Abstract
Eukaryotic organisms package DNA into chromatin for compact storage in the cell nucleus. However, this process promotes transcriptional repression of genes. To overcome the transcriptional repression, chromatin remodeling complexes have evolved that alter the configuration of chromatin packaging of DNA into nucleosomes by histones. The SWI/SNF chromatin remodeling complex uses energy from ATP hydrolysis to reposition nucleosomes and make DNA accessible to transcription factors. Recent studies showing mutations of BRG1, one of 2 mutually exclusive ATPase subunits, in human tumor cell lines and primary tissue samples have implicated a role for its loss in cancer development. While most of the mutations lead to complete loss of BRG1 protein expression, others result in single amino acid substitutions. To better understand the role of these BRG1 point mutations in cancer development, we characterized SWI/SNF function in human tumor cell lines with these mutations in the absence of BRM expression, the other ATPase component. We found that the mutant BRG1 proteins still interacted with the core complex members and appeared at the promoters of target genes. However, while these mutations did not affect CD44 and CDH1 expression, known targets of the SWI/SNF complex, they did abrogate Rb mediated cell cycle arrest. Therefore, our results implicate that these mutations disrupt the de novo chromatin remodeling activity of the complex without affecting the status of existing nucleosome positioning.
Keywords: Chromatin Remodeling, SWI/SNF complex, BRG1, CD44, CDH1
INTRODUCTION
Eukaryotic organisms package DNA into condensed solenoid chromatin for compact storage in the cell nucleus, resulting in transcriptional repression of genes. Therefore, mechanisms of chromatin remodeling have evolved to overcome this repressive nature of chromatin and make DNA accessible to sequence-specific transcription factors and transcription machinery (Clapier and Cairns, 2009). Chromatin remodeling results in an alteration of nucleosomes to allow the binding of transcription factors and initiation of transcription (Clapier and Cairns, 2009; Wang et al., 2007a; Wang et al., 2007b). Two classes of chromatin remodeling enzymes exist, histone modifying enzymes and ATP-dependent chromatin remodeling enzymes.
The SWI/SNF complex is an ATP-dependent chromatin remodeling complex that is conserved from humans to yeast (Clapier and Cairns, 2009). The complex is approximately 2-Mda in size and consists of 9–12 members varying in a tissue specific manner (Wang et al., 1996). To date, six SWI/SNF complexes have been isolated with each complex consisting of one catalytic subunit, BRG1 or BRM, plus 8–11 BAFs (BRG1 associated factors). A minimum catalytic core of BRG1 or BRM and BAF155 or BAF170 is required for disruption of nucleosome arrays in vitro, but all members of the complex are required for proper functioning in yeast (Laurent et al., 1991; Peterson and Herskowitz, 1992).
The main function of the SWI/SNF complex is transcriptional regulation. The complex has been shown to be required for expression of a variety of genes involved in cellular adhesion such as CD44 (Strobeck et al., 2001) and CDH1 (Banine et al., 2005), and genes involved in cellular proliferation such as cyclin A (Zhang et al., 2000) and cyclin E (Zhang et al., 2000), CSF1 (Liu et al., 2001), and p53-dependent target promoters (Lee et al., 2002). The complex has also been shown to be required for RB-mediated arrest by upregulating p21, which inhibits cyclin-dependent kinases and leads to RB hypophosphorylation (Hendricks et al., 2004; Kang et al., 2004). Due to the role of the complex in cellular adhesion and growth arrest, it is not surprising that evidence has linked loss of SWI/SNF complex members with human disease.
The first association between loss of a SWI/SNF complex member and human disease was established when loss of SNF5 expression was found to lead to development of malignant rhabdoid tumors (Versteege et al., 1998). Increasing evidence has also indicated a role for inactivation of other members of the SWI/SNF complex including BRG1, BRM, BAF155, and BAF57 in cancer development and/or cancer progression (Weissman and Knudsen, 2009). As expected, loss of the catalytic subunits, BRG1 and BRM, leads to loss of function of the complex. In fact, loss of BRG1 and BRM is found in human tumor cell lines and primary tumor of lung, breast, and prostate (Weissman and Knudsen, 2009). Wong et al screened a panel of tumor cell lines to determine if BRG1 is targeted for mutation (Wong et al., 2000). They identified 16/180 human tumor cell lines that possessed mutations in BRG1. We have found loss of expression of BRG1 and BRM in approximately 20% of NSCLC cell lines (Reisman et al., 2003). In most cases, loss of BRG1 protein expression arises from gene deletions or truncating point mutations (Wong et al., 2000). However, primary NSCLC tumors and several human tumor cell lines contain point mutations in BRG1 (Medina et al., 2004; Wong et al., 2000). Several groups, including our own, have found loss of BRG1 and/or BRM expression in primary NSCLC (Fukuoka et al., 2004; Reisman et al., 2003). Most importantly, loss of BRG1/BRM is an indicator of poor prognosis. NSCLC BRG1/BRM-negative tumor patients have a shorter survival time then NSCLC BRG1/BRM-positive tumor patients (Fukuoka et al., 2004; Reisman et al., 2003).
In a previous study, tumor cell lines lacking both BRG1 and BRM expression were analyzed for impaired RB-mediated growth arrest and for expression of CD44 and CDH1 (Banine et al., 2005; Reisman et al., 2002; Strobeck et al., 2001; Strobeck et al., 2002). It was found that either BRM or BRG1 was sufficient to restore RB-mediated growth arrest and CD44 and CDH1 expression, indicating that BRM can compensate for BRG1 loss. In contrast, Brm−/−;Brg1wt/wt mice failed to express CD44 in all tissues examined (Reisman et al., 2002). As a further characteristic of tumor suppressor genes, the stable reintroduction of BRG1 into the tumor cell lines resulted in replicative senescence (Hendricks et al., 2004; Kang et al., 2004).
In this study, we analyzed if point mutations in BRG1 found in human tumor cell lines resulted in altered functions. We suppressed BRM expression in each cell line by stable expression of shRNA and assessed them for impaired RB-mediated arrest, CD44 expression, and CDH1 expression. We found that 2 of the BRG1 mutations have lost the ability to promote RB-mediated cell cycle arrest and CD44 expression. We also showed that these mutant BRG1 proteins still associate with the other core members of the complex by co-immunoprecipitation and were present at target promoters by ChIPs analysis. Our results suggest that the point mutations found in these cell lines impair the function of BRG1 by altering the ability of the complex to remodel chromatin. The sites of these point mutations should provide insight into the structure and function of this important cellular regulatory protein.
