vitro transcribed mRNAs of eGFP and mCherry reporter constructs were coinjected with either control MASO or β-catenin miRNA TPs at 1000X (710 µM) molar excess of the estimated eGFP-β-catenin 3’UTR reporter construct. The early blastula (17 hpf) eGFP signals were normalized to the mCherry signals which serves as an injection control. (A) miRNA TPs result in blocking miRNA binding to the 3’UTR of β-catenin as supported by the significant increase in reporter construct expression in the presence of miRNA TPs (710 µM stock) compared to the control morpholino *Student T-Test, p < 0.0001. (B) Mesenchyme blastulae treated with miRNA TPs are morphologically normal compared to the control MASO-injected embryos. Arrows indicate uninjected embryos.
30 µg of 32-cell stage embryonic extracts were loaded onto the SDS-PAGE gel. Tenfold molecular excess of the β-catenin peptides was used as the antigen for the affinity-purified anti-β-catenin polyclonal antibodies, β-catenin 1 (β-1) and β-catenin55 (β-55). β-catenin bands were observed in the control lanes with β-catenin1 and β-catenin55 terminal peptides but not in preabsorbed antibodies. β-catenin (red); tubulin (green).
Highlights.
β-catenin is regulated by microRNAs.
Blocking miRNA regulation of β-catenin leads to defects in gut and esophageal muscle structures.
Blocking miRNA regulation of β-catenin leads to increased transcript levels of endodermal regulatory genes.
Acknowledgements
We thank the two anonymous reviewers for their comments. We thank Athula Wikramanayake for helpful discussions, reading of the manuscript, and the β-catenin antibodies. We thank David McClay for his 1D5 antibody against the PMCs. We thank Gary Wessel for his Endo1 and MHC antibodies. We also thank Dr. John McDonald for his consultation in statistics. We thank Santiago Suarez in cloning the β-catenin in situ probe. This work is funded by the University of Delaware Research Fund, the NIH NIGMS 5P20GM103653-02, and NIH NIGMS 8P20GM103446-12.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle H, Schwartz H, Kemler R. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61:514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- Andrikou C, Iovene E, Rizzo F, Oliveri P, Arnone MI. Myogenesis in the sea urchin embryo: the molecular fingerprint of the myoblast precursors. Evodevo. 2013;4:33. doi: 10.1186/2041-9139-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer LM, Yaguchi S, Angerer RC, Burke RD. The evolution of nervous system patterning: insights from sea urchin development. Development. 2011;138:3613–3623. doi: 10.1242/dev.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata R, Perillo M, Andrikou C, Cole AG, Martinez P, Arnone MI. Pattern and process during sea urchin gut morphogenesis: The regulatory landscape. Genesis. 2013 doi: 10.1002/dvg.22738. [DOI] [PubMed] [Google Scholar]
- Anton R, Chatterjee SS, Simundza J, Cowin P, DasGupta R. A Systematic Screen for Micro-RNAs Regulating the Canonical Wnt Pathway. Plos One. 2011;6 doi: 10.1371/journal.pone.0026257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Mena C, Cameron RA, Davidson EH. Hindgut specification and cell-adhesion functions of Sphox11/13b in the endoderm of the sea urchin embryo. Dev Growth Differ. 2006;48:463–472. doi: 10.1111/j.1440-169X.2006.00883.x. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Janknecht R, Hunter T. The kit receptor promotes cell survival via activation of PI 3-kinase and subsequent Akt-mediated phosphorylation of Bad on Ser136. Curr Biol. 1998;8:779–782. doi: 10.1016/s0960-9822(98)70302-1. [DOI] [PubMed] [Google Scholar]
- Borello U, Berarducci B, Murphy P, Bajard L, Buffa V, Piccolo S, Buckingham M, Cossu G. The Wnt/beta-catenin pathway regulates Gli-mediated Myf5 expression during somitogenesis. Development. 2006;133:3723–3732. doi: 10.1242/dev.02517. [DOI] [PubMed] [Google Scholar]
