Abstract
Proteoglycans control numerous normal and pathological processes, among which are morphogenesis, tissue repair, inflammation, vascularization and cancer metastasis. During tumor development and growth, proteoglycan expression is markedly modified in the tumor microenvironment. Altered expression of proteoglycans on tumor and stromal cell membranes affects cancer cell signaling, growth and survival, cell adhesion, migration and angiogenesis. Despite the high complexity and heterogeneity of breast cancer, the rapid evolution in our knowledge that proteoglycans are among the key players in the breast tumor microenvironment suggests their potential as pharmacological targets in this type of cancer. It has been recently suggested that pharmacological treatment may target proteoglycan metabolism, their utilization as targets for immunotherapy or their direct use as therapeutic agents. The diversity inherent in the proteoglycans that will be presented herein provides the potential for multiple layers of regulation of breast tumor behavior. This review summarizes recent developments concerning the biology of selected proteoglycans in breast cancer, and presents potential targeted therapeutic approaches based on their novel key roles in breast cancer.
Keywords: proteoglycans, versican, decorin, biglycan, syndecans, glypicans, heparanase, serglycin, signaling, breast cancer
1. Extracellular matrices in breast cancer: focus on the proteoglycans
1.1. Breast cancer: a complex disease
Breast cancer is a heterogeneous, tissue-specific disease, with substantial genotypic and phenotypic diversity. This type of cancer prevails in women, although male breast cancer is also observed. Estrogen receptor-alpha (ERα), progesterone receptor (PgR), and epidermal growth factor receptor-2 (HER2) are the three mandatory prognostic and predictive factors in invasive breast cancer used in routine clinical practice today [1]. Four main breast cancer subtypes drive treatment decisions: ERα-positive and HER2-negative with a low or intermediate differentiation grade (luminal A); ERα-positive and HER2-negative with a high differentiation grade (luminal B); aggressive type of HER2-positive and triple-negative breast cancer (ERα-, PgR- and HER2-negative). Two thirds of breast cancers are ERα-positive. ERα plays an important role in the development, progression and treatment of breast cancer and is of special interest because its protein level is elevated in premalignant and malignant breast lesions, but not in normal tissue. Therefore, ERα is a valuable predictive and prognostic factor in the clinical management of breast cancer. However, the majority of hormonally responsive breast cancers develop resistance to anti-estrogen treatment and progress to a more aggressive and hormonally independent phenotype. Several preclinical and clinical studies conducted until todays are mainly focused on genetic components involved in tumor progression and tumor microenvironment as to better understand the biology of breast tumor cells and improve breast cancer treatment.
1.2. Proteoglycans: key molecular effectors of breast cancer cell surface and pericellular microenvironments
Interactions of cancer cells with the tumor microenvironment are important determinants of cancer progression toward metastasis. The tumor microenvironment contains many distinct cell types, including endothelial cells and their precursors, pericytes, smooth muscle cells, fibroblasts, cancer/tumor-associated fibroblasts (CAFs/TAFs), myofibroblasts, and inflammatory cells [2]. These cells are immersed in highly dynamic and functional extracellular matrices (ECMs) composed by macromolecules, such as proteoglycans (PGs), collagen, laminin, fibronectin and proteinases. PGs are major components of ECMs as well as the cell surfaces. They are composed of a specific core protein substituted with one or more covalently linked glycosaminoglycan (GAG) chains resulting in high degree of structural and functional complexity. GAGs (chondroitin sulfate, CS; dermatan sulfate, DS; heparan sulfate, HS; heparin, HP) are linear heteropolysaccharides composed of repeating disaccharides of hexosamines (N-acetyl-galactosamine or N-acetyl-glucosamine) and uronic acids (D-glucuronic acid or L-iduronic acid) that are being sulfated at various positions. Keratan sulfate (KS) is composed of repeating disaccharides containing N-acetyl-glucosamine and galactose [3]. Notably, hyaluronan (HA) is the only GAG that is not covalently bound to PG core protein and its synthesis is epigenetically regulated [4]. The number and the type of GAG chains, as well as the specific structure of each GAG chain may differ greatly even within a certain PG molecule [3, 5]. These variations in the overall PG structure may not only be cell- and tissue-specific, but also may depend on the differentiation stage and the action of various stimuli on the cells. PGs assembly and modification involves the action of multiple enzymes, such as glycosyltransferases, sulfotransferases, epimerases, sulfatases, glycosidases, and heparanase, revealing multiple layers of regulation as well as the structural diversity and functional heterogeneity of these macromolecules.
According to their localization, PGs are categorized as ECM-secreted, cell surface-associated and intracellular. Each main group is further classified into subfamilies according to their gene homology, core protein properties, molecular size and modular composition [6, 7]. Secreted PGs involve large aggregating PGs, named hyalectans (aggrecan, versican, brevican, neurocan), small leucine-rich PGs (SLRPs; decorin, biglycan, lumican) and basement membrane PGs (perlecan, agrin, collagen XVIII). Cell-surface-associated PGs are divided into two main subfamilies (transmembrane syndecans and glycosylphosphatidylinositol (GPI)-anchored glypicans), whereas serglycin is the only intracellular PG characterized to date. PGs can interact with most of the proteins present in ECMs with different affinities. Their GAG chains are mainly implicated in these interactions, although their core proteins are sometimes involved. Apart from their participation in the organization of ECM and regulation of its mechanical properties, PGs interact with growth factors, cytokines and chemokines. Binding of these molecules to PGs restricts their diffusion along the surface of receiving cells forming effective gradients of these components in the ECM, preventing them from loss to the extracellular space or aberrant signaling, and protects them from degradation [3]. Moreover, PGs can provide a signaling platform for signaling molecules and morphogens to interact with other important components, because PGs are able to bind to many cell surface co-receptors and secreted proteins/proteinases thereby modulating their activities. In this context, PGs can finely tune the activity of multiple matrix effectors by forming concentration gradients and specify distinct cell fates in a concentration-dependent manner [8, 9].
There is an abundance of evidence relating PG/GAG expression levels and fine structures to breast cancer growth, invasion, and metastasis. CS/DSPGs are involved in mammary gland development and may, consequently, be involved in breast cancer development [10]. DSPGs expression was described to be increased in breast cancer fibroadenoma compared to healthy tissue [11]. A common finding is that matrix secreted CS/DSPGs such as decorin and versican are deposited in tumor stroma [12, 13] and are related to aggressive phenotype in breast cancer [14–16]. Relapse in women with node-negative breast cancer is related to the level of versican deposited in peritumoral stroma [14, 17]. In contrast, low levels of decorin in invasive breast carcinomas are associated with poor outcome[15], whereas chondroitinase ABC treatment, an enzymatic procedure used to degrade CS/DS chains, in tumors triggers metastasis [18]. Furthermore, it was recently shown that decorin has antiangiogenic activities [19], while it evokes mitochondrial autophagy (mitophagy) in breast carcinoma cells [20]. Biglycan, another DS/CSPG, acts as an endogenous danger signal and potently induces pro-inflammatory mediators actively participating in inflammatory processes. By binding to cell surface receptors, biglycan triggers innate immunity, but can also activate signaling pathways that bias oncogene activity, cell cycle, migration or survival [21–23].
Cell surface-associated HSPGs have been described as tumor biomarkers being differentially regulated during tumorigenesis [3, 24, 25]. Recently, a direct relationship between growth factor-mediated signaling, ERs and ECM components has been shown. Breast cancer cells that express ERα can be directly stimulated via estrogen, or indirectly stimulated via epidermal growth factor receptor (EGFR) or insulin-like growth factor receptor (IGFR). Activation of these pathways is crucial for tumor establishment and development and lead to specific modulation of HSPGs, such as syndecan-2 and syndecan-4 and glypican-1, in addition to other ECM-modulating molecules [26–28]. Review of data from patient studies has shown that elevated levels of syndecan-1 are associated with aggressive phenotype [29], whereas upregulation of syndecan-2 in breast cancer promotes the acquisition of an invasive phenotype through regulation of the cytoskeleton and GTPases [30]. In addition, by degrading HS chains, the heparanase enzyme alters PG function leading to the enhancement of tumor growth, angiogenesis, and metastasis. Growth factor binding specificity leads to different responses according to cell status and the type of HS chain presented by the cells and for that function, a balance between cell surface and shed HSPGs, such as syndecan-1, is crucial [31, 32]. Syndecan-1 shed by tumor cells binds to growth factors released into the tumor microenvironment. This protects growth factors from proteolytic attack and the syndecan-1/growth factor complex binds to and activates high affinity growth factor receptors on endothelial and other host cells [31, 32].
Recently it has been shown that serglycin promotes breast cancer cell anchorage-independent growth, migration and invasion of breast cancer cells and these properties are dependent on the expression and secretion of glycanated serglycin bearing CS chains [33].
Despite the high complexity and heterogeneity of breast cancer, the rapid evolution in our knowledge that PGs are among the key players in the breast tumor microenvironment suggests their potential as pharmacological targets. The key roles of the most important proteoglycans related to breast cancer progression and/or treatment are given in more details in the chapters below.
2. Versican: a tumor stroma-associated proteoglycan in breast cancer
2.1. Structural features and molecular interactions
Versican is present in the interstitial space of many tissues. Its core protein consists of two globular domains G1 and G3 present at the N-terminus and C-terminus, respectively, and a central part that may carry variable number of GAG chains. The G1 domain mediates the binding of versican to HA resulting in the formation of large aggregates in ECM. The G3 domain contains two epidermal growth factor repeats, a lectin binding domain and a complement regulatory region. The central domain that carries GAG chains consists of two discrete regions named as GAG-α and GAG-β, which are encoded by giant exon 7 of 3 kb and exon 8 of 5.3 kb size, respectively, in the human gene [3, 34]. At least four splice variants of versican exist that arise from the alternative splicing of these two exons encoding GAG-attachment region. The larger splice variants V0 can carry 17–23 CS/DS chains, whereas smaller variants V1 and V2, 12–15 and 5–8 CS/DS, respectively. The smallest variant V3 doesn’t carry GAG chains and exists as single protein [3, 34]. Versican is a multi-functional molecule that can interact with various ligands through its core protein and GAG chains. For example G3 domain binds PSGL-1, integrin β1, tenascin, fibulin-1 and -2, fibrillin-1, EGFR and fibronectin whereas G1 binds hyaluronan and link protein. The GAG chains mediate the binding to P- and L-selectin, CD44, chemokines, lipoproteins and most likely Toll-like receptors (TLR) [3].
2.2. Versican a tumor stroma modulator of breast cancer cell signaling and metastasis
Versican is accumulated in tumor stroma in various malignancies and its levels have been associated with cancer progression in various cancer types [3, 14, 35]. It is accumulated in the preclinical phase of breast cancer in non-palpable breast carcinomas and is associated with risk factors such as increased mammographic density and malignant appearing microcalcifications [16]. Versican is increased in fibroadenoma [11] and the elevated levels of stromal versican are associated with increased risk and rate of relapse in women with node-negative breast cancer [14, 17]. Although all versican splice variants are markedly accumulated in breast tumors, highly glycanated V0 and V1 variants predominate in tumor stroma. V2, V3 and a novel V4 splice variant are also expressed in tumor stroma [36]. V4 contains the first 1194 bp of exon 8 that encodes GAG-β domain that are sandwiched between the end of exon 6 and the beginning of exon 9. In this part of GAG-β domain several serine-glycine consensus sequences capable for carrying GAG chains are present and V4 may exist as true PG. The biological role of V4 variant in tumorigenesis is still unknown [36]. This alternative splice variant of versican may be also considered as a possible target for prognosis and/or therapeutic intervention with antibody-related agents. Apart from the variations occurred in the protein cores of versican due to alternative splicing, versican exhibits significant structural alterations on its glycosylation in various tumors [37–39]. In breast cancer, versican is differentially glycosylated, containing more sialic acid [40]. In most cases stromal cells are the main source of versican in tumor stroma although some cancer cells can synthesize versican themselves. Various stimuli such as platelet derived growth factor (PDGF), transforming growth factor β1 (TGF-β1), epidermal growth factor (EGF), insulin-like growth factor-I (IGF-I), interleukins (ILs) (IL-1β and IL-11), angiotensin II and steroid hormones affect versican synthesis in normal and cancer cell lines [3, 27, 34]. For example, TGF-β1 triggers the biosynthesis of versican in tumor cells and cancer associated fibroblasts [41, 42]. Versican derived from cancer-activated fibroblasts promotes the motility and invasion of ovarian cancer cells by activating the nuclear factor-κB (NF-κB) signaling pathway and by up-regulating expression of CD44, matrix metalloproteinase-9 (MMP-9), and the HA-mediated motility receptor [42]. Versican expressed by some tumor cells affects their growth and metastatic potential. For example, versican is highly expressed in sporadic clear cell renal cell carcinoma inhibiting cell death [43] and in sarcomas promoting tumor cell proliferation and migration and increasing HA production [44, 45]. Abrogation of versican expression in T-anaplastic large cell lymphoma results in decreased levels of membrane type 1-MMP (MT1-MMP) and CD44 and marked suppression of T-cell adhesion and invasion [46].
Versican also contributes to the formation of an inflammatory microenvironment in tumor stroma. Using mouse models of spontaneous breast cancer, it has been shown enhanced recruitment of bone marrow-derived CD11b(+)Gr1(+) myeloid progenitor cells in the pre-metastatic lungs. Versican secreted by these cells in the metastatic niche mediates suppression of the TGF-β/Smad2 pathway by stimulating mesenchymal to epithelial transition (MET), and increases breast cancer cell proliferation, which collectively promotes focal tumor outgrowth at the metastatic site [47]. Moreover, versican secreted by cancer cells interacts with TLR2 present on bone marrow derived macrophages. Versican activates TLR2/TLR6 complexes and induces TNF-alpha secretion enhancing the formation of lung metastasis [48]. Versican V1 secreted by ovarian cancer cells triggers TLR2 and vitamin D3 signaling and enhances hCAP18/LL-37 expression in macrophages. Subsequently, hCAP18/LL-37 secreted by macrophages stimulates growth and invasiveness of tumor cells in the co-culture experiments in vitro [49]. TLR2 signaling is directly involved in the growth of human breast cancers in vitro and in vivo and the inhibition of this pathway merits investigation as possible therapeutic and chemoprevention strategy [50]. Versican V1 variant is a direct transcriptional target of the transcription factor FoxQ1. Versican V1 over-expression stimulates the secretion of chemokine (C-C motif) ligand 2 (CCL2) from hepatocellular cancer (HCC) cells, infiltration of intra-tumoral tumor associated macrophages and augments the formation of metastases [51].
It is well established that G1 and G3 versican domains regulate cell proliferation in normal and tumor cells [3, 34]. The G1 domain of versican stimulates proliferation by creating a less adhesive microenvironment thus destabilizing cell adhesion. The G3 domain induces proliferation, at least in part, by activating EGFR via the action of EGF-like motifs. In breast cancer tissues, G1 and G3 versican levels are increased and they are localized in stromal tissue [52]. It has been shown that G3 via triggering EGFR signaling promotes breast cancer cell proliferation migration and invasion to bone with concordant inhibition of osteoblast differentiation and enhanced osteoblast apoptosis in vitro [53, 54] as well as the formation of spontaneous metastasis to bone in an orthotopic model [54]. EGF-like motifs present on G3 domain enhance EGFR/ERK or AKT signaling driving breast cancer cell invasion to bone stromal cells or osteoblast cells. These motifs are also responsible for the enhanced EGFR/JNK signaling that promotes osteoblast apoptosis and inhibits osteoblast differentiation as well as for repressed expression of GSK-3β (S9P) that contributes to inhibition of osteoblast growth [53]. G3 domain has a dual role in modulation breast cancer cell resistance to apoptosis against chemotherapeutic agents. It either enhances resistance to apoptosis in breast cancer cells cultured in serum free conditions, doxorubicin, or epirubicin by inducing pERK and GSK-3β or promotes apoptosis in cells treated with C2-ceramide or docetaxel by triggering pSAPK/JNK and decreasing expression of GSK-3β [55]. G3-induced EGFR/AKT/GSK-3β (S9P) signaling in breast cancer cells also enhances breast cancer cell self-renewal both in vitro and in vivo. In this model, versican is highly expressed in breast cancer progenitor cells and confers resistance to chemotherapeutic drugs [56]. It is obvious that accumulated versican in ECM is capable of stimulating several cell types through activation of various signaling pathways promoting the secretion of inflammatory mediators that augment tumor growth and metastasis.
It is notable that versican fragments liberated from the action of various proteases may also activate tumor or stromal cells at distant sites. Several protease families including a disintegrin and metalloproteinase domain with thrombospondin motifs (ADAMTS), MMPs and plasmin can cleave versican generating fragments containing the globular domains. The use of antibodies against an ADAMTS_specific versican cleavage site inhibits glioma cell migration [57]. The formation of neo-epitopes of versican fragments within tumor stroma may therefore be used as a potential targeted therapy [8].
3. The instructive role of decorin in autophagy and tumorigenesis
Decorin is a multifaceted PG and prototypical SLRP member that is rapidly evolving as a key factor in cell-matrix dynamics resulting in a multitude of cellular and biological phenotypes. Foremost, decorin is a pan-receptor tyrosine kinase (RTK) inhibitor [58, 59] that affects receptor function at multiple levels, including modulation and bioavailability of receptor ligands [60], for tumorigenic and metastatic suppression [61–65]. Perhaps the most striking evidence for decorin as “a guardian from the matrix” derives from the observation that decorin deficiency is permissive for tumorigenesis [66–68] and increases the basal activity of multiple receptor tyrosine kinases and is further permissive for progression of HCC [69]. Further, as decorin can integrate signaling over multiple receptors including EGFR and IGF-IR, it remains possible that decorin can affect the ERs as well in estrogen responsive breast carcinomas [27]. Decorin is over-expressed by stromal cells and is often accumulated in tumor stroma. Increased expression of decorin in breast cancer tissues is associated with lower tumor grade [70], reduced tumor size, reduced risk and rate of relapse and low survival in node-negative invasive breast cancer [15]. In contrast, high expression of decorin in malignant epithelial tissue is associated with increased lymph node metastasis, lower disease free survival in breast cancer [70]. High decorin expression in malignant epithelium is also correlated with decreased overall survival only in luminal B subtype of breast cancer tumors [70]. However, equally profound roles of decorin are quickly being elucidated and include the ultrastructure determinants of tendon and collagen biomechanics [71–74], a role in Lyme disease [75], maintaining the myogenic niche [76], a transcriptomic biomarker for HCC [77], keratinocyte function [78], fetal membrane regulation [79], and modulating the bone morphogenetic protein (BMP) and Wnt pathways [80, 81]. As a further indication concerning the functional diversity within the SLRP family, the closest relative of decorin, biglycan is primarily involved in orchestrating TLR2/4 as well as myeloid differentiation primary response gene 88 (MyD88) / toll-interleukin receptor-domain-containing adapter inducing interferon-beta (TRIF) mediated innate- immune responses as elegantly determined [23, 82]. Decorin also modulates TLR2/4 for immunomodulation and cancer progression [83].
The newly-discovered function of decorin in evoking protracted endothelial cell autophagy and tumor cell mitophagy, independent of nutrient deprivation and mediated by RTK modulation, is discussed below. Furthermore, decorin is part of an emerging subclass of matrix-derived effectors that engage the highly conserved autophagic machinery that will have profound effects on cell behavior and disease progression.
3.1. Extracellular matrix regulates autophagy
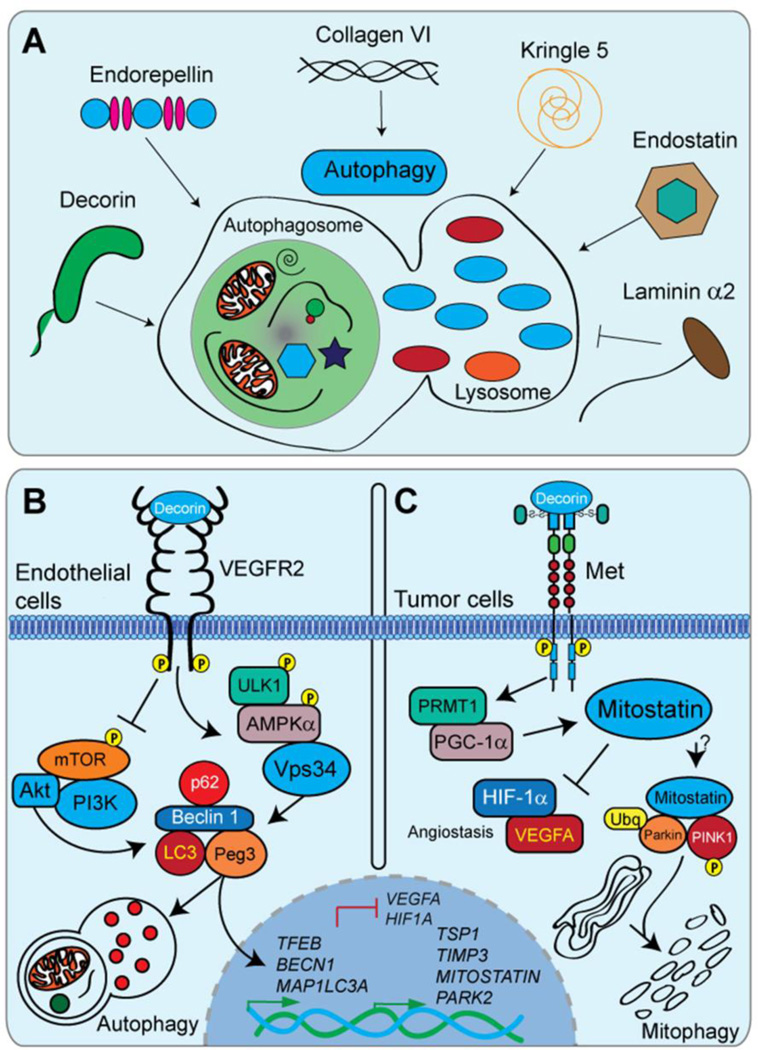
An emerging paradigm is the emerging concept regarding macroautophagic induction and regulation by a specific subset of multifunctional extracellular matrix constituents [84]. These constituents encompass diverse members including decorin, endorepellin, collagen VI, kringle 5, endostatin, and laminin α2 (Fig. 1A). Macroautophagy (hereafter, autophagy) is a tightly coordinated fundamental catabolic process responsible for the non-selective bulk degradation of cytosolic components and organelles [85, 86] following suboptimal metabolic conditions or nutritional dearth. Importantly, dysfunctional autophagy is increasingly being recognized as a key pathological mechanism responsible for several diseases including cancer [87, 88] as well as various forms of muscular dystrophy [89]. The multitude of biological processes orchestrated by the ECM parallels the progressive nature and recognition of autophagy in maintaining proper organismal homeostasis. Moreover, autophagic signaling via matrix components belies several well-established oncostatic and angiostatic functions of soluble matrix members such as decorin [59], endorepellin [90, 91] and endostatin [92]. When prolonged and unrestrained, autophagic induction is oncosuppressive [93] and can elicited by chemotherapeutic agents [94]
Figure 1.
The instructive roles of decorin in autophagy and tumorigenesis. A). Several matrix-derived molecules function as key modulators of autophagy. Each molecule engages a specific receptor for autophagic regulation while utilizing a common core of autophagic machinery. Decorin, endorepellin, collagen VI, kringle 5, and endostatin are pro-autophagic molecules. The laminin α2 chain from laminin 211 represses autophagic function. Please refer to section 3.1 for a more detailed analysis. B,C). Schematic representations of the mechanisms underlining decorin evoked endothelial cell autophagy via VEGFR2 and tumor cell mitophagy downstream of Met, respectively. Please refer to sections 3.2, 3.3, and 3.4 for additional information.
A crucial aspect of ECM-regulated autophagy is the wide functional variety and composition of the effector molecules, each engaging a distinct cell-surface receptor for proficient and differential signal transduction for autophagic regulation (Fig. 1A). Soluble decorin interacts with various RTKs including vascular endothelial growth factor receptor 2 (VEGFR2), for paternally expressed gene 3 (Peg3)-dependent endothelial cell autophagy [95, 96] (see section 3.2), and Met, for mitostatin-dependent tumor cell mitophagy and angiostasis [20] (see section 3.3 and 3.4) (Fig. 1B and C). Endorepellin, the C-terminal cleavage product of perlecan, commands a dual receptor antagonism by acting as a molecular bridge and simultaneously ligating the α2β1 integrin and VEGFR2 for angiostasis [90, 91]. Concurrent with the documented angiostatic properties of endorepellin, is the formation of Beclin 1 and LC3-positive autophagosomes (Fig. 1B) downstream of VEGFR2 in endothelial cells [97]. Molecular dissection of endorepellin into the bioactive (e.g. anti-angiogenic) N-terminal LG1/2 domains [98] was sufficient for autophagic induction, independent of the LG3/α2β1 integrin-binding module [98]. By analogy with endorepellin, several other proteolytically liberated, soluble pro-autophagic effectors such as endostatin (from the HSPG collagen XVIII) and kringle V (derived from an internal region of plasminogen) are also competent for autophagic induction [99, 100] (Fig. 1B).
Pertinent for maintaining skeletal muscle homeostasis [101], collagen VI has also been implicated in autophagic and mitochondrial regulation [102–104]. Loss of collagen VI (e.g. as seen in Ullrich and Bethlem muscular dystrophies) compromises AKT/FoxO3 signaling resulting in decreased autophagosome formation and disproportionate cytosolic levels of Beclin 1 and Bnip3 [103] (Fig. 1B). The above described ECM components function as pro-autophagic mediators for increased autophagy over basal levels. In contrast, laminin α2 (laminin 211), exerts anti-autophagic properties as mutations that arise in laminin α2 (as established in merosin-deficient congenital muscular dystrophy, MDC1A) manifest as loss of function alleles and consequent with a significant increase in autophagic markers (Beclin 1, p62,and LC3) [105]. Moreover, the intracellular signals and second messengers that are activated as a consequence of receptor recruitment and binding, seemingly converge upon a common core (Peg3, Beclin 1, LC3) of autophagic machinery required for an appropriate and germane autophagic response [84]. Characterization of the signals and relays necessary for this biological function are only beginning to be understood and elucidated. This unique collection of ECM molecules is quickly emerging as key regulators of autophagic programming in a wide array of tissues and microenvironments that appears independent of the prevailing nutrient concentrations.
Collectively, these candidate ECM molecules are pioneering a paradigmatic shift in understanding the complex determinants of intracellular behavior. The matrix provides soluble cues and embedded signals for the fine-tuning of this highly conserved intracellular process that factors markedly in the progression of complex pathologies.
