Abstract
Immunodominance describes a phenomenon whereby the immune system consistently targets only a fraction of the available antigen pool derived from a given pathogen. In the case of CD8+ T-cells, these constrained epitope targeting patterns are linked to human leukocyte antigen (HLA) class-I expression and determine disease progression. Despite the biological importance of these predetermined response hierarchies, however, little is known about the factors that control immunodominance in vivo. In this study, we conducted an extensive analysis of CD8+ T-cell responses restricted by a single HLA class-I molecule to evaluate the mechanisms that contribute to epitope targeting frequency and antiviral efficacy in HIV-1 infection. A clear immunodominance hierarchy was observed across 20 different epitopes restricted by HLA-B*42:01, which is highly prevalent in populations of African origin. Moreover, in line with previous studies, Gag-specific responses and targeting breadth were associated with lower viral load set-points. However, peptide-HLA-B*42:01 binding affinity and stability were not significantly linked with targeting frequencies. Instead, immunodominance correlated with epitope-specific usage of public TCRs, defined as amino acid residue-identical TRB sequences that occur in multiple individuals. Collectively, these results provide the first insights into a potential link between shared TCR recruitment, immunodominance and antiviral efficacy in a major human infection.
Keywords: CD8+ T-cell, clonotype, HIV-1, immunodominance, TCR
Introduction
CD8+ T-cells play a central role in the immune response to viruses (1). However, clear-cut differences exist between distinct specificities in terms of antiviral efficacy (2, 3). Moreover, epitope targeting patterns are often predetermined within a hierarchy of immunodominance across restriction elements (4-12). These biologically imposed limitations can therefore dictate the outcome of certain viral infections.
In the case of HIV-1, disease progression is strongly affected by the expression of particular HLA alleles (13, 14). Although several mechanisms have been proposed to explain this association (13), one key factor is the relative ability of epitope-specific CD8+ T-cell populations to kill HIV-infected cells. For example, Gag-specific responses typically contain HIV-1 more effectively in vivo (15-19) and in vitro (20, 21) compared to those that target other viral proteins. In addition, protective HLA class I molecules, such as HLA-B*27 and HLA-B*57 (22-24), restrict immunodominant Gag-specific responses that select viral escape variants with impaired replicative capacity (22, 25-27).
A number of factors can influence immunodominance (2, 4, 6, 11). Antigen presentation itself is the culmination of several upstream events, such as the kinetics of protein expression, the abundance of protein delivered into the cytoplasm, intra-cytoplasmic proteosomal cleavage, translocation by TAP, peptide loading onto MHC, ERAP1/2 trimming and transport to the cell surface (28). Peptide-MHC binding affinity and stability determine the subsequent availability of antigen over time, whereas TCR avidity and the frequency of naïve precursors govern the size of the potentially responsive T-cell pool (7, 9, 10, 12). The phenomenon of immunodomination, whereby certain responses are subordinated in the presence of particular high frequency responses (29), also plays a role. Despite the complexity of these multi-faceted processes, however, the end result is a largely predictable pattern of immunodominance for any given virus.
Emerging studies have highlighted a key role for the TCR repertoire as an independent determinant of antiviral efficacy in multiple systems (30-35). Although the process of V(D)J rearrangement can theoretically generate 1015-20 distinct TCRs (36), extreme biases exist during recombination, thymic selection, naïve T-cell recruitment and subsequent clonal expansion (6, 10, 36-38). These biases can ultimately generate identical or ‘public’ epitope-specific TCRs in multiple individuals (38). However, the role of TCR bias in relation to immunodominance remains ill-defined, with murine studies yielding apparently contradictory data (39, 40).
To assess the potential influence of TCR publicity on CD8+ T-cell immunodominance and antiviral efficacy in a human viral infection, we conducted an extensive analysis of HLA-B*42:01-restricted responses directed against an array of different epitopes derived from HIV-1. The prevalence of HLA-B*42:01 in most populations of African origin, combined with the substantial repertoire of associated viral peptides, enabled a large scale study controlled for the restriction element across multiple individuals with chronic HIV-1 infection. Consequently, we were able to detect statistically meaningful correlations between these parameters.
Materials and Methods
Study subjects
The study cohort comprised 2,093 female adults with chronic, antiretroviral therapy (ART)-naïve C-clade HIV-1 infection, recruited from five cohorts: (i) Durban, South Africa (14, 17, 25, 41); (ii) Gaborone, Botswana (42); (iii) Bloemfontein, South Africa (43); (iv) Kimberley, South Africa (44); and (v) Thames Valley, UK (45). A total of 246 HLA-B*42:01+ individuals with documented proviral DNA sequences, CD4+ T-cell counts, plasma viral loads and four-digit HLA genotyping data were identified within the entire cohort, from which 181 were screened with overlapping peptides (OLPs) to map HIV-specific CD8+ T-cell responses in IFNγ ELISpot assays. Informed consent was obtained from all participants. The following Institutional Review Boards approved the study: (i) University of KwaZulu-Natal, South Africa; (ii) University of the Free State, South Africa; (iii) Health Research Development Committee, Botswana Ministry of Health, Botswana; (iv) Office of Human Research Administration, Harvard School of Public Health, USA; and (v) University of Oxford, UK.
IFNγ ELISpot
Virus-specific CD8+ T-cell responses across the whole HIV-1 proteome were determined for 1,009 C-clade-infected subjects via direct ex vivo IFNγ ELISpot analysis. Antigens comprised 410 OLPs based on the C-clade consensus (2001) arranged in a matrix system with 11-12 peptides per pool. Responses to matrix pools were deconvoluted by subsequent testing with the individual 18mer peptides contained in each pool (15). Associations between HLA-B*42:01 expression and OLP targeting were calculated from a total of 181 HLA-B*42:01+ individuals. Percent targeting frequency (immunodominance rank) calculations for HLA-B*42:01-restricted epitopes required the exclusion of 27 individuals co-expressing HLA-B*07:02/39:10/42:02/81:01 to eliminate alternative presentation by other B7 superfamily members (46). Viral load set-point calculations versus targeting of HLA-B*42:01-restricted OLPs were based exclusively on data from the Durban cohort (n=126) to minimize the influence of external factors.
HLA class I typing
Four-digit HLA-A/B/C genotyping was performed using real-time reverse sequence-specific oligonucleotide (SSO) kits (Dynal) as described previously (47).
Epitope mapping and HLA restriction
Epitope mapping and recognition assays were performed as described previously (16, 45). Single allele-matched B-lymphoblastoid cell lines (BLCLs) were used to determine HLA restriction (16, 48).
Tetramers
Tetrameric peptide-HLA complexes were generated and used as described previously (16, 49). Samples were acquired using an LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo version 8.8.6 (TreeStar Inc.). Events were gated serially on singlets, lymphocytes, live cells and CD3+ T-cells prior to analysis in bivariate tetramer versus CD8 plots.
Proviral DNA sequencing
Sequences from Gag (n=1,857), Pol (n=1,052) and Nef (n=1,327) were generated by extraction of genomic DNA from peripheral blood mononuclear cells (PBMCs) and amplification via nested PCRs as described previously (17, 44, 47). Purified products were sequenced using the Big Dye Terminator Ready Reaction (Life Technologies) (41, 50). Vif, Rev and Env sequences were available from 255 subjects (51).
Peptide-HLA binding and stability assays
Peptide-HLA stability was measured as described previously (52). Peptide-HLA binding affinity was measured via AlphaScreen technology (Perkin Elmer) (53).
TCR clonotyping
Clonotypic analysis of antigen-specific CD8+ T-cell populations restricted by HLA-B*42:01 was performed as described previously (54). Briefly, viable HIV-specific tetramer+ CD8+ T-cell populations (n = 48) were sorted by flow cytometry at >98% purity directly into RNAlater. A median of 2,120 cells (25th percentile = 1,068 cells, 75th percentile = 4,340 cells) was sorted per population with a median response magnitude of 0.60% (25th percentile = 0.3%, 75th percentile = 1.4%). The number of sorted cells correlated with response magnitude (r = 0.8, P < 0.0001). Unbiased amplification of all expressed TRB gene products was then conducted using a template-switch anchored RT-PCR with a 3′ constant region primer. Amplicons were subcloned, sampled, sequenced (n = 3,592) and analyzed as described previously (55). The IMGT nomenclature is used in this report (56).
Statistical analysis
Associations between HLA-B*42:01 expression, HIV-1 polymorphisms and 18mer peptide (OLP) responses were determined as described previously (44, 57). The Dunn's multiple comparisons test was used to compare median viral loads between OLP responders and non-responders, the number of OLP responses between different HIV-1 proteins (breadth) and the number of different TCR clonotypes between responses. The two-tailed Mann-Whitney U-test was used to compare viral loads and CD4+ T-cell counts between individuals carrying wildtype sequences or escape polymorphisms within the Gag-RM9 epitope. P values were calculated using GraphPad Prism version 6.0c (GraphPad Software). Differences in Gag-RM9 sequence polymorphisms between HLA-B*42:01+ and HLA-B*42:01- subjects were calculated using Fisher's exact test. Correlations between % targeting frequencies (IFNγ OLP ELISpot responders) and either peptide-HLA binding affinities (IC50 KD value) or peptide-HLA binding half-lives (hours) were calculated using the Spearman rank test. Clonotypic data were normalized as described previously (58-60). Briefly, all samples were rarefied down to the lowest estimated coverage (88%) prior to calculations of TCR sharing. This process was repeated 10,000 times and mean publicity scores were used for statistical analysis.
Results
HLA-B*42:01-restricted responses conform to a strict immunodominance hierarchy
Initially, we used IFNγ ELISpot assays to screen 1,009 ART-naïve C-clade-infected female individuals, 181 (18%) of whom carried the HLA-B*42:01 allele, for responses to a panel of 410 OLPs (15, 16, 61) spanning the entire HIV-1 proteome (Table 1). This panel comprised seven HLA-B*42:01-restricted epitopes already listed in the Los Alamos Immunology Database (www.hiv.lanl.gov) (62). In addition, we identified 13 novel epitopes via statistical associations between recognition of a particular 18mer OLP and expression of HLA-B*42:01 (q < 0.05). This set of 20 HLA-B*42:01-restricted HIV-1 epitopes was used for further analysis.
Table 1. HLA-B*42:01 associations with IFNγOLP responses (n=1,009).
| Protein-OLP | Sequence OLP | Optimal | Epitope | HXB2 | Q-valued | Reference |
|---|---|---|---|---|---|---|
| Gag-3 | EKIRLRPGGKKHYMLKHL | RPGGKKHYMa | Gag-RM9 | Gag(22-30) | ns | This study |
| Gag-20 | QMVHQAISPRTLNAWVKV | SPRTLNAWVb | Gag-SV9 | Gag(148-156) | ns | This study |
| Gag-25 | GATPQDLNTMLNTVGGH | TPQDLNTML | Gag-TL9 | Gag(180-188) | 1.28E-124 | Llano 2008 |
| Gag-37 | WIILGLNKIVRMYSPVSIc | no | no | no | 1.28E-02 | This study |
| Gag-48 | ACQGVGGPSHKARVLAEA | GPSHKARVL | Gag-GL9 | Gag(355-363) | ns | Llano 2008 |
| Gag-52 | QRSNFKGPKRIVKCF | GPKRIVKCFb | Gag-GF9 | Gag(386-394) | 8.25E-03 | This study |
| RT-184 | PSINNETPGIRYQYNVL | TPGIRYQYNVLb | RT-TL11 | Pol(294-304) | 2.01E-12 | This study |
| RT-186 | QYNVLPQGWKGSPAIF | LPQGWKGSPAIb | RT-LI11 | Pol(304-314) | 3.90E-02 | This study |
| RT-187 | QGWKGSPAIFQSSMTKIL | SPAIFQSSMb | RT-SM9 | Pol(311-319) | ns | This study |
| RT-202 | SKLNWASQIYPGIKVRQL | YPGIKVRQL | RT-YL9 | Pol(426-434) | 7.17E-09 | Llano 2008 |
| Int-244 | MASEFNLPPIVAKEIVA | LPPIVAKEI | Int-LI9 | Pol(743-751) | 5.04E-49 | Llano 2008 |
| Int-275 | KVVPRRKAKIIKDYGKQM | IIKDYGKQMa | Int-IM9 | Pol(982-990) | 1.48E-98 | This study |
| Nef-73 | GALTSSNTDTTNADCAWLc | no | no | no | 3.45E-02 | This study |
| Nef-76 | EVGFPVRPQVPLRPMTFK | RPQVPLRPM | Nef-RM9 | Nef(71-79) | 8.24E-17 | Llano 2008 |
| Nef-78 | FKGAFDLSFFLKEKGGLc | no | no | no | 9.15E-03 | This study |
| Nef-84 | NYTPGPGVRYPLTFGWCF | TPGPGVRYPL | Nef-TL10 | Nef(382-411) | 3.26E-16 | Llano 2008 |
| Vpr-281 | ELKQEAVRHFPRPWLHGL | FPRPWLHGL | Vpr-FL9 | Vpr(34-42) | 1.61E-70 | Llano 2008 |
| Vif-407 | RHHYESRHPKVSSEVHI | HPKVSSEVHIa | Vif-HI10 | Vif(48-57) | 7.97E-69 | This study |
| Env-328 | VCTRPNNNTRKSIRI | RPNNNTRKSIb | Env-RI10 | Env(298-307) | 2.24E-03 | This study |
| Env-401 | NIPRRIRQGFEAALL | IPRRIRQGFa | Env-IF9 | Env(843-851) | ns | This study |
New optimal epitope (not listed in Los Alamos ‘A’ List Database) defined in this study.