MATERIAL AND METHODS
Cell Lines
MCF7, HeLa, SW13, NIH-OVCAR3, Hs578t, and HCT116 were obtained from ATCC. All cell lines were grown in RPMI with 10% FBS except HCT116 which was grown in DMEM with 10% FBS. Cell lines were routinely checked for mycoplasma contamination and were found to be negative.
Immunoblotting
Protein levels were determined as previously described (Chai et al., 2005). Briefly, cells were grown in 100 mm tissue-culture dishes, harvested in extraction buffer and collected with a scraper. After clarification by centrifugation, the supernatant was collected and protein concentration was determined by Bradford protein assay. Equal amounts of protein lysate (30 ug) were separated by SDS-polyacrylamide gel electrophoresis (either 7.5% or 4–20% gradient) for 1 hr at 100 V. Protein was then transferred to Immobilon-P membranes (Millipore) for 16–18 hr at 80 mA. Membranes were blocked for 1 hr in 5% non-fat dry milk/0.2% Tween 20 in PBS. Membranes were then incubated for 2 hr in primary antibody at room temperature. Primary antibodies included BRG1 (Santa Cruz, G7), BRM (Dr. Yaniv Moshe, Pasteur Institut or Abcam, 15597), CDH1 (Transduction Labs 610181), CD44 (H3) (Dr. Larry Sherman, Oregon Health Sciences University) and actin (Sigma, A2066). Membranes were washed and incubated in a 1:2000 dilution of either anti-mouse or anti-rabbit secondary antibody for 1 hr. Membranes were then washed and protein bands were detected with ECL chemiluminescence reagent (Amersham) on Biomax ML film (Kodak) or by Licor fluorescent imaging.
Nuclear Protein Extraction
1 × 109 cells were collected, washed and resuspended in hypotonic buffer (10 mM Hepes pH 7.9/10 mM NaCl/1.5 mM MgCl2/2 mM DTT) for 10 min. The pellet was the collected by centrifugation, resuspended in 2× original pellet volume with hypotonic buffer and extracted with a Dounce homogenizer. The nuclei were collected by centrifugation and resuspended in Nuclear Extract Buffer (20 mM Hepes ph 7.9/420 mM NaCl/1.5mM MgCl2/2.0 mM DTT/0.2 mM EDTA/25% glycerol). The nuclei were then sonicated and nuclear proteins extracted with a dounce homogenizer. Nuclear proteins were collected by centrifugation and then dialyzed against three changes of dialysis buffer (20 mM Hepes pH 7.9/50 mM KCl/1.0 mM DTT/0.2 mM EDTA/10% glycerol). Protein concentrations were determined by Bradford assay.
Immunoprecipitation
Nuclear extracts were precleared with Protein A and G sepharose beads (1:1) (Amersham) in RIPA buffer for 2 hr at 4°C. After removal of beads of centrifugation, equal amounts of protein were incubated with each antibody along with Protein A and G sepharose beads (1:1) (Amersham) saturated with BSA and salmon sperm in RIPA buffer at 4°C overnight. Primary antibodies included Normal Rabbit Serum, rabbit anti-BRG1 (J1, Weidong Wang, National Institute on Aging), rabbit anti-BAF155 (H-76, Santa Cruz) and rabbit anti-SNF5 (Tony Imbalzano, Univ. of Massachusetts). The next day, the beads were collected by centrifugation and washed once in RIPA buffer, IP wash buffer (100 mM Tris HCl pH 8.5/500 mM LiCl/1% NP40/1% deoxycholate), and in PBS. The proteins were then released directly from the beads by heating in loading buffer and loaded onto SDS-polyacrylamide gels for immunoblot analysis. Immunoblotting was carried out as above using the following antibodies- mouse anti-BRG1 (G7, Santa Cruz), rabbit anti-BAF155 (Weidong Wang, National Institute of Aging) and mouse anti-SNF5 (612111, BD Transduction Laboratories).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIPs) was carried out as described by Kuwahara et al (Kuwahara et al., 2010). Immunoprecipitation was performed with an antibody specific to SNF5 (Dr. Tony Imbalzano, University of Massachusetts School of Medicine), BRG-1 (J1; Dr. Weidong Wang, NIA), BAF155 (H76, Santa Cruz) or normal Rabbit IgG (sc-2027; Santa Cruz Biotechnology). DNA present in each IP was quantified by QT-PCR using gene-specific primers on an ABI 7000 sequence detection system (Table 1). All expression values were normalized against input DNA.
TABLE 1.
Sequences of QT-PCR primers used for ChIP
| GENE/POSITION | Forward Primer | Reverse Primer |
|---|---|---|
| CDH1/−354bp | 5' TCG AAC CCA GTG GAA TCA GAA 3' | 5' GGG TCT AGG TGG GTT ATG GGA C 3' |
| CD44/TSS1 | 5' TCA GCC TTT GGC CTC TCC TT 3' | 5' CTC CAG CCG GAT TCA GAG AA 3' |
| p21/−20bp | 5' TAT ATC AGG GCC GCG CTG 3' | 5' GGC TCC ACA AGG AAC TGA CTT C 3' |
| p21/−3kb | 5' CCG GCC AGT ATA TAT TTT TAA TTG AGA 3' | 5' AGT GGT TAG TAA TTT TCA GTT TGC TCA 3' |
TSS- transcription start site
Generation of Stable RNAi Clones
The generation and characterization of the BRM and BRG1 shRNA expression vectors were previously reported (Link et al., 2005; Rosson et al., 2005). Cell lines were transfected with these shRNAi vectors using Lipofectamine 2000 (Invitrogen) following the manufacturers instructions. Clones were selected using puromycin selection media based on death curves established for each cell line; 0.4ug/ml puromycin in RPMI for Hs578t cells, 1 ug/ml puromycin in RPMI for OVCAR3, and 2 ug/ml puromycin in DMEM for HCT116 cells. Messenger RNA and protein levels for each clone were screened by QT-PCR and Western blotting as described above.
Brdu Incorporation Assay
The assay was carried out as previously described (Betz et al., 2002). Briefly, cells were plated in eight well cover slips and transfected with 1 ug of pcDNA3 or p16/PSMRB and 0.2ug of GFP using lipofectamine 2000 (Invitrogen), following manufacturer's instructions. After 36 hr, 1 ug/ml Brdu was added for an additional 12 hr. The cells were then fixed in 10% formalin for 15–20 min. Brdu incorporation was detected using mouse anti-Brdu (Amersham) followed by alexa fluor 546 anti-mouse IgG (Molecular Probes). Fifty to one hundred GFP positive cells were counted for Brdu incorporation by fluorescent microscopy. The average of three experiments per sample was normalized to the parent cell line transfected with pcDNA3.