3.2. Decorin induces autophagy in normal endothelial cells
After successful establishment of tumor xenografts comprised of triple negative basal breast carcinoma cells, decorin was systemically administered and high-resolution transcriptomic profiling of the host Mus musculus stromal compartment and Homo sapiens tumor parenchyma was performed in parallel, on the same platform [106]. Bioinformatic analyses with this novel dataset unexpectedly revealed that decorin triggered significant and differential gene expression changes exclusively within the host microenvironment [106]. In striking contrast, no changes occurred within the human basal breast carcinoma [106]. Moreover, the stromal-specific genetic signature evoked by decorin decidedly disallows favorable tumorigenic growth and metastatic dissemination [59, 106].
Chronic decorin exposure permitted differential changes in a small, but robust, subset of genes operating wholly within the tumor stroma [106]. Of these, Peg3, a poorly understood genomically imprinted tumor suppressor [107, 108], emerged as a prime candidate. The biological activity of Peg3 aligns with the established oncostatic properties of decorin insofar as promoting the expression of an epigenetically silenced tumor suppressor gene [59, 109, 110] and modulation of the Wnt/β-catenin signaling axis [111]. Therefore, employing macrovascular and microvascular endothelial cells as the tumor microenvironment proxy, Peg3 distributed upon subcellular configurations reminiscent of autophagosomes in response to decorin [112]. Validating the identity of these structures with canonical autophagic markers, such as Beclin 1 and LC3, authenticated these Peg3-positive entities as autophagosomes (Fig. 1B). Functionally, Peg3 is necessary and sufficient for decorin-mediated transactivation of the BECN1 and MAP1LC3A genomic loci and eventual cytosolic accumulation of these proteins [112, 113]. Moreover, RNAi-mediated silencing of Peg3 results in a decrease of basal Beclin 1 mRNA and protein in endothelial cells (Fig. 1B) [112, 113].
Mechanistically, decorin induces Peg3-dependent endothelial cell autophagy downstream of VEGFR2 [113], the primary RTK responsible for coordinating endothelial cell behavior and homeostasis (Fig 1B). Intriguingly, decorin acts as a partial agonist via binding IgG modules 3–5 of the VEGFR2 ectodomain for competent autophagic induction (Fig. 1B) [113]. This activity stands in contrast with the well-documented role of decorin as a global RTK inhibitor [25, 114–116]. Upon decorin engagement of VEGFR2, the upstream signaling apparatus bifurcates and permits the simultaneous and protracted inhibition of the potently anti-autophagic PI3K/AKT/mTOR/p70S6K signalome with concurrent and sustained activation of the pro-autophagic ULK1/AMPKα/Vps34 pathway (Fig. 1B) [112, 117–119]. Consequently, the pro-autophagic signaling arm converges upon the physical assembly of a Peg3/Beclin 1/LC3/p62 supramolecular quaternary complex (Fig. 1B). The concerted formation of these Peg3-positive structures and the combinatorial disengagement of repressive Bcl2/Beclin 1 complexes are thereby permissive for competent isolation membrane formation, phagophore elongation, and autophagic gene target induction (Fig. 1B) [117]. Importantly, decorin promotes the rapid activation of the central energy sensor network via phosphorylation of AMPKα at Thr172 downstream of VEGFR2 and independent of prevailing nutrient requirements (Fig. 1B) [117]. As a step between VEGFR2 and phosphorylation of AMPKα (as no direct biochemical interaction was seen between VEGFR2 and AMPK), ULK1 may be recruited to AMPK and serve as an intermediary kinase for autophagic initiation and further attenuation of the anti-autophagic mTOR/Raptor/GβL/mLST/PRAS40 complex [120, 121].
Autophagy requires fusion between autophagosomes and lysosomes (autophagolysosomes) for engulfed target degradation by lysosomal hydrolases and nutrient recycling (Fig. 1B) [122, 123]; lysosomal biogenesis must be induced and maintained for continual and successful autophagic flux. Further, Peg3 functions as a master autophagic regulator and decorin may dynamically regulate transcription factor EB (TFEB) downstream of Peg3 activity [112, 124]. TFEB serves as a critical link for the synchronization of coordinated lysosomal-nuclear signaling and positive autophagic flux [125]. Phosphorylated TFEB is held in an inactive state in the cytosolic compartment upon the lysosomal membrane by positive mTOR signaling [126]. Since decorin staunchly inhibits mTOR activity in a VEGFR2 dependent manner, TFEB may become actively or passively dephosphorylated, translocate into the nucleus, and incorporate into transcriptionally competent pre-initiation complexes on the promoters of pro-autophagic targets downstream of Peg3 [124].
Collectively, the induction of endothelial cell autophagy proclaims a paradigmatic shift for elucidating not only the underlying molecular mechanisms of decorin, but also these findings could be applicable to the SLRP gene family as a whole. Autophagic induction in a tissue and organ specific manner may therefore represent heretofore unbeknownst, but evolutionarily conserved biological functions for matrix-derived cues, independent of nutrient conditions.
3.3. Decorin evokes mitophagy in breast carcinoma cells
Decorin has earned the title of “a guardian from the matrix” as decorin significantly disfavors tumorigenic growth [63, 127–129], circumvents rampant tumor neovascularization [19, 130], and suppresses bone metastasis [59, 131, 132]. In a mechanism analogous to the aforementioned activity of decorin-evoked endothelial cell autophagy, decorin acts as a partial Met agonist for the induction of tumor cell mitochondrial autophagy (Fig. 1C) [84, 117]. Mitophagic induction may, indeed, unify the classical tumoricidal functions of decorin [59]. Functioning at the core of this novel finding is a poorly studied decorin-inducible tumor suppressor known as mitostatin [133, 134]. Mitostatin, also known as trichoplein [135], localizes to mitochondria [133] as well as to highly specialized sites that exist in juxtaposition at endoplasmic reticulum-mitochondrial interfaces in conjunction with mitofusion-2 [135].
Downstream of Met, the regulatory scheme for mitostatin induction is dependent on PGC-1α, the molecular kingpin for mitochondrial biogenesis [136]. This is unique insofar as that PGC-1α has been implicated for BRAF-mediated oncogenesis [137] as well as metabolic reprogramming in several models of solid malignancies [138, 139]. However; in a Met tyrosine kinase dependent manner, decorin orchestrates rapid post-transcriptional stabilization of MITOSTATIN mRNA via direct binding of the C-terminal RNA recognition motif (RRM) of PGC-1α (Fig. 1C) [117]. Protein arginine methylation of the PGC-1α RRM is carried out by PRMT1 [130] and required for the formation of PGC-1α/MITOSTATIN-positive mRNP complexes (Fig. 1C) [117]. Genetically ablating the PGC-1α RRM disrupts mRNA binding and abrogates decorin-mediated stabilization of MITOSTATIN mRNA and downstream mitophagic induction in basal breast carcinoma cells (Fig. 1C).
RNAi-mediated suppression of mitostatin abolishes the response of breast carcinoma cells for canonically evoked (e.g. rapamycin, HBSS) or decorin-evoked mitophagy [117]. This manifests as a block in oxidative phosphorylation complex turnover, mitochondrial fragmentation, VDAC, and mtDNA depletion [117] (Fig. 1C). An early signaling event for the stimulation of mitophagic processes requires the loss of mitochondrial membrane potential [140]. Depolarization of the mitochondria outer membrane is a valid prognosticator of mitochondrial dysfunction and represents a “danger signal” [139] for degradation and / or apoptosis [141]. Depolarized mitochondria recruit a RING-between-RING (RBR) E3-ubiquitin ligase known as Parkin that executes the mitophagic cascade [142]. The importance of maintaining healthy mitochondria and their clearance via mitophagy is underscored in the development of several types of neurodegenerative diseases, such as recessive forms Parkinson’s, for which the eponym Parkin derives [140]. Over 18% of Parkinson’s disease patients harbor mutations in the PARK2 gene that encodes Parkin [142]. Moreover, this loss of membrane potential permits recognition of damaged versus healthy mitochondria for Parkin recruitment [142]. Therefore, as a very early event in the mitophagic pathway, decorin triggers mitochondrial depolarization to an extent that is analogous to the protonophore, FCCP [117]. The ability of decorin evoked mitochondrial depolarization may originate and succeed the calcium oscillations that occur upon decorin/RTK interactions [143].
Mechanistically, mitostatin may function as a molecular tether for Parkin recruitment to damaged, depolarized mitochondria and / or stimulate the activity of the PINK1/Parkin-mediated ubiquitination (Fig. 1C). The documented role of Parkin in evoking mitophagy [144] and respiratory chain turnover [145] functionally overlaps with the known roles of mitostatin signaling [117]. As such, mitostatin promotes the assembly of a pro-mitophagic signaling complex that includes PINK1, a master kinase necessary for mitophagic initiation and progression, and Parkin (Fig. 1C). This newly-formed ternary effector complex, downstream of positive decorin/Met signaling, may then permit activation, via PINK1 phosphorylation, of the Parkin RBR domain and downstream ubiquitination (Ubq) of mitochondrial targets, such as VDAC and p62/SQSTM1 [144, 146] (Fig. 1C). Tantalizingly, selective degradation of specific mitochondrial proteins in a PINK1/Parkin dependent manner [142] occurs primarily on the outer mitochondrial membrane, where mitostatin localizes [133, 134].
Therefore, soluble decorin engages Met in a positive fashion and evokes mitophagy in a mitostatin dependent manner within the tumor parenchyma. As will be discussed below, mitophagic induction may account for a classical hallmark of decorin bioactivity by suppressing tumor angiogenesis.
3.4. Anti-angiogenic function of decorin
A classic tenet of decorin is the innate ability of angiogenic suppression thereby preventing rampant tumor neovascularization and circumventing metastatic spread. In essence, decorin differentially modulates angiogenic effectors by inhibiting the transcription of pro-angiogenic angiokines [e.g. hypoxia inducible factor 1 α (HIF-1α) and vascular endothelial growth factor A (VEGFA)] with the concomitant induction and rapid secretion of potently anti-angiogenic molecules [tissue inhibitor of matrix metalloproteinase-3 (TIMP-3) and thrombospondin 1 (TSP1)] (Fig. 1C) [19, 130]. The induction of autophagic processes within the stroma and mitophagic activity within the tumor may underlie the molecular mechanism concerning this hallmark of decorin.
We have discussed above (section 3.2) that decorin binds VEGFR2 and positively signals for the induction of a macroautophagic program within the endothelial cells [112]. Endothelial cells, in turn, represent the fundamental cell type for being involved in both developmental and pathological vascularization. Indeed, migration, proliferation, tubulogenesis, and capillary plexus formation are chief angiogenic mechanisms by which a quickly developing tumor conciliates the need for nutrients, oxygen, and sustained growth and spreading. These properties are largely mediated by paracrine effects of VEGFA signaling, derived from the abnormal angiogenic stimulus (e.g. the tumor) and autocrine VEGFA effects stemming from the endothelial cells. Activation of the pro-autophagic VEGFR2 receptor stimulates the presumptive ULK1/AMPKα/Vps34/Peg3/TFEB signaling arm and may repress endothelial cell VEGFA or VEGFA responsiveness of the endothelial cells.
Intriguingly, upon loss of mitostatin, the ability decorin-mediated VEGFA suppression is wholly abrogated [117] (Fig. 1C). Therefore, mitophagic induction and angiogenic suppression may be inextricably and genetically linked. Several possible explanations that account for this connection exist. Turnover and degradation of electron transport chain components affect the production of reactive oxygen species [138, 147] which in turn drives HIF-1α/VEGFA signaling independent of oxygen tensions [148] in a manner akin to decorin [19]. Further, mitostatin-dependent mitophagy and recruitment of the PINK1/Parkin axis may ubiquitinate and trigger degradation of additional pro-angiogenic targets such as Myc, β-catenin, and HIF-1α [19, 127]. Importantly, as an associative partner of Parkin [149], the Skp1-Cul1-F-box (SCF)-containing E3 ubiquitin ligase, FBW7, may target HIF-1α and Myc for proteasomal degradation [150, 151] following mitophagic initiation. Therefore, activation of the mitophagic program, in a mitostatin and Parkin-dependent manner, under normoxic and nutrient rich conditions may provide a molecular link with the non-canonical, hypoxia-independent mechanism of decorin-mediated angiostasis (Fig. 1C) [19].
In conclusion, the ramification of decorin-mediated autophagy and mitophagy may have far-reaching consequences suppressing the overall integrity and viability of primary and metastatic solid neoplasms. As such, autophagic regulation may represent a generalized function for the surrounding matrix, and in particular for the multifunctional SLRP family, in the control of cell behavior.
4. Biglycan triggers inflammation and tumorigenesis
4.1 Biglycan as endogenous danger signal and its role in inflammatory diseases
Biglycan, another member of the class I family of SLRPs, consists of a 42 kDa protein core and up to two covalently-bound CS/DS side chains. This SLRP is ubiquitously expressed and acts as a structural component and stabilizer of the ECM via its interaction with numerous components of the ECM, e.g. collagens type I, II, III, and VI, and elastin [21, 22, 152]. Lessons learnt from biglycan-deficient mice that display an osteoporosis-like phenotype, established biglycan as an important regulator of bone formation and collagen fiber assembly [152, 153]. By interacting with tumor necrosis factor (TNF)-α, TGF-β1-3, BMP-4, Wnt (Wingless-type mouse mammary tumor virus integration site family) 1-induced secreted protein 1 (WISP-1) and VEGF, biglycan modifies a host of cellular processes [21, 22, 152].
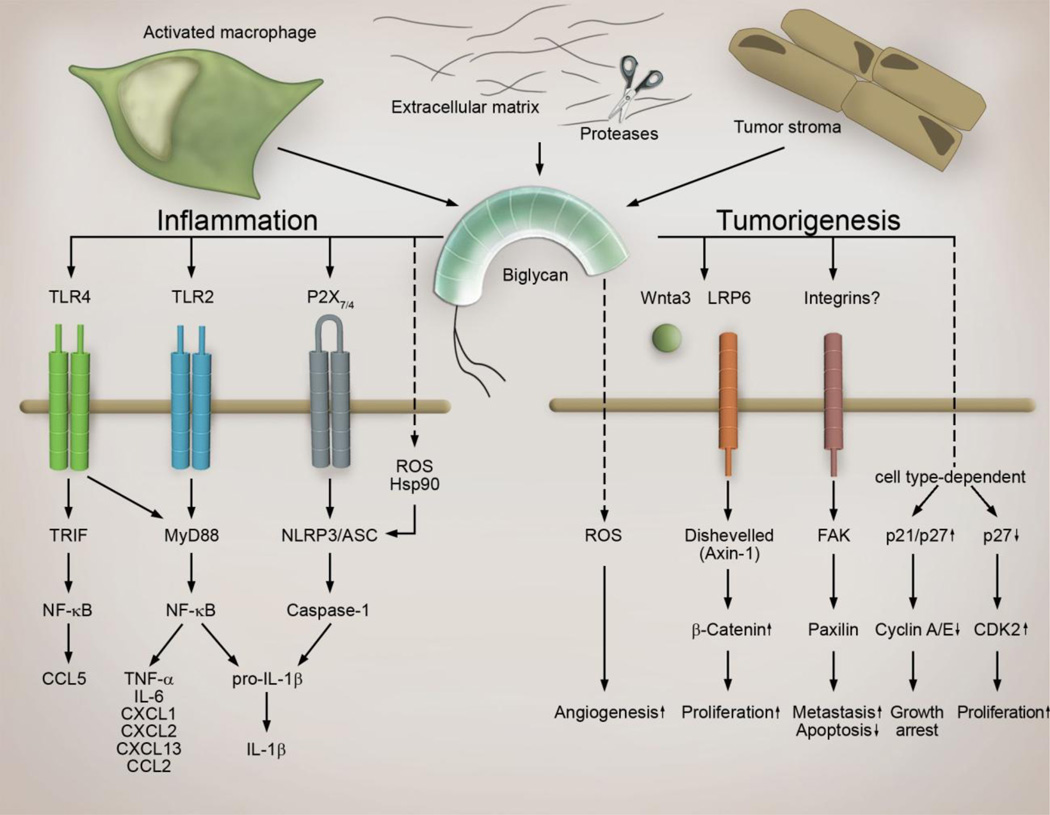
The most striking observation is that biglycan in its soluble form acts as a signaling molecule and “danger signal” by engaging the innate immunity TLR2 and TLR4 [154, 155] in macrophages (Fig. 2). Biglycan/TLR-mediated activation of the NF-κB leads to synthesis of proinflammatory TNF-α, IL-6 and pro-1β cytokines [82, 154] (Fig. 2). By clustering TLR2/4 with purinergic P2X7/P2X4 receptors along with induction of reactive oxygen species (ROS) and Heat shock protein (Hsp)90, biglycan triggers formation of NLRP3/ASC inflammasome (NLR pyrin domain containing 3/apoptosis-associated speck-like protein containing a carboxy-terminal caspase activation and recruitment domain) with subsequent activation of caspase-1 and processing of pro-IL-1β into mature IL-1β [3] (Fig. 2). Furthermore, an interplay of biglycan with either the adaptor molecule MyD88 or TRIF results in synthesis of various C-C and C-X-C motif ligands (CCL and CXCL), chemoattracting neutrophils (CXCL1, CXCL2), macrophages (CCL2), T-(CCL5), and B-lymphocytes (CXCL13) into the site of tissue injury [82, 156]. Consequently, studies in transgenic mice lacking or over-expressing soluble biglycan, have provided robust genetic evidence for the involvement of biglycan as an autonomous trigger in sterile inflammation (e.g. systemic lupus erythematosus, autoimmune perimyocarditis, diabetic nephropathy, ischemic kidney injury, and obesity) as well as a potentiator of pathogen-dependent inflammation (e. g. sepsis) [21, 22, 152, 154, 156].
Figure 2.
Multifunctional role of biglycan signaling in tumorigenesis. Soluble biglycan synthesized by host tissue, cancer and stromal cells (e. g. macrophages) or proteolytically released from the host- or tumor-derived extracellular matrix acts as a multifunctional signaling molecule. It stimulates angiogenesis by creating a reservoir of VEGF that can be released during tumor-associated ECM-degradation and presumably by interaction with TLR2 and ROS induction. It promotes cell cycle arrest but enhances development of metastases thereby promoting tumor progression. In stromal macrophages via TLR2/4 and NLRP3/ASC inflammasome biglycan triggers pro-inflammatory signaling thereby influencing tumorigenesis and metastasis. For details please refer to the text. (ASC, apoptosis-associated speck-like protein containing a carboxy-terminal Caspase activation and recruitment domain; CCL, chemokine (C-C motif) ligand; CXCL, chemokine (C-X-C motif) ligand; FAK, focal adhesion kinase; Hsp, Heat shock protein; IL, interleukin; MyD88, Myeloid differentiation primary response gene 88; NF-κB, nuclear factor 'kappa-light-chain-enhancer' of activated B-cells; NLRP3, Nod-like receptor pyrin domain containing 3; Nod, nucleotide-binding oligomerization domain; ROS, reactive oxygen species; TLR, Toll-like receptor; TNF, tumor-necrosis factor; TRIF, Toll-interleukin receptor-domain-containing adapter inducing interferon-β).
The ability of biglycan to create a pro-inflammatory milieu and to interfere with central signaling pathways operating in cancer (e.g. TGF-β- and Wnt- signaling) posits biglycan as a regulator of tumorigenesis. Below, we will review recent knowledge regarding the role of biglycan in cancer, metastasis and angiogenesis, and discuss potential therapeutic implications.
4.2 Biglycan expression in tumors
4.2.1 Biglycan: A prognostic marker for cancer progression and patients’ survival
There is a growing evidence for the over-expression of biglycan in various tumor types such as esophageal squamous cell carcinoma [157], intrahepatic cholangiocarcinoma [158], odontogenic cancer [159], melanoma [160],colorectal [161–163], endometrial [164] and gastric [165] that correlates with disease progression in some cases [162–165]. Interestingly, biglycan is also enriched in CD133-positive colon cancer stem cells, responsible for tumor motility and facilitation of drug resistance [166].
Notably, several studies correlate levels of biglycan in tumor tissue with a survival rate of patients. Patients suffering from esophageal squamous cell carcinoma with high tumor-associated biglycan expression possess a strongly reduced disease-specific survival rate [157]. Reduced survival of patients whose tumors had high expression of biglycan is also reported [167]. Accordingly, low biglycan levels tissue are beneficial and correspond to prolonged patients’ survival [164]. Whether these clinical effects reflect a role of biglycan in modulating the tumor stroma or the cancer needs to be further investigated.
A unique role for biglycan is reported in bladder cancer. In agreement with other clinical data, enhanced biglycan levels correlate with a high-grade human bladder cancer and muscle invasiveness. However, patients with high tumor-associated biglycan expression display the best survival rate [168]. This is in line with the in vitro and in vivo data showing increased proliferation of bladder cancer cells after knockdown of biglycan, indicating that biglycan may act as growth suppressor in urothelial neoplasms [168]. Furthermore, in diffuse large B-cell lymphomas biglycan expression is linked to improved success of therapies and patient survival by inducing a high intratumoral inflammatory reaction and an increased autologous tumor response [169]. In light of current knowledge regarding influence of inflammation on tumorigenesis, it is predictable that biglycan, similar to decorin, might inhibit tumor growth of established tumors by creating the TLR2/4-mediated pro-inflammatory environment [83]. However in early stages of tumor development biglycan-driven inflammation is expected rather to promote malignant growth.
Thus, cell type- and tumor stage-dependent expression of biglycan appears to be an important marker for prediction of tumor progression, development of metastases and for estimation of patients’ survival.
4.2.2 Triggers and sources of biglycan in cancer
In spite of the mounting evidence reporting enhanced biglycan expression in various malignant tumors not much is known about triggers and sources of biglycan in cancer. TGF-β is a major inducer of biglycan expression in the majority of cell types [156]. In fact, tumor-derived TGF-β has been shown to trigger biglycan expression in stromal fibroblasts via activation of growth arrest and DNA-damage inducible-beta (GADD45beta) and p38 [170, 171]. Furthermore, pro-inflammatory cytokines such as IL-1β and IL-6 are capable of inducing synthesis of biglycan in macrophages [154]. Therefore it is conceivable that pro-inflammatory factors secreted by stromal mononuclear cells will trigger de novo synthesis of biglycan in inflammatory and resident stromal cells. This in turn will cause TLR2/4-dependent synthesis of chemoattractants for neutrophils, macrophages, T- and B-lymphocytes recruiting these cells to the stroma (Fig. 2). Some of infiltrating mononuclear cells will contribute to a further synthesis of biglycan in the stroma, creating a feed-forward cycle driving an inflammatory response and influencing tumor growth in a cancer-stage dependent manner.
The majority of studies reporting enhanced biglycan levels in various cancers provide data generated in entire tumors. However it has to be considered that “biglycan pool” finally influencing tumor behavior originates from various sources of this SLRP. This “pool” consists of biglycan synthesized in cancer as well as in stromal cells of host and tumor (e. g. fibroblasts and macrophages) and of proteolytically released biglycan from host- and tumor-derived ECM (Fig. 2). Biglycan synthesized by various cells frequently differs in terms of type and length of its GAG chains. Therefore, it is conceivable that influence on tumor behavior in vivo caused by “biglycan pool” interfering with a crosstalk between host and tumor cells with the ECM, differs from those in vitro where single cell types and homogenous biglycan are used. Future studies identifying the cell type expressing biglycan at various stages of tumor progression are needed to provide a basis for the analysis of biglycan-mediated signaling crosstalk between tumor cells, stroma and the ECM. In particular, there is an urgent need to generate data regarding the soluble form of biglycan in cancer, as this is the form that is capable of acting as a receptor ligand and signaling molecule [154]. In fact, levels of soluble biglycan are markedly enhanced in sera from cancer patients [172, 173]. Furthermore, a gradual increase of circulating soluble biglycan is positively associated with tumor grade enhancement and lymph node metastases in patients suffering from endometrial cancer [173].
4.3 Biglycan-mediated signaling in tumorigenesis
In contrast to relative straightforward clinical data indicating enhancement of biglycan expression in various tumors, our understanding of biglycan signaling in tumorigenesis is quite sparse and controversial. Below, we critically analyze our current knowledge regarding biglycan effects on angiogenesis, malignant cell proliferation, growth arrest, innate immunity and inflammation as well as on development of metastases. Additionally, we anticipate biglycan-dependent signaling pathways known from non-carcinoma cells to be possibly operative in tumor cells as well.
4.3.1 Angiogenesis
There is a growing evidence for the importance of biglycan in promoting angiogenesis. Biglycan, constitutively expressed in normal endothelial cells, becomes markedly up-regulated under tumor condition and promotes endothelial cell migration and neovascularization of cancer [172]. Accordingly, biglycan-deficient mice exhibit extenuated neovascularization during healing of bone fractures [174]. In terms of underlying mechanisms triggers VEGF synthesis in carcinoma cells [175]. Additionally, biglycan has been shown to bind and sequester (VEGFA) in the ECM, thereby generating a reservoir of VEGF that can be released during tumor-associated ECM-degradation, enabling angiogenesis (Figure 2) [174]. Furthermore, neovascularization is also conveyed by TLR2 signaling and production of ROS [176]. Thus, it is conceivable that biglycan as a TLR2 ligand [154] and ROS-inducer [177] may trigger angiogenesis in a TLR2/ROS-dependent manner (Fig. 2).
4.3.2 Cell proliferation and breast cancer normalization
Anti-proliferative effects of biglycan are described in elaborated studies using human urothelial carcinoma cells either incubated with exogenous biglycan or over-expressing and lacking the biglycan gene, respectively [168]. Accordingly, in a model of subcutaneous mouse xenograft tumors, containing biglycan-depleted urothelial carcinoma cells, enhanced tumor growth is observed [168]. While mechanisms of anti-proliferative effects of biglycan are not clarified yet, activation of the P2X7 receptor and interference with TGF-β1-signaling can be considered as potential mechanisms of biglycan-dependent anti-proliferative effects in bladder cancer. In pancreatic cancer cells, biglycan-mediated cell cycle arrest due to up-regulation of the cyclin-dependent kinase inhibitor p27 and inhibition of cyclin A/E, provides further evidence that biglycan might act as a suppressor of tumor growth [170] (Figure 2). Additionally, biglycan inhibits cell proliferation in an in vitro model of HER-2/neu+ cell oncogenic transformation [178]. In renal mesangial cells, biglycan inhibits PDGF-mediated proliferation [179].