- Boyle MJ, Seaver EC. Developmental expression of foxA and gata genes during gut formation in the polychaete annelid, Capitella sp. I. Evol Dev. 2008;10:89–105. doi: 10.1111/j.1525-142X.2007.00216.x. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- Burke R. Structure of the digestive tract of the pluteus larva of Dendraster excentricus (Echinodermata: Echinoida) Zoomorphology. 1981;98:209–225. [Google Scholar]
- Burke RD, Alvarez CM. Development of the esophageal muscles in embryos of the sea urchin Strongylocentrotus purpuratus. Cell Tissue Res. 1988;252:411–417. doi: 10.1007/BF00214384. [DOI] [PubMed] [Google Scholar]
- Burtscher I, Lickert H. Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development. 2009;136:1029–1038. doi: 10.1242/dev.028415. [DOI] [PubMed] [Google Scholar]
- Cheers MS, Ettensohn CA. Rapid microinjection of fertilized eggs. Methods Cell Biol. 2004;74:287–310. doi: 10.1016/s0091-679x(04)74013-3. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Croce JC, McClay DR. Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development. 2010;137:83–91. doi: 10.1242/dev.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle S, Davidson EH. Precise cis-regulatory control of spatial and temporal expression of the alx-1 gene in the skeletogenic lineage of s. purpuratus. Dev Biol. 2011;357:505–517. doi: 10.1016/j.ydbio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de-Leon SB, Davidson EH. Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc Natl Acad Sci U S A. 2010;107:10103–10108. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drawbridge J. The color purple: analyzing alkaline phosphatase expression in experimentally manipulated sea urchin embryos in an undergraduate developmental biology course. Int J Dev Biol. 2003;47:161–164. [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Saudemont A, Bessodes N, Mekpoh F, Haillot E, Quirin M, Lepage T. Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development. 2010;137:223–235. doi: 10.1242/dev.042531. [DOI] [PubMed] [Google Scholar]
- Emily-Fenouil F, Ghiglione C, Lhomond G, Lepage T, Gache C. GSK3beta/shaggy mediates patterning along the animal-vegetal axis of the sea urchin embryo. Development. 1998;125:2489–2498. doi: 10.1242/dev.125.13.2489. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Wessel GM, Wray GA. Methods in Cell Biology. In: Ettensohn CA, Wessel GM, Wray GA, editors. Development of Sea Urchins, Ascidians, and other Invertebrate Deuterostomes: Experimental Approaches. San Diego: Elsevier Academic Press; 2004. pp. 1–13. [Google Scholar]
- Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7 doi: 10.1126/scisignal.2005189. re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Gross JM, McClay DR. The role of Brachyury (T) during gastrulation movements in the sea urchin Lytechinus variegatus. Dev Biol. 2001;239:132–147. doi: 10.1006/dbio.2001.0426. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Wessel GM. Exogenous RNA is selectively retained in the small micromeres during sea urchin embryogenesis. Mol Reprod Dev. 2010;77:836. doi: 10.1002/mrd.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood. 2009;114:404–414. doi: 10.1182/blood-2008-09-179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Degnan BM. Retinoic acid disrupts anterior ectodermal and endodermal development in ascidian larvae and postlarvae. Dev Genes Evol. 1998;208:336–345. doi: 10.1007/s004270050189. [DOI] [PubMed] [Google Scholar]
- Hiruta J, Mazet F, Yasui K, Zhang P, Ogasawara M. Comparative expression analysis of transcription factor genes in the endostyle of invertebrate chordates. Dev Dyn. 2005;233:1031–1037. doi: 10.1002/dvdy.20401. [DOI] [PubMed] [Google Scholar]
- Howard EW, Newman LA, Oleksyn DW, Angerer RC, Angerer LM. SpKrl: a direct target of beta-catenin regulation required for endoderm differentiation in sea urchin embryos. Development. 2001;128:365–375. doi: 10.1242/dev.128.3.365. [DOI] [PubMed] [Google Scholar]
