Abstract
Modern cancer treatment employs many effective chemotherapeutic agents originally discovered from natural sources. However, a significant challenge currently confronting clinical application is balancing systemic toxicity risk with therapeutic benefit. The cyclic depsipeptide didemnin B has demonstrated impressive anti-cancer activity in preclinical models. Clinical use has been approved but is limited by sparse patient responses combined with toxicity risk and an unclear mechanism of action. From a broad-scale effort to match antineoplastic natural products to their cellular activities, we found that didemnin B selectively induces rapid and wholesale apoptosis through dual inhibition of PPT1 and EEF1A1. Furthermore, empirical discovery of a small panel of exceptional responders to didemnin B allowed generation of a regularized regression model to extract a sparse-feature genetic biomarker capable of predicting sensitivity to didemnin B. This may facilitate patient selection that could enhance and expand therapeutic application of didemnin B against neoplastic disease.
Introduction
Natural products have contributed substantially to the arsenal of therapeutic compounds in use today, most notably as antibiotics and chemotherapy1. Their complex and varied chemistries confer potent and diverse bioactivities that have been honed and maintained by evolutionary pressure. Identifying the mechanisms of action of bioactive natural products has been a major challenge limiting our ability to harness their full therapeutic potential. To help address this challenge, we recently assembled a library of marine natural products and employed expression signature-based high-throughput screening to map the actions of these natural products to genetically-annotated functional space2. This strategy, Functional Signature Ontology (FUSION), has been demonstrated to effectively classify natural products that modulate a broad range of human cell biological systems, including nutrient homeostasis, extracellular matrix signaling, and oncogene signaling2,3.
Here we report the FUSION-inspired characterization of the chemotherapeutic agent didemnin B, a depsipeptide isolated from the marine tunicate Trididemnum solidum4,5. Didemnin B is of both biological and clinical interest as it targets cancer cell viability in vitro and in vivo through a mechanism that is not understood but is clearly distinct from that of other known antineoplastic agents6. The chemotherapeutic activity of didemnin B was first characterized in leukemia and the analog dehydrodidemnin B has been granted orphan drug status for treating acute lymphoblastic leukemia (ALL), though its therapeutic benefit does not appear to be limited to hematological malignancies4,6. Clinical trials of didemnin B and dehydrodidemnin B have documented responses in patients suffering from a wide array of solid tumors, including bronchial carcinoid, colon cancer, esophageal cancer, malignant melanoma, medullar thyroid carcinoma, metastatic breast cancer, non-small-cell lung cancer, renal cancer, and squamous cell cervical cancer7,8. However, the paucity of responders in each of these disease settings has precluded therapeutic application of didemnin analogs outside of ALL.
Through identification and characterization of multi-lineage tumor-derived cell lines that are exceptional responders to didemnin B, we find that the compound potently induces apoptosis, in an identifiable subset of human cancer cell lines, through dual inhibition of palmitoyl-protein thioesterase 1 (PPT1) and eukaryotic translation elongation factor 1 alpha 1 (EEF1A1). Furthermore, we present a quantitative sparse-feature expression biomarker, conserved in tumor samples, which can predict exceptional sensitivity to didemnin B in cell culture.
RESULTS
Didemnin B activates mTORC1 in vitro and in vivo
As part of a large-scale effort for unbiased mechanism of action annotation of genetic and chemical perturbations, we used functional signature-based ontology (FUSION) to cluster equivalent biological responses of HCT116 cells to 780 siRNA pools, 344 miRNA mimics, and 1186 natural product fractions2. From unsupervised hierarchical clustering2, we identified a dense clade heavily populated by reagents known to perturb AKT pathway activity (Fig. 1a; AKT2, AKT3, CNKSR19,10, RPS6KB211, WEE112, EEF2K13, miR-714,15, miR-49716,17, miR-38318, the miR-29 family19, and miR-193a20). Natural product fractions with FUSION signatures most similar to the genetic perturbations within this clade included UT-BA07-004-ETOAC from the tunicate Trididemnum solidum (Fig. 1b), an organism known to produce the antineoplastic compound didemnin B4,5. Indeed, structural determination revealed the most abundant compound in UT-BA07-004-ETOAC to be identical to didemnin B (Supplementary Results, Supplementary Fig. 1a). Guilt by association with the FUSION clade predicted activity of didemnin B against AKT pathway activation. Consistent with this, a 24-hour exposure of HCT116 cells to this compound inhibited AKT signaling in a dose-dependent manner, as indicated by reduced accumulation of activation site phosphorylation (S473) on AKT, on its direct substrate TSC2 (T1462), and on its downstream effector p70S6K(T389), an mTORC1 substrate (Fig. 1c). However, analysis of AKT signaling after short-term didemnin B exposure showed that increased phosphorylation of p70S6K (T389) occurred at lower concentrations and earlier time-points than any observable inhibition of AKT signaling (Supplementary Fig. 1b, c). Activation of mTORC1 is known to engage multiple negative feedback mechanisms that inhibit AKT signaling21–24. Indeed, didemnin B induced phosphorylation of the mTORC1 substrate site (T389) on p70S6K, with an EC50 of ~100 nM in HCT116 cells (Supplementary Fig. 1c), that was completely blocked by the mTORC1 inhibitor rapamycin (Fig. 1d). The mTORC1 substrate sites (T37/46) on 4E-BP1 responded similarly (Supplementary Fig. 1d). Activation of mTORC1 by didemnin B was conserved in all cell lines tested, including HCT116, U2OS, HeLa, primary human fibroblasts, telomerase-immortalized colonic epithelial cells, and mouse embryonic fibroblasts (Fig. 1d and Supplementary Fig. 1) as well as Vaco451 (see below). Importantly, intraperitoneal injection of didemnin B in mice induced a dose-dependent activation of mTORC1 in the liver, recapitulating the observations in tissue culture cells (Supplementary Fig. 1h).
Figure 1. Natural product FuSiOn identifies didemnin B as an activator of mTORC1.
(a) The FUSION clade enriched for known AKT pathway elements. Protein-coding genes and microRNAs known to participate in AKT signaling are highlighted in blue. ALDOC, aldolase C; NDRG1, N-myc downstream regulated 1; BNIP3L, BCL2/adenovirus E1B 19kDa interacting protein 3-like; BNIP3, BCL2/adenovirus E1B 19kDa interacting protein 3; ACSL5, acyl-CoA synthetase long-chain family member 5; LOXL2, lysyl oxidase-like 2. (b) The Euclidean rank sum values of the top 50 miRNA mimics and siRNA pools (of 1124 total) whose functional signatures correlate with that produced by UT-BA07-004-ETOAC are shown in rank order. Values corresponding to reagents from the AKT clade, shown in (a), are labeled. (c) 24 hour treatment of HCT116 cells with didemnin B inhibits AKT signaling. (d) 2.5 hour treatment of HCT116 cells with didemnin B activates P70S6K in a rapamycin-sensitive manner. (e) Didemnin B induces Redd-1 loss. (f) Didemnin B fails to activate mTORC1 in REDD1 knockout MEFs (REDD1 −/−). All immunoblots (c, d, e, f) are representative of two or more experiments.