However, there are several mechanisms in downstream signaling of biglycan that might suggest enhancement of proliferation in certain tumor cell types. In vascular smooth muscle cells, biglycan attenuates p27 levels with subsequent enhancement of cyclin-dependent kinase (CDK)2 expression and acceleration of mitosis [180]. Furthermore, biglycan interferes with Wnt/β-catenin-signaling, a central pathway involved in tumor progression. Biglycan binds to low-density lipoprotein receptor-related protein 6 (LRP6) and Wnt3a, an activator of the Wnt/β-catenin pathway, and increases β-catenin levels thereby supporting cell proliferation and differentiation [181].
Thus, it appears that there are several gaps in our knowledge regarding biglycan-dependent regulation of tumor growth. Besides not fully clarified effects of biglycan on carcinoma cell proliferation, data regarding biglycan-mediated regulation of tumor cell death is quite sparse (see below). Reports in non-carcinoma cells indicate biglycan-dependent inhibition of apoptosis in mesangial cells due to decreasing of caspase-3 activity [179] and pro-apoptotic effects in pre-adipocytes due to unknown mechanisms [182].
Despite being the most homologous relative of decorin, and in contrast to decorin, biglycan has been implicated in the development and progression of several genetically distinct cancers. Indeed, high levels of biglycan expression are associated with increased risk of esophageal squamous cell carcinoma [157], significant clinical outcome of pancreatic adenocarcinoma [167], enhanced gastric cancer invasion [183], and breast cancer normalization [184]. It is well established that breast cancer cells slow their growth and differentiate when associated with embryonic mesenchyme. Notably, when the matrix secreted by embryonic mammary mesenchyme was injected into fast-growing breast carcinoma in mice, there was a marked reduction of growth. Proteomics analysis of this mesenchyme ECM showed biglycan as a major constituent [184]. Moreover, addition of soluble biglycan was capable of evoking the tumor normalization response, and RNAi-mediated depletion of biglycan expression in cultured embryonic mesenchyme abolished the ECM’s inductive activity [184]. Thus, biglycan has a novel biological activity within the embryonic mammary mesenchyme that leads to partial breast cancer reversion. Additional studies in a broad-spectrum of carcinoma cell types and at various stages of tumor development are needed to provide a convincing proof for the inhibitory function of biglycan in tumorigenesis.
4.3.3 Development of metastases
In several human cancer types enhanced expression of biglycan is associated with the development of metastases. Furthermore, over-expression of biglycan in a mouse model of gastric xenograft tumors results in the development of metastases [183]. Mechanistically, biglycan triggers phosphorylation of the focal adhesion kinase (FAK) at Tyr576/577, Tyr925 and Tyr397 with subsequent induction of paxillin, resulting in enhanced migration and invasion [183] (Fig. 2). Accordingly, several reports describe biglycan-dependent induction of cell migration in various types of non-carcinoma cells [172, 178, 185]. In contrast, in osteosarcoma cells, biglycan reduces migratory capacity [186]. Interestingly, in lung fibroblasts biglycan activates the signaling pathways of RhoA and Rac1 thereby stimulating migration of these cells [185]. As phosphorylated paxillin is involved in Rac activation, it is conceivable that biglycan-FAK-paxillin-Rac1-signaling could be responsible for the biglycan-mediated induction of cell migration and development of metastases. In addition, anti-adhesive effects of biglycan [179] can further contribute to mechanisms of biglycan-dependent promotion of metastases.
4.4 Desensitization of tumors to chemotherapy
Of high therapeutic relevance appears the observation that biglycan expression in tumors correlates negatively with the cancer response to chemotherapy. A study that compared gene expression profiles of osteosarcoma biopsies either from patients with good or poor responses to chemotherapy, showed that biopsies from the non-responding group had twice as high biglycan levels as compared to responding patients [187]. Additionally, patients with ovarian cancer were chemotherapy-resistant when their tumors expressed enhanced levels of biglycan [188]. However, the mechanism of biglycan-dependent desensitization of tumors to chemotherapy remains elusive and should be addressed in future studies.
Taken together, the clinical message regarding biglycan and tumorigenesis is straightforward and shows over-expression of biglycan in various tumors in a positive correlation with the grade of tumor development and metastasis in cancer patients and experimental tumor models. However, the effects of biglycan on tumor growth still remain unclear. The majority of data underscores the role of biglycan as an inhibitor of cell proliferation and cell cycle suppressor. On the other hand biglycan promotes angiogenesis, cell migration and inflammation (Fig. 2).
Careful analysis of data published in this field, that appear in some cases to be controversial, reveals that these differences are mostly due to the usage of a wide variety of tumor cells with different histogenetic backgrounds and of tumor tissues at diverse stages of development and differentiation. Another critical point is the source and form of biglycan used in in vitro studies. We note that several commercial sources of biglycan do not provide a native form of this SLRP. Furthermore, it is frequently unclear whether effects of intact proteoglycan or protein core of biglycan on cell behavior are described. This might be essential for biglycan signaling as previously shown for inflammatory pathways [154, 156, 177]. Moreover, it is of importance whether soluble or immobilized biglycan was used in an experimental setting. Based on these variations, the underlying mechanisms and signaling pathways driving biglycan effects during the central steps in tumorigenesis are largely unknown. Thus, further studies are needed to unravel the biological roles of this SLRP in cancer progression and metastasis, and as potential therapeutic target for cancer.
5. Syndecans and their Roles in Breast Cancer
5.1. Syndecans as signaling receptors
Syndecans are a small family of type I transmembrane PGs. Mammals have four distinct genes encoding the core proteins, and with the exception of erythrocytes, all cells express at least one syndecan. Syndecan-4 is a ubiquitously expressed family member, while other family members are more tissue and spatio-temporally restricted [189]. For example, syndecan-1, the most studied of the family, is characteristic of simple and stratified epithelia. Syndecans are composed of a core protein bearing multiple GAG chains. These chains can be HS or CS/DS and the number and type of GAG chains vary depending on the syndecan core protein, although for the most part, glycosylation of syndecans in vivo is not well characterized. However, syndecan-1 and syndecan-3 can bear both HS and CS/DS chains whereas syndecan-2 and syndecan-4 predominantly have HS chains [189, 190]. HS chains are formed of repeating disaccharides of N-acetylglucosamine and glucuronic acid. These are extensively modified by sulfation and epimerization of the glucuronic acid to iduronic acid. The length and fine structure of GAG chains appear to be tissue and core protein specific, but generally there are between 50–150 disaccharides per chain. The structure of GAG chains has been discussed in detail recently [191, 192]. Mature HS chains are not uniformly modified by sulfation, but instead have regions of high sulfation interspersed among regions of low, or even no sulfation [191]. This patterning of HS chains encodes motifs that can interact with protein ligands. There are now over 100 potential ligands ranging from growth factors, cytokines, chemokines, ECM proteins and collagens, proteinases, to lipases and lipoproteins. As a result, syndecans are implicated in many cellular processes, but since many growth promoting ligands can bind HS, there is increasing focus on proliferative diseases, such as tumor progression [25].
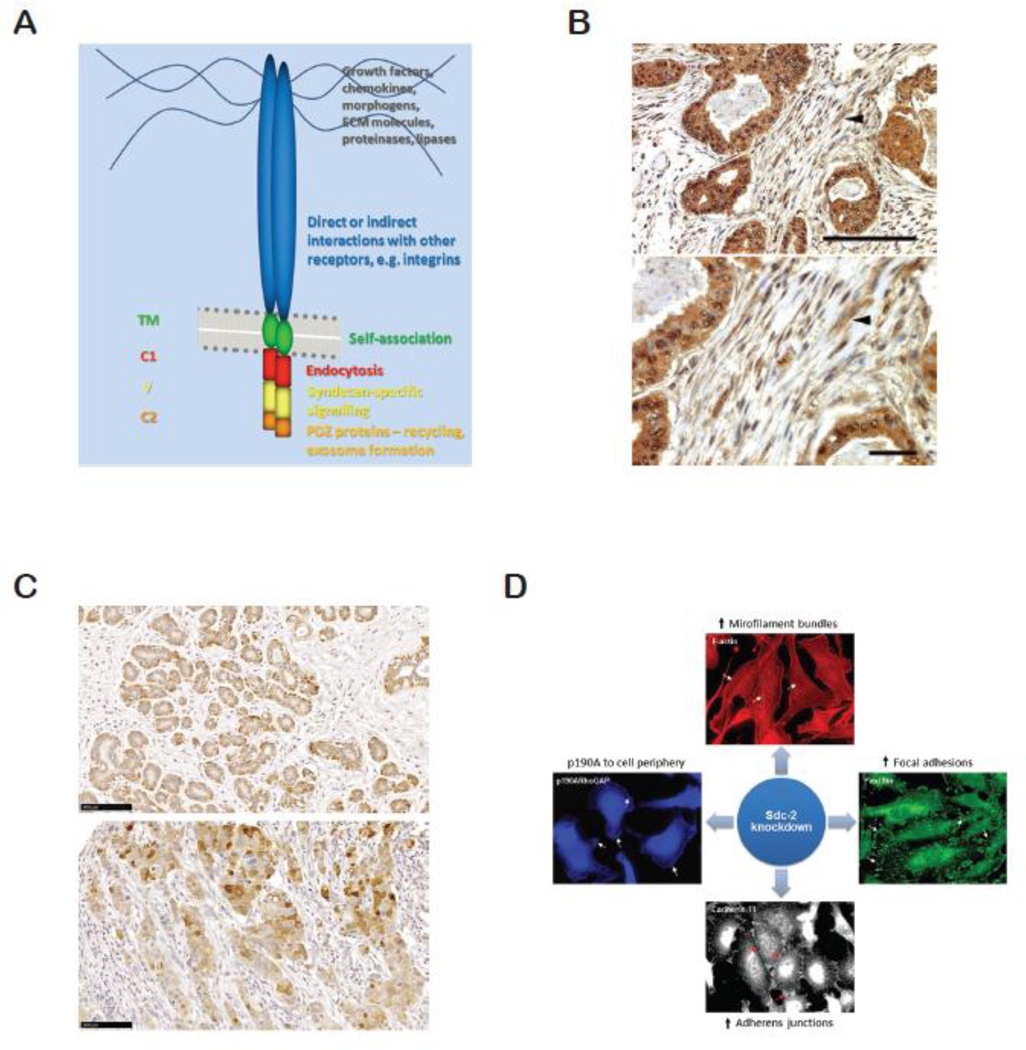
Syndecan core proteins are between 20–40kD can be divided into three domains; a large extracellular, single transmembrane and small cytoplasmic domain (Fig. 3A). While extracellular domains bear the GAG chains, transmembrane domains promote multimerization of the core protein, which appears necessary for signaling functions [193]. Cytoplasmic domains of syndecan can be further divided into two conserved (C1 and C2) with an intervening variable (V) region unique to each syndecan [189]. Though complete structure of syndecan core protein has not been elucidated, syndecan-4 cytoplasmic domain forms a twisted clamp dimeric structure [194]. The cytoplasmic domains have no intrinsic kinase activity, but can nevertheless signal through the docking of, for example, protein kinase Cα (PKCα) (in the case of syndecan-4; [195]). Broadly speaking, the C1 and C2 regions appear to be involved in trafficking of syndecans to or from the cell surface, together with subsequent formation of paracrine signaling organelles, exosomes [196–198]. V region interactions are quite poorly understood, with the exception of syndecan-4, where interactions with the cytoskeletal protein α–actinin and PKCα are documented [189, 195, 199, 200]. Downstream of these molecules is the regulation of Rho family GTPases and the actomyosin system, to control adhesion, migration and cellular morphology. For more details on signaling by syndecans, see reviews [189, 199, 201].
Figure 3.
A. Diagram of syndecan structure, showing some interactions and functions of the constituent domains. B. Intraductal invasive carcinoma grade III showing stromal staining (arrowheads) with mouse anti-syndecan-1 monoclonal antibody 11A9-14. Bar=100µm. C. Ductal hyperplasia (upper panel) and invasive ductal carcinoma grade III (lower panel) stained for syndecan-2 using monoclonal antibodies. Bar=100µm. D. Impact of syndecan-2 (Sdc-2) depletion on the behavior of triple negative MDA-MB231 cells. Cytoskeletal alterations include junction formation and microfilament bundle formation. Increased adhesion also results in decreased invasion and degradation of type I collagen gels.
5.2. Syndecans in the normal mammary gland
Considering the current interest in syndecans and breast cancer, it is surprising how little is known regarding their expression in the developing, lactating or involuting gland. The most studied member of syndecan family in normal mammary gland is syndecan-1. Through the development of the syndecan-1 knockout mouse, its function was addressed during mammary branching morphogenesis [202]. Syndecan-1 expression in the mouse mammary tissue is high in myoepithelial cells and ductal epithelial cells, notably on their lateral membrane [203]. Syndecan-1 null mice showed disrupted mammary gland development, as evidenced by hypomorphic glands and a sparse epithelial tree with 3 times less side branching than control mice. More importantly, absence of syndecan-1 conferred resistance to mammary hyperplasia and tumor development induced by constitutively active intracellular β-catenin expression [202]. The observed phenotype goes beyond the well-known syndecan-1 effect on the Wnt signaling complex. Rather, it was shown that syndecan-1 was essential to mammary epithelial cells responsiveness to β-catenin/TCF [202]. In contrast to syndecan-1, and even though syndecan-4 knockout mice have been reported [204, 205], there are no studies regarding its role during mammary gland development. In human breast tissue, syndecan-4 is expressed on luminal cells and weakly expressed on myoepithelial cells [29]. Stromal cell expression was not detected [29]. On the other hand, syndecan-2 expression in normal breast tissue was observed in myoepthelial cells (Fig. 3B). To the best of our knowledge, there is no report of how HSPGs are regulated during the different stages of mammary gland development. The available data regarding this aspect describes HS, CS and DS polysaccharide expression in virgin, lactating and involuting mouse mammary glands. Whereas HS chains are present at the basement membrane during all stages of development there is a shift between DS and CS expression. For instance, DS was highly expressed at the basement membrane during lactation stage while CS chains were the major GAG in mammary tissue during pregnancy [206].
5.3. Regulation of syndecan expression
The expression patterns of the four mammalian syndecans are distinct, suggesting that transcriptional regulation is an important feature. Despite this, little is currently understood regarding the regulation of the syndecan gene promoters. Soon after the identification of syndecan-1, there were some studies of its promoter [207, 208], indicating sites for Sp1 family (specifically Sp3 in more recent studies [209]), NF-kB, MyoD (Ebox) and Antennapedia [207] as well as Wilms’ tumor suppressor gene (WT1; [210]). However, syndecan-1 is not well known as an early response gene, unlike syndecan-4, where its expression has been well documented to be NF-kB and hypoxia sensitive [211, 212].
While none of the syndecan genes has been shown directly to be regulated by steroids, it is known that treatment of ERα+ breast carcinoma cells with estradiol (E2) exhibits significant increases in syndecan-2 transcriptional levels, but not syndecan-4 [26]. Moreover, the use of EGFR and IGF-IR inhibitors decrease the gene expression levels of syndecan-2 and -4, in contrast to E2-mediated treatment in the presence of inhibitors that also cause up-regulation of syndecan-2 and down-regulation of syndecan-4 gene expression levels [28]. The syndecan-2 promoter may be well worth characterizing, not least as it may be important in triple negative breast cancer [30].
Furthermore, treatment of breast cancer cells with pharmaceutical formulations or by other novel therapeutic approaches can affect syndecan expression levels. The bisphosphonate zoledronic acid suppresses syndecan-1 and syndecan-2 gene expression levels in human breast cancer cells, in contrast to significant increases in syndecan-4 mRNA levels [213]. Non-coding RNAs may also be important regulators since miR-10b, already implicated in breast cancer [214], regulates syndecan-1 levels in MDA-MB231 breast carcinoma, thereby promoting cell motility and invasiveness by a Rho-GTPase- and E-cadherin-dependent mechanism [215].
Syndecan-1 levels are also modified by omega-3 polyunsaturated fatty acids in human breast cancer cells and suggest that syndecan-1 mediated biological processes are modified through low-density lipoprotein delivery of n-3 polyunsaturated fatty acids [216]. In addition, syndecan-1 expression levels, shedding and localization in breast cancer cells are also enhanced by heparanase, an enzyme in current focus that promotes tumor progression and metastasis [217].
Very few studies have examined the genetic variation in syndecan genes and their association with malignancies. However, syndecan-1 and syndecan-4 polymorphic variations have been investigated in Australian breast cancer patients [218]. A single nucleotide polymorphism (SNP) in syndecan-1 (rs1131351) is associated with breast cancer in this population, in contrast to a syndecan-4 (rs67068737) polymorphism which has no association to the disease. This perspective is also enhanced by another study on European postmenopausal population, which shows that a syndecan-1 SNP is associated with breast cancer susceptibility [219]. The molecular implications of these findings remain to be investigated.
5.4. Syndecans and breast cancer
There have now been many studies on syndecans and breast cancer, although knowledge of mechanistic pathways is largely absent. Loss of syndecan-1 is associated in poor prognosis in many cancers such as lung cancer [220]. However, breast cancer research provides a different story. Several reports indicate that syndecan-1 is up-regulated in human breast cancer tissues compared to normal tissues, where it is correlated with higher histological tumor grading, increased mitotic index, increased tumor size, positive lymph node status and poor prognosis [29, 220–222].
Several studies confirmed the expression of syndecan-1 in both epithelial and stromal compartments of breast tumors [29, 223] (Fig. 3C). Epithelial syndecan-1 expression has been associated with negative ER status but stromal syndecan-1 expression with positive ER status. Moreover, triple negative breast carcinoma lines exhibit a higher expression of syndecan-1 compared to non-metastatic subtypes [224]. In addition, the HER2 positive and basal triple-negative carcinomas exhibit higher levels of syndecan-1 compared to luminal subtypes, though the latter may have higher expression than normal cells. Syndecan-1 expression in the reactive stroma cells has been proposed to create a favorable microenvironment for tumor cell growth and angiogenesis [225]. The source of stromal syndecan-1 is still debated, though some reports hold MT1-MMP mediated shedding responsible [226] while others detect the presence of syndecan-1 mRNA in the stroma [227]. In addition, a worse prognosis in breast carcinoma patients was reported where syndecan-1 expression extended to the stroma [223]. This was in agreement with earlier studies where stromal syndecan-1 promoted invasiveness of breast carcinomas [228]. In any case, distinct roles were suggested for soluble syndecan-1 in stroma and syndecan-1 in membrane bound form [229] and one study concluded that breast cancer-specific 10-year overall survival was reduced with higher expression of syndecan-1 in epithelium or stroma [223]. Several in vivo and in vitro models support the idea that syndecan-1 promotes tumorigenesis by promoting Wnt signaling [203], tumor cell adhesion, spreading [230], angiogenesis [231], proliferation [232] and ECM signaling [233]. Recently, Ibrahim et al. suggested that syndecan-1 promotes cancer stem cell properties in triple negative breast cancers [234], a factor that negatively impacts cancer therapies. The same study proposed that syndecan promotes stem cell properties via a pathway involving Wnt and IL-6/STAT3 signaling. Interestingly, administration of chemotherapy results in reduced syndecan-1 in cancers [235], but this treatment is less effective in patients with higher syndecan-1 expression [236].
Unlike syndecan-1, roles of syndecan-4 in breast cancer oncogenesis have been less studied, though syndecan-4 is known to be the second most abundant HSPG not only in normal mammary epithelium but also in breast carcinoma lines. Regardless of the expression, syndecan-4 was shown to mediate breast cancer cell adhesion, spreading [230] and growth factor signaling [224]. This might be important since receptor status is a key criterion for tumor classification and selection of treatment. However, syndecan-4 expression did not correlate with histological tumor type, age, lymph node status or grade of the tumor [29]. In contrast, a previous study suggested that syndecan-4 expression correlated significantly with high histological grade and negative estrogen receptor status [237], therefore a marker of poorer prognosis. These studies employed distinct methods and antibodies but suggest that the importance of syndecan-4 in breast cancer is not sufficiently resolved.
There are a few studies available concerning the roles of syndecan-2 and syndecan-3 in breast cancer progression. Our recent data from human tissue arrays suggest that syndecan-2 is up-regulated in breast tumors and in cases where the primary tumor and metastases from the same patient could be compared, syndecan-2 was expressed at higher levels in the latter [238]. Corresponding work in tissue culture suggested that syndecan-2 has an important role in regulating breast carcinoma cell morphology and invasive behavior [238]. A single report failed to correlate syndecan-3 expression mammary carcinoma outcome. It also indicated that syndecan-3 is not associated with lymph node metastasis and clinical stage, ruling out syndecan-3 as a possible prognostic marker [239].
5.5. Breast carcinoma in vitro
Breast tumors are characterized by loss of tissue architecture and tissue function, complex and altered patterns of gene expression and enormous heterogeneity [240, 241]. These factors make breast cancer a challenging disease to be studied. Syndecan roles include function as a receptor for ECM. According to the dynamic reciprocity model [242], organs and tissues are embedded in the ECM, a source of both biochemical and biophysical cues that control cell behavior. ECM cues are transduced by cell surface receptors through the cytoskeleton, which is connected to the nuclear matrix and chromatin. As a result of this intricate network, ECM information can decode change in gene expression and ultimately cell behavior. Syndecan HS chains interact with many ECM proteins such as collagen, fibronectin, laminins, and vitronectin [189, 190]. The triple negative and highly malignant MDA-MB-231 cells express many HSPGs, with syndecan-1 being dominant [230]. Cell spreading on vitronectin was achieved by a cooperative mechanism between syndecan-1 ectodomain and integrin αvβ3, since recombinant syndecan-1, syndecan-1 core protein-specific antibody or syndecan-1 down-regulation inhibited αvβ3 integrin-dependent spreading and migration [243]. Furthermore, through the use of syndecan-1 mutants lacking specific domains in the core protein, a peptide called synstatin (corresponding to amino acids 82–130 of mouse syndecan-1) was identified. Synstatin blocked interaction between syndecan-1 and αvβ3 and αvβ5 integrins [244]. Since these integrins are involved in tumor angiogenesis, synstatin was tested as an anti-angiogenic compound. Synstatin treatment inhibited xenograft tumor growth of human MDA-MB-231 breast cancer cells and tumor angiogenesis (11-fold reduction compared to untreated tumors), suggesting that syndecan-1 is a crucial regulator of integrin activation during angiogenesis and tumorigenesis [244]. The molecular mechanism by which syndecan-1 activated αvβ3 and αvβ5 integrins involved IGF-IR (insulin-like growth factor-I receptor) autophosphorylation mediated by syndecan-1 clustering. Indeed, IGF-IR inhibitors block mouse Sdc1-expressing breast cancer cell spreading and migration on vitronectin [245]. Studies using the S115 mouse mammary tumor cell line suggested that syndecan-1 expression inhibits tumor cell growth and supported epithelial morphology by inducing actin filament organization [246]. Similarly, targeting of syndecan-1 by the miR-10b or syndecan-1 knockdown in MDA-MB-231 cells induced increased cell migration and invasion [215]. The molecular mechanism that may explain cell phenotype upon syndecan-1 down regulation involves altered function of focal adhesion kinase, Rho-GTPases and E-cadherin [215].
Syndecan function in cell signaling induced by growth factors has also been addressed in breast cancer. Breast carcinoma tissue had an enhanced ability to promote assembly of fibroblast growth factor-2 (FGF-2) and fibroblast growth factor receptor 1 (FGFR1) complex when compared to normal tissue. In addition, syndecan-1 and syndecan-4 are the main proteoglycans responsible for FGF-2-FGFR1 complex formation in breast tumor samples [224].
Tumor cells and their microenvironment coexist in a relationship based on information exchanges. Stromal cells in the tumor microenvironment can also express syndecan-1, which contributes to tumor progression. Interestingly, fibroblast expression of syndecan-1 correlates with parallel stromal fiber organization in mammary tumors [247]. Through the use of syndecan-1 positive and syndecan-1 negative fibroblasts cultured on three dimensional ECM it was shown that syndecan-1 positive fibroblasts promoted ECM organization in a parallel fiber architecture. On the other hand, ECM in which syndecan-1 negative fibroblasts were cultured presented a random fiber arrangement [247]. Furthermore, fiber organization modulated by syndecan-1 positive fibroblasts controlled breast carcinoma cell migration since tumor cells preferentially migrate and invade along aligned collagen fibers [248].
It would therefore appear that syndecan-1 could influence the progression of breast cancer in several ways. Roles in supporting growth factor signaling are foremost, but if stromal syndecan-1, for example, influences integrin activity and the ECM, then it may also exert its effects through cell adhesion. This would be unsurprising since syndecans are bridges between the pericellular environment and the cytoskeleton. Syndecan-1 influences tumor cell behavior but also the stromal compartment and components of the immune system.
Recent data has unveiled novel roles for syndecan-2, which is more widely known as a mesenchymal HSPG, in breast cancer progression [30, 238]. Depletion of syndecan-2 in MDA-MB-231 cells led to profound impact on cytoskeletal organization in these cells. Cell spreading was enhanced with increased microfilament bundles, focal adhesions and cadherin-11 containing adherens junctions (Fig. 3D). Concomitantly, type I collagen invasion and degradation were blocked in the absence of syndecan-2 [238]. Mechanistically, syndecan-2 may signal through caveolin-2 to modulate breast carcinoma cell behavior since caveolin-2 formed a complex with syndecan-2 (but not syndecan-4). Depletion of caveolin-2 yielded the same phenotype as syndecan-2 depletion (unpublished data). In addition, our data also showed that protein levels of caveolin-2 were reduced upon syndecan-2 depletion, suggesting that syndecan-2 is a key player in maintaining caveolin-2 expression in these tumor cells. It would be interesting to investigate the fate of caveolin-2, for example proteasomal degradation, when syndecan-2 is depleted.
The cytoskeletal and behavioral consequences of syndecan-2 depletion were dependent on the Rho-GTPases [30]. A novel cross-talk between syndecan-2 and a negative regulator of Rho-GTPases, p190ARhoGAP, enabled spatiotemporal control of cytoskeletal rearrangement and cell migration in MDA-MB-231 cells. This GTPase activating protein was re-localized from cytoplasm to plasma membrane where RhoA is inactivated in the absence of syndecan-2. The re-localization of p190ARhoGAP appears to be syndcan-4 dependent. Consistent with this, Src-dependent tyrosine phosphorylation of p190ARhoGAP, which is a measure of its activity was increased upon syndecan-2 depletion, suggesting that syndecan-2 is a novel regulator of both distribution and activity of p190ARhoGAP in these tumor cells. A number of previous studies have indicated that syndecan-2 and -4 may have some overlapping roles since they are closely related in structure [189, 249]. However, in breast carcinoma, we found that syndecan-2 suppressed syndecan-4-induced focal adhesion formation [238] and cell surface levels of syndecan-4, on the other hand, were elevated by syndecan-2 depletion, suggesting that a compensatory up-regulation had occurred. However, further experiments are required to provide an answer on how syndecan-2 controls syndecan-4 leading to downstream effects on cytoskeletal rearrangement.
