Abstract
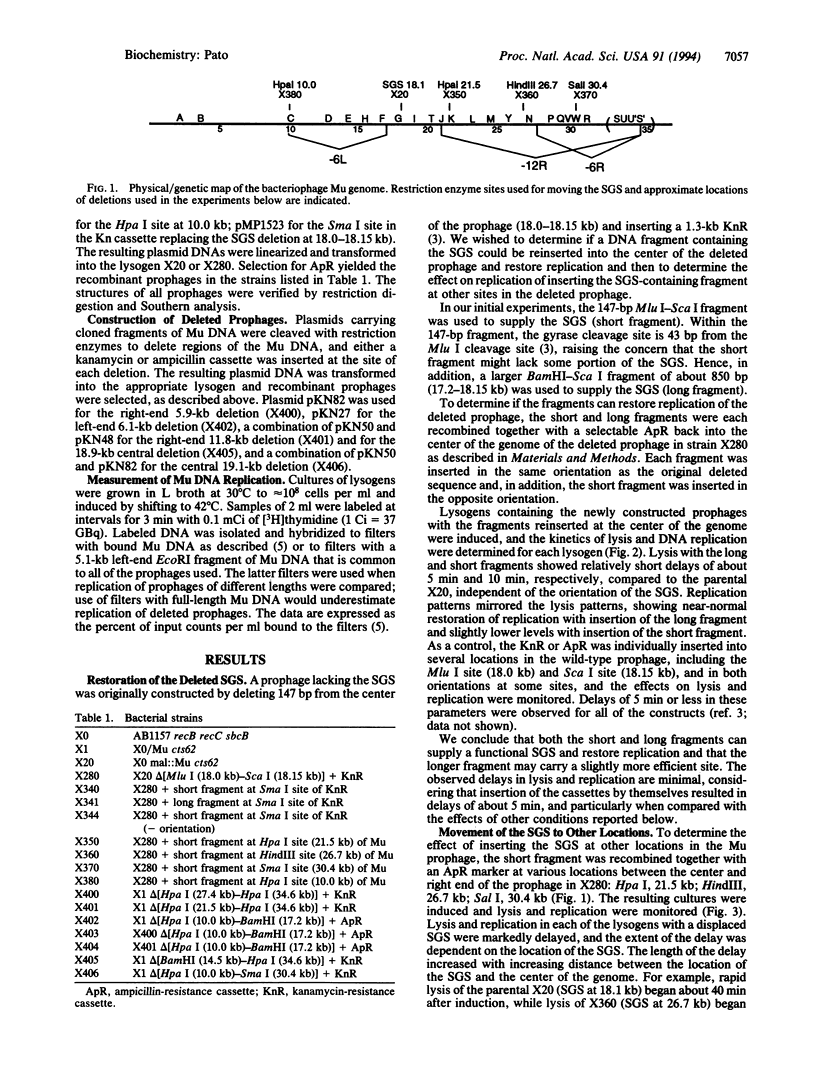
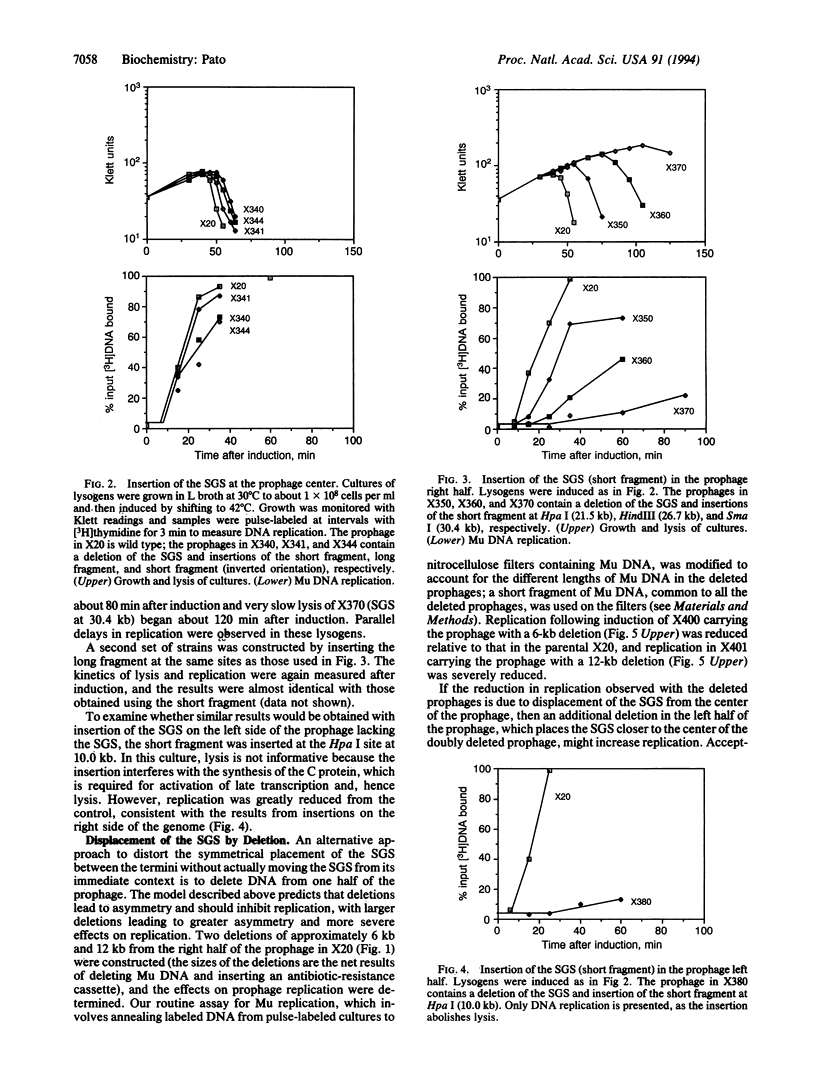
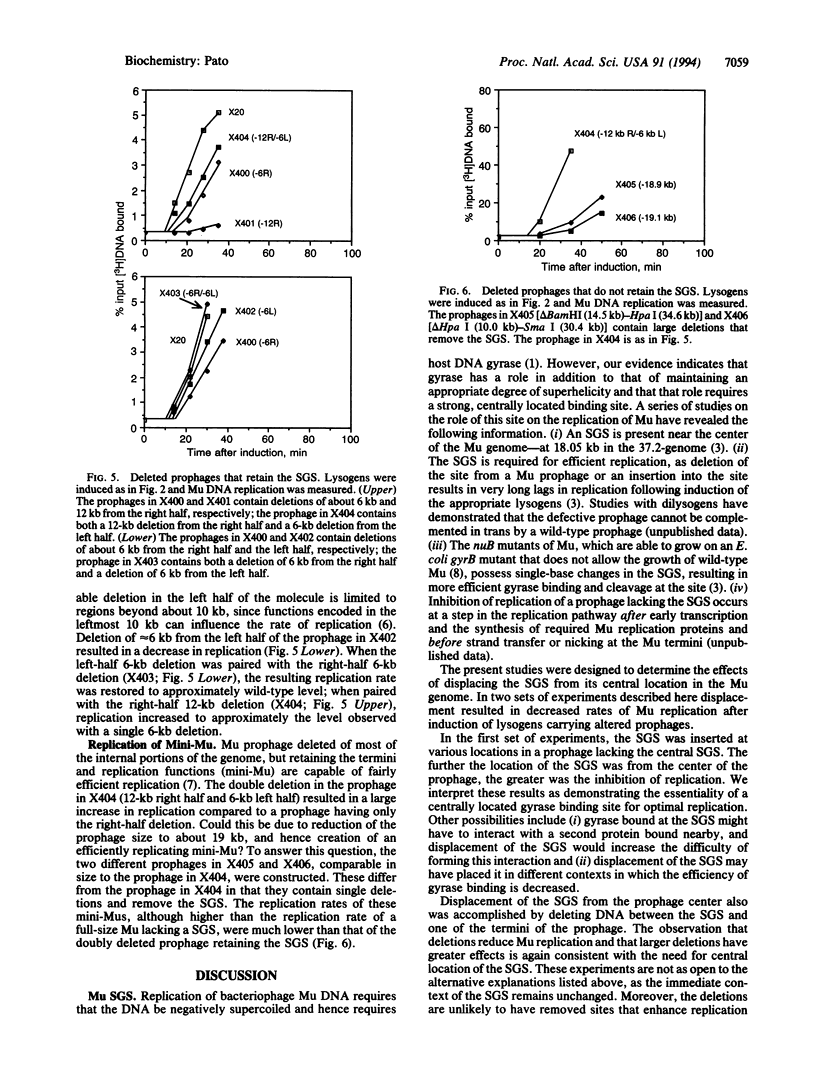
The bacteriophage Mu genome contains a strong DNA gyrase binding site (SGS) near its center, and disruption of the SGS by deletion or by insertion results in long delays in replication following induction of the appropriate lysogen. To determine if the central location of the SGS is obligatory for its function in Mu replication, we pursued two lines of investigation. First, fragments of Mu DNA containing the SGS were inserted into various locations in a Mu prophage lacking the central SGS. Replication following induction was restored in all of the lysogens constructed, but the observed rate of replication for different prophages decreased with increasing distance between the new location of the SGS and the center of the genome. We also deleted different lengths of DNA from within the right half of a wild-type prophage, retaining the SGS and displacing it from a central location. Replication rates of the deleted prophages were reduced, with larger deletions resulting in larger reductions. Pairing deletions in the right half of the prophage with a deletion in the left half resulted in substantially higher rates of replication than observed with the right half