Didemnin B activates mTORC1 by inhibition of Redd-1
Two direct protein targets of didemnin B have been identified: the eukaryotic translation elongation factor 1 alpha 1 (EEF1A1)25 and the palmitoyl-protein thioesterase 1 (PPT1)26. Didemnin B can inhibit protein translation by binding EEF1A1 and stabilizing aminoacyl-tRNA bound to the ribosomal A-site, thus preventing translocation27. Didemnin B is also a noncompetitive inhibitor of PPT1-dependent degradation of palmitoylated proteins in lysosomes28. To determine if didemnin B activity on the mTOR pathway could be ascribed to one of these known targets, we first examined if distinct chemical inhibitors of protein synthesis or PPT1 were sufficient to activate mTORC1. U2OS cells cultured under low nutrient conditions, to reduce basal mTORC1-S6K-S6 activity, were treated with cycloheximide (10 μg/ml) or puromycin (10 μg/ml) for 30 minutes. Both inhibitors of protein synthesis activated mTORC1 as indicated by increased phosphorylation of S6K and S6, while the PPT1 inhibitor HDSF29 did not (Supplementary Fig. 1i, j). In parallel assays, the extent of co-administered low-dose puromycin incorporation into nascent polypeptides was used as an indicator of protein synthesis, and confirmed translational inhibition by didemnin B (1 μM or 1.11 μg/ml) and cycloheximide (10 μg/ml) (Supplementary Fig. 1k). In contrast, hygromycin B, which impairs fidelity of translation without altering the rate of protein synthesis, neither activated mTORC1 nor impaired puromycin incorporation (Supplementary Fig. 1i, k). As expected, inhibition of protein synthesis was also accompanied by loss of the short-lived protein p53 (Supplementary Fig. 1k). Biological consequences of mTORC1 activation with either didemnin B or cycloheximide could be detected by accumulation of S6K phosphorylation within five minutes, accumulation of S6 phosphorylation within 10 minutes, and decreased autophagolysosome maturation within 120 minutes (Fig. 1e). Collectively, these observations indicate that translational inhibition activates mTORC1 and associated downstream signaling in a manner that is rapid, broad, and sustained. We considered that the mechanism underpinning the mTORC1 response to translational suppression might be accumulation of free amino acids and/or loss of a short-lived negative regulator30–32. Translational inhibition activated mTORC1 whether free amino acids were abundant (Fig. 1d and Supplementary Fig. 1b–h) or removed from the media (Fig. 1e, f and Supplementary Fig. 1i, l–n). However, among mTORC1 regulatory proteins, we found that loss of the mTORC1-inhibitor REDD1 strongly correlated with mTORC1-S6K-S6 activation induced by both cycloheximide and didemnin B (Fig. 1e and Supplementary Fig. 1l). Importantly, REDD1 knockout MEFs responded to amino acids by activation of mTORC1 signaling, but failed to respond to didemnin B or cycloheximide (Fig. 1f and Supplementary Fig. 1m, n). These findings indicate that REDD1 expression is required to couple mTORC1 activation to protein translation activity and that didemnin B activates mTORC1 through the same mechanism as cycloheximide (Supplementary Fig. 1o)30–32.
Selective induction of apoptosis by didemnin B
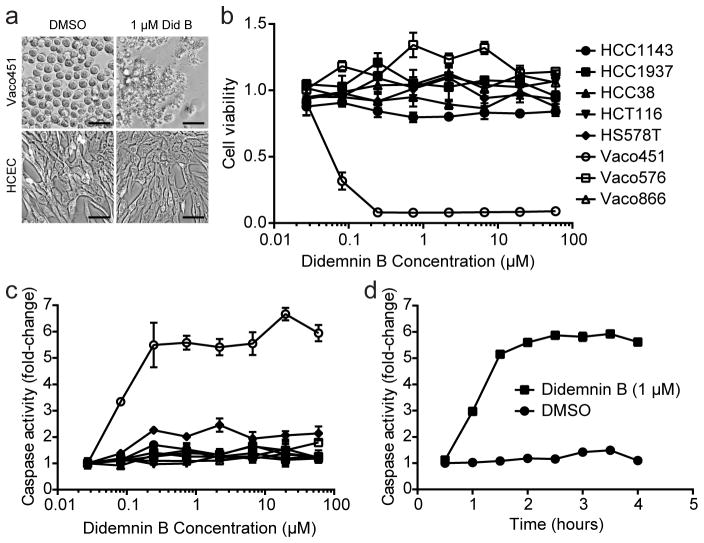
Although didemnin B and synthetic analogs have entered clinical trials for several oncological diseases, the anti-cancer mechanism of action is not yet understood6. Therefore we wished to test whether translational inhibition and mTORC1 activation contribute to the ability of didemnin B to induce cancer cell death. Among a small panel of breast cancer and colon cancer cell lines tested, we observed exceptionally selective toxicity in the colon cancer cell line Vaco451 (Fig. 2a, b and Supplementary Fig. 2a, b). Didemnin B toxicity in this colon cancer cell line achieved 100% kill at concentrations of 247 nM or higher within 96 hours of treatment, and displayed an LC50 of ~32 nM (Supplementary Fig. 2a) with no consequence on telomerase-immortalized colonic epithelial cells, HCT116 colon cancer cells, or any of several additional tumor-derived cell lines tested (Fig. 2a–c and Supplementary Fig. 2a, b). Remarkable selectivity for Vaco451 cells was observed at multiple time-points (ranging from 4–96 hours) and concentrations (ranging from 0.08 – 60 μM) of didemnin B treatment (Fig. 2a–c and Supplementary Fig. 2a, b). A four-hour exposure to didemnin B at concentrations of 247 nM or higher was sufficient to kill >90% of Vaco451 cells, without any reduction in viability of resistant cell lines at any concentration tested, up to 60 μM (Fig. 2b). The mode of cell death was apoptosis, with maximal induction of caspase activity observed within two hours of treatment in the exceptionally sensitive cell line (Fig. 2c, d).
Figure 2. An exceptional responder reveals context-specific induction of apoptosis by didemnin B.
(a) Didemnin B is selectively toxic in Vaco451 vs. HCEC. Indicated cell lines were treated with 1 μM didemnin B or equivalent volume DMSO for five hours before cells were imaged. Scale bar = 0.05 mm. (b) Vaco451 responds to low-nM concentrations of didemnin B. The indicated cell lines were treated with didemnin B at the indicated concentrations for six hours before cell viability was measured. Mean +/− SEM of triplicates is shown. (c) Didemnin B selectively induces apoptosis in Vaco451 cells. Indicated cell lines were treated with didemnin B at the indicated concentrations for three hours before caspase activity was measured. Mean +/− SEM of triplicates is shown. Legend from (b) applies also to (c). (d) Maximal caspase activation occurs within 2 hours of didemnin B exposure. Vaco451 cells were treated with 1 μM didemnin B or DMSO alone for the indicated time period before caspase activity was measured. Mean +/− SEM of triplicates is shown.
Didemnin B targets PPT1 and EEF1A1 to induce apoptosis
Notably, cell death was not rescued by pretreatment with rapamycin or Torin, indicating that activation of mTORC1 by didemnin B is not required for induction of cell death in Vaco451 cells (Supplementary Fig. 2c–h). Furthermore, although cycloheximide (10 μg/ml) inhibited protein synthesis in Vaco451 cells slightly better than didemnin B (1 μM or 1.11 μg/ml), it displayed minimal caspase activation in comparison to didemnin B (Supplementary Fig. 3a–c). Concordantly, translational inhibition via cycloheximide at this dose was insufficient to kill Vaco451 cells (Supplementary Fig. 3d). Concentrations of cycloheximide as high as 30 μg/ml (106.6 μM) were required to obtain modest reductions in cell viability (Supplementary Fig. 3e). These observations demonstrate that translational inhibition is insufficient to account for the remarkable response of Vaco451 cells to didemnin B, and indicate the participation of additional protein targets.