6. Heparanase, syndecan-1 shedding and exosomes facilitate intercellular communication that drives tumor progression
6.1. Heparanase acts as a master regulator of tumor-host crosstalk
Heparanase is a multifunctional molecule whose expression is closely associated with the aggressive behavior of many types of human cancers including breast cancer [250–254]. Heparanase binds to and enzymatically cleaves HS chains, thereby regulating HS availability and/or function both at the cell surface and within the ECM. The endoglucuronidase activity of heparanase may depend on the saccharide structures that surround the cleavage site of HS, thereby leading to variable substrate specificities and implying a complex role for heparanase in regulating HS biological activity [255]. Functionally, much of the impact of heparanase within the tumor microenvironment lies in its regulation of the bioavailability and activity of key factors that bind to HS including growth factors, chemokines, cytokines, enzymes and other effectors. These HS-binding factors represent a large number and broad range of functions [191], further underscoring the potential influence of heparanase in tumor-host cross-talk. Additionally, many factors utilize HS as a receptor or co-receptor on the surface of cells and modulation of HS by heparanase can impact this function. Heparanase function however is not limited solely to its enzymatic activity. Enzymatically inactive heparanase can activate signaling molecules such as AKT and p38 [256, 257] and promote transcription of several biologically important effectors [e.g., hepatocyte growth factor (HGF) and tissue factor] [258, 259]. This implies heparanase has broad functions beyond its impact on HS.
In breast cancer, analysis of clinical specimens led to early speculation that heparanase is associated with breast cancer metastasis. Heparanase expression is present in a high percent of patients having metastatic breast cancer as compared to patients without metastasis, where heparanase expression is rare [260]. Moreover, heparanase expression as determined by immunohistochemistry is associated with high-grade metastatic breast cancers [261] and with more invasive subtypes of human breast cancer as compared to less invasive subtypes [262]. Heparanase expression in breast cancer patients has also been associated with lymph node status, late clinical stages, a short overall survival and a short relapse-free survival [263].
Utilizing animal models of breast cancer, heparanase was shown to promote tumor growth, angiogenesis and survival apparently through its impact on generating a supportive tumor microenvironment [251, 264]. Much of this effect can be attributed to heparanase-mediated upregulation of VEGF and the downstream impact this has on enhancing angiogenesis [265]. Contributing to this effect is the ability of heparanase to enhance endothelial cell migration by stimulating AKT and PI3K [265]. In addition, heparanase has a major impact on promotion of the metastatic phenotype. Enhanced expression of heparanase in human breast cancer cell lines promotes tumor invasion, while knock-down of heparanase expression diminishes invasion capacity in vivo [264, 266, 267]. Heparanase plays important roles in breast cancer metastasis to the brain, an event that signals an exceptionally poor prognosis for the patient. Heparanase was found to regulate cytoskeletal dynamics of breast cancer cells and to mediate cross-talk between tumor and brain endothelial cells that together promote metastasis to the brain [268]. Stable expression of miR-1258 in metastatic cells inhibited heparanase expression and activity and diminished experimental metastasis to brain in vivo [269]. Moreover, isolation of circulating tumor cells from breast cancer patients and analysis of their protein signatures revealed that heparanase expression along with several other markers identified a population of circulating cells having a high probability of metastasizing to brain [270].
6.2. Shed syndecan-1 potentiates growth factor signaling that aids in establishing a supportive tumor microenvironment
Shedding of the transmembrane proteoglycan syndecan-1 from the surface of cells is elevated in many diseases and has a remarkable impact in tumor cell behavior [32, 271, 272]. Syndecan shedding is mediated by the action of a number of proteases that act at sites generally in the membrane-proximal region of the syndecan extracellular domain leading to release of an intact ectodomain with attached GAG (HS and CS) chains [273, 274]. Interestingly, heparanase also plays a role in increasing syndecan-1 shedding. In both myeloma and breast cancer, when heparanase expression was increased, syndecan-1 expression and shedding were substantially increased [217]. The increase was driven by heparanase-mediated stimulation of expression of sheddases MMP-9 and urokinase plasminogen activator and its receptor (uPA/uPAR) [275].
Because shed syndecan-1 retains its HS chains, it is free to bind to numerous effectors (growth factors, cytokines, chemokines and other HP-binding molecules) which can lead to diverse functional consequences both within the extracellular matrix and at the cell surface. These activities have been well-characterized within the myeloma tumor microenvironment where shed syndecan-1 potentiates the activity of factors such as VEGF and HGF [31, 258, 276]. Syndecan-1 shedding can influence FGF-2 mediated signaling in breast cancer cells. In the absence of shedding, syndecan-1 mediates FGF-2 signaling, but following induction of syndecan-1 shedding, FGF-2 signaling is mediated by the HSPG glypican-1 [277]. In breast cancer, shed syndecan-1 is derived predominantly from stromal fibroblasts that reside within the tumor [228]. This stromal-derived syndecan-1 stimulates breast cancer cell proliferation via activation of FGF-2 [272]. Together, these findings indicate differing roles exist for cell surface verses shed syndecan-1 in regulating breast cancer. This notion has been confirmed by other studies showing that shed syndecan-1 confers an invasive phenotype to breast cancer cells, whereas membrane syndecan-1 inhibits tumor cell invasion [229].
Interestingly, in addition to local interactions within the tumor microenvironment, shed syndecan-1 can regulate interactions with host cells that are distal to the tumor. When heparanase expression was enhanced in metastatic MDA-MD-231 breast cancer cells and these cells were implanted in the mammary fat pad of mice, a systemic bone resorption occurred even though tumor could not be detected within the bone [278]. This increased bone resorption was due to enhanced osteoclastogenesis stimulated, at least in part, by shed syndecan-1 released from the heparanase-expressing tumor cells growing in the mammary fat pad [279]. This suggests that the heparanase/syndecan-1 axis has broad impact on tumor-host behavior both within and beyond the immediate tumor microenvironment.
6.3. Heparanase and syndecans together regulate exosome secretion and composition
Exosomes are small (~30–100 nm) membrane vesicles that are produced within endosomal compartments and released at the cell surface. Following their release they can dock with recipient cells and deliver their cargo of signaling proteins, nucleic acids (DNA, mRNA and miRNA), carbohydrates and lipids thereby acting as powerful mediators of intercellular communication [280–282]. In cancer, this horizontal transfer of biological material can regulate the behavior of both tumor and host cells [283]. In addition to acting within the local tumor microenvironment, due to their small size, exosomes can escape the tumor, travel through the circulation and enter distal tissues where they can, for example, prepare metastatic niches prior to arrival of tumor cells [282, 283]. Emerging data also indicate that exosomes can act as barriers to anti-cancer therapy by interacting with tumor cells and enhancing their chemoresistance.
A number of publications over the last few years have begun to detail the impact of exosomes on breast cancer. Several of those indicate an important role for exosomes in breast cancer metastasis. For example, it was recently shown that breast cancer cell migration is stimulated by fibroblast-secreted exosomes that activate the protrusive activity and motility of breast cancer cells via Wnt-planer cell polarity signaling [284]. In vivo, when fibroblasts were co-injected with breast cancer cells, metastasis was dramatically enhanced and this was dependent on CD81, a well-known cargo present in exosomes. Breast cancer metastasis may also be mediated through miR-105, a microRNA found in breast cancer patients and associated with the occurrence of metastasis. Mechanistically, it was demonstrated that exosomes containing miR-105 carried by exosomes released from cancer cells target the tight junction protein ZO-1 [285]. This destroys the tight junctions of endothelial monolayers thereby compromising the integrity of this barrier and facilitating metastasis. Exosomes can also play an important regulatory role in breast cancer by enhancing chemoresistance. Exposure of drug-sensitive MCF-7 breast cancer cells to exosomes secreted by drug resistant variants of MCF-7 increased survival of the sensitive cells following their treatment with cytotoxic drugs [286]. This chemoresistant effect was traced to miR-100, miR-222 and miR-30a, a group of miRs previously associated with therapy failure. Additional studies have demonstrated a role for exosomal-delivered miRNAs in promoting resistance of breast cancer cells to docetaxel and tamoxifen [287, 288]. Interestingly, exosomes also play a role in dormancy of breast cancer within the bone marrow. This occurs through stroma-derived exosomes that deliver quiescence-inducing miRNAs to breast cancer cells [289].
Together, the studies above underscore the importance of understanding how exosome cargo and secretion are regulated. This is particularly important in cancer where it has been shown that the level of exosome secretion is significantly enhanced as tumors progress [290]. However, the mechanisms regulating exosome biogenesis are not well understood and may vary between cell types and within the context of their function [291]. There is considerable evidence that components of the Endosomal Sorting Complex Required for Transport (ESCRT) and members of the Rab family of GTPases play roles in mediating exosome secretion [292, 293]. In addition, there is emerging evidence that both syndecans and heparanase influence exosome secretion. Syndecans of MCF-7 breast cancer cells were recently shown to promote exosome formation through their binding to syntenin, a cytosolic adaptor protein [196]. Syntenin, through its LYPXX(n)L domains, also binds to ALIX, a component of the ESCRT machinery responsible for endosomal membrane budding and abscission. This syndecan-syntenin-ALIX complex segregates syndecans and their cargo (e.g., growth factors that are bound to syndecan HS chains) to budding endosomal membranes and supports the budding process resulting in formation of exosomes [196]. Interestingly, this syntenin-driven exosome formation is dependent on HS-mediated clustering of syndecans.
The finding that the status of HS influences exosome secretion raised the interesting possibility that physiologic modification of HS by heparanase would impact exosome secretion and molecular composition. This notion was confirmed by analysis of exosomes secreted by cells transfected with the cDNA for heparanase. In both myeloma and breast cancer cells, an elevation in heparanase expression led to a dramatic increase in exosome secretion [294]. This effect required the enzymatic activity of heparanase suggesting that exosome secretion was enhanced when syndecan-1 HS chains were remodeled by the enzyme. It is possible that heparanase-mediated shortening of the HS chains enhances formation of the syndecan-syntenin-ALIX complex thereby boosting the rate exosome formation. Enhanced heparanase expression in the tumor cells also led to alteration of the composition of the secreted exosomes including increased levels of heparanase, syndecan-1, HGF and VEGF [294]. This altered composition endowed these “heparanase exosomes” with an increased ability to promote tumor cell spreading and endothelial cell migration when compared to control exosomes. These findings indicate that as tumors progress and heparanase levels rise, it causes increased exosome secretion and alterations in exosome composition. This adds yet another mechanism whereby heparanase facilitates tumor-host crosstalk that helps drive aggressive tumor behavior and further validates heparanase as a target for anti-cancer therapy.
7. The role of Glypicans in breast cancer progression
7.1. The structure and function of glypicans
Glypicans are a family of proteoglycans that are linked to the plasma membrane through a GPI anchor [295]. Six members of the glypican family have been identified in mammals (glypican-1 to glypican-6) [295]. Structural features that are conserved across the family include the localization of 14 cysteine residues and of the insertion sites for GAG chains. All these insertion sites are close to the C-terminus, placing the GAG chains in proximity to the cell surface, and suggesting that these chains could mediate the interaction of glypicans with other cell surface proteins [295]. Most glypicans display HS chains. The number of GAG chains varies across the family (from two in glypican-3 to four in glypican-5), but the functional implications of this variation are unknown. Glypicans can be released from the cell surface by a lipase called Notum, which cleaves the GPI anchor [296]. These PGs can also be cleaved by furin-like convertases into two subunits that remain attached to each other by one or more disulfide bridges [297]. Notably, glypicans do not have domains with obvious homology to characterized domains found in other proteins, suggesting that they have unique functions. The crystal structure of glypican-1 lacking the GAG attachment domain has been recently reported [298]. The structure reveals that glypican-1 is a densely packed one-domain protein of cylindrical shape, consisting of 14 α-helices and three major loops.
Genetic and biochemical studies have demonstrated that glypicans can regulate several signaling pathways, including those triggered by Wnts [299–305], Hedgehogs (Hhs) [306–310], BMPs [311–314] and FGFs [315, 316]. In most cases this regulatory activity is based on the ability of glypicans to either inhibit or stimulate the interaction of these growth factors with their signaling receptors. It is now well established that the structural features of glypicans combine with the set of growth factors and growth factor receptors present in a given cell type to determine glypican function.
In addition to regulating signal reception at the cell membrane, glypicans have been shown to be involved in the secretion and/or transport of Hhs [307, 317–320]; Wnts [321–323], and BMPs [311, 312]. The studies that uncovered these functions have been mostly performed in the developing Drosophila wing. Similar functions of glypicans in a mammalian in vivo context remain to be demonstrated.
Glypicans also have specific functions in the nervous system. For example, glypicans have been shown to play a role in axon guidance [310, 324], and in the formation of excitatory synapses [325, 326].
7.2. Glypicans and breast cancer
It is well established that alterations of the signaling pathways regulated by glypicans contribute to malignant transformation. It is therefore not surprising that several studies have demonstrated that abnormal expression of members of the glypican family play a role in the progression of various tumor types, including breast cancer [308, 327–329].
The first study implicating a glypican in breast cancer progression reported the over-expression of glypican-1 in 10 out 20 tumors [330]. The levels of glypican-1 were assessed by Northern blot analysis. Notably, this study also showed that glypican-1 stimulates the mitogenic response of two breast cancer cell lines to -heparin binding epidermal growth factor (HB-EGF) and to FGF2, suggesting that the up-regulation of glypican-1 could play a role in breast cancer progression. It should be noted, however, that a more recent study of 23 breast tumor samples by qRT-PCR could not detect significant over-expression of glypican-1 [24].
The second investigation implicating glypicans in breast cancer progression showed a significant down-regulation of glypican-3 in the tumors compared to the surrounding non malignant tissue [329]. This study included 12 patients, and used in situ hybridization as a method to detect GPC3. The authors showed that the down-regulation of glypican-3 in breast cancer cell lines was due, at least in part, to the hypermethylation of the glypican-3 promoter. Furthermore, ectopic expression of glypican-3 inhibited the growth of eight out of ten breast cancer cell lines, suggesting that glypican-3 can act as an inhibitor of breast cancer growth [329]. The hypermethylation of the glypican-3 promoter in breast cancer was confirmed by a more extensive study that showed that this promoter was hypermethylated in 38 of 45 breast tumors [331]. Notably, this study reported that high levels of glypican-3 promoter methylation are more predominant in hormone receptor-negative patients. It should also be noted that the downregulation of glypican-3 in breast cancer has been recently confirmed by a study that included 23 patients [24]. Another investigation implicating glypican-3 in breast cancer showed that this glypican can inhibit experimental lung metastasis in a murine breast cancer cell line [332]. This finding is consistent with the previously reported glypican-3-induced inhibition of the growth of breast cancer cells.
Lastly, a recent study showed that glypican-6 stimulates the invasive migration of breast cancer cells [333]. This investigation also found that glypican-6 promotes invasiveness indirectly by stimulating Wnt5a expression leading to the activation of Jun N-terminal kinase (JNK) and p38 MAPK. It should be noted, however, that the authors of this study did not investigate whether glypican-6 is upregulated in breast cancer patients, and that a recent report found no difference in the glypican-6 mRNA levels of invasive breast cancer tissues compared to normal mammary gland [24].
Conclusively, the accumulated evidence strongly indicates that the glypican-3 is downregulated in most breast cancer patients, and that this down-regulation contributes to the progression of the disease. On the other hand, additional studies are required to confirm that the expression of glypican-1 and glypican-6 are deregulated in breast cancer, and that these glypicans play a role in this malignancy.
8. Serglycin: an inflammatory proteoglycan that is involved in tumorigenesis
Serglycin is the only characterized member of the family of intracellular PG and presents in intracellular secretory compartments. Serglycin is highly expressed in hematopoietic cells but recent studies demonstrated that it is also expressed by a variety of cell types and mediates crucial functions in both normal and pathological conditions [334]. The human serglycin gene is located in chromosome 10q.22. and consists of three exons. In human the small core protein of serglycin contains eight serine/glycine repeats, which are potential GAG attachment sites. The structure of serglycin differs between cell types due to variations of the number, the type and specific structure of GAGs attached on the core protein [334].
In hematopoietic cells serglycin is found in secretory granules and vesicles contributing in intracellular storage and secretion of bioactive molecules such as proteases, pore formation proteins, chemokines, growth factors and neurotransmitters. It has been shown that serglycin is secreted in the ECM in various cell types either constitutively or upon stimulation. In the ECM, serglycin forms complexes with bioactive molecules regulating their availability or transport to target sites [334].
8.1. Serglycin in inflammation
Serglycin is also synthesized by various stromal cells in tumor microenvironment for instance inflammatory cells, endothelial cells and activated fibroblasts [335, 336]. Serglycin is involved in the secretion of inflammatory mediators by these cells, which contribute to tumorigenesis [335, 336]. Serglycin plays crucial roles in the storage and secretion of various proteolytic enzymes in inflammatory cells but also regulates their functions upon secretion and may contribute to tumor progression. HP present on serglycin in mast cells forms complexes with chymase and promotes the binding of the enzyme to HP-binding substrates enhancing their proteolysis [337]. Furthermore, HP significantly blocks the inhibition of chymase by natural inhibitors such as α1-protease inhibitor, α1-antichymotrypsin, α2-macroglobulin and soybean trypsin inhibitor [338, 339]. HP present on serglycin is important for the formation of active tryptase tetramers [340, 341]. Chymase can activate various MMPs, whereas both tryptase and chymase directly degrade ECM components. Chymase cleaves vitronectin and procollagen, while tryptase degrades collagen type IV and both degrade fibronectin [334]. Serglycin is colocalized with MMP-13 in cytoplasmic granules in chondrocytes interacting with a fragment of MMP-13 that comprises the hinge and PEX domains [342]. Endogenous and exogenous added serglycin isolated from various sources forms complexes with the proform of MMP9 (proMMP9) in macrophages in vitro [343, 344]. The core protein interacts with both the hemopexin-like (PEX) domain and the fibronectin-like (FnII) module of proMMP-9. The formation of the complexes alters the mode of activation of proMMP9 and the interaction of the enzyme with its substrates [343, 345]. ProMMP-9 associated with PGs is activated in the presence of Ca2+ and it may be important for the activation of pro-enzyme in pathological situation such as breast cancer-induced bone disease [346].
8.2. Tumor-promoting role of serglycin in breast cancer
Serglycin is expressed in numerous human hematopoietic tumors including lymphoma, myeloma, mastocytoma, and thymoma but also in non-hematopoietic tumors [334]. Serglycin carrying CS side chains is highly expressed and constitutively secreted by multiple myeloma cells [347]. Serglycin levels are increased in bone marrow aspirates of patients with myeloma and inhibits bone mineralization through direct binding to hydroxyapatite, suggesting a potent correlation of serglycin accumulation with disease progression [347]. Serglycin knockdown in myeloma cells results in dramatically attenuated tumor growth in mice and impaired development of blood vessels, indicating that serglycin may affect tumor angiogenesis [348]. Serglycin is also localized on the cell surface of myeloma cells where it is attached through its CS-4S chains [347]. CD44 on myeloma cell surface may serve as a major ligand for serglycin promoting the adhesion of myeloma cells to collagen I and to bone marrow stromal cells [348, 349]. Binding of serglycin to collagen I enhances the biosynthesis and secretion of MMP2 and MMP9, which are involved in bone destruction [349].
Recently, increased expression of serglycin has been confirmed in nasopharyngeal and hepatocellular carcinoma. The elevated levels of serglycin in patients is correlated with unfavorable prognosis for overall survival and recurrence in nasopharyngeal cancer and for disease free and distant metastasis free survival in HCC [350, 351]. Serglycin secreted from metastatic nasopharyngeal carcinoma cells promotes EMT, motility, invasion, and metastasis [351]. Non-glycanated core protein of serglycin fails to induce cancer cell motility suggesting the involvement of GAG chains in tumor promoting properties of serglycin.
Serglycin is highly expressed in breast cancer tissues and cell lines [33]. The mRNA levels of serglycin are markedly up-regulated in aggressive breast cancer cells clustered into Basal B subgroup, which exhibit an EMT gene signature and resemble breast cancer stem cells being CD44highCD24low [33]. Basal-like breast cancers are correlated with increased risk of metastatic spread and poor patient prognosis. In contrast, serglycin is expressed in low levels in less aggressive subtypes of breast cancer cells [33]. Biochemical characterization of proteoglycans secreted by aggressive MDA-MB-231 breast cancer cells demonstrated that serglycin bearing CS chains is the major secreted proteoglycan and it is abundantly present in the cytoplasm and cell membrane showing both filamentous and granular distribution [33]. Serglycin promotes breast cancer cell anchorage-independent growth, migration and invasion when it is over-expressed in minimally invasive MCF-7 breast cancer cells. Interestingly, over-expression of a mutant form of serglycin lacking GAG attachment sites fails to induce breast cancer cell aggressiveness demonstrating that specific structure of CS-4S present on serglycin is important for its functions in breast cancer [33]. CHST11 gene that specifically mediates 4-O sulfation of CS is highly expressed in MDA-MB-231 breast cancer cells promoting their binding to P-selectin via CS-4S chains and facilitating the formation of metastasis [352]. It is also of great importance that CS-4 chains regulates the functional properties of proteolytic enzymes such as cathepsins, which are involved in ECM degradation and tumor metastases [8].
Serglycin also regulates immune system through its ability to inhibit complement system activity. Serglycin isolated from myeloma and breast cancer cells inhibits the classical and the lectin pathways of complement system via direct binding to C1q and MBL, respectively, and protects tumor cells from complement system attack [33, 353]. Only those CS-4S chains with a high proportion of 4-sulfated disaccharides interact efficiently with complement proteins [353]. CS-E and in a lower extent heparin compete with CS-4 chains of serglycin for binding to C1q, whereas only CS-E competes for binding to MBL. Binding of serglycin to C1q or/and C1 inhibits the cleavage of C4 in the classical pathway. In the lectin pathway, binding of serglycin to MBL either competes out MBL-associated proteases (MASPs) from the stalk region of MBL or sterically hinders cleavage of C2 and C4 by MASPs [353]. The inhibition of complement is a great limitation during immunotherapy against several types of cancer. These findings suggest a role for serglycin as a modulator of immune system response in tumor microenvironment.
9. Translational medicine: targeted therapeutic approaches based on the novel key roles of proteoglycans in breast cancer
Treating cancer poses a challenge because cancer cells have several inherent defense mechanisms. Not only do cancer cells originate from the host system, but they also use natural cellular metabolic pathways to grow. Additionally, due to the genetic errors that manifest cancer, tumors, including those of breast, are composed of heterogeneous populations of cells that respond differently to treatments and impart multi-drug resistance to tumors. In these cells, erroneous cellular machinery triggers abnormal signals, misinterpret incoming signals, and causes differentiation into several families of cancerous cells. The expanding repertoire of molecular interactions attributed to specific PGs emerges these molecules as powerful mediators that control a wide variety of processes and could represent novel therapeutic modalities against cancer as well as being targets themselves.
Importantly, most of these interactions are critically enhanced or inhibited by specific structural modules within GAG chains. Thus, therapeutics that target/modify specific PGs/GAGs will be able to attack cancer cells on multiple fronts because they can target their interactions such as growth factor binding, the coagulation cascade, proteinase activation and inhibition, heparanase and other GAG modifying enzymes activation and activity, and possibly tumor evolution/differentiation [354].
The use of modified GAGs or GAG mimetics to modulate GAG-protein interactions alone, or in conjunction with specific proteinases' exosites may introduce a new era in cancer therapeutics [8, 355]. One such approach could be the targeting of the exosites of specific cathepsins with negative charged inhibitors (such as poly-Asp and poly-Glu) with ionic properties similar to those of specific GAG moieties thereby modulating proteinase catalytic activities by interfering with the formation of cathepsin/GAG complexes [8]. It is possible to stimulate HS and CS biosynthesis by utilizing xylosides to prime GAG chains, however with no specific properties [356]. In another approach, it is possible to inhibit HS/CS biosynthesis by utilizing 4-deoxy-4-fluoro-xylosides [357]. Decreasing overall levels of HS and CS would affect HS/CS-matrix interactions and prevent tumor proliferation, invasion, metastasis, and angiogenesis by reducing for example FGF and VEGF signaling. Inhibition of HS production may also prevent heparanase activation and hence restrain heparanase activity by modulating the function of syndecans as the main mediators for heparanase uptake [358]. Preclinical and clinical studies have demonstrated that therapies targeting the heparanase/syndecan-1 axis hold promise for blocking the aggressive behavior of cancer since heparanase helps drive exosome secretion, alters exosome composition, and facilitates production of exosomes that impact both tumor and host cell behavior, thereby promoting tumor progression [31]. Notably, exosome secretion was markedly reduced by knocking down enzymes involved in HS synthesis or modification (EXT1/2 or NDST1/2) or by growing cells in the presence of heparitinase (heparinase III), a bacterial enzyme that degrades HS chains. Together these findings suggest that up or down regulation of syndecans in pathological processes could dramatically impact exosome formation and subsequent extent of intercellular communication. Similarly, this implies that therapeutic interventions designed to regulate the expression or abundance of syndecans could diminish the progression of diseases such as breast cancer. In addition to a role for HS in exosome formation, it was recently reported that HS on the surface of recipient cells plays an important role in exosome internalization [359]. It will be important to further explore this and to determine the full extent of HS function in the exosome docking and internalization process. Given the abundance of evidence that heparanase facilitates the progression of breast cancer, it will be important to eventually test heparanase inhibitors for their efficacy in breast cancer patients. Ongoing Phase I studies are now in progress testing three heparanase inhibitors including Roneparstat (SST0001) in myeloma patients [360], M402 in pancreatic cancer [361] and PG545 in patients with solid tumors [362, 363].
