Abstract
Background
The aim of this study was to evaluate the ability of 15 serotypes of Salmonella to form biofilm on polystyrene, polyvinyl chloride (PVC) and glass surfaces. .
Methods
Initially slime production was assessed on CRA agar and hydrophobicity of 20 Salmonella strains isolated from poultry and human and two Salmonella enterica serovar Typhimurium references strains was achieved by microbial adhesion to n-hexadecane. In addition, biofilm formation on polystyrene, PVC and glass surfaces was also investigated by using MTT and XTT colorimetric assay. Further, distribution of Salmonella enterotoxin (stn), Salmonella Enteritidis fimbrial (sef) and plasmid encoded fimbrial (pef) genes among tested strains was achieved by PCR.
Results
Salmonella strains developed red and white colonies on CRA and they are considered as hydrophilic with affinity values to n-hexadecane ranged between 0.29% and 29.55%. Quantitative biofilm assays showed that bacteria are able to form biofilm on polystyrene with different degrees and 54.54% of strains produce a strong biofilm on glass. In addition, all the strains form only a moderate (54.54%) and weak (40.91%) biofilm on PVC. PCR detection showed that only S. Enteritidis harbour Sef gene, whereas Pef and stn genes were detected in S. Kentucky, S. Amsterdam, S. Hadar, S. Enteritidis and S. Typhimurium.
Conclusion
Salmonella serotypes are able to form biofilm on hydrophobic and hydrophilic industrial surfaces. Biofilm formation of Salmonella on these surfaces has an increased potential to compromise food safety and potentiate public health risk.
Keywords: Salmonella, Hydrophobocity, Biofilm, Hydrophilic and hydrophobic surfaces, Fimbriae genes
Introduction
Salmonella enterica are important facultative intracellular pathogens that cause gastroenteritis in humans (1). The diverse Salmonella genus contains over 2500 serotypes (2), all of which are potentially pathogenic to humans (3). Specifically, Salmonella enterica serovar Typhimurium (S. Typhimurium) is implicated in human foodborne illnesses and often enters the human food supply via contamination of poultry, pork, beef and dairy products, and nuts such as peanuts and pistachios. Non-typhoidal salmonellosis is estimated to affect 1.4 million people each year in the United States, while more than 95% of cases of infections caused by these bacteria are foodborne. These infections caused account for about 30% of deaths resulting from foodborne illnesses (4).
“Biofilms are the predominant mode of bacterial growth, reflected in the observation that approximately 80% of all bacterial infections are related to biofilms (National Institutes of Health (USA)” (5–7). “Biofilms are defined as structured communities of bacterial cells enclosed in a self-produced polymeric matrix adherent to inert or living surfaces (8–9). Biofilm formation has serious implications in industrial, environmental, public health and medical situations” (10, 11). “In food industry, biofilms may create a persistent source of product contamination, leading to serious hygienic problems and also economic losses due to food spoilage” (12, 13). Improperly cleaned surfaces promote soil build-up, and, in the presence of water, contribute to the development of bacterial biofilms which may contain pathogenic microorganisms, such as Salmonella. Cross contamination occurs when cells detach from biofilm structure once food passes over contaminated surfaces or through aerosols originating from contaminated equipment. Bacteria in biofilms are generally well protected against environmental stresses, antibiotics (14), disinfectants and the host immune system (15) and as a consequence are extremely difficult to eradicate (16). Several reports have demonstrated the ability of Salmonella strains to form biofilms on abiotic surfaces such as plastic (17), rubber (18), cement (19), glass (20) and stainless steel (21).
The objective of the investigation was to study the ability of 15 serotypes of Salmonella originating from Tunisia to form biofilm on polystyrene, PVC and glass using quantitative calorimetric methods. Slime production and cell surface hydro-phobicity were also investigated. In addition the prevalence of Salmonella enterotoxin (stn), Salmonella Enteritidis fimbriae (sef) and plasmid encoded fimbriae (pef) genes was realised.
Materials and methods
Bacterial isolation and identification
Ten Salmonella isolated from human and ten isolates from poultry meat were used in this study (Table 1). Clinical isolates were delivered from Laboratory of Microbiology, University Hospital Fattouma Bourguiba, Monastir, Tunisia. Salmonella strains were isolated according to the standard procedure for Salmonella isolation. Isolates with typical cultural characteristics were further identified by conventional biochemical testing and sero-logic typing. In addition, two references strains, S. Typhimurium ATCC 14028s and S. Typhimurium LT2 DT104, provided from French Food Safety Agency, were also used in this study. These two species are part of S. enterica subspecies I, which colonizes mammals and birds and causes 99% of Salmonella infections in humans. All strains were maintained at −80°C in Luria-Bertani (LB) broth supplemented with glycerol (15%, vol/vol).
