Abstract
Non-muscle myosin 2 (NM2) is a major force-producing, actin-based motor in mammalian non-muscle cells, where it plays important roles in a broad range of fundamental biological processes, including cytokinesis, cell migration, and epithelial barrier function.. This breadth of function at the tissue and cellular levels suggests extensive diversity and differential regulation of NM2 bipolar filaments, the major, if not sole, functional form of NM2s in vivo. Previous in vitro, cellular and animal studies indicate that some of this diversity is supported by the existence of multiple NM2 isoforms. Moreover, two recent studies have shown that these isoforms can co-assemble to form heterotypic filaments, further expanding functional diversity. In addition to isoform co-assembly, cells may differentially regulate NM2 function via isoform-specific expression, RLC phosphorylation, MHC phosphorylation or regulation via binding partners. Here, we provide a brief summary of NM2 filament assembly, summarize the recent findings regarding NM2 isoform co-assembly, consider the mechanisms cells might utilize to differentially regulate NM2 isoforms, and review the data available to support these mechanisms.
Keywords: nonmuscle myosin II, isoform, filament assembly
Class 2 myosins consist of skeletal, cardiac, and smooth muscle myosins and nonmuscle myosin 2 (NM2). Consistent with their names, the expression of the three muscle myosins is restricted largely to their respective muscle tissue. In contrast, NM2 is present in most, if not all, differentiated mammalian cell types, where it assembles into bipolar filaments that engage actin filaments of opposing polarity. These NM2 filaments, although smaller than their muscle brethren, drive contractile events that are not unlike those that shorten the sarcomere in muscle. A plethora of studies have firmly established a core set of fundamental cell biological processes that are driven by NM2-dependent contractions. These include cell division (specifically cytokinesis [70]), cell adhesion (both cell: substrate [44] and cell: cell adhesion [18;36], cell migration [25;63], and tissue morphogenesis [11;27]). Sites where these myosins are critical for proper structure and/or function run the gamut from whole tissues (e.g. epithelia [23]) to specific subcellular compartments (e.g. the dendritic spines of neurons [34;81]). Moreover, NM2 exerts its influence at multiple organizational levels within the cell, from specific actin structures like stress fibers [75] and the contractile ring [70;71], to global structures like the viscoelastic cytoplasm [31]. Layered on top of these general functions are an expanding set of specialized functions, including roles in vesicle fission at the TGN [53], mitochondrial fission [43], and tumor progression via interactions with p53 [66]. This vast functional diversity begs the question: what variations in NM2 structure and function support this functional diversity? Relevant to this question, two recent studies have demonstrated in unequivocal fashion that the three NM2 isoforms expressed in mammals co-assemble to form mixed bipolar filaments, providing one mechanism by which filament diversity can be expanded. Here, we review these two studies and discuss their implications with regard to how cells might employ multiple NM2 isoforms to drive diverse cellular functions.
In mammals, three genes (MYH9, MYH10, and MYH14) encode three nonmuscle myosin heavy chain (NMHC) proteins (NMHC 2A, NMHC 2B, and NMHC 2C, respectively). Because NMHC dimerization during hexameric holoenzyme formation is strictly homophilic [29;39], cells contain up to three NM2 monomer pools (NM2A, NM2B, NM2C) from which bipolar filaments can be assembled. In contrast, many lower eukaryotes, including Dictyostelium discoideum, Drosophila melanogaster, and Saccaromyces cerevisiae, possess a single NM HC gene. This fact raises the possibility that the three mammalian isoforms are the product of simple gene duplications whose resultant proteins are performing redundant functions. Indeed, all three isoforms appear competent to function during cytokinesis [4]. However, studies in cell lines and mice have demonstrated isoform-specific, as well as isoform-redundant, functions for the three NM2 isoforms (reviewed in [78;79]). Additionally, while the biophysical properties of the three NM2 isoforms are more similar to one another than to muscle myosins, they still display significant differences between themselves in terms of actin-activated ATPase activity, duty ratio, and filament dynamics [12;29;58;80]. Together, these observations argue that multiple NM2 isoforms have evolved in mammalian cells to deal with the diversity of functions that must be supported by this class of myosin.
Full understanding of the physiological roles played by individual NM2 isoforms will require full understanding of the composition of the filaments they form. Until recently, no in vitro or in vivo study has resolved in definitive fashion the basic question of whether NM2 isoforms assemble in living cells to produce mixed (heterotypic) filaments, or whether filaments consist entirely of a single isoform (homotypic). Previous analyses of NM2 assembly have provided some insight into this question. NM2 filament assembly is regulated by the conversion of folded, inactive monomers into extended, assembly-competent molecules following phosphorylation of the myosin's regulatory light chain (RLC). Once in the assembly-competent state, the coiled-coil tail domains of individual NM2 monomers interact with each other in both parallel and anti-parallel fashion to drive the assembly of bipolar filaments. These tail: tail interactions are mediated largely by electrostatic attractions between a C-terminal, positively-charged “assembly-competent domain” and complementary, negatively-charged surfaces in upstream regions of the tail [61]. The bands of surface-exposed charges that drive assembly occur in repetitive, alternating fashion, making their patterns amenable to Fourier analyses. Such analyses indeed show similar patterns for all three NM2 isoforms, supporting the possibility that they form heterotypic filaments [12;72].