The didemnin B target enzyme palmitoyl-protein thioesterase 1 (PPT1)26,28 is considered generally dispensable for cell viability and function, as PPT1 knockout mice develop normally and remain healthy until neurodegeneration manifests with age33. However, selective vulnerability to PPT1 inhibition within tumorigenic regulatory contexts has not been examined. Transfection of a pool of siRNA oligos targeting PPT1 reduced PPT1 protein expression in Vaco451 cells below the limit of detection by western blot analysis (Supplementary Fig. 3f), but no consequences on cell viability were detected (Fig. 3a). However, siRNA-mediated depletion of PPT1 selectively sensitized Vaco451 cells to cycloheximide (Fig. 3a and Supplementary Fig. 3g). Four of five independent siRNA oligos targeting PPT1 were effective, arguing against the interpretation that sensitization was due to an off-target effect of the reagents employed (Supplementary Fig. 3f, h). Furthermore, an independent siRNA pool targeting PPT1 also sensitized to cycloheximide, as did three of four individual oligos within the second pool (Supplementary Fig. 3i, j). The sensitization was specific for PPT1 and was not observed with siRNAs targeting PPT2 (Supplementary Fig. 3i). We also did not observe sensitization to other potential inducers of apoptosis such as Smac mimetic or CFLAR RNAi upon PPT1 depletion (Fig. 3a and Supplementary Fig. 3k). Since PPT1 is a lysosomal thioesterase with optimal activity at pH of 4.034, we asked whether inhibition of lysosomal acidification via the vacuolar-ATPase inhibitor bafilomycin A1 would also sensitize Vaco451 cells to translational inhibition. Indeed, pretreatment with 50 nM bafilomycin A1 for 30 minutes sensitized to 10 μg/ml cycloheximide, resulting in >50% cell death after six hours (Fig. 3b). Sensitivity to combinatorial treatment with inhibitors of the vacuolar ATPase and protein synthesis was detected specifically in the didemnin B sensitive cell line Vaco451, but not didemnin B resistant cell lines Vaco866 or HCT116 even up to concentrations of 100 μg/ml cycloheximide and 200 nM bafilomycin A1 (Fig. 3b and Supplementary Fig. 3l, m). Rapamycin was not capable of substituting for cycloheximide in the combinatorial treatment, likely because it only inhibits translation of a specific subset of mRNAs, unlike cycloheximide, which broadly inhibits protein synthesis32 (Supplementary Fig. 3n). Finally, the PPT1 inhibitor hexadecylsulfonylfluoride (HDSF)29 selectively sensitized Vaco451, but not HCT116, to translational inhibition (Fig. 3c, d). Taken together, these observations indicate that didemnin B kills exceptional responders through combinatorial inhibition of PPT1 and protein synthesis.
Figure 3. Combinatorial inhibition of PPT1 and protein synthesis recapitulates selective induction of apoptosis by didemnin B.
(a) PPT1 RNAi sensitizes Vaco451 cells to translational inhibition. 72 hours post-transfection, cells were exposed to the indicated compounds for 6 hours. Mean + SEM of triplicates is shown. (b) Bafilomycin A1 sensitizes Vaco451 cells to translational inhibition. Indicated cells were pretreated with DMSO or 50 nM Bafilomycin A1 for 30 minutes before addition of DMSO, 10 μg/ml cycloheximide, or 1 μM didemnin B. Mean + SD of triplicates is shown. ns, p > 0.05; **, 0.001 < p < 0.01 by unpaired t test. (c) 24-hour pretreatment with HDSF sensitizes Vaco451 cells to caspase activation in response to translational inhibition. Mean + range of duplicates is shown. (d) HDSF sensitizes Vaco451 cells to translational inhibition. Mean +/− SD of triplicates is shown. (e) Didemnin B induces proteasome-dependent Mcl-1 loss. Vaco451 cells were pre-treated with 10 μM MG132 for 30 minutes prior to the addition of either 10 μg/ml cycloheximide, 1 μM didemnin B, or DMSO for 3 hours. Results are representative of two or more experiments. (f) Proteasome inhibition prevents didemnin B induced cell death. Mean +/− SEM of triplicates is shown. (g) Combinatorial inhibition of PPT1(12.5 μg/ml HDSF) and Mcl-1 (TW-37) selectively kills Vaco451 cells. Mean +/− SEM of triplicates is shown.
Mcl-1 accumulation is sensitive to EEF1A1 inhibition
We next tested whether translational inhibition might contribute to didemnin B-induced apoptosis in Vaco451 cells through depletion of one or more short-lived anti-apoptotic proteins such as Mcl-1 or c-Flip (encoded by the CFLAR gene). We first surveyed the effect of translational inhibition in Vaco451 cells on a handful of apoptosis-modulating proteins known to be rapidly degraded in other contexts (Supplementary Fig. 3n). We found that Mcl-1 and c-Myc were easily detected at baseline but disappeared rapidly upon translational inhibition in these cells, in a proteasome-dependent manner, with kinetics similar to those of caspase activation by didemnin B (Fig. 3e, Fig. 2d, and Supplementary Fig. 3n). On the other hand, c-Flip was barely detectable and disappeared with slightly slower kinetics (Supplementary Fig. 3o). Notably, didemnin B-induced cell death required proteasome function, as pretreatment with the proteasome inhibitor MG132 completely rescued viability in didemnin B-treated Vaco451 cells (Fig. 3f). This suggested that degradation of one or more proteasome substrates may be required for induction of apoptosis by didemnin B. Depletion of c-Flip using siRNA targeting CFLAR had no effect on viability of Vaco451 cells alone or in combination with PPT1 siRNA (Supplementary Fig. 3k). Depletion of c-Myc reduced viability by about half in Vaco451 cells, likely due to decreased proliferation, but did not sensitize to PPT1 siRNA (Supplementary Fig. 3p). Furthermore, neither gain nor loss of function of c-Myc modulated didemnin B-induced cell death (Supplementary Fig. 3q, r)35. In contrast, the Mcl-1 inhibitor TW-37 induced caspase activation and massive cell death selectively in Vaco451, but not HCT116 or Vaco866, pre-treated with the PPT1 inhibitor HDSF (Fig. 3g and Supplementary Fig. 3s–u). Together, these data suggest that didemnin B induces apoptosis in Vaco451 cells by combinatorial depletion of Mcl-1 and inhibition of PPT1.