Table 1.
Slime production, hydrophobicity and biofilm formation on polystyrene of Salmonella serotypes
| Strains | Origin | Biofilm phenotype CRA | Hydrophobicity (%) ±SD | Biofilm formation on polystyrene (XTT reduction mean OD492 ± SD) |
|---|---|---|---|---|
| S1: S. Enteritidis | Blood | white | 10.09± 0.32 | 0.256±0.17 |
| S2: S. Enteritidis | white | 13.75 ±0.2 | 0.74±0.05 | |
| S3: S. Enteritidis | white | 4.53± 0.1 | 0.50±0.27 | |
| S4: S. Enteritidis | Urine | white | 7.65± 0.2 | 0.65±0.21 |
| S5: S. Enteritidis | white | 18.51± 0.05 | 0.88±0.02 | |
| S6: S. Enteritidis | Pus | white | 0.29± 0.56 | 0.53±0.13 |
| S7: S. Amsterdam | Stool | Red | 9.24± 0.17 | 0.41±0.12 |
| S8: S. Muenster | white | 5.69± 0.21 | 0.73±0.03 | |
| S9: S. Kentucky | white | 7.05 ± 0.11 | 0.65±0.12 | |
| S10: S. Zanzibar | Red | 6.09± 0.14 | 1.05±0.1 | |
| S11: S. Arizona | Poultry meat | white | 17.09± 0.22 | 0.78±0.16 |
| S12: S. Wangata | white | 13.11± 0.15 | 1.08±0.05 | |
| S13: S. Braenderup | white | 1.59± 0.22 | 0.56±0.01 | |
| S14: S. Montevideo | white | 9.23± 0.19 | 0.63±0.18 | |
| S15: S. Cerro | white | 21.2± 0.42 | 0.58±0.02 | |
| S16: S. Agona | Red | 10± 0.2 | 0.60±0.07 | |
| S17: S. Hadar | white | 13.03 ± 0.15 | 0.61±0.03 | |
| S18 : S. Newport | white | 22.87± 0.33 | 0.82±0.05 | |
| S19: S. Altona | white | 15.58± 0.02 | 0.89±0.1 | |
| S20: S. Schwarzengrund | white | 29.55± 0.1 | 0.72±0.04 | |
| S21: S. Typhimurium 14028s | white | 28.66 ± 0.2 | 1.47±0.02 | |
| S22: S. Typhimurium LT2 DT104 | white | 25 ± 0.17 | 1.21±0.01 |
Phenotypic characterization of slime-producing bacteria
Qualitative detection of biofilm formation was studied by culturing the strains on Congo red agar (CRA) plates as described previously (22). Salmonella strains were inoculated into the surface of CRA plates, prepared by mixing 0.8 g Congo red with 36 g saccharose (Sigma-Aldrich, St Louis, MO) in 1 L of brain heart infusion agar, and were incubated for 24 h at 37°C under aerobic conditions and followed overnight at room temperature (23) Slime producing bacteria appeared as black colonies, whereas non-slime producers remained non pigmented.
Biofilm formation on polystyrene
The XTT assay was used to quantify bacterial biofilm (24). It measures the reduction of a tetrazolium salt (2, 3-bis (2-methyloxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) by metabolically active cells to a coloured water soluble formazan derivative that can be easily quantified colorimetrically (25). XTT (Sigma-Aldrich, Switzerland) solution (1 mg/ml) was prepared in PBS (7 mM Na2HPO4, 3 mM NaH2PO4 and 130 mM NaCl at pH 7.4), filter sterilized and stored at -80°C. Menadione (Sigma-Aldrich, Switzerland) solution (1 mM) was prepared in acetone and sterilized immediately before each assay.