Multiple studies have provided more direct evidence for isoform co-assembly. Specifically, Mitsuhashi and colleagues demonstrated heterotypic filament formation in vitro using purified tail fragments [54]. We note, however, that these tail fragments undergo several non-physiological forms of self-association, arguing that this data should be interpreted cautiously [61;62]. Moreover, expressing similar tail fragments as dominant negative constructs in cells provided data that actually favored homotypic filament formation [65]. To date, no evidence for co-assembly has been reported using purified, full-length NM2 isoforms. That said, early EM studies using purified platelet myosin (mostly NM2A), skeletal muscle myosin and smooth muscle myosin showed that these myosins could co-assemble into filaments in vitro [60], suggesting the three NM2 isoforms might do the same. With regard to studies in living cells, while a number of studies have provided data that is consistent with co-assembly, none have provided definitive proof. These data include the co-immunoprecipiation of NM2A and NM2B [51], the NM2B-dependent recruitment of the tail domain of NM2Ato the contractile ring [6], and the “co-localization” of different NM2 isoforms in confocal micrographs [7;23;41;51]. Importantly, such co-localization does not prove co-assembly into heterotypic filaments, as non-super resolution imaging typically cannot resolve in unequivocal fashion single bipolar filaments, whose length (∼300 nm) [12] is near the resolution limit of non-super resolution microscopy (although see [23]). Therefore, addressing NM2 isoform co-assembly in living cells requires the use of imaging techniques with resolutions beyond the limits of conventional light microscopy.
Recent advances in light microscopy, such as structured-illumination microscopy (SIM) [26], have enabled multi-color imaging in live cells at resolutions well beyond those attainable using standard light microscopy. Specifically, SIM can achieve a lateral resolution of ∼130 nm (∼100 nm with a high-NATIRF objective), sufficient to identify individual NM2 filaments and, with the appropriate tools, to discern their isoform composition. Immuno-electron microscopy has, of course, the ability to do the same at even higher resolution. Using these imaging modalities, two groups recently tackled the basic question posed above regarding the possible co-assembly of NM2 isoforms [9;68]. We next summarize the three main, shared findings presented in these two papers.
NM2 isoforms assemble into mixed bipolar filaments in living cells
To look for NM2 isoform co-assembly in living cells, Beach et al. [9] used exogenously-expressed, fluorescently-tagged versions of NM2A, NM2B and NM2C, isoform-specific antibodies against the endogenous proteins, and two-color TIRF-SIM imaging to visualize individual NM2 bipolar filaments. These efforts demonstrated unequivocally that NM2 isoforms (both expressed and endogenous) readily co-assemble into heterotypic filaments in a variety of settings within living cells. These settings included various types of stress fibers (e.g. ventral, sub nuclear, transverse arcs), individual filaments throughout the cytoplasm, and the contractile ring of dividing cells (see Figure 1A and 1B for an example and the legend for an explanation of the approach and techniques used). To look for NM2 isoform co-assembly in cells, Shutova et al. [68] used platinum replica electron microscopy (PREM) to image individual NM2 bipolar filaments in fixed, extracted cells, together with immuno-gold labeling to determine the isoform composition of individual filaments. As with the SIM images in Beach et al. [9], Shutova's electron micrographs showed clearly that endogenous NM2A and NM2B co-assemble into heterotypic filaments in living cells (see Figure 1C for examples).
Figure 1.
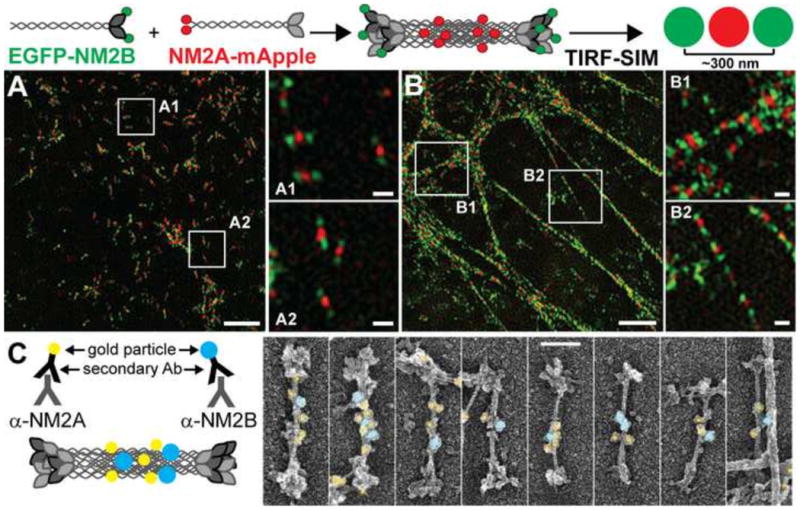
Evidence for NM2 isoform co-assembly. (A and B) The cartoon at the top depicts one experimental approach used by Beach et al. [9] to demonstrate NM2 isoform co-assembly. Briefly, NM2B was tagged on its N-terminus with EGFP, while NM2A was tagged on its C-terminus with mApple. Heterotypic filaments that formed in U20S cells expressing these two tagged myosins were identified in TIRF-SIM images as two green puncta ∼300 nm apart bifurcated by a single red punctum. These structures were readily apparent in TIRF-accessible regions, where their distribution varied from relatively random (A) to aligned within stress fibers (B). Images were reprinted with permission from [9] and correspond to Fig. 2B and Fig. 2D, respectively. Scale bars represent 2μm for the larger images and 300 nm for the insets. (C) The cartoon depicts the experimental approach used by Shutova et al. [68] to demonstrate NM2 isoform co-assembly. Briefly, detergent-extracted REF52 fibroblast cells were immuno-stained with primary antibodies that recognize specifically the C-terminus of NM2A or NM2B. Secondary antibodies conjugated to 12 nm (NM2A, pseudo-colored yellow) or 18 nm (NM2B, pseudo-colored blue) gold beads were then used to identify the respective NM2 isoform. Heterotypic filaments containing both NM2A and NM2B were clearly visible in platinum replica EM images. Images were reprinted with permission from [68] and correspond to Fig. 3A. The scale bar represents 100 nm.