deletions alone. We conclude that the SGS must be located centrally between the Mu termini for optimal rates of Mu replication. These results are discussed in terms of a model that proposes that the SGS is involved in organizing the topology of supercoiled prophage DNA to assist in synapsis of the Mu termini.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chandler M., Clerget M., Galas D. J. The transposition frequency of IS1-flanked transposons is a function of their size. J Mol Biol. 1982 Jan 15;154(2):229–243. doi: 10.1016/0022-2836(82)90062-6. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A., Waggoner B., Desmet L., Pato M. Transposition and replication of maxi-Mu derivatives of bacteriophage Mu. Virology. 1986 Aug;153(1):70–79. doi: 10.1016/0042-6822(86)90008-5. [DOI] [PubMed] [Google Scholar]
- Kavenoff R., Ryder O. A. Electron microscopy of membrane-associated folded chromosomes of Escherichia coli. Chromosoma. 1976 Mar 31;55(1):13–25. doi: 10.1007/BF00288323. [DOI] [PubMed] [Google Scholar]
- Laundon C. H., Griffith J. D. Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell. 1988 Feb 26;52(4):545–549. doi: 10.1016/0092-8674(88)90467-9. [DOI] [PubMed] [Google Scholar]
- Morisato D., Way J. C., Kim H. J., Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983 Mar;32(3):799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- Pato M. L., Howe M. M., Higgins N. P. A DNA gyrase-binding site at the center of the bacteriophage Mu genome is required for efficient replicative transposition. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8716–8720. doi: 10.1073/pnas.87.22.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26(3-4):335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci U S A. 1981 Jan;78(1):224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979 Jun 25;131(2):287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- Van Brunt J., Waggoner B. T., Pato M. L. Re-examination of F plasmid replication in a dnaC mutant of Escherichia coli. Mol Gen Genet. 1977 Feb 15;150(3):285–292. doi: 10.1007/BF00268127. [DOI] [PubMed] [Google Scholar]
- Waggoner B., Pato M., Toussaint A., Faelen M. Replication of mini-Mu prophage DNA. Virology. 1981 Aug;113(1):379–387. doi: 10.1016/0042-6822(81)90163-x. [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ames G. F. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R. K., Miller J. L., Miller H. I., Friedman D. I., Howe M. M. Isolation and mapping of Mu nu mutants which grow in him mutants of E. coli. Virology. 1982 Jul 15;120(1):269–272. doi: 10.1016/0042-6822(82)90027-7. [DOI] [PubMed] [Google Scholar]



