Abstract
To find the potential reasons for the discrepancies in the drug susceptibility test (DST) of M. tuberculosis isolates, twenty paired isolates with disputed drug susceptibilities to isoniazid (INH) were selected according to the MGIT960 testing and Löwenstein-Jensen (L-J) proportion methods. Their MICs were confirmed again by broth microdilution method and by L-J proportion method. The spoligotyping results showed that, of all the 20 paired strains, 11 paired isolates belonged to the Beijing genotype and 6 paired isolates belonged to SIT1634, and that each of the remaining 3 paired isolates had two genotypes, namely, SIT1 and SIT1634. Those 3 paired isolates with different intrapair spoligotypes were further confirmed as mixed infection by the results that those three pairs of isolates with different 12 locus MIRU intrapair types and one pair carried different base pair at codon 315 (AGC versus AAC). Totally mutations in the katG gene were identified in 13 paired isolates. No mutations were found in the regulatory sequences and open reading frames (ORF) of the inhA and ahpC genes in any of the tested isolates. Those results showed that the different test systems and the mixed infection with particular genotypes of M. tuberculosis strains contributed to the drug susceptibility discrepancies.
1. Introduction
Performance of drug susceptibility testing (DST) to measure drug resistance is important not only before treatment, but also in the course of therapy to identify acquired resistance, especially in the areas with a high incidence of MDR-TB [1]. Conventional DST methods rely on egg-based (Löwenstein-Jensen; L-J) or agar-based (Middlebrook) media, but these are laborious and time-consuming procedures requiring 3 to 8 weeks to obtain results [2]. A number of new methods for DST, including the mycobacterial growth indicator tube (MGIT) [3], E test [4], and Alamar blue [5] methods, have been introduced over the last decade to detect mycobacteria rapidly and to improve their growth rates [6, 7].
The BACTEC MGIT960 method has been assessed in many countries and its degree of agreement with conventional DST methods in M. tuberculosis has been assessed [8–10]. Meta-analysis of published results revealed high accuracy and high predictive value associated with the use of BACTEC MGIT960 [11]. However, there are still discrepancies in the DST results obtained for different anti-TB drugs between BACTEC MGIT960 and other DST methods. The discrepancies in INH susceptibility between the MGIT960 and L-J proportion methods, for example, varied from 0% to 1% [9]; however, few investigations have been reported that addressed the possible mechanisms underlying the discrepancies between the MGIT960 system and L-J proportion methods.
Discrepancies can arise from many reasons, for example, different DST systems used, mixed infection with different M. tuberculosis strains, and last but not least, contamination. In this study, 20 paired isolates with disputed drug susceptibilities to INH were selected according to the MGIT960 testing and L-J proportion methods. The name of the “paired isolates” referred to the two isolates obtained separately from the cultures after the DST by MGIT960 and L-J proportion methods from the same sputum of the patient. The reasons for the DST discrepancies were analyzed by the spoligotyping and VNTR genotyping methods and drug resistance-related mutations tested in INH resistance-related genes.
2. Materials and Methods
2.1. Strains and Antibiotics
A total of 20 paired M. tuberculosis isolates with DST discrepancies were collected in Tianjin Haihe Hospital in the year of 2006 from total 1412 isolates (Figure 1). “paired isolates” were from the the culture of the MGIT960 and L-J proportion method, respectively, which was mentioned above. Meanwhile, 96 randomly selected isolates, whose MGIT960 and agar proportion DST results were in agreement, were also collected from the same hospital. The 20 paired M. tuberculosis isolates were determined to be sensitive to INH using the conventional L-J proportion method (1 μg/mL) [12] but resistant to INH using the BACTEC MGIT960 method (0.1 μg/mL, Becton Dickinson Microbiology Systems, MD, USA) [9]. M. tuberculosis H37Rv (ATCC 27294) obtained from the Chinese National Reference Laboratory was used as a control.
Figure 1.