The differential distribution of NM2A and NM2B in well polarized cells is reflected in the composition of individual filaments
Using TIRF-SIM, Beach et al. [9] found that the fractional content of NM2A and NM2B within individual bipolar filaments in well polarized cells mirrored the differential distribution of these two isoforms typically seen in lower resolution images [41], with NM2A enriched peripherally and NM2B enriched centrally. Specifically, while NM2A dominated filaments in peripheral lamella, the ratio of NM2A to NM2B within filaments diminished progressively moving deeper into the cytoplasm, eventually favoring NM2B for interior filaments. Moreover, this result was seen for both endogenous NM2A/NM2B and exogenous, expressed NM2A/NM2B. Importantly, similar results for endogenous NM2A and NM2B were presented by Shutova et al. [68] based on correlation analyses of STED images of well polarized, detergent-extracted cells immuno-stained with isoform-specific antibodies to the C-termini of NM2A and NM2B.
The differential distribution of NM2A and NM2B is driven by a sorting mechanism acting over time
Interestingly, Beach et al. [9] found that the degree of “co-localization” between exogenously-expressed NM2A and NM2B in confocal micrographs was much stronger in freshly spread cells generating nascent lamella than in well-polarized cells. Importantly, TIRF-SIM revealed that this increase in signal overlap was reflected in the composition of individual bipolar filaments, with the relative ratio of NM2A to NM2B within filaments in freshly spread cells remaining much closer to 1 at nearly all distances from the cell edge. In other words, there was a much more even distribution of NM2A and NM2B within individual filaments in freshly-spread cells compared to well-polarized cells. Importantly, Shutova et al. [68] obtained similar results for endogenous NM2A and NM2B using correlation analyses of STED images of detergent-extracted, immuno-stained cells in which NM2 filament formation had been synchronized by treating cells with blebbistatin and then washing the drug out. Together, these observations indicate that both NM2A and NM2B readily co-assemble upon initiation of large-scale bipolar filament assembly, such as following cell attachment or release from blebbistatin treatment. These observations also argue that the differential distribution of NM2A and NM2B seen at steady state in well-polarized cells must arise from a sorting mechanism that functions over time and involves different rates of filament turnover for NM2A and NM2B, rather than from the sequential incorporation of these two isoforms into contractile arrays [77]. A number of significant biophysical differences between NM2A and NM2B, including differences in their stability within filaments and in their duty ratio, probably drive this sorting mechanism (see Beach et al. [9] and Shutova et al. [68]for further discussion). This model is consistent with the possibility that in well polarized cells, anterior NM2A-dominant filaments may be precursors to the NM2B-dominant filaments that form the posterior contractile apparatus [77].
The overarching observation made by these two studies - that individual NM2 isoforms form heterotypic filaments when they intermingle in the cytoplasm - has broad implications for efforts to assign specific cellular functions to specific NM2 isoforms. Most importantly, it argues that many functions attributed to NM2 may be being performed by heterotypic filaments. Critically, the properties of these mixed filaments are unknown and potentially complex. For example, do the properties of mixed filaments scale directly with fractional isoform content, or can one isoform dominate the properties of the mixed filament? Characterization of the biophysical properties of heterotypic filaments made using purified, full-length NM2 isoforms will be required to address these questions.
Layered over top of these issues are possible mechanisms used by cells to control the relative functional contributions made by NM2 isoforms. Specifically, while the two studies highlighted here demonstrate that NM2 isoforms co-assemble in cells, isoform-specific functions have been observed both in tissue culture settings and in the developing animal [5;79]. The realization that NM2 isoforms co-assemble into heterotypic filaments, yet remain capable of isoform-specific functions, poses an interesting dilemma. How do cells accomplish this feat? While it is certainly possible that these isoform-specific functions are accomplished by heterotypic filaments, it is also likely that cells possess mechanisms to differentially activate and assemble NM2 isoforms. We consider four such mechanisms below.
Select isoform expression
The first and most obvious mechanism involves the regulation of expression levels by controlling the transcription, translation, and/or turnover of the individual isoforms. Although NM2A and NM2B are expressed nearly ubiquitously [38], isoform specific-expression at the cellular level has been observed. For example, hematopoietic cells express almost exclusively NM2A (although knockout/replacement studies have not been performed to demonstrate that the NM2A isoform is specifically required for cell function) [56]. Additionally, isoform-specific expression was reported during epithelial-mesenchymal transition in mammary epithelial cells, in which epithelial cells expressing NM2A and NM2C switched their expression profile to NM2A and NM2B during their transition to a mesenchymal phenotype [7]. In this model system, the regulation of NM2 expression was downstream of TGFβ and hnRNPEl. Other reports have recognized isoform-specific expression of NM2 isoforms without exploring the underlying mechanism [13;79]. Various studies by Kawamoto and colleagues have made progress characterizing the promoter and enhancer elements for the human NMHC 2A gene (Myh9) [10;16;17] and have identified IRF-2 as a transcription factor capable of up-regulating NMHC 2A expression following phorbolester treatment [16]. Similar data are not available for NMHC 2B or NMHC 2C, and the general applicability of the reported mechanisms for NMHC 2A (TGFβ/hnRNPEl and IRF-2) have not been explored in other cells and tissues. Therefore, the mechanisms that regulate the isoform-specific expression of NM2 isoforms largely remain unknown.
