Abstract
During mitosis, the mitotic spindle is assembled to align chromosomes at the spindle equator in metaphase, and to separate the genetic material equally to daughter cells in anaphase. The spindle itself is a macromolecular machine composed of an array of dynamic microtubules and associated proteins that coordinate the diverse events of mitosis. Among the microtubule associated proteins are a plethora of molecular motor proteins that couple the energy of ATP hydrolysis to force production. These motors, including members of the kinesin superfamily, must function at the right time and in the right place to insure the fidelity of mitosis. Misregulation of mitotic motors in disease states, such as cancer, underlies their potential utility as targets for antitumor drug development and highlights the importance of understanding the molecular mechanisms for regulating their function. Here, we focus on recent progress about regulatory mechanisms that control the proper function of mitotic kinesins and highlight new findings that lay the path for future studies.
Keywords: Mitosis, Kinesin, Microtubule, Kinase, Spindle
Mitotic Motors are Controlled Temporally to Function during Mitosis
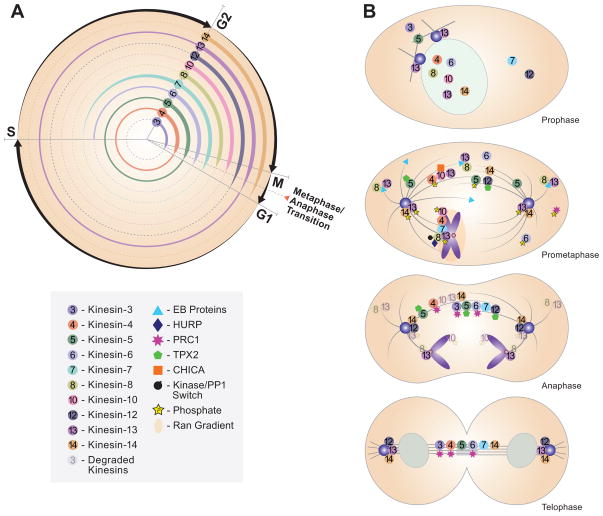
Many mitotic kinesins are regulated through temporal synthesis and degradation so that the protein is only present when needed during mitosis (Fig. 1A). Early studies of kinesin cell cycle expression came from simply analyzing protein expression and localization throughout the cell cycle. More recent studies include genomic approaches that have begun to uncover the transcriptional networks by which kinesin expression can be regulated. For example, binding of one or more mitosis-specific transcription factors, such as FOXM1, can promote expression of many cell cycle regulated genes at the G2/M transition [1]. This is interesting because FOXM1 is aberrantly expressed in many cancers and could contribute to the upregulation of kinesins seen in cancer cells [2]. Other important transcriptional networks controlling kinesin expression include those genes regulated by the DREAM/MMB (Myb-MuvB) complexes [3]. This is a multi-subunit complex that associates with repressive subunits, which inhibit gene expression during G1/S, and with activating subunits, which stimulate gene expression during G2/M. The promoters of the DREAM/MMB regulated genes often contain either a cell cycle homology region or a cell cycle dependent element, which can be bound by either the repressive or activating subunits of the DREAM/MMB complex. The DREAM/MMB complex is also highly integrated with the p53 control network, and p53 is mutated in a large number of cancers, highlighting the importance of the exquisite cell cycle control networks. In addition, the DREAM/MMB complex is important in controlling developmental expression of many proteins and may help tie together the understanding of the differential regulation of kinesin expression during development and in different cell types and how this relates to cell proliferation in general.
Figure 1.
Mechanisms that Control Mitotic Kinesin Localization. (A) Mitotic kinesins are regulated during the cell cycle through synthesis and degradation. The numbered circles represent different kinesin families and are in general representative of vertebrate kinesins. (B) Distinct kinesin family members are localized to different regions of the spindle, which is often controlled by binding partners and by regulatory proteins. Note that the precise localization of individual motors may vary between systems, and this provides a general overview of motor localization as it relates to motor function. Cartoon representations are modeled after [37].
