Abstract
Prostaglandin E2 (PGE2) is a potent lipid mediator involved in maintaining homeostasis but also promotion of acute inflammation or immune suppression in chronic inflammation and cancer. NLRP3 inflammasome plays an important role in host defense. Uncontrolled activation of NLRP3 inflammasome, due to mutations in the NLRP3 gene causes cryopyrin-associated periodic syndromes (CAPS). Here, we showed that NLRP3 inflammasome activation is inhibited by PGE2 in human primary monocyte-derived macrophages. This effect was mediated through prostaglandin E receptor 4 (EP4) and an increase in intracellular cAMP, independently of protein kinase A (PKA) or exchange protein directly activated by cAMP (Epac). A specific agonist of EP4 mimicked, while its antagonist or EP4 knockdown reversed PGE2-mediated NLRP3 inhibition. PGE2 caused an increase in intracellular cAMP. Blockade of adenylate cyclase by its inhibitor reversed PGE2-mediated NLRP3 inhibition. Increase of intracellular cAMP by an activator of adenylate cyclase or an analog of cAMP, or a blockade of cAMP degradation by phosphodiesterase inhibitor decreased NLRP3 activation. PKA or Epac agonists did not mimic and their antagonists did not reverse PGE2-mediated NLRP3 inhibition. In addition, constitutive IL-1β secretion from LPS-primed PBMCs of CAPS patients was substantially reduced by high doses of PGE2. Moreover, blocking cytosolic phospholipase A2α by its inhibitor or siRNA or inhibiting cyclooxygenase 2, resulting in inhibition of endogenous PGE2 production, caused an increase in NLRP3 inflammasome activation. Our results suggest that PGE2 might play a role in maintaining homeostasis during the resolution phase of inflammation and might serve as an autocrine and paracrine regulator.
Keywords: inflammasome, prostaglandin, PGE2, phospholipase, cAMP, macrophage, COX2, cPLA2
Introduction
The inflammasomes are a group of multimeric cytosolic protein complexes that consist of an inflammasome sensor molecule, adaptor protein ASC and caspase-1. Activation of caspase-1 by this complex leads to the processing of pro-IL-1β and pro-IL-18 to their mature active forms (1). Inflammasome forming sensor molecules encompass the family of the nucleotide binding domain, leucine rich repeat-containing proteins (NLRs) including NLRP1, NLRP3, NLRP6, NLRP7, NLRP12 and NLRC4 as well as two structurally different proteins, AIM2 and IFI16 (2). They have been shown to form inflammasomes upon activation by a variety of stimuli, through several different activation pathways, mainly in dendritic cells and macrophages (3). Among them, NLRP3 is best characterized and is connected to the pathogenesis of a variety of diseases (4) including inflammatory bowel disease and colon cancer (5), diabetes type II (6), gout (7), atherosclerosis (8) and Alzheimer disease (9). NLRP3 consists of an N-terminal pyrin domain, a central nucleotide binding domain (NBD), and a C-terminal leucin rich domain (LRR) (10). Gain-of-function mutations in the NLRP3 gene, mainly clustered in the NBD domain, result in its activation or predisposition for activation, and are associated with the cryopyrin-associated periodic fever syndromes (CAPS) including familial cold-induced autoinflammatory syndrome (FCAS), Muckle-Wells syndrome (MWS) and neonatal onset multisystem inflammatory disorder (NOMID) (11). In all three phenotypes the most common symptoms include periodic fever, arthralgia, rash and conjunctivitis (12). Both genetic and non-genetic diseases in which the inflammasome axis is dysregulated point to the importance of fine-tuning and modulation of its activity to maintain homeostasis. Since so many exogenous and endogenous factors are able to activate different inflammasomes, potent regulatory mechanisms must exist to allow the immune system to remove any sources of danger without causing excessive harm to the host. Recently, several factors and mechanisms have been identified to negatively regulate inflammasomes at different levels of their activation including autophagy (13), interferons type I (14), microRNAs (15), docosahexaenoic acid (16), nitric oxide (17) and cAMP (18). However the full spectrum, as well as downstream events involved in the regulation of inflammasome has not been elucidated.
Prostaglandin E2 (PGE2) belongs to a family of bioactive lipid mediators which have a broad range of effects (19). During the acute, initial stage of the inflammatory response PGE2 acts as a vasodilator and facilitates tissue influx of neutrophils (20), macrophages (21) and mast cells (22) as well as a regulator of nociception (23). However, PGE2 also has many potent immunosuppressive properties that contribute to the resolution phase of acute inflammation (24), facilitation of tissue regeneration (25) and the return to homeostasis (26). Yet in the context of many immunopathologies, those PGE2-mediated effects can lead to aggravation of the disease phenotype such as chronic inflammation or cancer (27). PGE2 regulates activities of both innate and adaptive immune cells. Its wide range of activities with often opposing effects depends on the species, cell and tissue types or context of action (28). PGE2 synthesis is initiated by phospholipases A2, catalyzing the hydrolysis of membrane phospholipids, liberating free fatty acids. Cytosolic phospholipase A2 group IVA (cPLA2α) is selective for arachidonate in the sn-2 position of membrane phospholipids, thus generating arachidonic acid (AA), the substrate of cyclooxygenases (COX1 and COX2), that convert AA to PGH2 (29). It is then converted to downstream active prostanoid by the terminal synthases. In many cells of innate immunity such as macrophages, cPLA2α is the rate-limiting enzyme in PGE2 production (30). The diverse effects of PGE2 may be also accounted for, at least in part, by the existence of four EP receptors, belonging to the family of G protein coupled receptors (GPCRs), differentially expressed in cells and by coupling to more than one G protein, initiating various signal-transduction pathways (31). While, EP1 mediates cytosolic Ca2+ mobilization (32), EP2 and EP4 couple primarily to Gsα, which activates adenylate cyclase (AC) to convert ATP to cyclic AMP (cAMP) (33, 34). Changes in cAMP levels are further translated into pleiotropic intracellular effects by a panel of cAMP binding effector proteins (35). The EP3 signaling pathway inhibits AC activity by coupling to Giα subunit and decreasing cAMP levels (36).
In macrophages, at the priming stage of NLRP3 inflammasome activation by TLR signaling, apart from induction of NLRP3 and pro-IL-1β expression, there is also an activation of cPLA2α, release of AA and production of PGE2 and other eicosanoids (37, 38). Moreover, aluminum salts and silica crystals (39), hyaluronan (40) as well as ATP (41) and other known activators of NLRP3 inflammasome further stimulate PGE2 production, although probably in an NLRP3 and caspase-1 independent manner (39). Furthermore, PGE2 and other prostanoids have been shown to be primarily responsible for several immediate reactions and sudden death in the “eicosanoid storm” after NLRC4 inflammasome activation by intracellular flagellin (42). In this case, this process appears to be NLRC4, casp-1 and COX1 dependent (42). However, the effects of exogenous or endogenous PGE2 on the NLRP3 inflammasome activation have not been studied. Due to the growing evidence of the co-existence of potent lipid mediators signaling and inflammasomes formation in response to infection, tissue damage or other cellular stress, we sought to analyze the effect of exogenous PGE2 on NLRP3 inflammasome activation. We also studied whether the endogenously produced lipid mediators might be involved in the modulation of NLRP3 inflammasome activation. We performed experiments in human primary monocyte-derived macrophages (MDM) and in peripheral blood mononuclear cells (PBMCs) from healthy donors as well as in PBMCs from CAPS patients. Here we found that PGE2 decreased NLRP3 inflammasome activation, triggered by aluminum crystals, ATP or nigericin. This effect was derived through the EP4 receptor and through an increase in intracellular cAMP, independently of PKA and Epac. In patients with CAPS, with mutations in the NBD domain of NLRP3, resulting in the decrease of cAMP binding (18), the inhibitory effect of PGE2 was observed only in high doses. Moreover, we found that blocking cPLA2α, or COX2 resulting in inhibition of endogenous PGE2 production, caused an increase in NLRP3 inflammasome activation. Our results suggest that PGE2 might play an important role in maintaining homeostasis during the resolution phase of inflammation by preventing excessive inflammasome activation in infiltrating macrophages as well as it might serve as a significant autocrine and paracrine regulator.
Materials and Methods
Reagents and antibodies
Aluminum potassium sulfate (alum crystals), monosodium urate crystals (MSU), and ultrapure LPS were purchased from Invivogen (San Diego, CA). ATP, nigericin sodium salt, 8-bromo-cAMP and IBMX were obtained from Sigma-Aldrich (St. Louis, MO); KH7 and forskolin from Tocris (Bristol, UK). Prostaglandin E2 (PGE2), PF-04418948 (EP2 inhibitor), GW 627368X (EP4 inhibitor), butaprost, free acid (EP2 agonist),CAY10598 (EP4 agonist) and NS-398 (COX2 inhibitor) were purchased from Cayman Chemical (Ann Arbor, MI). cPLA2α inhibitor (N-{(2S,4R)-4-(Biphenyl-2-ylmethyl-isobutyl-amino)-1-[2-(2,4-difluorobenzoyl)-benzoyl]-pyrrolidin-2-ylmethyl-3-[4-(2,4-dioxothiazolidin-5-ylidenemethyl)-phenyl] acrylamide, HCl) was obtained from Calbiochem, EMD Millipore (Gibbstown, NJ). 8-(4-Chlorophenylthio)-2'-O-methyladenosine 3',5'-cyclic Monophosphate . sodium salt (Epac agonist; activates Epac 1 and Epac 2) and 8-Bromoadenosine 3',5'-cyclic Monophosphothioate, Rp-Isomer . sodium salt (PKA inhibitor) were purchased from Enzo Life Science (Farmingdale, NY). N6-Benzoyl-8-piperidinoadenosine-3',5'-cyclic monophosphate . sodium salt (PKA agonist) and 4- Cyclopentyl- 2- (2, 5- dimethylbenzylsulfanyl)- 6- oxo- 1, 6- dihydropyrimidine- 5- carbonitrile (Epac inhibitor) were purchased from Axxora (Lausen, Switzerland). Antibody against IL-1β (AF-201-NA) was purchased from R&D Systems (Minneapolis, MN). Antibodies against caspase-1 (2225S) and total cPLA2α were purchased from Cell Signaling Technology (Danvers, MA); antibody against NLRP3 (AG-20B-0014-C100) from Adipogen (San Diego, CA); antibody against ASC (sc-22514-R) from Santa Cruz Biotechnology (Dallas, Texas). Monoclonal horseradish peroxidase (HRP)-conjugated beta actin antibody was obtained from GenScript (Piscataway, NJ) and Cell Signaling Technology. HRP-conjugated secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA).
Primary cells and cell lines
Human elutriated monocytes from healthy donors were obtained by an Institutional Review Board-approved protocol from the National Institutes of Health Blood Bank (Bethesda, MD). Monocytes were resuspended in RPMI 1640 medium with 2 mM of L-glutamine and supplemented with 10% heat-inactivated FBS (Life Technologies, Thermo Scientific, Waltham, MA) for overnight. The next day, 18×106 monocytes were seeded into a T75 tissue culture flask in 15 ml of Iscove's modified Dulbecco's medium (IMDM) (Life Technologies) with 10% heat-inactivated FBS and 50 ng/ml of M-CSF (Life Technologies) for 7 days. Half of the medium was replaced after 3 days of culture. 24 hours prior to experiments, macrophages were treated with trypsin (Lonza, Walkersville, MD) for 1 minute, scraped and plated in the IMDM medium without M-CSF into 12-well plates or 6-well plates at a density of 0.35×106 or 0.85×106, respectively.