Transient Transfection
Cell lines were transfected with shRNAi vectors using Lipofectamine 2000 (Invitrogen), following the manufacturers instructions. Transfected cells were harvested for protein 48 hours after transfection with detergent based extraction buffer (Banine et al., 2005).
RESULTS
Characterization of BRG1 mutant cell lines
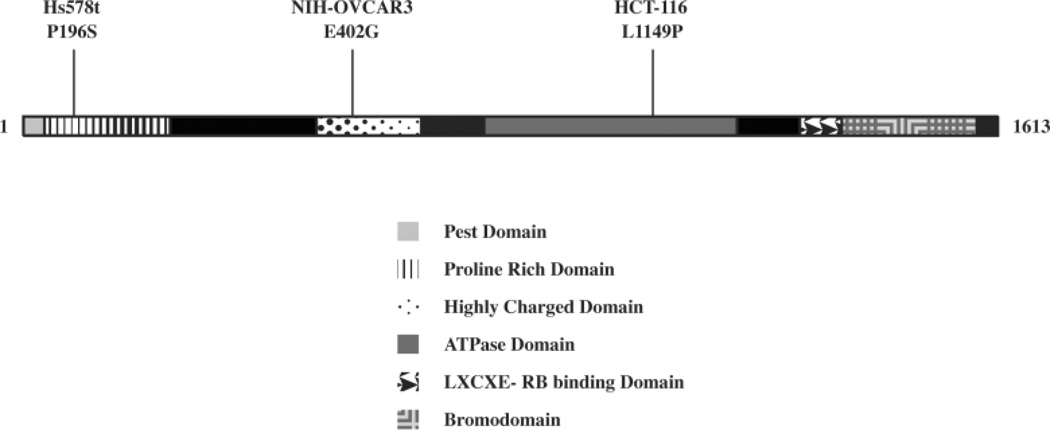
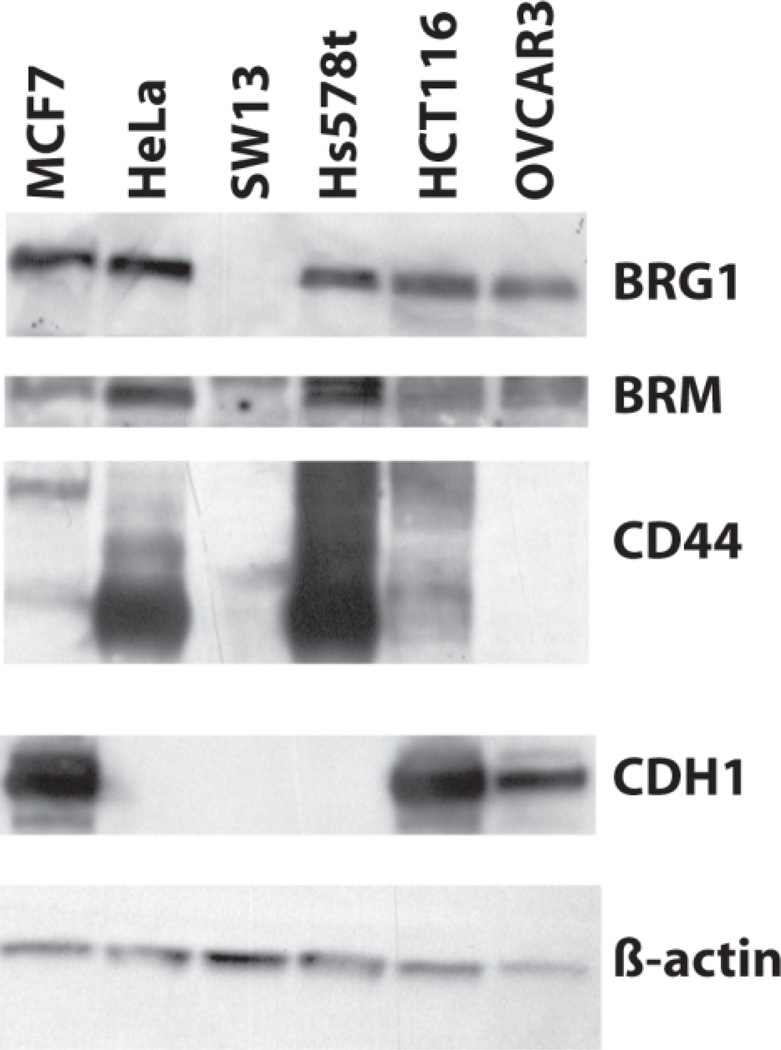
To determine whether point mutations in BRG1 affected its normal functions, we chose 3 human tumor cell lines with known mutations (Figure 1). These cell lines were chosen because the mutations appear in three different domains of BRG1. The Hs578t mutation contains a P196S mutation the proline rich domain I, near an area shown to be required for B-catenin signaling (Barker et al., 2001). The OVCAR3 E402G mutation is hemizygous, within the highly charged domain II, whose function remains unknown (Wong et al., 2000). The HCT116 L1149P mutation maps to the ATPase domain, which is responsible for hydrolyzing ATP (Wong et al., 2000). Previously, our lab determined a functional copy of either BRG1 or BRM was enough for CDH1 or CD44 expression and RB-mediated arrest (Banine et al., 2005; Reisman et al., 2002; Strobeck et al., 2001; Strobeck et al., 2000; Strobeck et al., 2002). These cell lines have previously been described for their CD44 expression and RB-sensitivity (Strobeck et al., 2002). Interestingly, a western-blot screen of our BRG1 mutant cell lines shows OVCAR3 lacks CD44 expression and Hs578t lacks CDH1 expression, even though they presumably contain at least a functional copy of BRM (Figure 2).
Figure 1. Location of BRG1 mutations.
The location of the three point mutations in human cancer cell lines Hs578t, NIH-OVCAR3, and HCT116 are indicated along with known functional domains of BRG1.
Figure 2. Western-blot analysis of BRG1 mutant cell lines.
Protein from BRG1 mutant cell lines was separated by SDS-PAGE and immunoblotted for BRG1, BRM, CD44, CDH1 and β-actin. MCF7 cells served as a positive control for CDH1 expression. β-actin served as a loading control.