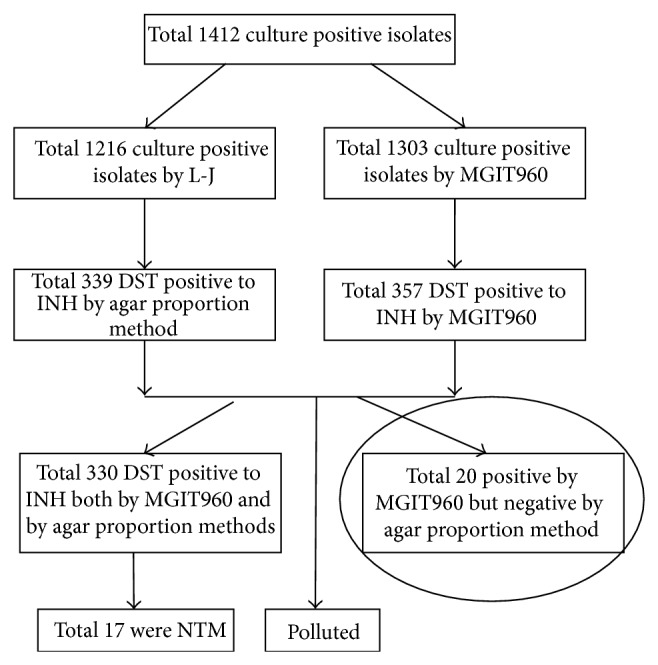
Strains selected in this experiment. A total of 1014 culture positive isolates were included in this study which were isolated in 2006. We focus on the INH as it is a very important antibiotic in curing tuberculosis. In this study of all the total 1412 culture positive isolates 1216 were positive on the L-J medium, of which 339 were resistant by L-J method to INH and 1303 isolates were positive by the MGIT960, of which 357 were resistant to INH by the MGIT960 system. Total 330 were DST positive to INH by both MGIT960 and agar proportion methods, of which 20 isolates with positive both by MGIT960 system but negative by agar proportion method were examined in this study.
2.2. Determination of the MIC of INH by Middlebrook 7H9 Broth Microdilution and L-J Agar Dilution
Resazurin was used as an indicator to test the MIC of INH in the Middlebrook 7H9 broth microdilution method [13]. Briefly, a 100 μL volume of Middlebrook 7H9 broth containing 0.05% Tween 80 and 10% OADC (Sigma, USA) was dispensed into the wells of a 96-well cell culture plate (Corning Coast). INH concentrations, in Middlebrook 7H9 medium, were as follows: 0.1, 0.2, 0.4, 0.8, 1.0, 1.2, 1.6, and 1.8 mg/L. Recovered isolates were collected from L-J slants and homogenized. Turbidity was adjusted to the number 1 McFarland standard (approximately 1 × 107 CFU/mL) and the suspension is diluted 1 : 10 and 100 μL of the dilution is added in each well that contains 100 μL of the appropriate INH dilution. The final inoculum concentration was 5 × 104 CFU/mL. The plates were sealed and incubated at 37°C for one week. Twenty-five microliter of 0.02% resazurin (Sigma Chem. Co., USA) solution was then added to each well and the plates were incubated for an additional 2 days. A change in color from blue to pink indicated the growth of bacteria and the MIC was read as the minimum INH concentration that prevented the color change in the presence of resazurin.
Determination of the MIC of INH using the L-J proportion method followed the protocol of the Chinese Anti-Tuberculosis Association [12]. INH concentrations used in the L-J medium were 2.0, 1.8, 1.6, 1.2, 1.0, 0.8, 0.6, 0.4, and 0.2 mg/L. About 105 CFU were inoculated on the INH-containing medium slants and results were recorded after 5-6 weeks.
2.3. Genomic DNA Isolation, Polymerase Chain Reaction (PCR), and Sequence Analysis
Colonies were first removed from the recovering slants by scraping, resuspended in 500 μL of TE (10 mM Tris, 1 mM EDTA (pH 8.0)), and killed by heating at 80°C for 30 min. The DNA extraction method, primers (from CyberSyn Co. Beijing, China), and PCR conditions were as described previously [14]. The primers were designed to amplify the katG gene, including the region around codon 315, the inhA regulatory region, the inhA ORF, and oxyR-ahpC regions (Table 1) [15, 16]. Both strands were sequenced for confirmation. Mutations were identified by BLAST comparisons with M. tuberculosis H37Rv as the reference (GenBank number NC_000962.3).
Table 1.
Primers used for PCR amplification in this study.