An overnight culture grown in TSB (Bio-Rad), of Salmonella cells, at 37°C was diluted to 1:100 in TSB supplement with 2% (wt/vol) glucose. A total of 200 μl of these cell suspensions was transferred in a U-bottomed 96-well microtiter plate. Each strain was tested in triplicate. Wells with sterile TSB alone was served as controls. Following incubation for 24 h at 37°C, the biofilms were first washed five times with 200 μl of PBS, and then 100 μl PBS and 12 μl XTT-menadione solution (12.5:1 v/v) were added to each of the pre-washed wells and the control wells. The plate was then incubated for 3 h in the dark at 37°C. Following incubation, 100 μl of the solution was transferred to fresh wells, and the colour change in the solution was measured with a multiscan reader (GIO, Rome, Italy) at 492 nm. The absorbance values for the controls were then subtracted from the values of the tested wells to eliminate spurious results due to background interference. Each assay was repeated three times.
Biofilm assessment on polyvinyl chloride (PVC) and on glass microscope slide covers surfaces
For thus, cells were grown for 18 h at 37°C in trypticase soy broth (TSB) supplemented with 2% of glucose. Batches of medium were inoculated with overnight cell cultures and incubated at 37°C in an orbital shaker operating at 150 rpm. Cells were harvested after 24 h (stationary growth phase), washed once with PBS (pH 7.2), and standardized to a density OD600 = 0.3. A volume of 80 μl of the standardized Salmonella cells suspensions was applied for all the tested strips (1cm2) placed in a 12-well tissue culture plate. The cells were allowed to adhere for 90 min at 37°C (adhesion phase). Non-adherent cells were removed from the strips by being gently washed with 5 ml PBS. Strips were then submerged in 4 ml of TSB containing 2% of glucose. Strips to which no cells were added served as negative controls. Control and experimental strips were incubated at 37°C for 90 min (adhesion step). The percentage of viable cells in biofilms was estimated with the bromure 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) colorimetric assay based on the ability of cells to metabolically reduce MTT to a water soluble formazan dye (26). Strips with biofilms were transferred to 12-well tissue culture plates containing 3 ml PBS/well. Fifty microliters of MTT (5 mg/ml in PBS) were added to each well. Plates were incubated for additional 24 h at 37°C. The supernatant was then removed and the formazan product was determined spectrophotometrically at 578 nm. Based on the O.D produced by bacterial films, strains were classified into the following categories: no biofilm producers, weak, moderate or strong biofilm producers, as previously described. Briefly, the cut-off O.D (ODc) was defined as three standard deviations above the mean O.D of the negative control. Strains were classified as follows: OD < ODc no biofilm producer, ODc < OD ≤ 2 × ODc weak biofilm producer, 2 × ODc < OD ≤ 4 × ODc moderate biofilm producer and 4 × ODc < OD strong biofilm producer. Experiments were performed in triplicate.
Cell surface hydrophobicity
Hydrophobicity was measured by the hexadecane partitioning method of van Loosdrecht et al. (27). Bacterial cells were grown overnight in TSB, and were washed three times by centrifugation at 6000 rpm for 10 min with PBS to remove the broth residues then resuspended in 4 ml of PBS, and the absorbance (DO540) was determined. One milliliter of hexadecane was added to each cell suspension and vortexed for 5 min. Each suspension was re-incubated at 37°C for 30 min. The aqueous layer was removed and aerated to remove all traces of hexadecane, and absorbance (DO540) was measured against a hexadecane-extracted PBS blank.
The hydrophobicity index was expressed as the ratio of absorbance of the hexadecane-extracted sample to absorbance of the sample before extraction.
Detection of stn, sef and pef genes by polymerase chain reaction
Bacterial chromosomal DNA was extracted using a Wizard Genomic Purification Kit (Promega, France). For all reactions, PCR were performed in 25 μl containing: 50 ng of extracted DNA, 5 μl green Go Taq buffer (5×), 0.25 μl of each deox-ynucleoside triphosphates (10 mM), 0.5 μl MgCl2 (50 mM), 1 μl of each primer (25 pM) and 1U of GO Taq DNA polymerase (Promega, USA).
Primers used for stn gene were: StnP1-5’-TTGTGTCGCTATCACTGGCAACC-3’ and Stn M13- 5’-ATTCGTAACCCGCTCTCGTCC-3’ which flank a 617 bp segment in the stn gene sequence; For sef gene, the primers used were, sefC 5’-GCGAAAACCAATGCGACTGTA-3’ and sefC 5’-CCCACCAGAAACATTCATCCC-3’ that flank a 1103 bp segment in the sef gene sequence. For pef gene the primers used were, pef A1 -5’-TGTTTCCGGGCTTGTGCT-3’ and pfA2-5’-CAGGGCATTTGCTGATTCTTCC-3’. These primers flank a 700 bp segment in the pef gene sequence (28).