The levels of kinesin proteins are also controlled by their regulated destruction at the end of mitosis. Several kinesins contain destruction box (D-box) or KEN box sequences that are targeted by APCCdc20 and by APCCdh1, which are ubiquitin ligases that tag proteins for destruction at the metaphase/anaphase transition and during early G1, respectively. The Kinesin-10, Kid, is one of the best-characterized examples of regulated destruction of a kinesin. Kid is a plus-end directed kinesin that is localized to chromosome arms and contributes to chromosome congression by mediating the polar ejection force [4]. It is destroyed at the metaphase/anaphase transition through APC to remove the force driving chromosomes to the spindle equator, which thus allows chromosomes to move poleward during anaphase [5]. The degradation of the budding yeast Kinesin-5, Cin8p, and the mammalian Kinesin-14, HSET, requires the activity of APCCdh1 instead of the early mitosis APCCdc20 for their regulated destruction during early G1. Although expression of a nondegradable Cin8p does not perturb cell cycle progression, there was a small increase in the percentage of mitotic cells with abnormal spindles [6]. The consequences of failing to degrade HSET have not been examined, although expression of mutant forms of HSET during interphase leads to excessive microtubule bundling and cellular toxicity [7].
In addition to cell cycle temporal control, some motors may need to act only at specific times during mitosis, and there are multiple cellular mechanisms that contribute to this type of motor regulation. The most notable examples are by controlling motor activity by modulating specific protein-protein interactions or through the control of the catalytic activity of the motor itself. This often occurs through the action of mitotic kinases, such as Aurora A/B, Plk1, and Cdk1, which phosphorylate multiple kinesins to regulate their specific localizations and activities in the spindle. These types of regulatory mechanisms are the focus of the rest of this review.
Spatial Control of Kinesin Activity Ensures Localized Function
The localization of mitotic kinesins can be as diverse as their functions, but the two go hand-in-hand because kinesins must be localized properly to complete their intended functions. Kinesins accomplish this task by directly binding to subsets of microtubules, to kinetochores, or to chromosomes. The affinities for these localizations are precisely controlled through gradients, intermolecular interactions with other proteins, and phosphorylation or other post-translational modifications (Fig. 1B).
Spatial Control by Gradients
Spindle assembly is under precise spatial regulation, which is controlled in part by the presence of gradients of signaling molecules in the spindle. The Ran gradient is found primarily around chromosomes and extends toward the spindle poles [8]. This gradient is formed because the guanine nucleotide exchange factor RCC1 is localized to chromosome arms, which causes a local increase in Ran-GTP around the chromosomes. The Aurora B/CPC gradient is controlled by the Chromosomal passenger complex (CPC), which consists of the Aurora B kinase along with its associated regulatory molecules INCENP and survivin [9]. The CPC targets to chromosome arms early in mitosis, focuses at the inner centromere during prometaphase and metaphase and then is left behind in the spindle midzone during anaphase and telophase. Both of these gradients have been shown to be important in the local regulation of motor activity.
The canonical example of spatial regulation by gradients in mitosis is illustrated by the Ran-GTP gradient, which regulates spindle assembly factors (SAFs) through binding to the nuclear transport receptors importin alpha and beta. During interphase, the Ran-GTP gradient is utilized to sequester proteins in the nucleus and then release them upon nuclear envelope breakdown. However, during mitosis the nuclear transport receptors are also involved directly in spatial control via the Ran-GTP gradient [8]. The importins bind specifically to the nuclear localization sequences (NLS) present in many mitotic kinesins. High levels of Ran-GTP result in the release of the importins from the kinesin, which can then target to its intended destination, such as microtubules or DNA. For the Kinesin-14s, this Ran regulation is important to regulate the microtubule cross-linking activity of the protein [10], and expression of NLS mutants results in aberrant microtubule cross-linking in interphase [7]. The Kinesin-10 Kid is interesting because it has a unique mechanism of importin regulation. Kid uses the binding to importins for its initial localization to chromatin but is also dependent upon Ran-GTP for continued association with chromosomes [11]. Thus for Kid, instead of the Ran-GTP gradient simply releasing the interaction between the importins and the cargo, it facilitates the loading of Kid to chromosome arms.