Isoform-specific RLC phosphorylation
Another mechanism by which cells might differentially regulate NM2 isoforms is via differential RLC phosphorylation, although how RLC kinases and/or phosphatase would differentially recognize NM2 isoforms is not clear. A previous report on myosin light chain kinase (MLCK), the most widely-studied RLC kinase, revealed MLCK from smooth muscle could phosphorylate SM2 but not skeletal muscle myosin 2 [73], setting a precedent for differential recognition of myosins by RLC kinases. Additional studies have demonstrated that MLCK primarily recognizes two regions: (1) the N-terminus of the RLC, where it phosphorylates the activating residue S19, and (2) the head-tail junction of MHC [28;69]. Analysis of these two regions may provide some insight into whether or not MLCK could differentially recognize NM2 isoforms. First, while the sequences immediately surrounding the activating residues (i.e. T18 and S19) are identical in the three smooth/nonmuscle myosin RLC isoforms, there is significant sequence divergence just upstream of this region near the very N-terminus, raising the possibility that different RLC isoforms may be differentially recognized. For this to affect NM2 isoforms, however, the different RLC isoforms would have to bind differentially to the NM HC isoforms, which has not been reported. That said, it remains a possibility given that the RLC-binding IQ2 domain is not entirely conserved between NMHC isoforms. As for the region of the head-tail junction recognized by MLCK, while the sequence immediately upstream of this junction is largely conserved, the sequence immediately downstream of this junction shows significant sequence divergence in NM2 isoforms. Therefore, if MLCK does differentially recognize NM2 isoforms, this region may play a critical role. Myosin phosphatase may also recognize both the RLC and portions of the MHC, providing an additional opportunity for isoform-specific regulation of RLC phosphorylation. The data on this is fairly ambiguous, however [32;52]. Unfortunately, interactions between other RLC kinases and myosin 2 (either SM2 or NM2) are largely unexplored. It is worth noting that the list of kinases capable of phosphorylating RLC at the activation sites continues to grow (rho kinase, citron kinase, zip kinase, MRCK, etc.). Some of these kinases may very well prove to be isoform-specific, or at least more active towards certain isoforms, while others may not discriminate. Finally, there is little data that addresses isoform-specific activation in living cells. That said, a study by Means and colleagues reported that thrombin treatment of MDA-MB-231 breast cancer cells resulted in the preferential ROCK-mediated phosphorylation of S19 on RLC bound to NM2A over NM2B [64].
The RLC can also be phosphorylated at S1, S2 and T9 by PKC. In vitro data shows that this phosphorylation directly decreases the myosin's actin-activated ATPase activity and inhibits the interaction between MCLK and RLC, further lowering the activation state. The effect of this putative inhibitory phosphorylation on NM2 in cells remains unclear [2;8;42]. Collectively, activating and inhibiting phosphorylations of the RLC represent potential but unproven mechanisms for isoform-specific NM2 regulation.
Isoform-specific MHC phosphorylation
Cells expressing multiple NM2 isoforms might utilize their functions differentially through MHC phosphorylation (see [20] for thorough review on MHC phosphorylation). Despite studies showing that MHC phosphorylation is the primary mechanism regulating filament assembly in the model organisms Dictyostelium discoideum and Acanthamoeba castelanii [24;50;76], as well as studies beginning in the late 1970's showing that mammalian MHCs could be phosphorylated [33;55], a deeper understanding of this regulatory mechanism in mammalian cells has been somewhat slow in coming. This is perhaps due in part to neglect following the discovery in the mid-1970's that the assembly of NM2 into filaments, as well as its actin-activated ATPase activity, were both dramatically activated by RLC phosphorylation [1;35]. Whatever the reason, the effects of NMHC phosphorylation have only recently been characterized and their in vivo significance remains incompletely understood. Studies using purified rod fragments have consistently shown that NMHC phosphorylation shifts the equilibrium toward the disassembled, monomeric state [22;57]. This is not surprising considering many of these phosphorylation sites are in or near regions in the tail necessary for bipolar filament formation [46;59;67]. While these in vitro studies argue that NMHC phosphorylation is inhibitory in nature, multiple cellular studies have shown that NMHC phosphorylation may be necessary for NM2 filament turnover and recycling [14;21]. Therefore, if NMHC kinases and/or phosphatases differentially recognize or act on NMHC isoforms, and if NMHC phosphorylation differentially alters the equilibrium between monomer and polymer for different isoforms, then this could represent a significant mechanism for regulating filament composition in an isoform-specific manner. At first glance, one might argue that this is not the simplest of regulatory mechanisms, as all three NM2 isoforms have putative phosphorylation sites for the three recognized mammalian NMHC kinases (PKC, CK2, and TRPM7) [20]. Similar to RLC phosphorylation, differential recognition by the kinases and phosphatases for the NMHC isoforms may prove critical. In the end, increased appreciation and investigation of NMHC phosphorylation may uncover mechanisms for isoform-specific regulation.