New optimal epitope (not listed in Los Alamos ‘A’ List Database) not defined in this study.
Optimal epitope not identified within the OLP sequence.
q-values for associations between HLA-B*42:01 expression (n=181) and OLP responses computed from analysis of IFNγ ELISpot data across 1,009 subjects. no, not optimized; ns, not significant.
Next, we ranked the response hierarchy among these 20 epitopes based on targeting frequency (immunodominance rank). A total of 154 HLA-B*42:01+ individuals were included in this analysis, stratified for lack of HLA-B*07:02/39:10/42:02/81:01 co-expression. Targeting frequency correlated with response magnitude (r = 0.96, P < 0.0001; Supplementary Fig S1) and conformed to a clear pattern of immunodominance (Fig 1A).
Figure 1. Identification of immunodominant and subdominant HLA-B*42:01-restricted epitopes.
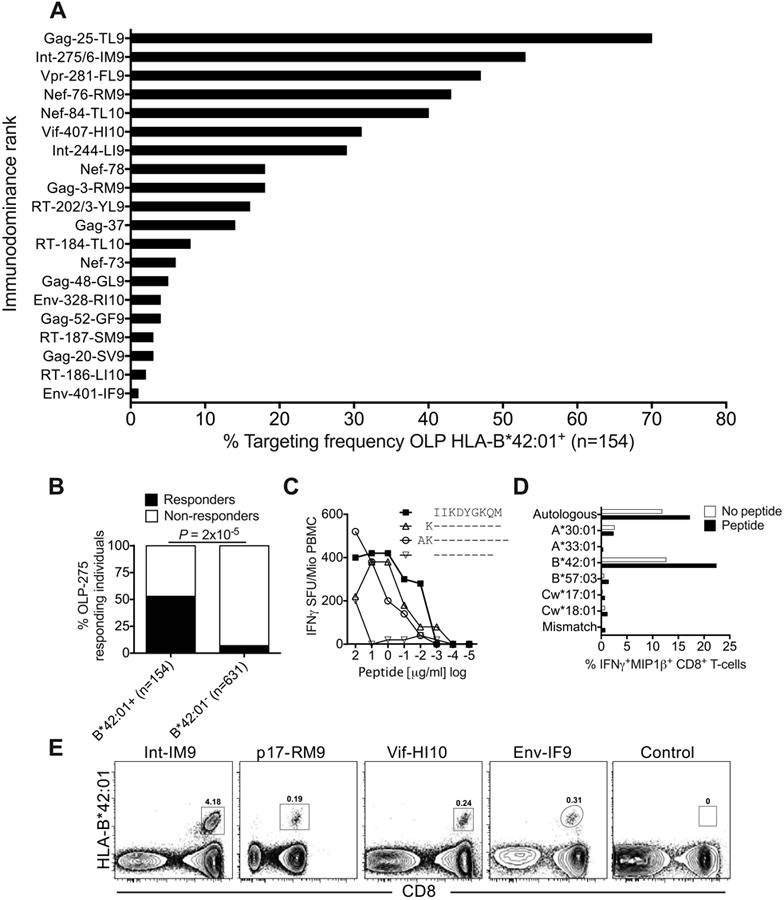
(A) Targeting frequencies for ‘protein-18mer peptide-optimal epitope name’ OLPs in HLA-B*42:01+ individuals, stratified for lack of HLA-B*07:02/39:10/42:02/81:01 co-expression (n=154). Responses were determined via IFNγ ELISpot assays. (B) Correlation between HLA-B*42:01 status and targeting of OLP-275 (KVVPRRKAKIIKDYGKQM) in IFNγ ELISpot assays. Significance was determined using Fisher's exact test. (C) Identification of the optimal HLA-B*42:01-restricted epitope Int-IM9 (IIKDYGKQM) via peptide truncations in IFNγ ELISpot assays. (D) Confirmation of HLA-B*42:01 as the restriction element for Int-IM9 via peptide pulsing of BLCLs partially HLA-matched to donor R014 (A*30:01/33:01, B*42:01/57:03, Cw17:01/18:01). Autologous or mismatched BLCLs were used as positive and negative controls, respectively. (E) Unequivocal confirmation of HLA-B*42:01 as the restriction element for Int-IM9 via cognate tetramer staining of responding PBMCs. Similar data are shown for three further novel epitopes: Gag-RM9 (RPGGKKHYM), Vif-HI10 (HPKVSSEVHI) and Env-IF9 (IPRRIRQGF). An HLA-mismatched tetramer was used as the negative control.
To validate the novel HLA-B*42:01-restricted CD8+ T-cell epitopes, we first mapped Int-IM9 (IIKDYGKQM) as the optimal peptide within OLP Int-275 (KVVPRRKAKIIKDYGKQM), which was targeted at a frequency of 53% (Fig 1B-D). Of note, Int-IM9 is the first example of a B7 superfamily-restricted epitope that does not contain proline at position 2, which acts as the primary anchor residue. We then used HLA-B*42:01 tetramers (16, 63) to rapidly and unequivocally identify an additional three optimal epitopes from OLP Gag-3 (Gag-RM9, RPGGKKHYM), OLP Vif-407 (Vif-HI10, HPKVSSEVHI) and OLP Env-401 (Env-IF9, IPRRIRQGF) (Fig 1E and Table 1). Finally, we demonstrated that 10 of the novel epitopes listed in Table 1 bound strongly to HLA-B*42:01 (KD, 2-82 nM; binding half-life, 1.1-22.4 hrs) (Table S1).
The identification of 20 HLA-B*42:01-restricted epitopes with predictable targeting patterns provided a unique opportunity to probe the biological impact and mechanistic basis of immunodominance patterns in HIV-1 infection.
Gag-specific responses are associated with lower viral load set-points
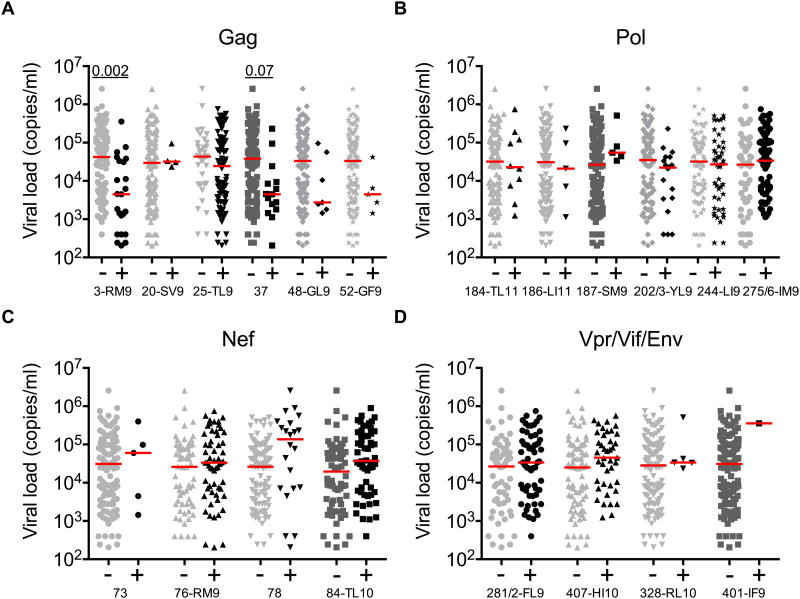
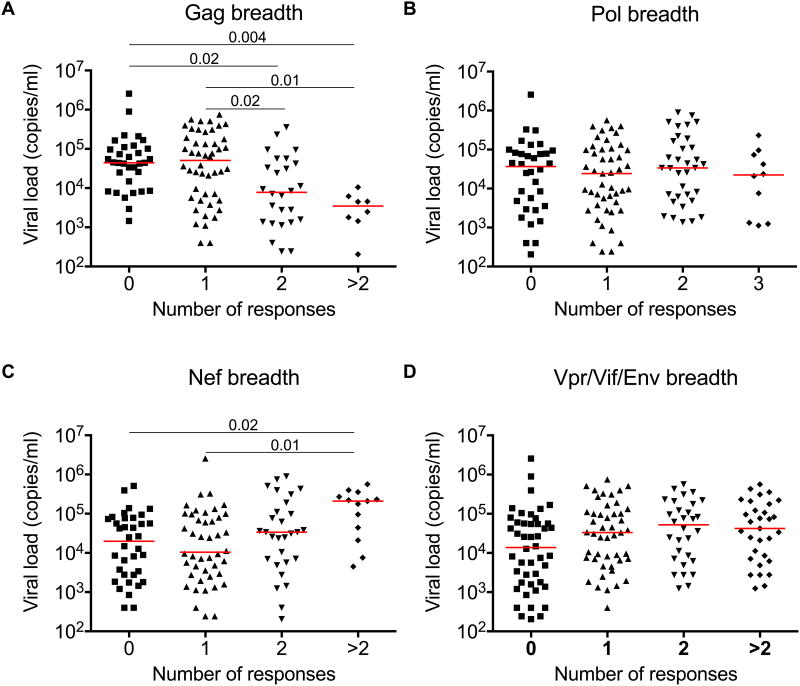
To establish the efficacy of each HLA-B*42:01-restricted response, we compared viral load set-points in HLA-B*42:01+ responders and non-responders (Fig 2). Five of the six Gag-specific responses showed a trend towards lower median viral loads, which was statistically significant for the Gag-RM9 epitope after multiple comparisons analysis (P = 0.002). In contrast, three of the six responses directed against accessory/regulatory proteins (two Nef-specific and one Vif-specific) showed a trend towards higher median viral loads, although no significant associations were observed after correction for multiple comparisons. Discordant associations with viral load set-point were also observed for response breadth (Fig 3). In particular, only Gag-specific response breadth was linked to immune control in the context of HLA-B*42:01 restriction, confirming previous analyses across the entire cohort (15). Epitope-specific differences therefore discriminate HLA-B*42:01-restricted responses with respect to immune control of HIV-1 replication.
Figure 2. Subdominant HLA-B*42:01-restricted Gag-specific responses are associated with lower viral loads.
Viral load set-points were compared across OLP-specific CD8+ T-cell responses targeting Gag (A), Pol (B), Nef (C) or Vpr/Vif/Env (D) in HLA-B*42:01+ responders and non-responders, stratified for lack of HLA-B*07:02/39:10/42:02/81:01 co-expression (n=126). Horizontal bars indicate median values. P values <0.1 by Dunn's multiple comparisons test are shown.
Figure 3. Discordant viral load associations with HLA-B*42:01-restricted protein-specific responses.
Viral load set-points were compared across groups making 0, 1, 2 and >2 protein-specific responses directed against Gag (A), Pol (B), Nef (C) or Vpr/Vif/Env (D) in the cohort of HLA-B*42:01+ individuals, stratified for lack of HLA-B*07:02/39:10/42:02/81:01 co-expression (n=126). Horizontal bars indicate median values. P values <0.05 by Dunn's multiple comparisons test are shown.
Gag-specific selection pressure is associated with loss of antiviral efficacy
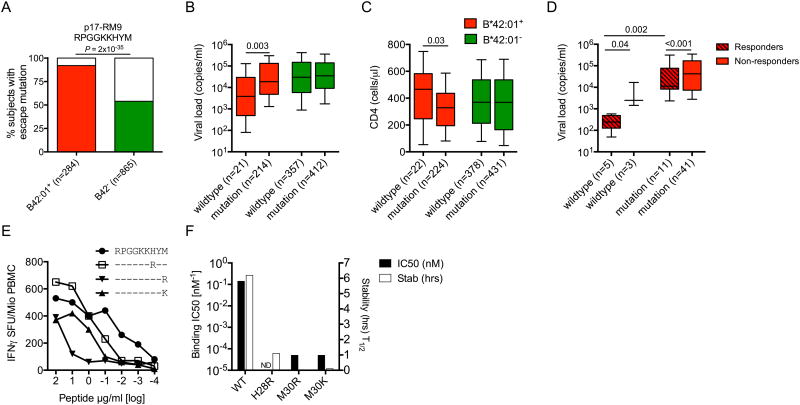
The ability of certain CD8+ T-cell responses to exert antiviral selection pressure has previously been associated with immune efficacy (15-17, 44). To examine this phenomenon across HLA-B*42:01-restricted specificities, we initially focused on the Gag-RM9 response, which exhibited the strongest association with viremic control (Fig 2A). Escape mutations were selected in 92% of HLA-B*42:01+ subjects compared to only 54% of HLA-B*42:01- individuals (P = 2×10-35) (Fig 4A). These polymorphisms were associated with higher viral load set-points (median HIV RNA copies/ml plasma = 3,860 vs 18,550, P = 0.003) (Fig 4B) and lower CD4+ T-cell counts (median CD4+ T-cells/μl = 467 vs 328, P = 0.03) (Fig 4C) in HLA-B*42:01+ individuals. Given the absence of such associations in HLA-B*42:01- subjects, it is likely that these effects operate via evasion of the Gag-RM9 response. Moreover, a beneficial effect on viral load set-point in Gag-RM9 responders carrying the wildtype epitope was observed across 60 HLA-B*42:01+ individuals for whom IFNγ ELISpot and viral sequence data were available from the same time point (median HIV RNA copies/ml plasma = 387 vs 2,470, P = 0.04) (Fig 4D). These findings suggest that the Gag-RM9 response contributes to viremic control most effectively in the absence of viral escape, but also to some extent in the presence of mutations that compromise CD8+ T-cell recognition (Fig 4E).