Intermolecular Protein Interactions
Another example of how kinesins are spatially localized is through intermolecular binding with other proteins. For example, several kinesins target to the plus ends of the microtubule through binding to EB (end binding) proteins, which interact with growing microtubule plus ends and act as a hub for recruiting proteins to this location [12]. Motor binding to EBs is primarily regulated through an SxIP motif within the kinesin sequence and has been shown for Kinesin-5s (Eg5), Kinesin-8s (Kif18B), and Kinesin-13s (MCAK) [13–17]. Mutation in the EB binding domain results in a deficiency in targeting to the microtubule plus end and a corresponding decrease in microtubule depolymerization activity for both Kif18B and MCAK [15, 16], suggesting that EB binding is functionally important for these motors. However, not all plus-end targeted kinesins require EBs for plus end localization. For example, the Kinesin-8, Kif18A localizes to the plus ends of K-fibers through an association with HURP (Hepatoma UpRegulated Protein). Overexpression of a HURP fragment that prevents Kif18A plus end localization leads to decreased association of Kif18A and an increased amplitude of kinetochore oscillations [18], ultimately leading to a delay in chromosome congression, similar to the phenotype of Kif18A knockdown. To highlight the complexity of these mitotic localization networks, HURP localization is also Ran-dependent [19], suggesting that the Ran gradient is a master control system that modulates the localization of many spindle motors both directly and indirectly by regulating other spindle MAPs.
The HURP complex may provide a key point of integration for the control of multiple mitotic motors. HURP was identified in a complex of proteins in Xenopus extracts that also contains the Kinesin-5, Eg5, the Kinesin-12 (KLP2) targeting protein, TPX2, and the MAP, XMAP215 [20]. Disruption of HURP results in defects in chromatin-mediated spindle assembly, suggesting that the effects of HURP are not limited to K-fiber associated motors. However, while the phenotypes of TPX2 disruption and HURP disruption are similar, they appear to act in independent pathways during spindle formation, suggesting that the HURP complex is not constitutively active [21]. Dissecting the contributions of each individual protein within these complexes as well as understanding the integration of activities will be important future endeavors.
The regulated binding of proteins to subsets of microtubules is also important in late mitosis. A major player in this process is PRC1, which serves as a scaffold for assembly of many proteins in the spindle midzone [22]. PRC1 is essential for central spindle assembly through its recruitment of the Kinesin-6 proteins MKLP-1 and MKLP-2, which are both essential for cytokinesis [22]. In vivo, the Kinesin-4, Kif4, is essential for targeting PRC1 to the central spindle [23]; however, elegant in vitro reconstitution experiments have demonstrated that PRC1 and XKlp1 (another Kinesin-4) coordinately regulate central spindle assembly by interacting specifically in regions of anti-parallel MT overlap and controlling the dynamics of the plus-ends of central spindle microtubules [24].
Phosphorylation by Mitotic Kinases
One major mechanism to control the localization of motors within the spindle is through protein phosphorylation, which has been shown to be critical for localization of several mitotic kinesins. The first example of this phosphoregulatory control was shown for the Kinesin-5s, where phosphorylation by Cdk1 of the Eg5 tail was shown to be required for localization of Eg5 to the spindle microtubules [25, 26]. It has been shown that Aurora kinases also contribute to Kinesin-5 targeting in C. elegans and X. laevis [27, 28], highlighting the complexity of these phosphoregulatory networks. The timing of the phosphorylation at individual sites is also important. While phosphorylation of the tail of Kinesin-5s early in mitosis is essential to target it to the spindle, phosphorylation of the yeast Kinesin-5 Cin8p weakens its microtubule association late in mitosis to control the rate and extent of anaphase spindle elongation. The localization of the Kinesin-6 proteins to the central spindle is also regulated by phosphorylation by multiple kinases. Phosphorylation of PRC1 by Cdk1 prevents its association with the spindle until late mitosis when dephosphorylated PRC1 binds to the spindle and recruits Kinesin-6 proteins [22].
