Abstract
Respiratory Complex I deficiency is implicated in numerous degenerative and metabolic diseases. In particular, mutations in several mitochondrial DNA (mtDNA)-encoded Complex I subunits including ND4, ND5 and ND6 have been identified in several neurological diseases. We previously demonstrated that these subunits played essential roles in Complex I assembly which in turn affected mitochondrial function. Here, we carried out a comprehensive study of the Complex I assembly pathway. We identified a new Complex I intermediate containing both membrane and matrix arms at an early assembly stage. We find that lack of the ND6 subunit does not hinder membrane arm formation; instead it recruits ND1 and ND5 enter the intermediate. While ND4 is important for the formation of the newly identified intermediate, the addition of ND5 stabilizes the complex and is required for the critical transition from Complex I to supercomplexes assembly. As a result, the Complex I assembly pathway has been redefined in this study.
Introduction
The mammalian NADH-ubiquinone dehydrogenase (Complex I) is the major entry point for electrons of the respiratory chain, yet it is the least understood complex in terms of its assembly, function and roles in different disorders. Complex I is a multi-subunit complex consisting of 44 subunits of which 7 subunits - ND1, -2, -3, -4, 4L, -5 and -6 - are encoded by the mitochondrial genome (1–3), (4). Electron microscopic (EM) studies indicate an L-shaped structure of Complex I consisting of a hydrophobic membrane arm embedded in the mitochondrial inner membrane and a hydrophilic peripheral arm that juts out into the matrix (5, 6). All of the mitochondrial DNA-encoded subunits are found in the membrane arm that participates in proton translocation (7), (3). The peripheral arm contains the nuclear-encoded subunits as well as Fe-S centers involved in transfer of electrons (8).
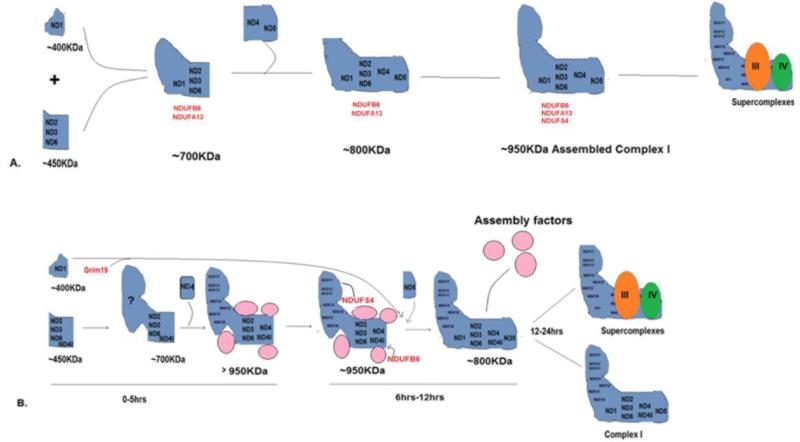
The currently favored model of the complex I assembly pathway consists of independent assembly of the membrane and the peripheral arm via sequential induction of subcomplexes which ultimately join together to form the L-shaped structure (9),(10). This model comprises of two starting subcomplexes: a 400KDa subcomplex containing ND1 and nuclear-encoded subunits such as NDUFS2, 3, 7, 8 and NDUFA9, and a ~460KDa subcomplex containing ND2, ND3 and ND4L. The additional presence of ND6 in the latter subcomplex has been debated. These two subcomplexes then combine to form a ~700KDa subcomplex that also contains NDUFB6 and NDUFA13. The subcomplex containing ND4 and ND5 then joins the Complex I assembly pathway and the assembly is completed with addition of the remaining nuclear-encoded subunits including NDUFS4 (10–15). Many assembly factors have been identified that help in the correct incorporation of Complex I subunits at different stages. Currently, approximately 11 assembly factors have been reported, including Ecsit, NDUFAF1-4, Aif, Ind1, ACAD9, C6ORF60, FOXRED1 and C20ORF7 (16–22).
Complex I deficiency has been associated with disorders such as Parkinson's disease, Leigh's syndrome, fatal infantile lactic acidosis, macrocephaly with progressive leukodystrophy, encephalomyopathy, and in particular, Leber's Hereditary Optic Neuropathy (LHON) (23–25). The ND1, ND4, ND5 and ND6 genes are also considered hotspots for pathological mutations (26). It has been observed that mutations in ND4 and ND6 obstruct complex I assembly, leading to a Complex I deficiency (27–29) while mutations in other mitochondrial subunits, such as ND1 and ND5, impair Complex I activity by causing a mild defect in Complex I assembly (30, 31).
In spite of this progress, details of the process of Complex I assembly, in particular those involving mtDNA-encoded subunits, are still largely unclear, with many contradictory observations. The entry point for ND6 had been thought to be as early as with ND2, ND3 and ND4L, but recently it was reported that ND6 might enter the assembly pathway later. Moreover, ND4 and ND5 had been thought to enter the assembly pathway later and in conjunction with each other, but this has also been debated. In addition, there are conflicting reports on the role of ND6 in Complex I assembly(12),28,32,33).
To help resolve these questions, we have developed an approach by isolating several mtDNA mutations based on resistance to a Complex I respiration inhibitor, Rotenone (33). In particular, cell lines carrying mutations in the ND4, ND5 and ND6 genes have been established (27, 33, 34). The availability of these mutant lines enables us to delineate the Complex I assembly pathway in mammalian cells. In this study, we provide evidence suggesting a different sequence of incorporation of mitochondrial encoded Complex I subunits than the model previously reported.
Materials and Methods
Cell lines
All the cell lines used in the present work were grown in monolayer culture. The cell line A9 (ATCC CCL-1.4) is a derivative of the mouse L fibroblast cell line (35). All cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). The rotenone-resistant clone 4A carrying the ND6 mutation was derived from A9 (35) and was grown in regular DMEM supplemented with 0.2 μM rotenone. Cybrids 4A-4 and 4A-6 are derived from the LL/2-m21 cell line with mitochondria from 4A (35). The 3A20-4 cells are mouse fibroblasts derived from the LL/2 cell line with a frameshift mutation in the ND5 subunit (33), while C4T cells are human osteosarcoma cells with frameshift mutation in the ND4 gene and are derived from the 143B.TK− cell line (27).
Mitochondrial Pulse-Chase
To test the stability of mitochondrial translational products, pulse-labeling experiments with [35]S methionine were performed according to a protocol described previously (36). Samples of 2 × 106 cells of the desired type were plated onto 10 cm dishes, incubated overnight, washed with methionine-free DMEM, and then incubated for 7 min at 37°C in 4 ml of the same medium containing 100 μg/ml of the cytoplasmic translational inhibitor cycloheximide. Thereafter, S[35] methionine (0.2 mCi [1,175 Ci/mmol]) was added, and the cells were incubated for 120 min, then washed and subjected to a 24 h chase in complete unlabeled medium in the absence of cycloheximide to allow incorporation of the labeled mtDNA-encoded subunits into the complexes. The labeled cells were treated with trypsin, washed and subjected to Blue native PAGE as described below.