Many of the previous studies of cell surface PGs and cancer progression are correlative. Two questions arise: (1) are the tumor-related changes in syndecan and glypican expression and function merely a consequence of the process, or active participants and (2) do these PGs make a relevant target? Syndecans and glypicans as potential targets in the wider cancer field has been the subject of recent analysis [3, 364, 365] and they are attractive in part because they are accessible on the cell surface. Most attention has been paid to syndecan-1, and it is both the most abundant member of the family in breast carcinoma and evidence suggests it supports growth and progression. However, there are no reports on the impact of targeting the core protein in breast carcinoma models. Evidence from knock-out mice suggests there may be redundancy between syndecan family members, in breast cancer at least there appears to be considerable specificity. Our very recent work with the MDA-MB-231 cell line suggests that syndecan-2 should also be further considered. It is only this syndecan that controls the poorly adhesive, highly migratory and invasive phenotype of this highly malignant cell line and once removed, cells become adherent and less motile, even though alternate syndecans remain on the cell surface. Moreover, it was found that the simple expedient of adding HS or HP to these cells was sufficient to alter behavior through competition with cell surface HSPGs. It will be interesting to determine whether targeting the syndecan-2 gene in invasive breast carcinoma renders them less metastatic in murine models.
The treatment with already existed pharmaceutical formulations in several in vitro and in vivo biological systems, suggests that they can regulate the expression levels of syndecans and glypicans, thus inhibiting their carcinogenic potential. According to that notion, the third generation bisphosphonate, zoledronate (zoledronic acid, Zometa®) is shown to down-regulate the expression levels of syndecan-1 -2 and glypican-1, in contrast with the up-regulation of syndecan-4 in human breast cancer cells with different metastatic potentials [213]. This effect is associated with the inhibition of cell growth, migration, adhesion, and invasion in correlation with the diminished levels of ανβ3, ανβ5, and α5β1 integrins [213]. Similar mode of action has the specific tyrosine kinase inhibitor imatinib (Glivec®), which targets PDGFRs, c-Kit and Bcr-Abl. It exerts a significant inhibitory effect on the expression of syndecan-2 -4 and glypican-1 on PDGF-BB-treated breast cancer cells, leading to suppressed cell growth ability, migration, and invasion [366]. Recent studies focus on exploring therapeutically approaches that are associated with syndecans ectodomain. As a result, monoclonal antibodies or peptides, which target specifically extracellular domain of syndecans, have been evaluated. For example, B-B4 (iodine-131-labeled anti-syndecan-1 antibody) was administrated to myeloma patients with success, promoting the notion of targeted radioimmunotherapy (RIT) [367]. Interestingly, recent studies indicate the importance of B-B4 antibody not only in multiple myeloma but also in triple-negative breast cancer in combination with immune-PET imaging and RIT [368]. Another approach in syndecan targeting involves the use of small peptides. For example, Synstatin was developed to the sequence between 82 and 130 amino acids of syndecan-1 ectodomain. In detail, this peptide antagonizes syndecan-1 domain, responsible for capturing and activating ανβ3 or ανβ5 integrins and IGF-IR. Synstatin’s action prevents the formation of the receptor complex, and in turn blocks tumor-induced angiogenesis and metastasis mediated by the initial complex [369].
It may be optimistic to expect that targeting a single receptor on the cell surface can provide a new opportunity for treating breast cancer. Syndecans and glypicans do not operate in isolation, but function alongside other receptors, including integrins and growth factor receptors. Moreover, the interplay with estrogen receptors may provide further complexity [29]. However, cell surface PGs are certainly worth pursuing to determine if they are important contributors to tumor progression that make them a viable target alongside other treatment options.
Versican deposition in the tumor stroma is associated with cancer relapse and poor patient outcome in several cancer types, including breast cancer [3, 25]. HA-versican pericellular matrices of cancer cells may be potential targets for tumor therapy due to their well-documented implication in cancer metastasis. Disruption of the HA–CD44 interaction with HA oligomers may be used for targeting tumor progression making HA oligomers promising inhibitors of cancer dissemination [370]. Furthermore, a novel versican isoform V4 is highly expressed in breast cancer [36], whereas versican is also differentially glycosylated in breast cancer because it contains more sialic acid [40]. This alternative splice variant of versican or the presence of unusual glycosylation may comprise possible targets for therapeutic intervention in breast cancer with antibody-related agents.
SLRPs such as decorin and biglycan have established roles in cancer progression and metastasis and thus, they constitute potential therapeutic targets for breast cancer treatment [3, 8, 371]. Adenoviral-mediated gene delivery of decorin or the systemic administration of human recombinant decorin or decorin core protein to various tumor xenograft models (breast and prostate carcinomas) suppresses tumor growth [62–64, 96, 372]. The recent discovery that decorin is pro-inflammatory and interacts with TLRs [83], together with the induction of autophagy in endothelial cells [95] and mitophagy in breast cancer cells [20], indicates that decorin can affect both the tumor stroma and the tumor itself in a variety of ways. Decorin-evoked endothelial cell autophagy reveals important therapeutic targets for augmenting autophagy and combating tumor angiogenesis. Induction of autophagic programs by decorin (and related autophagic matrikines) may represent a mechanism for tumorigenic and angiogenic suppression or for quelling homeostatic imbalances relevant for human pathologies.
On the other hand, the fact that biglycan is involved in numerous signaling cascades that strongly impact tumorigenesis harbors a great potential for targeting this molecule in therapeutic approaches. There are no doubts about the importance of innate immunity and inflammation for tumor growth. In this context lack of data regarding biglycan/TLR2/4-mediated inflammation [154] in tumorigenesis is surprising (Fig. 2). It is predictable that in developing cancer soluble biglycan promotes tumor growth by creating a pro-inflammatory environment in the stroma. Therefore, inhibitors of SLRP/TLR binding sites could be presumably effective in suppressing tumor growth. In contrast, in established tumors soluble biglycan potentially contributes to tumor growth retardation by boosting inflammation [83]. Thus, there is an urgent need for studies elucidating pro-inflammatory effects of biglycan in various stages of tumorigenesis in order to translate this knowledge into new cancer treatments.
Table 1.
Correlation of proteoglycans with clinicopathological characteristics and disease outcome in breast cancer
| Proteoglycan / Enzyme |
Expression mode | Correlation (References) |
|---|---|---|
| Versican | High stromal expression | Increased risk and rate of relapse in node-negative invasive breast cancer [14, 17]. Increased tumor grade, invasive disease and presence of malignant appearing microcalcifications [16]. |
| Decorin | High stromal expression | Lower tumor grade [70], reduced tumor size, reduced risk and rate of relapse and poor survival in node-negative invasive breast cancer [15]. |
| High expression in malignant epithelial tissue | Higher number of positive lymph nodes, increased lymph node metastasis, lower disease free survival in breast cancer [70]. Decreased overall survival only in luminal B subtype tumors [70]. | |
| Syndecan-1 | High expression in cancer cells | High tumor grade [29, 220, 237], large tumor size [220, 237], lymph node metastasis [237], reduced disease-free survival [220, 237] and poor overall survival [220, 222, 223, 237]. |
| Loss of expression in cancer cells | High tumor grade and reduced relapse-free survival in invasive ductal breast cancer [225]. | |
| High stromal expression | Increased blood vessel density and total vessel area [232], high tumor grade [225] and reduced survival [223]. | |
| Syndecan-4 | High expression in cancer cells | High tumor grade [237]. |
| Glypican-1 | High expression in cancer cells | High tumor size [237]. |
| Heparanase | High expression in cancer cells | Higher VNPI score in ductal in situ carcinoma [262], high grade [261], lymph node metastasis [260, 261, 263], tumor size [260, 263], clinical stage [263], reduced relapse-free and overall survival [263]. |
Highlights.
The biosynthesis of proteoglycans is dysregulated in breast cancer
Proteoglycans affect cancer cell signaling and phenotype
Targeting proteoglycans and modifying enzymes may provide novel therapeutic approaches
Acknowledgements
This research has been co-financed by the European Union (European Social Fund — ESF) and Greek National Funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) Research Funding Program: Thales. Investing in knowledge society through the European Social Fund.
This work was also supported in part by National Institutes of Health Grants RO1 CA39481, RO1 CA47282, RO1 CA164462 (R.V.I.) and Mizutani Foundation for Glycosciences (A.D.T.).
Original research on SLRP biology in the authors’ laboratories was supported by the German Research Council (SFB 815, project A5, SFB 1039, project B2, Excellence Cluster ECCPS to L.S.), LOEWE program Ub-Net (L.S.).
Support from the Danish Natural Science Research Council, Novo Nordisk Foundation, Lundbeck Foundation (J.R.C.) and Canadian Institute of Health Research (J.F.) to the authors is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Allred DC. Issues and updates: evaluating estrogen receptor-alpha, progesterone receptor, and HER2 in breast cancer. Mod Pathol. 2010;23(Suppl 2):S52–S59. doi: 10.1038/modpathol.2010.55. [DOI] [PubMed] [Google Scholar]
- 2.Theocharis AD, Gialeli C, Hascall VC, Karamanos NK. Extracellular matrix: a functional scaffold. In: Karamanos NK, editor. Extracellular matrix: pathobiology and signaling. Berlin/Boston: Walter de Gruyter GmbH & Co. KG; 2012. pp. 3–19. [Google Scholar]
- 3.Theocharis AD, Skandalis SS, Tzanakakis GN, Karamanos NK. Proteoglycans in health and disease: novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J. 2010;277:3904–3923. doi: 10.1111/j.1742-4658.2010.07800.x. [DOI] [PubMed] [Google Scholar]
- 4.Vigetti D, Viola M, Karousou E, De Luca G, Passi A. Metabolic control of hyaluronan synthases. Matrix Biol. 2014;35:8–13. doi: 10.1016/j.matbio.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, Pavao MS, Tzanakakis GN, Karamanos NK. Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J. 2012;279:1177–1197. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 6.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer L, Schaefer RM. Proteoglycans: from structural compounds to signaling molecules. Cell Tissue Res. 2010;339:237–246. doi: 10.1007/s00441-009-0821-y. [DOI] [PubMed] [Google Scholar]
- 8.Theocharis AD, Gialeli C, Bouris P, Giannopoulou E, Skandalis SS, Aletras AJ, Iozzo RV, Karamanos NK. Cell-matrix interactions: focus on proteoglycan-proteinase interplay and pharmacological targeting in cancer. FEBS J. 2014;281:5023–5042. doi: 10.1111/febs.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 2009;1:a002493. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson JE, Schor AM, Howell A, Ferguson MW. Changes in the extracellular matrix of the normal human breast during the menstrual cycle. Cell Tissue Res. 1992;268:167–177. doi: 10.1007/BF00338066. [DOI] [PubMed] [Google Scholar]
- 11.de Lima CR, de Arimatea dos Santos Junior J, Nazario AC, Michelacci YM. Changes in glycosaminoglycans and proteoglycans of normal breast and fibroadenoma during the menstrual cycle. Biochim Biophys Acta. 2012;1820:1009–1019. doi: 10.1016/j.bbagen.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Skandalis SS, Kletsas D, Kyriakopoulou D, Stavropoulos M, Theocharis DA. The greatly increased amounts of accumulated versican and decorin with specific post-translational modifications may be closely associated with the malignant phenotype of pancreatic cancer. Biochim Biophys Acta. 2006;1760:1217–1225. doi: 10.1016/j.bbagen.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Theocharis AD, Vynios DH, Papageorgakopoulou N, Skandalis SS, Theocharis DA. Altered content composition and structure of glycosaminoglycans and proteoglycans in gastric carcinoma. Int J Biochem Cell Biol. 2003;35:376–390. doi: 10.1016/s1357-2725(02)00264-9. [DOI] [PubMed] [Google Scholar]
- 14.Ricciardelli C, Brooks JH, Suwiwat S, Sakko AJ, Mayne K, Raymond WA, Seshadri R, LeBaron RG, Horsfall DJ. Regulation of stromal versican expression by breast cancer cells and importance to relapse-free survival in patients with node-negative primary breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:1054–1060. [PubMed] [Google Scholar]
- 15.Troup S, Njue C, Kliewer EV, Parisien M, Roskelley C, Chakravarti S, Roughley PJ, Murphy LC, Watson PH. Reduced expression of the small leucine-rich proteoglycans, lumican, and decorin is associated with poor outcome in node-negative invasive breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:207–214. [PubMed] [Google Scholar]
- 16.Skandalis SS, Labropoulou VT, Ravazoula P, Likaki-Karatza E, Dobra K, Kalofonos HP, Karamanos NK, Theocharis AD. Versican but not decorin accumulation is related to malignancy in mammographically detected high density and malignant-appearing microcalcifications in non-palpable breast carcinomas. BMC cancer. 2011;11:314. doi: 10.1186/1471-2407-11-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suwiwat S, Ricciardelli C, Tammi R, Tammi M, Auvinen P, Kosma VM, LeBaron RG, Raymond WA, Tilley WD, Horsfall DJ. Expression of extracellular matrix components versican, chondroitin sulfate, tenascin, and hyaluronan, and their association with disease outcome in node-negative breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:2491–2498. doi: 10.1158/1078-0432.ccr-03-0146. [DOI] [PubMed] [Google Scholar]
- 18.Prinz RD, Willis CM, Viloria-Petit A, Kluppel M. Elimination of breast tumor-associated chondroitin sulfate promotes metastasis. Genet Mol Res. 2011;10:3901–3913. doi: 10.4238/2011.December.8.9. [DOI] [PubMed] [Google Scholar]
- 19.Neill T, Painter H, Buraschi S, Owens RT, Lisanti MP, Schaefer L, Iozzo RV. Decorin antagonizes the angiogenic network: concurrent inhibition of Met, hypoxia inducible factor 1alpha, vascular endothelial growth factor A, and induction of thrombospondin-1 and TIMP3. The Journal of biological chemistry. 2012;287:5492–5506. doi: 10.1074/jbc.M111.283499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neill T, Torres A, Buraschi S, Owens RT, Hoek JB, Baffa R, Iozzo RV. Decorin induces mitophagy in breast carcinoma cells via peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha) and mitostatin. The Journal of biological chemistry. 2014;289:4952–4968. doi: 10.1074/jbc.M113.512566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frey H, Schroeder N, Manon-Jensen T, Iozzo RV, Schaefer L. Biological interplay between proteoglycans and their innate immune receptors in inflammation. FEBS J. 2013;280:2165–2179. doi: 10.1111/febs.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh LT, Nastase MV, Zeng-Brouwers J, Iozzo RV, Schaefer L. Soluble biglycan as a biomarker of inflammatory renal diseases. Int J Biochem Cell Biol. 2014;54:223–235. doi: 10.1016/j.biocel.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreth K, Frey H, Hubo M, Zeng-Brouwers J, Nastase MV, Hsieh LT, Haceni R, Pfeilschifter J, Iozzo RV, Schaefer L. Biglycan-triggered TLR-2- and TLR-4-signaling exacerbates the pathophysiology of ischemic acute kidney injury. Matrix Biol. 2014;35:143–151. doi: 10.1016/j.matbio.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Vega I, Garcia O, Crespo A, Castanon S, Menendez P, Astudillo A, Quiros LM. Specific genes involved in synthesis and editing of heparan sulfate proteoglycans show altered expression patterns in breast cancer. BMC Cancer. 2013;13:24. doi: 10.1186/1471-2407-13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J. Cell. Mol. Med. 2011;15:1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kousidou O, Berdiaki A, Kletsas D, Zafiropoulos A, Theocharis AD, Tzanakakis GN, Karamanos NK. Estradiol-estrogen receptor: a key interplay of the expression of syndecan-2 and metalloproteinase-9 in breast cancer cells. Mol Oncol. 2008;2:223–232. doi: 10.1016/j.molonc.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skandalis SS, Afratis N, Smirlaki G, Nikitovic D, Theocharis AD, Tzanakakis GN, Karamanos NK. Cross-talk between estradiol receptor and EGFR/IGF-IR signaling pathways in estrogen-responsive breast cancers: focus on the role and impact of proteoglycans. Matrix Biol. 2014;35:182–193. doi: 10.1016/j.matbio.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Tsonis AI, Afratis N, Gialeli C, Ellina MI, Piperigkou Z, Skandalis SS, Theocharis AD, Tzanakakis GN, Karamanos NK. Evaluation of the coordinated actions of estrogen receptors with epidermal growth factor receptor and insulin-like growth factor receptor in the expression of cell surface heparan sulfate proteoglycans and cell motility in breast cancer cells. FEBS J. 2013;280:2248–2259. doi: 10.1111/febs.12162. [DOI] [PubMed] [Google Scholar]
- 29.Lendorf ME, Manon-Jensen T, Kronqvist P, Multhaupt HA, Couchman JR. Syndecan-1 and Syndecan-4 are independent indicators in breast carcinoma. J Histochem Cytochem. 2011;59:615–629. doi: 10.1369/0022155411405057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim HC, Couchman JR. Syndecan-2 regulation of morphology in breast carcinoma cells is dependent on RhoGTPases. Biochim Biophys Acta. 2014;1840:2482–2490. doi: 10.1016/j.bbagen.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Ramani VC, Purushothaman A, Stewart MD, Thompson CA, Vlodavsky I, Au JL, Sanderson RD. The heparanase/Syndecan-1 axis in cancer: mechanisms and therapies. FEBS J. 2013;280:2294–2306. doi: 10.1111/febs.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanderson RD, Couchman JR. Targeting syndecan shedding in cancer. In: Karamanos NK, editor. Extracellular Matrix: Pathobiology and Signaling. de Gruyter GmbH; 2012. pp. 802–812. [Google Scholar]
- 33.Korpetinou A, Skandalis SS, Moustakas A, Happonen KE, Tveit H, Prydz K, Labropoulou VT, Giannopoulou E, Kalofonos HP, Blom AM, Karamanos NK, Theocharis AD. Serglycin is implicated in the promotion of aggressive phenotype of breast cancer cells. PloS one. 2013;8:e78157. doi: 10.1371/journal.pone.0078157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Theocharis AD. Versican in health and disease. Connect Tissue Res. 2008;49:230–234. doi: 10.1080/03008200802147571. [DOI] [PubMed] [Google Scholar]
- 35.Labropoulou VT, Theocharis AD, Ravazoula P, Perimenis P, Hjerpe A, Karamanos NK, Kalofonos HP. Versican but not decorin accumulation is related to metastatic potential and neovascularization in testicular germ cell tumours. Histopathology. 2006;49:582–593. doi: 10.1111/j.1365-2559.2006.02558.x. [DOI] [PubMed] [Google Scholar]
- 36.Kischel P, Waltregny D, Dumont B, Turtoi A, Greffe Y, Kirsch S, De Pauw E, Castronovo V. Versican overexpression in human breast cancer lesions: known and new isoforms for stromal tumor targeting. Int J Cancer. 2010;126:640–650. doi: 10.1002/ijc.24812. [DOI] [PubMed] [Google Scholar]
- 37.Skandalis SS, Theocharis AD, Papageorgakopoulou N, Vynios DH, Theocharis DA. The increased accumulation of structurally modified versican and decorin is related with the progression of laryngeal cancer. Biochimie. 2006;88:1135–1143. doi: 10.1016/j.biochi.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Theocharis AD. Human colon adenocarcinoma is associated with specific post-translational modifications of versican and decorin. Biochim Biophys Acta. 2002;1588:165–172. doi: 10.1016/s0925-4439(02)00161-8. [DOI] [PubMed] [Google Scholar]
- 39.Tsara ME, Theocharis AD, Theocharis DA. Compositional and structural alterations of proteoglycans in human rectum carcinoma with special reference to versican and decorin. Anticancer Res. 2002;22:2893–2898. [PubMed] [Google Scholar]
- 40.Tian Y, Esteva FJ, Song J, Zhang H. Altered expression of sialylated glycoproteins in breast cancer using hydrazide chemistry and mass spectrometry. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.011403. M111 011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikitovic D, Zafiropoulos A, Katonis P, Tsatsakis A, Theocharis AD, Karamanos NK, Tzanakakis GN. Transforming growth factor-beta as a key molecule triggering the expression of versican isoforms v0 and v1, hyaluronan synthase-2 and synthesis of hyaluronan in malignant osteosarcoma cells. IUBMB Life. 2006;58:47–53. doi: 10.1080/15216540500531713. [DOI] [PubMed] [Google Scholar]
- 42.Yeung TL, Leung CS, Wong KK, Samimi G, Thompson MS, Liu J, Zaid TM, Ghosh S, Birrer MJ, Mok SC. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013;73:5016–5028. doi: 10.1158/0008-5472.CAN-13-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dondeti VR, Wubbenhorst B, Lal P, Gordan JD, D'Andrea K, Attiyeh EF, Simon MC, Nathanson KL. Integrative genomic analyses of sporadic clear cell renal cell carcinoma define disease subtypes and potential new therapeutic targets. Cancer Res. 2012;72:112–121. doi: 10.1158/0008-5472.CAN-11-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keire PA, Bressler SL, Lemire JM, Edris B, Rubin BP, Rahmani M, McManus BM, van de Rijn M, Wight TN. A role for versican in the development of leiomyosarcoma. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.607168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wasa J, Nishida Y, Shinomura T, Isogai Z, Futamura N, Urakawa H, Arai E, Kozawa E, Tsukushi S, Ishiguro N. Versican V1 isoform regulates cell-associated matrix formation and cell behavior differentially from aggrecan in Swarm rat chondrosarcoma cells. Int J Cancer. 2012;130:2271–2281. doi: 10.1002/ijc.26230. [DOI] [PubMed] [Google Scholar]
- 46.Havre PA, Dang LH, Ohnuma K, Iwata S, Morimoto C, Dang NH. CD26 expression on Tanaplastic large cell lymphoma (ALCL) line Karpas 299 is associated with increased expression of versican and MT1-MMP and enhanced adhesion. BMC cancer. 2013;13:517. doi: 10.1186/1471-2407-13-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao D, Joshi N, Choi H, Ryu S, Hahn M, Catena R, Sadik H, Argani P, Wagner P, Vahdat LT, Port JL, Stiles B, Sukumar S, Altorki NK, Rafii S, Mittal V. Myeloid progenitor cells in the premetastatic lung promote metastases by inducing mesenchymal to epithelial transition. Cancer Res. 2012;72:1384–1394. doi: 10.1158/0008-5472.CAN-11-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Takahashi H, Lin WW, Descargues P, Grivennikov S, Kim Y, Luo JL, Karin M. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. 2009;457:102–106. doi: 10.1038/nature07623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D, Wang X, Wu JL, Quan WQ, Ma L, Yang F, Wu KY, Wan HY. Tumor-produced versican V1 enhances hCAP18/LL-37 expression in macrophages through activation of TLR2 and vitamin D3 signaling to promote ovarian cancer progression in vitro. PloS one. 2013;8:e56616. doi: 10.1371/journal.pone.0056616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheeren FA, Kuo AH, van Weele LJ, Cai S, Glykofridis I, Sikandar SS, Zabala M, Qian D, Lam JS, Johnston D, Volkmer JP, Sahoo D, van de Rijn M, Dirbas FM, Somlo G, Kalisky T, Rothenberg ME, Quake SR, Clarke MF. A cell-intrinsic role for TLR2-MYD88 in intestinal and breast epithelia and oncogenesis. Nat Cell Biol. 2014;16:1238–1248. doi: 10.1038/ncb3058. [DOI] [PubMed] [Google Scholar]
- 51.Xia L, Huang W, Tian D, Zhang L, Qi X, Chen Z, Shang X, Nie Y, Wu K. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology. 2014;59:958–973. doi: 10.1002/hep.26735. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi Y, Kuwabara H, Yoneda M, Isogai Z, Tanigawa N, Shibayama Y. Versican G1 and G3 domains are upregulated and latent transforming growth factor-beta binding protein-4 is downregulated in breast cancer stroma. Breast Cancer. 2012;19:46–53. doi: 10.1007/s12282-011-0264-7. [DOI] [PubMed] [Google Scholar]
- 53.Du WW, Fang L, Yang W, Sheng W, Zhang Y, Seth A, Yang BB, Yee AJ. The role of versican G3 domain in regulating breast cancer cell motility including effects on osteoblast cell growth and differentiation in vitro - evaluation towards understanding breast cancer cell bone metastasis. BMC cancer. 2012;12:341. doi: 10.1186/1471-2407-12-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du WW, Yang BB, Shatseva TA, Yang BL, Deng Z, Shan SW, Lee DY, Seth A, Yee AJ. Versican G3 promotes mouse mammary tumor cell growth, migration, and metastasis by influencing EGF receptor signaling. PloS one. 2010;5:e13828. doi: 10.1371/journal.pone.0013828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du WW, Yang BB, Yang BL, Deng Z, Fang L, Shan SW, Jeyapalan Z, Zhang Y, Seth A, Yee AJ. Versican G3 domain modulates breast cancer cell apoptosis: a mechanism for breast cancer cell response to chemotherapy and EGFR therapy. PloS one. 2011;6:e26396. doi: 10.1371/journal.pone.0026396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du WW, Fang L, Yang X, Sheng W, Yang BL, Seth A, Zhang Y, Yang BB, Yee AJ. The role of versican in modulating breast cancer cell self-renewal. Mol Cancer Res. 2013;11:443–455. doi: 10.1158/1541-7786.MCR-12-0461. [DOI] [PubMed] [Google Scholar]