| Gene | Forward primer, 5′-3′ | Reverse primer, 5′-3′ |
|---|---|---|
| katG | GCT GCT GTG GCC GGT CAA GA | CGT CCT TGG CGG TGT ATT GC |
| inhA reg | CCT CGC TGC CCA GAA AGG GA | ATC CCC CGG TTT CCT CCG GT |
| inhA ORF | GAA CTC GAC GTG CAA AAC | CAT CGA AGC ATA CGA ATA |
| oxyR-ahpC | CTG CGA CGG TGC TGG CACG | CAC GCT GCT GCG GGT GAT TGA T |
|
| ||
| MIRU and spoligotyping cluster for M. tuberculosis isolates | ||
| Spoligotyping | GGT TTT GGG TCT GAC GAC | CCG AGA GGG GAC GGA AAC |
| MIRU02 | TGG ACT TGC AGC AAT GGA CCA ACT | TAC TCG GAC GCC GGC TCA AAA T |
| MIRU04 | GCG CGA GAG CCC GAA CTG C | GCG CAG CAG AAA CGT CAG C |
| MIRU10 | GTT CTT GAC CAA CTG CAG TCG TCC | GCC ACC TTG GTG ATC AGC TAC CT |
| MIRU16 | TCG GAG AGA TGC CCT TCG AGT TAG | CCC GTC GTG CAG CCC TGG TAC |
| MIRU20 | TCG GAG AGA TGC CCT TCG AGT TAG | GGA GAC CGC GAC CAG GTA CTT GTA |
| MIRU23 | CTG TCG ATG GCC GCA ACA AAA CG | AGC TCA ACG GGT TCG CCC TTT TGT C |
| MIRU24 | CGA CCA AGA TGT GCA GGA ATA CAT | GGG CGA GTT GAG CTC ACA GAA |
| MIRU26 | TAG GTC TAC CGT CGA AAT CTG TGA C | CAT AGG CGA CCA GGC GAA TAG |
| MIRU27 | TCG AAA GCC TCT GCG TGC CAG TAA | GCG ATG TGA GCG TGC CAC TCA A |
| MIRU31 | ACT GAT TGG CTT CAT ACG GCT TTA | GTG CCG ACG TGG TCT TGA T |
| MIRU39 | CGC ATC GAC AAA CTG GAG CCA AAC | CGG AAA CGT CTA CGC CCC ACA CAT |
| MIRU40 | GGG TTG CTG GAT GAC AAC GTG T | GGG TGA TCT CGG CGA AAT CAG ATA |
2.4. Molecular Typing by Spoligotyping and the 12-Locus MIRU Method
Spoligotyping was performed with a commercial kit (Isogen Bioscience BV, Maarssen, The Netherlands) according to the manufacturer's instructions. Amplification of the direct variant regions for spoligotyping was performed essentially as described previously [17]. Interpretation of spoligotype patterns and assignment of octal codes were based on SITVIT2 database (Pasteur Institute of Guadeloupe, Parris, France), which is an updated version of the previously released SpolDB4 database (http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/tsSpoligo.jsp), as previously described [18].
The numbers of tandem repeats (TRs) at each locus in the isolates were determined on the basis of the number of whole repeats in a PCR product of the size estimated from the gel [19]. Polymerase chain reaction assays for the 12 chosen loci were repeated and compared within and between gels to ensure consistent estimation of size and TR copy number [20].
3. Results
3.1. Genotyping Analysis
Genotyping analysis can determine not only whether an infection results from transmission of the given tuberculosis isolate, but also whether the infection involves more than one strain of M. tuberculosis. Results from our genotyping analysis showed that 10 paired isolates belong to the Spoligotype International Type SIT1 (Beijing genotype, 000000000003771) and 6 paired isolates belong to the Spoligotype International Type SIT1634 (MANU2, 777777777723771) (Table 2), a spoligotype that was not found in the 96 randomly selected clinical isolates (Table 3). Three paired isolates were mixtures of the SIT1 and SIT1634 spoligotypes, and one pair was a mixture of SIT1 and the SIT269 (Beijing genotype, 000000000000771) spoligotypes. Compared with our set of 96 randomly selected isolates from Tianjin, only the Beijing and MANU genotypes were present and the percentage of the MANU genotype was extremely high (20 paired isolates: 15/40, 37.5%; 96 random clinical isolates: 3/96, 3.125%).
Table 2.
Genotypes of the 20 isolates with discrepancies in their INH DST as determined by the Middlebrook 7H9 broth microdilution and L-J agar dilution methods.