Rare and diverse representation of didemnin B sensitivity
To develop leads for molecular contexts that may account for or correlate with exceptional response to didemnin B, we searched for additional sensitive cell lines. From NCI-60 profiling efforts, the acute promyelocitic leukemia cell line HL-60 was found to undergo rapid and complete apoptosis in response to 1 μM didemnin B exposure36. One commonality between the colorectal cancer cell line Vaco451 and the acute promyelocitic leukemia cell line HL-60 is substantial amplification of the Myc locus37,38. This led us to test whether Myc amplification might predict sensitivity to didemnin B within a small panel of cancer cell lines of known Myc copy number status39 (Supplementary Fig. 4a). However, we found no association between Myc amplification status and sensitivity to didemnin B among this panel (Supplementary Fig. 4a). While these efforts eliminated Myc amplification as a predictive marker, they successfully uncovered two additional didemnin B sensitive cell lines: the breast cancer cell line HCC1187 and the small cell lung cancer cell line NCIH211 (Supplementary Fig. 4a). Among the cell line panel tested, selective sensitivity of HCC1187 and NCIH211 was observed at concentrations of didemnin B ranging from 0.08–60 μM and treatment times ranging from 4–72 hours (Fig. 4a and Supplementary Fig. 4b, c). Like Vaco451, both HCC1187 and NCIH211 responded to didemnin B with rapid activation of caspases (Fig. 4b and Supplementary Fig. 4d), were resistant to single agent administration of cycloheximide (Supplementary Fig. 4e), and were selectively sensitive to siRNA-mediated co-depletion of PPT1 and Mcl-1 (Fig. 4c). These observations suggest that a subset of epithelially-derived human cancer cells spanning multiple tissues of origin exhibit selective sensitivity to didemnin B-induced apoptotic cell death through the same mechanism of action.
Figure 4. A subset of breast, colon, and lung cancer cell lines are selectively sensitive to didemnin B.
(a) The indicated cell lines were treated with didemnin B at the indicated concentrations for six hours before cell viability was measured. Mean +/− SEM of triplicates is shown. (b) The indicated cell lines were treated with 1 μM didemnin B or equivalent volume of DMSO for three hours. Caspase activity was measured using Caspase-Glo and expressed as fold-change vs. DMSO-treated cells. Mean + SEM of triplicates is shown. (c) The indicated cell lines were transfected with siRNA pools targeting LONRF1 (negative control), UBB (positive control), or PPT1 and Mcl-1. 72 hours later, cell viability was measured and normalized to control. Mean + SEM of triplicates is shown.
Multi-feature biomarker predicts didemnin B sensitivity
We therefore leveraged this cohort to generate a data-driven molecular response predictor using the elastic net penalized linear regression model40. Genome-wide mRNA expression data obtained using a common technical platform is publicly available from three of the four sensitive lines, as well as ten of the cell lines we found to be resistant to didemnin B (Fig. 2b, c, Fig. 4a and Supplementary Fig. 4a). From a binary indication of “sensitive” or “resistant” assigned to each cell line, a four-gene biomarker emerged, such that high expression of LOC101927886, HNRNPM, BCL11A, and TP53BP2 correlated with sensitivity to didemnin B among the 13 cell lines (Fig. 5a). This multi-feature biomarker was used to predict sensitivity among 1022 additional cell lines for which microarray data were available (Fig. 5b and Supplementary Data Set 1). Normalized predictive values ranged from 0.141 (predicted to be most sensitive) to −0.080 (predicted to be most resistant). Cancer cell lines from the “haematopoietic and lymphoid tissue” lineage were highly enriched among top-ranked predicted sensitive lines (indicated in purple in Fig. 5b and Supplementary Data Set 1). Cell lines derived from patients with ALL were particularly enriched among top-ranked lines, which is notable given that a synthetic analog of didemnin B has been granted orphan drug status in this disease setting (Supplementary Data Set 1)6. Of note, rare cell lines from other lineages such as “breast” (yellow) and “lung” (blue) were also among top-ranked predicted sensitive lines (Fig. 5b and Supplementary Data Set 1). To ask whether this was an artifact of cell culture or, alternatively, might indicate the presence of a rare, distinct subset of epithelially-derived cancer patients, we applied the multi-feature biomarker to publicly available expression profiles from patient-derived tumor samples assembled by The Cancer Genome Atlas (http://cancergenome.nih.gov/). Indeed, patient-derived tumor samples spanned a larger range of predicted sensitivity than that represented by the cell line panel, with three colorectal adenocarcinoma patient samples in particular displaying a stronger match to the multi-feature biomarker predicting sensitivity to didemnin B than any cell line in the panel (Supplementary Data Set 2, Fig. 5c, and Supplementary Fig. 5a, b). To evaluate the predictive potential of the multi-feature biomarker among epithelially-derived cancers, five non-hematopoietic lines spanning the entire range were obtained and tested for sensitivity to didemnin B (Fig. 5d, e). The breast cancer cell line HCC1599, ranked # 13 of 1022, had the highest predicted sensitivity among cell lines to which we had access, and was indeed a remarkable responder to didemnin B (Fig. 5d, e and Supplementary Fig. 5c–e). The lung cancer cell line NCIH82, ranked # 35 of 1022, displayed partial sensitivity that required either higher concentrations of didemnin B or longer treatment times to exhibit responses comparable to those seen with fully sensitive lines (Fig. 5d, e and Supplementary Fig. 5c–e). The remaining cell lines tested, ranked # 192, # 337, and # 1022 of 1022, were all resistant to didemnin B (Fig. 5d, e and Supplementary Fig. 5c–e). Thus the multi-feature biomarker could successfully predict sensitivity to didemnin B among epithelially-derived human cancer cell lines propagated in culture and could identify a subset of colorectal, breast, and lung cancer patients that might be expected to respond to didemnin B or its analogs.
Figure 5. Molecular response predictor for exceptional sensitivity to didemnin B.
(a) Expression values of the indicated genes in the indicated cell lines are displayed as a heatmap. For compound sensitivity (top), a binary call was made with a value of “1” assigned to cell lines exhibiting sensitivity to didemnin B and a value of “0” assigned to resistant cell lines. Respective weights for each gene from the elastic net algorithm are displayed in yellow on the left. (b) The ranked predicted sensitivity for 1022 lines are displayed, with five experimentally tested cell lines indicated by name with dotted lines at the corresponding predicted sensitivity value. Cell lines are color-coded by anatomical site of origin in graph (left) and by rank (right). (c) The predicted sensitivities for 185 colorectal adenocarcinoma patients, computed using the elastic net-derived multi-feature biomarker from (a), are displayed in ranked order. (d) The indicated cell lines were treated with 1 μM didemnin B or equivalent volume of DMSO for six hours. Cell viability was measured using Cell TiterGlo, normalized to DMSO alone, and displayed as mean + SEM of n= 6–9 replicates. (e) The indicated cell lines were treated with 1 μM didemnin B or equivalent volume DMSO for three hours. Caspase activity was measured using CaspaseGlo and displayed as fold-change versus DMSO-treated cells. Mean + SEM of triplicates are shown. In (d and e), * indicates P value < 0.05; ns indicates P value > 0.05.
GSEA further indicates polypharmacology of mechanism
A total of four sensitive lines and ten resistant lines with associated expression data were used in an S2N analysis to identify the top 2% of differentially expressed genes between didemnin B-sensitive and -resistant classes (Fig. 6a, Supplementary Fig. 6a and Supplementary Data Set 3). Pathway enrichment analysis identified “KEGG_LYSOSOME” as one of the top pathways enriched in the resistant versus sensitive class (p=0.0014, hypergeometric test, Supplementary Data Set 4). The leading-edge genes included CLN5, which similarly to the didemnin B target PPT1 (also called CLN1), is mutated in some forms of neuronal ceroid lipofuscinoses41 (Fig. 6b, Supplementary Data Sets 3 and 4). Conversely, the top pathway enriched in the sensitive versus resistant class was “1060_Retromer complex (SNX1 SNX2 VPS35 VPS29 VPS26B)” with p=0.0010 by the hypergeometric test (Supplementary Data Set 5). Given that CLN5 is required for recruitment of the retromer to the lysosomal sorting receptor42, these findings suggest a coherent biological process which may be impaired specifically in the didemnin B sensitive class of cancer cells. In addition, EEF1A2, which is 98% similar (92% identical) to the didemnin B target EEF1A1 and functions similarly in translational elongation43, was one of the top 2% of genes expressed more highly in the resistant than the sensitive class by S2N analysis (Fig. 6c and Supplementary Data Set 3).
