All living organisms must transmit genetic information through successive generations, and all six eukaryotic supergroups utilize a mitotic spindle to accomplish this task. The mitotic spindle is the cellular machinery responsible for chromosome segregation during mitosis, and it is comprised of hundreds of proteins. During evolution, eukaryotic cells have developed different mechanisms and different kinds of mitotic spindles to segregate the chromosomes (Drechsler and McAinsh, 2012). Regardless of the differences, the common characteristic of all these different mitotic spindles is the utilization of microtubules and kinesins, as all known eukaryotes studied to this date possess kinesins (Wickstead et al., 2010).
The mitotic spindle is a bipolar array of microtubules of varied lengths that continuously grow and shrink. These highly dynamic microtubules are nucleated by centrosomes and contact the chromosomes in the centromeric region to facilitate chromosome attachment and segregation. Although chromosome movement is powered in part by changes in microtubule assembly (Shelden and Wadsworth, 1992), kinesins associated with microtubules and other spindle structures refine the movement of chromosomes in the spindle. Kinesins participate in chromosome attachment, influence microtubule dynamics and contribute to anaphase spindle elongation (reviewed in (Cross and McAinsh, 2014)). Thus, kinesins, in conjunction with dynamic microtubules, ensure the proper distribution of genetic material between the two daughter cells to avoid aneuploidy.
Kinesins are a class of molecular motors that use the energy from hydrolysis of ATP to translocate along the microtubule or control microtubule end dynamics (Vale et al., 1985, 1996; Desai et al., 1999). They have been identified in members of all six eukaryotic supergroups including extremely deep-rooted members (Fig. 1). Processive kinesins are able to perform successive work-producing cycles of ATP hydrolysis without detaching from the substrate microtubule whereas non-processive kinesins readily detach from microtubules but can be quite effective working in ensembles. There are 14 families of kinesins, and most members possess two distinct functional domains: an ATP-hydrolyzing motor domain and a tail domain that can associate with cellular structures or cargo (Lawrence et al., 2004; Miki et al., 2005). The motor domain is very well conserved among the different kinesins families while the tail domains are more divergent. Most kinesins translocate to the plus ends of the microtubule and possess an N-terminal motor domain. There are kinesins with the motor at the C-terminus that translocate to the minus end of the microtubule. Besides this hand over hand “walking” activity (Yildiz et al., 2004), there are some kinesins that are able to control microtubule dynamics through promoting polymerization, promoting depolymerization or pausing polymerization activity (Desai et al., 1999; Bringmann et al., 2004; Cui et al., 2005).
Figure 1.
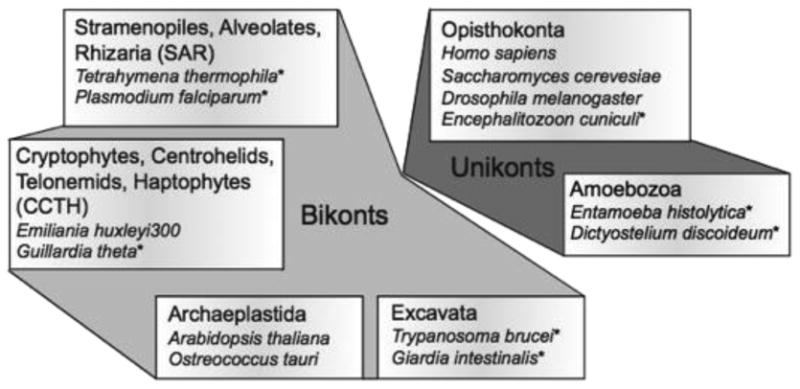
Six eukaryotic supergroups including, in each case, a few members in which kinesins have been unambiguously identified. Organisms thought to be more deeply rooted are indicated (*).
In Opisthokont metazoans there appear to be Kinesins involved in every step of mitosis, belonging to the families 4, 5, 6, 7, 8, 10, 12, 13, 14 (reviewed in (Cross and McAinsh, 2014)). Here we are going to review the kinesins involved in each phase of mitosis with an emphasis on the stages of mammalian cell division. Additionally, we will consider the role of kinesins in deep-rooted eukaryotes.