Blood specimens from patients with CAPS were drawn after obtaining informed consent under a protocol approved by the NIAMS/NIDDK Institutional Review Board. Blood specimens from healthy volunteers were obtained under a protocol approved by the NHLBI Institutional Review Board. Blood samples were processed immediately using lymphocyte separation medium (LSM) (Lonza), according to the manufacturer's protocol to obtain PBMCs. 2×106 PBMCs were seeded, allowed to attach to a 12-well plate containing RPMI medium without serum for 20 min, immediately followed by the inflammasome activation/inhibition experiments. THP-1 cells were obtained from ATCC (Manassas, VA) and cultured in RPMI medium containing 10% heat-inactivated FBS and 0.05 mM 2-mercaptoethanol. 0.5×106 THP-1cells were plated into a 12-well plate in the presence of 50 nM PMA (Sigma-Aldrich) for 12 hours before the experiment was performed.
Inflammasome activation and inhibition
Human primary MDM were treated with/without LPS (100 ng/ml) for 4 h in IMDM with 10% FBS, followed by 30 min treatment with PGE2 (0.1 μM or as indicated), EP2 agonist (butaprost, free acid; 0.5 μM), EP4 agonist (CAY10598; 0.1 μM), 8-bromo-cAMP (5-20 μM), PKA selective agonist (6-Bnz-8-PIP-cAMP; 20 or 50 μM) or EPAC selective agonist (8-(4-Chlorophenylthio)-2'-O-methyladenosine 3',5'-cyclic Monophosphate . sodium salt, 20 or 50 μM), followed by stimulation with alum (400 μg/ml for 5 h), ATP (2 mM for 1 h) or nigericin (4 μM for 1 h) in Opti-MEM medium (Life Technologies). EP2 inhibitor (PF-04418948, 0.5 μM), EP4 inhibitor (GW627368X, 2 μM), PKA inhibitor (8-Bromoadenosine 3',5'-cyclic Monophosphothioate, Rp-Isomer . sodium salt, 50 μM), EPAC inhibitor (4- Cyclopentyl- 2- (2, 5- dimethylbenzylsulfanyl)- 6- oxo- 1, 6- dihydropyrimidine- 5- carbonitrile, 10 μM) or vehicle (DMSO) were added 30 min prior treatment with PGE2 or vehicle (EtOH). KH7 (25 μM) or IBMX (200 μM) or forskolin (50 μM) were added 1 h prior to treatment with PGE2 or vehicle (EtOH). In the experiments with cPLA2α inhibitor (2 μM) in human primary MDM, the inhibitor or DMSO were added before and after priming or only after priming, followed by treatment with alum (400 μg/ml for 5 h) in Opti-MEM. In the experiments with COX2 inhibitor (NS398, 5 μM) in human primary MDM, the inhibitor or DMSO were added before and after priming, followed by treatment with alum (400 μg/ml for 5 h) or ATP (2 mM for 1 h) in Opti-MEM. PBMCs from CAPS patients or healthy volunteers were primed for 3 h with LPS (1 μg/ml) in RPMI 1640 with 10% FBS and followed by indicated doses of PGE2 (in CAPS patients) or PGE2 with alum (400 μg/ml) or ATP (2 mM) (in healthy volunteers) for 30 min in RPMI 1640 without serum. PMA-treated THP-1 cells were treated with cPLA2α inhibitor for 30 min, then primed with 100 ng/ml of LPS and stimulated by MSU (100 μg/ml for 5 h), alum (400 μg/ml for 5 h), ATP (2 mM for 1 h) or nigericin (4 μM for 45 min) in Opti-MEM medium. Supernatants, lysates and cell pellets were collected for the following analyses.
NLRP3, EP2, EP4 and PLA2G4A knockdown
ON-TARGETplus SMART pool small interfering RNA (siRNA) (Dharmacon, Thermo Scientific, Lafayette, CO) against human NLRP3 (L-017367-00-0005), or sc-45469, Santa Cruz), EP2 (PTGER2) (L-005712-00-0005), EP4 (PTGER4) (L-005714-00-0005) and cPLA2α (PLA2G4A) (L-009886-00-005) together with ON-TARGETplus Control Non-targeting pool (D-00181970-05) were used to perform knockdown experiments in human primary MDM or THP-1 cells. Human primary MDM (0.5×106 cells per 100 μl cuvette) or THP-1 cells (5×106 per 100 μl cuvette) were transfected with siRNA pools (1 μM or 100 nM) using a P3 Primary Cell 4D-Nucleofector X Kit L (Amaxa, Cologne, Germany) according to the manufacturer's protocols for human macrophages and monocytes, respectively. The silencing of gene expression was confirmed by RT-PCR. All experiments on transfected cells were performed after 48 hours.
Real-Time PCR
Total RNA was extracted from cells using QIAshredder columns and RNeasy mini kit and treated with DNase (Qiagen, Valencia, CA). Reverse transcription was performed using an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Gene expression was assessed using RT-PCR performed on an ABI Prism ViiA7 sequence detection system (Applied Biosystems, Foster City, CA) using commercially available probe and primers sets (Applied Biosystems) as follows: GAPDH, Hs02758991_g1; IL-1β, Hs01555410_m1; IL-6, Hs00174131_m1; TNF, Hs99999043_m1; cPLA2α (PLA2G4A), Hs00233352_m1; NLRP3, Hs00918082_m1; caspase-1, Hs00354836_m1; EP1 (PTGER1), Hs00168752_m1, EP2, Hs04183523_m1, EP3, Hs00168755_m1, EP4, Hs00168761_m1 and iTaq Universal Probes Supermix (Bio-Rad). Gene expression was normalized to GAPDH transcripts and represented as a relative quantification (RQ) compared with control.
ELISA
IL-1β and IL-18 in cell culture supernatants were measured using Human IL-1 beta/IL-1F2 Quantikine Kit (SLB50) from R&D Systems or Human IL-18 ELISA kit from MBL (Nagoya, Japan) respectively, according to the manufacturer's directions. There were substantial differences in the IL-1β and IL-18 release between the donors, as depicted in the Supplementary Figure 1 A and B. Therefore in order to combine similar experiments and calculate statistical significance, the values in all experiments are presented as the percentage of the response after LPS/alum treatment. The range of the raw values from combined experiments is included in each figure legend. PGE2 in cell culture supernatants was measured using EIA kit from Cayman Chemical Co. TNF in cell culture supernatants was measured using Human TNF-alpha ELISA Kit (DTA00C) from R&D Systems. Intracellular cAMP quantification was performed using EIA kit from Enzo (Farmingdale, NY). Human primary MDM seeded on a 6-well plate were stimulated with PGE2 (0.1 μM), or forskolin (50 μM) in the presence of IBMX (200 μM) for 15 min, followed by 10 min cell lysis in 0.1 N HCl containing 0.1 % triton X-100 (Sigma-Aldrich). For the cAMP quantification in the experiments with NS398, primary MDM were treated with LPS (100 ng/ml) with/without NS398 (5 μM), NS398 alone or DMSO for 4 h in complete medium and then stimulated with Alum for 15 min in serum-free medium followed by 10 min cell lysis in 0.1 N HCl containing 0.1 % triton X-100 and IBMX (200 μM). Cell lysates were collected, centrifuged and stored at −80 °C before cAMP detection according to the manufacturer's protocol.
Western Blot
Supernatants were collected and cells were lysed with RIPA buffer (Thermo Scientific, Rockford, IL) containing protease inhibitor cocktails (Roche, Basel, Switzerland). Supernatants were precipitated with equal volume of methanol and ¼ volumes of chloroform (Sigma-Aldrich) and centrifuged at room temperature at 12,000 rpm for 5 min. The upper layer was aspirated, double volume of methanol added and centrifuged again. The liquid phase was discarded and the pellet was dried at 50°C for 5 min. The pellets were dissolved in 1X NuPage Sample Buffer (Life Technologies) and heated at 70°C for 10 min and then subjected to electrophoresis using the 4-12% NuPAGE Bis-Tris gels using MES buffer (Life Technologies). The cell lysates were placed on ice for 15 min and then centrifuged at 13,000 rpm for 20 min. Protein concentration of cell lysates was determined by BCA kit (Pierce, Rockford, IL). Equal amounts of lysate proteins (20 μg) were resolved by 4–12% NuPAGE Bis-Tris gels using MOPS running buffer (Life Technologies) and then transblotted to the nitrocellulose membranes using the IBlot Dry Blotting System (Life Technologies). For ASC pyroptosome, cell pellets were cross-linked using 2 mM disuccinimidyl suberate (DSS) (Thermo Scientific) for 30 min in PBS in RT; 6x SDS sample buffer was added and samples were processed as cell lysates. The membranes were blocked with 5% nonfat skim milk in PBST (phosphate buffer saline containing 0.1% Tween 20) for 2 h and then incubated with primary antibodies overnight at 4°C. Subsequently, the membranes were washed and then incubated with specific secondary antibodies conjugated with horseradish peroxidase (HRP) for 1 h. The blots were developed using the SuperSignal West Femto Chemiluminescent Substrate kit (Thermo Scientific) and visualized on a Bio-Rad Image Station (Chemi-Doc-MP) (Hercules, CA).
Statistical analysis
Data were analyzed by one-way ANOVA with Holm-Sidak post hoc test, Kruskall-Wallis ANOVA on ranks with Dunn's post hoc test, RM (repeated measurements)-ANOVA, followed by Sidak post hoc test or unpaired Student t test, as appropriate. Differences were considered significant when p < 0.05. The data are presented as the mean ± SEM from the indicated number of independent experiments, each performed in duplicate.