In addition, the Kinesin-6 proteins are phosphorylated by spindle midzone-associated Aurora B, which positively influences their association with the central spindle [29]. In addition to targeting motors to spindle microtubules, phosphorylation is also important for localization of some kinesins to other spindle structures, such as chromosomes, kinetochores, or spindle poles. The best characterized example of this type of regulation is for the Kinesin-13 MCAK (Mitotic Centromere-Associated Kinesin), which is a microtubule depolymerizing kinesin that localizes to at least three distinct places in the cell during mitosis [30]. Aurora B positively influences MCAK localization to centromeres by phosphorylating S110 in its kinetochore-targeting domain, whereas phosphorylation at T95 promotes association with chromosome arms. Aurora B also indirectly facilitates MCAK association with centromeres by phosphorylation of Sgo2, which then interacts with MCAK and targets it to centromeres [31]. Aurora-B dependent phosphorylation is physiologically important in error correction, as phosphorylated MCAK is enriched at merotelically attached kinetochores [32]. Additionally, MCAK phosphorylation by Aurora A within its C-terminus regulates its association with spindle poles and contributes to Ran-regulated spindle bipolarity [33]. In contrast, Aurora A phosphorylation of the Kinesin-13 Kif2a decreases its microtubule association, whereas Plk1 phosphorylation of Kif2a increases its microtubule and pole localization [33, 34]. Together these studies highlight the intricate phosphorylation networks that control the localization of just one family of motors and thus influence localized spindle microtubule dynamics.
The spatial localization of Kid is also controlled by Cdk1 phosphorylation, which regulates the binding affinity of Kid to microtubules, which in turn influences the pool of available Kid to bind to chromosome arms [35]. The binding of Kid to spindle microtubules is further regulated by CHICA, a novel spindle protein required for chromosome congression [36]. Thus, Kid is regulated spatially and temporally by the Ran-GTP gradient, phosphorylation, and protein-protein interaction providing an example of how the cell uses multiple mechanisms to ensure the precise localization of mitotic motors to sub-spindle structures. Understanding the mechanisms that coordinate Kid function on the spindle versus the chromosome arms will provide significant new insights into how the polar ejection force contributes to chromosome congression.
Modulation of Motor Activity is Critical to Proper Spindle Function
While the proper expression and physical localization of mitotic kinesins are essential for mitosis, ensuring that the activities of motors are precisely tuned is also an important regulatory mechanism. Three kinesin families (Kinesin-5, Kinesin-7, and Kinesin-13) provide excellent examples for how the activities of motors can be regulated.
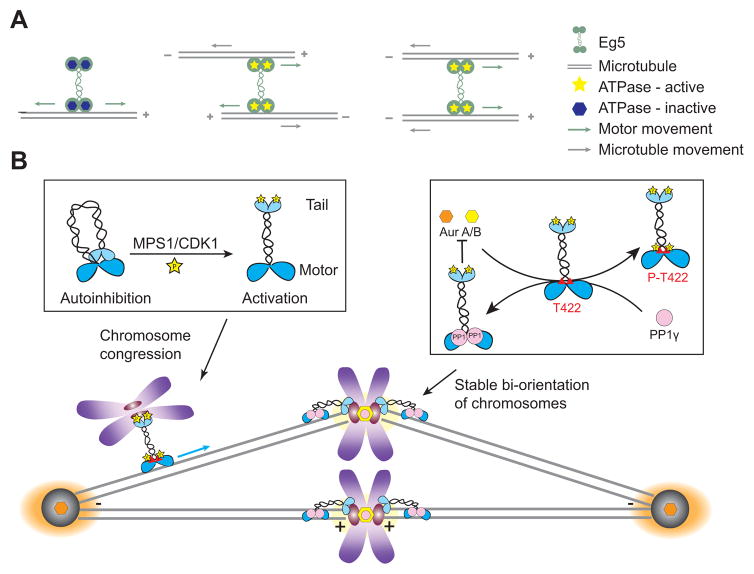
It is well-established that the motility of conventional kinesin, Kinesin-1, is regulated by cargo binding to the tail domain, which relieves kinesin autoinhibition allowing for vesicle motility during interphase [37]. Likewise, cargo binding of Kinesin-5 also regulates motility but by a different mechanism (Fig. 2A). The Kinesin-5, Eg5, is a plus-end directed motor that has two motor dimers oriented in an anti-parallel fashion with a pair of motor domains (heads) on either end (Fig. 2A) [38]. This dumbbell structure allows Eg5 to crosslink and slide apart two microtubules [39]. Eg5 utilizes ATP-independent diffusion when it binds to single microtubules, whereas binding to a second microtubule activates ATP hydrolysis activity and switches Eg5 from diffusive to directed movement [40]. This allows Eg5 to only slide microtubules when it is actively engaged in microtubule