Kinesins in Mitosis
Prophase: Centrosome separation
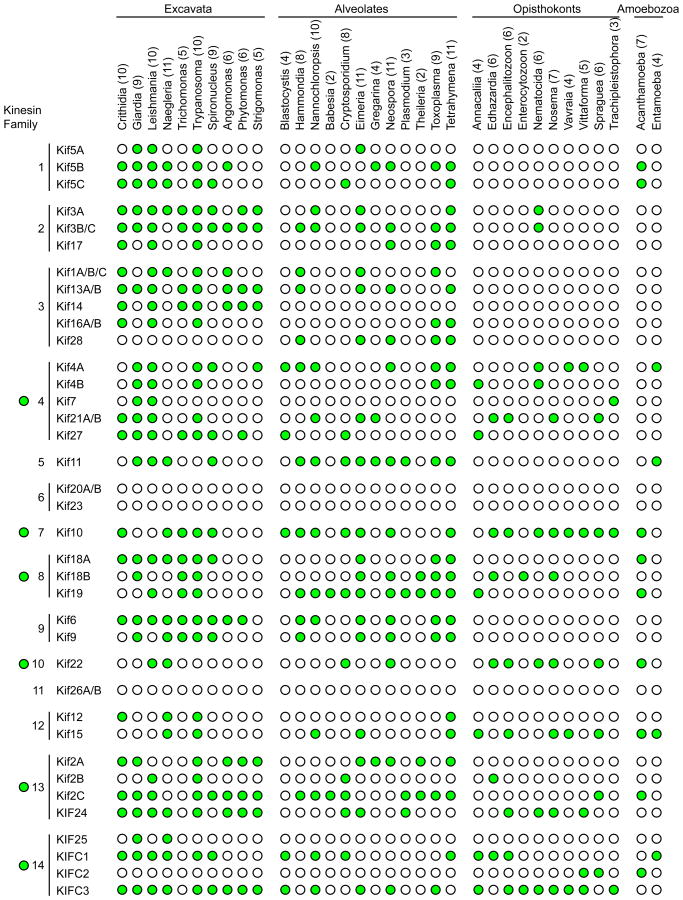
During prophase, the two centrosomes close to the nuclear envelope separate and travel to opposites sides of the cell to form a bipolar spindle. At the same time the chromatin begins to condense to form chromosomes. The most important kinesin family involved in the formation of the bipolar spindle is the kinesin-5 family (Kashina et al., 1997). Kif11 (also known as Eg5) is the Kinesin-5 family member involved in the bipolar spindle formation in humans (Slangy et al., 1995). Kif11 acts as a tetramer with two kinesin heads contacting one microtubule and the other pair of heads contacting a parallel or antiparallel microtubule. Thus, Kif11 acts as a microtubule crosslinker that is able to force two microtubules to glide with respect to each other, and uses this activity to separate the centrosomes during the beginning of mitosis (Kapitein et al., 2005). A representative Kif11 member is present in many deep-rooted eukaryotes and lacking in others, even across those (such as T. brucei vs. S. cerevisiae) with closed mitosis (Fig. 2).
Figure 2.
Distribution of kinesins in deep-rooted eukaryotes. We manually analyzed the genome of several protists from the groups Excavata, Alveolates, Opisthokonts and Amoebozoa searching for kinesins. Presence of a kinesin member in the genome is indicated with a green filled circle. The number in parenthesis after the name indicates the number of kinesin families for each organism.
We have checked the genomes of the next eukaryotes: Acanthamoeba (A castellanii), Angomonas (A deanei), Anncaliia (A algerae), Babesia (B. bovis, B. microti), Blastocystis (B. hominis), Crithidia (C. fasciculate), Cryptosporidium (C. hominis, C. muris, C. parvum), Edhazardia (E. aedis), Eimeria (E. acervulina, E. brunetti, E. falciformis, E. maxima, E. mitis, E. necatrix, E. praecox, E. tenella), Encephalitozoon (E. cuniculi, E. hellem, E. intestinalis, E. romaleae), Entamoeba (E. dispar, E. histolytica, E. invadens, E. moshkovskii, E. nuttalli), Enterocytozoon (E. bieneusi), Giardia (G. intestinalis), Gregarina (G. niphandrodes), Hammondia (H. hammondi), Leishmania (L braziliensis, L donovani, L infantum, L major, L mexicana, L tarentolae), Naegleria (N. gruberi), Nannochloropsis (N. gaditana), Nematocida (N. parisii), Neospora (N. caninum), Nosema (N. bombycis, N. ceranae), Phytomonas spp (different isolates), Plasmodium (P. berghei, P. chabaudi, P. cynomolgi, P. falciparum, P. gallinaceum, P. knowlesi, P. reichenowi, P. vivax, P. yoelii), Spironucleus (S. salmonicida), Spraguea (S. lophii), Strigomonas (S. culicis), Tetrahymena (T. thermophila), Theileria (T. annulata, T. equi, T. orientalis, T. parva), Toxoplasma (T. gondii), Trachipleistophora (T. hominis), Trichomonas (T. vaginalis), Trypanosoma (T. brucei, T. congolense, T. cruzi, T. evansi, T. grayi, T. rangeli, T. vivax), Vavraia (V. culicis), and Vittaforma (V. corneae).
Prometaphase and Metaphase: Organization of a bipolar spindle and chromosome congression
During prometaphase the centrosomes localize to opposites sides of the cell to form a bipolar spindle. There is evidence that Kif15 (a Kinesin-12 family member) functionally overlaps Kif11 in the separation of the centrosomes and in the formation of a bipolar spindle (Tanenbaum et al., 2009; Sturgill and Ohi, 2013). KifC1 (a kinesin-14 motor) is a minus-end directed motor that generates an inward force during the formation of the spindle. The outward force created by Kif11 and Kif15 compensates this inward force to help maintain the spindle length (Mountain et al., 1999). Interestingly, some organisms that lack Kif11, such as P. tetraurelia and T. brucei, do possess Kif15 (Wickstead et al., 2010).
During this phase, microtubules contact the chromosomes in the kinetochores and move them to the middle of the spindle forming the metaphase plate. Several kinesin families are involved in the capture and congression of the chromosomes: Kinesin-4 (Kif4), Kinesin-7 (Kif10), Kinesin-8 (Kif18A), Kinesin-10 (Kif22), Kinesin-13 (Kif2B, Kif2C) and Kinesin-14 (KifC1).