| Pairs | Isolates | Spoligotyping pattern | MIRU pattern |
|---|---|---|---|
| 1 | 2235 | 777777777723771 | 1241 2728 3422 |
| 3010 | 777777777723771 | 1241 2728 3422 | |
|
| |||
| 2 | 3195 | 000000000003771 | 1261 2718 3322 |
| 2986 | 000000000003771 | 1261 2718 3322 | |
|
| |||
| 3 | 3184 | 777777777723771 | 2261 2425 3322 |
| 3255 | 777777777723771 | 2261 2425 3322 | |
|
| |||
| 4 | 2577 | 000000000003771 | 1261 2718 3322 |
| 549 | 000000000003771 | 1261 2718 3322 | |
|
| |||
| 5 | 3478 | 000000000003771 | 1361 2618 3322 |
| 3972 | 000000000003771 | 1361 2618 3322 | |
|
| |||
| 6 | 322 | 777777777723771 | 1241 2728 3422 |
| 501 | 000000000003771 | 1261 2718 3322 | |
|
| |||
| 7 | 2671 | 000000000000771 | 1261 2719 3312 |
| 1182 | 000000000003771 | 1261 2719 3312 | |
|
| |||
| 8 | 2851 | 000000000003771 | 1241 2728 3422 |
| 1563 | 000000000003771 | 1241 2728 3422 | |
|
| |||
| 9 | 2566 | 777777777723771 | 1241 2728 3322 |
| 497 | 777777777723771 | 1241 2728 3322 | |
|
| |||
| 10 | 3079 | 777777777723771 | 1241 2728 3322 |
| 2435 | 777777777723771 | 1241 2728 3322 | |
|
| |||
| 11 | 3995 | 000000000003771 | 1261 2728 3322 |
| 4835 | 000000000003771 | 1261 2728 3322 | |
|
| |||
| 12 | 4394 | 000000000003771 | 1261 2718 3322 |
| 4396 | 777777777723771 | 1241 2728 3422 | |
|
| |||
| 13 | 4124 | 000000000003771 | 1361 2615 3322 |
| 4198 | 000000000003771 | 1361 2615 3322 | |
|
| |||
| 14 | 4192 | 000000000003771 | 2261 2615 3322 |
| 4199 | 000000000003771 | 2261 2615 3322 | |
|
| |||
| 15 | 4348 | 000000000003771 | 1261 2628 3321 |
| 4355 | 000000000003771 | 1261 2628 3321 | |
|
| |||
| 16 | 4482 | 777777777723771 | 1241 2618 3322 |
| 1901 | 777777777723771 | 1241 2618 3322 | |
|
| |||
| 17 | 4484 | 777777777723771 | 2261 2631 3321 |
| 1914 | 777777777723771 | 2261 2631 3321 | |
|
| |||
| 18 | 2098 | 777777777723771 | 2261 2631 3321 |
| 2099 | 000000000003771 | 1241 2648 3322 | |
|
| |||
| 19 | 2785 | 000000000003771 | 1241 2648 3422 |
| 1554 | 000000000003771 | 1241 2648 3422 | |
|
| |||
| 20 | 2789 | 000000000003771 | 1241 2648 3422 |
| 1344 | 000000000003771 | 1241 2648 3422 | |
Note: order of 12 MIRU loci is 2, 4, 10, 16, 20, 23, 24, 26, 27, 31, 39, and 40.
Table 3.
Spoligotyping patterns of the 96 randomly selected M. tuberculosis isolates.
| Number of isolates | Shared types | Spoligotyping pattern |
|---|---|---|
| 85 | Beijing (SIT1) | 000000000003771 |
| 2 | Beijing-like (SIT269) | 000000000000771 |
| 1 | Beijing-like (SIT585) | 000000000000031 |
| 2 | T1 (SIT261) | 737777773760771 |
| 1 | T1 (SIT5) | 000677777760771 |
| 1 | T1 (SIT353) | 777777774760771 |
| 1 | MANU2 (SIT53) | 777777777760771 |
| 1 | Manu_ancestor (SIT523) | 777777777777771 |
| 1 | MANU2 (SIT1195) | 777767477763771 |
| 1 | U (SIT1200) | 703777747777771 |
Results obtained by using the 12-locus MIRU method [19] showed that 20 pairs of isolates had 14 MIRU patterns. Both the spoligotyping and the MIRU patterns were different in the isolates named as 6, 12, and 18 pairs, individually. The isolates named as 7 pairs had different spoligotypes, but the same MIRU type (Table 2).
3.2. MICs of the Tested Strains
To identify the differences between the liquid Middlebrook 7H9 and L-J proportion methods in DST, we tested the MICs of each of the 16 paired INH-resistant isolates and 4 pairs of isolates which consisted of different genotypes using both Middlebrook 7H9 broth microdilution and L-J proportion methods. The MICs of all the 24 tested isolates were determined to be greater than 0.1 μg/mL (0.1 to 0.6 μg/mL) using the Middlebrook 7H9 broth microdilution method and greater than 0.3 μg/mL (0.4 to 1.8 μg/mL) using the L-J proportion method (Table 4). The MICs of 5 pairs of the tested isolates using the L-J proportion method were higher than 1 μg/mL, the cutoff concentration for determining drug susceptibility in the L-J agar proportion method in this study (Table 4).