- Huang K, Zhang J-X, Han L, You Y-P, Jiang T, Pu P-Y, Kang C-S. MicroRNA roles in beta-catenin pathway. Molecular Cancer. 2010;9 doi: 10.1186/1476-4598-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Takada N, Satoh N, Satou Y. (beta)-catenin mediates the specification of endoderm cells in ascidian embryos. Development. 2000;127:3009–3020. doi: 10.1242/dev.127.14.3009. [DOI] [PubMed] [Google Scholar]
- Incassati A, Chandramouli A, Eelkema R, Cowin P. Key signaling nodes in mammary gland development and cancer: beta-catenin. Breast Cancer Res. 2010;12:213. doi: 10.1186/bcr2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci U S A. 2008;105:15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Kim M, Jho EH. Wnt/beta-catenin signalling: from plasma membrane to nucleus. Biochem J. 2013;450:9–21. doi: 10.1042/BJ20121284. [DOI] [PubMed] [Google Scholar]
- Koga H, Matsubara M, Fujitani H, Miyamoto N, Komatsu M, Kiyomoto M, Akasaka K, Wada H. Functional evolution of Ets in echinoderms with focus on the evolution of echinoderm larval skeletons. Development Genes and Evolution. 2010;220:107–115. doi: 10.1007/s00427-010-0333-5. [DOI] [PubMed] [Google Scholar]
- Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano G, Nishida H. Maternal and zygotic expression of the endoderm-specific alkaline phosphatase gene in embryos of the ascidian, Halocynthia roretzi. Dev Biol. 1998;198:245–252. [PubMed] [Google Scholar]
- Kurokawa D, Kitajima T, Mitsunaga-Nakatsubo K, Amemiya S, Shimada H, Akasaka K. HpEts, an ets-related transcription factor implicated in primary mesenchyme cell differentiation in the sea urchin embryo. Mech Dev. 1999;80:41–52. doi: 10.1016/s0925-4773(98)00192-0. [DOI] [PubMed] [Google Scholar]
- Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, Kimelman D, Moon RT. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Liu YC, Lai WC, Chuang KA, Shen YJ, Hu WS, Ho CH, Chen YB, Hsu MF, Hsu HC, Lieu CH. Blockade of JAK2 activity suppressed accumulation of beta-catenin in leukemic cells. J Cell Biochem. 2010;111:402–411. doi: 10.1002/jcb.22714. [DOI] [PubMed] [Google Scholar]
- Livi CB, Davidson EH. Expression and function of blimp1/krox, an alternatively transcribed regulatory gene of the sea urchin endomesoderm network. Dev Biol. 2006;293:513–525. doi: 10.1016/j.ydbio.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Logan CY, McClay DR. The allocation of early blastomeres to the ectoderm and endoderm is variable in the sea urchin embryo. Development. 1997;124:2213–2223. doi: 10.1242/dev.124.11.2213. [DOI] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Davidson EH. A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev Biol. 2012;364:77–87. doi: 10.1016/j.ydbio.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Oliveri P. A protocol for unraveling gene regulatory networks. Nat Protoc. 2008;3:1876–1887. doi: 10.1038/nprot.2008.187. [DOI] [PubMed] [Google Scholar]
- McClay DR, Cannon GW, Wessel GM, Fink RD, Marchase RB. Patterns of Antigenic Expression in Early Sea Urchin Development. In: Raff WRJaRA., editor. Time, Space, and Pattern in Embryonic Development. New York: A.R. Liss; 1983. pp. 157–169. [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Moon RT. Wnt/beta-catenin pathway. Sci STKE 2005, cm1. 2005 doi: 10.1126/stke.2712005cm1. [DOI] [PubMed] [Google Scholar]
- Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nature reviews. Genetics. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas FE. Experimental validation of microRNA targets using a luciferase reporter system. Methods Mol Biol. 2011;732:139–152. doi: 10.1007/978-1-61779-083-6_11. [DOI] [PubMed] [Google Scholar]