The first contact is usually a “lateral connection”, where the kinetochores contact the microtubule via the lattice rather than the microtubule tip. In these cases, the molecular motors Dynein and Kif10 (also known as CenpE, a Kinesin-7 member) are the players involved in the transport of the chromosomes to the plus-end tip of the microtubule to establish a stronger connection kinetochore-microtubule (Schaar et al., 1997; Wood et al., 1997; Kapoor et al., 2006; Cai et al., 2009). Kinesin-7 is widely represented in deep-rooted organisms.
Members of the Kinesin-8 family are important for the correct chromosome alignment in metaphase. For example, the deletion of Kinesin-8 members Klp5 and Klp6 in S. pombe and Kip3 in S. cerevisiae, alters the alignment of chromosomes in metaphase (Garcia et al., 2002; West et al., 2002; Wargacki et al., 2010). In human cells the depletion of Kif18A generates a congression defect with chromosomes dispersed through all the spindle (Mayr et al., 2007; Stumpff et al., 2008, 2012). Kinesin-13 member's role in chromosome alignment at the metaphase plate is not as well understood as Kinesin-8, but it is known that the lack of Kif2C (MCAK) affects attachment (Domnitz et al., 2012) and congression in human cells (Zhu et al., 2005) possibly by limiting microtubule length within the spindle.
Another activity influenced by kinesins is chromosome oscillation around the metaphase plate. Once two sister chromatids are attached to microtubule tips emerging from respective opposite poles, the chromosome is bi-oriented. Stable, bi-oriented attachment of the chromosomes to the mitotic spindle is a requirement to turn off the mitotic checkpoint. The checkpoint assures that both daughter cells receive the same amount of genetic material by inhibiting the initiation of anaphase chromosome segregation in the presence of improperly attached chromosomes. Bi-oriented chromosomes establish a meta-stable position at the metaphase plate by oscillating back and forth across the spindle midpoint. It is hypothesized that in addition to properly positioning chromosomes for anaphase, oscillations may facilitate the shedding of improper microtubule attachments during prometaphase and metaphase (Holt et al., 2005; Wordeman et al., 2007). These oscillations are possible thanks to forces derived from changes in microtubule polymerization rates that are controlled in part by kinesins. There are several kinesins implicated as participating in the oscillation movements: Kif2B, Kif2C, Kif4, Kif10, Kif18A and Kif22.
Polar ejection forces help to push chromosomes away from the spindle poles and relocate them to the metaphase plate if they have moved far away from it (Rieder et al., 1986). This is achieved thanks to a group of kinesins that interact with chromosomes called chromokinesins. Kif22 (also known as Kid, member of the Kinesin-10 family) is a chromokinesin that uses solely plus-end directed motility to facilitate chromosome congression (Stumpff et al., 2012). Kif4 (Kinesin-4 family) is a plus-end directed chromokinesin that can also regulate microtubule dynamics and microtubule length to influence congression (Oh et al., 2000; Samejima et al., 2012; Stumpff et al., 2012; Wandke et al., 2012). This unusual kinesin family member can be detected in a number of deep-rooted eukaryotes (Fig. 2).
The Kinesin-8 family member, Kif18A, is able to control microtubule dynamics (Roostalu and Surrey, 2013; Su et al., 2013) and chromosome oscillations probably by pausing or reducing the polymerization rates of microtubules ends close to the kinetochores (Du et al., 2010; Stumpff et al., 2012). Studies in human cells have shown that Kif18A localizes in the spindle with a higher concentration at the tip of the plus-end microtubule close to kinetochores (Mayr et al., 2007; Stumpff et al., 2008). Kinetochores closer to the metaphase plate have longer k-fibers, and therefore accumulate more Kif18A molecules. Growth suppression of these longer k-fibers would expected to be enhanced, preventing the chromosomes from moving farther from the metaphase plate. In contrast, the loss of Kif18A allows chromosomes to stray farther from the metaphase plate during each oscillation. This would explain why there are chromosomes dispersed through all the spindle in the Kif18A knockdown (Mayr et al., 2007; Stumpff et al., 2008, 2011, 2012). There are very few eukaryotes that do not possess either a kinesin-13 member, a kinesin-8 member or both, which suggests microtubule length modulation by kinesins is an essential activity. Curiously, one primitive eukaryote, Cyanidioschyzon merolae, that lacks kinesin-8 and -13 family members accomplishes much of its G1 activities without utilizing assembled microtubules (Imoto et al., 2011). In this organism MTs are only present in the mitotic spindle, and the authors suggest the idea that MTs first evolved associated to mitosis, and that the cytoskeleton and transport functions evolved later.