Table 4.
MIC of INH and the katG, inhA, and oxyR-ahpC mutations of the 20 pairs of M. tuberculosis isolates with DST discrepancies.
| Pairs | Isolate* | 7H9 Middlebrook (μg/mL) | L-J agar (μg/mL) | katG315 | inhA reg | inhA ORF | oxyR-ahpC |
|---|---|---|---|---|---|---|---|
| 1 | 2235 | 0.6 | 1 | AAC | None | None | None |
| 3010 | 0.6 | 1 | AAC | None | None | None | |
| 2 | 3195 | 0.1 | 1 | AGC | None | None | None |
| 2986 | 0.1 | 1 | AGC | None | None | None | |
| 3 | 3184 | 0.4 | 0.6 | ACC | None | None | None |
| 3255 | 0.4 | 0.6 | ACC | None | None | None | |
| 4 | 2577 | 0.2 | 0.4 | AGC | None | None | None |
| 549 | 0.2 | 0.4 | AGC | None | None | None | |
| 5 | 3478 | 0.6 | 1.2 | ACC | None | None | None |
| 3972 | 0.6 | 1.2 | ACC | None | None | None | |
| 6 | 322 | 0.4 | 1 | ACC | None | None | None |
| 501 | 0.2 | 0.6 | ACC | None | None | None | |
| 7 | 2671 | 0.6 | 1.2 | AGC | None | None | None |
| 1182 | 0.4 | 1 | AGC | None | None | None | |
| 8 | 2851 | 0.4 | 1 | AAC | None | None | None |
| 1563 | 0.4 | 1 | AAC | None | None | None | |
| 9 | 2566 | 0.4 | 1 | ACC | None | None | None |
| 497 | 0.4 | 1 | ACC | None | None | None | |
| 10 | 3079 | 0.6 | 1.4 | AGC | None | None | None |
| 2435 | 0.6 | 1.4 | AGC | None | None | None | |
| 11 | 3995 | 0.4 | 1 | ACC | None | None | None |
| 4835 | 0.4 | 1 | ACC | None | None | None | |
| 12 | 4394 | 0.4 | 0.8 | ACC | None | None | None |
| 4396 | 0.4 | 0.8 | ACC | None | None | None | |
| 13 | 4124 | 1 | 1.4 | AAC | None | None | None |
| 4198 | 1 | 1.4 | AAC | None | None | None | |
| 14 | 4192 | 0.4 | 1 | ACC | None | None | None |
| 4199 | 0.4 | 1 | ACC | None | None | None | |
| 15 | 4348 | 0.2 | 0.8 | ACC | None | None | None |
| 4355 | 0.2 | 0.8 | ACC | None | None | None | |
| 16 | 4482 | 0.4 | 1 | AGC | None | None | None |
| 1901 | 0.4 | 1 | AGC | None | None | None | |
| 17 | 4484 | 1 | 1.8 | ACC | None | None | None |
| 1914 | 1 | 1.8 | ACC | None | None | None | |
| 18 | 2098 | 0.4 | 1 | AGC | None | None | None |
| 2099 | 0.4 | 1 | AAC | None | None | None | |
| 19 | 2785 | 0.4 | 1 | AAC | None | None | None |
| 1554 | 0.4 | 1 | AAC | None | None | None | |
| 20 | 2789 | 0.2 | 0.6 | AGC | None | None | None |
| 1344 | 0.2 | 0.6 | AGC | None | None | None |
Note: katG315 is the predominant mutation. The wild type is AGC.
*16 isolates with consistent genotype in pair and 4 pairs of isolates (bold) with different genotypes in pair.
3.3. Sequence Analysis of the Putative INH-Target Genes
Mutations in the katG gene were identified in 13 paired isolates, of which each of 12 paired isolates carried the same mutations and one pair which showed a DST discrepancy by MGIT960 and L-J proportion methods carried different base pair at codon 315 (AGC versus AAC). The AGC315AAC mutation was found in 4 paired isolates, while 9 paired isolates carried the mutation AGC315ACC. The AGC315AAC and AGC 315ACC mutations were not associated with specificity to the Beijing or MANU2 genotypes among the tested isolates. Seven paired isolates did not contain mutations in the katG gene and no mutations were found in the regulatory sequences and open reading frames (ORF) of the inhA and ahpC genes in any of the tested isolates (Table 4).