Figure 4. HIV-1 sequence polymorphisms in Gag-RM9 are CD8+ T-cell escape mutations associated with loss of immune control.
(A) HLA-B*42:01+ (red) and HLA-B*42:01- (green) associations with mutations in Gag-RM9 (departure from wildtype). Significance was determined using Fisher's exact test. (B) Viral load set-points and (C) CD4+ T-cell counts in HLA-B*42:01+ (red) and HLA-B*42:01- (green) individuals carrying either wildtype Gag-RM9 (RPGGKKHYM) or any mutation in this epitope. The x-axis indicates the number of sequences analyzed in each case. (D) Viral load set-points in 60 HLA-B*42:01+ individuals carrying either wildype Gag-RM9 or any mutation in this epitope stratified for responder (crossed boxes) or non-responder (open boxes) status. (E) Impact of the commonly selected H28R, M30R and M30K variants on CD8+ T-cell recognition in IFNγ ELISpot assays. Data from subject R019 (A*02:01/30:01, B*35:01/42:01, Cw*16:01/17:01) are shown. (F) HLA-B*42:01 binding affinities and half-lives (stability) for the peptides shown in (E). In (B-D), horizontal bars indicate median values presented by 25th percentile boxes and ranges (standard deviation); significance was determined using the Mann-Whitney U-test.
In further analyses, we examined the relationship between viral sequences across the whole HIV-1 genome and HLA-B*42:01 expression in a total of 1,867 individuals. Four polymorphisms in the Gag-RM9 epitope were selected by HLA-B*42:01 (H28R, H28S, M30R and M30K) (Table 2). These mutations resulted in reduced CD8+ T-cell recognition (Fig 4E), at least in part through decreased peptide-HLA-B*42:01 binding (Fig 4F). In total, we identified 20 different polymorphic sites located in nine of the 20 HLA-B*42:01-restricted epitopes (q < 0.05), including the novel Gag-GF9 epitope (Table 2). Of the 10 top-ranking immunodominant HLA-B*42:01-restricted responses, all six targeting epitopes in Gag, Pol or Vpr showed evidence of selection pressure on the virus. In contrast, only one of the four responses targeting epitopes in Nef or Vif showed evidence of selection pressure. These data provide further evidence of variable antiviral efficacy across the 20 HLA-B*42:01-restricted responses, most notably those targeting immunodominant epitopes.
Table 2.
HLA-B*42:01-associated HIV-1 polymorphisms.
| Protein | HXB2 location | Escape variantselecteda | Sequenceb | % B*42:01+ | % B*42:01- | P | Qc | N |
|---|---|---|---|---|---|---|---|---|
| Gag-p17 | 20 | S | IRLRPGGKKHYMLKH | 5 | 2 | 6.8E-03 | 4.6E-02 | 1767 |
| 28 | Q | IRLRPGGKKHYMLKH | 29 | 13 | 8.0E-12 | 2.9E-10 | 1765 | |
| 28 | S | IRLRPGGKKHYMLKH | 6 | 2 | 8.8E-05 | 8.7E-04 | 1765 | |
| 28 | R | IRLRPGGKKHYMLKH | 22 | 13 | 2.3E-04 | 1.9E-03 | 1765 | |
| 30 | R | IRLRPGGKKHYMLKH | 25 | 12 | 9.8E-10 | 2.7E-08 | 1765 | |
| 30 | K | IRLRPGGKKHYMLKH | 14 | 7 | 1.0E-04 | 9.2E-04 | 1765 | |
| Gag-p24 | 182 | T | EGATPQDLNTMLNTV | 9 | 1 | 2.6E-08 | 5.8E-07 | 1857 |
| 386 | S | NFKGPKRIVKCFNCG | 46 | 29 | 6.4E-06 | 8.8E-05 | 1122 | |
| 387 | R | NFKGPKRIVKCFNCG | 43 | 34 | 2.6E-03 | 1.9E-02 | 1131 | |
| Pol-RT | 427 | S | SQIYPGIKVRQLCKL | 24 | 5 | 2.5E-15 | 4.8E-14 | 1052 |
| 427 | A | SQIYPGIKVRQLCKL | 31 | 11 | 8.5E-12 | 7.1E-11 | 1052 | |
| 427 | Q | SQIYPGIKVRQLCKL | 4 | 1 | 2.4E-03 | 9.4E-03 | 1052 | |
| 429 | V | SQIYPGIKVRQLCKL | 10 | 1 | 5.8E-10 | 3.9E-09 | 1051 | |
| 433 | N | SQIYPGIKVRQLCKL | 5 | 2 | 9.8E-03 | 3.1E-02 | 1049 | |
| 436 | R | SQIYPGIKVRQLCKL | 18 | 7 | 2.3E-05 | 1.2E-04 | 1049 | |
| Pol-Int | 746 | V | EFNLPPIVAKEIVAS | 51 | 17 | 5.9E-13 | 6.3E-12 | 565 |
| 749 | R | EFNLPPIVAKEIVAS | 28 | 2 | 6.8E-17 | 1.7E-15 | 565 | |
| 984 | R | KVKIIKDYGKQMAGA | 79 | 41 | 8.7E-15 | 1.1E-13 | 562 | |
| 985 | E | KVKIIKDYGKQMAGA | 7 | 1 | 1.6E-03 | 6.7E-03 | 562 | |
| 990 | V | KVKIIKDYGKQMAGA | 7 | 1 | 1.3E-03 | 5.6E-03 | 562 | |
| Vpr | 42 | I | VRHFPRPWLHSLGQY | 12 | 3 | 1.1E-02 | 3.2E-02 | 255 |
| 45 | H | VRHFPRPWLHSLGQY | 61 | 42 | 5.6E-03 | 2.2E-02 | 254 | |
| Vif | 48 | N | ESRHPKVSSEVHIPLG | 32 | 9 | 4.7E-05 | 7.5E-04 | 255 |
| Env | 843 | V | IRNIPRRIRQGFEAA | 17 | 4 | 2.0E-03 | 2.0E-02 | 248 |
| 845 | T | IRNIPRRIRQGFEAA | 41 | 22 | 1.1E-03 | 1.9E-02 | 245 |
Escape polymorphism shows the amino acid selected in that particular HXB2 location, also indicated by bold face.
Optimal epitope is underlined with the consensus sequence shown for +/- 3 amino acids; bold face indicates the site of polymorphism.
Only q values <0.05 are included.
TCR bias in the HLA-B*42:01-restricted repertoire
The presence of public TCR clonotypes, defined on the basis of TRB amino acid sequence identity across multiple individuals responding to the same epitope (36, 38, 64), has previously been linked with CD8+ T-cell efficacy in SIV (33, 34), HIV (30, 65) and CMV (35) infection. To examine this phenomenon in the setting of HLA-B*42:01-restricted responses to HIV-1, we sorted viable tetramer+ CD8+ T-cell populations (n = 48) specific for six of the seven most frequently targeted epitopes (Gag-TL9, Int-IM9, Vpr-FL9, Nef-TL10, Vif-HI10 and Int-LI9) (Supplementary Fig S2) and sequenced a total of 3,592 constituent TCR clones across eight different donors per specificity (Tables 3-8). Clonotypic data for the Gag-RM9 response were only available from one subject (Table S2) and therefore could not be included in further analyses.
Table 3.
Clonal analysis of HLA-B*42:01-restricted TL9-specific CD8+ T-cell populations.
| Subject (ddmmyy) | Epitope | CD8+Tet+ | pVL | CD4 | TRBV | CDR3 | TRBJ | Freq (%) |
|---|---|---|---|---|---|---|---|---|
| N021 (040211) | TL9 | 1.4 | 8249 | 510 | 4-1 | CASSQEGGGQGQPQH | 1-5 | 60.00 |
| HLA-A*30:01/34:02 | 5-6 | CASSLATDGYT | 1-2 | 9.41 | ||||
| HLA-B*42:01/44:03 | 4-1 | CASSQEGGGDGQPQH | 1-5 | 8.24 | ||||
| HLA-C*07:01/17:01 | 12-3/4 | CASSFGLDEAF | 1-1 | 5.88 | ||||
| 4-1 | CASSQEGGGEGQPQH | 1-5 | 4.71 | |||||
| 12-3/4 | CASSFSKNTEAF | 1-1 | 3.53 | |||||
| 5-5 | CASSLEGTSGPQETQY | 2-5 | 2.35 | |||||
| 12-3/4 | CASSVGPNEQF | 2-1 | 2.35 | |||||
| 4-1 | CASSQEGGGDGQPPH | 1-5 | 1.18 | |||||
| 4-1 | CASRQEGGGDGQPQH | 1-5 | 1.18 | |||||
| 4-1 | CAPTPEGGGQGQPQH | 1-5 | 1.18 | |||||
| Number of cells sorted | 5000 | count | (85) | |||||
| R014 (130907) | TL9 | 2.7 | 1177 | 300 | 12-3/4 | CASSLGPTEAF | 1-1 | 31.25 |
| HLA-A*30:01/33:01 | 12-3/4 | CASSLDPEKGAF | 1-1 | 21.25 | ||||
| HLA-B*42:01/57:03 | 12-3/4 | CASSLGLNTIY | 1-3 | 20.00 | ||||
| HLA-C*17:01/18:01 | 12-3/4 | CASSFSKNTEAF | 1-1 | 10.00 | ||||
| 12-3/4 | CASSLGVNTIY | 1-3 | 7.50 | |||||
| 12-3/4 | CASSQGPTEAF | 1-1 | 6.25 | |||||
| 6-2/3 | CATHAGTGELF | 2-2 | 2.50 | |||||
| 12-3/4 | CASSLSFTEAF | 1-1 | 1.25 | |||||
| Number of cells sorted | 4701 | count | (80) | |||||
| H022 (170807) | TL9 | 6.1 | 125688 | 430 | 12-3/4 | CASSLNGADGYT | 1-2 | 76.83 |
| HLA-A*02:02/03:01 | 12-3/4 | CASSLGLNTIY | 1-3 | 13.41 | ||||
| HLA-B*08:01/42:01 | 7-9 | CASSSQTSGLFANTGELF | 2-2 | 9.76 | ||||
| HLA-C*07:02/17:01 | count | (82) | ||||||
| Number of cells sorted | 5000 | |||||||
| R081 (070909) | TL9 | 1.1 | 503 | 130 | 12-3/4 | CASSLGANTIY | 1-3 | 52.50 |
| HLA-A*30:01/30:02 | 5-1 | CASSLAFGTSGGEQY | 2-7 | 11.25 | ||||
| HLA-B*35:01/42:01 | 4-1 | CASSQEGGGGGQPQH | 1-5 | 11.25 | ||||
| HLA-C*04:01/17:01 | 5-1 | CASSLSDVSWNTEAF | 1-1 | 8.75 | ||||
| 12-3/4 | CASSREGYSNQPQH | 1-5 | 3.75 | |||||
| 12-3/4 | CASSLSKNTEAF | 1-1 | 2.50 | |||||
| 12-3/4 | CASSPGNTEAF | 1-1 | 2.50 | |||||
| 12-3/4 | CASRDPYEQY | 2-7 | 2.50 | |||||
| 12-3/4 | CASDKGTGNYGYT | 1-2 | 1.25 | |||||
| 12-3/4 | CASSHSKNTEAF | 1-1 | 1.25 | |||||
| 12-3/4 | CASSFGGTTEAF | 1-1 | 1.25 | |||||
| 12-3/4 | CASGLGANTIY | 1-3 | 1.25 | |||||
| Number of cells sorted | 2008 | count | (80) | |||||
| N033 (100707) | TL9 | 3.7 | 415 | 620 | 7-9 | CASSSTITGMGDSGNTIY | 1-3 | 96.63 |
| HLA-A*30:01/36:01 | 7-9 | CTSSSTITGMGDSGNTIY | 1-3 | 1.12 | ||||
| HLA-B*42:01/53:01 | 7-9 | CASSSTITGMGVSGNTIY | 1-3 | 1.12 | ||||
| HLA-C*04:01/17:01 | 7-9 | CASSSAITGMGDSGNTIY | 1-3 | 1.12 | ||||
| Number of cells sorted | 5000 | count | (89) | |||||
| N086 (181208) | TL9 | 0.7 | 2794 | 370 | 6-1 | CASRASTGSGNTIY | 1-3 | 93.33 |
| HLA-A*33:03/33:03 | 27 | CASSLRHLASDYNSPLH | 1-5 | 2.22 | ||||
| HLA-B*42:01/53:01 | 6-1 | CAGRASTGSGNTIY | 1-3 | 2.22 | ||||
| HLA-C*03:02/04:01 | 6-1 | CASRARRPSNTIY | 1-3 | 2.22 | ||||
| Number of cells sorted | 4340 | count | (45) | |||||
| N106 (030310) | TL9 | 1.8 | 14014 | 350 | 5-5 | CASSLVFGTAGGQQF | 2-1 | 63.10 |
| HLA-A*02:01/02:05 | 12-3/4 | CASSFSKNTEAF | 1-1 | 33.33 | ||||
| HLA-B*42:01/53:01 | 6-5 | CASSWTETGELF | 2-2 | 2.38 | ||||
| HLA-C*04:01/17:01 | 5-5 | CTSSLVFGTAGGRQF | 2-1 | 1.19 | ||||
| Number of cells sorted | 4160 | count | (84) | |||||
| SK191 | TL9 | 2.7 | 76900 | 12-3/4 | CASSFSKNTEAF | 1-1 | 80.95 | |
| HLA-A*23:01/30:01 | 4-1 | CASSQEGGGQGQPQH | 1-5 | 5.95 | ||||
| HLA-B*08:01/42:01 | 5-1 | CASSLMGASGANVLT | 2-6 | 4.76 | ||||
| HLA-C*16:01/17:01 | 12-3/4 | CASSLGANTIY | 1-3 | 4.76 | ||||
| 12-3/4 | CASSLSKNTEAF | 1-1 | 2.38 | |||||
| 29-1 | CSVRTHQGPTNEKLF | 1-4 | 1.19 | |||||
| Number of cells sorted | 3674 | count | (84) |
Table 8.