Results
PGE2 inhibits NLRP3-triggered inflammasome activity in human macrophages
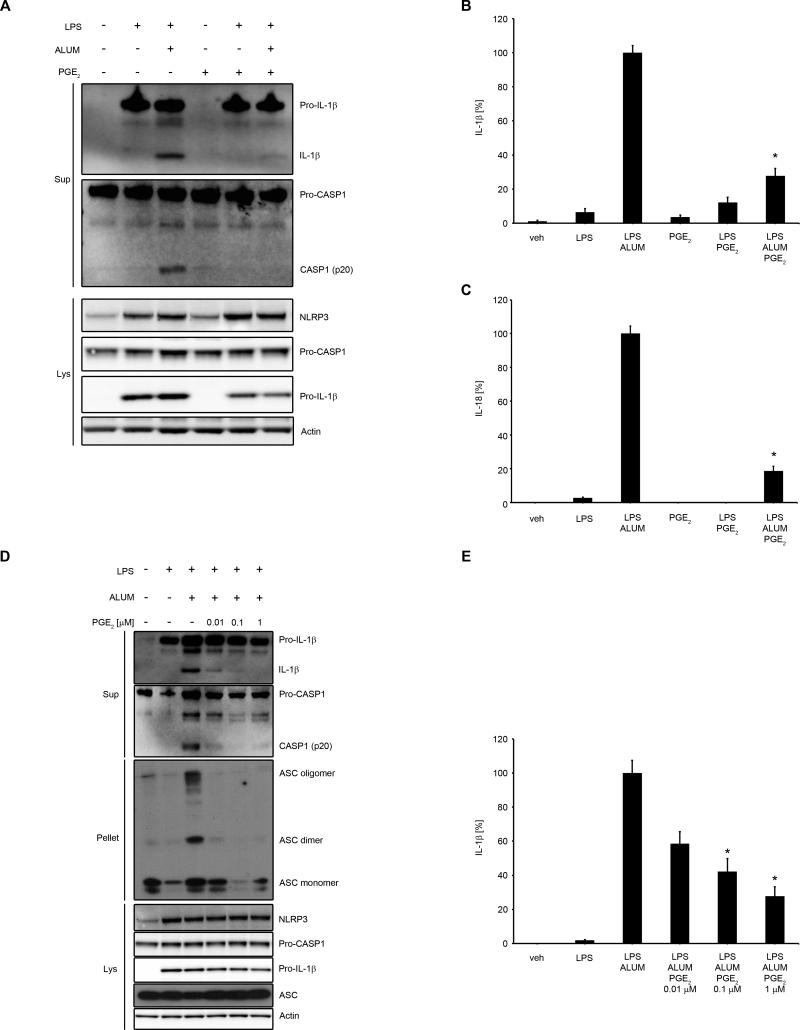
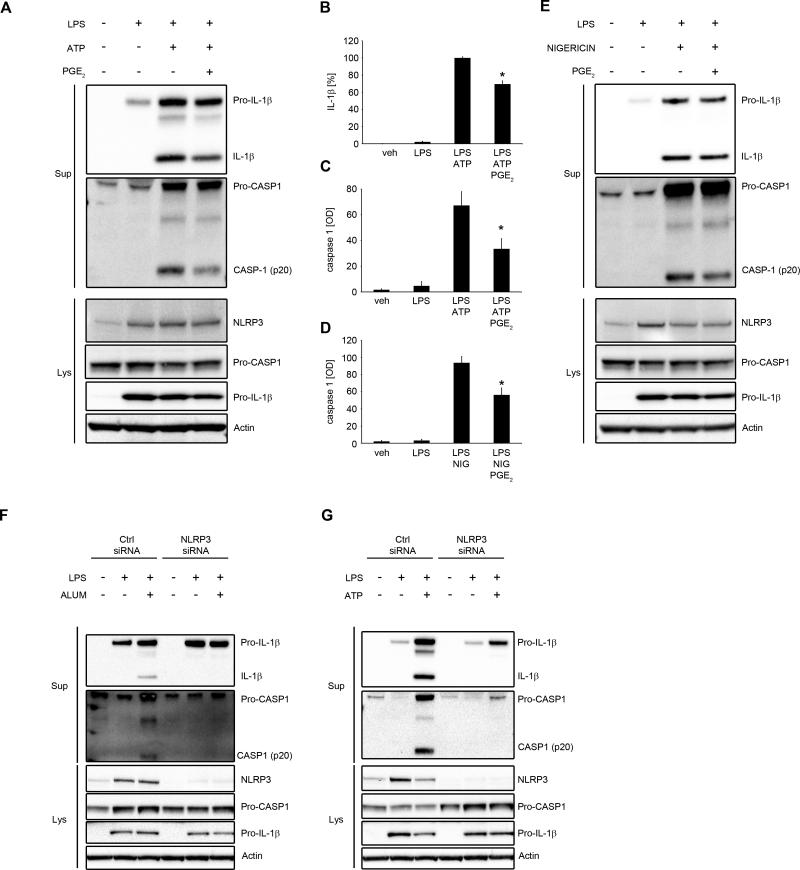
To assess the effect of PGE2 on NLRP3 inflammasome activation, we first examined whether PGE2 could inhibit caspase-1 cleavage and IL-1β secretion. Human primary MDM were primed with LPS followed by treatment with PGE2 and subsequent stimulation with alum, to activate IL-1β maturation via NLRP3 inflammasome. We found that PGE2 blocked alum-induced activation of caspase-1 and production of mature IL-1β and IL-18 (Fig. 1A-C; Supplementary Fig. 1). We also observed that PGE2, in these experimental conditions, slightly decreased the expression of pro-IL-1β, without affecting NLRP3 or caspase-1 (Fig. 1A). The inhibitory effect of PGE2 on caspase-1 cleavage, IL-1β maturation, ASC oligomerization (Fig. 1D) and IL-1β secretion (Fig. 1E) was dose-dependent and began with very low doses of PGE2 (0.01 μM-0.1 μM) in human primary MDM. The effectiveness of these doses points to a physiologic and receptor-specific mode of PGE2-mediated inhibition of NLRP3 activation (31). We also tested other activators of NLRP3 inflammasome, such as ATP and nigericin (43) to determine whether the suppression exerted by PGE2 specifically affected alum-dependent NLRP3 inflammasome activity, a phagocytosis-dependent model of NLRP3 activation (44). We found that caspase-1 cleavage and IL-1β secretion was also decreased by PGE2 in an ATP- (Fig. 2A-C) or nigericin- (Fig. 2D, E) induced NLRP3 inflammasome activation model. To control for NLRP3 specificity of studied stimuli in human macrophages we performed NLRP3 siRNA experiments, with two different sources of siRNAs, confirming that both alum and ATP specifically induced NLRP3-dependent caspase-1 and IL-1β cleavage (Fig. 2F, G). Thus, PGE2 specifically inhibited the activity of NLRP3 inflammasome.
Figure 1. PGE2 inhibits alum-induced NLRP3 inflammasome activation and IL-1β and IL-18 secretion.
(A-E) human primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by 30 min treatment with PGE2 (0.1 μM) or vehicle (EtOH) (A, B and C) or indicated doses of PGE2 (D and E) then stimulated with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants, lysates and cell pellets were collected for WB (A and D) or ELISA (B, C and E). (A and D) the WB are representative of 3 independent experiments from 3 healthy donors, each showing similar results. (B) IL-1β release data represent the mean ± SEM from 7 independent experiments from 7 healthy donors, performed in duplicate. IL-1β data are presented as the percentage of the response after LPS/alum treatment, ranging from 61.6 to 831.6 pg/ml. (C) IL-18 release data represent the mean ± SEM from 5 independent experiments from 5 healthy donors, performed in duplicate. IL-18 data are presented as the percentage of the response after LPS/alum treatment, ranging from 18.2 to 821.3 pg/ml. (E) IL-1β release data represent the mean ± SEM from 6 independent experiments from 6 healthy donors, performed in duplicate. IL-1β data are presented as the percentage of the response after LPS/alum treatment, which ranged from 11.4 to 299 pg/ml.* P < .05 as compared to LPS/alum treatment, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test.
Figure 2. PGE2-induced inhibition of IL-1β production is driven through NLRP3 inflammasome.
(A-E) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by 30 min treatment with PGE2 (0.1 μM) or vehicle (EtOH), then stimulated with ATP (2 mM for 1 h) (A, B and C) or nigericin (4 μM for 1 h) (D and E) in serum-free medium. Supernatants and lysates were collected for WB or ELISA. (A and E) the WB are representative of 3 independent experiments from 3 healthy donors, each showing similar results. (B) IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, performed in duplicate. Data are presented as the percentage of the response after LPS/ATP treatment, ranging from 226.6 to 5851.4 pg/ml. (C and D) mature caspase-1 is presented in optical density units [OD] after subtraction of background from 3 independent experiments from 3 healthy donors. (F and G) MDM were transfected with NLRP3 siRNA (1 μM; L-017367-00-0005). After 48 h cells were treated with/without LPS (100 ng/ml) for 4 h in complete culture medium, followed by stimulation with alum (400 μg/ml for 5 h) (F) or ATP (2 mM for 1 h) (G) in serum-free medium. The WB are representative of 3 independent experiments from 3 healthy donors, each showing similar results.
PGE2 differentially influences the gene expression of NLRP3 inflammasome complex components
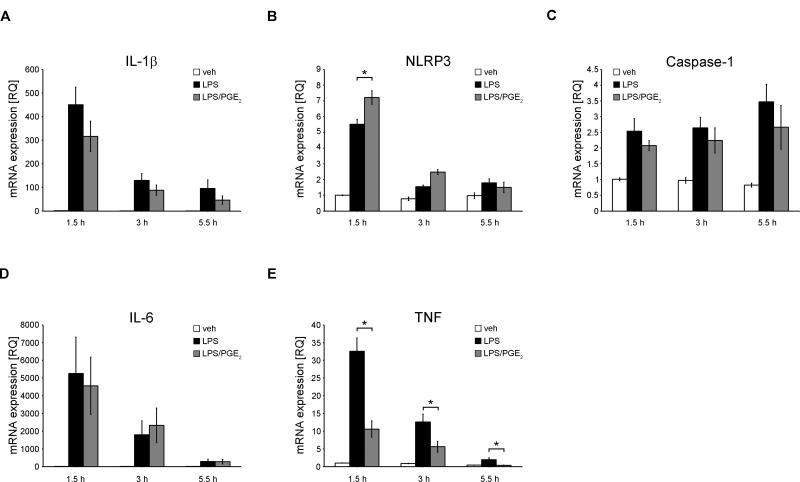
To evaluate the role of PGE2 on the expression of NLRP3 inflammasome components, we performed a time course analysis of PGE2 on LPS-induced gene expression. Human primary MDM were treated with/without LPS, followed by treatment with PGE2 for 1.5 h, 3 h and 5.5 h. We found that LPS-induced IL-1β mRNA expression was not significantly altered by PGE2 in either of the time points, though there was a downward trend (Fig. 3A). NLRP3 mRNA expression was significantly increased by PGE2 at 1.5 h time point (Fig 3B), while caspase-1 mRNA expression was unchanged (Fig. 3C). TNF and IL-6 usually serve as controls for specific inflammasome and non-priming effecting mechanisms. However, both of these cytokines have been reported to be regulated by PGE2 and cAMP in opposite directions in macrophages and through mechanisms not affecting intracellular canonical TLR signaling (45, 46). Interestingly, we found that LPS-induced IL-6 mRNA expression was not changed by the addition of PGE2 after LPS stimulation (Fig. 3D), as opposed to TNFα, in which mRNA expression was significantly decreased at each time-point studied (Fig. 3E). Taken together, the lack of the effect of PGE2 on the transcription of genes encoding proteins involved in the NLRP3 inflammasome complex formation, as well as its inhibitory effect on mature caspase-1, ASC oligomerization and constitutively expressed IL-18, as shown in the previous paragraph, suggests that PGE2 acts on NLRP3 inflammasome assembly.
Figure 3. PGE2 does not decrease gene expression of proteins involved in NLRP3 inflammasome complex formation.
(A-E) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by treatment with PGE2 (0.1 μM) or vehicle (EtOH) for indicated time in serum-free medium. At each time point, cells were lysed and total RNA was extracted. Gene expression was assessed using RT PCR, normalized to GAPDH transcripts and represented as a relative quantification (RQ) compared with vehicle treated cells from the first time-point. Data represent the mean ± SEM from 4 independent experiments from 4 healthy donors, each performed in duplicate. * P < .05 as indicated, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test.