Blue Native gel electrophoresis/2D SDS PAGE
Complex assembly analysis was carried out by Blue native gel electrophoresis (37). Sample preparation and Blue native gel electrophoresis were carried out as described previously (38). Briefly, the cells were suspended in lysis buffer and subjected to homogenization using a Potter-Elvehjem homogenizer with a rotating pestle until 80% of the cells were broken. The homogenate was then centrifuged at 700 g at 4°C to remove nuclear and cytoplasmic debris. The resulting supernatant was again centrifuged at 3000g to pellet the mitochondria. The mitochondrial protein concentration was determined using a BSA protein assay from Thermo-Fisher (Waltham, MA). For Western blotting and immunodetection, antibodies against complex I subunits NDUFA13 (Abcam, Cambridge, GBN), NDUFB6 (Abcam), NDUFS4 (Abcam) and NDUFA9 (Santa Cruz Biochemicals Dallas, TX) were utilized. The Western blots were carried out according to the protocol provided by Mitosciences. In-gel activity assays for Complex I and Complex V were conducted as described before (39). 2D-SDS electrophoresis was performed as previously described. Lanes from the Blue native gel were cut out and laid onto 10-16% gradient Tricine SDS PAGE gel as published previously (40).
Mass spectroscopy
Protein peptide identification by mass spectroscopy was carried out at the Mass Spectroscopy Institutional Core at UTHSCSA. The samples were processed as described above for Blue native gels and equal amounts of protein were loaded. Complex I bands were cut from the A9 and 4A sample lanes, respectively. The bands were subjected to digestion and protein identification was carried out using Liquid chromatography–mass spectrometry (LC/MS) at the UTHSCSA Core. Each experiment was conducted twice.
Results
We and others have previously shown that ND6, ND4 and ND5 are essential for Complex I assembly (27–29, 33, 34). With the development of approaches to delineate the dynamics of the respiratory complexes’ assembly process and generation of more cell lines carrying various mutations in mtDNA encoded Complex I subunits, we carried out a careful investigation into the roles of several key subunits in Complex I assembly.
Complex I assembly in cells without mtDNA encoded ND6 subunit
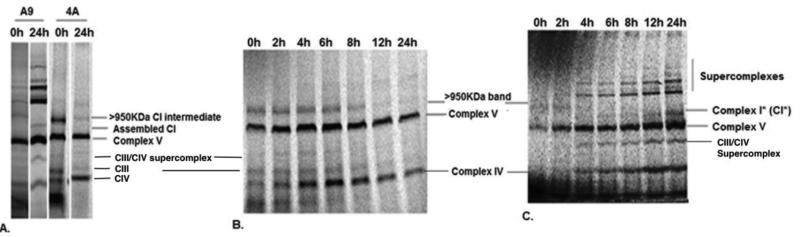
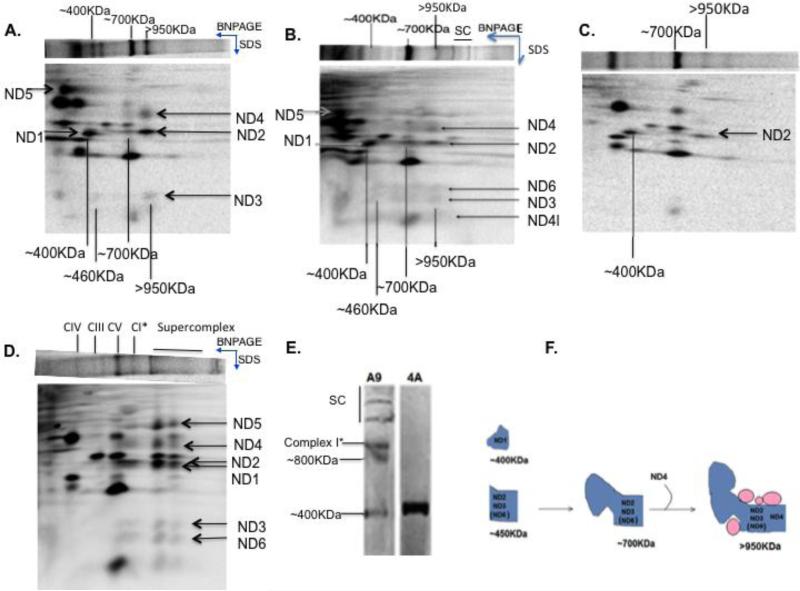
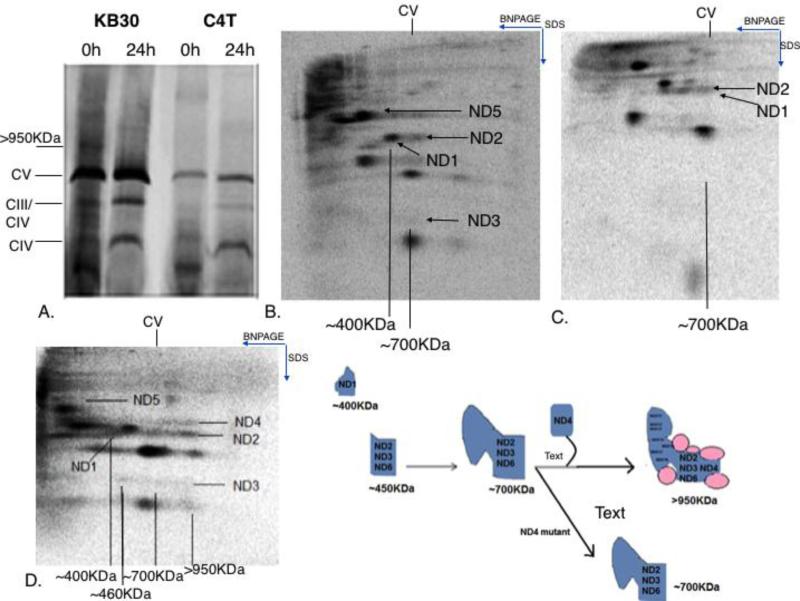
Cell line 4A is one that we established which carries a near-homoplasmic frameshift mutation in the mtDNA-encoded ND6 gene (28). To characterize Complex I assembly in the absence of ND6 subunit, 4A cells were grown along with the control A9 cells in the presence of a mitochondrial protein synthesis inhibitor, chloramphenicol, for 24 h. This leads to an enrichment of nuclear-encoded respiratory chain subunits in the mitochondria and blocks mtDNA-encoded protein synthesis. Cells were subsequently labeled for 2 h with [35]S methionine and chased for 24 h in complete unlabeled medium to allow incorporation of labeled, mtDNA-encoded respiratory subunits into respiratory complexes. Cells were collected at different time points and solubilized with digitonin for native gel electrophoresis analysis. As shown in Fig. 1A, at the 0h time point, i.e., 2h after addition of labeled [35]S, both A9 and 4A showed a band corresponding to MW greater than 950KDa, which is close to the complete Complex I. After 24 h of chase, this band shifted to a slightly lower molecular weight band in A9 cells, while it significantly decreased in mutant 4A cells. To more carefully follow the changes of this putative early assembled Complex I, we carried out the chase analysis with more time points between 0h and 24 h. In 4A cells, where ND6 is absent, the putative Complex I band started to decrease around ~6 h (Fig.1B). In contrast to 4A cells, this putative Complex I intermediate band in A9 cells was rapidly incorporated into supercomplexes by 4-6 h (Fig.1C), and at about the same time, a