Clonal analysis of HLA-B*42:01-restricted TL10-specific CD8+ T-cell populations.
| Subject (ddmmyy) | Epitope | CD8+Tet+ | pVL | CD4 | TRBV | CDR3 | TRBJ | Freq (%) |
|---|---|---|---|---|---|---|---|---|
| N086 (181208) | TL10 | 1.4 | 2794 | 370 | 5-4 | CASSPRGGGETQY | 2-5 | 89.23 |
| HLA-A*33:03/33:03 | 14 | CASSQRGADTEAF | 1-1 | 3.08 | ||||
| HLA-B*42:01/53:01 | 7-9 | CASRLLGPSTFFYGYT | 1-2 | 1.54 | ||||
| HLA-C*03:02/04:01 | 9 | CASSVVGGGAADTQY | 2-3 | 1.54 | ||||
| 15 | CATSERGGMETQY | 2-5 | 1.54 | |||||
| 5-4 | CPSSPRGGGETQY | 2-5 | 1.54 | |||||
| 5-4 | CASSPRGGGETQH | 2-5 | 1.54 | |||||
| Number of cells sorted | 5000 | count | (65) | |||||
| R081 (070909) | TL10 | 1.4 | 503 | 130 | 9 | CASSVSGGQVTDTQY | 2-3 | 48.00 |
| HLA-A*30:01/30:02 | 4-1 | CASSQDRATQETQY | 2-5 | 12.00 | ||||
| HLA-B*42:01/44:03 | 4-3 | CASSQYRATQETQY | 2-5 | 9.33 | ||||
| HLA-C*04:01/17:01 | 14 | CASYRSDRPTEAF | 1-1 | 8.00 | ||||
| 27 | CASRKGIQETQY | 2-5 | 6.67 | |||||
| 4-3 | CASSQEGRRDTQY | 2-3 | 4.00 | |||||
| 7-9 | CASSPPIPGTADTIY | 1-3 | 2.67 | |||||
| 10-2 | CASSESGRGDTQY | 2-3 | 2.67 | |||||
| 7-9 | CASTTAADTQY | 2-3 | 2.67 | |||||
| 9 | CASSVAGGQVTDTQY | 2-3 | 1.33 | |||||
| 14 | CASSQAASGNTIY | 1-3 | 1.33 | |||||
| 4-1 | CASSQEGRRDTQY | 2-3 | 1.33 | |||||
| Number of cells sorted | 2079 | count | (75) | |||||
| N003 (110507) | TL10 | 0.2 | 12802 | 180 | 5-1 | CASSTFGATQETQY | 2-5 | 98.63 |
| HLA-A*03:01/33:03 | 5-1 | CASSTFGATQEIQY | 2-5 | 1.37 | ||||
| HLA-B*15:10/42:01 | count | (73) | ||||||
| HLA-C*03:02/17:01 | ||||||||
| Number of cells sorted | 1020 | |||||||
| SK178 | TL10 | 0.5 | 220000 | 27 | CASSLSRQGTGELF | 2-2 | 31.88 | |
| HLA-A*34:02/34:02 | 6-2/3 | CASSYRVTEAF | 1-2 | 30.43 | ||||
| HLA-B*42:01/44:03 | 6-2/3 | CASSYYSGGSSYNEQF | 2-1 | 8.70 | ||||
| HLA-C*04:01/17:01 | 6-2/3 | CASSYSRTDLKNIQY | 2-4 | 8.70 | ||||
| 29-1 | CSVKLTEFGYT | 1-2 | 7.25 | |||||
| 19 | CASMGLVGGTDTQY | 2-3 | 4.35 | |||||
| 6-2/3 | CASSYSRAPKLENIQY | 2-4 | 2.90 | |||||
| 4-3 | CASSQFTGTQETQY | 2-5 | 2.90 | |||||
| 6-2/3 | CASSYYSGGTTYNEQF | 2-1 | 1.45 | |||||
| 4-3 | CASSPVGRGTEAF | 1-1 | 1.45 | |||||
| Number of cells sorted | 1478 | count | (69) | |||||
| SK040 | TL10 | 0.1 | 164000 | 5-1 | CASSPRGTRTDTQY | 2-3 | 48.05 | |
| HLA-A*30:01/33:03 | 27 | CASRRGFHQPQH | 1-5 | 36.36 | ||||
| HLA-B*42:01/53:01 | 20-1 | CSAPEPTSGRWSGELF | 2-2 | 7.79 | ||||
| HLA-C*04:01/17:01 | 14 | CASSLSPTEAF | 1-1 | 2.60 | ||||
| 6-1 | CASSGRDTSTDTQY | 2-3 | 1.30 | |||||
| 5-1 | CASSPRETRTDTQY | 2-3 | 1.30 | |||||
| 15 | CATSPRGGAVEQF | 2-1 | 1.30 | |||||
| Number of cells sorted | 325 | count | (61) | |||||
| SK191 | TL10 | 0.2 | 76900 | 9 | CASSVWGDPSYEQY | 2-7 | 97.22 | |
| HLA-A*01:02/30:01 | 5-1 | CASSPNTIANEQF | 2-1 | 2.78 | ||||
| HLA-B*15:03/42:01 | count | (36) | ||||||
| HLA-C*02:10/17:01 | ||||||||
| Number of cells sorted | 1043 | |||||||
| SK046 | TL10 | 0.6 | 4850 | 5-1 | CASRPIGGAQETQY | 2-5 | 100.00 | |
| HLA-A*02:02/30:01 | count | (40) | ||||||
| HLA-B*15:16/42:01 | ||||||||
| HLA-C*14:02/17:01 | ||||||||
| Number of cells sorted | 842 | |||||||
| SK075 | TL10 | 0.6 | 25900 | 5-1 | CASSPRGTRTDTQY | 2-3 | 48.05 | |
| HLA-A*23:01/30:01 | 27 | CASRRGFHQPQH | 1-5 | 36.36 | ||||
| HLA-B*42:01/58:01 | 20-1 | CSAPEPTSGRWSGELF | 2-2 | 7.79 | ||||
| HLA-C*06:02/17:01 | 14 | CASSLSPTEAF | 1-1 | 2.60 | ||||
| 6-1 | CASSGRDTSTDTQY | 2-3 | 1.30 | |||||
| 5-1 | CASSPRETRTDTQY | 2-3 | 1.30 | |||||
| 15 | CATSPRGGAVEQF | 2-1 | 1.30 | |||||
| 7-9 | CASSLASDTQY | 2-3 | 1.30 | |||||
| Number of cells sorted | 2070 | count | (77) |
Columns show: HLA genotyping data below each subject identifier with sample date in brackets; targeted epitope; frequency of tetramer+ (Tet+) cells in the total CD8+ T-cell population; plasma viral load (pVL, HIV RNA copies/ml of plasma); CD4+ T-cell count (CD4, cells/μl blood); TRBV usage; CDR3 amino acid sequence; TRBJ usage; clonotype frequency (%). Public clonotypes are color-coded. Data represent one time point per subject.
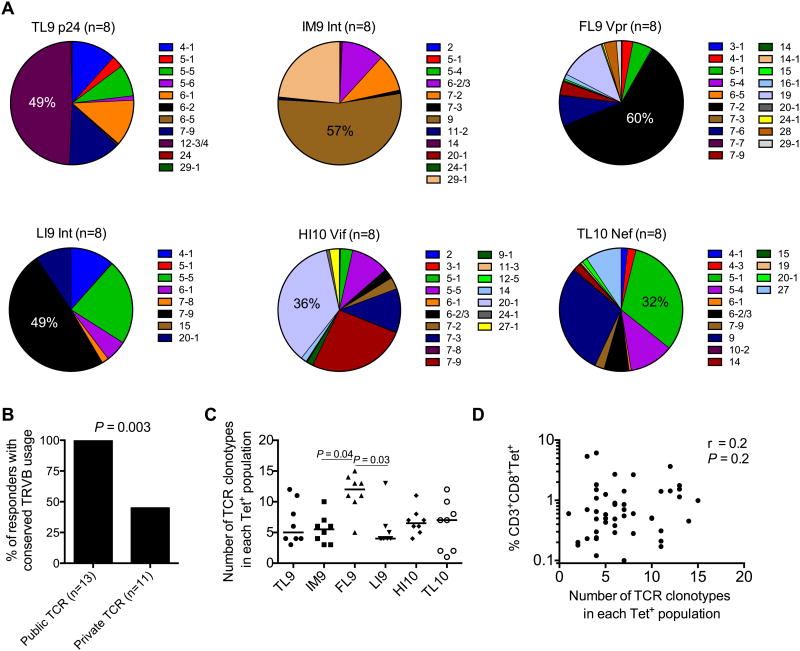
For the top two ranking immunodominant responses, Gag-TL9 (Table 3) and Int-IM9 (Table 4), public TCRs were identified in 6/8 and 4/8 subjects, respectively. The most prevalent examples in terms of recurrence were TRBV12-3/4/CASSFSKNTEAF/TRBJ1-1 (Gag-TL9) and TRBV9/CASSVDKGGTDTQY/TRBJ2-3 (Int-IM9), each of which were shared by 4/8 individuals. Public clonotypes were also identified for Vpr-FL9, ranked third in the immunodominance hierarchy (Table 5). Each specificity was characterized by diverse but distinct patterns of TRBV usage (Fig 5A). Moreover, within each specificity, all public clonotypes used the same TRBV gene (Fig 5B). For example, the four different Int-IM9-specific public clonotypes all expressed the TRBV9 (Table 4), suggesting ‘hard-wired’ germline-encoded antigen recognition.
Table 4.
Clonal analysis of HLA-B*42:01-restricted IM9-specific CD8+ T-cell populations.