PGE2 inhibits NLRP3 inflammasome through EP4 receptor
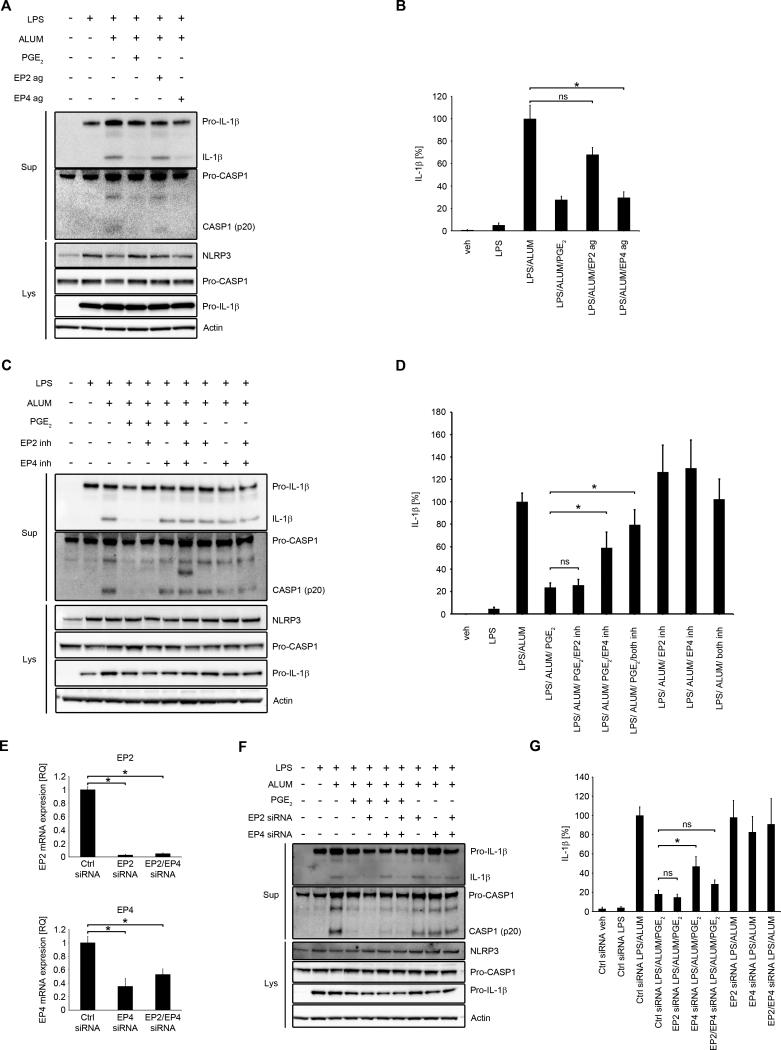
PGE2 is known to exert various effects through at least four different receptors EP1, EP2, EP3 and EP4 (PTGER1-4), all belonging to the GPCR family (31). Therefore, we first analyzed which receptors are expressed on human primary MDM. In agreement with the literature on mouse macrophages, cell lines and human macrophages (34) we found that EP1 basal mRNA expression was at the edge of the detection level; EP3 receptor expression was very low, while EP2 and EP4 were abundantly expressed (Supplementary Fig. 2A). Furthermore, both EP2 and EP4 mRNA expression was significantly increased by LPS priming (Supplementary Fig. 2B, 2C). To determine which receptor is responsible for the PGE2 effect on NLRP3 inflammasome, we used specific pharmacological agonists and antagonists of each receptor as well as a pool of siRNAs to knockdown each receptor. The EP4 agonist, CAY10598, but not the EP2 agonist, butaprost, blocked alum-induced secretion of mature IL-1β and caspase-1 (Fig. 4A, 4B). Moreover, we found that the EP4 antagonist, GW627368X, blocked PGE2-induced inhibition of NLRP3 inflammasome activation, while the EP2 antagonist, PF-04418948, had no effect (Fig. 4 C, 4D). Both inhibitors applied simultaneously had similar effect to EP4 antagonist alone (Fig. 4C, 4D). The function of EP2 and EP4 agonists and antagonists was controlled by measuring TNF (Supplementary Fig. 3 A, B), which inhibition was reported to be driven through both of these receptors in human macrophages (47). Knockdown of EP2 or EP4 receptors was performed by electroporation with the specific pools of siRNAs. The knockdown efficiency for EP2 and EP4 was 97% and 65%, respectively. When both siRNA were applied together, then EP2 was reduced by 93% and EP4 by 50% as assessed by RT-PCR (Fig. 4 E). Similar to the antagonists results, EP4 knockdown, as opposed to EP2 knockdown reduced PGE2-induced inhibition of NLRP3 inflammasome activation (Fig. 4F, 4G). Simultaneous knockdown did not have an effect on PGE2-induced inhibition of NLRP3 inflammasome activation, probably because of the low efficiency of the knockdown of EP4 (Fig. 4 E, 4F, 4G). Altogether, these data suggest that PGE2 decreases NLRP3-inflammasome activity through EP4 receptor.
Figure 4. PGE2 inhibits NLRP3 inflammasome through EP4 receptor.
(A and B) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by 30 min treatment with EP2 agonist (butaprost, free acid, 0.5 μM), EP4 agonist (CAY10598, 0.1 μM), PGE2 (0.1 μM) or vehicle (EtOH), then stimulated with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for WB (A) or IL-1β ELISA (B). IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 111.5 to 139.9 pg/ml.* P < .05 as indicated, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test (B). (C and D) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete culture medium, then 30 min with EP2 inhibitor (PF-04418948, 0.5 μM), EP4 inhibitor (GW627368X, 2 μM), both inhibitors or vehicle (DMSO), followed by 30 min treatment with PGE2 (0.1 μM) or vehicle (EtOH), and stimulation with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for WB (C) or IL-1β ELISA (D). IL-1β release data represent the mean ± SEM from 5 independent experiments from 5 healthy donors, each performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 36.6 to 372.6 pg/ml (D). (E, F and G), MDM were transfected with EP2 siRNA (1 μM), EP4 siRNA (1 μM) or both. After 48 h cells were treated with/without LPS (100 ng/ml) for 4 h in complete culture medium, followed by 30 min treatment with PGE2 (0.1 μM) or vehicle (EtOH), and then stimulated with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for RT-PCR (E), WB (F) or IL-1β ELISA (G). Gene expression was normalized to GAPDH transcripts and represented as a relative quantification (RQ) compared with vehicle-treated control siRNA transfected cells. Data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. * P < .05 as indicated, as assessed Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test (E). IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 34.6 to 491.5 pg/ml (G). * P < .05 as indicated, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test (D and G). The WB are representative of 3 independent experiments from 3 healthy donors, each showing similar results (A, C and F).
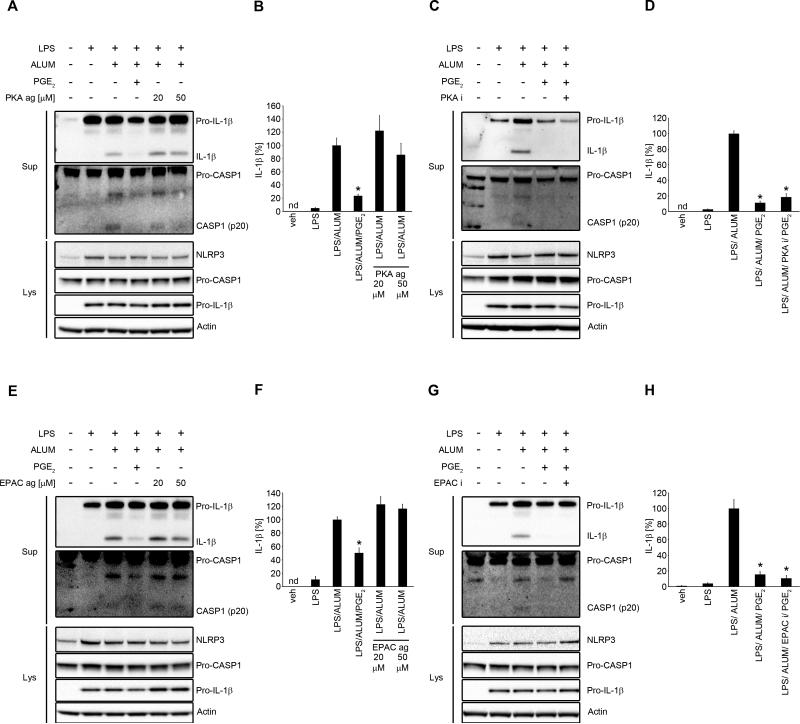
cAMP, independent of PKA and Epac, is involved in PGE2-mediated NLRP3 inflammasome inhibition
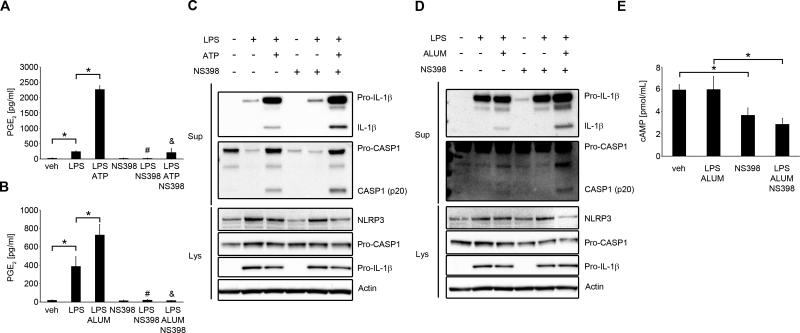
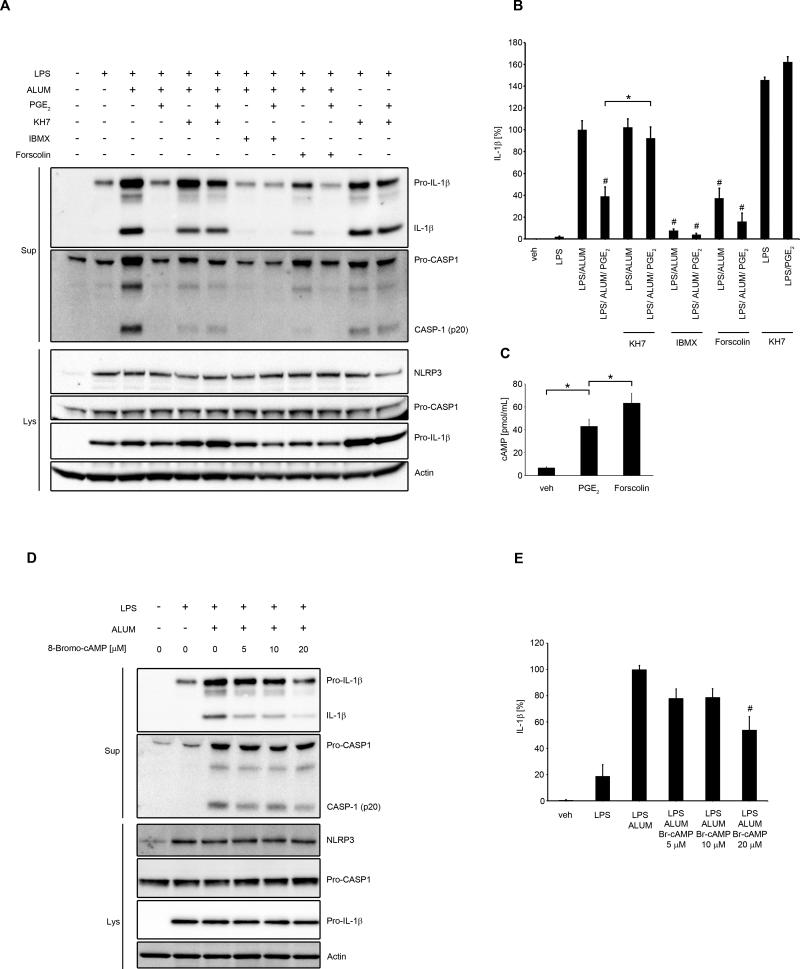
The most widely described signaling pathway through EP4 in macrophages is coupling with Gsα subunit and activation of adenylate cyclase (34). Adenylate cyclase converts ATP to cAMP, which is an important second messenger. Intracellular cAMP levels are further controlled by the actions of phosphodiesterase, which converts cAMP to inactive 5’-AMP, by degrading the phosphodiester bond (48). Signaling through EP4 increases intracellular cAMP levels (34). It was previously demonstrated that cAMP inhibits NLRP3 inflammasome through direct interaction with NLRP3 protein in mouse BMDM and in human PBMCs (18). To determine, if this is the major mechanism of the observed effect of PGE2-induced NLRP3 inhibition in human primary MDM, we applied several approaches that involved the key molecules in the regulation of intracellular cAMP levels. We used KH7, an adenylate cyclase inhibitor which decreases basal or activated cAMP levels (49); forskolin, a direct adenylate cyclase activator, that increases intracellular cAMP levels (50) and IBMX, a phosphodiesterase inhibitor, that increases cAMP levels by blocking its degradation (51). First, we confirmed that in human primary MDM, 15 min treatment with PGE2 stimulated cAMP production (Fig. 5C). Next, we found that blocking adenylate cyclase with KH7 in the presence or absence of alum significantly reduced PGE2-mediated NLRP3-inflammasome inhibition (Fig. 5A, 5B) and IL-1β secretion (Fig. 5B). Forskolin treatment significantly decreased NLRP3 activation (Fig. 5A) and IL-1β secretion (Fig. 5B), which was further decreased by adding PGE2, suggesting additive PGE2-mediated mechanisms of adenylate cyclase activation. Not surprisingly, prolonged IBMX treatment completely abrogated NLRP3 activation (Fig. 5A) and IL-1β secretion (Fig. 5B). To further confirm cAMP-mediated NLRP3 inflammasome inhibition, we used 8-Bromo-cAMP, a cell permeable analog of cAMP. We found that 8-Bromo-cAMP in a dose-dependent manner suppressed activation of caspase-1 and production of mature IL-1β (Fig. 5D) and in the dose of 20 μM significantly decreased IL-1β secretion (Fig. 5E). In macrophages, cAMP might act through ubiquitously expressed intracellular cAMP receptors, the