For all genes, PCR were performed in 25 μl containing: 50 ng of extracted DNA, 5 μl green Go Taq buffer (5×), 0.25 μl of each deoxynucleoside triphosphates (10 mM), 0.5 μl MgCl2 (50 mM), 1 μl of each primer (25 pM) and 1U of GO Taq DNA polymerase (Promega, USA). Reaction mixtures were incubated for 5 min at 94°C, followed by 25 cycles at 94°C for 1min, annealing at 55°C for 1min for Pef and Sef, 72°C for 1 min and a final extension at 72°C for 10 min. The annealing temperature for the detection of the stn gene was 59°C.
PCR products (5 μl) were analysed on 1% agarose gels stained with ethidium bromide (0.5 mg/ml) at 90V for 1 h and visualized under ultraviolet transillumination and photographed using Gel doc (Bio-Rad). All PCR positive strains indicated the presence of genes was confirmed by repeating the PCR three times.
Statistical analysis
Each analysis was performed using the S.P.S.S. 13.0 statistics package for windows. Differences in the degree of biofilm formation were examined by the Friedman test, followed by the Wilcoxon signed ranks test. P-values of < 0.05 were considered significant.
Results
Determination of Slime production
Phenotypic slime production, presented in table 1, was assessed by culturing the investigated strains on CRA plates. Our results showed that the 22 Salmonella strains were considered as non-slime producer since they showed white or red colonies on CRA plates.
Cell surface hydrophobicity
The results of microbial adhesion to hexadecane are summarized in Table 1. We have found that the affinity to hexadecane (apolar solvent) was low with values between 0.29±0.56 and 29.55±0.1 suggesting a hydrophilic character for the human as well as for the poultry meat isolates and for the two reference strains.
Biofilm formation on polystyrene
Table 1 presents the results of the XTT quantitative adherence assay. Based on the oxidative activity we noted that most of the serotypes were adhesive to polystyrene at different degrees. The optical density values of XTT reduction estimated at 492 nm were ranged from 0.256±0.17 to 1.05±0.1 for the strains isolated from human. Whereas, these values were ranged from 0.56±0.01 to 1.08±0.05 for poultry meat isolates. In addition, we noted that S. Typhimurium ATCC 14028s and S. Typhimurium LT2 DT104 displayed the high oxidative activity (1.47±0.02 and 1.21±0.01 respectively) therefore they are considered strongly biofilm formation.
Biofilm formation on PVC and glass
The results of the biofilm formation on PVC and glass were realized by MTT assay. Using this method, we quantify the viable cells in biofilm. At the beginning we noted that all the tested strains are not able to form a strong biofilm on PVC. In addition, all serotypes form only a moderate (70% and 50% for human and poultry meat isolates respectively) and weak (30% and 50% for human and poultry meat isolates respectively) biofilms on PVC. Contrarily, 80% of human Salmonella isolates produce a strong biofilm on glass. Further, we noted that 70% of poultry meat isolates produce a moderate biofilm and only S. Zanzibar isolated from stool was considered as weakly biofilm producer on glass (Table 2).
Table 2.