| Subject (ddmmyy) | Epitope | CD8+ Tet+ | pVL | CD4 | TRBV | CDR3 | TRBJ | Freq (%) |
|---|---|---|---|---|---|---|---|---|
| R020 (150607) | IM9 | 0.4 | 5723 | 600 | 9 | CASSDNPLVGGFTDTQY | 2-3 | 85.37 |
| HLA-A*29:01/29:02 | 29-1 | CSDDGGQEGYGYT | 1-2 | 9.76 | ||||
| HLA-B*42:01/51:01 | 29-1 | CSVEEETNYGYT | 1-2 | 3.66 | ||||
| HLA-C*16:01/17:01 | 9 | CASSDNPLVGGFTDMQY | 2-3 | 1.22 | ||||
| Number of cells sorted | 1068 | count | (82) | |||||
| N003 (110507) | IM9 | 0.3 | 12802 | 180 | 7-2 | CASSLALRGGDQETQY | 2-5 | 57.89 |
| HLA-A*03:01/33:03 | 7-3 | RASSSLRGTAADTQY | 2-3 | 35.79 | ||||
| HLA-B*15:10/42:01 | 7-2 | CAGSLALRLADQETQY | 2-5 | 2.11 | ||||
| HLA-C*03:02/17:01 | 2 | CASLEGGSYT | 1-2 | 2.11 | ||||
| 7-3 | CASGLALRGGDQETQY | 2-5 | 1.05 | |||||
| 6-2/3 | CAIRTSGDYEQY | 2-7 | 1.05 | |||||
| Number of cells sorted | 1813 | count | (95) | |||||
| H022 (170807) | IM9 | 5.4 | 125688 | 430 | 29-1 | CSVEGMRDYGYT | 1-2 | 98.51 |
| HLA-A*02:02/03:01 | 29-1 | CSVEGMREYGYT | 1-2 | 1.49 | ||||
| HLA-B*08:01/42:01 | count | (68) | ||||||
| HLA-C*07:02/17:01 | ||||||||
| Number of cells sorted | 5000 | |||||||
| R094 (030810) | IM9 | 0.8 | 69724 | 230 | 6-2/3 | CASRGSGVYEQY | 2-7 | 80.00 |
| HLA-A*29:02/30:01 | 5-1 | CASSLVDPTGFGLETQY | 2-5 | 11.76 | ||||
| HLA-B*42:01/42:01 | 9 | CASSVDKGGADEQF | 2-1 | 3.53 | ||||
| HLA-C*17:01/17:01 | 14 | CASSPRDFSPTDTQY | 2-3 | 1.18 | ||||
| 9 | CASSVDKGGTDTQY | 2-3 | 1.18 | |||||
| 20-1 | CSAREDEGWGGYT | 1-2 | 1.18 | |||||
| 24-1 | CATSDSYEQY | 2-7 | 1.18 | |||||
| Number of cells sorted | 1940 | count | (85) | |||||
| N033 (100707) | IM9 | 0.2 | 415 | 620 | 6-2/3 | CASRGTGVHEQY | 2-7 | 56.96 |
| HLA-A*30:01/36:01 | 9 | CASSVDKGGADTQY | 2-3 | 29.11 | ||||
| HLA-B*42:01/53:01 | 6-2/3 | CASRTSGGHEQF | 2-1 | 12.66 | ||||
| HLA-C*04:01/17:01 | 11-2 | CASSLDPRMNTEAF | 1-1 | 1.27 | ||||
| Number of cells sorted | 2788 | count | (79) | |||||
| N052 (171007) | IM9 | 0.5 | 14935 | 270 | 6-2/3 | CASRTSGEETQY | 2-5 | 36.84 |
| HLA-A*30:02/30:02 | 9 | CASSVDKGGTDTQY | 2-3 | 34.21 | ||||
| HLA-B*08:01/42:01 | 9 | CASSEDKGGGDTQY | 2-3 | 7.89 | ||||
| HLA-C02:10/17:01 | 9 | CASSVDKGGVDEQF | 2-1 | 6.58 | ||||
| 5-1 | CASGDSGDEQF | 2-1 | 5.36 | |||||
| 6-2/3 | CASRTSGDYEQY | 2-7 | 3.95 | |||||
| 5-4 | CASSFLTGARSKNIQY | 2-4 | 1.32 | |||||
| 9 | CASSVDKGGPDTQY | 2-3 | 1.32 | |||||
| 9 | CASSEDKGGADTQY | 2-3 | 1.32 | |||||
| 6-2/3 | CASRTGGEETQY | 2-5 | 1.32 | |||||
| Number of cells sorted | 1828 | count | (76) | |||||
| N073 (210508) | IM9 | 0.2 | 1302 | 430 | 9 | CASALEQGGYNEQF | 2-1 | 34.83 |
| HLA-A*23:01/26:01 | 9 | CASSVDKGGTDTQY | 2-3 | 32.58 | ||||
| HLA-B*41:01/42:01 | 9 | CASSEDKGGGDTQY | 2-3 | 29.21 | ||||
| HLA-C-17:01/17:01 | 9 | CASSVDKGGTDAQY | 2-3 | 1.12 | ||||
| 9 | CTSSEDKGGGDTQY | 2-3 | 1.12 | |||||
| 9 | CVSSVDKGGTDTQY | 2-3 | 1.12 | |||||
| Number of cells sorted | 581 | count | (89) | |||||
| N058 (270509) | IM9 | 0.4 | 4534 | 510 | 9 | CASSVDKGGTDTQY | 2-3 | 81.82 |
| HLA-A*01:02/30:01 | 9 | CASSVDKGGTDEQF | 2-1 | 9.09 | ||||
| HLA-B*15:03/42:01 | 9 | CASSVDKGGADEQF | 2-1 | 6.49 | ||||
| HLA-C*02:10/17:01 | 9 | CVSSVDKGGTDTQY | 2-3 | 2.60 | ||||
| Number of cells sorted | 3070 | Count | (77) |
Table 5.
Clonal analysis of HLA-B*42:01-restricted FL9-specific CD8+ T-cell populations.
| Subject (ddmmyy) | Epitope | CD8+Tet+ | pVL | CD4 | TRBV | CDR3 | TRBJ | Freq (%) |
|---|---|---|---|---|---|---|---|---|
| N037 (061009) | FL9 | 1.3 | 3170 | 360 | 5-1 | CASSHLDSGLAVDTEAF | 1-1 | 40.26 |
| HLA-A*29:02/30:02 | 28 | CASSFRQGLGHTGELF | 2-2 | 25.97 | ||||
| HLA-B*42:01/42:01 | 7-2 | CASSLWSGASNEQF | 2-1 | 24.68 | ||||
| HLA-C*17:01/17:01 | 7-2 | CASSLYSGADQPQH | 1-5 | 7.79 | ||||
| 7-2 | CASSLWPGASNEQF | 2-1 | 1.30 | |||||
| Number of cells sorted | 4059 | count | (77) | |||||
| N021 (040211) | FL9 | 0.5 | 8249 | 510 | 7-2 | CASSLWGGDFSQEQF | 2-1 | 40.00 |
| HLA-A*30:01/34:02 | 7-2 | CASSLWSGVGDGYT | 1-2 | 35.00 | ||||
| HLA-B*42:01/44:03 | 7-2 | CASSLYGGPEQPQH | 1-5 | 6.25 | ||||
| HLA-C*07:01/17:01 | 7-9 | CASSPISDRSGNTIY | 1-3 | 5.00 | ||||
| 7-2 | CASSLWAGVSDTQY | 2-3 | 2.50 | |||||
| 4-1 | CASSQDMKGGSFTGELF | 2-2 | 1.25 | |||||
| 7-2 | CASSLWGGGFSQEQF | 2-1 | 1.25 | |||||
| 7-2 | CASSLWGGDFSQERF | 2-1 | 1.25 | |||||
| 7-2 | CASSLYSGGDQPQH | 1-5 | 1.25 | |||||
| 7-2 | CASSLYSGLDQPQH | 1-5 | 1.25 | |||||
| 7-2 | CASSLYGGGEQPQH | 1-5 | 1.25 | |||||
| 29-1 | CSVAGTGMTDTQY | 2-3 | 1.25 | |||||
| 6-5 | CASRSGRTNEKLF | 1-4 | 1.25 | |||||
| 5-1 | CASSLEAPTDTQY | 1-3 | 1.25 | |||||
| Number of cells sorted | 2753 | count | (80) | |||||
| R014 (130907) | FL9 | 1.0 | 1177 | 300 | 7-2 | CASSLFSGPSQTQY | 2-5 | 23.75 |
| HLA-A*30:01/33:01 | 7-2 | CASSLYSGGGQPQH | 1-5 | 22.50 | ||||
| HLA-B*42:01/57:03 | 7-2 | CASSLYSGGDKEQY | 2-7 | 10.00 | ||||
| HLA-C*17:01/18:01 | 7-2 | CASSLYGSPDQPQH | 1-5 | 10.00 | ||||
| 7-2 | CASSLYGGGDQPQH | 1-5 | 8.75 | |||||
| 29-1 | CSGGGYGSGTYEQY | 2-7 | 6.25 | |||||
| 7-2 | CASSLWAGETEAF | 1-1 | 6.25 | |||||
| 7-2 | CASSLYSGGDQPQH | 1-5 | 3.75 | |||||
| 7-2 | CASSLYHSPTDQPQH | 1-5 | 1.25 | |||||
| 7-2 | CASRLYSGGDKEQY | 2-7 | 1.25 | |||||
| 7-2 | CASSLFSGASQTQY | 2-5 | 1.25 | |||||
| 7-2 | CASSLYGRGDQPQH | 1-5 | 1.25 | |||||
| 7-2 | CASSLYLAPNEKLF | 1-4 | 1.25 | |||||
| 4-1 | CASSQDHGGGTEAF | 1-1 | 1.25 | |||||
| 20-1 | CSARGGQLQETQY | 2-5 | 1.25 | |||||
| Number of cells sorted | 1892 | count | (80) | |||||
| R094 (030810) | FL9 | 0.2 | 69724 | 230 | 19 | CASSIKGYNEQF | 2-17890-/* | 82.76 |
| HLA-A*29:02/30:01 | 19 | CASSIQTGNSPLH | 1-6 | 5.75 | ||||
| HLA-B*42:01/42:01 | 19 | CASSIKGYNERF | 2-1 | 2.30 | ||||
| HLA-C*17:01/17:01 | 19 | CTSSIKGYNEQF | 2-1 | 2.30 | ||||
| 5-4 | CASSFYPTDEQF | 2-1 | 2.30 | |||||
| 19 | CASSIQTVNSPLH | 1-6 | 1.15 | |||||
| 4-1 | CASSQEGGPAEQF | 2-1 | 1.15 | |||||
| 29-1 | CSAGDWANNEQF | 2-1 | 1.15 | |||||
| 19 | CASSIKGYNELF | 2-1 | 1.15 | |||||
| 19 | CANSIKGYNEQF | 2-1 | 1.15 | |||||
| 14 | CASSQDRREQY | 2-7 | 1.15 | |||||
| Number of cells sorted | 590 | count | (87) | |||||
| N114 (080211) | FL9 | 1.5 | 2382 | 570 | 7-2 | CASSLWSGIADTQY | 2-3 | 75.90 |
| HLA-A*30:01/30:02 | 7-2 | CASSLWAGGSNEQF | 2-1 | 6.02 | ||||
| HLA-B*42:01/57:03 | 15 | CATSRDRETGGDYGYT | 1-2 | 3.61 | ||||
| HLA-C*17:01/18:01 | 24-1 | CATRDRDRENQPQH | 1-5 | 3.61 | ||||
| 7-2 | CASSLWGGGDREQY | 2-7 | 1.20 | |||||
| 7-2 | CASSLFSGGEETQY | 2-5 | 1.20 | |||||
| 7-2 | CASSLWSGRADTQY | 2-3 | 1.20 | |||||
| 7-2 | CASSLWSGGADTQY | 2-3 | 1.20 | |||||
| 7-2 | CAGSLWSGIADTQY | 2-3 | 1.20 | |||||
| 7-2 | CASSLWGGIADTQY | 2-3 | 1.20 | |||||
| 7-2 | CASSLWAGVSDTQY | 2-3 | 1.20 | |||||
| 7-2 | CASSLYSGYDQPQH | 1-5 | 1.20 | |||||
| 7-9 | CASSLEGVPVEF | 2-1 | 1.20 | |||||
| Number of cells sorted | 5000 | count | (83) | |||||
| N080 (180808) | FL9 | 1.8 | 1520 | 610 | 7-2 | CASSLFGSPEQPQH | 1-5 | 38.81 |
| HLA-A*02:02/03:01 | 7-2 | CASSLYTGSDQPQH | 1-5 | 19.40 | ||||
| HLA-B*15:03/42:01 | 7-2 | CASQLYSGGDQPQH | 1-5 | 13.43 | ||||
| HLA-C*02:01/17:01 | 7-2 | CASSLYSGPDQPQH | 1-5 | 7.46 | ||||
| 7-2 | CASSLWDQETQY | 2-5 | 4.48 | |||||
| 7-2 | CASSLYTGGDQPQH | 1-5 | 2.99 | |||||
| 7-2 | CASSLYSGAEQPQH | 1-5 | 2.99 | |||||
| 7-2 | CASKLYTGGDQPQH | 1-5 | 2.99 | |||||
| 7-2 | CVSSLFGSPEQPQH | 1-5 | 1.49 | |||||
| 7-2 | CASSVFGGPDQPQH | 1-5 | 1.49 | |||||
| 7-2 | CASSLYAGPDQPQH | 1-5 | 1.49 | |||||
| 7-2 | CASSLFGSPGQPQH | 1-5 | 1.49 | |||||
| 7-2 | CANSLYTGSDQPQH | 1-5 | 1.49 | |||||
| Number of cells sorted | 3223 | count | (67) | |||||
| R066 (260610) | FL9 | 0.2 | 127 | NA | 7-2 | CASSLWGGPTNEQY | 2-7 | 50.00 |
| HLA-A*30:01/74:01 | 7-9 | CASSSVDRSSYEQY | 2-7 | 16.00 | ||||
| HLA-B*42:01/53:01 | 7-2 | CASSLWGGSSNEQF | 2-1 | 10.00 | ||||
| HLA-C*04:01/17:01 | 7-2 | CASSLFSGGDQPQH | 1-5 | 6.00 | ||||
| 7-9 | CASSSVDRNSYEQY | 2-7 | 4.00 | |||||
| 7-2 | CASSLWAGGSNEQF | 2-1 | 4.00 | |||||
| 7-9 | CASSSVDRNSYEQY | 2-7 | 2.00 | |||||
| 7-2 | CASSLWGGASNEQY | 2-7 | 2.00 | |||||
| 7-2 | CASSLWAGGPETQY | 2-5 | 2.00 | |||||
| 7-2 | CASSLWAGPSNEQF | 2-1 | 2.00 | |||||
| 3-1 | CASSQGQSSYEQY | 2-7 | 2.00 | |||||
| Number of cells sorted | 1000 | count | (50) | |||||
| N033 (100707) | FL9 | 415 | 620 | 7-6 | CASSLERSSEQY | 2-7 | 62.71 | |
| HLA-A*30:01/36:01 | 4-1 | CASSQDRGPDTQY | 2-3 | 18.64 | ||||
| HLA-B*42:01/53:01 | 16-1 | CASSEGRDQETQY | 2-5 | 6.78 | ||||
| HLA-C*04:01/17:01 | 20-1 | CSARTYAGGTDTQY | 2-3 | 1.69 | ||||
| 16-1 | CTSSEGRDQETQY | 2-5 | 1.69 | |||||
| 16-1 | CAGSEGRDQETQY | 2-5 | 1.69 | |||||
| 7-7 | CASSPARGTDTQY | 2-3 | 1.69 | |||||
| 7-3 | CASSKDRGTDTQY | 2-3 | 1.69 | |||||
| 29-1 | CSVEDSLVNEQF | 2-1 | 1.69 | |||||
| 14-1 | CASRGTGESPLH | 1-6 | 1.69 | |||||
| Number of cells sorted | ND | count | (59) |
Figure 5. Clonality and TCR bias in HLA-B*42:01-restricted epitope-specific responses.