Isoform-specific binding partners
Finally, it remains possible that isoform-specific binding partners regulate NM2 isoform localization and assembly levels. S100A4, or metastasin (mtsl), is the best characterized NM2 binding partner (reviewed in [20]) and has been shown to enhance cell migration and metastasis in a number of cell types [30;47;48;74]. S100A4 preferentially binds NM2A, and possibly NM2C, in a Ca2+-dependent manner and drives filament depolymerization, probably by unwinding the coiled-coil of individual monomers and destabilizing intermolecular interactions within the filament [3;40;47;49;54]. More recently, another S100 family member (S100P), which has a similar -10-fold preference for NM2A and NM2C over NM2B, was shown to dissociate preformed filaments in vitro, and to enhance cell migration [19]. Another recent study suggested that the Arf GEFs BIG1 and BIG2 can form a multimolecular complex with myosin phosphatase and with NM2A, but not with NM2B or NM2C [45]. In this role, BIG1/BIG2 appear to be negative regulators of NM2A, as suppression of BIG proteins resulted in a decrease in NM2A: myosin phosphatase interaction and an increase in RLC-T18/S19 phosphorylation. It is not clear if other NM2 binding partners (anillin, supervillin, sept2, etc.) [15;37;71] have any preference for NM2 isoforms, and no regulatory proteins have been identified for NM2B. Collectively, NM2 binding partners represent an attractive mechanism for regulating NM2 isoform-specific functions in vivo.
Conclusion
The recent, unequivocal demonstration that NM2 isoforms co-assemble into mixed bipolar filaments in cells [9;68] will further complicate efforts to parse out the unique and redundant functions performed by the three mammalian NM2 isoforms. Further characterization of the mechanochemical properties of these heterotypic filaments, and more extensive efforts to define the locations within cells where they form, are required. These efforts will be further bolstered by in vitro and in vivo studies focused on clarifying the pathways by which the assembly and activity of NM2 isoforms can be differentially regulated. Together, these efforts should lead to a deeper, mechanistic understanding of how NM2 isoforms are capable of supporting such a vast array of biological processes.
Highlights.
-
-
The breadth of NM2-supported processes infers functional diversity of NM2 filaments
-
-
The ability of NM2 isoforms to co-assemble expands this functional diversity
-
-
Cells may also differentially regulate NM2 isoforms using a variety of mechanisms
Acknowledgments
The authors would like to thank Kirsten Remmert (NHLBI) for artistic contributions to the figure, Jim Sellers (NHLBI) for helpful discussions, and Tom Egelhoff (CCF) for critical reading of the manuscript. This work was supported by NIH DIR funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Adelstein RS, Conti MA. Phosphorylation of platelet myosin increases actin-activated myosin ATPase activity. Nature. 1975;256:597–598. doi: 10.1038/256597a0. [DOI] [PubMed] [Google Scholar]
- 2.Asokan SB, Johnson HE, Rahman A, King SJ, Rotty JD, Lebedeva IP, Haugh JM, Bear JE. Mesenchymal Chemotaxis Requires Selective Inactivation of Myosin II at the Leading Edge via a Noncanonical PLCgamma/PKCalpha Pathway. Dev Cell. 2014;31:747–760. doi: 10.1016/j.devcel.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badyal SK, Basran J, Bhanji N, Kim JH, Chavda AP, Jung HS, Craig R, Elliott PR, Irvine AF, Barsukov IL, Kriajevska M, Bagshaw CR. Mechanism of the Ca(2)+-dependent interaction between S100A4 and tail fragments of nonmuscle myosin heavy chain MA. J Mol Biol. 2011;405:1004–1026. doi: 10.1016/j.jmb.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao J, Jana SS, Adelstein RS. Vertebrate nonmuscle myosin II isoforms rescue small interfering RNA-induced defects in COS-7 cell cytokinesis. J Biol Chem. 2005;280:19594–19599. doi: 10.1074/jbc.M501573200. [DOI] [PubMed] [Google Scholar]
- 5.Bao J, Ma X, Liu C, Adelstein RS. Replacement of nonmuscle myosin II-B with II-A rescues brain but not cardiac defects in mice. J Biol Chem. 2007;282:22102–22111. doi: 10.1074/jbc.M702731200. [DOI] [PubMed] [Google Scholar]
- 6.Beach JR, Egelhoff TT. Myosin II recruitment during cytokinesis independent of centralspindlin-mediated phosphorylation. J Biol Chem. 2009;284:27377–27383. doi: 10.1074/jbc.M109.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beach JR, Hussey GS, Miller TE, Chaudhury A, Patel P, Monslow J, Zheng Q, Keri RA, Reizes O, Bresnick AR, Howe PH, Egelhoff TT. Myosin II isoform switching mediates invasiveness after TGF-beta-induced epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108:17991–17996. doi: 10.1073/pnas.1106499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beach JR, Licate LS, Crish JF, Egelhoff TT. Analysis of the role of Serl/Ser2/Thr9 phosphorylation on myosin II assembly and function in live cells. BMC Cell Biol. 2011;12:52. doi: 10.1186/1471-2121-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beach JR, Shao L, Remmert K, Li D, Betzig E, Hammer JA., III Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beohar N, Kawamoto S. Transcriptional regulation of the human nonmuscle myosin II heavy chain-A gene. Identification of three clustered cis-elements in intron-1 which modulate transcription in a cell type- and differentiation state-dependent manner. J Biol Chem. 1998;273:9168–9178. doi: 10.1074/jbc.273.15.9168. [DOI] [PubMed] [Google Scholar]