Biofilm formation on PVC, glass and genes detection in Salmonella serotypes
| Strains | Biofilm formation on PVC | Biofilm formation on glass | Presence of genes | ||||
|---|---|---|---|---|---|---|---|
| MTT reduction (mean OD578 ± SD) | Biofilm state | MTT reduction (mean OD578 ± SD) | Biofilm state | stn | sef | pef | |
| S1: S. Enteritidis | 2.13±0.21 | moderate | 4.35±0.01 | strong | + | ||
| S2: S. Enteritidis | 1.89±0.17 | weak | 4.11±0.04 | strong | + | + | |
| S3: S. Enteritidis | 2.70±0.09 | moderate | 5.06±0.01 | strong | + | ||
| S4: S. Enteritidis | 2.19±0.15 | moderate | 4.24±0.13 | strong | + | ||
| S5: S. Enteritidis | 2.10±0.14 | moderate | 4.7±0.02 | strong | + | + | |
| S6: S. Enteritidis | 1.96±0.06 | weak | 4.17±0.12 | strong | + | ||
| S7: S. Amsterdam | 1.98±0.05 | weak | 4.27±0.05 | strong | + | ||
| S8: S. Muenster | 2.17±0.13 | moderate | 4.07±0.08 | strong | |||
| S9: S. Kentucky | 2.33±0.18 | moderate | 3.65±0.15 | moderate | + | ||
| S10: S. Zanzibar | 2.64±0.17 | moderate | 1.68±0.06 | weak | |||
| S11: S. Arizona | 1.68±0.001 | weak | 5.50±0.01 | strong | |||
| S12: S. Wangata | 2.34±0.12 | moderate | 5.6±0.08 | strong | |||
| S13: S. Braenderup | 1.97±0.04 | weak | 4.15±0.01 | strong | |||
| S14: S. Montevideo | 2.03±0.03 | moderate | 2.82±0.13 | moderate | |||
| S15: S. Cerro | 2.46±0.23 | moderate | 3.77±0.01 | moderate | |||
| S16: S. Agona | 2.10±0.20 | moderate | 2.60±0.06 | moderate | |||
| S17: S. Hadar | 1.58±0.07 | weak | 3.02±0.17 | moderate | + | ||
| S18 : S. Newport | 1.74±0.14 | weak | 3.29±0.06 | moderate | |||
| S19: S. Altona | 2.23±0.20 | moderate | 3.10±0.13 | moderate | |||
| S20: S. Schwarzengrund | 1.39±0.08 | weak | 2.88±0.11 | moderate | |||
| S21: S. Typhimurium 14028s | 1.41±0.09 | weak | 3.64±0.03 | moderate | + | + | |
| S22: S. Typhimurium LT2 DT104 | 1.87±0.14 | weak | 4.01±0.08 | strong | + | + | |
Detection of stn, sef and pef genes
Among the isolates we noted that only S. Enter-itidis strains isolated from blood, urine and pus were positive for sef gene giving a 617-bp band (Figure 1). In addition S. Kentucky and S. Amsterdam isolated from stool and S. Hadar isolated from poultry meat harbour Salmonella enterotoxin gene (stn). However plasmid encoded fimbriae (pef) was detected in two human S. Enteritidis isolates (S2 and S5). Further, S. Typhimurium ATCC 14028s and S. Typhimurium LT2 DT104 were positive for stn and pef genes (Table 2).
Fig. 1.
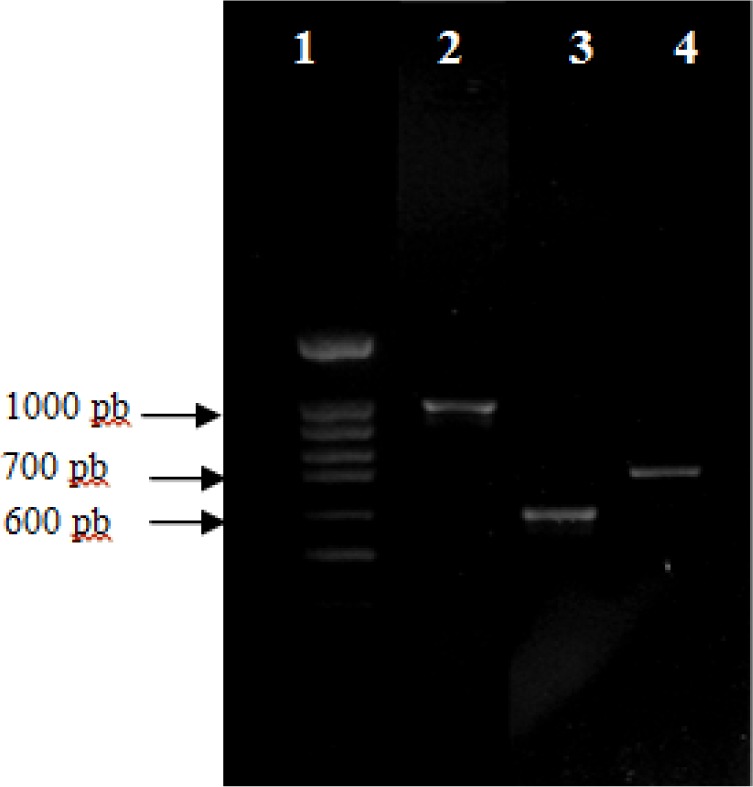
Agarose gel electrophoresis of polymerase chain reaction (PCR) amplification of pef, sef and stn genes. Lane 1, 100 bp DNA molecular size marker, Lane 2 through 4: pef, sef and stn genes respectively.