(A) TRBV usage is depicted for six different HLA-B*42:01-restricted epitope-specific responses. The number of individuals analyzed is indicated in each case. (B) Percentage of responders to Gag-TL9, Int-IM9 or Vpr-FL9 stratified for the presence of public TCRs. Significance was determined using Fisher's exact test. (C) Number of different TCR clonotypes detected in each of the six epitope-specific responses. P values <0.1 by Dunn's multiple comparisons test are shown. (D) Correlation between clonality and response magnitude. Significance was determined using the Spearman rank test.
Overall, we identified 11 different public clonotypes, all of which were confined exclusively to the top three ranked immunodominant specificities (Gag-TL9, Int-IM9 and Vpr-FL9) (Tables 3-5). In contrast, no public clonotypes were present within the Nef-TL10, Vif-HI10 and Int-LI9 specificities, ranked fifth to seventh in the immunodominance hierarchy (Tables 6-8). These observations suggest that clonotypic publicity is a feature of immunodominant HIV-specific CD8+ T-cell responses restricted by HLA-B*42:01. The Vpr-FL9-specific CD8+ T-cell populations were the most polyclonal (Fig 5C), but no correlation was found between clonality and response magnitude (P = 0.2) (Fig 5D). Almost identical results were obtained after normalization procedures that account for differences in sampling depth between individuals (data not shown).
Table 6. Clonal analysis of HLA-B*42:01-restricted LI9-specific CD8+ T-cell populations.
| Subject (ddmmyy) | Epitope | CD8+Tet+ | pVL | CD4 | TRBV | CDR3 | TRBJ | Freq (%) |
|---|---|---|---|---|---|---|---|---|
| N037 (061009) | LI9 | 0.2 | 3170 | 360 | 5-5 | CASSLVGPPGELF | 2-2 | 73.40 |
| HLA-A*29:02/30:02 | 6-1 | CASSRSGLYEQY | 2-7 | 24.47 | ||||
| HLA-B*42:01/42:01 | 5-5 | CASSSLGPPGEQY | 2-7 | 1.06 | ||||
| HLA-C*17:01/17:01 | 5-5 | CVSSLVGPPGELF | 2-7 | 1.06 | ||||
| Number of cells sorted | 876 | count | (94) | |||||
| N086 (181208) | LI9 | 0.7 | 2794 | 370 | 7-9 | CASSSRDGQEQY | 2-7 | 96.77 |
| HLA-A*33:03/33:03 | 7-9 | CASSPRDRDFNYGYT | 1-2 | 1.08 | ||||
| HLA-B*42:01/53:01 | 7-9 | CASSSRGGQEQY | 2-7 | 1.08 | ||||
| HLA-C*03:02/04:01 | 5-1 | CASSGMNTEAF | 1-1 | 1.08 | ||||
| Number of cells sorted | 4156 | count | (93) | |||||
| R020 (150607) | LI9 | 0.6 | 5723 | 600 | 4-1 | CASSHGMGASTSGYT | 1-2 | 90.59 |
| HLA-A*29:01/29:02 | 7-9 | CASSFPQNTEAF | 1-1 | 5.88 | ||||
| HLA-B*42:01/51:01 | 7-9 | CASRDGQGHEQY | 2-7 | 2.35 | ||||
| HLA-C*16:01/17:01 | 4-1 | CASGHGMGASTSGYT | 1-2 | 1.18 | ||||
| Number of cells sorted | ND | count | (85) | |||||
| N033 (100707) | LI9 | 0.5 | 415 | 620 | 7-9 | CASSDRQSLVQF | 2-1 | 86.84 |
| HLA-A*30:01/36:01 | 7-8 | CASSKPLYEQY | 2-7 | 7.89 | ||||
| HLA-B*42:01/53:01 | 7-9 | CTSSDRQSLVQF | 2-1 | 2.63 | ||||
| HLA-C*04:01/17:01 | 7-9 | CASEIGNSGQETQY | 2-5 | 1.32 | ||||
| 7-9 | CVSSDRQSLVQF | 2-1 | 1.32 | |||||
| Number of cells sorted | 3337 | count | (76) | |||||
| N080 (180808) | LI9 | 1.1 | 1520 | 610 | 7-9 | CASSPIQGSEQY | 2-7 | 56.36 |
| HLA-A*02:02/03:01 | 7-9 | CASSSKDGQSQY | 2-3 | 25.45 | ||||
| HLA-B*15:03/42:01 | 5-5 | CASSWTGPPGEQF | 2-1 | 10.91 | ||||
| HLA-C*02:01/17:01 | 7-9 | CASSPRQGKEQF | 2-1 | 7.27 | ||||
| Number of cells sorted | 2120 | count | (55) | |||||
| N021 (040211) | LI9 | 0.1 | 8249 | 510 | 7-9 | CASSLAQSREQY | 2-7 | 25.71 |
| HLA-A*30:01/34:02 | 7-9 | CASSSRQGKEAF | 1-1 | 25.71 | ||||
| HLA-B*42:01/44:03 | 7-9 | CASSPRQGHEQY | 2-7 | 22.86 | ||||
| HLA-C*07:01/17:01 | 7-9 | CASSPRTGGTEAF | 1-1 | 7.14 | ||||
| 7-9 | CASSSRQSKEAF | 1-1 | 4.29 | |||||
| 7-9 | CASSLAQSRERY | 2-7 | 2.86 | |||||
| 5-5 | CASSSVGPPGELF | 2-2 | 2.86 | |||||
| 15 | CATSRSGLAGKDTQY | 2-3 | 1.43 | |||||
| 7-9 | CASSSRQGKEQY | 2-7 | 1.43 | |||||
| 7-9 | CASSPRQGQEQY | 2-7 | 1.43 | |||||
| 7-9 | CASSPGQGREQY | 2-7 | 1.43 | |||||
| 7-9 | CASSPGQGQEQY | 2-7 | 1.43 | |||||
| 7-9 | CASSLVQSREQY | 2-7 | 1.43 | |||||
| Number of cells sorted | 1085 | count | (70) | |||||
| SK178 | LI9 | 1.5 | 222000 | 5-5 | CASSLVGPPGEAF | 1-1 | 76.79 | |
| HLA-A*34:02/34:02 | 6-1 | CASRDRQSHEQY | 2-7 | 17.86 | ||||
| HLA-B*42:01/44:03 | 7-9 | CASSFTSGVITGELF | 2-2 | 3.57 | ||||
| HLA-C*04:01/17:01 | 6-1 | CASSASVIAGKLF | 1-4 | 1.79 | ||||
| Number of cells sorted | 4762 | count | (56) | |||||
| SK075 | LI9 | 0.9 | 25900 | 1-1 | CSARGLGVNTEAF | 1-1 | 75.32 | |
| HLA-A*23:01/30:01 | 2-1 | CASIPNLGNEQF | 2-1 | 12.99 | ||||
| HLA-B*42:01/58:01 | 1-1 | CASSLWGAKMNTEAF | 1-1 | 6.49 | ||||
| HLA-C*06:02/17:01 | 1-1 | CASSPRQGKEAF | 1-1 | 2.60 | ||||
| 2-6 | CASSPRQGLEGANVLT | 2-6 | 1.30 | |||||
| 2-1 | CASSVSAIYNEQF | 2-1 | 1.30 | |||||
| Number of cells sorted | 5182 | count | (77) |
Public clonotypes are involved in immunodominant CD8+ T-cell responses
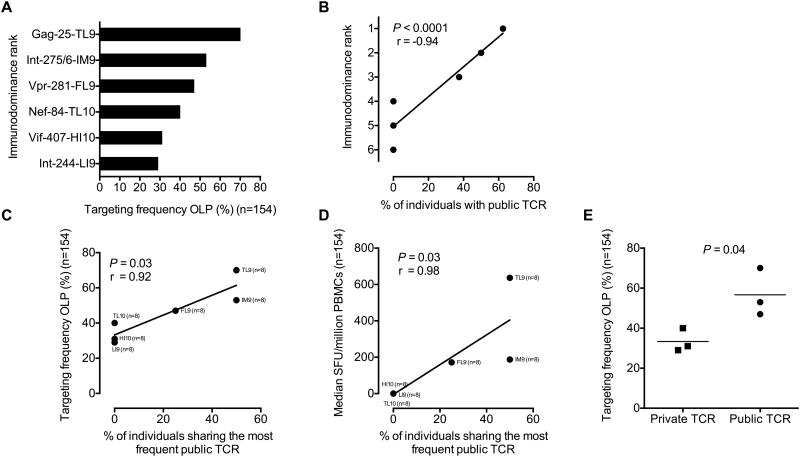
A stable peptide-MHC interaction is a prerequisite for immunogenicity, but other factors also contribute (12). Based on our observations above, we hypothesized that TCR bias may play a role in immunodominance. Accordingly, we compared measures of public TCR occurrence with epitope targeting across all HLA-B*42:01+ individuals (n=154) (Fig 6A). Strong correlations were observed for immunodominance rank (r = -0.94, P < 0.0001) (Fig 6B) and targeting frequency (r = 0.92, P = 0.03) (Fig 6C), both of which held whether we compared the frequency of individuals sharing any public TCR or the frequency of individuals sharing the most common public TCR. The latter correlation was even more marked after normalization for sampling depth (r = 0.941, P = 0.005; data not shown). Although we deliberately analyzed an identical number of CD8+ T-cell populations from an identical number of individuals for each specificity (n = 8), this result is important because it indicates that the detected association is not an artefact of differential sequence coverage. An additional correlation was observed for response magnitude (Fig 6D). Moreover, exclusively private responses were significantly less dominant than responses incorporating public TCRs (P = 0.04) (Fig 6E).
Figure 6. Epitope targeting frequency correlates with shared TCR recruitment.
(A) Targeting frequencies for six optimal epitopes in HLA-B*42:01+ individuals, stratified for lack of HLA-B*07:02/39:10/42:02/81:01 co-expression (n=154). (B) Immunodominance rank, shown as most targeted ranked 1, versus the percentage of individuals sharing any public TCR. Significance was determined using the Spearman rank test. (C) Targeting frequencies for the same six epitopes versus the percentage of individuals sharing the most frequent public TCR. Significance was determined using the Spearman rank test. (D) Response magnitude, including responders and non-responders (mean), versus the percentage of individuals sharing the most frequent public TCR. Significance was determined using the Spearman rank test. (E) Targeting frequencies for all six epitopes stratified for the presence or absence of public TCRs. Significance was determined using the Mann-Whitney U-test.
Type 1 bias, defined by TRBV sharing alone, was not directly linked to immunodominance (P = 0.2; data not shown). Similarly, there was no association between type IV bias, defined by near-identical (disparity <2 amino acids) CDR3 sequences (36), and targeting frequency (P = 0.7; data not shown). It is also notable that response magnitude did not correlate with the frequency of public TCRs (P = 0.74; data not shown). Thus, public TCR recruitment is linked to increased targeting frequency independently of non-identical bias and response magnitude.
Peptide-HLA-B*42:01 binding contributes minimally to immunodominance
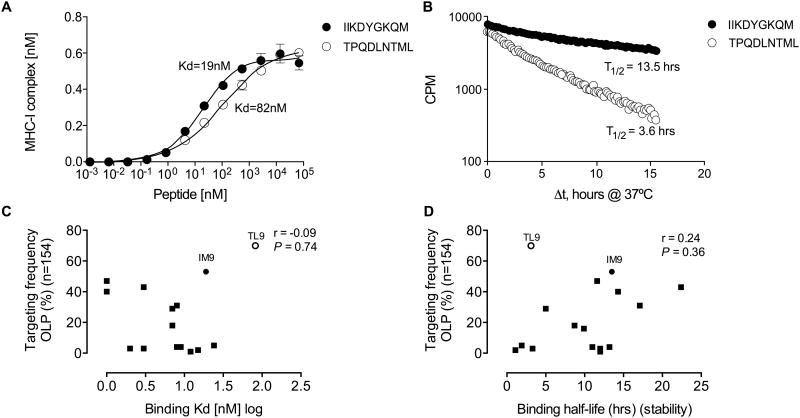
To assess the relative impact of TCR bias with respect to other factors that shape immunodominance (6), (16, 66), we examined peptide-HLA-B*42:01 binding affinity and stability (52, 53) for 16 of the epitopes described in this study (Table S1). The two immunodominant epitopes, Gag-TL9 and Int-IM9, bound HLA-B*42:01 with affinities of KD = 82 nM and KD = 19 nM, respectively, and corresponding half-lives of 3.6 hrs and 13.5 hrs (Fig 7A and B). Across all epitopes, however, no significant correlations were detected between these binding parameters and immunodominance (Fig 7C and D). Nonetheless, peptide-HLA-B*42:01 binding half-life correlated positively with targeting frequency when Gag-TL9 was excluded from the analysis (r = 0.53, P = 0.04; data not shown). Neither binding affinity nor stability correlated with response magnitude (P = 0.42 and P = 0.32, respectively; data not shown). Thus, peptide binding alone cannot explain the observed patterns of immunodominance in this system.
Figure 7. Peptide-HLA-B*42:01 binding affinities and half-lives for 16 optimal epitopes.
HLA-B*42:01 binding affinities (A) and half-lives (B) for the Gag-TL9 (TPQDLNTML) and Int-IM9 (IIKDYGKQM) peptides. Peptide binding affinities (C) and half-lives (D) for the optimal epitopes versus targeting frequencies for the 16 corresponding OLPs in HLA-B*42:01+ individuals, stratified for lack of HLA-B*07:02/39:10/42:02/81:01 co-expression (n=154). Significance was determined using the Spearman rank test.