4. Discussion
Different DST methods have been developed and are used in routine clinical practice such as the conventional L-J methods and the automated MB/BacT (Organon Teknika, Turnhout, Belgium), ESPII (Difco Laboratories, Detroit, Michigan), BACTEC 9000MB (Becton Dickenson Microbiology System, Sparks, MD), and BACTEC MGIT 960 (BBL Becton Dickinson Microbiology Systems, Cockeysville, MD) systems [5, 21–23]. The DST results would be influenced by many steps of the protocol, including the culture and the DST methods. In this study, we analyzed the discrepancy of the drug susceptibility test by the MGIT and L-J methods for the isolates collected from the culture by MGIT and L-J, respectively.
Except for the median time to report the DST results the M. tuberculosis complex culture positivity rates were also greatly different in MGIT and L-J [24], which indicated the possible culture preference to somewhat. And the detection time, accuracy, and performance capacity are also variable by different DST methods. Studies reported that the reasons for the different performance capacity among these methods mainly resulted from the different DST systems [23, 25]. The most obvious difference is the drug concentrations used for the DST. In MGIT system, the sensitive strains were susceptible to the INH less than the 0.1 μg/mL, while the concentration of the INH was 1 μg/mL in L-J system in this study [12, 26, 27]. Of all the 20 paired cases 15 cases had MIC in borderlines between the MGIT and the DST methods, which was a usual reason for the discordant.
Many reports showed that there was a good concordance between DST on L-J and MGIT for INH in DST [25–27]. In this study, we still found that 20 paired isolates with the same genotypes individually showed the discrepancy in the drug susceptibilities to INH according to the MGIT960 testing and L-J proportion methods. Lawson et al. demonstrated that there was a substantial degree of agreement between the two methods, with similar INH and rifampicin DST patterns, but more frequent detection of streptomycin resistance and less frequent detection of ethambutol with L-J than MGIT-960. However, the differences were not statistically significant [25]. A multiple center evaluation showed that the discrepancies in INH susceptibility between the MGIT960 and L-J proportion methods varied from 0% to 1% [9].
Mixed infection with the different genotypes of M. tuberculosis in the same patient also affected the DST results even by the same testing systems [28, 29]. In this study heterogeneous genotypes were found in the isolates from each of the 4 patients. Three patients were infected by the different stains with Spoligotype International Type SIT1634 (Manu2) and Beijing genotypes and 1 patient was infected by the strains with two different Beijing genotypes. And also our test on the mutations of the putative INH-target genes, katG, inhA, and ahpC further confirmed one patient (number 18) with mixed infection by the heterogeneous genotypes (Table 4).
Some mycobacterial characteristics might be associated with particular genotypes. A well-known but controversial example is that the Beijing family strains of M. tuberculosis are often associated with relapse [30], drug resistance [31], and an increased ability to cause disease, to be transmitted within certain geographic settings [32, 33]. The isolates with particular genotypes, such as Spoligotype International Type SIT1634 (Manu2) in this study, showed higher rate of resistance in MGIT960 system than in L-J system. In this study, we found that the percentage of “MANU” genotype strains was markedly increased in paired isolates whose DST results showed discrepancies (37.5%) compared to the randomly selected clinical isolates (3.125%). An unusually high proportion of strains belonging to the “Manu” clade (27.15%) were also reported by Helal et al. [18]. Interestingly, Manu2 strains (SIT1634) have rarely been reported in Tianjin or even in China as a whole [34, 35] or in the SPOLDB4 database (excluding this study, n = 3, 1, from India and 2 from the USA).
In this study, all the 40 isolates were determined as resistant by MGIT and sensitive by L-J, of which twenty-seven isolates were found with mutations in katG315 and 13 isolates were found with no mutations in katG315 (Table 4). Those results of the mutations found in the INH-targeted genes supported that the DST result by the MGIT was more accurate than that by the L-J, and we also found that the MICs of some isolates by L-J agar method were very higher than those in the first execution in clinic, which indicated, to some extent, that the operation needs to be improved in proportion method on L-J agar.
5. Conclusion
Our study confirmed that the discrepancies of the DST in M. tuberculosis clinical isolates did exist for INH. One of the reasons for the discrepancy is the different test systems between the BACTEC MGIT960 system and the traditional L-J proportion method. Mixed infection by the strains with MANU2 and Beijing genotype patterns could also contributed to drug discrepancies.