slightly lower molecular weight band, labeled Complex I* (CI*), started to appear and accumulate. The CI* band appeared simultaneously with the supercomplexes. To confirm that the >950KDa band seen at 0 h in 4A cells is indeed an intermediate of Complex I, a second dimension tricine SDS gel electrophoresis was carried out. The presence of mtDNA-encoded Complex I subunits was detected in the >950KDa band (Fig. 2A). Furthermore, immediately after labeling, a similar pattern of CI assembly was observed in both the wild type as well as the ND6 mutant line (Fig. 2A and 2B). Within 2 h of labeling, incorporation of ND4, ND2, ND3 and ND4L (and ND6 in A9 cells) was seen in the relatively high molecular weight >950KDa Complex I intermediate in both A9 and 4A cells while ND1 and ND5 remained in the ~400KDa and ~200KDa subcomplexes, respectively. ND2, ND3 and ND6 were also seen in ~450KDa and ~700KDa subcomplexes in A9 and 4A cells at early time of assembly. However, the levels of the subunits in the early assembled >950KDa complex I intermediate in 4A cells decreased over a period of 24 h and only ND1 was seen in the ~400KDa subcomplex, while supercomplex assembly was seen in A9 cells. Also observed in A9 cells at 24hs is a ~750-800 Kodak Complex I band, which is probably not seen in 1D gel due to the strong Complex V signal, and the CI* band (Fig. 2C and 2D). Steady-state levels of Complex I assembly intermediates in 4A cells were detected with multiple antibodies against its subunits including anti-NDUFA13 antibody, and results revealed the presence of the ~400KDa band (Figure 2E), confirming the observations in 24h [35]S-labeled gel in 4A cells.
Figure 1. Early Complex I assembly intermediate in ND6 mutant cells.
A) shows the presence of a >950KDa putative Complex I band in A9 and ND6 mutant 4A cells at 0h after [35]S methionine labeling. The >950KDa band can also be seen in 4A cells at 24 h, while in A9 cells, the assembled Complex I band along with supercomplexes is seen. B) the chase experiment shows the gradual disappearance of the >950KDa Complex I intermediate band in 4A cells over time. C) [35]S labeled pulse chase of wild type A9 cells showing the >950KDa complex at early times and the assembled Complex I, indicated as Complex I*, and supercomplexes at later time point. Complexes III, IV and V are marked for molecular weight reference.
Figure 2. Presence of Complex I subunits in ND6 mutant cells.
A) shows the 2D SDS PAGE profile of ND6 mutant 4A and B) shows wild type A9 cells after 2h of [35]S labeling and C) shows mutant 4A and D) shows wild type A9 profile after 24 h of [35]S labeling. The samples were run on a BNPAGE in the direction indicated and were subsequently run on SDSPAGE in the second dimension. The molecular weights of all subcomplexes observed have been indicated. Complex III, IV and V have been indicated for molecular weight reference. E) shows the formation of a steady state ~400KDa band in 4A cells but no complex I assembly when probed with anti-NDUFA13 antibody. F) The cartoon shows the three subcomplexes observed in 4A and A9 cells and their progress into the >950KDa Complex I intermediate, pink circles indicating presence of assembly factors.
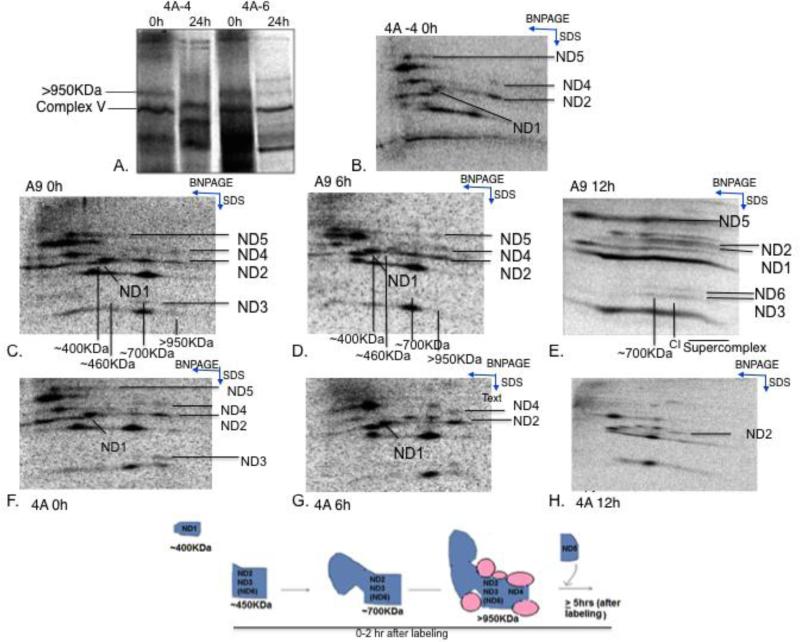
To confirm that these findings were not characteristic of only the 4A cell line, due to some specific alterations in nuclear background, we transferred 4A mtDNA carrying the ND6 mutation to a new nuclear background, mouse Lewis Lung cancer line LL/2 derived, mtDNA-less rho zero cells (33). We then conducted similar experiments in two such cybrids, 4A-4 and 4A-6. We observed Complex I assembly pattern at an early time point as observed in the 4A cells (Figure 3A), and the high molecular weight Complex I band immediately after labeling also contained mtDNA-encoded Complex I subunits as in 4A cells, as represented by 4A-4 cells (Figure 3B).
Figure 3. Assembly of mtDNA subunits into Complex I in ND6 mutant cells.