Figure 6. Predictive power in hematological cancer.
(a) The normalized expression of the top 2% of genes differentially expressed between the sensitive and resistant class of cell lines, as determined by S2N analysis, is shown as a heat map. (b, c) the expression of two genes from (a), CLN5 (b), and EEF1A2 (c), in sensitive versus resistant cell lines is displayed. (d) Selective response of hematologic cell lines to didemnin B. The indicated cell lines were treated with didemnin B at the indicated concentrations for 24, 48, or 72 hours before cell viability was measured. Mean +/− range of duplicates is shown.
Biomarker enrichment in hematologic cancers
Although 12 of the 13 cell lines used to generate the multi-feature biomarker were derived from solid tumors of epithelial origin, CCLE lines predicted to be sensitive to didemnin B were highly enriched for those from hematopoietic lineages (Fig. 5a, b, Supplementary Data Set 1, and discussed above). Analysis of biomarker-derived sensitivity scores among several leukemia and lymphoma patient cohorts exhibited a wide range of predicted sensitivity (Supplementary Fig. 6b). Therefore, we acquired eight hematopoietic lines from the CCLE panel that spanned the range of predicted sensitivity and tested the ability of didemnin B to induce apoptosis. Three of four predicted responders were indeed sensitive to didemnin B by this criterion (Supplementary Fig. 6c). Sensitive lines from the hematopoietic lineages responded to didemnin B by rapid apoptotic cell death that was similar with respect to magnitude (<5% viability), active dose range (0.02 – 60 μM) and timing/persistence (4 – 72 hours) to that of sensitive lines from epithelial lineages (Fig. 6d and Supplementary Fig. 6d, e).
DISCUSSION
Using quantitative gene expression signatures as a pattern-matching tool for detection of mechanistically equivalent chemical and genetic perturbations, the cyclic depsipeptide didemnin B was identified as a persistent mTORC1 pathway agonist in vitro and in vivo. Mode of action studies indicated this activity was the consequence of release of REDD1-dependent inhibition of mTORC1 via the well-described suppression of protein translation by didemnin B. These observations revealed REDD1 as a short-lived protein that acts as a self-sufficient brake on latent mTORC1 activity. Loss of REDD1 accumulation resulted in rapid and full mTORC1 activation in the absence of instructive signals such as amino acids, growth factors, or serum. This relationship suggests that, within the complex hierarchy of mTORC1 pathway control mechanisms, coupling of mTORC1 activity to dynamic rates of protein synthesis is dominant to signal transduction mechanisms that monitor nutrient availability or mitogen pathway activity.
REDD1-mediated mTORC1 inhibition therefore likely serves to maintain equilibrium between cellular rates of protein synthesis and degradation across ever-changing conditions. The robustness of this system is perhaps highlighted by the fact that humans can develop and thrive even when protein synthesis is substantially perturbed by mutations in genes encoding core translational machinery. Notably, a sequela of such genetic diseases, termed ribosomopathies, is an increased incidence of cancer44. This could be due to sustained activation of mTORC1 in response to impaired protein translation through the mechanism described here.
Didemnin B has long been recognized as an antiproliferative agent, in a variety of inflammatory and neoplastic settings, with a cryptic mechanism of action that is not accounted for by targeting EEF1A18. From a cohort of molecularly characterized breast, colon, and lung tumor-derived cell lines, we identified exceptional responders to didemnin B-induced apoptosis. Mechanistic evaluation of this activity indicated an obligate combinatorial suppression of both protein targets of didemnin B, EEF1A125,27 and PPT126,28. This adds didemnin B to a growing list of anti-neoplastic compounds in which the bioactivity of interest results from plural mechanisms of action of a single molecule.
We found that didemnin B causes rapid loss of the anti-apoptotic protein Mcl-1, and that selective induction of apoptosis in sensitive cancer cell lines by didemnin B could be reproduced by combinatorial treatment with the PPT1 inhibitor HDSF and the Mcl-1 inhibitor TW-37. This mechanistic insight raises two distinct therapeutic possibilities warranting further investigation. First, in clinical settings where didemnin B analogs are found to be effective, combinatorial treatment with specific inhibitors of PPT1 and Mcl-1 might potentially maintain effectiveness while reducing toxicity otherwise caused by blunt translational inhibition7,45. Second, in clinical settings where Mcl-1 represents a resistance mechanism to an otherwise promising therapeutic option (or a bona fide target itself), didemnin B analogs may be effective in combination with other therapies (or as a single agent)46,47.
Public efforts facilitating accumulation of molecular annotations for both tumor-derived cell lines and tumor tissues allowed identification of conserved genomic features that predict exceptional response to didemnin B. Specifically, a multi-feature quantitative gene expression profile was sufficient to predict tumor-derived cell lines with an apoptotic response to low-dose didemnin B. By ranking the diverse human cancer cell lines represented in the CCLE, we noted a significant enrichment of hematological malignancies as compared to other disease lineages. This is notable given that dehydrodidemnin B was granted orphan drug status for treating ALL and is currently in phase 1, 2, and 3 clinical trials for other hematological malignancies. When applied to patient cohorts across multiple solid tumor disease settings, this biomarker identifies a distinct population of patients that we believe may be most likely to respond to didemnin B or its analogs. Though genome-wide expression data are not routinely used diagnostically, this biomarker may correlate with more easily detected features such as translocations. Intriguingly, both breast cancer cell lines identified as sensitive to didemnin B in this study (HCC1187 and HCC1599) harbor activating Notch rearrangements, a relatively rare event in this disease setting48. Furthermore, predicted sensitivity to didemnin B positively correlated with presence of the TEL-AML fusion in two independent preB-ALL patient cohorts (GSE12995, p=1.1e-05, and GSE13425, p=7.3e-08). If verified by clinical studies assessing patient outcome, these biomarkers could enable guided therapeutic use of didemnin B or an analog in patients most likely to respond, thus expanding the clinical benefit without unnecessarily exposing large numbers of patients to the associated toxicities.