cross-links, which contributes to spindle pole separation. Eg5 can also tether to microtubule plus-ends, which may be important in parallel microtubule cross-links in the half spindle. Furthermore, it was shown that Eg5 possesses another microtubule binding site in its tail that enhances motor processivity, microtubule association, and microtubule crosslinking [41], but it is still unknown how microtubule-binding events regulate the motor activity. It has been proposed that the tail-motor and/or tail-microtubule interaction triggers allosteric changes in the protein that mediate the switch of Eg5 motor activity between the two microtubule-binding states [41, 42]. The regulation of microtubule cross-linking activity has also been shown for members of the Kinesin-14 family, which are minus-end directed motors. Like Eg5, the Kinesin-14s are activated by microtubule cross-linking [43] to slide anti-parallel microtubules, but they tether parallel microtubules [44]. Given that Kinesin-5 and Kinesin-14 proteins play antagonistic roles in spindle length control and that this antagonism can be recapitulated in vitro [45, 46], this system will be ideal to begin to look at the integration of motor activity and regulation in more complex motor ensembles.
Figure 2.
Regulation of Mitotic Motor Activity. (A) Eg5 diffuses on a single microtubule in an ATP-independent manner. When Eg5 binds to a second microtubule, its ATP hydrolysis activity is stimulated, and Eg5 switches from diffusive to directed movement toward the microtubule plus end. Eg5 can either crosslink and slide apart antiparallel microtubules or crosslink and generate force on parallel microtubules. (B) The motor activity of Cenp-E is autoinhibited by direct binding of the tail to the motor. This autoinhibition is reversed by phosphorylation of the tail by either MPS1 or Cdk1 kinase. Phosphorylation by Aurora A/Aurora B at T422 also contributes to regulation of Cenp-E during chromosome congression. After chromosome alignment, the activity of Cenp-E is fine-tuned by a balance between Aurora kinases and PP1γ phosphatase to maintain chromosome bi-orientation. Type 1 phosphatase (PP1γ) can bind to a docking motif adjacent to T422, blocking the binding of Aurora kinases and allowing Cenp-E to deliver PP1 to kinetochores, where it contributes to the stable bi-orientation of chromosomes.
Motor protein activity can also be regulated through intramolecular interactions. Cenp-E, a member of the Kinesin-7 family of plus-end directed motor proteins, acts at the kinetochore-microtubule interface to direct chromosome congression by moving chromosomes towards the spindle equator [47, 48]. Like the canonical transport protein Kinesin-1, the motor activity of Cenp-E is autoinhibited by direct binding of the C-terminal tail to the motor domain [49, 50] (Fig. 2B). Phosphorylation of the tail by either MPS1-or Cdk1- reverses the autoinhibition, which provides a mechanism to activate Cenp-E motility during early mitosis when it is needed for chromosome congression.
Not all intramolecular interactions of kinesins are autoinhibitory. For example, the microtubule depolymerization activity of MCAK is regulated by Aurora B-dependent phosphorylation of S196 in the neck domain of MCAK. Phosphorylation at this site could potentially disfavor electrostatic interactions of the positively charged neck with the negatively charged microtubule lattice [51]. However, this regulation is more complex because it was shown that Aurora B phosphorylation at S196 changes MCAK from a closed to open conformation, which results in a decrease in MCAK affinity for the microtubule that contributes to reduced microtubule depolymerization activity [52]. This means that for at least some Kinesin-13s, a closed conformation is the high-affinity binding state to microtubule ends to stimulate microtubule depolymerization.