classic protein kinase A (PKA) and exchange protein directly activated by cAMP (Epac) or might directly bind to the NLRP3 protein (18). To study whether PGE2-dependent cAMP-mediated effect on NLRP3 is derived through PKA or Epac, we used their specific agonists and antagonists. We found that PKA selective agonist (6-Bnz-8-PIP-cAMP) did not mimic PGE2-dependent NLRP3 inflammasome inhibition (Fig. 6A, B). Also, we observed that PKA specific inhibitor (8-Bromoadenosine 3',5'-cyclic Monophosphothioate) did not block PGE2-mediated NLRP3 inflammasome inhibition (Fig. 6C, D). Similarly, Epac selective agonist (8-(4-Chlorophenylthio)-2'-O-methyladenosine 3',5'-cyclic Monophosphate) did not inhibit caspase-1 cleavage and production of mature IL-1β (Fig. 6E, F) and Epac selective inhibitor (4- Cyclopentyl- 2- (2, 5- dimethylbenzylsulfanyl)-6- oxo- 1, 6- dihydropyrimidine- 5- carbonitrile) did not reverse PGE2-mediated NLRP3 inflammasome inhibition (Fig. 6G, H). At the same time, these agonists and antagonists had an effect on TNF production (Supplementary Fig. 3C-F), as previously reported (52, 53), proving that they were effective and more importantly suggesting separate pathways of cAMP-dependent NLRP3 inflammasome and TNF inhibition. Altogether, these data suggest that PGE2-mediated NLRP3 inflammasome inhibition is exerted through cAMP, but not through PKA or Epac.
Figure 5. cAMP is involved in PGE2-mediated NLRP3 inhibition.
(A, B) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by treatment for 1 h with KH7 (25 μM) or IBMX (200 μM) or forskolin (50 μM), and followed by PGE2 (0.1 μM) or vehicle (EtOH) for 30 min and stimulated with alum for 5 h (400 μg/ml) in serum-free medium. Supernatants and lysates were collected for WB (A) or IL-1β ELISA (B). IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 76.7 to 562.8 pg/ml. # P < .05 as compared to LPS/alum treatment, * P < .05 as indicated, as assessed by one-way ANOVA, followed by Holm-Sidak post hoc test (B). (C) primary MDM were treated with IBMX (200 μM) with either vehicle (EtOH), PGE2 (0.1 μM) or forskolin (50 μM) for 15 min. cAMP measured in cell lysates represents the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in triplicate. * P < .05 as indicated, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test. (D and E) MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by 30 min treatment with indicated doses of 8-Bromo-cAMP or vehicle (Tris) and then stimulated with alum for 5 h (400 μg/ml) in serum-free medium. Supernatants and lysates were collected for WB (D) or IL-1β ELISA (E). IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 70.2 to 1100.9 pg/ml.* P < .05 as compared to LPS/alum treatment, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test (E). The WB are representative of 3 independent experiments from 3 healthy donors, each showing similar results (A and D).
Figure 6. PGE2-derived NLRP3 inflammasome inhibition does not require PKA or EPAC.
(A and B) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by 30 min treatment with PKA selective agonist (6-Bnz-8-PIP-cAMP, 20 or 50 μM), PGE2 (0.1 μM) or vehicle, then stimulated with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for WB (A) or IL-1β ELISA (B). IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 44.7 to 103 pg/ml.* P < .05 as compared to LPS/alum, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test (B). (C and D) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete culture medium, then 30 min with PKA inhibitor (8-Bromoadenosine 3',5'-cyclic Monophosphothioate, Rp-Isomer . sodium salt, 50 μM) or vehicle (DMSO), followed by 30 min treatment with PGE2 (0.1 μM) or vehicle (EtOH), and stimulation with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for WB (C) or IL-1β ELISA (D). IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 43.9 to 167.7 pg/ml. * P < .05 as compared to LPS/alum, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test (D). (E and F) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by 30 min treatment with EPAC selective agonist (8-(4-Chlorophenylthio)-2'-O-methyladenosine 3',5'-cyclic Monophosphate . sodium salt, 20 or 50 μM), PGE2 (0.1 μM) or vehicle, then stimulated with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for WB (E) or IL-1β ELISA (F). IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 82.9 to 245.6 pg/ml.* P < .05 as compared to LPS/alum, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test (F). (G and H) primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete culture medium, then 30 min with EPAC inhibitor (4- Cyclopentyl- 2- (2, 5- dimethylbenzylsulfanyl)- 6- oxo- 1, 6- dihydropyrimidine- 5- carbonitrile, 10 μM) or vehicle (DMSO), followed by 30 min treatment with PGE2 (0.1 μM) or vehicle (EtOH), and stimulation with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for WB (G) or IL-1β ELISA (H). IL-1β release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. Data are presented as the percentage of the response after LPS/alum treatment, which ranged from 206.2 to 226.4 pg/ml * P < .05 as compared to LPS/Alum, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test (H).
PGE2-mediated NLRP3 inflammasome inhibition in CAPS patients requires higher doses
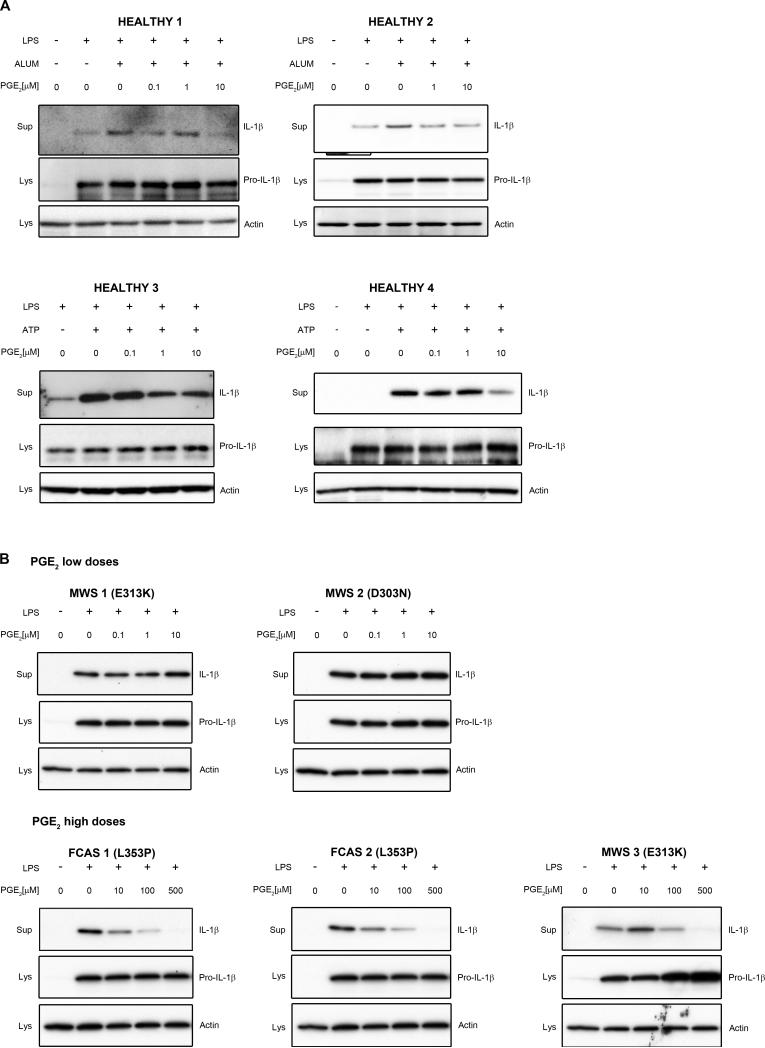
As mentioned above, it has been previously reported that cAMP binds directly to the nucleotide-binding domain (NBD or NACHT) of NLRP3 protein and this binding is impaired in cases of the most common mutations of NLRP3 gene, present in patients with CAPS (18). Therefore, to further analyze whether PGE2-mediated NLRP3 inhibition is cAMP-NBD specific, we studied the effect of PGE2 on IL-1β secretion in PBMCs from mutation-positive CAPS patients. PBMCs of these patients are especially prone to NLRP3 inflammasome activation and abundant secretion of mature IL-1β in response to LPS priming only, without other NLRP3 stimuli (54). Monocytes from healthy subjects are also reported to respond to LPS signal only, probably by intrinsic release of ATP, but the second signal makes their response much more robust (55). We found here that in monocytes from healthy subjects PGE2 decreased alum or ATP-dependent mature IL-1β secretion in similar doses as in monocyte-derived macrophages (0.1-10 μM) (Fig 7A). However, in patients with familial cold autoinflammatory syndrome (FCAS) or Muckle-Wells syndrome (MWS) with known mutations in NLRP3 (Fig. 7B), these doses were less effective and PGE2 inhibitory effect was visible in doses 10-100 times higher than in healthy subjects (Fig. 7B). These results suggest that PGE2-mediated NLRP3 inhibition is diminished in patients carrying an NLRP3 mutation in NBD domain.
Figure 7. PGE2-induced NLRP3 inflammasome inhibition requires higher doses in CAPS patients.