ONLINE METHODS
Cell lines
Cell lines are as described49–51. They were kind gifts from Adi Gazdar (breast cancer lines), James Willson and Sandy Markowitz (Vaco colon cancer lines), Jerry Shay (human colonic epithelial cells), John Minna (lung cancer lines), Fred Grinnell (BR5 human fibroblasts), Ryan Potts (U2OS), Xiaodong Wang (U2OS GFP-LC3 and Jurkat), Josh Mendell (P493), and James Brugarolas (REDD1 wild-type and knockout MEFs), or were purchased from the ATCC (HCT116) or the DSMZ (TALL-1 and RCH-ACV). The identities of all human cancer cell lines from epithelial origin were verified using the Promega Powerplex 1.2 system and an in-house database of more than 500 reference fingerprints52. Growth media was DMEM supplemented with 10% fetal bovine serum (FBS) for HCT116, U2OS, HeLa, MEF, and BR5 fibroblast cells. Growth media was MEM2+ for Vaco576 and Vaco871 cells. Growth media was Iscove’s supplemented with 20% FBS for OCI-M1, OCI-Ly3, OCI-Ly10, and OCI-Ly19. Growth media was AlphaMEM supplemented with 10% FBS for OCI-AML3. Growth media was RPMI supplemented with 5–15% FBS (as directed by supplier) for all remaining cell lines, except for the normal human colonic epithelial cells (HCECs), which were maintained and passaged in S+2% media under low oxygen conditions, but were assayed in RPMI supplemented with 10% FBS under normoxia conditions. The U2OS cell line stably expressing GFP-LC3 was used to assess the effect of translational inhibition via didemnin B or cycloheximide on autophagy53.
Antibodies and other materials
Antibodies recognizing pan-AKT, p-AKT (S473), GSK3β, p-GSK3β (S9), TSC2, p-TSC2 (T1462), S6K, p-S6K (T389), S6, p-S6 (S235/6), PRAS40, ULK1, Rag A, Rag B, Rag C, AMPK, cleaved PARP, and Mcl-1 were from Cell Signaling Technology. β-tubulin antibody was from Sigma-Aldrich. XPB (TFIIH p89 S-19), p53, c-Myc, and GFP antibodies were from Santa Cruz. Puromycin antibody was from KeraFAST. REDD1/DDIT4 antibodies were from Novus and Bethyl. C-FLIP antibody was from Enzo Life Sciences. All antibodies were used at final concentrations of 1:1000 – 1:2000 except for cleaved PARP, c-Myc, p-TSC2, ULK1, and XPB (1:500), AMPK, P70S6K, and TSC2 (1:750), and β-tubulin, S6, and p-S6 (1:4000). DMSO, cycloheximide, puromycin, rapamycin, bafilomycin A1, and MG-132 were from Sigma-Aldrich. Hygromycin B was from Corning. Hexadecylsulfonylfluoride (HDSF) was from Santa Cruz. TW-37 was from Selleckchem. Torin1 was a kind gift from David Sabatini and Nathanael Gray. Smac mimetic was a kind gift from Xiaodong Wang.
Natural products
Generation and functional mapping of the natural product library was described previously2. Tunicate sample UTSW-BA07-004 was collected at Sweetings Cay, Bahamas in 2008. Based on morphology, the tunicate was identified as Trididemnum solidum. A type sample is preserved in the MacMillan lab at UTSW. An additional Trididemnum solidum specimen was kindly provided by Tadeusz F. Molinski (UC – San Diego). A sample of T. solidum (60 g wet wt.) was lyophilized, extracted with CH2Cl2:MeOH (2:1) for 24 hours and concentrated. Subsequent partitioning between aq. MeOH (10%) and n-hexane, aq. MeOH (30%) and CH2Cl2. Didemnin B was found to be present in large quantities in the CH2Cl2 fraction, which was subsequently purified using flash SiO2 chromatography with a gradient of CH2Cl2:MeOH (100:0 to 80:20) to give UTBA07-didemnin B (10 mg). Didemnin B, white solid, [α]D25 −81.4 (c 0.5, CH2Cl2); 1H NMR (Supplementary Fig. 1a, 500 MHz, CDCl3) and 13C NMR (125 MHz, CD3Cl3) are consistent with previously reported NMR data54; HRESIMS [M + H]+m/z 1112.6492 (calcd for C57H90N7O15, 1112.6495).
Compound treatment, viability assays, caspase activity assays, and western blots
Cells were plated in 96-well plates for viability assays and caspase activity assays, and in 96-, 24-, 12-, or 6-well plates for lysate collection for western blot experiments. Compounds in DMSO were added to achieve the indicated final concentration, for the indicated length of time. The same volume of DMSO was added as a negative control. Viability was measured using CellTiter Glo (Promega) following the manufacturer’s protocols, or caspase activity was measured using Caspase-Glo reagent (Promega) following manufacturer’s protocols, or cells were collected in hot 2x sample buffer, boiled, and sonicated for immunoblot analysis using standard western blot protocols. XPB (TFIIH p89 S-19, Santa Cruz) was used as a loading control.
Mice
Didemnin B was formulated in 90% D5W + 5% cremophor + 5% DMSO for animal delivery. Three female SCID-NOD mice per condition (6–8 weeks of age) were treated with high concentration didemnin B (1 mg/kg), low concentration didemnin B (0.11 mg/kg) or vehicle alone, by two intraperitoneal injections 96 hours and 7 hours before harvesting livers for flash freezing in liquid nitrogen. Lysates were generated from small sections of mouse liver in M-PER Mammalian Protein Extraction Reagent (Thermo Scientific) supplemented with Phosphatase Inhibitor Cocktails #2 and #3 (Sigma-Aldrich) using a TissueLyser II (Qiagen), following the manufacturer’s protocols. 10–40 μg per sample was loaded on each of four SDS-PAGE gels and analyzed by standard western blot protocols. These animals were cared for according to guidelines set forth by the American Association for Accreditation of Laboratory Animal Care and the U.S. Public Health Service policy on Human Care and Use of Laboratory Animals. All mouse studies were approved and supervised by the UT Southwestern Institutional Animal Care and Use Committee.
Puromycin incorporation assay
Cells were plated in 6-well plates and pre-treated with the indicated compounds at the indicated concentrations for 30 minutes. Puromycin (Sigma-Aldrich) was added to the media at a final concentration of 1 μM (0.471 μg/ml). Thirty minutes after addition of puromycin, cells were pelleted, resuspended in hot 2X sample buffer, boiled, sonicated, and loaded on a 5–15% SDS-PAGE gel (Bio-Rad). After transfer to PVDF membrane, standard western blot protocols were used to probe with the antibody recognizing puromycin (KeraFAST) and other indicated antibodies.
Amino acid stimulation
Cells were plated in 6-well plates in growth media. Two days later, media was removed, cells were rinsed once with EBSS, and fresh EBSS was added. Two hours later, L-glutamine was added to a final concentration of 2 mM only in wells destined for amino acid stimulation. One hour later, amino acids were added to a final concentration of 1x for the 30 minute time-point. Twenty minutes later, amino acids were added to a final concentration of 1x for the 10 minute time-point. Ten minutes later, lysates were collected and analyzed by western blot.
RNA interference
RNAi was performed by transfecting siRNA oligos via reverse transfection using RNAiMax (Life Technologies) according to the manufacturer’s instructions. 96-well plate format was used. A pool of four siRNA oligos targeting each gene was used to dilute off-target effects, except where use of an individual siRNA oligo was specifically indicated. Pools of four siRNAs targeting either LONRF1 or UBB were used as negative or positive transfection controls, respectively.