A) shows the presence of the >950KDa Complex I band in 1D native gel in 4A-4 and 4A-6 cells at 0 h and 24 h after [35]S labeling. B) shows the 2D SDS PAGE profile of 4A-6 cells similar to 4A cells after 2 h of labeling with the >950KDa band containing Complex I subunits. (C-E) shows a 2D SDS PAGE profile of A9 cells and (F-H) shows 2D SDS PAGE profile of 4A cells at the 0 h, 4h and 10 h time points (i.e., 2 h, 6 h and 12 h after [35]S labeling begins) respectively. Each observed subcomplex in marked on the SDS PAGE as well as the BNPAGE. The direction for each gel dimension is also indicated by the blue arrows. I) The cartoon shows the kinetic progression of different subcomplexes as observed on the gel.
Incorporation of complex I subunits depending on the mtDNA-encoded ND6 subunit
To map out the sequential incorporation of the different mtDNA subunits in the presence and absence of ND6, we first labeled the mtDNA-encoded subunits, and then carried out pulse-chase experiments at different time points. In mutant 4A cells, we observed that the ND5 subunit remained in the premature ~200KDa sub-complexes and disappeared after 6h of chase (Figure 3F-H), while ND1 did not progress to the>950KDa band but was still detectable in the ~400KDa subcomplexes. In both cell lines, ND2 and ND3 (ND6 in A9) were consistently seen in the ~450KDa, ~700KDa and >950KDa subcomplexes. These molecular weights were tentatively assigned with respect to other respiratory complexes; Complex III, IV and V. In the wild type cells, by 6h of labeling, ND5 had already started accumulating in the higher Complex I band and by 12h, the subcomplexes had assembled into supercomplexes (Figure 3C-E).
To more carefully determine the composition of the nuclear-encoded subunits in this early >950KDa Complex I intermediate band, we allowed for de novo Complex I assembly in A9 and 4A cells by treating them with chloramphenicol for 6 days to allow for existing Complex I to get turned over (41). A9 and 4A cells were collected at the 0h time point (i.e., 2h removal of block) and mitochondria were isolated and subjected to Blue native gel electrophoresis. This Coomassie Blue stained band was subjected to mass spectrometry analysis. Assembled Complex I band from A9 cells was used as a control. Table 1 provides a list of Complex I subunits incorporated in the presence and in absence of ND6 as they were identified in 4A and A9 cells. Among the subunits identified in both A9 and 4A cells, there were NDUFS2, NDUFS3, NDUFA9 and others which previously reported as early assembled subunits (10, 15). We also detected NDUFV1, NDUFV2, NDUFS1, 5 and 6 which were incorporated at a later stage of in complex I assembly (42) indicating that ND6 was not required to incorporate these subunits and that in the absence of ND6, the assembly of Complex I could still carry on to a certain extent.
Table 1.
Mass spectroscopy detects presence of nuclear encoded Complex I subunits in the >950KDa intermediate in ND6 mutant 4A cells and A9 cells. Steady state Complex I band from A9 cells was used as control.
| Proteins Identified | A9 (Control) | A9 (>950KDa band) | 4A (>950KDa band) |
|---|---|---|---|
| NDUFA9 | + | + | + |
| NDUFS1 | + | + | + |
| NDUFV1 | + | + | + |
| NDUFB10 | + | + | + |
| NDUFS2 | + | + | + |
| NDUFS3 | + | + | + |
| NDUFV2 | + | + | + |
| NDUFA12 | + | + | + |
| NDUS7 | + | + | + |
| NDUFS8 | + | - | + |
| NDUFA5 | + | - | - |
| NDUFA13 | + | - | - |
| NDUFA7 | + | - | - |
| NDUFA10 | + | - | - |
| NDUFS6 | + | - | - |
| NDUFA8 | + | - | - |
| NDUFB5 | + | - | - |
Complex I assembly in cells without mtDNA-encoded ND4 or ND5 subunit
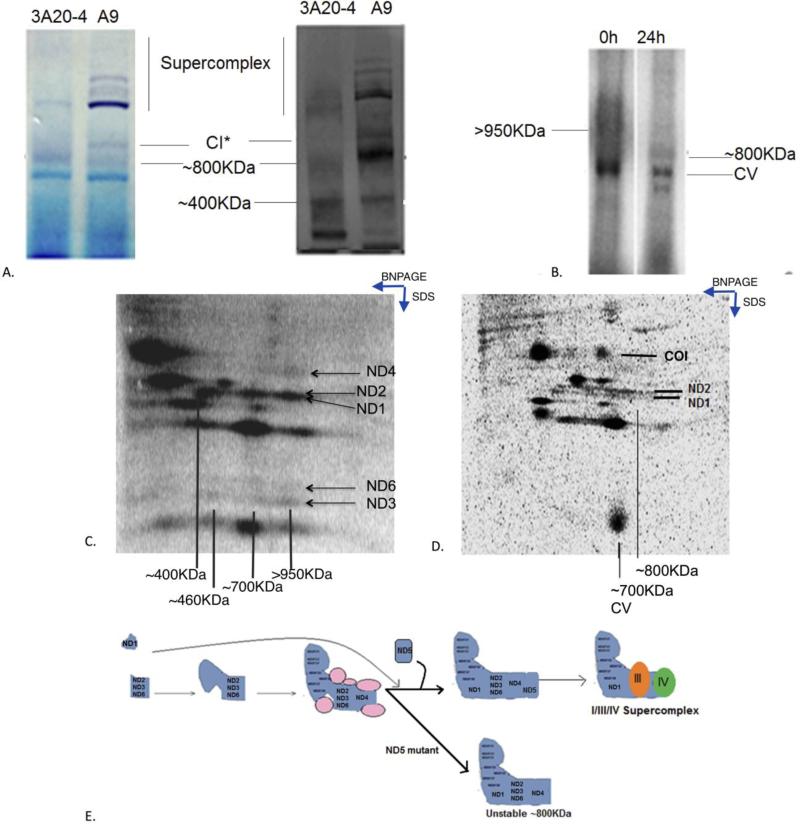
To extend the study to the role of other mtDNA-encoded subunits in complex I assembly, we took advantage of human and mouse cell lines carrying various mutations in ND5 and ND4 with different nuclear backgrounds. To determine the role that ND5 plays in the Complex I assembly pathway, we used mouse 3A20-4 cells isolated previously with a near homoplasmic nonsense mutation in the ND5 gene (33). We followed a similar approach as before wherein we labeled the mitochondrial translational products and observed the incorporation of Complex I subunits into different subcomplexes at 2h and 24h post-labeling.
We confirmed that Complex I assembly was disrupted in the absence of ND5 subunit with both in-gel assay and western blotting, showing almost no Complex I in 3A20-4 cells even though we could see a faint ~800KDa band (Figure 4A). We assigned this molecular weight w.r.t the ~800KDa band observed earlier in control cells, just above the ~750KDa Complex V band. In the 2D SDS PAGE after [35]S labeling we observed ND2, ND3 and ND6 in the ~450KDa and the ~700KDa subcomplex. We also detected a similar >950KDa upper band immediately after labeling and 2D SDS PAGE showed the presence of all mtDNA-encoded subunits except for ND5. However, the intensity of this band significantly decreased with the concurrent appearance of a ~800KDa band after 24h (Figure 4B, C), which was visible even at steady-state levels (Figure 4A). After 24 h, ND1, ND2 and ND4 were still seen in the faint ~800KDa band (Figure 4D).