(A) PBMCs from four healthy subjects were treated with LPS (1 μg/ml) for 3 h in complete medium, followed by treatment with the indicated doses of PGE2 or vehicle (EtOH) and ATP (2 mM) or alum (400 μg/ml) for 30 min in serum-free medium. Supernatants and lysates were collected for WB. (B) PBMCs from five CAPS patients with designated mutations in NLRP3 were treated with LPS (1 μg/ml) for 3 h in complete medium, followed by treatment with indicated doses of PGE2 or vehicle (EtOH) for 30 min in serum-free medium. Supernatants and lysates were collected for WB.
cPLA2α and COX2 are involved in endogenous NLRP3 inflammasome regulation
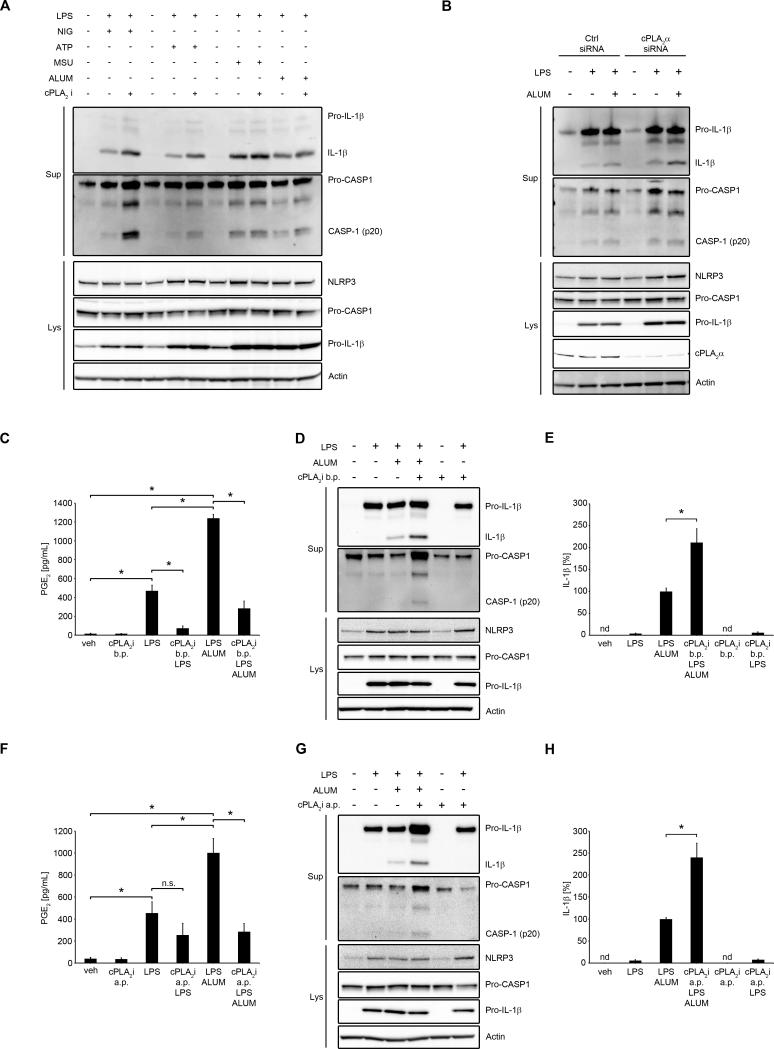
Having found that exogenous PGE2 inhibits NLRP3 inflammasome activation in human primary MDM, we wondered whether endogenous PGE2 production might serve as a regulatory autocrine or paracrine mechanism against excessive NLRP3 activation. Along with others, we have previously shown that LPS induces cPLA2α activation, AA release and downstream PGE2 production (37, 38). Moreover, ATP or crystals can lead to cPLA2α-dependent PGE2 production (39, 41). Therefore, we studied whether blockade of AA release and PGE2 production would influence NLRP3 activation. PMA-induced THP-1 macrophages, human primary MDM and cPLA2α siRNA-transfected THP-1 cells were used. We found that inhibition of cPLA2α enhanced NLRP3 inflammasome activation in THP-1 cells (Fig. 8A). Similarly, knockdown of cPLA2α in THP-1 cells resulted in an increase of NLRP3 inflammasome activation (Fig. 8B). We also studied primary human MDMs, which were treated with cPLA2 inhibitor before the priming step (b.p.), or after priming (a.p.) to block cPLA2α-dependent signals during inflammasome activation. We first confirmed that PGE2 production was induced by LPS treatment and further increased by alum (Fig. 8C, 8F). PGE2 production induced by LPS alone was blocked in the presence of cPLA2 inhibitor added before priming (Fig. 8C) and was significantly decreased when added before alum treatment (Fig. 8F). Under the same experimental conditions we found that cPLA2α inhibitor added before or after priming increased NLRP3 inflammasome activation in human primary MDM (Fig. 8D, 8E, 8G, 8H). To more selectively block only PGE2 production, we also inhibited COX2, by using its selective inhibitor NS398. First, we confirmed that NS398 blocked LPS/ATP- (Fig. 9A) as well as LPS/alum-induced PGE2 (Fig. 9B) production. Similar to cPLA2 inhibition, inhibition of COX2 led to an increase of NLRP3 inflammasome activation (Fig. 9C, D). Additionally, we also noted that treatment with NS398 decreased intracellular cAMP (Fig. 9E). Altogether, these data suggest that cPLA2α and COX2, through PGE2 production, are involved in the regulation of NLRP3 inflammasome activation.
Figure 8. cPLA2α is involved in endogenous NLRP3 inflammasome regulation.
(A) THP-1 cells were treated with cPLA2 inhibitor (pyrrolidone derivative, 2 μM, for 30 min) or vehicle (DMSO) then primed with LPS (100 ng/ml) for 4 h in serum-free medium, followed by treatment with nigericin (4 μM for 45 min), ATP (1 mM, for 1 h), monosodium urate crystals (MSU, 100 μg/ml, for 5h) or alum (200 μg/ml, for 5h). Supernatants and lysates were collected for WB. The WB are representative of 2 independent experiments, each showing similar results. (B) THP-1 cells were transfected with cPLA2α siRNA or control siRNA (1 μM). After 48 h transfected cells were primed with LPS (100 ng/ml) for 4 h in serum-free medium, followed by stimulation with alum (200 μg/ml, for 5h). Supernatants and lysates were collected for WB. The WB are representative of 2 independent experiments, each showing similar results. (C, D and E) human primary MDM were treated with/without LPS (100 ng/ml) with cPLA2 inhibitor (2 μM) or DMSO for 4 h in complete medium, followed by subsequent treatment with cPLA2 inhibitor or DMSO for 30 min, and then stimulated with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for PGE2 ELISA (C), WB (D) or IL-1β ELISA (E). PGE2 (C) and IL-1β (E) release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. IL-1β data are presented as the percentage of the response after LPS/alum treatment, which ranged from 409.4 to 647.6 pg/ml (E). (F, G and H) human primary MDM were treated with/without LPS (100 ng/ml) for 4 h in complete medium, followed by treatment with cPLA2 inhibitor or DMSO for 30 min, and then stimulated with alum (400 μg/ml for 5 h) in serum-free medium. Supernatants and lysates were collected for PGE2 ELISA (F), WB (G) or IL-1β ELISA (H). PGE2 (F) and IL-1β (H) release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. IL-1β data are presented as the percentage of the response after LPS/alum treatment, which ranged from 261.2 to 516.7 pg/ml (E). The WB are representative of 3 independent experiments from 3 healthy donors, each showing similar results (D, G). * P < .05 as indicated, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test. b.p. (before priming), a.p. (after priming)
Figure 9. COX2 is involved in endogenous NLRP3 inflammasome regulation.
(A-D) primary MDM were treated with/without LPS (100 ng/ml) with NS398, a specific COX2 inhibitor (5 μM) or DMSO for 4 h in complete medium, followed by subsequent treatment with NS398 (5 μM) or DMSO for 30 min, and then stimulated with ATP (2 mM for 1 h) (A and C) or alum (400 μg/ml for 5 h) (B and D) in serum-free medium. Supernatants and lysates were collected for PGE2 ELISA (A and B) or WB (C and D). The WB are representative of 3 independent experiments from 3 healthy donors, each showing similar results. PGE2 release data represent the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in duplicate. * P < .05 as indicated, # P < .05 as compared to LPS, & P < .05 as compared to LPS/ATP or LPS/alum, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test. (E) Primary MDM were treated with LPS (100 ng/ml) with/without NS398 (5 μM), NS398 alone or DMSO for 4 h in complete medium and then stimulated with alum for 15 min in serum-free medium. cAMP measured in cell lysates represents the mean ± SEM from 3 independent experiments from 3 healthy donors, each performed in triplicate. * P < .05 as indicated, as assessed by Kruskal-Wallis ANOVA on ranks, followed by Dunn's post hoc test.
Discussion
Monocytes and macrophages are among the first cells involved in the acute phase of inflammation. They infiltrate the tissue in response to chemotactic agents and respond to the invading pathogen or tissue trauma by several mechanisms including inflammasome activation. The inflammatory response should subside once it has carried out its function to maintain homeostasis. Activated NLRP3 inflammasome needs to be counterbalanced by the existence of local mediators or mechanisms that quickly quiet the response and lead to the resolution of acute inflammation. Here we have found that PGE2 inhibited NLRP3 inflammasome activation in human primary MDM. We have demonstrated that this effect depends on the EP4 receptor and an increase in intracellular cAMP, but not on PKA or Epac-mediated pathways. We also found that PGE2 decreased IL-1β secretion in patients with CAPS, but in higher doses than in healthy subjects. Moreover, we have demonstrated that this mechanism is involved in an autocrine or paracrine loop, possibly controlling the extent of NLRP3 inflammasome activation.
Our results are consistent with the recent findings that cAMP is directly involved in the endogenous control of NLRP3 inflammasome activation (18). Calcium sensing receptor (CASR), belonging to the GPCRs family in response to the extracellular Ca2+ increase activates phospholipase C (PLC) and inhibits adenylate cyclase (AC). As a result there is an increase in intracellular Ca2+ and decrease in cAMP, both of which can independently activate NLRP3 inflammasome (18). Here we also provided three lines of evidence showing that PGE2-mediated inhibition of NLRP3 inflammasome occurred by an increase in intracellular cAMP. First, KH7, an AC inhibitor, reversed PGE2-mediated NLRP3 inflammasome inhibition. Second, forskolin itself as an AC activator decreased NLRP3 activation and this effect was enhanced by cotreatment with PGE2. Moreover, IMBX, a phosphodiesterase inhibitor totally blocked alum-induced NLRP3 activation. Finally, 8-Bromo-cAMP, a cell permeable analog of cAMP in a dose-dependent manner mimicked PGE2-mediated NLRP3 inflammasome inhibition. Another recent study in which the authors used relatively low concentrations of forskolin (up to 20 μM) reports extracellular Ca2+ sensing by both CASR and GPRC6A in NLRP3 activation; however, cAMP involvement was not demonstrated (56). We have shown that even when forskolin was used in a concentration of 50 μM, there was still an additive effect of PGE2, suggesting that AC might require stronger activation to elicit its full cAMP-producing potential. Interestingly, it has been demonstrated that this cAMP effect is not dependent on protein kinase A (PKA), a major cAMP binding protein, but is driven by direct binding of cAMP to NBD domain of NLRP3 protein (18). We have confirmed these findings and extended them, showing that cAMP-mediated inhibition of NLRP3 is also not dependent on Epac, the second major intracellular cAMP binding protein. Similarly, we have also found that in patients with CAPS, carrying mutations in NBD domain of NLRP3 gene that result in the impairment of cAMP binding (18), the PGE2-mediated NLRP3 inflammasome inhibition was less efficient and required 10-100 times higher doses of PGE2 to elicit similar effect as in healthy subjects. This suggests that patients with CAPS might be more resistant to signals leading to resolution of inflammation resulting in prolonged inflammation. They might also generate more PGE2 as a derivative of an excessive need to extinguish NLRP3 inflammasome and increased IL-1β signaling giving the positive feedback for activation of cPLA2α and expression of COX2 and mPGES1 (57). The pleiotropic and detrimental effects of prolonged, excessive PGE2 signaling in various tissues might lead to secondary pathologies (27). Indeed, this has been demonstrated in patients with NOMID, the most severe phenotype among CAPS variants, who suffer from severe arthropathy and develop bulky masses in their long bones (58). There is an increase in PGE2, cAMP and PKA activity in NOMID tumor cells, leading to abnormal Wnt signaling and proliferation, while inhibition of both PGE2 and caspase-1 leads to a decrease in NOMID cell proliferation (58).