Besides chromosome congression and positioning, another process regulated by kinesins is the turnover of the kinetochore microtubules. The kinesin-13 family members (Kif2A, Kif2B and Kif2C) are involved in this process likely because they are capable of using the energy of ATP to directly disassemble microtubules (Desai et al., 1999; Hunter et al., 2003; Cooper et al., 2010). During the first contacts of the microtubule with the kinetochores, erroneous connections are common (Cimini et al., 2003). Kinetochore-associated Kif2C is implicated in the correction of MT-KT attachments (Kline-Smith et al., 2004; Wordeman et al., 2007). Kif2C is found both on kinetochores and also as a complex with EB1 on microtubule plus-ends. It facilitates end-on attachment of kinetochores to microtubule tips by suppressing plus-end microtubule length within the spindle (Domnitz et al., 2012). Kif2A also controls spindle microtubule length but its centrosomal position suggests control of microtubule minus ends so its overarching role may be to set overall spindle length (Wilbur and Heald, 2013).
Anaphase, Telophase and Cytokinesis
During anaphase the separated sister chromosomes move from the center of the spindle in the metaphase plate toward opposite poles of the cell (anaphase A) and the mitotic spindle is elongated (anaphase B). Microtubule depolymerization is the principle driving force for anaphase chromosome segregation (Gorbsky et al., 1987; Shelden and Wadsworth, 1992). In Drosophila the Kinesin-13 members KLP59C and KLP10A may influence microtubule depolymerization to facilitate chromosome segregation as well (Rogers et al., 2004). Spindle elongation is controlled by kinesin-5 members in flies and yeast (Straight et al., 1998; Brust-Mascher et al., 2009). Interestingly, it is not well-understood what controls anaphase B spindle elongation in mammals.
The central spindle is a structure of antiparallel microtubules formed between the two sets of segregating chromosomes important for the regulation cytokinesis. Kif2A and Kif4 control the size of the central spindle through the control of microtubule dynamics (Bieling et al., 2010; Hu et al., 2011; Uehara et al., 2013). There are other kinesins important for cytokinesis like Kif10, Kif14, Kif20A, Kif20B and Kif23 (reviewed in (Lee et al., 2012) and chapter xxx in this same number).
Kinesins and the control of Microtubule dynamics
The classical or conventional role of kinesins is the transport of cargo from one place in the cell to another, e.g. the transport of cargo down the axon of a neuron. But as described above, some kinesins are able to influence MT dynamics. Microtubules are dynamic biological polymers that constantly grow and shrink in an assembly/disassembly cycle known as dynamic instability (Mitchison and Kirschner, 1984).
Microtubule dynamics varies depending on the cell cycle, cell events and cellular structures encountered by the microtubule end. During interphase the half-life of MT is around 5 min, and in the mitotic spindle it is of around 5 seconds (Saxton et al., 1984). But even inside of the spindle there are differences between microtubule subpopulations. The connection of MT with the KT (K-fibers) is very, stable, resulting in higher half-life than in the rest of the mitotic spindle (around 5 minutes) (Gorbsky and Borisy, 1989; Zhai et al., 1995). The turnover of microtubules in the kinetochore fiber is an important contributor to error correction during cell division (Bakhoum et al., 2009; Ertych et al., 2014).
Microtubule dynamics is highly regulated by a complex of proteins like MAPs (microtubule associated proteins) and kinesins (Vaart et al., 2009). Based on existing studies, we can differentiate three classes of kinesins regulating MT dynamics:
Kinesins that promote or enhance the addition of subunit to the MT: kinesin-7 (CenpE), Kinesin-10 (Nod)
Kinesins that enhance or promotes tubulin subunit loss: kinesin-8 (Kip3, Klp5/6, Kif19), Kinesin-13 (Kif2A, 2B and 2C/MCAK), Kinesin-14 (Kar3)
Kinesins that suppress the dynamics at the MT ends: Kinesin-4 (Kif4/Xklp-1), Kinesin-8 (Kif18A).
Kinesins proteins are the only cytoskeletal motors (kinesins, dyneins and myosins) that have been found in all eukaryotes studied to date (Richards and Cavalier-Smith, 2005; Wickstead and Gull, 2006, 2007; Wickstead et al., 2010). This would put the apparition of the first kinesins between 1.6 and 2.2 billion years ago, with the apparition of the first eukaryotic cells. But, when did kinesins acquire the ability to modulate microtubule dynamics? Is that function previous or subsequent to translocation along the microtubule lattice? It is possible that regulation of microtubule assembly is an ancient and perhaps the original function of the earliest kinesins, with the transport function evolving later. With this in mind, what would be the set of kinesins possessed by the FECA (First Eukaryotic Common Ancestor) and the LECA (Last Eukaryotic Common Ancestor)? In a 2010 paper Wickstead et al. studied kinesin diversity through a wide range of eukaryotes organisms concluding that is probably the LECA had a whole set of kinesins comprising members of families 1, 2, 3, 4, 5, 8, 9, 10, 13, 14, and 17 (the Kinesin-17 family is a new family only found in bikonts (Wickstead and Gull, 2006)). In addition, it appears that most eukaryotic organisms have kinesins controlling MT dynamics and most of them have some kind of MT-depolymerizing kinesin from families Kinesin-8, Kinesin-13 or Kinesin-14 (Wickstead and Gull, 2006; Wickstead et al., 2010). Even the primitive red alga C. merolae, which lacks kinesin-8 and -13 family members, possesses a kinesin-14. We have performed a simple manual BLAST analysis of kinesins in eukaryote protists from the groups Excavata, Alveolates, Opisthokonts and Amoebozoa (Fig. 1). Some of these organisms are pathogens, and some of them are known as deep-rooted eukaryotes because they are usually positioned close to the root of the eukaryotic tree of life. We have not found kinesins from families 6 and 11, and our results fit with the results from previous papers.