The effects of phosphorylation on motor activity are counteracted by the opposing roles of serine-threonine phosphatases, such as PP1 and PP2A. For example, the motor processivity and the microtubule binding of Cenp-E are reduced by Aurora A and Aurora B-mediated phosphorylation at T422, which lies close to the motor domain (Fig 2B). This results in the ability of Cenp-E to drive mono-oriented chromosomes near the pole toward the spindle equator. However, this activity needs to be modulated in order for chromosomes to become stably attached to spindle microtubules once they are bi-oriented. Adjacent to T422 is a docking motif for type 1 phosphatase (PP1γ), which blocks the direct binding of Aurora kinases and allows Cenp-E to deliver PP1 to kinetochores, where it also helps contribute to the stable bi-orientation of chromosomes [53]. These activities are also spatially controlled because an Aurora A gradient emanates from the spindle poles whereas Aurora B and PP1γ are primarily localized at the kinetochores. Thus, this Aurora/PP1 switch is critical for spatially modulating Cenp-E’s role in chromosome congression and stable bi-orientation. An Aurora/PP1 regulatory network also plays a role in the bi-orientation of chromosomes in fission yeast by its regulation of the plus-end directed Kinesin-8 proteins, Klp5 and Klp6. The non-motor tail domains of Klp5 and Klp6 bind to PP1Dis, and the association of PP1Dis is required for chromosome bi-orientation and for efficient spindle checkpoint silencing [54].
Integration of Phosphoregulatory Mechanisms
Emerging evidence suggests there is intense crosstalk between different regulatory pathways. For example, although Aurora A and Aurora B have distinct localizations and fundamentally different functions, they share some substrates, such as CENP-E, Kif18B, MCAK, Kif2a, and TPX2 to cooperatively regulate mitotic events [55]. In addition, some kinases and phosphatases simultaneously hit the same kinesins to tune the balance between phosphorylation and dephosphorylation to modulate activity or spindle targeting [56, 57]. For instance, centrosomal Plk1 and Aurora A antagonistically regulate the spatial targeting and depolymerase activity of Kif2a. In early mitosis, as the spindle starts to form, Aurora A phosphorylation of Kif2a inhibits its enzymatic activity and decreases the targeting of Kif2a on spindle microtubules, which allows for stabilization of the minus ends of microtubules that are emanating from the chromatin. After these microtubule minus ends are embedded in the spindle poles, Plk1 then activates Kif2a, which results in depolymerization from the microtubule minus ends to exert the pulling force for microtubule poleward flux [58]. Multi-kinase regulation has also been demonstrated for MCAK, Eg5, and MKLP-1, suggesting that this is likely a common mechanism of action.
The importance of the kinase/phosphatase balance at the kinetochore has also been shown for Aurora B/PP2A, which controls the phosphorylation levels of multiple substrates at the kinetochore [57]. This type of balanced phosphorylation/dephosphorylation system is going to be critical to maintain the fine spatial gradients of Aurora B phosphorylation at the kinetochore, wherein tension is thought to allow substrate movement into and out of the regions of highest Aurora B kinase activity [59]. Together these studies highlight the fine-tuned control that is needed to locally modulate spindle protein function.
The above studies provide an example of how multiple kinase and phosphatase pathways can be integrated. However, an additional complexity will be to elucidate how the more global gradients of Ran-GTP and the Aurora B/CPC around the chromosomes and centromeres coordinately regulate these mitotic proteins. While it is known that both pathways contribute to spindle assembly [60], their local concentrations and activation could differentially affect the activity of their downstream targets, some of which are identical between the two pathways.
Conclusions
In order to ensure the fidelity of mitosis, mitotic kinesins need to be highly regulated in both space and time and to locally control their activity. These distinct temporal and spatial regulatory mechanisms can affect the same motors at different points in the cell cycle and different motors at the same point in the cell cycle. Although the past few decades have resulted in large advances in our understanding of the general mechanisms by which these motors are regulated, there is still much to learn about individual pathways of regulation. In addition, newer proteomics approaches are identifying novel protein-protein interactions and putative sites of phosphoregulation and other post-translational modifications, which should provide a complete roadmap of the mitotic spindle [61, 62]. However, lists are simply lists, until the functional and mechanistic analyses help elucidate how these individual changes modulate the localization and activity of each substrate and how these individual regulatory events are integrated into the global network of the spindle itself. Then we will be able to build a more detailed roadmap of mitosis.