Mutant BRG1 proteins still interact with core complex members
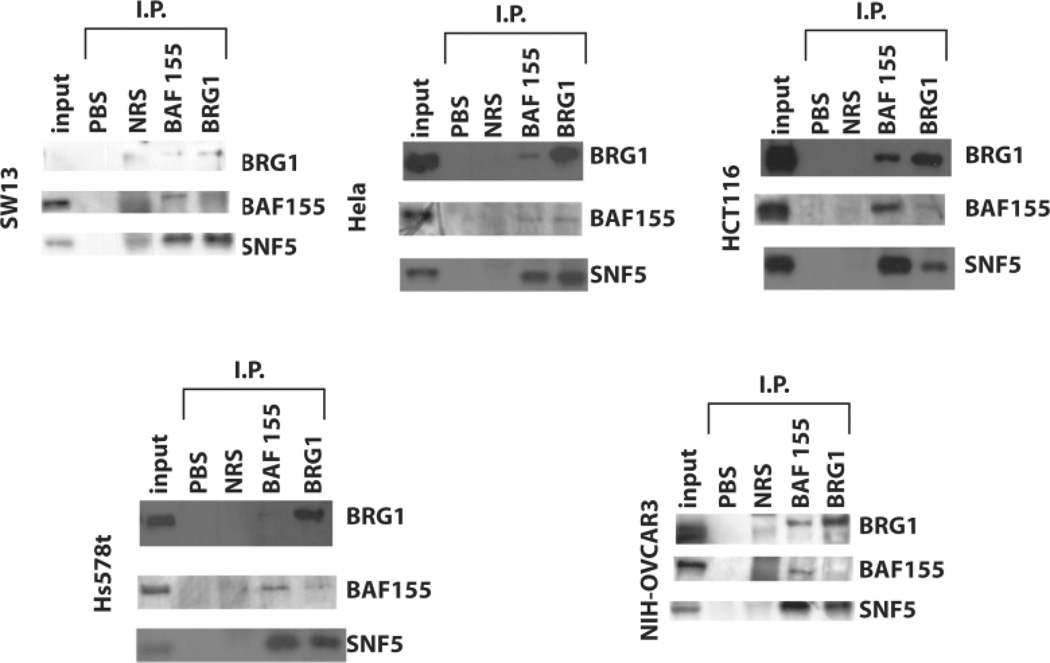
Point mutations in proteins can affect their folding or their binding properties. To determine if the BRG1 mutations in HCT116, Hs578t, and OVCAR3 inhibited its interaction with other members of the SWI/SNF complex, we performed immunoprecipitation of nuclear extracts with antibodies against BRG1 and BAF155. Immunoprecipitated protein was analyzed by western-blot analysis (Figure 3). Both of the other core complex members, BAF155 and SNF5, co-immunoprecipitated from HeLa, our positive control for wild-type BRG1 (Figure 3). However, the stoichiometry of the complex appears to vary depending upon which complex member we used for immunoprecipitation. In contrast, only SNF5 co-immunoprecipitated with BAF155 in the BRG1 deficient SW13 (Figure 3). We did observe a small amount of both BAF155 and SNF5 protein co-precipitating with BRG1 due to a small sub-population of BRG1-positive cells within this cell line (Yamamichi-Nishina et al., 2003). In general, the results with the HCT116 cell line appear similar to those observed with the HeLa cell line indicating that the BRG1 mutation did not apparently affect the ability of the protein to interact with these core proteins (Figure 3). However, the immunoprecipitation results with the Hs578t suggest a reduced interaction with the BAF155 subunit (Figure 3). The results with the NIH-OVCAR3 cell line also indicate altered protein interactions. These cells express both wtBRG1 and mutant BRG1 proteins. Therefore, the absence of BAF155 protein in the BRG1 pull-down lane but not the BAF155 pull-down lane may indicate a reduced binding of BAF155 by the BRG1 mutant protein.
Figure 3. Immunoprecipitation of SWI/SNF complex core components.
Nuclear proteins from BRG1 mutant cell lines were immunoprecipitated for BRG1 (J1 antibody) of BAF155 (H76, Santa Cruz) and then analyzed by immunoblot for expression of the other core complex members of the SWI/SNF complex. Input samples were run as positive controls. PBS precipitated protein served as a negative control. Protein precipitated with normal rabbit serum (NRS) indicated any background protein precipitated by IgG. HeLa cells were precipitated as a positive control for an intact complex while SW13 serves as a negative control for complex formation because only a minor population expresses BRG1.
Analysis of BRM-deficient HCT116 and Hs578t cell lines
Because the BRG1 mutant protein appeared to be in a complex with at least one other core member of the SWI/SNF complex, we wanted to determine if the BRG1 mutant complexes remained functional. To test this possibility, we decided to abrogate BRM function to negate its ability to compensate for BRG1 loss. To create HCT116, NIH-OVCAR3 and Hs578t stable knockdowns of BRM, we transfected each cell line with shRNAi vectors against BRM and selected stable clones. We also isolated cell lines with stable expression of a BRG1 shRNA or vector alone as controls.
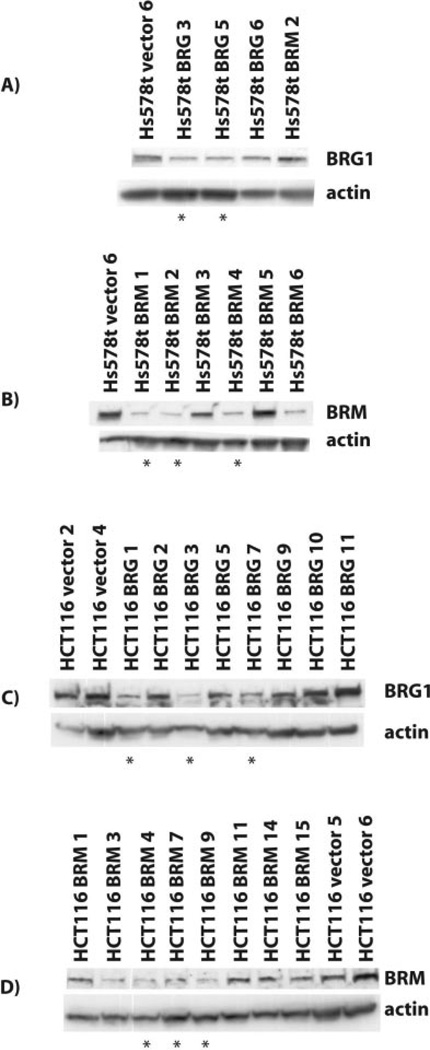
Western-blot analysis confirmed that stable expression of the RNAi vectors in the HCT116 and Hs578t cell lines resulted in downregulation of BRG1 by the BRG1 RNAi (Figure 4a and 4c) and downregulation of BRM by the BRM RNAi vector (Figure 4b and 4d). Attempts to generate NIH-OVCAR3 stable clones expressing a BRM shRNAi vector were unsuccessful. Although we were able to obtain colonies for vector alone and BRG1 shRNAi transfected cells, no colonies were able to grow out in BRM shRNAi transfected cells. Transient transfection with BRM shRNAi confirmed initial reduction of BRM expression in NIH-OVCAR-3 cells but no stable colonies with reduced expression proliferated (data not shown). Therefore, we chose the cell lines indicated by a star in Figure 4 and tested each one for three phenotypes associated with SWI/SNF complex function: RB-mediated cell cycle arrest and expression of CD44 and CDH1 mRNA (Table 2).