Polymerizing kinesins
There are several kinesins in different systems that promote MT polymerization or MT nucleation. Kif10 (also known as Cenp-E) is a plus-end directed kinesin from the Kinesin-7 family (Wood et al., 1997). It localizes to MT ends, is able to stabilize GTP-microtubules and promote the elongation of the stabilized MTs (Sardar et al., 2010). The Drosophila Nod (Kinesin-10 family) is a non-motile kinesin that plays an important role in chromosome segregation during meiosis. It localizes to MT ends and promotes MT polymerization (Cui et al., 2005). Additionally, there is the special case of the S. pombe KLP5/6 (Kinesin-8) motors that promote both MT nucleation and catastrophe (Cui et al., 2005). In our analysis we have found proteins similar to Kif10 (CenpE, kinesin-7) in most of the eukaryotes analyzed (Fig. 2).
Depolymerizing Kinesins
The most studied group of kinesins controlling MT dynamics is the depolymerases, especially the Kinesin-8 and Kinesin-13 families.
Kinesin-13 family
Kinesins-13 members have the catalytic domain in the center of the protein, and do not walk over the MT, but use their ATPase activity to remove tubulin subunits from both ends of the MT (Desai et al., 1999; Walczak, 2003), arriving to the ends mainly by diffusion (Helenius et al., 2006). The first kinesins with depolymerization activity identified in mammals were Kif2A and Kif2C (MCAK) from the Kinesin-13 family (Noda et al., 1995; Wordeman and Mitchison, 1995). Kif2A and 2C have been studied in a lot of detail, but the first studies showing the depolymerization activity of MCAK came studying the orthologous in Xenopus: XKCM1. The depletion of XKCM1 in Xenopus egg extracts causes an excessive growth of the MT (spindle and astral MT) and the prevention of mitotic spindle formation (Walczak et al., 1996; Kline-Smith and Walczak, 2002). Cells overexpressing XKCM1 do not form a bipolar spindle because the MTs are too small and not able to grow (Ohi et al., 2007). In human cells overexpression of MCAK cause similar defects, which MCAK loss promoted kinetochore attachment errors and spindle positioning defects (Maney et al., 1998; Kline-Smith and Walczak, 2002; Wordeman et al., 2007; Rankin and Wordeman, 2010; Domnitz et al., 2012). S. cerevisiae doesn't possess members of the Kinesin-13 family (Wickstead et al., 2010). However, both Kar3 (a Kinesin-14) and Kip3 (a kinesin-8) have the ability to destabilize MT ends (Sproul et al., 2005), so it is possible that they may have acquired the depolymerization activity to compensate for the lack of Kinesin-13. Alternatively, the kinesin-14, Kar3, may supply depolymerizing activity to functionally subsidize activities controlled by kinesin-13 in other organisms (Saunders et al., 1997).
Kinesin-13 members are present in many deep-rooted eukaryotes. In Giardia intestinalis Kinesin-13 localizes to the median body and flagellum, affecting flagellum length, median body behavior and mitotic MT dynamics (Dawson et al., 2007). Kinesin-13 in Leishmania major is involved in flagellar length control (Blaineau et al., 2007). Trypanosoma brucei Kinesin-13 localizes to the flagellar tip but it seems that its role in regulating flagellar length is very modest (Chan and Ersfeld, 2010). And in the single cell green alga Chlamydomonas reinhardtii Kinesin-13 is involved in flagellum assembly/disassembly cycle (Piao et al., 2009; Wang et al., 2013).
Kinesin-8 family
Kinesin-8 proteins possess an N-terminal motor domain and use highly processive motility to reach the plus-end of the MT where they can influence MT dynamics (Gupta et al., 2006; Varga et al., 2006; Su et al., 2012). Kinesin-8 has two MT interacting sites, one of them in the C-terminal of the protein, that allows the kinesin to keep attached to the MT and exhibit high processivity (Mayr et al., 2011; Stumpff et al., 2011; Su et al., 2011; Weaver et al., 2011). Thanks to this high processivity Kinesin-8 motors can accumulate at the end of the plus-side of the long MT creating a gradient of kinesin motors along the length of the MT that is more pronounced in longer MT fibers. In other words, greater numbers of motors can accumulate at the ends of longer MTs imparting more kinesin-8 activity at the ends of longer microtubules (Varga et al., 2006, 2009).