- 57.Arslan F, Bosserhoff AK, Nickl-Jockschat T, Doerfelt A, Bogdahn U, Hau P. The role of versican isoforms V0/V1 in glioma migration mediated by transforming growth factor-beta2. Br J Cancer. 2007;96:1560–1568. doi: 10.1038/sj.bjc.6603766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morrione A, Neill T, Iozzo RV. Dichotomy of decorin activity on the insulin-like growth factor-I system. FEBS J. 2013;280:2138–2149. doi: 10.1111/febs.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neill T, Schaefer L, Iozzo RV. Decorin, a guardian from the matrix. Am. J. Pathol. 2012;181:380–387. doi: 10.1016/j.ajpath.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morcavallo A, Buraschi S, Xu S-Q, Belfiore A, Schaefer L, Iozzo RV, Morrione A. Decorin differentially modulates the activity of insulin receptor isoform A ligands. Matrix Biol. 2014;35:82–90. doi: 10.1016/j.matbio.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldoni S, Seidler DG, Heath J, Fassan M, Baffa R, Thakur ML, Owens RT, McQuillan DJ, Iozzo RV. An antimetastatic role for decorin in breast cancer. Am J Pathol. 2008;173:844–855. doi: 10.2353/ajpath.2008.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu Y, Sun H, Owens RT, Wu J, Chen YQ, Berquin IM, Perry D, O'Flaherty JT, Edwards IJ. Decorin suppresses prostate tumor growth through inhibition of epidermal growth factor and androgen receptor pathways. Neoplasia. 2009;11:1042–1053. doi: 10.1593/neo.09760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reed CC, Gauldie J, Iozzo RV. Suppression of tumorigenicity by adenovirus-mediated gene transfer of decorin. Oncogene. 2002;21:3688–3695. doi: 10.1038/sj.onc.1205470. [DOI] [PubMed] [Google Scholar]
- 64.Reed CC, Waterhouse A, Kirby S, Kay P, Owens RT, McQuillan DJ, Iozzo RV. Decorin prevents metastatic spreading of breast cancer. Oncogene. 2005;24:1104–1110. doi: 10.1038/sj.onc.1208329. [DOI] [PubMed] [Google Scholar]
- 65.Seidler DG, Goldoni S, Agnew C, Cardi C, Thakur ML, Owens RA, McQuillan DJ, Iozzo RV. Decorin protein core inhibits in vivo cancer growth and metabolism by hindering epidermal growth factor receptor function and triggering apoptosis via caspase-3 activation. J. Biol. Chem. 2006;281:26408–26418. doi: 10.1074/jbc.M602853200. [DOI] [PubMed] [Google Scholar]
- 66.Bi X, Pohl NM, Qian Z, Yang GR, Gou Y, Guzman G, Kajdacsy-Balla A, Iozzo RV, Yang W. Decorin-mediated inhibition of colorectal cancer growth and migration is associated with E-cadherin in vitro and in mice. Carcinogenesis. 2012;33:326–330. doi: 10.1093/carcin/bgr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bi X, Tong C, Dockendorff A, Bancroft L, Gallagher L, Guzman G, Iozzo RV, Augenlicht LH, Yang W. Genetic deficiency of decorin causes intestinal tumor formation through disruption of intestinal cell maturation. Carcinogenesis. 2008;29:1435–1440. doi: 10.1093/carcin/bgn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iozzo RV, Chakrani F, Perrotti D, McQuillan DJ, Skorski T, Calabretta B, Eichstetter I. Cooperative action of germline mutations in decorin and p53 accelerates lymphoma tumorigenesis. Proc. Natl. Acad. Sci. USA. 1999;96:3092–3097. doi: 10.1073/pnas.96.6.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horvath Z, Kovalszky I, Fullar A, Kiss K, Schaff Z, Iozzo RV, Baghy K. Decorin deficiency promotes hepatic carcinogenesis. Matrix Biol. 2014;35:194–205. doi: 10.1016/j.matbio.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cawthorn TR, Moreno JC, Dharsee M, Tran-Thanh D, Ackloo S, Zhu PH, Sardana G, Chen J, Kupchak P, Jacks LM, Miller NA, Youngson BJ, Iakovlev V, Guidos CJ, Vallis KA, Evans KR, McCready D, Leong WL, Done SJ. Proteomic analyses reveal high expression of decorin and endoplasmin (HSP90B1) are associated with breast cancer metastasis and decreased survival. PloS one. 2012;7:e30992. doi: 10.1371/journal.pone.0030992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson PS, Huang TF, Kazam E, Iozzo RV, Birk DE, Soslowsky LJ. Influence of decorin and biglycan on mechanical properties of multiple tendons in knockout mice. J. Biomechanical Eng. 2005;127:181–185. doi: 10.1115/1.1835363. [DOI] [PubMed] [Google Scholar]
- 72.Dunkman AA, Buckley MR, Mienaltowski MJ, Adams SM, Thomas SJ, Satchell L, Kumar A, Pathmanathan L, Beason DP, Iozzo RV, Birk DE, Soslowsky LJ. Decorin expression is important for age-related changes in tendon structure and mechanical properties. Matrix Biol. 2013;32:3–13. doi: 10.1016/j.matbio.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reese SP, Underwood CJ, Weiss JA. Effects of decorin proteoglycan on fibrillogenesis, ultrastructure, and mechanics of type I collagen gels. Matrix Biol. 2013;32:414–423. doi: 10.1016/j.matbio.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen S, Young MF, Chakravarti S, Birk DE. Interclass small leucine-rich repeat proteoglycan interactions regulate collagen fibrillogenesis and corneal stromal assembly. Matrix Biol. 2014;35:103–111. doi: 10.1016/j.matbio.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown EL, Wooten RM, Johnson BJ, Iozzo RV, Smith A, Dolan MC, Guo BP, Weis JJ, Hook M. Resistance to Lyme disease in decorin-deficient mice. The Journal of clinical investigation. 2001;107:845–852. doi: 10.1172/JCI11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brandan E, Gutierrez J. Role of skeletal muscle proteoglycans during myogenesis. Matrix Biol. 2013;32:289–297. doi: 10.1016/j.matbio.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 77.Duncan MB. Extracellular matrix transcriptome dynamics in hepatocellular carcinoma. Matrix Biol. 2013;32:393–398. doi: 10.1016/j.matbio.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 78.Nikolovska K, Renke JK, Jungmann O, Grobe K, Iozzo RV, Zamfir AD, Seidler DG. A decorin-deficient matrix affects skin chondroitin/dermatan sulfate levels and keratinocyte function. Matrix Biol. 2014;35:91–102. doi: 10.1016/j.matbio.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu Z, Horgan CE, Carr O, Owens RT, Iozzo RV, Lechner BE. Biglycan and decorin differentially regulate signaling in the fetal membranes. Matrix Biol. 2014;35:266–275. doi: 10.1016/j.matbio.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zoeller JJ, Pimtong W, Corby H, Goldoni S, Iozzo AE, Owens RT, Ho S-Y, Iozzo RV. A central role for decorin during vertebrate convergent extension. J. Biol. Chem. 2009;284:11728–11737. doi: 10.1074/jbc.M808991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ichii M, Frank MB, Iozzo RV, Kincade PW. The canonical Wnt pathway shapes niches supportive of hematopoietic stem/progenitor cells. Blood. 2012;119:1683–1692. doi: 10.1182/blood-2011-07-369199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng-Brouwers J, Beckmann J, Nastase MV, Iozzo RV, Schaefer L. De novo expression of circulating biglycan evokes an innate inflammatory tissue response via MyD88/TRIF pathways. Matrix Biol. 2014;35:132–142. doi: 10.1016/j.matbio.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhao JG, Lemarchand P, Pfeilschifter J, Schaefer RM, Iozzo RV, Schaefer L. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA-21. Sci Signal. 2011;4:ra75. doi: 10.1126/scisignal.2001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Neill T, Schaefer L, Iozzo RV. Instructive roles of extracellular matrix on autophagy. Am. J. Pathol. 2014;184:2146–2153. doi: 10.1016/j.ajpath.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koren I, Kimchi A. Promoting tumorigenesis by suppressing autophagy. Science. 2012;338:889–890. doi: 10.1126/science.1230577. [DOI] [PubMed] [Google Scholar]
- 88.Chen P, Cescon M, Bonaldo P. Collagen VI in cancer and its biological mechanisms. Trends Mol. Med. 2013;19:410–417. doi: 10.1016/j.molmed.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 89.Bonaldo P, Sandri M. Cellular and molecular mechanisms of muscle atrophy. Disease Models Mechan. 2013;6:25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goyal A, Pal N, Concannon M, Paulk M, Doran M, Poluzzi C, Sekiguchi K, Whitelock JM, Neill T, Iozzo RV. Endorepellin, the angiostatic module of perlecan, interacts with both the α2β1 integrin and vascular endothelial growth factor receptor 2 (VEGFR2) J. Biol. Chem. 2011;286:25947–25962. doi: 10.1074/jbc.M111.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goyal A, Poluzzi C, Willis AC, Smythies J, Shellard A, Neill T, Iozzo RV. Endorepellin affects angiogenesis by antagonizing diverse VEGFR2- evoked signaling pathways: transcriptional repression of HIF-1α and VEGFA and concurrent inhibition of NFAT1 activation. J. Biol. Chem. 2012;287:43543–43556. doi: 10.1074/jbc.M112.401786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hood BL, Lucas DA, Kim G, Chan KC, Blonder J, Issaq HJ, Veenstra TD, Conrads TP, Pollet I, Karsan A. Quantitative analysis of the low molecular weight serum proteome using (18)O stable isotope labeling in a lung tumor xenograft mouse model. J. Am. Soc. Mass Spectrom. 2005;16:1221–1230. doi: 10.1016/j.jasms.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 93.Morselli E, Galluzzi L, Kepp O, Marino G, Michaud M, Vitale I, Maiuri MC, Kroemer G. Oncosuppressive functions of autophagy. Antioxid. Redox. Signal. 2011;14:2251–2269. doi: 10.1089/ars.2010.3478. [DOI] [PubMed] [Google Scholar]
- 94.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Kepp O, Scoazec M, Mignot G, Rello-Varona S, Tailler M, Menger L, Vacchelli E, Galluzzi L, Ghiringhelli F, diVirgilio F, Zitvogel L, Kroemer G. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 95.Buraschi S, Neill T, Goyal A, Poluzzi C, Smythies J, Owens RT, Schaefer L, Torres A, Iozzo RV. Decorin causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci U S A. 2013;110:E2582–E2591. doi: 10.1073/pnas.1305732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Buraschi S, Neill T, Owens RT, Iniguez LA, Purkins G, Vadigepalli R, Evans B, Schaefer L, Peiper SC, Wang ZX, Iozzo RV. Decorin protein core affects the global gene expression profile of the tumor microenvironment in a triple-negative orthotopic breast carcinoma xenograft model. PloS one. 2012;7:e45559. doi: 10.1371/journal.pone.0045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Poluzzi C, Casulli J, Goyal A, Mercer TJ, Neill T, Iozzo RV. Endorepellin evokes autophagy in endothelial cells. J. Biol. Chem. 2014;289:16114–16128. doi: 10.1074/jbc.M114.556530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Willis CD, Poluzzi C, Mongiat M, Iozzo RV. Endorepellin laminin-like globular repeat 1/2 domains bind Ig3–5 of vascular endothelial growth factor(VEGF) receptor 2 and block pro-angiogenic signaling by VEGFA in endothelial cells. FEBS J. 2013;280:2271–2294. doi: 10.1111/febs.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nguyen TMB, Subramanian IV, Xiao X, Ghosh G, Nguyen P, Kelekar A, Ramakrishnan S. Endostatin induces autophagy in endothelial cells by modulating Beclin 1 and β-catenin levels. J. Cell. Mol. Med. 2009;13:3687–3698. doi: 10.1111/j.1582-4934.2009.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nguygen TMB, Subramanian IV, Kelekar A, Ramakrishnan S. Kringle 5 of human plasminogen, an angiogenesis inhibitor, induces both autophagy and apoptotic death in endothelial cells. Blood. 2007;109:4793–4802. doi: 10.1182/blood-2006-11-059352. [DOI] [PubMed] [Google Scholar]
- 101.Urciuolo A, Quarta A, Morbidoni V, gattazzo F, Molon S, Grumati P, Montemurro F, Tedesco FS, Blaauw B, Cossu G, Vozzi G, Rando TA, Bonaldo P. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun. 2013;4:1964. doi: 10.1038/ncomms2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Irwin WA, Bergamin N, Sabatelli P, Reggiani C, Megighian A, Merlini L, Braghetta P, Columbaro M, Volpin D, Bressan GM, Bernardi P, Bonaldo P. Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat. Genet. 2003;35:367–371. doi: 10.1038/ng1270. [DOI] [PubMed] [Google Scholar]
- 103.Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, Blaauw B, Urciuolo A, Tiepolo T, Merlini L, Maraldi NM, Bernardi P, Sandri M, Bonaldo P. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med. 2011;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 104.Grumati P, Bonaldo P. Autophagy in skeletal muscle homeostasis and in muscular dystrophies. Cells. 2012;1:325–345. doi: 10.3390/cells1030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carmignac V, Svensson M, Kφrner Z, Elowsson L, Matsumura C, Gawlik KI, Allamand V, Durbeej M. Autophagy is increased in laminin α2 chain-deficient muscle and its inhibition improves muscle morphology in a mouse model of MDC1A. Human Mol. Gen. 2011;20:4891–4902. doi: 10.1093/hmg/ddr427. [DOI] [PubMed] [Google Scholar]
- 106.Buraschi S, Neill T, Owens RT, Iniguez LA, Purkins G, Vadigepalli R, Evans B, Schaefer L, Peiper SC, Wang Z, Iozzo RV. Decorin protein core affects the global gene expression profile of the tumor microenvironment in a triple-negative orthotopic breast carcinoma xenograft model. PloS one. 2012;7:e45559. doi: 10.1371/journal.pone.0045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li L-L, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, Barton SC, Ishino F, Surani MA. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nature Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- 108.Yamaguchi A, Taniguchi M, Hori O, Ogawa S, Tojo N, Matsuoka N, Miyake S-i, Kasai K, Sugimoto H, Tamatani M, Yamashita T, Yamashita T, Tohyama M. Peg3/Pw1 is involved in p53-mediated cell death pathway in brain ischemia/hypoxia. J. Biol. Chem. 2002;277:623–629. doi: 10.1074/jbc.M107435200. [DOI] [PubMed] [Google Scholar]
- 109.Kohda T, Asai A, Kuroiwa Y, Kobayashi S, Aisaka K, Nagashima G, Yoshida MC, Kondo Y, Kagiyama N, Kirino T, Kaneko-Ishino T, Ishino F. Tumour suppressor activity of human imprinted gene PEG3 in a glioma cell line. Genes Cells. 2001;6:237–247. doi: 10.1046/j.1365-2443.2001.00412.x. [DOI] [PubMed] [Google Scholar]
- 110.Dowdy SC, Gostout BS, Shridhar V, Wu X, Smith DI, Podratz KC, Jiang S-W. Biallelic methylation and silencing of paternally expressed gene 3 (PEG3) in gynecologic cancer cell lines. Gynecol. Oncol. 2005;99:126–134. doi: 10.1016/j.ygyno.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 111.Jiang X, Yu Y, Yang HW, Agar NYR, Frado L, Johnson MD. The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. J. Biol. Chem. 2010;285:8472–8480. doi: 10.1074/jbc.M109.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Buraschi S, Neill T, Goyal A, Poluzzi C, Smythies J, Owens RT, Schaefer L, Torres AT, Iozzo RV. Decorin causes autophagy in endothelial cells via Peg3. Proc. Natl. Acad. Sci. USA. 2013;110:E2582–E2591. doi: 10.1073/pnas.1305732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Neill T, Torres AT, Buraschi S, Iozzo RV. Decorin has an appetite for endothelial cell autophagy. Autophagy. 2013;9:1626–1628. doi: 10.4161/auto.25881. [DOI] [PubMed] [Google Scholar]
- 114.Iozzo RV, Goldoni S, Berendsen A, Young MF. Small leucine-rich proteoglycans. In: Mecham RP, editor. Extracellular Matrix: An overview. Springer; 2011. pp. 197–231. [Google Scholar]
- 115.Merline R, Nastase MV, Iozzo RV, Schaefer L. Small Leucine-rich proteoglycans: multifunctional signaling effectors. In: Karamanos N, editor. Extracellular Matrix: Pathobiology and signaling. Berlin: Walter de Gruytier Gmbh and Co.; 2012. pp. 185–196. [Google Scholar]
- 116.Schaefer L, Iozzo RV. Small leucine-rich proteoglycans, at the crossroad of cancer growth and inflammation. Curr. Opin. Genet. Dev. 2012;22:56–57. doi: 10.1016/j.gde.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 117.Goyal A, Neill T, Owens RT, Schaefer L, Iozzo RV. Decorin activates AMPK, an energy sensor kinase, to induce autophagy in endothelial cells. Matrix Biol. 2014;34:46–54. doi: 10.1016/j.matbio.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex- at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phopshorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee JW, Park S, Takahashi Y, Wang H-G. The association of AMPK with ULK1 regulates autophagy. PloS one. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Choi AMK, Ryter SW, Levine B. Autophagy in human health and disease. New Engl. J. Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 124.Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-lysosome signaling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buraschi S, Pal N, Tyler-Rubinstein N, Owens RT, Neill T, Iozzo RV. Decorin antagonizes Met receptor activity and downregulates β-catenin and Myc levels. J. Biol. Chem. 2010;285:42075–42085. doi: 10.1074/jbc.M110.172841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Goldoni S, Humphries A, Nystrom A, Sattar S, Owens RT, McQuillan DJ, Ireton K, Iozzo RV. Decorin is a novel antagonistic ligand of the Met receptor. J Cell Biol. 2009;185:743–754. doi: 10.1083/jcb.200901129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Goldoni S, Iozzo RV. Tumor microenvironment: Modulation by decorin and related molecules harboring leucine-rich tandem motifs. Int. J. Cancer. 2008;123:2473–2479. doi: 10.1002/ijc.23930. [DOI] [PubMed] [Google Scholar]
- 130.Neill T, Jones HR, Crane-Smith Z, Owens RT, Schaefer L, Iozzo RV. Decorin induces rapid secretion of thrombospondin-1 in basal breast carcinoma cells via inhibition of Ras homolog gene family, member A/Rho-associated coiled-coil containing protein kinase 1. FEBS J. 2013;280:2353–2368. doi: 10.1111/febs.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu W, Neill T, Yang Y, Hu Z, Cleveland E, Wu Y, Hutten R, Xiao X, Stock SR, Shevrin D, Kaul K, Brendler C, Iozzo RV, Seth P. The systemic delivery of an oncolytic adenovirus expressing decorin inhibits bone metastasis in a mouse model of human prostate cancer. Gene Therapy. 2014 doi: 10.1038/gt.2014.110. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Reed CC, Waterhouse A, Kirby S, Kay P, Owens RA, McQuillan DJ, Iozzo RV. Decorin prevents metastatic spreading of breast cancer. Oncogene. 2005;24:1104–1110. doi: 10.1038/sj.onc.1208329. [DOI] [PubMed] [Google Scholar]
- 133.Fassan M, D'Arca D, Letko J, Vecchione A, Gardiman MP, McCue P, Wildemore B, Rugge M, Shupp-Byrne D, Gomella LG, Morrione A, Iozzo RV, Baffa R. Mitostatin is down-regulated in human prostate cancer and suppresses the invasive phenotype of prostate cancer cells. PloS one. 2011;6:e19771. doi: 10.1371/journal.pone.0019771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vecchione A, Fassan M, Anesti V, Morrione A, Goldoni S, Baldassarre G, Byrne D, D'Arca D, Palazzo JP, Lloyd J, Scorrano L, Gomella LG, Iozzo RV, Baffa R. MITOSTATIN a putative tumor suppressor on chromosome 12q24.1, is downregulated in human bladder and breast cancer. Oncogene. 2009;28:257–269. doi: 10.1038/onc.2008.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cerqua C, Anesti V, Pyakurel A, Liu D, Naon D, Wiche G, Baffa R, Dimmer KS, Scorrano L. Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Reports. 2010;11:854–860. doi: 10.1038/embor.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ventura-Clapier R, Garnier A, Veksler W. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1α. Cardiovasc. Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 137.Vazquez F, Lim J-H, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, Puigserver P. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 139.Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat. Rev. Mol. Cell Biol. 2012;13:780–788. doi: 10.1038/nrm3479. [DOI] [PubMed] [Google Scholar]
- 140.Dagda R, Cherra SJI, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 142.Narendra D, Walker JE, Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Csordas G, Santra M, Reed CC, Eichstetter I, McQuillan DJ, Gross D, Nugent MA, Hajnoczky G, Iozzo RV. Sustained down-regulation of the epidermal growth factor receptor by decorin. A mechanism for controlling tumor growth in vivo. J. Biol. Chem. 2000;275:32879–32887. doi: 10.1074/jbc.M005609200. [DOI] [PubMed] [Google Scholar]
- 144.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 145.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl. Acad. Sci U.S.A. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not appreciate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cenki B, Sephton CF, Cenik BK, Herz J, Yu G. Progranulin: A proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J. Biol. Chem. 2012;287:32298–32306. doi: 10.1074/jbc.R112.399170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- 150.Flugel D, Gorlach A, Kietzmann T. GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood. 2012;119:1292–1301. doi: 10.1182/blood-2011-08-375014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 2004;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 152.Nastase MV, Young MF, Schaefer L. Biglycan: a multivalent proteoglycan providing structure and signals. J Histochem Cytochem. 2012;60:963–975. doi: 10.1369/0022155412456380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xu T, Bianco P, Fisher LW, Longenecker G, Smith E, Goldstein S, Bonadio J, Boskey A, Heegaard AM, Sommer B, Satomura K, Dominguez P, Zhao C, Kulkarni AB, Robey PG, Young MF. Targeted disruption of the biglycan gene leads to an osteoporosis-like phenotype in mice. Nat Genet. 1998;20:78–82. doi: 10.1038/1746. [DOI] [PubMed] [Google Scholar]
- 154.Schaefer L, Babelova A, Kiss E, Hausser HJ, Baliova M, Krzyzankova M, Marsche G, Young MF, Mihalik D, Gotte M, Malle E, Schaefer RM, Grone HJ. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. The Journal of clinical investigation. 2005;115:2223–2233. doi: 10.1172/JCI23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schaefer L. Complexity of danger: the diverse nature of damage-associated molecular patterns. The Journal of biological chemistry. 2014;289:35237–35245. doi: 10.1074/jbc.R114.619304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Moreth K, Brodbeck R, Babelova A, Gretz N, Spieker T, Zeng-Brouwers J, Pfeilschifter J, Young MF, Schaefer RM, Schaefer L. The proteoglycan biglycan regulates expression of the B cell chemoattractant CXCL13 and aggravates murine lupus nephritis. The Journal of clinical investigation. 2010;120:4251–4272. doi: 10.1172/JCI42213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu YH, Yang F, Zhang SS, Zeng TT, Xie X, Guan XY. High expression of biglycan is associated with poor prognosis in patients with esophageal squamous cell carcinoma. International journal of clinical and experimental pathology. 2013;6:2497–2505. [PMC free article] [PubMed] [Google Scholar]
- 158.Nishino R, Honda M, Yamashita T, Takatori H, Minato H, Zen Y, Sasaki M, Takamura H, Horimoto K, Ohta T, Nakanuma Y, Kaneko S. Identification of novel candidate tumour marker genes for intrahepatic cholangiocarcinoma. J Hepatol. 2008;49:207–216. doi: 10.1016/j.jhep.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 159.Modolo F, Biz MT, Martins MT, Machado de Sousa SO, de Araujo NS. Expression of extracellular matrix proteins in adenomatoid odontogenic tumor. J Oral Pathol Med. 2010;39:230–235. doi: 10.1111/j.1600-0714.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- 160.Jaeger J, Koczan D, Thiesen HJ, Ibrahim SM, Gross G, Spang R, Kunz M. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:806–815. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 161.Galamb O, Sipos F, Spisak S, Galamb B, Krenacs T, Valcz G, Tulassay Z, Molnar B. Potential biomarkers of colorectal adenoma-dysplasia-carcinoma progression: mRNA expression profiling and in situ protein detection on TMAs reveal 15 sequentially upregulated and 2 downregulated genes. Cell Oncol. 2009;31:19–29. doi: 10.3233/CLO-2009-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gu X, Ma Y, Xiao J, Zheng H, Song C, Gong Y, Xing X. Up-regulated biglycan expression correlates with the malignancy in human colorectal cancers. Clin Exp Med. 2012;12:195–199. doi: 10.1007/s10238-011-0155-4. [DOI] [PubMed] [Google Scholar]
- 163.Mikula M, Rubel T, Karczmarski J, Goryca K, Dadlez M, Ostrowski J. Integrating proteomic and transcriptomic high-throughput surveys for search of new biomarkers of colon tumors. Funct Integr Genomics. 2011;11:215–224. doi: 10.1007/s10142-010-0200-5. [DOI] [PubMed] [Google Scholar]
- 164.Liu Y, Li W, Li X, Tai Y, Lu Q, Yang N, Jiang J. Expression and significance of biglycan in endometrial cancer. Arch Gynecol Obstet. 2014;289:649–655. doi: 10.1007/s00404-013-3017-3. [DOI] [PubMed] [Google Scholar]
- 165.Wang B, Li GX, Zhang SG, Wang Q, Wen YG, Tang HM, Zhou CZ, Xing AY, Fan JW, Yan DW, Qiu GQ, Yu ZH, Peng ZH. Biglycan expression correlates with aggressiveness and poor prognosis of gastric cancer. Exp Biol Med (Maywood) 2011;236:1247–1253. doi: 10.1258/ebm.2011.011124. [DOI] [PubMed] [Google Scholar]