Discussion
The ability of Salmonella to attach to abiotic surfaces and form biofilms is a cause of concern for many industries, including the food ones (29). Poor sanitation of food-contact surfaces is believed to be an essential contributing factor in foodborne disease outbreaks, especially those involving Salmonella. This is because the attachment of bacterial cells to such surfaces is the first step of a process which can ultimately lead to the contamination of food products. Accordingly, in this work, we have demonstrated that Salmonella, food-borne pathogen, is able to form biofilm on industrial surfaces such as plastic (PVC and polystyrene) and glass but with different degrees. Thus, biofilms formed in these surfaces are of special importance since they may act as a persistent source of Salmonella contamination which may lead to food spoilage or/and transmission of diseases (12-30).
In this work, results showed that human and poultry meat Salmonella isolates and the two references strains were categorized as non slime-producer on CRA plate according to Chaieb et al. (23) and Ben Abdallah et al. (31), developing red and white colonies. Indeed, slime production by Salmonella plays an important role in its pathogenesis of infections (32). Slimes are generally polysaccharidic materials, although other polymers may also be present. They are probably involved in the protection of microbial cells (33). However, these molecules are also important in the formation of biofilms on surfaces and they have been considered to be in volved in the first steps of biofilm formation (34). Statistical analysis showed no correlation between the biofilm formation and slime production by Salmonella isolated from human and poultry meat. On the other hand, biofilm of Salmonella is mainly composed of curli and cellulose, and Salmonella strains were grouped into distinct morphotypes according to Congo red (35, 36). In our study S. Agona, S. Zanzibar and S. Amsterdam showed red, dry and rough colonies indicating curli and cellulose production (RDAR) thereby they were categorized as biofilm producers. Whereas the others serotypes without biofilm-forming ability showed nearly smooth and white colonies indicating a lack of both curli and cellulose production (SAW).
Evaluation of biofilm formation by Salmonella serotypes revealed that these hydrophilic strains possess a capacity for biofilm formation on plastic (PVC and polystyrene) hydrophobic surfaces. These results confirm previous findings, which showed that Salmonella spp. are able to form biofilm on plastic surfaces (37, 38). In general, it is assumed that glass is hydrophilic material while PVC and plastic are hydrophobic materials (39, 40). It has been previously shown that micro-organisms, including Salmonella spp., adhere in higher numbers to more hydrophobic than hydro-philic materials (39–41). However, obtained results showed that Salmonella serotypes, isolated from human and poultry meat, form a strong biofilm on “hydrophilic material” such as glass microscope slide covers than hydrophilic support. Statistical analysis revealed no correlation between cell surface hydrophobicity and biofilm formation. According to Crawford et al. (42) cellulose, an intact LPS, a functional type III secretion system (TTSS) apparatus and flagellar motility are of crucial importance for biofilms grown of Salmonella on hydrophilic glass coverslips. Statistical analysis revealed a correlation between the biofilm formation on PVC and on glass (P<0.05).
In general the process of biofilm formation by microorganisms is influenced by various factors including nutrients level, pH, temperature, incubation period, ionic strength, culture concentration, etc., but the contact surface characteristics and the bacterial cell surface appendages, such fimbriae, flagella, curli, exopolysaccharides, outer membrane proteins, are the most important among all of them Agrawal et al. (43). Thereby we searched by PCR for the presence of sef and pef genes involved in adhesion and invasion and Salmonella enterotoxin gene (stn) and statistical analysis showed no correlation between the presence of genes and biofilm formation. In term of biofilm formation ability, no correlation is drawn when we compared clinical and poultry meat strains.
Conclusion
In summary, biofilms have great importance for public health because of their role in certain infectious diseases and importance in a variety of device-related infections. In this work we showed that Salmonella serotypes isolated from poultry meat and human are able to adhere and form biofilm on industrial surfaces such as polystyrene, PVC and glass. We have also observed a variability inter and intraspecies. These may explain the higher prevalence and persistence of Salmonella on food product and certainly this prevalence leads to food spoilage or/and transmission of diseases. In addition, Salmonella biofilms have great significance for public health, since biofilm-associated microorganisms exhibit dramatically decreased susceptibility to antimicrobial agents and treatments.
Ethical considerations
Ethical issues (Including plagiarism, Informed Consent, misconduct, data fabrication, double publication and/or submission, redundancy, etc.) have been completely checked by the authors.
Acknowledgments
Financial source of the study was provided by Ministry of Higher Education, Scientific Research and Information and Communication Technologies, Tunisia. We are grateful to Dr. Maha Mastouri (Laboratory of Microbiology, University Hospital Fattouma Bourguiba, Monastir, Tunisia) for his help in clinical strains. The authors declare that there is no conflict of interests.