Figure 4. Early Complex I assembly in ND5 mutant 3A 20-4 cells.
A) shows the lack of Complex I in gel activity in 3A 20-4 cells, using A9 cells as wild type controls (left panel) and the lack of assembled Complex I in 3A 20-4 cells using anti-NDUFA13 antibody (right panel). B) shows the 1D native gel profile in 3A20-4 cells at the 0 h time and 24 h and C) shows the 2D SDS PAGE profile and presence of mtDNA-encoded Complex I subunits at the 0 h time point in the >950KDa Complex I intermediate. D) shows the 2D SDS gel profile at 24 h time point of labeling where the presence of the a faint ~800KDa complex can be detected. Complex V has been labeled for molecular weight reference. E) The cartoon shows the presence of the ~400KDa, 700KDa , >950KDa and ~800KDa subcomplexes seen in the 2D SDS profile and 1D Blue native gel in ND5 mutant cells.
We then used C4T cells derived from the 143B.TK− cell line, which is a homoplasmic ND4 mutant human osteosarcoma cell line (34) to examine the role of ND4 subunits in Complex I assembly . Within 2 h of labeling, a Complex I band was seen in wild type KB30 cells which incorporated into supercomplex by 24 h, while in C4T cells, no Complex I band was seen either early or 24h after labeling (Figure 5A). 2D SDS analysis of these subunits revealed the presence of ND2, ND3 and some ND6 (very faint) in the ~450KDa and ~700KDa subcomplex in C4T cells during the early assembly stages while ND1 was seen in the ~400KDa subcomplex and ND5 was observed in the early ~200KDa subcomplex (Figure 5B). The early assembled >950KDa Complex I intermediate as seen in ND5 and ND6 mutant cells was not observed in these cells, indicating that ND4 is incorporated into Complex I assembly earlier than ND5 and ND6. Almost all subunits except ND2 disappeared after 24 h of assembly and we saw maturation of some portion of ND1 subcomplex to the upper molecular weight band, possibly due to the presence of some ND6, but the majority of the ~700KDa complex was degraded (Figure 5C). In wild type cells, on the other hand, a pattern similar to A9 cells was observed at early times after labeling and after 24 h (Figure 5D, E).
Figure 5. Lack of assembly of Complex I in ND4 mutant cells.
A) shows the 1D BNPAGE of KB30 and ND4 mutant C4T cells at the 0 h and 24 h time points of [35]S labeling. An early Complex I band is seen in KB30 at 0 h but not in C4T cells. B) Shows the 2D SDS PAGE of C4T cells and D) KB30 cells at 0 h time point after [35]S labeling and C) shows the 2DSDS PAGE of C4T cells and E) KB30 cells at the 24 h time point after [35]S labeling. The cartoon shows the presence of the ~400KDa and ~700KDa subcomplexes seen in the 2DSDS profile in ND4 mutant cells.
Incorporation of nuclear encoded subunits in Complex I assembly
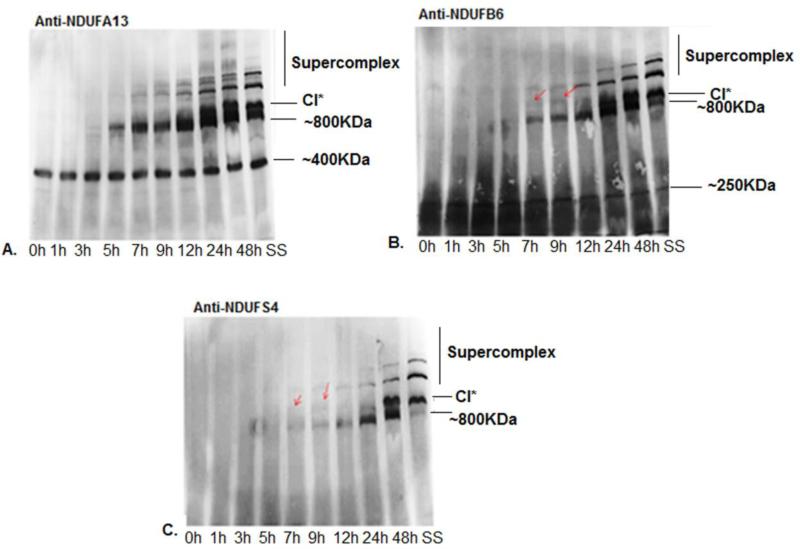
To obtain a complementary picture of Complex I assembly associated with the alteration of mtDNA-encoded subunits described above, we then analyzed the incorporation of nuclear DNA-encoded subunits. We chose three subunits NDUFB6, NDUFS4 and NDUFA13 as they are incorporated at different stages in the assembly pathway, as reviewed previously (42). Mouse fibroblast A9 cells were treated with chloramphenicol, an inhibitor of mitochondrial translation, at 40μg/ml, for 6 days (41). At 6 days of treatment, there was an almost complete turnover of existing Complex I with almost none present immediately after removal of the inhibitor (Figure 6). De novo Complex I assembly was then followed at different time points to observe the incorporation of these four subunits into Complex I (Figure 6A-C). While it is possible that detection of different bands may be affected by different antibody sensitivities, we chose these three antibodies as they were the most sensitive and gave the strongest signal on a blue native gel. Since we corroborated these results with our [35]S results and previously established subcomplex modules we believe the bands we observe to be genuine representation of Complex I subcomplexes. We observed that NDUFA13 was present consistently in a ~400KDa subcomplex, closer to the Complex III band and progressed to the ~800KDa subcomplex almost 5h after the removal of the CAP block (Figure 6A). It then entered into a supercomplex starting at 7h after the assembly begins; at the same time, the CI* band also started accumulating. This was also seen with the other two nuclear encoded subunits. NDUFB6 is a nuclear-encoded subunit that forms a part of the membrane arm. NDUFB6 was a part of a low molecular weight subcomplex approximately ~200KDa near the Complex IV band (Figure 6B). NDUFB6 was observed in the ~800KDa subcomplex at around 7h after removal of inhibitor, indicating it was incorporated after the assembly of NDUFA13 into Complex I. At the same time, NDUFB6 was also seen in a band that was slightly larger than the Complex I band which likely corresponds to the early >950KDa Complex I intermediate seen in both A9 and 4A cells immediately after labeling of mtDNA-encoded subunits with [35]S (Figure 1B, C).
Figure 6. Incorporation of nuclear encoded subunits in Complex I assembly.