PGE2 acts through four different receptors designated EP1, EP2, EP3 and EP4, with expression variations occurring in different cells and tissue types. Using multiple pharmacological and molecular approaches, we have determined that PGE2 acted specifically through EP4 to decrease NLRP3 inflammasome activation in human primary MDM. Recently, two other GPCRs have been shown to be involved in the negative regulation of NLRP1 and NLRP3 inflammasomes. Yan et al have determined that ω-3 fatty acids such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) acting through GPR120 and GPR40 and at least partially by their downstream scaffolding protein β-arrestin-2 decrease NLRP1 and NLRP3 activation (16). β-arrestin-2 was shown to bind to full-length NLRP3 and its LRR and NBD (NACHT) domains as well as to NLRP1 in a HEK293T overexpression system, but in Arrb2−/− knockout mice the reversal of DHA or EPA-mediated inhibition was incomplete. However, DHA via this axis is able to suppress high fat diet-induced NLRP3 inflammasome activation and the prevention of NLRP3 inflammasome-dependent insulin resistance in vivo (16). We have recently determined that DHA also through GRP120 activates cPLA2α and induces PGE2 production in RAW 264.7 cells and human primary macrophages (59). This suggests that the proposed PGE2-mediated inhibition of NLRP3 by increase in cAMP might be an additional mechanism of ω-3 derived effects.
The role of PGE2 on IL-1β production in monocytes and macrophages has been previously suggested; however, several contradictory results have been reported and the mechanisms involved have not been described. Early studies suggested that PGE2 inhibits IL-1β secretion in macrophages (60-62) and monocytes (63, 64), not at the level of gene transcription but by a posttranscriptional mechanism which was not well defined (63). Others reported that in monocytes, PGE2 enhances IL-1β mRNA expression and protein production via cAMP (65-67) and PKA-CREB-dependent transcriptional events (68). With these differences in mind, we studied the effect of PGE2 on inflammasome activation. We found that PGE2 added after LPS priming decreased NLRP3 inflammasome activation via a cAMP-dependent mechanism as assessed by the decrease in mature IL-1β and mature caspase-1 release, and IL-18 release, as well as a decrease in ASC oligomerization. This might account for previous observations that were noted before the development of the inflammasome concept. Nonetheless, having controlled for IL-1β mRNA and protein expression, we also noted a non-significant decrease at the mRNA level, though reflected by the slight decreases in total pro-IL-1β levels in the cells, pointing out the complexity of PGE2 in the control of IL-1β release.
Our findings that cPLA2α or COX2 inhibition resulting in the decrease of PGE2 led to an increase in NLRP3 inflammasome activation and IL-1β secretion in macrophages are consistent with the several early works studying effects of various NSAIDs on IL-1β production. Indomethacin (61-64, 69) and piroxicam (69) were reported to increase IL-1β production. On the other hand, it was reported that inhibition of cPLA2 or COX2 leads to a decrease in mature IL-1β secretion (55, 70, 71). The differences with our findings might result from the dose and type of inhibitors used in those studies, as well as the type of cells, their redox state and EP receptors expression. Our results of pharmacological inhibition with a low dose of a highly specific COX2 inhibitor, cPLA2α inhibitor and cPLA2α knockdown by siRNA, in conjunction with the observations of the exogenous PGE2 effects on NLRP3 inflammasome activation suggest that this axis might be play a regulatory role in human macrophages.
In summary, our results suggest that PGE2 might serve as a local inhibitor of NLRP3 inflammasome in monocytes and macrophages at the site of acute inflammation. In general this mechanism might serve as a switch to resolution of inflammation and return to homeostasis, but once dysregulated, it might lead to detrimental effects and pathology.
Supplementary Material
Acknowledgements
We are grateful to Drs. Chong-Shan Shi and Jehad Edwan for their input and helpful discussions.
Source of funding: NIH intramural program
Abbreviations used
- AA
Arachidonic acid
- AC
Adenylate cyclase
- AIM2
Absent in melanoma 2
- CAPS
Cryopyrin-associated periodic fever syndromes
- COX
Cyclooxygenase
- cPLA2α
Cytosolic phospholipase A2 group IVA
- DHA
Docosahexaenoic acid
- dsDNA
double-stranded DNA
- Epac
Exchange protein directly activated by cAMP
- FCAS
Familial cold-induced autoinflammatory syndrome
- GPCR
G protein-coupled receptors
- IFI16
Interferon, gamma-inducible protein 16
- NBD
Nucleodtide binding domain
- LRR
C-terminal leucin rich domain
- MDM
Monocyte-derived macrophages
- MWS
Muckle-Wells syndrome
- NLR
Nucleotide-binding domain, leucine rich repeat-containing proteins
- NOMID
Neonatal onset multisystem inflammatory disorder
- PG
Prostaglandin
- PKA
Protein kinase A
- PLC
Phospholipase C
- siRNA
Small interfering RNA
References
- 1.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 2.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nature Reviews. Immunology. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Nardo D, De Nardo CM, Latz E. New Insights into Mechanisms Controlling the NLRP3 Inflammasome and Its Role in Lung Disease. The American Journal of Pathology. 2014;184:42–54. doi: 10.1016/j.ajpath.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annual review of immunology. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. The Journal of Experimental Medicine. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT, Brickey WJ, Ting JP. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nature Immunology. 2011;12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 8.Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nunez G, Schnurr M, Espevik T, Lien E, Fitzgerald KA, Rock KL, Moore KJ, Wright SD, Hornung V, Latz E. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heneka MT, Kummer MP, Stutz A, Delekate A, Schwartz S, Vieira-Saecker A, Griep A, Axt D, Remus A, Tzeng TC, Gelpi E, Halle A, Korte M, Latz E, Golenbock DT. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nature Genetics. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*). Annual Review of Immunology. 2009;27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldbach-Mansky R, Kastner DL. Autoinflammation: the prominent role of IL-1 in monogenic autoinflammatory diseases and implications for common illnesses. The Journal of Allergy and Clinical Immunology. 2009;124:1141–1149. doi: 10.1016/j.jaci.2009.11.016. quiz 1150-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nature Immunology. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, Farlik M, Decker T, Du Pasquier RA, Romero P, Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, Hornung V, Latz E. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Y, Jiang W, Spinetti T, Tardivel A, Castillo R, Bourquin C, Guarda G, Tian Z, Tschopp J, Zhou R. Omega-3 fatty acids prevent inflammation and metabolic disorder through inhibition of NLRP3 inflammasome activation. Immunity. 2013;38:1154–1163. doi: 10.1016/j.immuni.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez-Cuellar E, Tsuchiya K, Hara H, Fang R, Sakai S, Kawamura I, Akira S, Mitsuyama M. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. J Immunol. 2012;189:5113–5117. doi: 10.4049/jimmunol.1202479. [DOI] [PubMed] [Google Scholar]
- 18.Lee GS, Subramanian N, Kim AI, Aksentijevich I, Goldbach-Mansky R, Sacks DB, Germain RN, Kastner DL, Chae JJ. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalinski P. Regulation of immune responses by prostaglandin E2. J Immunol. 2012;188:21–28. doi: 10.4049/jimmunol.1101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desouza IA, Franco-Penteado CF, Camargo EA, Lima CS, Teixeira SA, Muscara MN, De Nucci G, Antunes E. Inflammatory mechanisms underlying the rat pulmonary neutrophil influx induced by airway exposure to staphylococcal enterotoxin type A. British Journal of Pharmacology. 2005;146:781–791. doi: 10.1038/sj.bjp.0706393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tajima T, Murata T, Aritake K, Urade Y, Hirai H, Nakamura M, Ozaki H, Hori M. Lipopolysaccharide induces macrophage migration via prostaglandin D(2) and prostaglandin E(2). The Journal of Pharmacology and Experimental Therapeutics. 2008;326:493–501. doi: 10.1124/jpet.108.137992. [DOI] [PubMed] [Google Scholar]
- 22.Weller CL, Collington SJ, Hartnell A, Conroy DM, Kaise T, Barker JE, Wilson MS, Taylor GW, Jose PJ, Williams TJ. Chemotactic action of prostaglandin E2 on mouse mast cells acting via the PGE2 receptor 3. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11712–11717. doi: 10.1073/pnas.0701700104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natura G, Bar KJ, Eitner A, Boettger MK, Richter F, Hensellek S, Ebersberger A, Leuchtweis J, Maruyama T, Hofmann GO, Halbhuber KJ, Schaible HG. Neuronal prostaglandin E2 receptor subtype EP3 mediates antinociception during inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13648–13653. doi: 10.1073/pnas.1300820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellmann J, Zhang MJ, Tang Y, Rane M, Bhatnagar A, Spite M. Increased saturated fatty acids in obesity alter resolution of inflammation in part by stimulating prostaglandin production. J Immunol. 2013;191:1383–1392. doi: 10.4049/jimmunol.1203369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvirkvelia N, McMenamin M, Chaudhary K, Bartoli M, Madaio MP. Prostaglandin E2 promotes cellular recovery from established nephrotoxic serum nephritis in mice, prosurvival, and regenerative effects on glomerular cells. American Journal of Physiology. Renal Physiology. 2013;304:F463–470. doi: 10.1152/ajprenal.00575.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKenzie KF, Clark K, Naqvi S, McGuire VA, Noehren G, Kristariyanto Y, van den Bosch M, Mudaliar M, McCarthy PC, Pattison MJ, Pedrioli PG, Barton GJ, Toth R, Prescott A, Arthur JS. PGE(2) induces macrophage IL-10 production and a regulatory-like phenotype via a protein kinase A-SIK-CRTC3 pathway. J Immunol. 2013;190:565–577. doi: 10.4049/jimmunol.1202462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakanishi M, Rosenberg DW. Multifaceted roles of PGE2 in inflammation and cancer. Seminars in Immunopathology. 2013;35:123–137. doi: 10.1007/s00281-012-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirata T, Narumiya S. Prostanoids as regulators of innate and adaptive immunity. Advances in Immunology. 2012;116:143–174. doi: 10.1016/B978-0-12-394300-2.00005-3. [DOI] [PubMed] [Google Scholar]
- 29.Dennis EA, Cao J, Hsu YH, Magrioti V, Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chemical Reviews. 2011;111:6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murakami M, Taketomi Y, Miki Y, Sato H, Hirabayashi T, Yamamoto K. Recent progress in phospholipase A(2) research: from cells to animals to humans. Progress in Lipid Research. 2011;50:152–192. doi: 10.1016/j.plipres.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Hirata T, Narumiya S. Prostanoid receptors. Chemical reviews. 2011;111:6209–6230. doi: 10.1021/cr200010h. [DOI] [PubMed] [Google Scholar]