Acknowledgments
Work in the Walczak Lab is supported by NIH R01GM059618. We thank Stephanie Ems-McClung and Lesley Weaver for critical comments on the manuscript. We thank all members of the Walczak lab for insightful discussions about mitotic motors. We apologize to colleagues whose original work was not cited due to strict limitations on the number of references.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alvarez-Fernandez M, Medema RH. Novel functions of FoxM1: from molecular mechanisms to cancer therapy. Front Oncol. 2013;3:30. doi: 10.3389/fonc.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozielski F, editor. Kinesins in Cancer. Springer; in press. [Google Scholar]
- 3.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanneste D, Ferreira V, Vernos I. Chromokinesins: localization-dependent functions and regulation during cell division. Biochem Soc Trans. 2011;39:1154–1160. doi: 10.1042/BST0391154. [DOI] [PubMed] [Google Scholar]
- 5.Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 6.Hildebrandt ER, Hoyt MA. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol Biol Cell. 2001;12:3402–3416. doi: 10.1091/mbc.12.11.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai S, Weaver LN, Ems-McClung SC, Walczak CE. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell. 2009;20:1348–1359. doi: 10.1091/mbc.E08-09-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller BG. Self-organization of intracellular gradients during mitosis. Cell Div. 2010;5:5. doi: 10.1186/1747-1028-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ems-McClung SC, Zheng Y, Walczak CE. Importin alpha/beta and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol Biol Cell. 2004;15:46–57. doi: 10.1091/mbc.E03-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahara K, Takagi M, Ohsugi M, Sone T, Nishiumi F, Maeshima K, Horiuchi Y, Tokai-Nishizumi N, Imamoto F, Yamamoto T, et al. Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J Cell Biol. 2008;180:493–506. doi: 10.1083/jcb.200708003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 13.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 14.Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell. 2011;22:3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Montenegro Gouveia S, Leslie K, Kapitein LC, Buey RM, Grigoriev I, Wagenbach M, Smal I, Meijering E, Hoogenraad CC, Wordeman L, et al. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr Biol. 2010;20:1717–1722. doi: 10.1016/j.cub.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Jiang K, Toedt G, Montenegro Gouveia S, Davey NE, Hua S, van der Vaart B, Grigoriev I, Larsen J, Pedersen LB, Bezstarosti K, et al. A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol. 2012;22:1800–1807. doi: 10.1016/j.cub.2012.07.047. [DOI] [PubMed] [Google Scholar]
- 18.Ye F, Tan L, Yang Q, Xia Y, Deng LW, Murata-Hori M, Liou YC. HURP regulates chromosome congression by modulating kinesin Kif18A function. Curr Biol. 2011;21:1584–1591. doi: 10.1016/j.cub.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 19.Sillje HH, Nagel S, Korner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 20.Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 21.Casanova CM, Rybina S, Yokoyama H, Karsenti E, Mattaj IW. Hepatoma up-regulated protein is required for chromatin-induced microtubule assembly independently of TPX2. Mol Biol Cell. 2008;19:4900–4908. doi: 10.1091/mbc.E08-06-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douglas ME, Mishima M. Still entangled: assembly of the central spindle by multiple microtubule modulators. Semin Cell Dev Biol. 2010;21:899–908. doi: 10.1016/j.semcdb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 25.Blangy A, Lane HA, d’Hérin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 26.Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci USA. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell. 2005;16:742–756. doi: 10.1091/mbc.E04-08-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giet R, Uzbekov R, Cubizolles F, Le Guellec K, Prigent C. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J Biol Chem. 1999;274:15005–15013. doi: 10.1074/jbc.274.21.15005. [DOI] [PubMed] [Google Scholar]