Using chloramphenicol treatment for 6 days followed by recovery at different time points, indicated below each image, to document the incorporation of the following Complex I subunits in Complex I assembly A) anti- NDUFA13 antibody, B) anti-NDUFB6 antibody, C) anti-NDUFS4. Red arrows indicate the position of the >950KDa complex I intermediate while other Complex I subcomplexes have been indicated by their molecular weights. SS denotes steady state controls i.e. Complex I bands observed at steady state only. These samples were not treated with chloramphenicol.
We expected NDUFS4 to appear in Complex I at a very late stage, as reported recently (43) but our observations showed that NDUFS4 is incorporated in Complex I assembly intermediates earlier than 24 h (Figure 6C). While NDUFS4 is seen in the Complex I and supercomplexes bands around 24h, it was seen earlier in the ~800KDa subcomplex and the >950KDa band which was also observed in the NDUFB6 pattern of assembly at about 5h. It is interesting to note that the ~800KDa subcomplex was seen around ~7h using all three subunits. Since we saw the >950KDa intermediate well before that in our [35]S-labeled cells, we propose that the ~800KDa subcomplex appears after the >950KDa subcomplex in the Complex I assembly pathway. This is also corroborated by our data on ND5 mutant cells which show a ~800KDa band at steady-state levels and also in the 24hs [35]S-labeled gel but not immediately after labeling.
Discussion
Using cybrid cell lines with mutations in ND6, we showed a Complex I assembly intermediate, that has not been observed before, that is an almost complete Complex I which, however, is degraded over a period of 24h. We also showed that this early complex I intermediate contains all the mtDNA-encoded subunits except ND1 and ND5, indicating that the incorporation of ND1 and ND5 into Complex I depends on the presence of ND6. This agrees with a previous report suggesting a role for ND6 in the maturation of ND1- and ND5-containing subcomplexes. Our results thus suggest that in ND6 and ND5 mutant cells, the inability of the >950KDa band to progress to supercomplex leads to a Complex I deficiency. It was previously predicted that Complex III and Complex IV associate with Complex I through the distal part of the membrane arm and the supercomplex formation is required to stabilize Complex I (44–46). It is therefore possible that, as a result of lack of ND5 or ND6, association of the other respiratory complexes through the membrane arm may be hindered, in turn causing the >950KDa intermediate to be unstable.
Unlike previous reports, our results show that in ND4, ND5 and ND6 mutant cells, the ND1 signal remains strong in the ~400KDa subcomplex and does not enter the assembling Complex I until much later. While this was also observed in ND6 mutant cells in a previous study (12) we also showed this in wild type cells as well as in ND4 mutant cells. We also demonstrate that in the absence of ND1, a peripheral arm is still formed in the >950KDa intermediate in ND6 mutant cells, and that the presence of the ~450KDa subcomplex containing ND2, ND3 and ND6 is enough for addition of peripheral arm subunits.
Analysis of mouse as well as human cybrids with ND5 mutation showed a pattern of assembly similar to the ND6 mutant cells, i.e., the presence of an almost complete Complex I intermediate band at an early time point. In ND5 mutant cells, this band dissociates to form the ~800KDa subcomplex over time. However, in these cells we also see ND1 in the final ~800KDa band, indicating that ND5 may be one of the last mtDNA-encoded subunits incorporated into Complex I. The presence of a faint ~800KDa subcomplex as well as a supercomplex band (Figure 4A) at steady-state indicates that without ND5, these cells can still process Complex I assembly all the way through until the last step.
We also show that the ND4 mutation completely abolishes Complex I assembly. We do not see the almost complete Complex I intermediate band in ND4 mutant cells, indicating that the ND4-containing subcomplex may be necessary to push the subcomplexes into the >950KDa band seen in ND5 and ND6 mutant cells. Recently, a study showed that lack of ND6 in ND4 mutant cells indicates the assembly of ND4 earlier than ND6 (12). Even in the wild type cells, ND6 is seen to migrate with ND2 and ND3, making it a part of the earliest Complex I subcomplex and hence it is incorporated into Complex I assembly earlier than ND4. This is corroborated by the recent findings (47) of the mammalian Complex I architecture study wherein supernumerary subunits form protective layers around ND2, ND4l, ND3 and ND6 indicating that they may be incorporated together in the assembly pathway. We therefore propose that because ND4 is also seen in the almost complete complex I intermediate in absence of ND6, indicating that the assembly of ND4 into Complex I may follow ND6 but does not depend on the presence or absence of ND6, thus making our assembly pathway model partially sequential in contrast to other models.
Our model suggests that the >950KDa intermediate lasts until ~6h post de novo assembly before assembling into supercomplexes via the ~800KDa subcomplex. In wild type A9 cells, as early as 2 h after labeling we observed that the >950KDa intermediate progress into supercomplexes, explaining the lesser intensity of the >950KDa band in A9 cells as compared to ND6 mutant 4A cells. This slight shift in molecular weight may be interpreted as assembly and stability factors dissociating from a matured, assembled Complex I. The presence of ND4, and not ND5, in the early upper band also disproves previous reports that suggest ND4 is incorporated later and in conjunction with ND5 (42, 48, 49), and we also show that ND4 is needed to generate the >950KDa intermediate. Our study also reveals that the ND1-containing subcomplex does not join the assembling Complex until much later than previously thought.
Our data thus suggest a sequential pathway for assembly of mtDNA-encoded subunits into Complex I assembly. ND2 and ND3 (ND6 in wild type cells) appear to be the earliest incorporated subunits, appearing in all three mutant cell lines as well as wild type cells at the earliest hour after labeling. ND2, ND3 and ND6 are seen in the ~450KDa, ~700KDa and >950KDa subcomplex 2 h after labeling of mtDNA-encoded subunits, while ND1 remains in a slightly lower ~400KDa subcomplex but does not incorporate into the other subcomplexes. The ~450KDa subcomplex then assembles into a ~700KDa subcomplex, presumably with addition of peripheral arm subunits and containing ND2, ND3, ND6 and ND4L. This is followed by ND4-containing subcomplex that joins the ~700KDa subcomplex. Thus, in our results we observe the entry of ND6 prior to ND4 into the Complex I assembly pathway. Addition of the ND4 subcomplex propels the Complex I intermediates into the >950KDa intermediate, presumably with assembly factors still associated. The presence of NDUFS4 in the >950KDa band in our Blue Native gel and the presence of other nuclear-encoded subunits detected by mass spectroscopy confirm the existence of a peripheral arm in our >950KDa intermediate. In ND6 mutant cells, we see all the mtDNA-encoded subunits except ND1 and ND5, suggesting that these incorporate after ND6 assembles into Complex I. The assembly of ND1 and ND5 also does not appear to be sequential, but merely depends on the presence of ND6 in the assembling Complex I. Addition of ND1- and ND5-containing subcomplexes signals the end of Complex I assembly. At this point, ~6h after the de novo assembly begins, the >950KDa complex I shifts in molecular weight to a ~800KDa intermediate with subcomplexes added from ND1 and ND5. While the ~400 Kodak, ~700KDa and ~800KDa subcomplexes have been described before to some extent (10, 50, 51), the dynamic and kinetic progression of these subcomplexes as well as the presence of the >950KDa intermediate have not been described before. This may be partly because most of these studies are in patients’ cells and as such it is hard to determine whether the presence and composition of these subcomplexes in these cells is physiologically relevant or an artifact of defective Complex I assembly.