Members of this family can regulate MT dynamics by removing tubulin subunits or by blocking the addition of new tubulin. The mammalian Kinesin-8 Kif18A appears to suppress microtubule assembly at microtubule plus ends (Du et al., 2010; Stumpff et al., 2011, 2012). A pause in tubulin addition could allow GTP hydrolysis to reach the microtubule plus-end and increase catastrophes, but this mechanism has yet to be proven. Thus, these motors could induce MT depolymerization without actually removing tubulin subunits. S. cerevisiae Kip3p, in contrast, promotes the loss of tubulin subunits from the microtubule plus end (Varga et al., 2006; Su et al., 2011). To remove tubulin subunits, Kip3p accumulates on the plus-side of the MT and when new Kip3p arrives to the plus-end it pushes the previous molecules removing the motor and tubulin from the MT fiber (Varga et al., 2009). In keeping with the controversial nature of kinesin-8 activity, it is not clear if Klp5/6 from S. pombe has depolymerizing activity or not, as there are studies showing depolymerizing activity (Erent et al., 2012), and others showing the opposite (Grissom et al., 2009). While yeast do not have a Kinesin-13 member, Giardia and some other deep eukaryotes have Kinesin-13, so it is possible that yeast has lost Kinesin-13 during evolution. However they have members of the Kinesin-8 family, so they still have kinesins with depolymerizing function. It is possible that organisms with simpler genomes and fewer kinesins have combined the depolymerizing functions in just one family.
Evolution of Kinesins
From our analysis we have reached similar conclusions to those in Wickstead. et al. There are organisms like Crithidia, Eimeria, Leishmania, Neospora, Trypanosoma or Naegleria that possess kinesins from 10 different families (Fig. 2). This would support the idea that the LECA was fully equipped with a complete set of kinesins for different cellular functions. However there are some other organisms like Babesia (Kinesin-8 and 13), Enterocytozoon (Kinesin-8 and 14) and Theileria (Kinesin-8 and 13) that have just two kinesins in their genomes (also previously reported by (Wickstead and Gull, 2006)). Plasmodium has just three kinesins (Kinesin-5, 8 and 13). And there are several other organisms with just three or four kinesins (Fig. 2). Importantly, in all the cases of organisms with just two, three or four kinesins, all of them possess representatives from kinesin families implicated in controlling MT dynamics, i.e. Kinesin-4, 7, 8, 13 and 14. So it is possible that the earliest eukaryotic cells (not the LECA) were equipped principally with a set of kinesins in charge of controlling MT dynamics and that kinesins specific for transport functions did not appear until later in evolution through gene duplication and specialization. It is possible that the organisms with only 2-3 kinesins have lost some kinesins members during evolution and would, therefore, not exemplify the most ancient eukaryotes. However, it is also possible that organisms with more than four kinesins have gained kinesins during evolution. Most of these deep-rooted eukaryotes possessing a broad range of kinesins are pathogens, so it is possible that horizontal gene transfer occurred between the host and the pathogen, or even between different pathogens. There are some reported cases of gene transfer between eukaryotic pathogens, and between the pathogen and a host (Andersson, 2005, 2009; Keeling and Palmer, 2008; Alsmark et al., 2009; Bar, 2011; Selman et al., 2011). For these reasons, the protist that epitomizes this putative ancient eukaryote remains to be unambiguously identified.
One characteristic common to most of these deep-rooted eukaryotes is the existence of flagella, and also a cytoskeleton composed of unusually complex MT structures (e.g. Giardia, Trypanosoma, Plasmodium, etc) (Wickstead and Gull, 2011; Dawson and Paredez, 2013). In some cases their life cycle depends strongly on the proper assembly and function of these flagella, for which they need to tightly control MT length and dynamics. It is difficult to separate the existence of a flagellum from transport. The IFT (IntraFlagellar Transport) is a system to build and maintain the eukaryotic flagellum that relies on the kinesins' transport activity. As the LECA is likely to have possessed a full 9+2 flagellar apparatus, which would presumably require transport activity, it is probable that it had several kinesins in charge of the transport of cargo from the cytoplasm to the end of the flagellum (Yubuki and Leander, 2013). The origin of the flagellum is not yet clear (Carvalho-Santos et al., 2011; Yubuki and Leander, 2013), but one hypothesis proposes an autogenous origin of the flagellum from an MTOC organizing the mitotic spindle (Pickett-Heaps, 1974). C. merolae presents assembled MTs only during the formation of a mitotic spindle, suggesting that MTs first evolved to facilitate mitosis (Imoto et al., 2011). If the first eukaryotic cells with a flagellum did organize this structure from a mitotic spindle, it is possible that they were able to do it without the transport-kinesins required for IFT. For example, the pathogen Plasmodium doesn't have an IFT system and it is able to build a flagellum (Briggs et al., 2004). Moreover, the flagellar machinery is often co-opted to build the mitotic spindle during cell division in some of these deep-rooted eukaryotes.
It is possible that the very earliest eukaryotic cells, not the LECA, had just a minimum number of two or three kinesins in charge of MT dynamics to control mitotic spindle, cytoskeleton and an ancient flagellum. Other kinesins dedicated to transport may have appeared later in evolution as cells became larger and more specialized. More evolutionary cell biology research and more studies centering on these deep-rooted eukaryotes will be necessary to understand the nature and evolution of the kinesins controlling MT dynamics.
Acknowledgments
This work was supported by National Institutes of Health grant to L. Wordeman (GM69429).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Juan Jesus Vicente, Email: jjvr@uw.edu.
Linda Wordeman, Email: worde@uw.edu.