Acknowledgments
The authors thank the Beijing Bio-Bank of Clinical Resources on Tuberculosis and the Outpatient Department of Tianjin Haihe Hospital for supplying M. tuberculosis H37Rv and clinical isolates. They also thank the National Special Key Project of China on Major Infectious Diseases (2013ZX10003009), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (ZYLX201304), and the Beijing High-level Technical Personnel Training Project in Health (2011-3-069) for financial support.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Zaoxian Mei and Zhaogang Sun contributed equally to this work.
References
- 1.Pasipanodya J. G., Srivastava S., Gumbo T. Meta-analysis of clinical studies supports the pharmacokinetic variability hypothesis for acquired drug resistance and failure of antituberculosis therapy. Clinical Infectious Diseases. 2012;55(2):169–177. doi: 10.1093/cid/cis353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kent P. T., Kubica G. P. Public Health Mycobacteriology. A Guide for Level III Laboratory. Atlanta, Ga, USA: Centers for Diseases Control; 1985. [Google Scholar]
- 3.Walters S. B., Hanna B. A. Testing of susceptibility of Mycobacterium tuberculosis to isoniazid and rifampin by mycobacterium growth indicator tube method. Journal of Clinical Microbiology. 1996;34(6):1565–1567. doi: 10.1128/jcm.34.6.1565-1567.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wanger A., Mills K. Testing of Mycobacterium tuberculosis susceptibility to ethambutol, isoniazid, rifampin, and streptomycin by using Etest. Journal of Clinical Microbiology. 1996;34(7):1672–1676. doi: 10.1128/jcm.34.7.1672-1676.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins L. A., Franzblau S. G. Microplate Alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium . Antimicrobial Agents and Chemotherapy. 1997;41(5):1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piersimoni C. TB control strategies: present and future in diagnostic methods: what the clinician can ask to the lab. Monaldi Archives for Chest Disease. 2002;57(5-6):306–310. [PubMed] [Google Scholar]
- 7.Woods G. L., Fish G., Plaunt M., Murphy T. Clinical evaluation of Difco ESP culture system II for growth and detection of mycobacteria. Journal of Clinical Microbiology. 1997;35(1):121–124. doi: 10.1128/jcm.35.1.121-124.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adegbola R. A., Hill P., Baldeh I., et al. Surveillance of drug-resistant Mycobacterium tuberculosis in The Gambia. International Journal of Tuberculosis and Lung Disease. 2003;7(4):390–393. [PubMed] [Google Scholar]
- 9.Giampaglia C. M. S., Martins M. C., De Oliveira Vieira G. B., et al. Multicentre evaluation of an automated BACTEC 960 system for susceptibility testing of Mycobacterium tuberculosis . International Journal of Tuberculosis and Lung Disease. 2007;11(9):986–991. [PubMed] [Google Scholar]
- 10.Heifets L., Cangelosi G. A. Drug susceptibility testing of Mycobacterium tuberculosis: a neglected problem at the turn of the century. International Journal of Tuberculosis and Lung Disease. 1999;3(7):564–581. [PubMed] [Google Scholar]
- 11.Piersimoni C., Olivieri A., Benacchio L., Scarparo C. Current perspectives on drug susceptibility testing of Mycobacterium tuberculosis complex: the automated nonradiometric systems. Journal of Clinical Microbiology. 2006;44(1):20–28. doi: 10.1128/JCM.44.1.20-28.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinese Anti-Tuberculosis Association. Protocols for Tuberculosis Diagnosis in Laboratory. 1st. Beijing, China: Chinese Educational and Cultural Publisher; 2006. [Google Scholar]
- 13.Palomino J.-C., Martin A., Camacho M., Guerra H., Swings J., Portaels F. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis . Antimicrobial Agents and Chemotherapy. 2002;46(8):2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddiqi N., Shamim M., Hussain S., et al. Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrobial Agents and Chemotherapy. 2002;46(2):443–450. doi: 10.1128/AAC.46.2.443-450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipin M. Y., Stepanshina V. N., Shemyakin I. G., Shinnick T. M. Association of specific mutations in katG, rpoB, rpsL and rrs genes with spoligotypes of multidrug-resistant Mycobacterium tuberculosis isolates in Russia. Clinical Microbiology and Infection. 2007;13(6):620–626. doi: 10.1111/j.1469-0691.2007.01711.x. [DOI] [PubMed] [Google Scholar]