It is interesting to note that our study indicates that it would take 12-24 hours for a newly synthesized complex I subunit to incorporate into functional respiratory machinery. It might be reasonable to assume that in order to maintain a stable respiratory phenotype, time required for Complex I assembly should not exceed cell doubling time. Together, the rate of assembly of respiratory complexes, in particular Complex I, might be subjected to tissue-specific regulation, and that in turn could be the base for the tissue-specific presentation of some mitochondrial diseases which has been shown to associate with the regulatory factors of Complex I assembly. ND4 and ND5 genes are among the most affected diseases-associated ones for mitochondrial diseases. Our results, indicating that they likely play an important role in stabilizing Complex I and facilitating supercomplex assembly, provide some insights for the pathogenesis for the related diseases. Another important implication of this study is although specific mutations in ND1, ND4 and ND6 genes are reported as the primary ones for LHON, the different roles played by these subunits in Complex I assembly may be responsible for differences in severity seen and as such different treatment approaches might be taken as these three subunits play distinctive roles in Complex I dynamics. In this study, we combined mass spectroscopy, blue native and 2D gels approaches to show a kinetic progression of Complex I assembly, and these techniques have allowed us to discern the assembly pattern to the greatest detail. Our results also showed the formation of a new Complex I intermediate that may be important for progression of Complex I into supercomplexes. Meanwhile, our data gives rise to other questions: what role does ND6 play in assembly of ND1 and ND5, two subunits that are situated at the opposite ends on the membrane arm (47) that warrants future studies. Nevertheless, these new findings may provide a new basis for understanding how Complex I assembly progresses into supercomplex and the role of these supercomplexes in mitochondrial disorders with ND4, ND5 and ND6 mutations.
Highlights.
We find that ND6 involves in recruitment of ND1 and ND5 enter the intermediates.
The addition of ND5 stabilizes the complex and is required for the critical transition from Complex I to supercomplexes assembly.
We identified a new Complex I intermediate containing both membrane and matrix arms at an early assembly stage.
The Complex I assembly pathway has been redefined.
Figure 7. Model for Complex I assembly.
A) shows the current hypothesized model for Complex I assembly with the nuclear encoded subunits used in this study B) Stepwise and kinetic assembly of the mitochondrial encoded subunits and some of the nuclear encoded subunits during Complex I assembly. The subunits labeled in red are nuclear encoded subunits while the pink circles are assembly factors.
Acknowledgements
This work was supported by grants from National Institute of Health (1 R01 GM109434-01A1 and a pilot grant from M01RR01346-8UL1TR000149) to YB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine Complex I Is a Complex of 45 Different Subunits. J. Biol. Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 2.Chomyn A, Mariottini P, Cleeter MWJ, Ragan CI, Matsuno-Yagi A, Hatefi Y, Doolittle RF, Attardi G. Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Publ. Online 18 April 1985 Doi101038314592a0. 1985;314:592–597. doi: 10.1038/314592a0. [DOI] [PubMed] [Google Scholar]
- 3.Chomyn A, Cleeter MW, Ragan CI, Riley M, Doolittle RF, Attardi G. URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit. Science. 1986;234:614–618. doi: 10.1126/science.3764430. [DOI] [PubMed] [Google Scholar]
- 4.Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landázuri MO, Enríquez JA. NDUFA4 Is a Subunit of Complex IV of the Mammalian Electron Transport Chain. Cell Metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Friedrich T, Weidner U, Nehls U, Fecke W, Schneider R, Weiss H. Attempts to define distinct parts of NADH:ubiquinone oxidoreductase (complex I). J. Bioenerg. Biomembr. 1993;25:331–337. doi: 10.1007/BF00762458. [DOI] [PubMed] [Google Scholar]
- 6.Hofhaus G, Weiss H, Leonard K. Electron microscopic analysis of the peripheral and membrane parts of mitochondrial NADH dehydrogenase (Complex I). J. Mol. Biol. 1991;221:1027–1043. doi: 10.1016/0022-2836(91)80190-6. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich T, Weiss H. Modular evolution of the respiratory NADH:ubiquinone oxidoreductase and the origin of its modules. J. Theor. Biol. 1997;187:529–540. doi: 10.1006/jtbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 8.Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell. Proteomics MCP. 2003;2:117–126. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Thorburn DR, Ryan MT, McKenzie M, Lazarou Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta. 2009;1793:78–88. doi: 10.1016/j.bbamcr.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the Assembly Profiles for Mitochondrial- and Nuclear-DNA-Encoded Subunits into Complex I. Mol. Cell. Biol. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardol P, Boutaffala L, Memmi S, Devreese B, Matagne RF, Remacle C. In Chlamydomonas, the loss of ND5 subunit prevents the assembly of whole mitochondrial complex I and leads to the formation of a low abundant 700 kDa subcomplex. Biochim. Biophys. Acta. 2008;1777:388–396. doi: 10.1016/j.bbabio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Perales-Clemente E, Fernández-Vizarra E, Acín-Pérez R, Movilla N, Bayona-Bafaluy MP, Moreno-Loshuertos R, Pérez-Martos A, Fernández-Silva P, Enríquez JA. Five entry points of the mitochondrially encoded subunits in mammalian complex I assembly. Mol. Cell. Biol. 2010;30:3038–3047. doi: 10.1128/MCB.00025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadava N, Houchens T, Potluri P, Scheffler IE. Development and characterization of a conditional mitochondrial complex I assembly system. J. Biol. Chem. 2004;279:12406–12413. doi: 10.1074/jbc.M313588200. [DOI] [PubMed] [Google Scholar]
- 14.Bourges I, Ramus C, Mousson de Camaret B, Beugnot R, Remacle C, Cardol P, Hofhaus G, Issartel J-P. Structural organization of mitochondrial human complex I: role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem. J. 2004;383:491–499. doi: 10.1042/BJ20040256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel RO, Dieteren CEJ, van den Heuvel LPWJ, Willems PHGM, Smeitink JAM, Koopman WJH, Nijtmans LGJ. Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J. Biol. Chem. 2007;282:7582–7590. doi: 10.1074/jbc.M609410200. [DOI] [PubMed] [Google Scholar]