Discussion
In this study, we conducted an extensive analysis of CD8+ T-cell responses restricted by a single HLA class I molecule to evaluate the mechanisms that contribute to immunodominance and antiviral efficacy in HIV-1 infection. The key findings were: (i) targeting frequencies conformed to a clear hierarchy across 20 different HLA-B*42:01-restricted epitope-specific responses; (ii) Gag-specific responses and targeting breadth were associated with lower viral load set-points; (iii) epitope-specific public TCR usage correlated with immunodominance; and (iv) peptide-HLA-B*42:01 binding affinity and stability were not significantly linked with targeting frequencies. These results suggest that the available TCR repertoire can influence immunodominance patterns and CD8+ T-cell efficacy in chronic HIV-1 infection.
Initially, we used OLPs spanning the entire viral proteome to screen a large cohort of individuals with chronic C-clade infection for HIV-specific CD8+ T-cell responses. This comprehensive and unbiased strategy allowed the identification of novel and previously defined HLA-B*42:01-restricted epitopes that adhered to a strict pattern of immunodominance, thereby enabling more detailed downstream analyses. It is notable that the accurate quantification of certain responses can be compromised by this approach, especially when the optimal epitope resides in the central part of the corresponding OLP (18, 67). Nonetheless, we found a strong correlation between optimal peptide-specific and OLP-defined responses (r = 0.85, P < 0.0001; data not shown). Although this finding justifies the use of OLP screening, it remains conceivable that minor discrepancies could negatively influence the observed correlation between response magnitude and targeting frequency.
Subsequent analyses revealed a significant association between HLA-B*42:01-restricted Gag-specific CD8+ T-cell responses and lower viral load set-points, consistent with the notion that protein targeting patterns influence immune efficacy. Mechanistically, this may be a function of the rapid processing kinetics and relative abundance of Gag-derived epitopes on the cell surface, enabling the elimination of infected targets prior to the production of viral progeny (57, 68, 69). In addition, Gag targeting may be beneficial due to the fitness costs incurred by viral escape mutations within highly conserved, functionally constrained regions of the viral proteome (25-27, 70, 71). However, not all escape mutations reduce viral replicative capacity, as exemplified by the Gag-RM9 variants associated with higher viral loads in our study. This observation is consistent with a recent analysis of the M30R polymorphism, which was shown to increase viral replicative capacity and precipitate disease progression in association with HLA-B*42:01 (72).
The detected association between TCR publicity and immunodominance is particularly striking given the vast recombinatorial diversity that shapes the available repertoire in each individual (36, 38, 64, 65). Across a total of 48 ex vivo datasets, however, TCR bias was apparent at multiple levels. In particular, each epitope-specific CD8+ T-cell population displayed distinct patterns of TRBV usage. Moreover, the extent of TCR sharing varied as a function of epitope specificity. In line with previous studies, clonal diversity per se did not correlate with immunodominance (73). However, the most frequently targeted epitopes mobilized cognate repertoires characterized by the presence of high frequency public TCRs. It is established that public TCRs arise in the naïve repertoire as a function of convergent recombination, whereby production frequency is dictated on a probabilistic basis by the number of rearrangements, nucleotide additions and amino acid codons that can generate a particular sequence(64, 74). The subsequent recruitment of these TCRs into the epitope-specific memory repertoire is dictated by antigen avidity and clonal proliferation (10, 55). Our observations therefore suggest that commonly targeted epitopes are structurally compatible with frequently generated TCRs, at least in the system described here. In this scenario, public TCRs populate a recurrence ‘hot-spot’ in the naïve repertoire, enabling immunodominant epitopes to initiate and maintain CD8+ T-cell responses in multiple individuals. Consistent with this proposition, naive precursor frequencies are known to influence immunodominance patterns (6, 7, 9, 75, 76). However, the extent to which frequently produced TCRs contribute to the overall precursor pool for any given epitope remains to be determined.
It is noteworthy that the public TCRs detected for each of the three immunodominant specificities were constrained by a distinct TRBV segment. This suggests a determinative role for the germline-encoded CDR1 and CDR2 loops. One possibility is that specific residues encoded by these TRBV genes interact with HLA-B*42:01, potentially influencing thymic selection to skew naïve CD8+ T-cell frequencies towards particular specificities (77). Alternatively, germline-encoded recognition of the bound peptide may contribute to immunodominance patterns (78). Immunodominant epitope-specific TCRs constructed almost entirely from germline DNA have been described previously and may represent an evolutionarily conserved mechanism to combat ancient pathogens that have co-evolved with the human race (37, 64, 79, 80). Although this is unlikely to apply directly in the case of HIV-1, it is intriguing to speculate that structural homology with such epitopes may inadvertently underlie the immunodominance patterns described here.
In contrast to the association between immunodominance and TCR bias controlled within the framework of a single HLA class I molecule, peptide binding to HLA-B*42:01 contributed little to the observed epitope targeting frequencies in this study. However, it is important to note that other factors, such as antigen processing efficiency (81, 82), kinetics (57, 69, 83) and protein abundance (84, 85), play a key role alongside a requirement for sufficient peptide-MHC binding (9, 86). Further studies will therefore be necessary to assess the contribution of TCR recruitment in relation to these well defined determinants of immunodominance.
In summary, we present the first evidence linking epitope targeting frequencies to TCR bias. Although the extent to which this phenomenon applies across other systems remains to be defined, our data suggest that antigen-specific repertoire studies will be important for a full understanding of both natural and vaccine-induced immune responses against intracellular pathogens.
Supplementary Material
Table 7.
Clonal analysis of HLA-B*42:01-restricted HI10-specific CD8+ T-cell populations.
| Subject (ddmmyy) | Epitope | CD8+ Tet+ | pVL | CD4 | TRBV | CDR3 | TRBJ | Freq (%) |
|---|---|---|---|---|---|---|---|---|
| H022 (170807) | HI10 | 0.3 | 125688 | 430 | 20-1 | CSATDRASGIEQY | 2-7 | 43.66 |
| HLA-A*02:02/03:01 | 20-1 | CSATSRDSALEQY | 2-7 | 33.80 | ||||
| HLA-B*08:01/42:01 | 14 | CASSQEGWDGRYEQY | 2-7 | 9.86 | ||||
| HLA-C*07:02/17:01 | 20-1 | CSATSRQGGLEQY | 2-7 | 2.82 | ||||
| 6-2/3 | CASNPVFAAGGETQY | 2-5 | 1.41 | |||||
| 20-1 | CSATSRDRALEQY | 2-7 | 1.41 | |||||
| 20-1 | CSATSRASAIEQY | 2-7 | 1.41 | |||||
| 20-1 | CSATGRESGIEQY | 2-7 | 1.41 | |||||
| 20-1 | CSATARQGGMEQY | 2-7 | 1.41 | |||||
| 20-1 | CSAADRASGIEQY | 2-7 | 1.41 | |||||
| 20-1 | CSARGGTSIFYT | 1-2 | 1.41 | |||||
| Number of cells sorted | 2608 | count | (71) | |||||
| N106 (030310) | HI10 | 0.9 | 14014 | 350 | 20-1 | CSATDRDGGLEQY | 2-7 | 48.31 |
| HLA-A*02:01/02:05 | 7-9 | CASSLGPVEAF | 1-1 | 37.08 | ||||
| HLA-B*42:01/53:01 | 20-1 | CSATSRDRGLEQY | 2-7 | 5.62 | ||||
| HLA-C*04:01/17:01 | 14 | CASSQENRQGRYEQY | 2-7 | 2.25 | ||||
| 7-2 | CASRRDRDMNTEAF | 1-1 | 2.25 | |||||
| 20-1 | CSATHRDRGLEQY | 2-7 | 2.25 | |||||
| 20-1 | CSATDRDGGPEQY | 2-7 | 1.12 | |||||
| 20-1 | CSARRFPEAF | 1-1 | 1.12 | |||||
| Number of cells sorted | 2190 | count | (89) | |||||
| N080 (180808) | HI10 | 0.5 | 1520 | 610 | 5-5 | CASSLSRTGFYEQY | 2-7 | 48.15 |
| HLA-A*02:02/03:01 | 27-1 | CASSLGADQPQH | 1-5 | 20.37 | ||||
| HLA-B*15:03/42:01 | 9-1 | CASGAAMNTEAF | 1-1 | 12.96 | ||||
| HLA-C*02:01/17:01 | 6-2/3 | CASSLGPAAGYT | 1-2 | 11.11 | ||||
| 6-2/3 | CASRRGSTYNEQF | 2-1 | 5.56 | |||||
| 7-9 | CASSLGPTVQGNYGYT | 1-2 | 1.85 | |||||
| Number of cells sorted | 923 | count | (54) | |||||
| N058 (270509) | HI10 | 0.6 | 4534 | 510 | 7-3 | CASRPPDTGELF | 2-2 | 48.68 |
| HLA-A*01:02/30:01 | 20-1 | CSATSRDGDNEQF | 2-1 | 32.89 | ||||
| HLA-B*15:03/42:01 | 7-9 | CASSLGPLTGLGPEAF | 1-1 | 13.16 | ||||
| HLA-C*02:10/17:01 | 7-9 | CASGLGPLTGLGPEAF | 1-1 | 1.32 | ||||
| 7-8 | CASSLAGQGNGYT | 1-2 | 1.32 | |||||
| 7-3 | CASRSPDTGELF | 2-2 | 1.32 | |||||
| 6-2/3 | GASRTSGEETQY | 2-5 | 1.32 | |||||
| Number of cells sorted | 4729 | count | (76) | |||||
| SK178 | HI10 | 0.3 | 220000 | 20-1 | CSATNRDRGLEQY | 2-7 | 68.57 | |
| HLA-A*34:02/34:02 | 7-2 | CASSFDKGYEQY | 2-7 | 22.86 | ||||
| HLA-B*42:01/44:03 | 24-1 | CATRGRGSEETQY | 2-5 | 7.14 | ||||
| HLA-C*04:01/17:01 | 11-3 | CASSSTWGTGELF | 2-2 | 1.43 | ||||
| Number of cells sorted | 1316 | count | (70) | |||||
| SK191 | HI10 | 0.5 | 76900 | 7-9 | CASSLGPAIPGNTIY | 1-3 | 50.68 | |
| HLA-A*01:02/30:01 | 7-3 | CASRGADTGELF | 2-2 | 42.47 | ||||
| HLA-B*15:03/42:01 | 20-1 | CSATSRAGDNEQF | 2-1 | 4.11 | ||||
| HLA-C*02:10/17:01 | 12-5 | CASGLAVPVDGYT | 1-2 | 1.37 | ||||
| 6-1 | CASTLDRLAF | 1-1 | 1.37 | |||||
| Number of cells sorted | 1306 | count | (73) | |||||
| R020 (150607) | HI10 | 0.7 | 5723 | 600 | 7-9 | CASSLGPRYEQY | 2-7 | 62.34 |
| HLA-A*29:01/29:02 | 5-5 | CASSFTRQSPYNEQF | 2-1 | 31.17 | ||||
| HLA-B*42:01/51:01 | 5-5 | CASSSTRQSPYNEQF | 2-1 | 1.30 | ||||
| HLA-C*16:01/17:01 | 3-1 | CASSQDRTSGNTIY | 1-3 | 1.30 | ||||
| 20-1 | CSATRRDRGLEQY | 2-7 | 1.30 | |||||
| 7-9 | CASSTTTGNTEAF | 1-1 | 1.30 | |||||
| 2 | CASSRGNTIY | 1-3 | 1.30 | |||||
| Number of cells sorted | 2068 | count | (77) | |||||
| N037 (061009) | HI10 | 0.3 | 3170 | 360 | 7-9 | CASSLGPTVPGNTIY | 1-3 | 34.62 |
| HLA-A*29:02/30:02 | 20-1 | CSATSRQGGREQY | 2-7 | 32.05 | ||||
| HLA-B*42:01/42:01 | 5-1 | CASSSFRDGGTDTQY | 2-3 | 25.64 | ||||
| HLA-C*17:01/17:01 | 7-9 | CASRGGPLTEAF | 1-1 | 5.13 | ||||
| 7-9 | CASSLGPTVPGNAIY | 1-3 | 1.28 | |||||
| 20-1 | CSATSRRGGREQY | 2-7 | 1.28 | |||||
| Number of cells sorted | 963 | count | (78) |
Acknowledgments
This work was supported by the Wellcome Trust (D.A.P. and P.J.G.) and the National Institutes of Health (grant #R01 AI46995). H.N.K is funded by the Danish Agency for Science, Technology and Innovation (grant #12-132295), The Lundbeck Foundation (grant #R151-2013-14624) and The MAERSK Foundation for Medical Improvement. D.A.P. is a Wellcome Trust Senior Investigator.