- 11.Bertet C, Sulak L, Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- 12.Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bond JE, Ho TQ, Selim MA, Hunter CL, Bowers EV, Levinson H. Temporal spatial expression and function of non-muscle myosin II isoforms MA and MB in scar remodeling. Lab Invest. 2011;91:499–508. doi: 10.1038/labinvest.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breckenridge MT, Dulyaninova NG, Egelhoff TT. Multiple regulatory steps control mammalian nonmuscle myosin II assembly in live cells. Mol Biol Cell. 2009;20:338–347. doi: 10.1091/mbc.E08-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Takizawa N, Crowley JL, Oh SW, Gatto CL, Kambara T, Sato O, Li XD, Ikebe M, Luna EJ. F-actin and myosin II binding domains in supervillin. J Biol Chem. 2003;278:46094–46106. doi: 10.1074/jbc.M305311200. [DOI] [PubMed] [Google Scholar]
- 16.Chung MC, Kawamoto S. IRF-2 is involved in up-regulation of nonmuscle myosin heavy chain 11-A gene expression during phorbol ester-induced promyelocytic HL-60 differentiation. J Biol Chem. 2004;279:56042–56052. doi: 10.1074/jbc.M404791200. [DOI] [PubMed] [Google Scholar]
- 17.Chung MC, Kim HK, Kawamoto S. TFEC can function as a transcriptional activator of the nonmuscle myosin II heavy chain-A gene in transfected cells. Biochemistry. 2001;40:8887–8897. doi: 10.1021/bi002847d. [DOI] [PubMed] [Google Scholar]
- 18.Conti MA, Even-Ram S, Liu C, Yamada KM, Adelstein RS. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J Biol Chem. 2004;279:41263–41266. doi: 10.1074/jbc.C400352200. [DOI] [PubMed] [Google Scholar]
- 19.Du M, Wang G, Ismail TM, Gross S, Fernig DG, Barraclough R, Rudland PS. S100P dissociates myosin MA filaments and focal adhesion sites to reduce cell adhesion and enhance cell migration. J Biol Chem. 2012;287:15330–15344. doi: 10.1074/jbc.M112.349787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dulyaninova NG, Bresnick AR. The heavy chain has its day: regulation of myosin-II assembly. Bioarchitecture. 2013;3:77–85. doi: 10.4161/bioa.26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18:3144–3155. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulyaninova NG, Malashkevich VN, Almo SC, Bresnick AR. Regulation of myosin-IIA assembly and Mtsl binding by heavy chain phosphorylation. Biochemistry. 2005;44:6867–6876. doi: 10.1021/bi0500776. [DOI] [PubMed] [Google Scholar]
- 23.Ebrahim S, Fujita T, Millis BA, Kozin E, Ma X, Kawamoto S, Baird MA, Davidson M, Yonemura S, Hisa Y, Conti MA, Adelstein RS, Sakaguchi H, Kachar B. NMII forms a contractile transcellular sarcomeric network to regulate apical cell junctions and tissue geometry. Curr Biol. 2013;23:731–736. doi: 10.1016/j.cub.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- 25.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin MA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 26.Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MG. Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc Natl Acad Sci U S A. 2012;109:5311–5315. doi: 10.1073/pnas.1119262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franke JD, Montague RA, Kiehart DP. Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol. 2005;15:2208–2221. doi: 10.1016/j.cub.2005.11.064. [DOI] [PubMed] [Google Scholar]
- 28.Gallagher PJ, Herring BP, Stull JT. Myosin light chain kinases. J Muscle Res Cell Motil. 1997;18:1–16. doi: 10.1023/a:1018616814417. [DOI] [PubMed] [Google Scholar]
- 29.Golomb E, Ma X, Jana SS, Preston YA, Kawamoto S, Shoham NG, Goldin E, Conti MA, Sellers JR, Adelstein RS. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J Biol Chem. 2004;279:2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- 30.Grum-Schwensen B, Klingelhofer J, Berg CH, EI-Naaman C, Grigorian M, Lukanidin E, Ambartsumian N. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mtsl) gene. Cancer Res. 2005;65:3772–3780. doi: 10.1158/0008-5472.CAN-04-4510. [DOI] [PubMed] [Google Scholar]
- 31.Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, Lippincott-Schwartz J, Mackintosh FC, Weitz DA. Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell. 2014;158:822–832. doi: 10.1016/j.cell.2014.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartshorne DJ, Ito M, Erdodi F. Role of protein phosphatase type 1 in contractile functions: myosin phosphatase. J Biol Chem. 2004;279:37211–37214. doi: 10.1074/jbc.R400018200. [DOI] [PubMed] [Google Scholar]
- 33.Hesketh JE, Virmaux N, Mandel P. Evidence for a cyclic nucleotide-dependant phosphorylation of retinal myosin. FEBS Lett. 1978;94:357–360. doi: 10.1016/0014-5793(78)80976-4. [DOI] [PubMed] [Google Scholar]