References
- Alsmark UC, Sicheritz-Ponten T, Foster PG, Hirt RP, Embley TM. Horizontal gene transfer in eukaryotic parasites: a case study of Entamoeba histolytica and Trichomonas vaginalis. Methods Mol Biol Clifton NJ. 2009;532:489–500. doi: 10.1007/978-1-60327-853-9_28. [DOI] [PubMed] [Google Scholar]
- Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO. Horizontal gene transfer between microbial eukaryotes. Methods Mol Biol Clifton NJ. 2009;532:473–487. doi: 10.1007/978-1-60327-853-9_27. [DOI] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar DZ. Evidence of Massive Horizontal Gene Transfer Between Humans and Plasmodium vivax. Nat Preced 2011 [Google Scholar]
- Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pagès M, Bastien P. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr Biol CB. 2007;17:778–782. doi: 10.1016/j.cub.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Briggs LJ, Davidge JA, Wickstead B, Ginger ML, Gull K. More than one way to build a flagellum: comparative genomics of parasitic protozoa. Curr Biol. 2004;14:R611–R612. doi: 10.1016/j.cub.2004.07.041. [DOI] [PubMed] [Google Scholar]
- Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303:1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell. 2009;20:1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, O'Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibres. Nat Cell Biol. 2009;11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Ersfeld K. The role of the Kinesin-13 family protein TbKif13-2 in flagellar length control of Trypanosoma brucei. Mol Biochem Parasitol. 2010;174:137–140. doi: 10.1016/j.molbiopara.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol. 2010;17:77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross RA, McAinsh A. Prime movers: the mechanochemistry of mitotic kinesins. Nat Rev Mol Cell Biol. 2014;15:257–271. doi: 10.1038/nrm3768. [DOI] [PubMed] [Google Scholar]
- Cui W, Sproul LR, Gustafson SM, Matthies HJG, Gilbert SP, Hawley RS. Drosophila Nod protein binds preferentially to the plus ends of microtubules and promotes microtubule polymerization in vitro. Mol Biol Cell. 2005;16:5400–5409. doi: 10.1091/mbc.E05-06-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SC, Paredez AR. Alternative cytoskeletal landscapes: cytoskeletal novelty and evolution in basal excavate protists. Curr Opin Cell Biol. 2013;25:134–141. doi: 10.1016/j.ceb.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SC, Sagolla MS, Mancuso JJ, Woessner DJ, House SA, Fritz-Laylin L, Cande WZ. Kinesin-13 regulates flagellar, interphase, and mitotic microtubule dynamics in Giardia intestinalis. Eukaryot Cell. 2007;6:2354–2364. doi: 10.1128/EC.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Domnitz SB, Wagenbach M, Decarreau J, Wordeman L. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J Cell Biol. 2012;197:231–237. doi: 10.1083/jcb.201108147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McAinsh AD. Exotic mitotic mechanisms. Open Biol. 2012;2:120140. doi: 10.1098/rsob.120140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol CB. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Erent M, Drummond DR, Cross RA. S. pombe kinesins-8 promote both nucleation and catastrophe of microtubules. PloS One. 2012;7:e30738. doi: 10.1371/journal.pone.0030738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertych N, Stolz A, Stenzinger A, Weichert W, Kaulfuβ S, Burfeind P, Aigner A, Wordeman L, Bastians H. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol. 2014;16:779–791. doi: 10.1038/ncb2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol CB. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ, Borisy GG. Microtubules of the kinetochore fiber turn over in metaphase but not in anaphase. J Cell Biol. 1989;109:653–662. doi: 10.1083/jcb.109.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ, Sammak PJ, Borisy GG. Chromosomes move poleward in anaphase along stationary microtubules that coordinately disassemble from their kinetochore ends. J Cell Biol. 1987;104:9–18. doi: 10.1083/jcb.104.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- Holt SV, Vergnolle MAS, Hussein D, Wozniak MJ, Allan VJ, Taylor SS. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J Cell Sci. 2005;118:4889–4900. doi: 10.1242/jcs.02614. [DOI] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Field CM, Mitchison TJ. KIF4 regulates midzone length during cytokinesis. Curr Biol CB. 2011;21:815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto Y, Yoshida Y, Yagisawa F, Kuroiwa H, Kuroiwa T. The cell cycle, including the mitotic cycle and organelle division cycles, as revealed by cytological observations. Microscopy. 2011;60:S117–S136. doi: 10.1093/jmicro/dfr034. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Rogers GC, Scholey JM. The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim Biophys Acta BBA - Mol Cell Res. 1997;1357:257–271. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith SL, Walczak CE. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol Biol Cell. 2002;13:2718–2731. doi: 10.1091/mbc.E01-12-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Davies T, Mishima M. Cytokinesis microtubule organisers at a glance. J Cell Sci. 2012;125:3495–3500. doi: 10.1242/jcs.094672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hümmer S, Bormann J, Grüner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol CB. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Mayr MI, Storch M, Howard J, Mayer TU. A non-motor microtubule binding site is essential for the high processivity and mitotic function of kinesin-8 Kif18A. PloS One. 2011;6:e27471. doi: 10.1371/journal.pone.0027471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda Y, Sato-Yoshitake R, Kondo S, Nangaku M, Hirokawa N. KIF2 is a new microtubule-based anterograde motor that transports membranous organelles distinct from those carried by kinesin heavy chain or KIF3A/B. J Cell Biol. 1995;129:157–167. doi: 10.1083/jcb.129.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Burbank K, Liu Q, Mitchison TJ. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr Biol CB. 2007;17:953–959. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Oh S, Hahn H, Torrey TA, Shin H, Choi W, Lee YM, Morse HC, Kim W. Identification of the human homologue of mouse KIF4, a kinesin superfamily motor protein. Biochim Biophys Acta. 2000;1493:219–224. doi: 10.1016/s0167-4781(00)00151-2. [DOI] [PubMed] [Google Scholar]