Elastic net for predictive biomarker discovery
The elastic net is a penalized linear regression model that is used to select features that are associated with a drug response, measured, in this case, as a binary vector of sensitivity. The elastic net is member of a class of machine learning procedures that are specialized for the case when there are many more input features (i.e. gene expression measurements) than samples (i.e. cancer cell lines or tissues). It is advantageous in that it allows for the identification of multi-feature biomarker signatures whose additive patterns across the cell line panel act to predict a response vector. The elastic net also allows for the weighting of identified features to enable predictions of sensitivities in untested cell lines. We specifically chose the elastic net because it combines the advantages of an L1-norm (Lasso) regression and an L2-norm (Ridge) regression model. Candidate predictive features were selected from 21,087 measures of copy number expression profiles from Affymetrix SNP 6.0 arrays, 14,569 measures of gene expression from Affymetrix Human Genome U133 Plus 2.0 microarray platforms, and 1,667 measures of binary mutation statuses from massively parallel sequencing across 13 cell lines. Normalized values for each dataset were downloaded from the CCLE website (http://www.broadinstitute.org/ccle/), and duplicated measurements for the same gene were compressed into a single value by taking the median value between the duplicates.
Let X ∈ ℝnxp be the matrix of predictive features where n is the number of cell lines included in the training dataset and p is the number of features, and let y ∈ ℝn be the vector of binary sensitivity values for the same cell line panel. Columns of the predictive features matrix and y were normalized to have a mean of zero and a standard deviation of 1. The elastic net attempts to find which weighted linear combination of the columns of the predictive features matrix can best approximate y, or it solves the following equation for w:
The elastic net solves the above by enforcing a penalty to the solution that makes the solution both unique and sparse so that only the features that best approximate y are left with non-zero weight values. It does this by combining L1-norm and L2-norm regularization parameters so that the elastic net formulation to the above problem is given by:
where λ, α, are two adjustable parameters such that λ controls the degree of the overall penalty and α controls the degree to which the L1 norm and L2 norm constraints are applied so that when α=0, only the L1 penalty is applied and when α=1, only the L2 penalty is applied. In order to determine the optimal values of alpha and lambda to use in the model, we did 100 iterations of 5 fold cross-validation where, in each iteration, the cells were randomly re-sampled into different groups. The values of alpha and lambda were chosen to be those that resulted in the minimum mean squared error for each fold. Features were then chosen to be those with the highest weights that were selected as features in at least 70% of the cross-validation permutations.
Predicting sensitivities in untested cell lines
Weights were calculated for each of the features selected from the elastic net using the original 13 cell lines as a training set. Normalized predictive sensitivity values for untested cell lines were then calculated for each of the 1022 cell lines in the cancer cell line encyclopedia that were not included in the original training set with the following formula:
where wi is the weight determined from the elastic net for feature i, and xij is the normalized expression value of feature i in cell line j. The range of si values predicts the degree of sensitivity where a high value of si predicts sensitive and a low value of si predicts resistance. The described four feature gene signature was used to predict sensitivities in cells from the CCLE panel. The probe for ‘LOC101927886’ was not present in the tumor datasets, so a three feature gene signature was used to predict tumor sensitivities.
Supplementary Material
Acknowledgments
We thank Tadeusz Molinski for providing additional Trididemnum solidum material, James Willson and Sanford Markowitz for the Vaco colon cancer cell lines, Kenneth Huffman, Michael Peyton, and John Minna for the lung and breast cancer cell lines, Jerry Shay for the human colonic epithelial cells, Xiaodong Wang for the U2OS GFP-LC3 cells, James Brugarolas for the REDD1 knockout MEFs, Frederick Grinnell for the BR5 fibroblasts, and Joshua Mendell for the P493 cells. We also thank Noelle Williams for formulating didemnin B for animal delivery. This study was supported by the Welch Foundation (I-1414, I-1689), the National Cancer Institute (CA071443, CA176284, CA149833) and CPRIT (RP120718, RP110708). M.B.P. was supported by a Komen for the Cure Postdoctoral Fellowship. E.A.M. was supported in part by the NIH (2T32GM008203). Y-H.O. and H.S.K. were supported by fellowships from the Cancer Interventions and Discoveries Program (RP101496).
Footnotes
Author contributions
J.B.M. produced UT-BA07-004-ETOAC, purified didemnin B, and solved its structure. M.B.P., H.S.K., and M.A.W. performed the analyses that led to the prediction that UT-BA07-004-ETOAC may inhibit AKT signaling. M.B.P., T.I.R., and Y-H.O. performed the experiments that confirmed this prediction and elucidated the underlying mechanism. R.A.B., J.E.T., and M.B.P. designed and performed the mouse experiment. M.B.P. and T.I.R. identified cancer cell lines exhibiting exceptional sensitivity to didemnin B and determined the underlying mechanism. E.A.M. performed the elastic net analysis and the biomarker-based prediction analyses, and T.I.R. and M.B.P. tested the resulting predictions experimentally. M.D.M. provided the OCI cell lines. M.A.W. supervised the research. M.B.P. and M.A.W. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potts MB, et al. Using functional signature ontology (FUSION) to identify mechanisms of action for natural products. Sci Signal. 2013;6:ra90. doi: 10.1126/scisignal.2004657. 6/297/ra90 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Y, et al. Discoipyrroles A-D: isolation, structure determination, and synthesis of potent migration inhibitors from Bacillus hunanensis. J Am Chem Soc. 2013;135:13387–13392. doi: 10.1021/ja403412y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinehart KL, Jr, et al. Didemnins: antiviral and antitumor depsipeptides from a caribbean tunicate. Science. 1981;212:933–935. doi: 10.1126/science.7233187. [DOI] [PubMed] [Google Scholar]
- 5.Rinehart KL, Jr, Gloer JB, Cook JC, Jr, Mizsak SA, Scahill TA. Structures of the didemnins antiviral and cytotoxic depsipeptides from a Caribbean tunicate. J Am Chem Soc. 1981;103:1857–1859. [Google Scholar]
- 6.Lee J, Currano JN, Carroll PJ, Joullie MM. Didemnins, tamandarins and related natural products. Nat Prod Rep. 2012;29:404–424. doi: 10.1039/c2np00065b. [DOI] [PubMed] [Google Scholar]
- 7.Le Tourneau C, Raymond E, Faivre S. Aplidine: a paradigm of how to handle the activity and toxicity of a novel marine anticancer poison. Current pharmaceutical design. 2007;13:3427–3439. [PubMed] [Google Scholar]
- 8.Vera MD, Joullie MM. Natural products as probes of cell biology: 20 years of didemnin research. Medicinal research reviews. 2002;22:102–145. doi: 10.1002/med.10003. [DOI] [PubMed] [Google Scholar]
- 9.Fritz RD, Varga Z, Radziwill G. CNK1 is a novel Akt interaction partner that promotes cell proliferation through the Akt-FoxO signalling axis. Oncogene. 2010;29:3575–3582. doi: 10.1038/onc.2010.104. onc2010104 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Lim J, Zhou M, Veenstra TD, Morrison DK. The CNK1 scaffold binds cytohesins and promotes insulin pathway signaling. Genes Dev. 2010;24:1496–1506. doi: 10.1101/gad.1904610. 24/14/1496 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sridharan S, Basu A. S6 kinase 2 promotes breast cancer cell survival via Akt. Cancer research. 2011;71:2590–2599. doi: 10.1158/0008-5472.CAN-10-3253. 0008-5472.CAN-10-3253 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katayama K, Fujita N, Tsuruo T. Akt/protein kinase B-dependent phosphorylation and inactivation of WEE1Hu promote cell cycle progression at G2/M transition. Mol Cell Biol. 2005;25:5725–5737. doi: 10.1128/MCB.25.13.5725-5737.2005. 25/13/5725 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaul G, Pattan G, Rafeequi T. Eukaryotic elongation factor-2 (eEF2): its regulation and peptide chain elongation. Cell Biochem Funct. 2011;29:227–234. doi: 10.1002/cbf.1740. [DOI] [PubMed] [Google Scholar]