- Nishida H, Kumano G. Analysis of the temporal expression of endoderm-specific alkaline phosphatase during development of the ascidian Halocynthia roretzi. Dev Growth Differ. 1997;39:199–205. doi: 10.1046/j.1440-169x.1997.t01-1-00008.x. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Developmental Biology. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci. 1999;112(Pt 8):1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- Paul I, Bhattacharya S, Chatterjee A, Ghosh MK. Current Understanding on EGFR and Wnt/beta-Catenin Signaling in Glioma and Their Possible Crosstalk. Genes Cancer. 2013;4:427–446. doi: 10.1177/1947601913503341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi A, Careccia S, Lulli V, Romania P, Marziali G, Testa U, Lavorgna S, Lo-Coco F, Petti MC, Calabretta B, Levrero M, Piaggio G, Rizzo MG. miRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene. 2013;32:3648–3654. doi: 10.1038/onc.2012.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CF, Wikramanayake AH. Differential regulation of Disheveled in a novel vegetal cortical domain in sea urchin eggs and embryos: Implications for the localized activation of canonical Wnt signaling. PLoS One in press. 2013 doi: 10.1371/journal.pone.0080693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2010;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. A gene regulatory network controlling the embryonic specification of endoderm. Nature. 2011;474:635–639. doi: 10.1038/nature10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056–1068. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Range RC, Angerer RC, Angerer LM. Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior-posterior axis of sea urchin embryos. PLoS Biol. 2013;11:e1001467. doi: 10.1371/journal.pbio.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. Late specification of Veg1 lineages to endodermal fate in the sea urchin embryo. Dev Biol. 1998;195:38–48. doi: 10.1006/dbio.1997.8814. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Revilla-I-Domingo R, Oliver P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeser T, Stein S, Kessel M. Nuclear beta-catenin and the development of bilateral symmetry in normal and LiCl-exposed chick embryos. Development. 1999;126:2955–2965. doi: 10.1242/dev.126.13.2955. [DOI] [PubMed] [Google Scholar]
- Rowning BA, Wells J, Wu M, Gerhart JC, Moon RT, Larabell CA. Microtubule-mediated transport of organelles and localization of beta-catenin to the future dorsal side of Xenopus eggs. Proc Natl Acad Sci U S A. 1997;94:1224–1229. doi: 10.1073/pnas.94.4.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Samanta MP, Tongprasit W, Istrail S, Cameron RA, Tu Q, Davidson EH, Stolc V. Report -The transcriptome of the sea urchin embryo. Science. 2006;314:960–962. doi: 10.1126/science.1131898. [DOI] [PubMed] [Google Scholar]
- Saurat N, Andersson T, Vasistha NA, Molnar Z, Livesey FJ. Dicer is required for neural stem cell multipotency and lineage progression during cerebral cortex development. Neural Dev. 2013;8:14. doi: 10.1186/1749-8104-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saydam O, Shen Y, Wurdinger T, Senol O, Boke E, James MF, Tannous BA, Stemmer-Rachamimov AO, Yi M, Stephens RM, Fraefel C, Gusella JF, Krichevsky AM, Breakefield XO. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol. 2009;29:5923–5940. doi: 10.1128/MCB.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Steinbeisser H, Warga RM, Hausen P. Beta-catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhaeusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sharma T, Ettensohn CA. Activation of the skeletogenic gene regulatory network in the early sea urchin embryo. Development. 2010;137:1149–1157. doi: 10.1242/dev.048652. [DOI] [PubMed] [Google Scholar]
- Sharma T, Ettensohn CA. Regulative deployment of the skeletogenic gene regulatory network during sea urchin development. Development. 2011;138:2581–2590. doi: 10.1242/dev.065193. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, McClay DR. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126:1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Hagen JW, Okamura K, Perrimon N, Lai EC. Functional screening identifies miR-315 as a potent activator of Wingless signaling. Proc Natl Acad Sci U S A. 2007;104:18151–18156. doi: 10.1073/pnas.0706673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev Biol. 2008;313:863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, Coffman JA, Dean M, Elphick MR, Ettensohn CA, Foltz KR, Hamdoun A, Hynes RO, Klein WH, Marzluff W, McClay DR, Morris RL, Mushegian A, Rast JP, Smith LC, Thorndyke MC, Vacquier VD, Wessel GM, Wray G, Zhang L, Elsik CG, Ermolaeva O, Hlavina W, Hofmann G, Kitts P, Landrum MJ, Mackey AJ, Maglott D, Panopoulou G, Poustka AJ, Pruitt K, Sapojnikov V, Song X, Souvorov A, Solovyev V, Wei Z, Whittaker CA, Worley K, Durbin KJ, Shen Y, Fedrigo O, Garfield D, Haygood R, Primus A, Satija R, Severson T, Gonzalez-Garay ML, Jackson AR, Milosavljevic A, Tong M, Killian CE, Livingston BT, Wilt FH, Adams N, Belle R, Carbonneau S, Cheung R, Cormier P, Cosson B, Croce J, Fernandez-Guerra A, Geneviere A-M, Goel M, Kelkar H, Morales J, Mulner-Lorillon O, Robertson AJ, Goldstone JV, Cole B, Epel D, Gold B, Hahn ME, Howard-Ashby M, Scally M, Stegeman JJ, Allgood EL, Cool J, Judkins KM, McCafferty SS, Musante AM, Obar RA, Rawson AP, Rossetti BJ, Gibbons IR, Hoffman MP, Leone A, Istrail S, Materna SC, Samanta MP, Stolc V, Tongprasit W, Tu Q, Bergeron K-F, Brandhorst BP, Whittle J, Berney K, Bottjer DJ, Calestani C, Peterson K, Chow E, Yuan QA, Elhaik E, Graur D, Reese JT, Bosdet I, Heesun S, Marra MA, Schein J, Anderson MK, Brockton V, Buckley KM, Cohen AH, Fugmann SD, Hibino T, Loza-Coll M, Majeske AJ, Messier C, Nair SV, Pancer Z, Terwilliger DP, Agca C, Arboleda E, Chen N, Churcher AM, Hallbook F, Humphrey GW, Idris MM, Kiyama T, Liang S, Mellott D, Mu X, Murray G, Olinski RP, Raible F, Rowe M, Taylor JS, Tessmar-Raible K, Wang D, Wilson KH, Yaguchi S, Gaasterland T, Galindo BE, Gunaratne HJ, Juliano C, Kinukawa M, Moy GW, Neill AT, Nomura M, Raisch M, Reade A, Roux MM, Song JL, Su Y-H, Townley IK, Voronina E, Wong JL, Amore G, Branno M, Brown ER, Cavalieri V, Duboc V, Duloquin L, Flytzanis C, Gache C, Lapraz F, Lepage T, Locascio A, Martinez P, Matassi G, Matranga V, Range R, Rizzo F, Rottinger E, Beane W, Bradham C, Byrum C, Glenn T, Hussain S, Manning G, Miranda E, Thomason R, Walton K, Wikramanayke A, Wu S-Y, Xu R, Brown CT, Chen L, Gray RF, Lee PY, Nam J, Oliveri P, Smith J, Muzny D, Bell S, Chacko J, Cree A, Curry S, Davis C, Dinh H, Dugan-Rocha S, Fowler J, Gill R, Hamilton C, Hernandez J, Hines S, Hume J, Jackson L, Jolivet A, Kovar C, Lee S, Lewis L, Miner G, Morgan M, Nazareth LV, Okwuonu G, Parker D, Pu L-L, Thorn R, Wright R Sea Urchin Genome Sequencing C. The genome of the sea urchin Strongylocentrotus purpuratus. Science (New York, N.Y.) 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JL, Stoeckius M, Maaskola J, Friedlander M, Stepicheva N, Juliano C, Lebedeva S, Thompson W, Rajewsky N, Wessel GM. Select microRNAs are essential for early development in the sea urchin. Dev Biol. 2012;362:104–113. doi: 10.1016/j.ydbio.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton AA, Giraldez AJ. Use of target protector morpholinos to analyze the physiological roles of specific miRNA-mRNA pairs in vivo. Nat Protoc. 2011;6:2035–2049. doi: 10.1038/nprot.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepicheva NA, Song JL. High throughput microinjections of sea urchin zygotes. J Vis Exp. 2014 doi: 10.3791/50841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet HC, Gehring M, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- Sweet HC, Hodor PG, Ettensohn CA. The role of micromere signaling in Notch activation and mesoderm specification during sea urchin embryogenesis. Development. 1999;126:5255–5265. doi: 10.1242/dev.126.23.5255. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, Buckingham M, Cossu G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 1998;125:4155–4162. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci U S A. 2008;105:18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA. Wnt signaling in myogenesis. Trends Cell Biol. 2012;22:602–609. doi: 10.1016/j.tcb.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonica A, Weng W, Gumbiner BM, Venuti JM. TCF is the nuclear effector of the beta-catenin signal that patterns the sea urchin animal-vegetal axis. Dev Biol. 2000;217:230–243. doi: 10.1006/dbio.1999.9551. [DOI] [PubMed] [Google Scholar]