- 32.Katoh H, Watabe A, Sugimoto Y, Ichikawa A, Negishi M. Characterization of the signal transduction of prostaglandin E receptor EP1 subtype in cDNA-transfected Chinese hamster ovary cells. Biochimica et Biophysica Acta. 1995;1244:41–48. doi: 10.1016/0304-4165(94)00182-w. [DOI] [PubMed] [Google Scholar]
- 33.Fujino H, Salvi S, Regan JW. Differential regulation of phosphorylation of the cAMP response element-binding protein after activation of EP2 and EP4 prostanoid receptors by prostaglandin E2. Molecular Pharmacology. 2005;68:251–259. doi: 10.1124/mol.105.011833. [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama U, Iwatsubo K, Umemura M, Fujita T, Ishikawa Y. The prostanoid EP4 receptor and its signaling pathway. Pharmacological Reviews. 2013;65:1010–1052. doi: 10.1124/pr.112.007195. [DOI] [PubMed] [Google Scholar]
- 35.Lefkimmiatis K, Zaccolo M. cAMP signaling in subcellular compartments. Pharmacology & Therapeutics. 2014 doi: 10.1016/j.pharmthera.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimoto Y, Namba T, Honda A, Hayashi Y, Negishi M, Ichikawa A, Narumiya S. Cloning and expression of a cDNA for mouse prostaglandin E receptor EP3 subtype. The Journal of Biological Chemistry. 1992;267:6463–6466. [PubMed] [Google Scholar]
- 37.Qi HY, Shelhamer JH. Toll-like receptor 4 signaling regulates cytosolic phospholipase A2 activation and lipid generation in lipopolysaccharide-stimulated macrophages. The Journal of Biological Chemistry. 2005;280:38969–38975. doi: 10.1074/jbc.M509352200. [DOI] [PubMed] [Google Scholar]
- 38.Buczynski MW, Stephens DL, Bowers-Gentry RC, Grkovich A, Deems RA, Dennis EA. TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264.7 cells. The Journal of Biological Chemistry. 2007;282:22834–22847. doi: 10.1074/jbc.M701831200. [DOI] [PubMed] [Google Scholar]
- 39.Kuroda E, Ishii KJ, Uematsu S, Ohata K, Coban C, Akira S, Aritake K, Urade Y, Morimoto Y. Silica crystals and aluminum salts regulate the production of prostaglandin in macrophages via NALP3 inflammasome-independent mechanisms. Immunity. 2011;34:514–526. doi: 10.1016/j.immuni.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Sokolowska M, Chen LY, Eberlein M, Martinez-Anton A, Liu Y, Alsaaty S, Qi HY, Logun C, Horton M, Shelhamer JH. Low molecular weight hyaluronan activates cytosolic phospholipase A2alpha and eicosanoid production in monocytes and macrophages. The Journal of Biological Chemistry. 2014;289:4470–4488. doi: 10.1074/jbc.M113.515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbera-Cremades M, Baroja-Mazo A, Gomez AI, Machado F, Di Virgilio F, Pelegrin P. P2X7 receptor-stimulation causes fever via PGE2 and IL-1beta release. FASEB Journal. 2012;26:2951–2962. doi: 10.1096/fj.12-205765. [DOI] [PubMed] [Google Scholar]
- 42.von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, Vance RE. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 2012;490:107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 44.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature Immunology. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koga K, Takaesu G, Yoshida R, Nakaya M, Kobayashi T, Kinjyo I, Yoshimura A. Cyclic adenosine monophosphate suppresses the transcription of proinflammatory cytokines via the phosphorylated c-Fos protein. Immunity. 2009;30:372–383. doi: 10.1016/j.immuni.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 46.Ma W, Quirion R. Up-regulation of interleukin-6 induced by prostaglandin E from invading macrophages following nerve injury: an in vivo and in vitro study. Journal of Neurochemistry. 2005;93:664–673. doi: 10.1111/j.1471-4159.2005.03050.x. [DOI] [PubMed] [Google Scholar]
- 47.Ratcliffe MJ, Walding A, Shelton PA, Flaherty A, Dougall IG. Activation of E-prostanoid4 and E-prostanoid2 receptors inhibits TNF-alpha release from human alveolar macrophages. The European Respiratory Journal. 2007;29:986–994. doi: 10.1183/09031936.00131606. [DOI] [PubMed] [Google Scholar]
- 48.Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacological Reviews. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 49.Kumar S, Kostin S, Flacke JP, Reusch HP, Ladilov Y. Soluble adenylyl cyclase controls mitochondria-dependent apoptosis in coronary endothelial cells. The Journal of Biological Chemistry. 2009;284:14760–14768. doi: 10.1074/jbc.M900925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwatsubo K, Tsunematsu T, Ishikawa Y. Isoform-specific regulation of adenylyl cyclase: a potential target in future pharmacotherapy. Expert Opinion on Therapeutic Targets. 2003;7:441–451. doi: 10.1517/14728222.7.3.441. [DOI] [PubMed] [Google Scholar]
- 51.Fawcett L, Baxendale R, Stacey P, McGrouther C, Harrow I, Soderling S, Hetman J, Beavo JA, Phillips SC. Molecular cloning and characterization of a distinct human phosphodiesterase gene family: PDE11A. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3702–3707. doi: 10.1073/pnas.050585197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frei R, Ferstl R, Konieczna P, Ziegler M, Simon T, Rugeles TM, Mailand S, Watanabe T, Lauener R, Akdis CA, O'Mahony L. Histamine receptor 2 modifies dendritic cell responses to microbial ligands. The Journal of Allergy and Clinical Immunology. 2013;132:194–204. doi: 10.1016/j.jaci.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 53.Aronoff DM, Canetti C, Serezani CH, Luo M, Peters-Golden M. Cutting edge: macrophage inhibition by cyclic AMP (cAMP): differential roles of protein kinase A and exchange protein directly activated by cAMP-1. J Immunol. 2005;174:595–599. doi: 10.4049/jimmunol.174.2.595. [DOI] [PubMed] [Google Scholar]
- 54.Gattorno M, Tassi S, Carta S, Delfino L, Ferlito F, Pelagatti MA, D'Osualdo A, Buoncompagni A, Alpigiani MG, Alessio M, Martini A, Rubartelli A. Pattern of interleukin-1beta secretion in response to lipopolysaccharide and ATP before and after interleukin-1 blockade in patients with CIAS1 mutations. Arthritis and Rheumatism. 2007;56:3138–3148. doi: 10.1002/art.22842. [DOI] [PubMed] [Google Scholar]
- 55.Piccini A, Carta S, Tassi S, Lasiglie D, Fossati G, Rubartelli A. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1beta and IL-18 secretion in an autocrine way. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8067–8072. doi: 10.1073/pnas.0709684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossol M, Pierer M, Raulien N, Quandt D, Meusch U, Rothe K, Schubert K, Schoneberg T, Schaefer M, Krugel U, Smajilovic S, Brauner-Osborne H, Baerwald C, Wagner U. Extracellular Ca2+ is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nature Communications. 2012;3:1329. doi: 10.1038/ncomms2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park JY, Pillinger MH, Abramson SB. Prostaglandin E2 synthesis and secretion: the role of PGE2 synthases. Clin Immunol. 2006;119:229–240. doi: 10.1016/j.clim.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 58.Almeida MQ, Tsang KM, Cheadle C, Watkins T, Grivel JC, Nesterova M, Goldbach-Mansky R, Stratakis CA. Protein kinase A regulates caspase-1 via Ets-1 in bone stromal cell-derived lesions: a link between cyclic AMP and pro-inflammatory pathways in osteoblast progenitors. Human Molecular Genetics. 2011;20:165–175. doi: 10.1093/hmg/ddq455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Chen LY, Sokolowska M, Eberlein M, Alsaaty S, Martinez-Anton A, Logun C, Qi HY, Shelhamer JH. The fish oil ingredient, docosahexaenoic acid, activates cytosolic phospholipase A via GPR120 receptor to produce prostaglandin E and plays an anti-inflammatory role in macrophages. Immunology. 2014;143(1):81–95. doi: 10.1111/imm.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brandwein SR. Regulation of interleukin 1 production by mouse peritoneal macrophages. Effects of arachidonic acid metabolites, cyclic nucleotides, and interferons. The Journal of Biological Chemistry. 1986;261:8624–8632. [PubMed] [Google Scholar]
- 61.Shirahama M, Ishibashi H, Tsuchiya Y, Kurokawa S, Hayashida K, Okumura Y, Niho Y. Kupffer cells may autoregulate interleukin 1 production by producing interleukin 1 inhibitor and prostaglandin E2. Scandinavian Journal of Immunology. 1988;28:719–725. doi: 10.1111/j.1365-3083.1988.tb01505.x. [DOI] [PubMed] [Google Scholar]
- 62.Goss JA, Mangino MJ, Flye MW. Kupffer cell autoregulation of IL-1 production by PGE2 during hepatic regeneration. The Journal of Surgical Research. 1992;52:422–428. doi: 10.1016/0022-4804(92)90306-k. [DOI] [PubMed] [Google Scholar]
- 63.Knudsen PJ, Dinarello CA, Strom TB. Prostaglandins posttranscriptionally inhibit monocyte expression of interleukin 1 activity by increasing intracellular cyclic adenosine monophosphate. J Immunol. 1986;137:3189–3194. [PubMed] [Google Scholar]
- 64.Hart PH, Whitty GA, Piccoli DS, Hamilton JA. Control by IFN-gamma and PGE2 of TNF alpha and IL-1 production by human monocytes. Immunology. 1989;66:376–383. [PMC free article] [PubMed] [Google Scholar]
- 65.Sung SS, Walters JA. Increased cyclic AMP levels enhance IL-1 alpha and IL-1 beta mRNA expression and protein production in human myelomonocytic cell lines and monocytes. The Journal of Clinical Investigation. 1991;88:1915–1923. doi: 10.1172/JCI115515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohmori Y, Strassman G, Hamilton TA. cAMP differentially regulates expression of mRNA encoding IL-1 alpha and IL-1 beta in murine peritoneal macrophages. J Immunol. 1990;145:3333–3339. [PubMed] [Google Scholar]
- 67.Oshima H, Hioki K, Popivanova BK, Oguma K, Van Rooijen N, Ishikawa TO, Oshima M. Prostaglandin E(2) signaling and bacterial infection recruit tumor-promoting macrophages to mouse gastric tumors. Gastroenterology. 2011;140:596–607. e597. doi: 10.1053/j.gastro.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 68.Chandra G, Cogswell JP, Miller LR, Godlevski MM, Stinnett SW, Noel SL, Kadwell SH, Kost TA, Gray JG. Cyclic AMP signaling pathways are important in IL-1 beta transcriptional regulation. J Immunol. 1995;155:4535–4543. [PubMed] [Google Scholar]
- 69.Otterness IG, Bliven ML, Eskra JD, Reinke M, Hanson DC. The pharmacologic regulation of interleukin-1 production: the role of prostaglandins. Cellular Immunology. 1988;114:385–397. doi: 10.1016/0008-8749(88)90330-9. [DOI] [PubMed] [Google Scholar]
- 70.Andrei C, Margiocco P, Poggi A, Lotti LV, Torrisi MR, Rubartelli A. Phospholipases C and A2 control lysosome-mediated IL-1 beta secretion: Implications for inflammatory processes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9745–9750. doi: 10.1073/pnas.0308558101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hua KF, Chou JC, Ka SM, Tasi YL, Chen A, Wu SH, Chiu HW, Wong WT, Wang YF, Tsai CL, Ho CL, Lin CH. Cyclooxygenase-2 Regulates NLRP3 Inflammasome-Derived IL-1beta Production. J Cell Physiol. 2014;230:863–874. doi: 10.1002/jcp.24815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.