References
- Finlay BB, Falkow S (1989). Salmonella as an intracellular parasite. Mol Microbiol, 3(12): 1833–1841. [DOI] [PubMed] [Google Scholar]
- Swaminathan B, Barrett TJ, Fields P (2006). Surveillance for human Salmonella infections in the United States. J AOAC Int, 89(2): 553–559. [PubMed] [Google Scholar]
- Tauxe RV, Pavia AT (1998) Salmonellosis: nontyphoidal. In Bacterial infections of humans: epidemiology and control Edited by: Evans A, Brachman P. New York: Plenum, pp. 613–628. [Google Scholar]
- Mead PS, Slutsker L, Dietz V,. McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV (1999). Food-related illness and death in the United States. Emerg Infect Dis, 5(5): 607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D (2003). Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov, 2(2): 114–122. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Stoodley P (2009). Evolving concepts in biofilm infections. Cell Microbiol, 11(7): 1034–1043. [DOI] [PubMed] [Google Scholar]
- Raza A, Sarwar Y, Ali A, Jamil A, Haque A, Haque A (2011). Effect of biofilm formation on the excretion of Salmonella enterica serovar Typhi in feces. Int J Infect Dis, 15:(11) e747–e752. [DOI] [PubMed] [Google Scholar]
- Homoe P, Bjarnsholt T, Wessman M, Sorensen HC, Johansen HK (2009). Morphological evidence of biofilm formation in Greenlanders with chronic suppurative otitis media. Eur Arch Oto-Rhino-L, 266(10): 1533–1538. [DOI] [PubMed] [Google Scholar]
- Dunne WM (2002). Bacterial Adhesion: Seen Any Good Biofilms Lately? Clin Microbiol Rev, 15(2): 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol, 2(2): 95–108. [DOI] [PubMed] [Google Scholar]
- Biswas R, Agarwal RK, Bhilegaonkar KN, Kumar A, Nambiar P, Rawat S, Singh M (2011). Cloning and sequencing of biofilm-associated protein (bapA) gene and its occurrence in different serotypes of Salmonella. Lett Appl Microbiol, 52: (2) 138–143. [DOI] [PubMed] [Google Scholar]
- Brooks JD, Flint SH (2008). Biofilms in the food industry: problems and potential solutions. Inter J Food Sci Tech, 43(12): 2163–2176. [Google Scholar]
- Giaouris E, Chorianopoulos N, Skandamis PN, Nychas GJ (2012). Attachment and biofilm formation by Salmonella in food processing environments, p157–180 In Barakat SMM (ed), Salmonella-a dangerous foodborne pathogen. InTech, Rijeka, Croatia. [Google Scholar]
- Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O (2010). Antibiotic resistance of bacterial biofilms. Inter J Antimicrob Ag, 35(4): 322–332. [DOI] [PubMed] [Google Scholar]
- Jensen PO, Givskov M, Bjarnsholt T, Moser C (2010). The immune system vs. Pseudomonasaeruginosa biofilms. FEMS Immunol Med Mic, 59(3): 292–305. [DOI] [PubMed] [Google Scholar]
- Burmolle M, Bahl MI, Jensen LB, Sorensen SJ, Hansen LH (2008). Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology, 154 (Pt1): 187–195. [DOI] [PubMed] [Google Scholar]
- Hurrell E, Kucerova E, Loughlin M, Caubilla-Barron J, Forsythe SJ (2009). Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Int J Food Microbiol, 136(2): 227–231. [DOI] [PubMed] [Google Scholar]
- Arnold JW, Yates IE (2009). Interventions for control of Salmonella: clearance of microbial growth from rubber picker fingers. Poultry Science, 88(6): 1292–1298. [DOI] [PubMed] [Google Scholar]
- Joseph B, Otta SK, Karunasagar I, Karunasagar I (2001). Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int J Food Microbiol, 64(3): 367–372. [DOI] [PubMed] [Google Scholar]
- Prouty AM, Gunn JS (2003). Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect Immun, 71(12): 7154–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretro T, Vestby LK, Nesse LL, Storheim SE, Kotlarz K, Langsrud S (2009). Evaluation of efficacy of disinfectants against Salmonella from the feed industry. J Appl Microbiol, 106(3): 1005–1012. [DOI] [PubMed] [Google Scholar]
- Freeman DJ, Falkiner FR, Keane CT (1989). New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol, 42(8): 872–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaieb K, Chehab O, Zantar TRouabhia M, Mahdouani K, Bakhrouf A (2007). In vitro effect of pH and ethanol on biofilm formation by clinical ica-positive Staphylococcus epidermidis strains. Ann Microbiol, 57(3): 431–437. [Google Scholar]