- Piao T, Luo M, Wang L, Guo Y, Li D, Li P, Snell WJ, Pan J. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc Natl Acad Sci U S A. 2009;106:4713–4718. doi: 10.1073/pnas.0808671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J. The evolution of mitosis and the eukaryotic condition. Biosystems. 1974;6:37–48. doi: 10.1016/0303-2647(74)90009-4. [DOI] [PubMed] [Google Scholar]
- Rankin KE, Wordeman L. Long astral microtubules uncouple mitotic spindles from the cytokinetic furrow. J Cell Biol. 2010;190:35–43. doi: 10.1083/jcb.201004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Roostalu J, Surrey T. The multiple talents of kinesin-8. Nat Cell Biol. 2013;15:889–891. doi: 10.1038/ncb2820. [DOI] [PubMed] [Google Scholar]
- Samejima K, et al. Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIα. J Cell Biol. 2012;199:755–770. doi: 10.1083/jcb.201202155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar HS, Luczak VG, Lopez MM, Lister BC, Gilbert SP. Mitotic kinesin CENP-E promotes microtubule plus-end elongation. Curr Biol CB. 2010;20:1648–1653. doi: 10.1016/j.cub.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar BT, Chan GK, Maddox P, Salmon ED, Yen TJ. CENP-E function at kinetochores is essential for chromosome alignment. J Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman M, Pombert JF, Solter L, Farinelli L, Weiss LM, Keeling P, Corradi N. Acquisition of an animal gene by microsporidian intracellular parasites. Curr Biol CB. 2011;21:R576–R577. doi: 10.1016/j.cub.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelden E, Wadsworth P. Microinjection of biotin-tubulin into anaphase cells induces transient elongation of kinetochore microtubules and reversal of chromosome-to-pole motion. J Cell Biol. 1992;116:1409–1420. doi: 10.1083/jcb.116.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slangy A, Lane HA, d' Hérin P, Harper M, Kress M, Niggt EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol CB. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A Tethering Mechanism Controls the Processivity and Kinetochore-Microtubule Plus-End Enrichment of the Kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev Cell. 2012;22:1017–1029. doi: 10.1016/j.devcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Ohi R. Kinesin-12 Differentially Affects Spindle Assembly Depending on Its Microtubule Substrate. Curr Biol. 2013;23:1280–1290. doi: 10.1016/j.cub.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Arellano-Santoyo H, Portran D, Gaillard J, Vantard M, Thery M, Pellman D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat Cell Biol. 2013;15:948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ohi R, Pellman D. Move in for the kill: motile microtubule regulators. Trends Cell Biol. 2012;22:567–575. doi: 10.1016/j.tcb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macůrek L, Janssen A, Geers EF, Alvarez-Fernández M, Medema RH. Kif15 Cooperates with Eg5 to Promote Bipolar Spindle Assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Uehara R, Tsukada Y, Kamasaki T, Poser I, Yoda K, Gerlich DW, Goshima G. Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J Cell Biol. 2013;202:623–636. doi: 10.1083/jcb.201302123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaart B van der, Akhmanova A, Straube A. Regulation of microtubule dynamic instability. Biochem Soc Trans. 2009;37:1007. doi: 10.1042/BST0371007. [DOI] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Walczak CE. The Kin I kinesins are microtubule end-stimulated ATPases. Mol Cell. 2003;11:286–288. doi: 10.1016/s1097-2765(03)00067-4. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Wandke C, et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J Cell Biol. 2012;198:847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Piao T, Cao M, Qin T, Huang L, Deng H, Mao T, Pan J. Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. J Cell Sci. 2013;126:1531–1540. doi: 10.1242/jcs.124255. [DOI] [PubMed] [Google Scholar]
- Wargacki MM, Tay JC, Muller EG, Asbury CL, Davis TN. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle Georget Tex. 2010;9:2581–2588. doi: 10.4161/cc.9.13.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, LeBlanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol CB. 2011;21:1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Gull K. A “Holistic” Kinesin Phylogeny Reveals New Kinesin Families and Predicts Protein Functions. Mol Biol Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic Cph Den. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. The evolution of the cytoskeleton. J Cell Biol. 2011;194:513–525. doi: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K, Richards TA. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol. 2010;10:110. doi: 10.1186/1471-2148-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife. 2013;2:e00290. doi: 10.7554/eLife.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood KW, Sakowicz R, Goldstein LS, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks handover-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- Yubuki N, Leander BS. Evolution of microtubule organizing centers across the tree of eukaryotes. Plant J Cell Mol Biol. 2013;75:230–244. doi: 10.1111/tpj.12145. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]