Figure 4. Analysis of RNAi stable clones by protein expression.
Hs578t and HCT116 cells were transfected with shRNAi vectors and selected in growth medium supplemented with puromycin. Total cellular protein was extracted from colonies using urea, separated by SDSPAGE and immunoblotted for BRG1 (G-7, Santa Cruz) in Hs578t cells (A) and HCT116 cells (C) and BRM (Dr. Yaniv, Pasteur Institut) in Hs578t cells (B) and HCT116 cells (D). β-actin served as a loading control.
TABLE 2.
Characteristics of the BRG1/BRM-deficient Cell Lines
| Cell Line | RB Sensitivity* | CD44 Expression | CDH1 Expression |
|---|---|---|---|
| HCT116 | S | + | + |
| Hs578t | S | + | − |
| NIH-OVCAR3 | R | − | + |
- S- sensitive; R- resistant.
Loss of BRM Expression Abrogates Rb Sensitivity
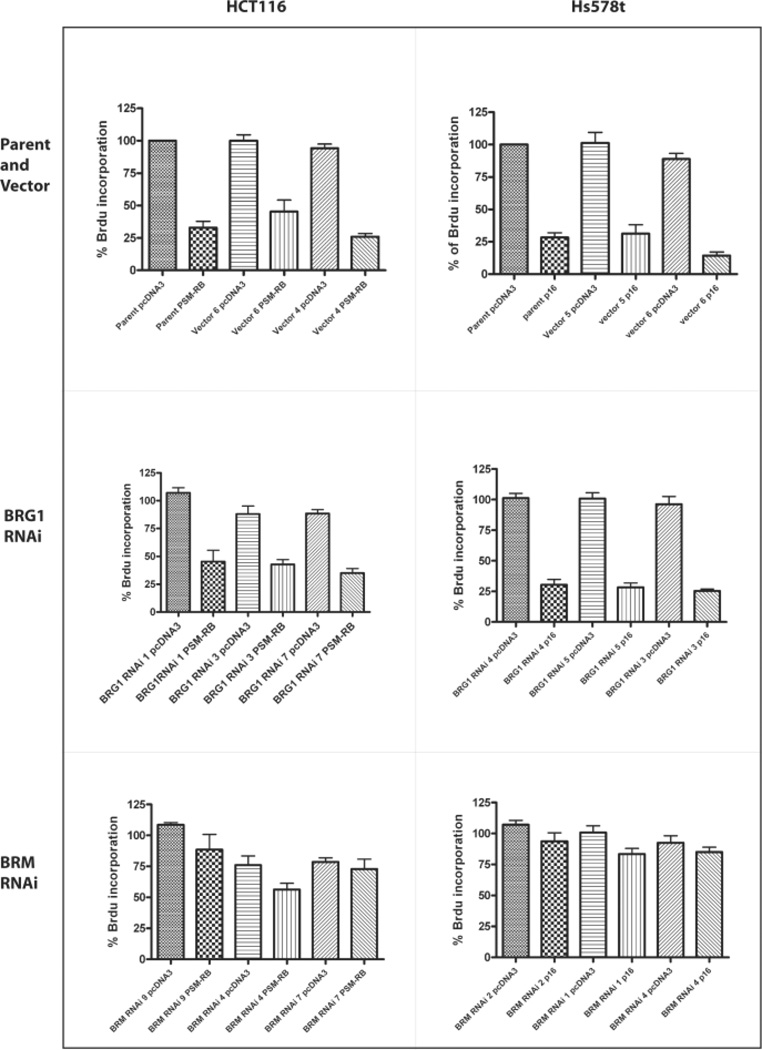
Normally, cells transfected with p16INK4A or a constitutively active form of Rb, PSM-RB, undergo growth arrest (Strobeck et al., 2002). However, in previous studies, we and others have shown that cell lines without a functional BRM and BRG1 were insensitive to RB-mediated cell cycle arrest (Strobeck et al., 2002). Previously, Hs578t was found to be sensitive to RB-mediated arrest due to the presence of functional BRM. In a similar fashion, the p16-deficient HCT116 cell line undergoes cell-cycle arrest when transfected with PSM-RB. To determine if the BRG1 mutations affected RB sensitivity, we analyzed the RNAi stable clones of HCT116 and Hs578t for incorporation of Brdu during the S phase of the cell cycle. In parent cell lines, stable vector clones, and BRG1 RNAi stable clones, transfection of Hs578t with p16, and transfection of HCT116 cells with PSM-RB caused reduction of Brdu incorporation due to growth arrest of the cells (Figure 5). In contrast, the BRM RNAi stable clones became resistant to Rb-mediated growth arrest and continued to incorporate Brdu (Figure 5). These data indicate that the mutations in BRG1 abrogated its ability to regulate the cell cycle.
Figure 5. RB sensitivity of HCT116 and Hs578t RNAi stable clones.
HCT116 and Hs578t RNAi stable clones were transfected with either pcDNA3 or p16 for Hs578t cells, or PSM-RB for HCT116 cells. Brdu incorporation was assessed for Hs578t and HCT116 parent cell lines and vector alone stable clones (A), BRG1 RNAi stable clones (B), and BRM RNAi stable clones (C) as described in the Material and Methods.