- 34.Hodges JL, NeweII-Litwa K, Asmussen H, Vicente-Manzanares M, Horwitz AR. Myosin IIb activity and phosphorylation status determines dendritic spine and post-synaptic density morphology. PLoS One. 2011;6:e24149. doi: 10.1371/journal.pone.0024149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikebe M, Hartshorne DJ, Elzinga M. Phosphorylation of the 20,000-dalton light chain of smooth muscle myosin by the calcium-activated, phospholipid-dependent protein kinase. Phosphorylation sites and effects of phosphorylation. J Biol Chem. 1987;262:9569–9573. [PubMed] [Google Scholar]
- 36.Ivanov AI, Hunt D, Utech M, Nusrat A, Parkos CA. Differential roles for actin polymerization and a myosin II motor in assembly of the epithelial apical junctional complex. Mol Biol Cell. 2005;16:2636–2650. doi: 10.1091/mbc.E05-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joo E, Surka MC, Trimble WS. Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev Cell. 2007;13:677–690. doi: 10.1016/j.devcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Kawamoto S, Adelstein RS. Chicken nonmuscle myosin heavy chains: differential expression of two mRNAs and evidence for two different polypeptides. J Cell Biol. 1991;112:915–924. doi: 10.1083/jcb.112.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley CA, Sellers JR, Gard DL, Bui D, Adelstein RS, Baines IC. Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J Cell Biol. 1996;134:675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiss B, Duelli A, Radnai L, Kekesi KA, Katona G, Nyitray L. Crystal structure of the S100A4-nonmuscle myosin MA tail fragment complex reveals an asymmetric target binding mechanism. Proc Natl Acad Sci U S A. 2012;109:6048–6053. doi: 10.1073/pnas.1114732109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolega J. Cytoplasmic dynamics of myosin MA and MB: spatial 'sorting' of isoforms in locomoting cells. J Cell Sci. 1998;111(Pt 15):2085–2095. doi: 10.1242/jcs.111.15.2085. [DOI] [PubMed] [Google Scholar]
- 42.Komatsu S, Ikebe M. The phosphorylation of myosin M at the Ser1 and Ser2 is critical for normal platelet-derived growth factor induced reorganization of myosin filaments. Mol Biol Cell. 2007;18:5081–5090. doi: 10.1091/mbc.E06-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korobova F, Gauvin TJ, Higgs HN. A role for myosin M in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo JC, Han X, Hsiao CT, Yates JR, III, Waterman CM. Analysis of the myosin-M-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le K, Li CC, Ye G, Moss J, Vaughan M. Arf guanine nucleotide-exchange factors BIG1 and BIG2 regulate nonmuscle myosin MA activity by anchoring myosin phosphatase complex. Proc Natl Acad Sci U S A. 2013;110:E3162–E3170. doi: 10.1073/pnas.1312531110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee RJ, Egelhoff TT, Spudich JA. Molecular genetic truncation analysis of filament assembly and phosphorylation domains of Dictyostelium myosin heavy chain. J Cell Sci. 1994;107(Pt 10):2875–2886. doi: 10.1242/jcs.107.10.2875. [DOI] [PubMed] [Google Scholar]
- 47.Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- 48.Li ZH, Dulyaninova NG, House RP, Almo SC, Bresnick AR. S100A4 regulates macrophage Chemotaxis. Mol Biol Cell. 2010;21:2598–2610. doi: 10.1091/mbc.E09-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li ZH, Spektor A, Varlamova O, Bresnick AR. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42:14258–14266. doi: 10.1021/bi0354379. [DOI] [PubMed] [Google Scholar]
- 50.Luck-Vielmetter D, Schleicher M, Grabatin B, Wippler J, Gerisch G. Replacement of threonine residues by serine and alanine in a phosphorylatable heavy chain fragment of Dictyostelium myosin II. FEBS Lett. 1990;269:239–243. doi: 10.1016/0014-5793(90)81163-i. [DOI] [PubMed] [Google Scholar]
- 51.Marini M, Bruschi M, Pecci A, Romagnoli R, Musante L, Candiano G, Ghiggeri GM, Balduini C, Seri M, Ravazzolo R. Non-muscle myosin heavy chain MA and MB interact and co-localize in living cells: relevance for MYH9-related disease. Int J Mol Med. 2006;17:729–736. [PubMed] [Google Scholar]
- 52.Matsumura F, Hartshorne DJ. Myosin phosphatase target subunit: Many roles in cell function. Biochem Biophys Res Commun. 2008;369:149–156. doi: 10.1016/j.bbrc.2007.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12:645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- 54.Mitsuhashi M, Sakata H, Kinjo M, Yazawa M, Takahashi M. Dynamic assembly properties of nonmuscle myosin II isoforms revealed by combination of fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy. J Biochem. 2011;149:253–263. doi: 10.1093/jb/mvq134. [DOI] [PubMed] [Google Scholar]
- 55.Muhlrad A, Oplatka A. Phosphorylation of fibroblast myosin. FEBS Lett. 1977;77:37–40. doi: 10.1016/0014-5793(77)80188-9. [DOI] [PubMed] [Google Scholar]
- 56.Murakami N, Mehta P, Elzinga M. Studies on the distribution of cellular myosin with antibodies to isoform-specific synthetic peptides. FEBS Lett. 1991;278:23–25. doi: 10.1016/0014-5793(91)80074-d. [DOI] [PubMed] [Google Scholar]
- 57.Murakami N, Singh SS, Chauhan VP, Elzinga M. Phospholipid binding, phosphorylation by protein kinase C, and filament assembly of the COOH terminal heavy chain fragments of nonmuscle myosin II isoforms MIIA and MIIB. Biochemistry. 1995;34:16046–16055. doi: 10.1021/bi00049a019. [DOI] [PubMed] [Google Scholar]