- 166.Fang DD, Kim YJ, Lee CN, Aggarwal S, McKinnon K, Mesmer D, Norton J, Birse CE, He T, Ruben SM, Moore PA. Expansion of CD133(+) colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br J Cancer. 2010;102:1265–1275. doi: 10.1038/sj.bjc.6605610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Aprile G, Avellini C, Reni M, Mazzer M, Foltran L, Rossi D, Cereda S, Iaiza E, Fasola G, Piga A. Biglycan expression and clinical outcome in patients with pancreatic adenocarcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:131–137. doi: 10.1007/s13277-012-0520-2. [DOI] [PubMed] [Google Scholar]
- 168.Niedworok C, Rock K, Kretschmer I, Freudenberger T, Nagy N, Szarvas T, Vom Dorp F, Reis H, Rubben H, Fischer JW. Inhibitory role of the small leucine-rich proteoglycan biglycan in bladder cancer. PloS one. 2013;8:e80084. doi: 10.1371/journal.pone.0080084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rydstrom K, Joost P, Ehinger M, Eden P, Jerkeman M, Cavallin-Stahl E, Linderoth J. Gene expression profiling indicates that immunohistochemical expression of CD40 is a marker of an inflammatory reaction in the tumor stroma of diffuse large B-cell lymphoma. Leuk Lymphoma. 2012;53:1764–1768. doi: 10.3109/10428194.2012.666541. [DOI] [PubMed] [Google Scholar]
- 170.Weber CK, Sommer G, Michl P, Fensterer H, Weimer M, Gansauge F, Leder G, Adler G, Gress TM. Biglycan is overexpressed in pancreatic cancer and induces G1-arrest in pancreatic cancer cell lines. Gastroenterology. 2001;121:657–667. doi: 10.1053/gast.2001.27222. [DOI] [PubMed] [Google Scholar]
- 171.Takekawa M, Tatebayashi K, Itoh F, Adachi M, Imai K, Saito H. Smad-dependent GADD45beta expression mediates delayed activation of p38 MAP kinase by TGF-beta. EMBO J. 2002;21:6473–6482. doi: 10.1093/emboj/cdf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yamamoto K, Ohga N, Hida Y, Maishi N, Kawamoto T, Kitayama K, Akiyama K, Osawa T, Kondoh M, Matsuda K, Onodera Y, Fujie M, Kaga K, Hirano S, Shinohara N, Shindoh M, Hida K. Biglycan is a specific marker and an autocrine angiogenic factor of tumour endothelial cells. Br J Cancer. 2012;106:1214–1223. doi: 10.1038/bjc.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liu T, Du X, Sheng X. Gene expression changes after ionizing radiation in endothelial cells derived from human endometrial cancer-preliminary outcomes. Arch Gynecol Obstet. 2014;289:1315–1323. doi: 10.1007/s00404-013-3136-x. [DOI] [PubMed] [Google Scholar]
- 174.Berendsen AD, Pinnow EL, Maeda A, Brown AC, McCartney-Francis N, Kram V, Owens RT, Robey PG, Holmbeck K, de Castro LF, Kilts TM, Young MF. Biglycan modulates angiogenesis and bone formation during fracture healing. Matrix Biol. 2014;35:223–231. doi: 10.1016/j.matbio.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xing X, Gu X, Ma T, Ye H. Biglycan up-regulated vascular endothelial growth factor (VEGF) expression and promoted angiogenesis in colon cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014 doi: 10.1007/s13277-014-2779-y. [DOI] [PubMed] [Google Scholar]
- 176.West XZ, Malinin NL, Merkulova AA, Tischenko M, Kerr BA, Borden EC, Podrez EA, Salomon RG, Byzova TV. Oxidative stress induces angiogenesis by activating TLR2 with novel endogenous ligands. Nature. 2010;467:972–976. doi: 10.1038/nature09421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Babelova A, Moreth K, Tsalastra-Greul W, Zeng-Brouwers J, Eickelberg O, Young MF, Bruckner P, Pfeilschifter J, Schaefer RM, Grone HJ, Schaefer L. Biglycan, a danger signal that activates the NLRP3 inflammasome via toll-like and P2X receptors. The Journal of biological chemistry. 2009;284:24035–24048. doi: 10.1074/jbc.M109.014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Recktenwald CV, Leisz S, Steven A, Mimura K, Muller A, Wulfanger J, Kiessling R, Seliger B. HER-2/neu-mediated down-regulation of biglycan associated with altered growth properties. The Journal of biological chemistry. 2012;287:24320–24329. doi: 10.1074/jbc.M111.334425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schaefer L, Beck KF, Raslik I, Walpen S, Mihalik D, Micegova M, Macakova K, Schonherr E, Seidler DG, Varga G, Schaefer RM, Kresse H, Pfeilschifter J. Biglycan, a nitric oxide-regulated gene, affects adhesion, growth, and survival of mesangial cells. The Journal of biological chemistry. 2003;278:26227–26237. doi: 10.1074/jbc.M210574200. [DOI] [PubMed] [Google Scholar]
- 180.Shimizu-Hirota R, Sasamura H, Kuroda M, Kobayashi E, Hayashi M, Saruta T. Extracellular matrix glycoprotein biglycan enhances vascular smooth muscle cell proliferation and migration. Circ Res. 2004;94:1067–1074. doi: 10.1161/01.RES.0000126049.79800.CA. [DOI] [PubMed] [Google Scholar]
- 181.Berendsen AD, Fisher LW, Kilts TM, Owens RT, Robey PG, Gutkind JS, Young MF. Modulation of canonical Wnt signaling by the extracellular matrix component biglycan. Proc Natl Acad Sci U S A. 2011;108:17022–17027. doi: 10.1073/pnas.1110629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ward M, Ajuwon KM. Regulation of pre-adipocyte proliferation and apoptosis by the small leucine-rich proteoglycans, biglycan and decorin. Cell Prolif. 2011;44:343–351. doi: 10.1111/j.1365-2184.2011.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hu L, Duan YT, Li JF, Su LP, Yan M, Zhu ZG, Liu BY, Yang QM. Biglycan enhances gastric cancer invasion by activating FAK signaling pathway. Oncotarget. 2014;5:1885–1896. doi: 10.18632/oncotarget.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bischof AG, Yuksel D, Mammoto T, Mammoto A, Krause S, Ingber DE. Breast cancer normalization induced by embryonic mesenchyme is mediated by extracellular matrix biglycan. Integrative biology : quantitative biosciences from nano to macro. 2013;5:1045–1056. doi: 10.1039/c3ib40103k. [DOI] [PubMed] [Google Scholar]
- 185.Tufvesson E, Westergren-Thorsson G. Biglycan and decorin induce morphological and cytoskeletal changes involving signalling by the small GTPases RhoA and Rac1 resulting in lung fibroblast migration. J Cell Sci. 2003;116:4857–4864. doi: 10.1242/jcs.00808. [DOI] [PubMed] [Google Scholar]
- 186.Datsis GA, Berdiaki A, Nikitovic D, Mytilineou M, Katonis P, Karamanos NK, Tzanakakis GN. Parathyroid hormone affects the fibroblast growth factor-proteoglycan signaling axis to regulate osteosarcoma cell migration. FEBS J. 2011;278:3782–3792. doi: 10.1111/j.1742-4658.2011.08300.x. [DOI] [PubMed] [Google Scholar]
- 187.Mintz MB, Sowers R, Brown KM, Hilmer SC, Mazza B, Huvos AG, Meyers PA, Lafleur B, McDonough WS, Henry MM, Ramsey KE, Antonescu CR, Chen W, Healey JH, Daluski A, Berens ME, Macdonald TJ, Gorlick R, Stephan DA. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005;65:1748–1754. doi: 10.1158/0008-5472.CAN-04-2463. [DOI] [PubMed] [Google Scholar]
- 188.Pan S, Cheng L, White JT, Lu W, Utleg AG, Yan X, Urban ND, Drescher CW, Hood L, Lin B. Quantitative proteomics analysis integrated with microarray data reveals that extracellular matrix proteins, catenins, and p53 binding protein 1 are important for chemotherapy response in ovarian cancers. OMICS. 2009;13:345–354. doi: 10.1089/omi.2009.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Couchman JR. Transmembrane signaling proteoglycans. Annu Rev Cell Dev Biol. 2010;26:89–114. doi: 10.1146/annurev-cellbio-100109-104126. [DOI] [PubMed] [Google Scholar]
- 190.Xian X, Gopal S, Couchman JR. Syndecans as receptors and organizers of the extracellular matrix. Cell Tissue Res. 2010;339:31–46. doi: 10.1007/s00441-009-0829-3. [DOI] [PubMed] [Google Scholar]
- 191.Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kreuger J, Kjellen L. Heparan sulfate biosynthesis: regulation and variability. J Histochem Cytochem. 2012;60:898–907. doi: 10.1369/0022155412464972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lee D, Oh ES, Woods A, Couchman JR, Lee W. Solution structure of a Syndecan-4 cytoplasmic domain and its interaction with phosphatidylinositol 4,5-bisphosphate. The Journal of biological chemistry. 1998;273:13022–13029. doi: 10.1074/jbc.273.21.13022. [DOI] [PubMed] [Google Scholar]
- 194.Choi S, Lee E, Kwon S, Park H, Yi JY, Kim S, Han IO, Yun Y, Oh ES. Transmembrane domain-induced oligomerization is crucial for the functions of Syndecan-2 and Syndecan-4. The Journal of biological chemistry. 2005;280:42573–42579. doi: 10.1074/jbc.M509238200. [DOI] [PubMed] [Google Scholar]
- 195.Oh ES, Woods A, Couchman JR. Syndecan-4 proteoglycan regulates the distribution and activity of protein kinase C. The Journal of biological chemistry. 1997;272:8133–8136. doi: 10.1074/jbc.272.13.8133. [DOI] [PubMed] [Google Scholar]
- 196.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 197.Morgan MR, Hamidi H, Bass MD, Warwood S, Ballestrem C, Humphries MJ. Syndecan-4 phosphorylation is a control point for integrin recycling. Dev Cell. 2013;24:472–485. doi: 10.1016/j.devcel.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen K, Williams KJ. Molecular mediators for raft-dependent endocytosis of Syndecan-1, a highly conserved, multifunctional receptor. The Journal of biological chemistry. 2013;288:13988–13999. doi: 10.1074/jbc.M112.444737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Morgan MR, Humphries MJ, Bass MD. Synergistic control of cell adhesion by integrins and syndecans. Nat Rev Mol Cell Biol. 2007;8:957–969. doi: 10.1038/nrm2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Greene DK, Tumova S, Couchman JR, Woods A. Syndecan-4 associates with alpha-actinin. The Journal of biological chemistry. 2003;278:7617–7623. doi: 10.1074/jbc.M207123200. [DOI] [PubMed] [Google Scholar]
- 201.Lambaerts K, Wilcox-Adelman SA, Zimmermann P. The signaling mechanisms of syndecan heparan sulfate proteoglycans. Curr Opin Cell Biol. 2009;21:662–669. doi: 10.1016/j.ceb.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu BY, Kim YC, Leatherberry V, Cowin P, Alexander CM. Mammary gland development requires Syndecan-1 to create a beta-catenin/TCF-responsive mammary epithelial subpopulation. Oncogene. 2003;22:9243–9253. doi: 10.1038/sj.onc.1207217. [DOI] [PubMed] [Google Scholar]
- 203.Alexander CM, Reichsman F, Hinkes MT, Lincecum J, Becker KA, Cumberledge S, Bernfield M. Syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice. Nat Genet. 2000;25:329–332. doi: 10.1038/77108. [DOI] [PubMed] [Google Scholar]
- 204.Ishiguro K, Kadomatsu K, Kojima T, Muramatsu H, Tsuzuki S, Nakamura E, Kusugami K, Saito H, Muramatsu T. Syndecan-4 deficiency impairs focal adhesion formation only under restricted conditions. The Journal of biological chemistry. 2000;275:5249–5252. doi: 10.1074/jbc.275.8.5249. [DOI] [PubMed] [Google Scholar]
- 205.Echtermeyer F, Streit M, Wilcox-Adelman S, Saoncella S, Denhez F, Detmar M, Goetinck P. Delayed wound repair and impaired angiogenesis in mice lacking Syndecan-4. The Journal of clinical investigation. 2001;107:R9–R14. doi: 10.1172/JCI10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Beck JC, Lekutis C, Couchman J, Parry G. Stage-specific remodeling of the mammary gland basement membrane during lactogenic development. Biochem Biophys Res Commun. 1993;190:616–623. doi: 10.1006/bbrc.1993.1093. [DOI] [PubMed] [Google Scholar]
- 207.Hinkes MT, Goldberger OA, Neumann PE, Kokenyesi R, Bernfield M. Organization and promoter activity of the mouse Syndecan-1 gene. The Journal of biological chemistry. 1993;268:11440–11448. [PubMed] [Google Scholar]
- 208.Vihinen T, Maatta A, Jaakkola P, Auvinen P, Jalkanen M. Functional characterization of mouse Syndecan-1 promoter. The Journal of biological chemistry. 1996;271:12532–12541. doi: 10.1074/jbc.271.21.12532. [DOI] [PubMed] [Google Scholar]
- 209.Iguchi-Ishiguro H, Ouchi Y, Watanabe S, Numabe Y. Analysis of Syndecan-1 gene promoter during mouse tooth development. Arch Oral Biol. 2012;57:531–538. doi: 10.1016/j.archoralbio.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 210.Cook DM, Hinkes MT, Bernfield M, Rausche FJ., 3rd Transcriptional activation of the Syndecan-1 promoter by the Wilms' tumor protein WT1. Oncogene. 1996;13:1789–1799. [PubMed] [Google Scholar]
- 211.Okuyama E, Suzuki A, Murata M, Ando Y, Kato I, Takagi Y, Takagi A, Murate T, Saito H, Kojima T. Molecular mechanisms of Syndecan-4 upregulation by TNF-alpha in the endothelium-like EAhy926 cells. J Biochem. 2013;154:41–50. doi: 10.1093/jb/mvt024. [DOI] [PubMed] [Google Scholar]
- 212.Fujita N, Hirose Y, Tran CM, Chiba K, Miyamoto T, Toyama Y, Shapiro IM, Risbud MV. HIF-1-PHD2 axis controls expression of syndecan 4 in nucleus pulposus cells. FASEB J. 2014;28:2455–2465. doi: 10.1096/fj.13-243741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Dedes PG, Gialeli C, Tsonis AI, Kanakis I, Theocharis AD, Kletsas D, Tzanakakis GN, Karamanos NK. Expression of matrix macromolecules and functional properties of breast cancer cells are modulated by the bisphosphonate zoledronic acid. Biochim Biophys Acta. 2012;1820:1926–1939. doi: 10.1016/j.bbagen.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 214.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 215.Ibrahim SA, Yip GW, Stock C, Pan JW, Neubauer C, Poeter M, Pupjalis D, Koo CY, Kelsch R, Schule R, Rescher U, Kiesel L, Gotte M. Targeting of Syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. Int J Cancer. 2012;131:E884–E896. doi: 10.1002/ijc.27629. [DOI] [PubMed] [Google Scholar]
- 216.Sun H, Berquin IM, Edwards IJ. Omega-3 polyunsaturated fatty acids regulate Syndecan-1 expression in human breast cancer cells. Cancer Res. 2005;65:4442–4447. doi: 10.1158/0008-5472.CAN-04-4200. [DOI] [PubMed] [Google Scholar]
- 217.Yang Y, Macleod V, Miao HQ, Theus A, Zhan F, Shaughnessy JD, Jr, Sawyer J, Li JP, Zcharia E, Vlodavsky I, Sanderson RD. Heparanase enhances Syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis. The Journal of biological chemistry. 2007;282:13326–13333. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 218.Okolicsanyi RK, Buffiere A, Jacinto JM, Chacon-Cortes D, Chambers SK, Youl PH, Haupt LM, Griffiths LR. Association of heparan sulfate proteoglycans SDC1 and SDC4 polymorphisms with breast cancer in an Australian Caucasian population. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014 doi: 10.1007/s13277-014-2774-3. [DOI] [PubMed] [Google Scholar]
- 219.Menashe I, Maeder D, Garcia-Closas M, Figueroa JD, Bhattacharjee S, Rotunno M, Kraft P, Hunter DJ, Chanock SJ, Rosenberg PS, Chatterjee N. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 2010;70:4453–4459. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Barbareschi M, Maisonneuve P, Aldovini D, Cangi MG, Pecciarini L, Angelo Mauri F, Veronese S, Caffo O, Lucenti A, Palma PD, Galligioni E, Doglioni C. High Syndecan-1 expression in breast carcinoma is related to an aggressive phenotype and to poorer prognosis. Cancer. 2003;98:474–483. doi: 10.1002/cncr.11515. [DOI] [PubMed] [Google Scholar]
- 221.Lofgren L, Sahlin L, Jiang S, Von Schoultz B, Fernstad R, Skoog L, Von Schoultz E. Expression of Syndecan-1 in paired samples of normal and malignant breast tissue from postmenopausal women. Anticancer Res. 2007;27:3045–3050. [PubMed] [Google Scholar]
- 222.Tsanou E, Ioachim E, Briasoulis E, Charchanti A, Damala K, Karavasilis V, Pavlidis N, Agnantis NJ. Clinicopathological study of the expression of Syndecan-1 in invasive breast carcinomas. correlation with extracellular matrix components. J Exp Clin Cancer Res. 2004;23:641–650. [PubMed] [Google Scholar]
- 223.Leivonen M, Lundin J, Nordling S, von Boguslawski K, Haglund C. Prognostic value of Syndecan-1 expression in breast cancer. Oncology. 2004;67:11–18. doi: 10.1159/000080280. [DOI] [PubMed] [Google Scholar]
- 224.Mundhenke C, Meyer K, Drew S, Friedl A. Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 receptor binding in breast carcinomas. Am J Pathol. 2002;160:185–194. doi: 10.1016/S0002-9440(10)64362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Loussouarn D, Campion L, Sagan C, Frenel JS, Dravet F, Classe JM, Pioud-Martigny R, Berton-Rigaud D, Bourbouloux E, Mosnier JF, Bataille FR, Campone M. Prognostic impact of Syndecan-1 expression in invasive ductal breast carcinomas. Br J Cancer. 2008;98:1993–1998. doi: 10.1038/sj.bjc.6604400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Su G, Blaine SA, Qiao D, Friedl A. Membrane type 1 matrix metalloproteinase-mediated stromal Syndecan-1 shedding stimulates breast carcinoma cell proliferation. Cancer Res. 2008;68:9558–9565. doi: 10.1158/0008-5472.CAN-08-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mennerich D, Vogel A, Klaman I, Dahl E, Lichtner RB, Rosenthal A, Pohlenz HD, Thierauch KH, Sommer A. Shift of Syndecan-1 expression from epithelial to stromal cells during progression of solid tumours. Eur J Cancer. 2004;40:1373–1382. doi: 10.1016/j.ejca.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 228.Stanley MJ, Stanley MW, Sanderson RD, Zera R. Syndecan-1 expression is induced in the stroma of infiltrating breast carcinoma. Am J Clin Pathol. 1999;112:377–383. doi: 10.1093/ajcp/112.3.377. [DOI] [PubMed] [Google Scholar]
- 229.Nikolova V, Koo CY, Ibrahim SA, Wang Z, Spillmann D, Dreier R, Kelsch R, Fischgrabe J, Smollich M, Rossi LH, Sibrowski W, Wulfing P, Kiesel L, Yip GW, Gotte M. Differential roles for membrane-bound and soluble Syndecan-1 (CD138) in breast cancer progression. Carcinogenesis. 2009;30:397–407. doi: 10.1093/carcin/bgp001. [DOI] [PubMed] [Google Scholar]
- 230.Beauvais DM, Rapraeger AC. Syndecan-1-mediated cell spreading requires signaling by alphavbeta3 integrins in human breast carcinoma cells. Exp Cell Res. 2003;286:219–232. doi: 10.1016/s0014-4827(03)00126-5. [DOI] [PubMed] [Google Scholar]
- 231.Maeda T, Alexander CM, Friedl A. Induction of Syndecan-1 expression in stromal fibroblasts promotes proliferation of human breast cancer cells. Cancer Res. 2004;64:612–621. doi: 10.1158/0008-5472.can-03-2439. [DOI] [PubMed] [Google Scholar]
- 232.Maeda T, Desouky J, Friedl A. Syndecan-1 expression by stromal fibroblasts promotes breast carcinoma growth in vivo and stimulates tumor angiogenesis. Oncogene. 2006;25:1408–1412. doi: 10.1038/sj.onc.1209168. [DOI] [PubMed] [Google Scholar]
- 233.Burbach BJ, Ji Y, Rapraeger AC. Syndecan-1 ectodomain regulates matrix-dependent signaling in human breast carcinoma cells. Exp Cell Res. 2004;300:234–247. doi: 10.1016/j.yexcr.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 234.Ibrahim SA, Hassan H, Vilardo L, Kumar SK, Kumar AV, Kelsch R, Schneider C, Kiesel L, Eich HT, Zucchi I, Reinbold R, Greve B, Gotte M. Syndecan-1 (CD138) modulates triple-negative breast cancer stem cell properties via regulation of LRP-6 and IL-6-mediated STAT3 signaling. PloS one. 2013;8:e85737. doi: 10.1371/journal.pone.0085737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tokes AM, Szasz AM, Farkas A, Toth AI, Dank M, Harsanyi L, Molnar BA, Molnar IA, Laszlo Z, Rusz Z, Kulka J. Stromal matrix protein expression following preoperative systemic therapy in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:731–739. doi: 10.1158/1078-0432.CCR-08-1523. [DOI] [PubMed] [Google Scholar]
- 236.Gotte M, Kersting C, Ruggiero M, Tio J, Tulusan AH, Kiesel L, Wulfing P. Predictive value of Syndecan-1 expression for the response to neoadjuvant chemotherapy of primary breast cancer. Anticancer Res. 2006;26:621–627. [PubMed] [Google Scholar]
- 237.Baba F, Swartz K, van Buren R, Eickhoff J, Zhang Y, Wolberg W, Friedl A. Syndecan-1 and Syndecan-4 are overexpressed in an estrogen receptor-negative, highly proliferative breast carcinoma subtype. Breast Cancer Res Treat. 2006;98:91–98. doi: 10.1007/s10549-005-9135-2. [DOI] [PubMed] [Google Scholar]
- 238.Lim H, Multhaupt H, Couchman JR. Cell surface heparan sulfate proteoglycans control adhesion and invasion of breast carcinoma cells. Molecular cancer. 2015;14:15. doi: 10.1186/s12943-014-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wu ZS, Pandey V, Wu WY, Ye S, Zhu T, Lobie PE. Prognostic significance of the expression of GFRalpha1, GFRalpha3 and Syndecan-3, proteins binding ARTEMIN, in mammary carcinoma. BMC cancer. 2013;13:34. doi: 10.1186/1471-2407-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 241.Polyak K. Heterogeneity in breast cancer. The Journal of clinical investigation. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 243.Beauvais DM, Burbach BJ, Rapraeger AC. The Syndecan-1 ectodomain regulates alphavbeta3 integrin activity in human mammary carcinoma cells. J Cell Biol. 2004;167:171–181. doi: 10.1083/jcb.200404171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Beauvais DM, Ell BJ, McWhorter AR, Rapraeger AC. Syndecan-1 regulates alphavbeta3 and alphavbeta5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J Exp Med. 2009;206:691–705. doi: 10.1084/jem.20081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Beauvais DM, Rapraeger AC. Syndecan-1 couples the insulin-like growth factor-1 receptor to inside-out integrin activation. J Cell Sci. 2010;123:3796–3807. doi: 10.1242/jcs.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Leppä S, Mali M, Miettinen HM, Jalkanen M. Syndecan expression regulates cell morphology and growth of mouse mammary epithelial tumor cells. Proc Natl Acad Sci U S A. 1992;89:932–936. doi: 10.1073/pnas.89.3.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yang N, Mosher R, Seo S, Beebe D, Friedl A. Syndecan-1 in breast cancer stroma fibroblasts regulates extracellular matrix fiber organization and carcinoma cell motility. Am J Pathol. 2011;178:325–335. doi: 10.1016/j.ajpath.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, Friedl A, Keely PJ. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Oh ES, Couchman JR. Syndecans-2 and -4; close cousins, but not identical twins. Mol Cells. 2004;17:181–187. [PubMed] [Google Scholar]
- 250.Barash U, Cohen-Kaplan V, Dowek I, Sanderson RD, Ilan N, Vlodavsky I. Proteoglycans in health and disease: new concepts for heparanase function in tumor progression and metastasis. FEBS J. 2010;277:3890–3903. doi: 10.1111/j.1742-4658.2010.07799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Arvatz G, Shafat I, Levy-Adam F, Ilan N, Vlodavsky I. The heparanase system and tumor metastasis: is heparanase the seed and soil? Cancer Metast Rev. 2011;30:253–268. doi: 10.1007/s10555-011-9288-x. [DOI] [PubMed] [Google Scholar]
- 252.Hammond E, Khurana A, Shridhar V, Dredge K. The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front Oncol. 2014;4:195. doi: 10.3389/fonc.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 254.Gomes AM, Stelling MP, Pavao MS. Heparan sulfate and heparanase as modulators of breast cancer progression. Biomed Res Int. 2013;2013:852093. doi: 10.1155/2013/852093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Peterson SB, Liu J. Multi-faceted substrate specificity of heparanase. Matrix Biol. 2013;32:223–227. doi: 10.1016/j.matbio.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 256.Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. The Journal of biological chemistry. 2004;279:23536–23541. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- 257.Sotnikov I, Hershkoviz R, Grabovsky V, Ilan N, Cahalon L, Vlodavsky I, Alon R, Lider O. Enzymatically quiescent heparanase augments T cell interactions with VCAM-1 and extracellular matrix components under versatile dynamic contexts. J Immunol. 2004;172:5185–5193. doi: 10.4049/jimmunol.172.9.5185. [DOI] [PubMed] [Google Scholar]
- 258.Ramani VC, Yang Y, Ren Y, Nan L, Sanderson RD. Heparanase plays a dual role in driving hepatocyte growth factor (HGF) signaling by enhancing HGF expression and activity. The Journal of biological chemistry. 2011;286:6490–6499. doi: 10.1074/jbc.M110.183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Nadir Y, Brenner B, Gingis-Velitski S, Levy-Adam F, Ilan N, Zcharia E, Nadir E, Vlodavsky I. Heparanase induces tissue factor pathway inhibitor expression and extracellular accumulation in endothelial and tumor cells. Thromb Haemost. 2008;99:133–141. [PubMed] [Google Scholar]
- 260.Maxhimer JB, Quiros RM, Stewart R, Dowlatshahi K, Gattuso P, Fan M, Prinz RA, Xu X. Heparanase-1 expression is associated with the metastatic potential of breast cancer. Surgery. 2002;132:326–333. doi: 10.1067/msy.2002.125719. [DOI] [PubMed] [Google Scholar]
- 261.Gawthorpe S, Brown JE, Arif M, Nightingale P, Nevill A, Carmichael AR. Heparanase and COX-2 expression as predictors of lymph node metastasis in large, high-grade breast tumors. Anticancer Res. 2014;34:2797–2800. [PubMed] [Google Scholar]
- 262.Maxhimer JB, Pesce CE, Stewart RA, Gattuso P, Prinz RA, Xu X. Ductal carcinoma in situ of the breast and heparanase-1 expression: a molecular explanation for more aggressive subtypes. J Am Coll Surg. 2005;200:328–335. doi: 10.1016/j.jamcollsurg.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 263.Tang D, Zhang Q, Zhao S, Wang J, Lu K, Song Y, Zhao L, Kang X, Wang J, Xu S, Tian L. The expression and clinical significance of microRNA-1258 and heparanase in human breast cancer. Clin Biochem. 2013;46:926–932. doi: 10.1016/j.clinbiochem.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 264.Cohen I, Pappo O, Elkin M, San T, Bar-Shavit R, Hazan R, Peretz T, Vlodavsky I, Abramovitch R. Heparanase promotes growth, angiogenesis and survival of primary breast tumors. Int J Cancer. 2006;118:1609–1617. doi: 10.1002/ijc.21552. [DOI] [PubMed] [Google Scholar]
- 265.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66:1455–1463. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 266.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- 267.Zhang ZH, Chen Y, Zhao HJ, Xie CY, Ding J, Hou YT. Silencing of heparanase by siRNA inhibits tumor metastasis and angiogenesis of human breast cancer in vitro and in vivo. Cancer Biol Ther. 2007;6:587–595. doi: 10.4161/cbt.6.4.3888. [DOI] [PubMed] [Google Scholar]