Loss of BRM Expression Does Not Alter CD44 or CDH1 Expression
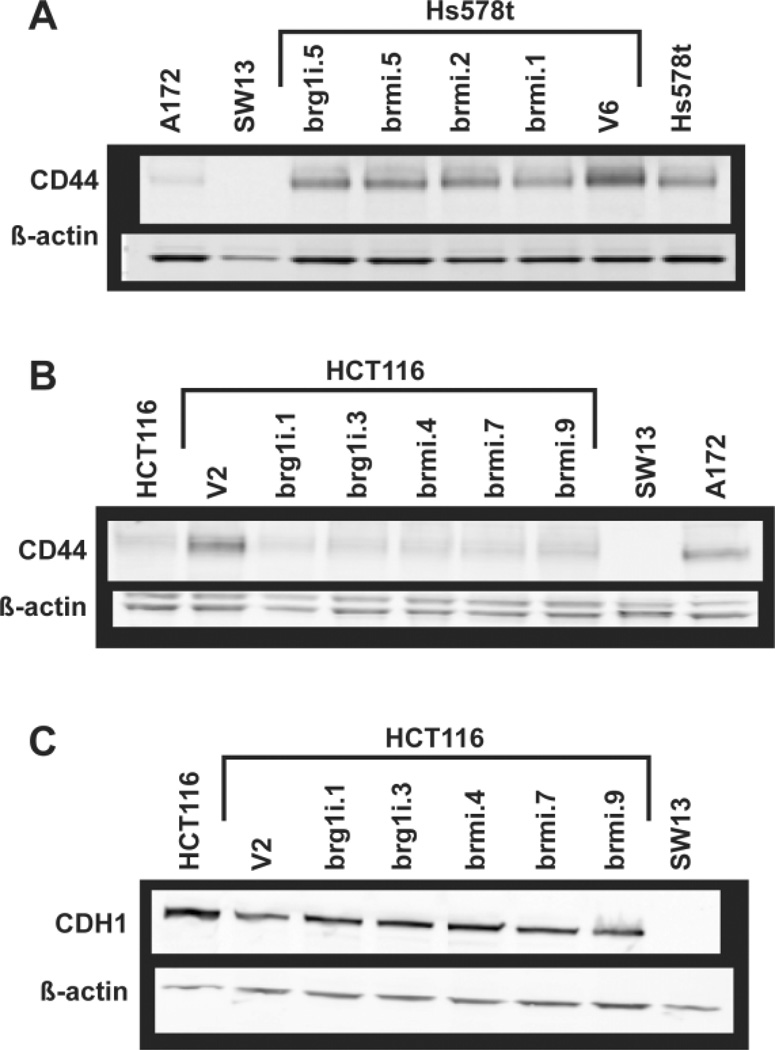
We also assessed if the BRM and BRG1 knockdown cell lines showed altered expression of cell adhesion genes previously associated with BRG1 and BRM regulation. Using Western blotting, we examined CD44 and CDH1 expression in a subset of these cell lines compared to BRG1 and BRM expression. As shown in Figure 6, we did not observe a correlation between BRG1 mutations and reduced CD44 or CDH1 expression. Therefore, in contrast to responsiveness to RB-mediated growth arrest, transcription of these genes does not seem affected by either of these mutations.
Figure 6. CD44 and CDH1 protein expression in Hs578t and HCT116 RNAi stable clones.
Total cellular protein was extracted from the Hs578t and HCT116 parental cell lines and representative vector and RNAi stable clones using either urea (CD44) or high salt RIPA (CDH1) buffers, separated by SDS-PAGE and immunoblotted for CD44 (A & B) or CDH1 (C). β-actin served as a loading control.
BRG1 mutant proteins still bind to the CD44 and CDH1 promoters
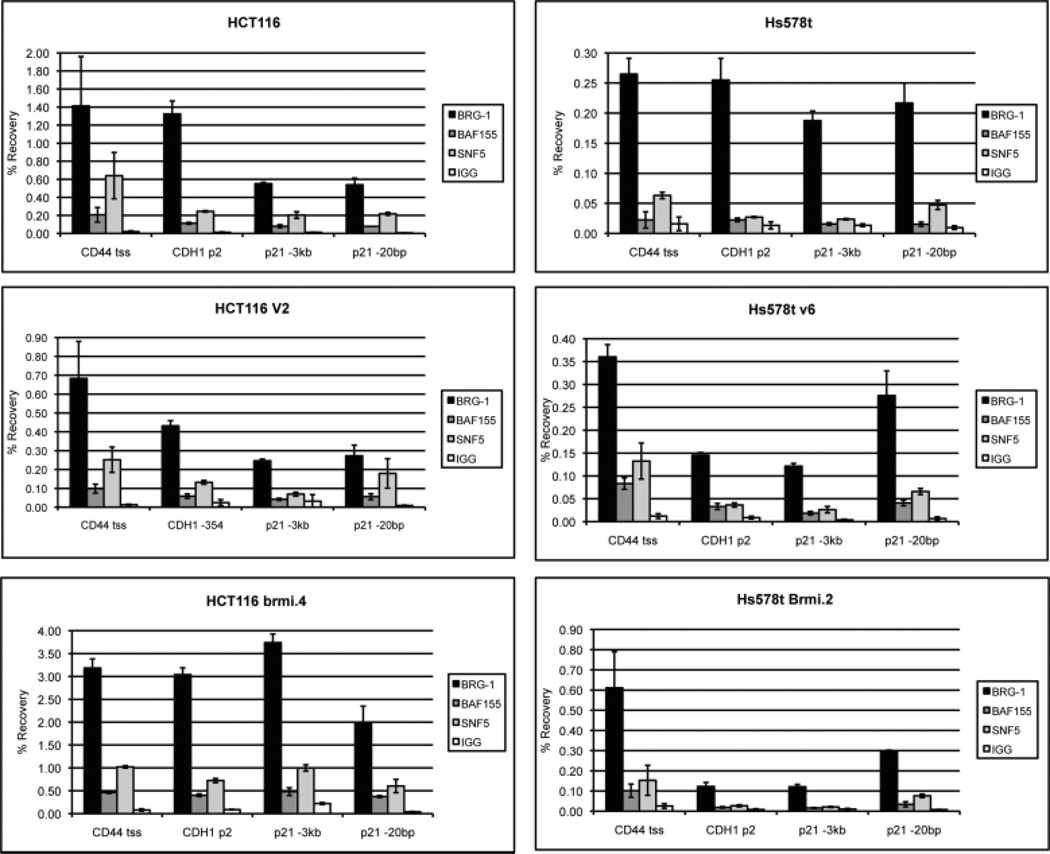
Although the mutant BRG1 proteins could still associate with the core complex members (Figure 3), they may still compromise the ability of the complex to bind to chromatin. To assess this possibility, we examined the binding of BRG1, BAF155 and SNF5 at the CD44, CDH1 and p21 promoters by ChIPs. We and others had previously shown the presence of the complex at these 3 promoters (Banine et al., 2005; Hendricks et al., 2004; Kang et al., 2004). We looked at binding in the parental cell lines and representative vector control and BRMrnai cell lines. As shown in Figure 7 A and B, we observed similar levels of BRG1 and SNF5 binding at all 3 promoters in all Hs578t cell lines. In contrast, an increase in BRG1 and SNF5 binding occurred at all promoters in the HCT116BRMrnai cell line compared to the parent and vector control (Figure 7C & D). However, this appears to result from a non-specific increase in binding because it occurs at both the low binding (−3kb) and high binding (−20bp) p21 sites (Kuwahara et al., 2010).