- 58.Nagy A, Takagi Y, Billington N, Sun SA, Hong DK, Homsher E, Wang A, Sellers JR. Kinetic characterization of nonmuscle myosin IIb at the single molecule level. J Biol Chem. 2013;288:709–722. doi: 10.1074/jbc.M112.424671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakasawa T, Takahashi M, Matsuzawa F, Aikawa S, Togashi Y, Saitoh T, Yamagishi A, Yazawa M. Critical regions for assembly of vertebrate nonmuscle myosin II. Biochemistry. 2005;44:174–183. doi: 10.1021/bi048807h. [DOI] [PubMed] [Google Scholar]
- 60.Pollard TD. Electron microscopy of synthetic myosin filaments. Evidence for cross-bridge. Flexibility and copolymer formation. J Cell Biol. 1975;67:93–104. doi: 10.1083/jcb.67.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ricketson D, Johnston CA, Prehoda KE. Multiple tail domain interactions stabilize nonmuscle myosin II bipolar filaments. Proc Natl Acad Sci U S A. 2010;107:20964–20969. doi: 10.1073/pnas.1007025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenberg M, Straussman R, Ben-Ya'acov A, Ronen D, Ravid S. MHC-IIB filament assembly and cellular localization are governed by the rod net charge. PLoS One. 2008;3:e1496. doi: 10.1371/journal.pone.0001496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandquist JC, Means AR. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Mol Biol Cell. 2008;19:5156–5167. doi: 10.1091/mbc.E08-05-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sandquist JC, Swenson KI, Demali KA, Burridge K, Means AR. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem. 2006;281:35873–35883. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- 65.Sato MK, Takahashi M, Yazawa M. Two regions of the tail are necessary for the isoform-specific functions of nonmuscle myosin IIB. Mol Biol Cell. 2007;18:1009–1017. doi: 10.1091/mbc.E06-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, Reva B, Fuchs E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shoffner JD, De LA. Sequences in the myosin II tail required for self-association. Biochem Biophys Res Commun. 1996;218:860–864. doi: 10.1006/bbrc.1996.0153. [DOI] [PubMed] [Google Scholar]
- 68.Shutova MS, Spessott WA, Giraudo CG, Svitkina T. Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr Biol. 2014;24:1958–1968. doi: 10.1016/j.cub.2014.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silver DL, Vorotnikov AV, Watterson DM, Shirinsky VP, Sellers JR. Sites of interaction between kinase-related protein and smooth muscle myosin. J Biol Chem. 1997;272:25353–25359. doi: 10.1074/jbc.272.40.25353. [DOI] [PubMed] [Google Scholar]
- 70.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin M Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 71.Straight AF, Field CM, Mitchison TJ. Anillin binds nonmuscle myosin M and regulates the contractile ring. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Straussman R, Squire JM, Ben-Ya'acov A, Ravid S. Skip residues and charge interactions in myosin II coiled-coils: implications for molecular packing. J Mol Biol. 2005;353:613–628. doi: 10.1016/j.jmb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 73.StuII JT, Nunnally MH, Moore RL, Blumenthal DK. Myosin light chain kinases and myosin phosphorylation in skeletal muscle. Adv Enzyme Regul. 1985;23:123–140. doi: 10.1016/0065-2571(85)90043-3. [DOI] [PubMed] [Google Scholar]
- 74.Takenaga K, Nakamura Y, Endo H, Sakiyama S. Involvement of S100-related calcium-binding protein pEL98 (or mts1) in cell motility and tumor cell invasion. Jpn J Cancer Res. 1994;85:831–839. doi: 10.1111/j.1349-7006.1994.tb02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tojkander S, Gateva G, Lappalainen P. Actin stress fibers-assembly, dynamics and biological roles. J Cell Sci. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- 76.Vaillancourt JP, Lyons C, Cote GP. Identification of two phosphorylated threonines in the tail region of Dictyostelium myosin II. J Biol Chem. 1988;263:10082–10087. [PubMed] [Google Scholar]
- 77.Vicente-Manzanares M, NeweII-Litwa K, Bachir AI, Whitmore LA, Horwitz AR. Myosin MA/MB restrict adhesive and protrusive signaling to generate front-back polarity in migrating cells. J Cell Biol. 2011;193:381–396. doi: 10.1083/jcb.201012159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang A, Ma X, Conti MA, Adelstein RS. Distinct and redundant roles of the non-muscle myosin II isoforms and functional domains. Biochem Soc Trans. 2011;39:1131–1135. doi: 10.1042/BST0391131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang A, Ma X, Conti MA, Liu C, Kawamoto S, Adelstein RS. Nonmuscle myosin II isoform and domain specificity during early mouse development. Proc Natl Acad Sci U S A. 2010;107:14645–14650. doi: 10.1073/pnas.1004023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang F, Kovacs M, Hu A, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of nonmuscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 81.Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