- 268.Ridgway LD, Wetzel MD, Ngo JA, Erdreich-Epstein A, Marchetti D. Heparanase-induced GEF-H1 signaling regulates the cytoskeletal dynamics of brain metastatic breast cancer cells. Mol Cancer Res. 2012;10:689–702. doi: 10.1158/1541-7786.MCR-11-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zhang L, Sullivan PS, Goodman JC, Gunaratne PH, Marchetti D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res. 2011;71:645–654. doi: 10.1158/0008-5472.CAN-10-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, Goodman JC, Groves MD, Marchetti D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5:180ra148. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Manon-Jensen T, Itoh Y, Couchman JR. Proteoglycans in health and disease: the multiple roles of syndecan shedding. FEBS J. 2010;277:3876–3889. doi: 10.1111/j.1742-4658.2010.07798.x. [DOI] [PubMed] [Google Scholar]
- 272.Su G, Blaine SA, Qiao D, Friedl A. Shedding of Syndecan-1 by stromal fibroblasts stimulates human breast cancer cell proliferation via FGF2 activation. The Journal of biological chemistry. 2007;282:14906–14915. doi: 10.1074/jbc.M611739200. [DOI] [PubMed] [Google Scholar]
- 273.Manon-Jensen T, Multhaupt HA, Couchman JR. Mapping of matrix metalloproteinase cleavage sites on Syndecan-1 and Syndecan-4 ectodomains. FEBS J. 2013;280:2320–2331. doi: 10.1111/febs.12174. [DOI] [PubMed] [Google Scholar]
- 274.Teng YH, Aquino RS, Park PW. Molecular functions of Syndecan-1 in disease. Matrix Biol. 2012;31:3–16. doi: 10.1016/j.matbio.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Purushothaman A, Chen L, Yang Y, Sanderson RD. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. The Journal of biological chemistry. 2008;283:32628–32636. doi: 10.1074/jbc.M806266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Purushothaman A, Uyama T, Kobayashi F, Yamada S, Sugahara K, Rapraeger AC, Sanderson RD. Heparanase-enhanced shedding of Syndecan-1 by myeloma cells promotes endothelial invasion and angiogenesis. Blood. 2010;115:2449–2457. doi: 10.1182/blood-2009-07-234757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ding K, Lopez-Burks M, Sanchez-Duran JA, Korc M, Lander AD. Growth factor-induced shedding of Syndecan-1 confers glypican-1 dependence on mitogenic responses of cancer cells. J Cell Biol. 2005;171:729–738. doi: 10.1083/jcb.200508010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kelly T, Suva LJ, Huang Y, Macleod V, Miao HQ, Walker RC, Sanderson RD. Expression of heparanase by primary breast tumors promotes bone resorption in the absence of detectable bone metastases. Cancer Res. 2005;65:5778–5784. doi: 10.1158/0008-5472.CAN-05-0749. [DOI] [PubMed] [Google Scholar]
- 279.Kelly T, Suva LJ, Nicks KM, MacLeod V, Sanderson RD. Tumor-derived Syndecan-1 mediates distal cross-talk with bone that enhances osteoclastogenesis. J Bone Mineral Res. 2010;25:1295–1304. doi: 10.1002/jbmr.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clinical & developmental immunology. 2011;2011:842849. doi: 10.1155/2011/842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Record M, Subra C, Silvente-Poirot S, Poirot M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochemical pharmacology. 2011;81:1171–1182. doi: 10.1016/j.bcp.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 282.Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184:28–41. doi: 10.1016/j.ajpath.2013.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar CM, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 285.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, Tang JH. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PloS one. 2014;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y, Li H, Zhu X, Yao L, Zhang J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res Treat. 2014;147:423–431. doi: 10.1007/s10549-014-3037-0. [DOI] [PubMed] [Google Scholar]
- 288.Chen WX, Cai YQ, Lv MM, Chen L, Zhong SL, Ma TF, Zhao JH, Tang JH. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:9649–9659. doi: 10.1007/s13277-014-2242-0. [DOI] [PubMed] [Google Scholar]
- 289.Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, Greco SJ, Bryan M, Patel PS, Rameshwar P. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 2011;71:1550–1560. doi: 10.1158/0008-5472.CAN-10-2372. [DOI] [PubMed] [Google Scholar]
- 290.Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Harding CV, Heuser JE, Stahl PD. Exosomes: looking back three decades and into the future. J Cell Biol. 2013;200:367–371. doi: 10.1083/jcb.201212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 293.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 294.Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, Sanderson RD. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. The Journal of biological chemistry. 2013;288:10093–10099. doi: 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Traister A, Shi W, Filmus J. Mammalian Notum induces the release of glypicans and other GPI-anchored proteins from the cell surface. Biochem. J. 2008;410:503–511. doi: 10.1042/BJ20070511. [DOI] [PubMed] [Google Scholar]
- 297.De Cat B, Muyldermans SY, Coomans C, Degeest G, Vanderschueren B, Creemers J, Biemar F, Peers B, David G. Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J. Cell Biol. 2003;163:625–635. doi: 10.1083/jcb.200302152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Svensson G, Awad W, Hakansson M, Mani K, Logan DT. Crystal structure of N-glycosylated human glypican-1 core protein: structure of two loops evolutionary conserved in vertebrate glypican-1. J. Biol. Chem. 2012;287:14040–14051. doi: 10.1074/jbc.M111.322487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Fico A, de Chevigny A, Egea J, Bosl MR, Cremer H, Maina F, Dono R. Modulating Glypican4 suppresses tumorigenicity of embryonic stem cells while preserving self-renewal and pluripotency. Stem Cells. 2012;30:1863–1874. doi: 10.1002/stem.1165. [DOI] [PubMed] [Google Scholar]
- 300.Gao W, Kim H, Feng M, Phung Y, Xavier CP, Rubin JS, Ho M. Inactivation of Wnt signaling by a human antibody that recognizes the heparan sulfate chains of glypican-3 for liver cancer therapy. Hepatology. 2014;60:576–587. doi: 10.1002/hep.26996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Shiba K, Hu N, Bronner-Fraser M. Altering glypican-1 levels modulates canonical Wnt signaling during trigeminal placode development. Dev. Biol. 2010;348:107–118. doi: 10.1016/j.ydbio.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Song HH, Shi W, Xiang Y, Filmus J. The loss of Glypican-3 induces alterations in Wnt signaling. J. Biol. Chem. 2005;280:2116–2125. doi: 10.1074/jbc.M410090200. [DOI] [PubMed] [Google Scholar]
- 303.Topczewsky J, Sepich DS, Myers DC, Walker C, Amores A, Lele Z, Hammerschmidt M, Postlethwait J, Solnica-Krezel L. The Zebrafish Glypican Knypek controls cell polarity during gastrulation movements of convergent extension. Dev. Cell. 2001;1:251–264. doi: 10.1016/s1534-5807(01)00005-3. [DOI] [PubMed] [Google Scholar]
- 304.Tsuda M, Kamimura K, Nakato H, Archer M, Staatz W, Fox B, Humphrey M, Olson S, Futch T, Kaluza V, Siegfried E, Stam L, Selleck SB. The cell-surface proteoglycan Dally regulates Wingless signaling in Drosophila. Nature. 1999;400:276–280. doi: 10.1038/22336. [DOI] [PubMed] [Google Scholar]
- 305.Yan D, Wu Y, Feng Y, Lin SC, Lin X. The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Dev. Cell. 2009;17:470–481. doi: 10.1016/j.devcel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits hedgehog signaling during development by competing with Patched for Hedgehog binding. Dev. Cell. 2008;14:700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 307.Desbordes SC, Sanson B. The glypican Dally-like is required for hedgehog signalling in the embryonic epidermis of Drosophila. Development. 2003;130:6245–6255. doi: 10.1242/dev.00874. [DOI] [PubMed] [Google Scholar]
- 308.Li F, Shi W, Capurro M, Filmus J. Glypican-5 stimulates rhabdomyosarcoma cell proliferation by activating Hedgehog signaling. J. Cell Biol. 2011;192:691–704. doi: 10.1083/jcb.201008087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 310.Rawson JM, Dimitroff B, Johnson KG, Rawson JM, Ge X, Van Vactor D, Selleck SB. The heparan sulfate proteoglycans Dally-like and Syndecan have distinct functions in axon guidance and visual-system assembly in Drosophila. Curr. Biol. 2005;15:833–838. doi: 10.1016/j.cub.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 311.Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, Liu H, Lin X. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 312.Fujise M, Takeo S, Kamimura K, Matsuo T, Aigaki T, Izumi S, Nakato H. Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development. 2003;130:1515–1522. doi: 10.1242/dev.00379. [DOI] [PubMed] [Google Scholar]
- 313.Kuo WJ, Digman MA, Lander AD. Heparan sulfate acts as a bone morphogenetic protein coreceptor by facilitating ligand-induced receptor hetero-oligomerization. Mol. Biol. Cell. 2010;21:4028–4041. doi: 10.1091/mbc.E10-04-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Taneja-Bageshwar S, Gumienny TL. Two functional domains in C. elegans glypican LON-2 can indepepndently inhibit BMP-like signaling. Dev. Biol. 2012;371:66–76. doi: 10.1016/j.ydbio.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Gutierrez J, Brandan E. A novel mechanism of sequestering FGF-2 by glypican in lipid rafts, allowing skeletal muscle differentiation. Mol. Cell. Biol. 2010;30:1634–1649. doi: 10.1128/MCB.01164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Okamoto K, Tokunaga K, Doi K, Fujita T, Suzuki H, Katoh T, Watanabe T, Nishida N, Mabuchi A, Takahashi A, Kubo M, Maeda S, Nakamura Y, Noiri E. Common variation in GPC5 is associated with acquired nephrotic syndrome. Nature Genet. 2011;43:459–463. doi: 10.1038/ng.792. [DOI] [PubMed] [Google Scholar]
- 317.Ayers KL, Gallet A, Staccini-Lavenant L, Therond PP. The long-range activity of hedgehog is regulated in the apical extracellular space by the glypican Dally and the hydrolase Notum. Dev. Cell. 2010;18:605–620. doi: 10.1016/j.devcel.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 318.Eugster C, Panakova D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev. Cell. 2007;13:57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 319.Vyas N, Goswami D, Manonmani A, Sharma P, Ranganath HA, VijayRaghavan K, Shashidhara LS, Sowdhamini R, Mayor S. Nanoscale organization of hedgehog is essential for long-range signaling. Cell. 2008;133:1214–1227. doi: 10.1016/j.cell.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 320.Yan D, Wu Y, Yang Y, Belenkaya TY, Tang X, Lin X. The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development. 2010;137:2033–2044. doi: 10.1242/dev.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Franch-Marro X, Marchand O, Piddini E, Ricardo S, Alexandre C, Vincent JP. Glypicans shunt the wingless signal between local signalling and further transport. Development. 2005;132:659–666. doi: 10.1242/dev.01639. [DOI] [PubMed] [Google Scholar]
- 322.Gallet A, Staccini-Lavenant L, Therond PP. Cellular trafficking of the glypican Dally-like is required for full-strength hedgehog signaling and wingless transcytosis. Dev. Cell. 2008;14:712–725. doi: 10.1016/j.devcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 323.Kreuger J, Perez L, Giraldez AJ, Cohen SM. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev. Cell. 2004;7:503–512. doi: 10.1016/j.devcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 324.Smart AD, Course MM, Rawson J, Selleck SB, Van Vactor D, Johnson KG. Heparan sulfate proteoglycan specificity during axon pathway formation in Drosophila embryo. Dev. Neurobiol. 2011;71:608–618. doi: 10.1002/dneu.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.de Wit J, O'sullivan ML, Savas JN, Condomitti G, Caccese MC, Vennekens KM, Yates JR, Ghosh A. Unbiased discovery of glypican as a receptor for LRRTM4 in regulating excitatory synapse development. Neuron. 2013;79:1–16. doi: 10.1016/j.neuron.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Capurro M, Xiang YY, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65:6245–6254. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- 328.Murthy SS, Shen T, De Rienzo A, Lee WC, Ferriola PC, Jhanwar SC, Mossman BT, Filmus J, Testa JR. Expression of GPC3, an X-linked recessive overgrowth gene, is silenced in malignant mesothelioma. Oncogene. 2000;19:410–416. doi: 10.1038/sj.onc.1203322. [DOI] [PubMed] [Google Scholar]
- 329.Xiang YY, Ladeda V, Filmus J. Glypican-3 expression is silenced in human breast cancer. Oncogene. 2001;20:7408–7412. doi: 10.1038/sj.onc.1204925. [DOI] [PubMed] [Google Scholar]
- 330.Matsuda K, Maruyama H, Guo F, Kleeff J, Itakura J, Matsumoto Y, Lander AD, Korc M. Glypican-1 is overexpressed in human breast cancer and modulates the mitogenic effects of multiple heparin-binding growth factors in breast cancer cells. Cancer Res. 2001;61:5562–5569. [PubMed] [Google Scholar]
- 331.Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang THM. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- 332.Peters MG, Farias E, Colombo L, Filmus J, Puricelli L, Bal de Kier Joffe E. Inhibition of invasion and metastasis by glypican-3 in a syngeneic breast cancer model. Breast Cancer Res. Treat. 2003;80:221–232. doi: 10.1023/A:1024549729256. [DOI] [PubMed] [Google Scholar]
- 333.Yiu GK, Kaunisto A, Chin R, Toker A. NFAT promotes carcinoma invasive migration through glypican-6. Biochem. J. 2011;440:157–166. doi: 10.1042/BJ20110530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Korpetinou A, Skandalis SS, Labropoulou VT, Smirlaki G, Noulas A, Karamanos NK, Theocharis AD. Serglycin: at the crossroad of inflammation and malignancy. Front Oncol. 2014;3:327. doi: 10.3389/fonc.2013.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Meen AJ, Oynebraten I, Reine TM, Duelli A, Svennevig K, Pejler G, Jenssen T, Kolset SO. Serglycin is a major proteoglycan in polarized human endothelial cells and is implicated in the secretion of the chemokine GROalpha/CXCL1. The Journal of biological chemistry. 2011;286:2636–2647. doi: 10.1074/jbc.M110.151944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Tyan SW, Hsu CH, Peng KL, Chen CC, Kuo WH, Lee EY, Shew JY, Chang KJ, Juan LJ, Lee WH. Breast cancer cells induce stromal fibroblasts to secrete ADAMTS1 for cancer invasion through an epigenetic change. PloS one. 2012;7:e35128. doi: 10.1371/journal.pone.0035128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Pejler G, Sadler JE. Mechanism by which heparin proteoglycan modulates mast cell chymase activity. Biochemistry. 1999;38:12187–12195. doi: 10.1021/bi991046b. [DOI] [PubMed] [Google Scholar]
- 338.Lindstedt L, Lee M, Kovanen PT. Chymase bound to heparin is resistant to its natural inhibitors and capable of proteolyzing high density lipoproteins in aortic intimal fluid. Atherosclerosis. 2001;155:87–97. doi: 10.1016/s0021-9150(00)00544-x. [DOI] [PubMed] [Google Scholar]
- 339.Pejler G, Berg L. Regulation of rat mast cell protease 1 activity. Protease inhibition is prevented by heparin proteoglycan. Eur J Biochem. 1995;233:192–199. doi: 10.1111/j.1432-1033.1995.192_1.x. [DOI] [PubMed] [Google Scholar]
- 340.Hallgren J, Spillmann D, Pejler G. Structural requirements and mechanism for heparin-induced activation of a recombinant mouse mast cell tryptase, mouse mast cell protease-6: formation of active tryptase monomers in the presence of low molecular weight heparin. The Journal of biological chemistry. 2001;276:42774–42781. doi: 10.1074/jbc.M105531200. [DOI] [PubMed] [Google Scholar]
- 341.Sakai K, Ren S, Schwartz LB. A novel heparin-dependent processing pathway for human tryptase. Autocatalysis followed by activation with dipeptidyl peptidase I. The Journal of clinical investigation. 1996;97:988–995. doi: 10.1172/JCI118523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Zhang L, Yang M, Yang D, Cavey G, Davidson P, Gibson G. Molecular interactions of MMP-13 C-terminal domain with chondrocyte proteins. Connect Tissue Res. 2010;51:230–239. doi: 10.3109/03008200903288902. [DOI] [PubMed] [Google Scholar]
- 343.Malla N, Berg E, Theocharis AD, Svineng G, Uhlin-Hansen L, Winberg JO. In vitro reconstitution of complexes between pro-matrix metalloproteinase-9 and the proteoglycans serglycin and versican. FEBS J. 2013;280:2870–2887. doi: 10.1111/febs.12291. [DOI] [PubMed] [Google Scholar]
- 344.Winberg JO, Kolset SO, Berg E, Uhlin-Hansen L. Macrophages secrete matrix metalloproteinase 9 covalently linked to the core protein of chondroitin sulphate proteoglycans. J Mol Biol. 2000;304:669–680. doi: 10.1006/jmbi.2000.4235. [DOI] [PubMed] [Google Scholar]
- 345.Winberg JO, Berg E, Kolset SO, Uhlin-Hansen L. Calcium-induced activation and truncation of promatrix metalloproteinase-9 linked to the core protein of chondroitin sulfate proteoglycans. Eur J Biochem. 2003;270:3996–4007. doi: 10.1046/j.1432-1033.2003.03788.x. [DOI] [PubMed] [Google Scholar]
- 346.Labropoulou VT, Theocharis AD, Symeonidis A, Skandalis SS, Karamanos NK, Kalofonos HP. Pathophysiology and pharmacological targeting of tumor-induced bone disease: current status and emerging therapeutic interventions. Curr Med Chem. 2011;18:1584–1598. doi: 10.2174/092986711795471275. [DOI] [PubMed] [Google Scholar]
- 347.Theocharis AD, Seidel C, Borset M, Dobra K, Baykov V, Labropoulou V, Kanakis I, Dalas E, Karamanos NK, Sundan A, Hjerpe A. Serglycin constitutively secreted by myeloma plasma cells is a potent inhibitor of bone mineralization in vitro. The Journal of biological chemistry. 2006;281:35116–35128. doi: 10.1074/jbc.M601061200. [DOI] [PubMed] [Google Scholar]
- 348.Purushothaman A, Toole BP. Serglycin proteoglycan is required for multiple myeloma cell adhesion, in vivo growth, and vascularization. The Journal of biological chemistry. 2014;289:5499–5509. doi: 10.1074/jbc.M113.532143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Skliris A, Labropoulou VT, Papachristou DJ, Aletras A, Karamanos NK, Theocharis AD. Cell-surface serglycin promotes adhesion of myeloma cells to collagen type I and affects the expression of matrix metalloproteinases. FEBS J. 2013;280:2342–2352. doi: 10.1111/febs.12179. [DOI] [PubMed] [Google Scholar]
- 350.He L, Zhou X, Qu C, Tang Y, Zhang Q, Hong J. Serglycin (SRGN) overexpression predicts poor prognosis in hepatocellular carcinoma patients. Med Oncol. 2013;30:707. doi: 10.1007/s12032-013-0707-4. [DOI] [PubMed] [Google Scholar]
- 351.Li XJ, Ong CK, Cao Y, Xiang YQ, Shao JY, Ooi A, Peng LX, Lu WH, Zhang Z, Petillo D, Qin L, Bao YN, Zheng FJ, Chia CS, Iyer NG, Kang TB, Zeng YX, Soo KC, Trent JM, Teh BT, Qian CN. Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res. 2011;71:3162–3172. doi: 10.1158/0008-5472.CAN-10-3557. [DOI] [PubMed] [Google Scholar]
- 352.Cooney CA, Jousheghany F, Yao-Borengasser A, Phanavanh B, Gomes T, Kieber-Emmons AM, Siegel ER, Suva LJ, Ferrone S, Kieber-Emmons T, Monzavi-Karbassi B. Chondroitin sulfates play a major role in breast cancer metastasis: a role for CSPG4 and CHST11 gene expression in forming surface P-selectin ligands in aggressive breast cancer cells. Breast Cancer Res. 2011;13:R58. doi: 10.1186/bcr2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Skliris A, Happonen KE, Terpos E, Labropoulou V, Borset M, Heinegard D, Blom AM, Theocharis AD. Serglycin inhibits the classical and lectin pathways of complement via its glycosaminoglycan chains: implications for multiple myeloma. Eur J Immunol. 2011;41:437–449. doi: 10.1002/eji.201040429. [DOI] [PubMed] [Google Scholar]
- 354.Raman K, Kuberan B. Chemical Tumor Biology of Heparan Sulfate Proteoglycans. Curr Chem Biol. 2010;4:20–31. doi: 10.2174/187231310790226206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Casu B, Naggi A, Torri G. Heparin-derived heparan sulfate mimics to modulate heparan sulfate-protein interaction in inflammation and cancer. Matrix Biol. 2010;29:442–452. doi: 10.1016/j.matbio.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Kuberan B, Ethirajan M, Victor XV, Tran V, Nguyen K, Do A. "Click" xylosides initiate glycosaminoglycan biosynthesis in a mammalian cell line. Chembiochem. 2008;9:198–200. doi: 10.1002/cbic.200700494. [DOI] [PubMed] [Google Scholar]
- 357.Garud DR, Tran VM, Victor XV, Koketsu M, Kuberan B. Inhibition of heparan sulfate and chondroitin sulfate proteoglycan biosynthesis. The Journal of biological chemistry. 2008;283:28881–28887. doi: 10.1074/jbc.M805939200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Gingis-Velitski S, Zetser A, Kaplan V, Ben-Zaken O, Cohen E, Levy-Adam F, Bashenko Y, Flugelman MY, Vlodavsky I, Ilan N. Heparanase uptake is mediated by cell membrane heparan sulfate proteoglycans. The Journal of biological chemistry. 2004;279:44084–44092. doi: 10.1074/jbc.M402131200. [DOI] [PubMed] [Google Scholar]
- 359.Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci U S A. 2013;110:17380–17385. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Ritchie JP, Ramani VC, Ren Y, Naggi A, Torri G, Casu B, Penco S, Pisano C, Carminati P, Tortoreto M, Zunino F, Vlodavsky I, Sanderson RD, Yang Y. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/Syndecan-1 axis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:1382–1393. doi: 10.1158/1078-0432.CCR-10-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Zhou H, Roy S, Cochran E, Zouaoui R, Chu CL, Duffner J, Zhao G, Smith S, Galcheva-Gargova Z, Karlgren J, Dussault N, Kwan RY, Moy E, Barnes M, Long A, Honan C, Qi YW, Shriver Z, Ganguly T, Schultes B, Venkataraman G, Kishimoto TK. M402, a novel heparan sulfate mimetic, targets multiple pathways implicated in tumor progression and metastasis. PloS one. 2011;6:e21106. doi: 10.1371/journal.pone.0021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Dredge K, Hammond E, Handley P, Gonda TJ, Smith MT, Vincent C, Brandt R, Ferro V, Bytheway I. PG545, a dual heparanase and angiogenesis inhibitor, induces potent anti-tumour and anti-metastatic efficacy in preclinical models. Br J Cancer. 2011;104:635–642. doi: 10.1038/bjc.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Hammond E, Brandt R, Dredge K. PG545, a heparan sulfate mimetic, reduces heparanase expression in vivo, blocks spontaneous metastases and enhances overall survival in the 4T1 breast carcinoma model. PloS one. 2012;7:e52175. doi: 10.1371/journal.pone.0052175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Barbouri D, Afratis N, Gialeli C, Vynios DH, Theocharis AD, Karamanos NK. Syndecans as modulators and potential pharmacological targets in cancer progression. Front Oncol. 2014;4:4. doi: 10.3389/fonc.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Choi S, Kang DH, Oh ES. Targeting syndecans: a promising strategy for the treatment of cancer. Expert Opin Ther Targets. 2013;17:695–705. doi: 10.1517/14728222.2013.773313. [DOI] [PubMed] [Google Scholar]
- 366.Malavaki CJ, Roussidis AE, Gialeli C, Kletsas D, Tsegenidis T, Theocharis AD, Tzanakakis GN, Karamanos NK. Imatinib as a key inhibitor of the platelet-derived growth factor receptor mediated expression of cell surface heparan sulfate proteoglycans and functional properties of breast cancer cells. FEBS J. 2013;280:2477–2489. doi: 10.1111/febs.12163. [DOI] [PubMed] [Google Scholar]
- 367.Rousseau C, Ferrer L, Supiot S, Bardies M, Davodeau F, Faivre-Chauvet A, Baumgartner P, Wijdenes J, Lacombe M, Barbet J, Guillaume T, Moreau P, Harousseau JL, Kraeber-Bodere F, Cherel M. Dosimetry results suggest feasibility of radioimmunotherapy using anti-CD138 (B-B4) antibody in multiple myeloma patients. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:679–688. doi: 10.1007/s13277-012-0362-y. [DOI] [PubMed] [Google Scholar]
- 368.Rousseau C, Ruellan AL, Bernardeau K, Kraeber-Bodere F, Gouard S, Loussouarn D, Sai-Maurel C, Faivre-Chauvet A, Wijdenes J, Barbet J, Gaschet J, Cherel M, Davodeau F. Syndecan-1 antigen, a promising new target for triple-negative breast cancer immuno-PET and radioimmunotherapy. A preclinical study on MDA-MB-468 xenograft tumors. EJNMMI Res. 2011;1:20. doi: 10.1186/2191-219X-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Rapraeger AC. Synstatin: a selective inhibitor of the Syndecan-1-coupled IGF1R-alphavbeta3 integrin complex in tumorigenesis and angiogenesis. FEBS J. 2013;280:2207–2215. doi: 10.1111/febs.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Ween MP, Hummitzsch K, Rodgers RJ, Oehler MK, Ricciardelli C. Versican induces a pro-metastatic ovarian cancer cell behavior which can be inhibited by small hyaluronan oligosaccharides. Clin Exp Metastasis. 2011;28:113–125. doi: 10.1007/s10585-010-9363-7. [DOI] [PubMed] [Google Scholar]
- 371.Nastase MV, Iozzo RV, Schaefer L. Key roles for the small leucine-rich proteoglycans in renal and pulmonary pathophysiology. Biochim Biophys Acta. 2014;1840:2460–2470. doi: 10.1016/j.bbagen.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Tralhao JG, Schaefer L, Micegova M, Evaristo C, Schonherr E, Kayal S, Veiga-Fernandes H, Danel C, Iozzo RV, Kresse H, Lemarchand P. In vivo selective and distant killing of cancer cells using adenovirus-mediated decorin gene transfer. FASEB J. 2003;17:464–466. doi: 10.1096/fj.02-0534fje. [DOI] [PMC free article] [PubMed] [Google Scholar]