- 14.Kefas B, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer research. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. 68/10/3566 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Li ZY, Na HM, Peng G, Pu J, Liu P. Alteration of microRNA expression correlates to fatty acid-mediated insulin resistance in mouse myoblasts. Mol Biosyst. 2011;7:871–877. doi: 10.1039/c0mb00230e. [DOI] [PubMed] [Google Scholar]
- 16.Creevey L, et al. MicroRNA-497 increases apoptosis in MYCN amplified neuroblastoma cells by targeting the key cell cycle regulator WEE1. Mol Cancer. 2013;12:23. doi: 10.1186/1476-4598-12-23. 1476-4598-12-23 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo ST, et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2013;32:1910–1920. doi: 10.1038/onc.2012.214. onc2012214 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Z, et al. Downregulation of miR-383 promotes glioma cell invasion by targeting insulin-like growth factor 1 receptor. Med Oncol. 2013;30:557. doi: 10.1007/s12032-013-0557-0. [DOI] [PubMed] [Google Scholar]
- 19.Gong JN, et al. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ. 2014;21:100–112. doi: 10.1038/cdd.2013.133. cdd2013133 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121:499–509. doi: 10.1182/blood-2012-07-444729. blood-2012-07-444729 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. S0960982204006116 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Briaud I, et al. Insulin receptor substrate-2 proteasomal degradation mediated by a mammalian target of rapamycin (mTOR)-induced negative feedback down-regulates protein kinase B-mediated signaling pathway in beta-cells. J Biol Chem. 2005;280:2282–2293. doi: 10.1074/jbc.M412179200. M412179200 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. 332/6035/1317 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. 332/6035/1322 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crews CM, Collins JL, Lane WS, Snapper ML, Schreiber SL. GTP-dependent binding of the antiproliferative agent didemnin to elongation factor 1 alpha. J Biol Chem. 1994;269:15411–15414. [PubMed] [Google Scholar]
- 26.Crews CM, Lane WS, Schreiber SL. Didemnin binds to the protein palmitoyl thioesterase responsible for infantile neuronal ceroid lipofuscinosis. Proc Natl Acad Sci U S A. 1996;93:4316–4319. doi: 10.1073/pnas.93.9.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SirDeshpande BV, Toogood PL. Mechanism of protein synthesis inhibition by didemnin B in vitro. Biochemistry. 1995;34:9177–9184. doi: 10.1021/bi00028a030. [DOI] [PubMed] [Google Scholar]
- 28.Meng L, Sin N, Crews CM. The antiproliferative agent didemnin B uncompetitively inhibits palmitoyl protein thioesterase. Biochemistry. 1998;37:10488–10492. doi: 10.1021/bi9804479. bi9804479 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Das AK, et al. Structural basis for the insensitivity of a serine enzyme (palmitoyl-protein thioesterase) to phenylmethylsulfonyl fluoride. J Biol Chem. 2000;275:23847–23851. doi: 10.1074/jbc.M002758200. M002758200 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Blenis J, Chung J, Erikson E, Alcorta DA, Erikson RL. Distinct mechanisms for the activation of the RSK kinases/MAP2 kinase/pp90rsk and pp70-S6 kinase signaling systems are indicated by inhibition of protein synthesis. Cell Growth Differ. 1991;2:279–285. [PubMed] [Google Scholar]
- 31.Kimball SR, Do AN, Kutzler L, Cavener DR, Jefferson LS. Rapid turnover of the mTOR complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following inhibition of protein synthesis. J Biol Chem. 2008;283:3465–3475. doi: 10.1074/jbc.M706643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terada N, et al. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci U S A. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta P, et al. Disruption of PPT1 or PPT2 causes neuronal ceroid lipofuscinosis in knockout mice. Proc Natl Acad Sci U S A. 2001;98:13566–13571. doi: 10.1073/pnas.251485198. 98/24/13566 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voznyi YV, et al. A new simple enzyme assay for pre- and postnatal diagnosis of infantile neuronal ceroid lipofuscinosis (INCL) and its variants. J Med Genet. 1999;36:471–474. [PMC free article] [PubMed] [Google Scholar]
- 35.Pajic A, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. International journal of cancer Journal international du cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 36.Grubb DR, Wolvetang EJ, Lawen A. Didemnin B induces cell death by apoptosis: the fastest induction of apoptosis ever described. Biochem Biophys Res Commun. 1995;215:1130–1136. doi: 10.1006/bbrc.1995.2580. S0006-291X(85)72580-6 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Collins S, Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982;298:679–681. doi: 10.1038/298679a0. [DOI] [PubMed] [Google Scholar]
- 38.Leary RJ, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. 0808041105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou H, Hastie T. Regularization and variable selection via the elastic net. J Roy Stat Soc B. 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 41.Savukoski M, et al. Defined chromosomal assignment of CLN5 demonstrates that at least four genetic loci are involved in the pathogenesis of human ceroid lipofuscinoses. American journal of human genetics. 1994;55:695–701. [PMC free article] [PubMed] [Google Scholar]
- 42.Mamo A, Jules F, Dumaresq-Doiron K, Costantino S, Lefrancois S. The role of ceroid lipofuscinosis neuronal protein 5 (CLN5) in endosomal sorting. Mol Cell Biol. 2012;32:1855–1866. doi: 10.1128/MCB.06726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soares DC, Abbott CM. Highly homologous eEF1A1 and eEF1A2 exhibit differential post-translational modification with significant enrichment around localised sites of sequence variation. Biology direct. 2013;8:29. doi: 10.1186/1745-6150-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stumpf CR, Ruggero D. The cancerous translation apparatus. Current opinion in genetics & development. 2011;21:474–483. doi: 10.1016/j.gde.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ribrag V, et al. Multicenter phase II study of plitidepsin in patients with relapsed/refractory non-Hodgkin’s lymphoma. Haematologica. 2013;98:357–363. doi: 10.3324/haematol.2012.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suryani S, et al. Cell and molecular determinants of in vivo efficacy of the BH3 mimetic ABT-263 against pediatric acute lymphoblastic leukemia xenografts. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:4520–4531. doi: 10.1158/1078-0432.CCR-14-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belmar J, Fesik SW. Small molecule Mcl-1 inhibitors for the treatment of cancer. Pharmacology & therapeutics. 2015;145C:76–84. doi: 10.1016/j.pharmthera.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson DR, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nature medicine. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gazdar AF, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. International journal of cancer. Journal international du cancer. 1998;78:766–774. doi: 10.1002/(SICI)1097-0215(19981209)78:6<766::AID-IJC15>3.0.CO;2-L. [pii] [DOI] [PubMed] [Google Scholar]
- 50.Hackett AJ, et al. Two syngeneic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines. J Natl Cancer Inst. 1977;58:1795–1806. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- 51.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. 1133427 [pii] [DOI] [PubMed] [Google Scholar]
- 52.Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–1321. doi: 10.1093/jnci/djq279. djq279 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukimoto M, et al. Bacterial production of the tunicate-derived antitumor cyclic depsipeptide didemnin B. J Nat Prod. 2011;74:2329–2331. doi: 10.1021/np200543z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.