Figure 7. ChIP analysis of BRG1 binding at cellular promoters.
The parent as well as representative cell lines from the vector and BRM RNAi stable clones were harvested and protein was extracted for ChIPs assays as described in the Material and Methods. ChIPs assays were performed using antibodies directed against 3 core members of the SWI/SNF complex including hSNF5, BAF155 and BRG-1. ChIP analyses were carried out at 3 promoters where the SWI/SNF complex had previously shown binding. Normal rabbit IgG served as a control for non-specific antibody binding. Values are the mean of triplicates.
DISCUSSION
Research focused on the SWI/SNF complex has increased recently due to expanding evidence that aberrations in the complex lead to cancer development (Weissman and Knudsen, 2009). Several lines of evidence implicate loss of catalytic subunits in cancer development. Loss of BRG1 is found in human tumor cell lines, and primary tumors (Medina et al., 2004; Reisman et al., 2003; Weissman and Knudsen, 2009). Similarly, mutations in BRG1 are found in human tumor cell lines and primary tumors (Medina et al., 2004; Wong et al., 2000). Interestingly, the presence of point mutations and internal deletions shows that BRG1 loss is not due to the presence of a larger genomic deletion. Loss of BRG1 or BRM most likely contributes to cancer development by altering gene expression on a global scale and/or disrupting RB-mediated growth arrest. Importantly, our results show that mutations that alter BRG1 function do not a priori affect expression of known target genes i.e. CD44 and CDH1. Presumably, nucleosome positioning remained in place in the absence of SWI/SNF complex function. Rather, the effects of the mutations appeared upon the requirement for active nucleosome repositioning. In this study, the mutant BRG1 proteins lost the ability to respond to Rb-mediated growth arrest, presumably through the absence of chromatin remodeling activity at the necessary promoter regions. Further studies examining nucleosome positioning at promoters such as p21WAF1/CIP1 will address this possibility.
Previous examination of human tumor cell lines revealed that cell lines deficient for both BRG1/BRM were impaired for their ability to express CD44 protein and were unable to growth arrest through RB indicating compensatory function between the two (Strobeck et al., 2002). Expression of BRG1 or BRM has been shown to rescue these defects. HCT116 cells contain a mutation in the ATPase domain in Motif V (Medina et al., 2004; Smith and Peterson, 2005; Wong et al., 2000). A screen of 20 NSCLC found two mutations in BRG1, both of which are in Motif V of the ATPase domain (Medina et al., 2004). These findings indicate, although rare, BRG1 mutations may contribute to tumor development. Additional research has indicated how this mutation may impair BRG1 function. Mutations in Motif V don’t impair nucleosomal binding but do appear to reduce ATPase activity (Smith and Peterson, 2005), so mutations in Motif V are believed to alter the coupling of ATPase activity to the specific function of BRG1 (Smith and Peterson, 2005). The BRG1 mutation in breast cancer cell Hs578t is in domain I (Wong et al., 2000). Part of domain I has been shown to interact with B-catenin (Barker et al., 2001). Previous studies have shown that these cell lines are capable of CD44 protein expression and undergo RB-mediated arrest due to presence of functional BRM (Strobeck et al., 2002). In this study we wanted to determine if the function of BRG1 mutants in human tumor cell lines was impaired in the absence of BRM using BRM shRNAi. We found that both the mutations found in HCT116 and Hs578t were impaired in terms of RB sensitivity. Neither mutation showed consistent effects on protein expression.
CDH1 is not expressed in BRG1/BRM deficient cells but can be upregulated by transfection of BRG1 or BRM into deficient cells. In a western-blot screen of Hs578t and HCT116 we found Hs578t doesn’t express CDH1. Apparently, BRM is unable to compensate for the BRG1 mutation in this cell line and/or CDH1 remains silent due to other mechanisms. The BRG1 mutation in breast cancer cell Hs578t is in domain I, a domain that is partially required to interact with B-catenin and is required for transcriptional activation of Tcf dependent promoters (Barker et al., 2001). CDH1 is regulated by this pathway, and aberrations in B-catenin signaling and loss of CDH1 commonly occurs in breast cancer (Graff et al., 1995). The Hs578t CDH1 promoter is methylated (Graff et al., 1995), but treatment with 5-azacytidine does not restore CDH1 expression, indicating the requirement of other transcriptional factors (Hajra et al., 1999). We found transfection of Hs578t cells with BRG1 doesn’t upreguate CDH1 expression (data not shown). Other labs have demonstrated inducible expression of K789R, a DNBRG1 with no ATPase activity, reduces expression of Tcf target genes (Barker et al., 2001). The BRG1 mutation in Hs578t may also be acting as a dominant negative and inhibiting BRM from compensating in the absence of functional BRG1. Reduction of BRM in HCT116 stable RNAi clones didn’t result in consistent reduction of CDH1 expression. BRG1 and BRM are approximately 76% homologous but they lack homology in the N-terminal region of their proteins including the area of the Hs578t mutation. So alternatively, Hs578t may not express CDH1 with functional BRM because it may be functionally unique to BRG1. Its induction by BRM may result from the high non-physiological levels used in previous experiments.