- 16.Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 2005;115:2784–2792. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nouws J, Nijtmans L, Houten SM, van den Brand M, Huynen M, Venselaar H, Hoefs S, Gloerich J, Kronick J, Hutchin T, Willems P, Rodenburg R, Wanders R, van den Heuvel L, Smeitink J, Vogel RO. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 2010;12:283–294. doi: 10.1016/j.cmet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Saada A, Edvardson S, Rapoport M, Shaag A, Amry K, Miller C, Lorberboum-Galski H, Elpeleg O. C6ORF66 is an assembly factor of mitochondrial complex I. Am. J. Hum. Genet. 2008;82:32–38. doi: 10.1016/j.ajhg.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunning CJR, McKenzie M, Sugiana C, Lazarou M, Silke J, Connelly A, Fletcher JM, Kirby DM, Thorburn DR, Ryan MT. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 2007;26:3227–3237. doi: 10.1038/sj.emboj.7601748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheftel AD, Stehling O, Pierik AJ, Netz DJA, Kerscher S, Elsässer H-P, Wittig I, Balk J, Brandt U, Lill R. Human Ind1, an Iron-Sulfur Cluster Assembly Factor for Respiratory Complex I. Mol. Cell. Biol. 2009;29:6059–6073. doi: 10.1128/MCB.00817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugiana C, Pagliarini DJ, McKenzie M, Kirby DM, Salemi R, Abu-Amero KK, Dahl H-HM, Hutchison WM, Vascotto KA, Smith SM, Newbold RF, Chistodoulou J, Calvo S, Mootha VK, Ryan MT, Thorburn DR. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am. J. Hum. Genet. 2008;83:468–478. doi: 10.1016/j.ajhg.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saada A, Vogel RO, Hoefs SJ, van den Brand MA, Wessels HJ, Willems PH, Venselaar H, Shaag A, Barghuti F, Reish O, Shohat M, Huynen MA, Smeitink JAM, van den Heuvel LP, Nijtmans LG. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am. J. Hum. Genet. 2009;84:718–727. doi: 10.1016/j.ajhg.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19:802–815. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winklhofer KF, Haass C. Mitochondrial dysfunction in Parkinson’s disease. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 2010;1802:29–44. doi: 10.1016/j.bbadis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Sharma LK, Lu J, Bai Y. Mitochondrial respiratory complex I: structure, function and implication in human diseases. Curr. Med. Chem. 2009;16:1266–1277. doi: 10.2174/092986709787846578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofhaus G, Attardi G. Lack of assembly of mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase and loss of enzyme activity in a human cell mutant lacking the mitochondrial ND4 gene product. EMBO J. 1993;12:3043–3048. doi: 10.1002/j.1460-2075.1993.tb05973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai Y, Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai Y, Hu P, Park JS, Deng J-H, Song X, Chomyn A, Yagi T, Attardi G. Genetic and functional analysis of mitochondrial DNA-encoded complex I genes. Ann. N. Y. Acad. Sci. 2004;1011:272–283. doi: 10.1007/978-3-662-41088-2_26. [DOI] [PubMed] [Google Scholar]
- 30.Hofhaus G, Attardi G. Efficient selection and characterization of mutants of a human cell line which are defective in mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase. Mol. Cell. Biol. 1995;15:964–974. doi: 10.1128/mcb.15.2.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malfatti E, Bugiani M, Invernizzi F, de Souza CF-M, Farina L, Carrara F, Lamantea E, Antozzi C, Confalonieri P, Sanseverino MT, Giugliani R, Uziel G, Zeviani M. Novel mutations of ND genes in complex I deficiency associated with mitochondrial encephalopathy. Brain J. Neurol. 2007;130:1894–1904. doi: 10.1093/brain/awm114. [DOI] [PubMed] [Google Scholar]
- 32.Chomyn A. Mitochondrial genetic control of assembly and function of complex I in mammalian cells. J. Bioenerg. Biomembr. 2001;33:251–257. doi: 10.1023/a:1010791204961. [DOI] [PubMed] [Google Scholar]
- 33.Bai Y, Shakeley RM, Attardi G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell. Biol. 2000;20:805–815. doi: 10.1128/mcb.20.3.805-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai Y, Hájek P, Chomyn A, Chan E, Seo BB, Matsuno-Yagi A, Yagi T, Attardi G. Lack of complex I activity in human cells carrying a mutation in MtDNA-encoded ND4 subunit is corrected by the Saccharomyces cerevisiae NADH-quinone oxidoreductase (NDI1) gene. J. Biol. Chem. 2001;276:38808–38813. doi: 10.1074/jbc.M106363200. [DOI] [PubMed] [Google Scholar]
- 35.Bai Y, Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 37.Wittig I, Braun H-P, Schägger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 38.Fernández-Vizarra E, Ferrín G, Pérez-Martos A, Fernández-Silva P, Zeviani M, Enríquez JA. Isolation of mitochondria for biogenetical studies: An update. Mitochondrion. 2010;10:253–262. doi: 10.1016/j.mito.2009.12.148. [DOI] [PubMed] [Google Scholar]
- 39.Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- 40.Schägger H. Tricine-SDS-PAGE. Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 41.Storrie B, Attardi G. EXPRESSION OF THE MITOCHONDRIAL GENOME IN HELA CELLS. J. Cell Biol. 1973;56:819–831. doi: 10.1083/jcb.56.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta BBA - Bioenerg. 2012;1817:851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Moreno-Lastres D, Fontanesi F, García-Consuegra I, Martín MA, Arenas J, Barrientos A, Ugalde C. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 2012;15:324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Althoff T, Mills DJ, Popot J-L, Kühlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schäfer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J. Architecture of Active Mammalian Respiratory Chain Supercomplexes. J. Biol. Chem. 2006;281:15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- 47.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gershoni M, Fuchs A, Shani N, Fridman Y, Corral-Debrinski M, Aharoni A, Frishman D, Mishmar D. Coevolution predicts direct interactions between mtDNA-encoded and nDNA-encoded subunits of oxidative phosphorylation complex i. J. Mol. Biol. 2010;404:158–171. doi: 10.1016/j.jmb.2010.09.029. [DOI] [PubMed] [Google Scholar]
- 49.Mckenzie M, Ryan MT. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life. 2010;62:497–502. doi: 10.1002/iub.335. [DOI] [PubMed] [Google Scholar]
- 50.Lazarou M, Smith SM, Thorburn DR, Ryan MT, McKenzie M. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. FEBS J. 2009;276:6701–6713. doi: 10.1111/j.1742-4658.2009.07384.x. [DOI] [PubMed] [Google Scholar]
- 51.Ugalde C, Vogel R, Huijbens R, Heuvel B, van den, Smeitink J, Nijtmans L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum. Mol. Genet. 2004;13:2461–2472. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]