- Pettit RK, Weber CA, Kean MJ, Hoffmann H, Pettit GR, Tan R, Franks KS, Horton ML (2005). Microplate Alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob Agents Chemother, 49(7): 261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunney MM, Ramage G, Field TR, Moriarty TF, Storey DG (2004). Rapid colorimetric assay for antimicrobial susceptibility testing of Pseudomonas aeruginosa. Antimicrob Agents Chemother, 48(5): 1879–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods, 65 (1–2): 55–63. [DOI] [PubMed] [Google Scholar]
- van Loosdrecht MC, Lyklema J, Norde W, Schraa G, Zehnder A J (1987). The role of bacterial cell wall hydrophobicity in adhesion. Appl Environ Microbiol, 53(8): 1893–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugkar HV, Rahman H, Dutta PK (2003). Distribution of virulence genes in Salmonella serovars isolated from man & animals. Indian J Med Res, 117(2): 66–70 [PubMed] [Google Scholar]
- Mahdavi M, Jalali M, Kermanshahi RK (2008). Biofilm formation by Salmonellaenteritidis on food contact surfaces. J Biol Sci, 8(2): 502–505. [Google Scholar]
- Zottola EA, Sasahara KC (1994). Microbial biofilms in the food processing industry –Should they be a concern? Int J Food Microbiol, 23(2): 125–148. [DOI] [PubMed] [Google Scholar]
- Ben Abdallah F, Chaieb K, Zmanta T, Kallel H, Bakhrouf A (2009). Adherence assays and slime production of Vibrio alginolyticus and Vibrio parahaemolyticus. Br J Microbiol, 40(2): 393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaráz LE, Satorres SE, Lucero RM, Puig de centorli ON (2003). Species identification, slime production and oxacillin susceptibility in coagulase-negative staphylococci isolated from nosocomial specimens. Br J Microbiol, 34(1): 45–51. [Google Scholar]
- Ophir T, Gutnick DL (1994). A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl Environ Microbiol, 60(2): 740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E, Hübner J, Gutierrez N, Takeda S, Goldmann DA, Pier GB (1993). Isolation and characterisation of transposon mutants of Staphylococcus epidermidis deficient in capsular polysaccharide/adhesion and slime. Infect Immun, 61(2): 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Dong H, Chen S, Chen Y, Peng D, Liu X (2011). Characterization of biofilm formation by Salmonella enterica serovar Pullorum strains. Afr J Microbiol Res, 5(17): 2428–2437. [Google Scholar]
- Malcova M, Hradecka H, Karpiskova R, Rychlik I (2008). Biofilm formation in field strains of Salmonella enterica serovar Typhimurium: identification of a new colony morphology type and the role of SGI1 in biofilm formation. Vet Microbiol, 129 (3–4): 360–366. [DOI] [PubMed] [Google Scholar]
- Djordjevic D, Wiedmann M, McLandsborough LA (2002). Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol, 68(6): 2950–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanović S, Cirković I, Ranin L, Svabić-Vlahović M (2004). Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol, 38(5): 428–432. [DOI] [PubMed] [Google Scholar]
- Sinde E, Carballo J (2000). Attachment of Salmonella spp. and Listeria monocytogenes to stainless steel, rubber and polytetrafluorethylene: the influence of free energy and the effect of commercial sanitizers. Food Microbiol, 17(4): 439–447. [Google Scholar]
- Donlan RM (2002). Biofilms: microbial life on surfaces. Emerg Infect Dis, 8(9): 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe D, Smart CA, Alexander C, Vulfson EN (1999). Bacterial adhesion at synthetic surfaces. Appl Environ Microbiol, 65(11): 4995–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Gibson DL, Kay WW, Gunn JS (2008). Identification of a bile induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect Immu, 76(11): 5341–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal RK, Singh S, Bhilegaonkar KN, Singh VP (2011). Optimization of microtitre plate assay for the testing of biofilm formation ability in different Salmonella serotypes. Inter Food Res J, 18(4): 1493–1498. [Google Scholar]