- 16.Sun Z., Zhang J., Zhang X., Wang S., Zhang Y., Li C. Comparison of gyrA gene mutations between laboratory-selected ofloxacin-resistant Mycobacterium tuberculosis strains and clinical isolates. International Journal of Antimicrobial Agents. 2008;31(2):115–121. doi: 10.1016/j.ijantimicag.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Kamerbeek J., Schouls L., Kolk A., et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. Journal of Clinical Microbiology. 1997;35(4):907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helal Z. H., Ashour M. S. E.-D., Eissa S. A., et al. Unexpectedly high proportion of ancestral manu genotype Mycobacterium tuberculosis strains cultured from tuberculosis patients in Egypt. Journal of Clinical Microbiology. 2009;47(9):2794–2801. doi: 10.1128/JCM.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Supply P., Mazars E., Lesjean S., Vincent V., Gicquel B., Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Molecular Microbiology. 2000;36(3):762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 20.Boniotti M. B., Goria M., Loda D., et al. Molecular typing of Mycobacterium bovis strains isolated in Italy from 2000 to 2006 and evaluation of variable-number tandem repeats for geographically optimized genotyping. Journal of Clinical Microbiology. 2009;47(3):636–644. doi: 10.1128/JCM.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamin W. H., Jr., Waites K. B., Beverly A., et al. Comparison of the MB/BacT system with a revised antibiotic supplement kit to the BACTEC 460 system for detection of mycobacteria in clinical specimens. Journal of Clinical Microbiology. 1998;36(11):3234–3238. doi: 10.1128/jcm.36.11.3234-3238.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organisation. The Use of Liquid Medium for Culture and DST in Middle- and Low-Income Countries. 2007. http://www.who.int/tb/dots/laboratory/policy/en/index3.htm. [Google Scholar]
- 23.Yew W. W., Tong S. C. W., Lui K. S., Leung S. K. F., Chau C. H., Wang E. P. Comparison of MB/BacT system and agar proportion method in drug susceptibility testing of Mycobacterium tuberculosis . Diagnostic Microbiology and Infectious Disease. 2001;39(4):229–232. doi: 10.1016/S0732-8893(01)00237-1. [DOI] [PubMed] [Google Scholar]
- 24.Balabanova Y., Drobniewski F., Nikolayevskyy V., et al. An integrated approach to rapid diagnosis of tuberculosis and multidrug resistance using liquid culture and molecular methods in Russia. PLoS ONE. 2009;4(9) doi: 10.1371/journal.pone.0007129.e7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawson L., Emenyonu N., Abdurrahman S. T., et al. Comparison of Mycobacterium tuberculosis drug susceptibility using solid and liquid culture in Nigeria. BMC Research Notes. 2013;6(1, article 215) doi: 10.1186/1756-0500-6-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S. J. Drug-susceptibility testing in tuberculosis: methods and reliability of results. European Respiratory Journal. 2005;25(3):564–569. doi: 10.1183/09031936.05.00111304. [DOI] [PubMed] [Google Scholar]
- 27.World Health Organization. Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. 2008, http://www.who.int/tb/features_archive/xdr_mdr_policy_guidance. [PubMed]
- 28.Shamputa I. C., Jugheli L., Sadradze N., et al. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respiratory Research. 2006;7, article 99 doi: 10.1186/1465-9921-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamputa I. C., Rigouts L., Eyongeta L. A., et al. Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. Journal of Clinical Microbiology. 2004;42(12):5528–5536. doi: 10.1128/JCM.42.12.5528-5536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huyen M. N. T., Buu T. N., Tiemersma E., et al. Tuberculosis relapse in vietnam is significantly associated with Mycobacterium tuberculosis Beijing genotype infections. The Journal of Infectious Diseases. 2013;207(10):1516–1524. doi: 10.1093/infdis/jit048. [DOI] [PubMed] [Google Scholar]
- 31.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerging Infectious Diseases. 2006;12(5):736–743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Jong B. C., Hill P. C., Aiken A., et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in the Gambia. Journal of Infectious Diseases. 2008;198(7):1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanekom M., van der Spuy G. D., Streicher E., et al. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. Journal of Clinical Microbiology. 2007;45(5):1483–1490. doi: 10.1128/JCM.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong Y., Cave M. D., Zhang L., et al. Association between Mycobacterium tuberculosis Beijing/W lineage strain infection and extrathoracic tuberculosis: Insights from epidemiologic and clinical characterization of the three principal genetic groups of M. tuberculosis clinical isolates. Journal of Clinical Microbiology. 2007;45(2):409–414. doi: 10.1128/JCM.01459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang Y., Zhou Y., Zhao B., et al. Spoligotyping and drug resistance analysis of Mycobacterium Tuberculosis strains from national survey in China. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0032976.e32976 [DOI] [PMC free article] [PubMed] [Google Scholar]