- Wahl ME, Hahn J, Gora K, Davidson EH, Oliveri P. The cis-regulatory system of the tbrain gene: Alternative use of multiple modules to promote skeletogenic expression in the sea urchin embryo. Dev Biol. 2009;335:428–441. doi: 10.1016/j.ydbio.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang H, Li M, Frid MG, Flockton AR, McKeon BA, Yeager ME, Fini MA, Morrell NW, Pullamsetti SS, Velegala S, Seeger W, McKinsey TA, Sucharov CC, Stenmark KR. MicroRNA-124 controls the proliferative, migratory, and inflammatory phenotype of pulmonary vascular fibroblasts. Circ Res. 2014;114:67–78. doi: 10.1161/CIRCRESAHA.114.301633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Xu Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun. 2010;402:186–189. doi: 10.1016/j.bbrc.2010.08.031. [DOI] [PubMed] [Google Scholar]
- Wessel GM, McClay DR. Sequential expression of germ-layer specific molecules in the sea urchin embryo. Dev Biol. 1985;111:451–463. doi: 10.1016/0012-1606(85)90497-x. [DOI] [PubMed] [Google Scholar]
- Wessel GM, Zhang W, Klein WH. Myosin heavy chain accumulates in dissimilar cell types of the macromere lineage in the sea urchin embryo. Dev Biol. 1990;140:447–454. doi: 10.1016/0012-1606(90)90093-x. [DOI] [PubMed] [Google Scholar]
- Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- Whittaker JR. Determination of Alkaline Phosphatase Expression in Endodermal Cell Lineages of an Ascidian Embryo. Biol Bulletin. 1990;178:222–230. doi: 10.2307/1541823. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Huang L, Klein WH. beta-Catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryo. Proc Natl Acad Sci U S A. 1998;95:9343–9348. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- Xia H, Ng SS, Jiang S, Cheung WK, Sze J, Bian XW, Kung HF, Lin MC. miR-200a–mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 2010;391:535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Angerer RC, Angerer LM. A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell. 2008;14:97–107. doi: 10.1016/j.devcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Zeng G, Germinaro M, Micsenyi A, Monga NK, Bell A, Sood A, Malhotra V, Sood N, Midda V, Monga DK, Kokkinakis DM, Monga SP. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8:279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J-X, Yue W-F, Zhu M-J, Du M. AMP-activated Protein Kinase Regulates beta-Catenin Transcription via Histone Deacetylase 5. Journal of Biological Chemistry. 2011;286 doi: 10.1074/jbc.M110.199372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reporter constructs were used to test the binding of miRNA TPs to β-catenin 3’UTR. eGFP reporter constructs were fused to the β-catenin 3’UTR containing wild type miRNA binding sites and mCherry constructs were fused to the Xenopus β-globin 3’UTR. In vitro transcribed mRNAs of eGFP and mCherry reporter constructs were coinjected with either control MASO or β-catenin miRNA TPs at 1000X (710 µM) molar excess of the estimated eGFP-β-catenin 3’UTR reporter construct. The early blastula (17 hpf) eGFP signals were normalized to the mCherry signals which serves as an injection control. (A) miRNA TPs result in blocking miRNA binding to the 3’UTR of β-catenin as supported by the significant increase in reporter construct expression in the presence of miRNA TPs (710 µM stock) compared to the control morpholino *Student T-Test, p < 0.0001. (B) Mesenchyme blastulae treated with miRNA TPs are morphologically normal compared to the control MASO-injected embryos. Arrows indicate uninjected embryos.
30 µg of 32-cell stage embryonic extracts were loaded onto the SDS-PAGE gel. Tenfold molecular excess of the β-catenin peptides was used as the antigen for the affinity-purified anti-β-catenin polyclonal antibodies, β-catenin 1 (β-1) and β-catenin55 (β-55). β-catenin bands were observed in the control lanes with β-catenin1 and β-catenin55 terminal peptides but not in preabsorbed antibodies. β-catenin (red); tubulin (green).