Redundant functions of BRG1/BRM suggest that cells must lose expression of both genes for tumorigenic effects. Although redundant functions have been shown for the catalytic subunits in in vitro experiments, some assays have shown differential functions for the two proteins. Staining of MEFs from BRM knockout mice reveals that CD44 expression is lost even in the presence of BRG1 (Reisman et al., 2002). This is surprising since transfection of BRG1 into deficient cells upregulates CD44 protein expression. Again overexpression of BRG1, at non-physiological levels, may be able to regulate CD44 in cells deficient for BRG1 and BRM. Kadam and Emerson have shown BRG1 and BRM exhibit transcriptional specificity. They found BRG1 binds zinc finger protein transcription factors while BRM binds ankyrin repeat transcription factors involved in notch signaling (Kadam and Emerson, 2003). Examination of normal human tissues shows differential expression of the proteins, implicating distinct roles for the catalytic subunits in differentiated tissue (Reisman et al., 2005). In fact, homozygous knockout mice indicate differential functions of the two catalytic subunits. Homozygous knockout mice for BRG1 are embryonic lethal and BRG1 heterozygous mice are prone to tumor development by haploinsufficiency (Bultman et al., 2000; Bultman et al., 2008) while knockout mice for BRM are viable with no significant phenotype (Reyes et al., 1998). Due to evidence indicating differential functions of BRG1 and BRM, inactivating mutations of BRG1 could lead to cancer development or progression.
Further characterization of BRG1 and BRM mutations will provide important insights into the normal functions of these proteins, their unique properties and how their loss leads to cancer progression or development. Investigating the differential expression of the catalytic subunits may also lead to information about the role of the complex in different cellular environments. It will also be important to study the temporal expression of the catalytic subunits during development and differentiation to see if alterations in BRG1 during these times result in tumor development. Loss of the catalytic subunits may prove more catastrophic during these developmental and differentiation windows leading to tumor initiation while loss of the catalytic subunits in differentiated tissues may contribute more to cancer progression. Our studies also emphasize the need for careful mutational analysis of BRG1 in primary tumor material because single amino acid changes can lead to substantial alterations in protein function.
ACKNOWLEDGEMENTS
We wish to thank Mr. Nisarg Desai for excellent technical assistance.
Contract grant sponsor: NIH/NCI; Contract grant number: CA138841. Contract grant sponsor: NIH/NIEHS; Contract grant number: T32 ES007126.
LITERATURE CITED
- Banine F, Bartlett C, Gunawardena R, Muchardt C, Yaniv M, Knudsen ES, Weissman BE, Sherman LS. SWI/SNF chromatin-remodeling factors induce changes in DNA methylation to promote transcriptional activation. Cancer Res. 2005;65(9):3542–3547. doi: 10.1158/0008-5472.CAN-04-3554. [DOI] [PubMed] [Google Scholar]
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. Embo J. 2001;20(17):4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21(34):5193–5203. doi: 10.1038/sj.onc.1205706. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27(4):460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65(22):10192–10198. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, Hong K, Settnek S, Gupta A, Buetow K, Hewitt S, Travis WD, Jen J. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10(13):4314–4324. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55(22):5195–5199. [PubMed] [Google Scholar]
- Hajra KM, Ji X, Fearon ER. Extinction of E-cadherin expression in breast cancer via a dominant repression pathway acting on proximal promoter elements. Oncogene. 1999;18(51):7274–7279. doi: 10.1038/sj.onc.1203336. [DOI] [PubMed] [Google Scholar]
- Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24(1):362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. Transcriptional Specificity of Human SWI/SNF BRG1 and BRM Chromatin Remodeling Complexes. Mol Cell. 2003;11(2):377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol. 2004;24(3):1188–1199. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara Y, Charboneau A, Knudsen ES, Weissman BE. Reexpression of hSNF5 in malignant rhabdoid tumor cell lines causes cell cycle arrest through a p21(CIP1/WAF1)-dependent mechanism. Cancer Res. 2010;70(5):1854–1865. doi: 10.1158/0008-5472.CAN-09-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002;277(25):22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- Link KA, Burd CJ, Williams E, Marshall T, Rosson G, Henry E, Weissman B, Knudsen KE. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25(6):2200–2215. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106(3):309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Medina PP, Carretero J, Fraga MF, Esteller M, Sidransky D, Sanchez-Cespedes M. Genetic and epigenetic screening for gene alterations of the chromatin-remodeling factor, SMARCA4/BRG1, in lung tumors. Genes Chromosomes Cancer. 2004;41(2):170–177. doi: 10.1002/gcc.20068. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Bouldin TW, Weissman BE, Funkhouser WK. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl Immunohistochem Mol Morphol. 2005;13(1):66–74. doi: 10.1097/00129039-200503000-00011. [DOI] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63(3):560–566. [PubMed] [Google Scholar]
- Reisman DN, Strobeck MW, Betz BL, Sciariotta J, Funkhouser W, Jr, Murchardt C, Yaniv M, Sherman LS, Knudsen ES, Weissman BE. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002;21(8):1196–1207. doi: 10.1038/sj.onc.1205188. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2a) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosson GB, Bartlett C, Reed W, Weissman BE. BRG1 loss in MiaPaCa2 cells induces an altered cellular morphology and disruption in the organization of the actin cytoskeleton. J Cell Physiol. 2005;205(2):286–294. doi: 10.1002/jcp.20397. [DOI] [PubMed] [Google Scholar]
- Smith CL, Peterson CL. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol Cell Biol. 2005;25(14):5880–5892. doi: 10.1128/MCB.25.14.5880-5892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck MW, DeCristofaro MF, Banine F, Weissman BE, Sherman LS, Knudsen ES. The BRG-1 Subunit of the SWI/SNF Complex Regulates CD44 Expression. J Biol Chem. 2001;276(12):9273–9278. doi: 10.1074/jbc.M009747200. [DOI] [PubMed] [Google Scholar]
- Strobeck MW, Knudsen KE, Fribourg AF, DeCristofaro MF, Weissman BE, Imbalzano AN, Knudsen ES. BRG-1 is required for RB-mediated cell cycle arrest. Proc Natl Acad Sci U S A. 2000;97(14):7748–7753. doi: 10.1073/pnas.97.14.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck MW, Reisman DN, Gunawardena RW, Betz BL, Angus SP, Knudsen KE, Kowalik TF, Weissman BE, Knudsen ES. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J Biol Chem. 2002;277(7):4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007a;13(9):363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007b;13(9):373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes and Development. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69(21):8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, Ghaffari S, Iliev D, Penn B, Woodland AM, Smith R, Salada G, Carillo A, Laity K, Gupte J, Swedlund B, Tavtigian SV, Teng DH, Lees E. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60(21):6171–6177. [PubMed] [Google Scholar]
- Yamamichi-Nishina M, Ito T, Mizutani T, Yamamichi N, Watanabe H, Iba H. SW13 cells can transition between two distinct subtypes by switching expression of BRG1 and Brm genes at the post-transcriptional level. J Biol Chem. 2003;278(9):7422–7430. doi: 10.1074/jbc.M208458200. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean dC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]