- 29.White EA, Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskeleton (Hoboken) 2012;69:882–892. doi: 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ems-McClung SC, Walczak CE. Kinesin-13s in mitosis: Key players in the spatial and temporal organization of spindle microtubules. Semin Cell Dev Biol. 2010;21:276–282. doi: 10.1016/j.semcdb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanno Y, Kitajima TS, Honda T, Ando Y, Ishiguro K, Watanabe Y. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 2010;24:2169–2179. doi: 10.1101/gad.1945310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Ems-McClung SC, Walczak CE. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol Biol Cell. 2008;19:2752–2765. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang CY, Coppinger JA, Seki A, Yates JR, 3rd, Fang G. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J Cell Sci. 2009;122:1334–1341. doi: 10.1242/jcs.044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohsugi M, Tokai-Nishizumi N, Shiroguchi K, Toyoshima YY, Inoue J, Yamamoto T. Cdc2-mediated phosphorylation of Kid controls its distribution to spindle and chromosomes. EMBO J. 2003;22:2091–2103. doi: 10.1093/emboj/cdg208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santamaria A, Nagel S, Sillje HH, Nigg EA. The spindle protein CHICA mediates localization of the chromokinesin Kid to the mitotic spindle. Curr Biol. 2008;18:723–729. doi: 10.1016/j.cub.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 37.Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 38.Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 40.Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJG. Microtubule cross-linking triggers the directional motility of kinesin-5. The J Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinger JS, Qiu MH, Yang G, Kapoor TM. A Nonmotor Microtubule Binding Site in Kinesin-5 Is Required for Filament Crosslinking and Sliding. Curr Biol. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thiede C, Lakamper S, Wessel AD, Kramer S, Schmidt CF. A chimeric kinesin-1 head/kinesin-5 tail motor switches between diffusive and processive motility. Biophys J. 2013;104:432–441. doi: 10.1016/j.bpj.2012.11.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furuta K, Toyoshima YY. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr Biol. 2008;18:152–157. doi: 10.1016/j.cub.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 44.Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- 45.Hentrich C, Surrey T. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J Cell Biol. 2010;189:465–480. doi: 10.1083/jcb.200910125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 47.Wood KW, Sakowicz R, Goldstein LSB, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- 48.Wood KW, Sakowicz R, Goldstein LS, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- 49.Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 50.Espeut J, Gaussen A, Bieling P, Morin V, Prieto S, Fesquet D, Surrey T, Abrieu A. Phosphorylation relieves autoinhibition of the kinetochore motor Cenp-E. Mol Cell. 2008;29:637–643. doi: 10.1016/j.molcel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol. 2010;17:77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ems-McClung SC, Hainline SG, Devare J, Zong H, Cai S, Carnes SK, Shaw SL, Walczak CE. Aurora B Inhibits MCAK Activity through a Phosphoconformational Switch that Reduces Microtubule Association. Curr Biol. 2013;23:2491–2499. doi: 10.1016/j.cub.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora Kinases and Protein Phosphatase 1 Mediate Chromosome Congression through Regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meadows JC, Shepperd LA, Vanoosthuyse V, Lancaster TC, Sochaj AM, Buttrick GJ, Hardwick KG, Millar JBA. Spindle Checkpoint Silencing Requires Association of PP1 to Both Spc7 and Kinesin-8 Motors. Dev Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hochegger H, Hegarat N, Pereira-Leal JB. Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open Biol. 2013;3 doi: 10.1098/rsob.120185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lens SMA, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 57.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang CY, Coppinger JA, Seki A, Yates JR, Fang GW. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J Cell Sci. 2009;122:1334–1341. doi: 10.1242/jcs.044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maresca TJ, Groen AC, Gatlin JC, Ohi R, Mitchison TJ, Salmon ED. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr Biol. 2009;19:1210–1215. doi: 10.1016/j.cub.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Syred HM, Welburn J, Rappsilber J, Ohkura H. Cell cycle regulation of microtubule interactomes: multi-layered regulation is critical for the interphase/mitosis transition. Mol Cell Proteomics. 2013;12:3135–3147. doi: 10.1074/mcp.M113.028563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maliga Z, Junqueira M, Toyoda Y, Ettinger A, Mora-Bermudez F, Klemm RW, Vasilj A, Guhr E, Ibarlucea-Benitez I, Poser I, et al. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol. 2013;15:325–334. doi: 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]