References
- 1.Zinkernagel RM. Immunology taught by viruses. Science. 1996;271:173–178. doi: 10.1126/science.271.5246.173. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Appay V, Douek DC, Price DA. CD8+ T cell efficacy in vaccination and disease. Nature medicine. 2008;14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 4.Boulanger DS, Oliveira R, Ayers L, Prior SH, James E, Williams AP, Elliott T. Absence of tapasin alters immunodominance against a lymphocytic choriomeningitis virus polytope. J Immunol. 2010;184:73–83. doi: 10.4049/jimmunol.0803489. [DOI] [PubMed] [Google Scholar]
- 5.Crotzer VL, Christian RE, Brooks JM, Shabanowitz J, Settlage RE, Marto JA, White FM, Rickinson AB, Hunt DF, Engelhard VH. Immunodominance among EBV-derived epitopes restricted by HLA-B27 does not correlate with epitope abundance in EBV-transformed B-lymphoblastoid cell lines. J Immunol. 2000;164:6120–6129. doi: 10.4049/jimmunol.164.12.6120. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins MK, Chu HH, McLachlan JB, Moon JJ. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol. 2010;28:275–294. doi: 10.1146/annurev-immunol-030409-101253. [DOI] [PubMed] [Google Scholar]
- 7.Jenkins MK, Moon JJ. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J Immunol. 2012;188:4135–4140. doi: 10.4049/jimmunol.1102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koelle DM, Liu Z, McClurkan CL, Cevallos RC, Vieira J, Hosken NA, Meseda CA, Snow DC, Wald A, Corey L. Immunodominance among herpes simplex virus-specific CD8 T cells expressing a tissue-specific homing receptor. Proc Natl Acad Sci U S A. 2003;100:12899–12904. doi: 10.1073/pnas.2131705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.La Gruta NL, Rothwell WT, Cukalac T, Swan NG, Valkenburg SA, Kedzierska K, Thomas PG, Doherty PC, Turner SJ. Primary CTL response magnitude in mice is determined by the extent of naive T cell recruitment and subsequent clonal expansion. J Clin Invest. 2010;120:1885–1894. doi: 10.1172/JCI41538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddiqui S, Basta S. CD8+ T cell immunodominance in lymphocytic choriomeningitis virus infection is modified in the presence of toll-like receptor agonists. J Virol. 2011;85:13224–13233. doi: 10.1128/JVI.05996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, Cerundolo V, Bonneville M. Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. J Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 13.Goulder PJ, Walker BD. HIV and HLA Class I: An Evolving Relationship. Immunity. 2012;37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 15.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nature medicine. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 16.Kloverpris HN, Stryhn A, Harndahl M, van der Stok M, Payne RP, Matthews PC, Chen F, Riddell L, Walker BD, Ndung'u T, Buus S, Goulder P. HLA-B*57 micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure and HIV immune control. J Virol. 2012 doi: 10.1128/JVI.06150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews PC, Prendergast A, Leslie A, Crawford H, Payne R, Rousseau C, Rolland M, Honeyborne I, Carlson J, Kadie C, Brander C, Bishop K, Mlotshwa N, Mullins JI, Coovadia H, Ndung'u T, Walker BD, Heckerman D, Goulder PJ. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol. 2008;82:8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mothe B, Llano A, Ibarrondo J, Daniels M, Miranda C, Zamarreno J, Bach V, Zuniga R, Perez-Alvarez S, Berger CT, Puertas MC, Martinez-Picado J, Rolland M, Farfan M, Szinger JJ, Hildebrand WH, Yang OO, Sanchez-Merino V, Brumme CJ, Brumme ZL, Heckerman D, Allen TM, Mullins JI, Gomez G, Goulder PJ, Walker BD, Gatell JM, Clotet B, Korber BT, Sanchez J, Brander C. Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med. 2011;9:208. doi: 10.1186/1479-5876-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews PC, Koyanagi M, Kloverpris HN, Harndahl M, Stryhn A, Akahoshi T, Gatanaga H, Oka S, Juarez Molina C, Valenzuela Ponce H, Avila Rios S, Cole D, Carlson J, Payne RP, Ogwu A, Bere A, Ndung'u T, Gounder K, Chen F, Riddell L, Luzzi G, Shapiro R, Brander C, Walker B, Sewell AK, Reyes Teran G, Heckerman D, Hunter E, Buus S, Takiguchi M, Goulder PJ. Differential Clade-Specific HLA-B*3501 Association with HIV-1 Disease Outcome Is Linked to Immunogenicity of a Single Gag Epitope. J Virol. 2012;86:12643–12654. doi: 10.1128/JVI.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Piechocka-Trocha A, Miura T, Brockman MA, Julg BD, Baker BM, Rothchild AC, Block BL, Schneidewind A, Koibuchi T, Pereyra F, Allen TM, Walker BD. Differential neutralization of human immunodeficiency virus (HIV) replication in autologous CD4 T cells by HIV-specific cytotoxic T lymphocytes. J Virol. 2009;83:3138–3149. doi: 10.1128/JVI.02073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saez-Cirion A, Sinet M, Shin SY, Urrutia A, Versmisse P, Lacabaratz C, Boufassa F, Avettand-Fenoel V, Rouzioux C, Delfraissy JF, Barre-Sinoussi F, Lambotte O, Venet A, Pancino G. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with Gag-specific CD8 T cell responses. J Immunol. 2009;182:7828–7837. doi: 10.4049/jimmunol.0803928. [DOI] [PubMed] [Google Scholar]
- 22.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. HIV evolution: CTL escape mutation and reversion after transmission. Nature medicine. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien SJ, Gao X, Carrington M. HLA and AIDS: a cautionary tale. Trends Mol Med. 2001;7:379–381. doi: 10.1016/s1471-4914(01)02131-1. [DOI] [PubMed] [Google Scholar]
- 24.Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, Yu XG, Draenert R, Johnston MN, Strick D, Allen TM, Feeney ME, Kahn JO, Sekaly RP, Levy JA, Rockstroh JK, Goulder PJ, Walker BD. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS. 2003;17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 25.Crawford H, Prado JG, Leslie A, Hue S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins JI, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker BD, Goulder PJ. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA, Kuntzen T, Tung CS, LaBute MX, Mueller SM, Harrer T, McMichael AJ, Goulder PJ, Aiken C, Brander C, Kelleher AD, Allen TM. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peaper DR, Cresswell P. Regulation of MHC class I assembly and peptide binding. Annu Rev Cell Dev Biol. 2008;24:343–368. doi: 10.1146/annurev.cellbio.24.110707.175347. [DOI] [PubMed] [Google Scholar]
- 29.Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, Burgett N, Swartz ME, Yang A, Alter G, Yu XG, Meier A, Rockstroh JK, Allen TM, Jessen H, Rosenberg ES, Carrington M, Walker BD. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iglesias MC, Almeida JR, Fastenackels S, van Bockel DJ, Hashimoto M, Venturi V, Gostick E, Urrutia A, Wooldridge L, Clement M, Gras S, Wilmann PG, Autran B, Moris A, Rossjohn J, Davenport MP, Takiguchi M, Brander C, Douek DC, Kelleher AD, Price DA, Appay V. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood. 2011;118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladell K, Hashimoto M, Iglesias MC, Wilmann PG, McLaren JE, Gras S, Chikata T, Kuse N, Fastenackels S, Gostick E, Bridgeman JS, Venturi V, Arkoub ZA, Agut H, van Bockel DJ, Almeida JR, Douek DC, Meyer L, Venet A, Takiguchi M, Rossjohn J, Price DA, Appay V. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity. 2013;38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 33.Price DA, Asher TE, Wilson NA, Nason MC, Brenchley JM, Metzler IS, Venturi V, Gostick E, Chattopadhyay PK, Roederer M, Davenport MP, Watkins DI, Douek DC. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG. T Cell Receptor alphabeta Diversity Inversely Correlates with Pathogen-Specific Antibody Levels in Human Cytomegalovirus Infection. Sci Transl Med. 2012;4:128ra142. doi: 10.1126/scitranslmed.3003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89:375–387. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 37.Miles JJ, Bulek AM, Cole DK, Gostick E, Schauenburg AJ, Dolton G, Venturi V, Davenport MP, Tan MP, Burrows SR, Wooldridge L, Price DA, Rizkallah PJ, Sewell AK. Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus infection. PLoS Pathog. 2010;6:e1001198. doi: 10.1371/journal.ppat.1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 39.Billam P, Bonaparte KL, Liu J, Ruckwardt TJ, Chen M, Ryder AB, Wang R, Dash P, Thomas PG, Graham BS. T Cell receptor clonotype influences epitope hierarchy in the CD8+ T cell response to respiratory syncytial virus infection. J Biol Chem. 2011;286:4829–4841. doi: 10.1074/jbc.M110.191437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kedzierska K, Guillonneau C, Gras S, Hatton LA, Webby R, Purcell AW, Rossjohn J, Doherty PC, Turner SJ. Complete modification of TCR specificity and repertoire selection does not perturb a CD8+ T cell immunodominance hierarchy. Proc Natl Acad Sci U S A. 2008;105:19408–19413. doi: 10.1073/pnas.0810274105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feeney ME, Tang Y, Roosevelt KA, Leslie AJ, McIntosh K, Karthas N, Walker BD, Goulder PJ. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J Virol. 2004;78:8927–8930. doi: 10.1128/JVI.78.16.8927-8930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro RL, Hughes MD, Ogwu A, Kitch D, Lockman S, Moffat C, Makhema J, Moyo S, Thior I, McIntosh K, van Widenfelt E, Leidner J, Powis K, Asmelash A, Tumbare E, Zwerski S, Sharma U, Handelsman E, Mburu K, Jayeoba O, Moko E, Souda S, Lubega E, Akhtar M, Wester C, Tuomola R, Snowden W, Martinez-Tristani M, Mazhani L, Essex M. Antiretroviral regimens in pregnancy and breast-feeding in Botswana. N Engl J Med. 2010;362:2282–2294. doi: 10.1056/NEJMoa0907736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang KH, Goedhals D, Carlson JM, Brockman MA, Mishra S, Brumme ZL, Hickling S, Tang CS, Miura T, Seebregts C, Heckerman D, Ndung'u T, Walker B, Klenerman P, Steyn D, Goulder P, Phillips R, van Vuuren C, Frater J. Progression to AIDS in South Africa Is Associated with both Reverting and Compensatory Viral Mutations. PLoS One. 2011;6:e19018. doi: 10.1371/journal.pone.0019018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, Walker BD, Ndung'u T, Shapiro R, Frater J, Brumme ZL, Goulder PJ, Heckerman D. Widespread Impact of HLA Restriction on Immune Control and Escape Pathways in HIV-1. J Virol. 2012 doi: 10.1128/JVI.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Payne RP, Kloverpris H, Sacha JB, Brumme Z, Brumme C, Buus S, Sims S, Hickling S, Riddell L, Chen F, Luzzi G, Edwards A, Phillips R, Prado JG, Goulder PJ. Efficacious Early Antiviral Activity of HIV Gag- and Pol-Specific HLA-B*2705-Restricted CD8+ T Cells. J Virol. 2010;84:10543–10557. doi: 10.1128/JVI.00793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leslie A, Price DA, Mkhize P, Bishop K, Rathod A, Day C, Crawford H, Honeyborne I, Asher TE, Luzzi G, Edwards A, Rousseau CM, Mullins JI, Tudor-Williams G, Novelli V, Brander C, Douek DC, Kiepiela P, Walker BD, Goulder PJ. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J Immunol. 2006;177:4699–4708. doi: 10.4049/jimmunol.177.7.4699. [DOI] [PubMed] [Google Scholar]
- 47.Honeyborne I, Prendergast A, Pereyra F, Leslie A, Crawford H, Payne R, Reddy S, Bishop K, Moodley E, Nair K, van der Stok M, McCarthy N, Rousseau CM, Addo M, Mullins JI, Brander C, Kiepiela P, Walker BD, Goulder PJ. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J Virol. 2007;81:3667–3672. doi: 10.1128/JVI.02689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goulder PJ, Addo MM, Altfeld MA, Rosenberg ES, Tang Y, Govender U, Mngqundaniso N, Annamalai K, Vogel TU, Hammond M, Bunce M, Coovadia HM, Walker BD. Rapid definition of five novel HLA-A*3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by elispot and intracellular cytokine staining assays. J Virol. 2001;75:1339–1347. doi: 10.1128/JVI.75.3.1339-1347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 50.Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, Pillay T, Hilton L, Thobakgale C, Ramduth D, Draenert R, Le Gall S, Luzzi G, Edwards A, Brander C, Sewell AK, Moore S, Mullins J, Moore C, Mallal S, Bhardwaj N, Yusim K, Phillips R, Klenerman P, Korber B, Kiepiela P, Walker B, Goulder P. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rousseau CM, Birditt BA, McKay AR, Stoddard JN, Lee TC, McLaughlin S, Moore SW, Shindo N, Learn GH, Korber BT, Brander C, Goulder PJ, Kiepiela P, Walker BD, Mullins JI. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J Virol Methods. 2006;136:118–125. doi: 10.1016/j.jviromet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Harndahl M, Rasmussen M, Roder G, Buus S. Real-time, high-throughput measurements of peptide-MHC-I dissociation using a scintillation proximity assay. J Immunol Methods. 2010 doi: 10.1016/j.jim.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harndahl M, Justesen S, Lamberth K, Roder G, Nielsen M, Buus S. Peptide binding to HLA class I molecules: homogenous, high-throughput screening, and affinity assays. J Biomol Screen. 2009;14:173–180. doi: 10.1177/1087057108329453. [DOI] [PubMed] [Google Scholar]
- 54.Quigley MF, Almeida JR, Price DA, Douek DC. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. In: Coligan John E, et al., editors. Current protocols in immunology. Chapter 10. 2011. Unit10 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Developmental and comparative immunology. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 57.Kloverpris HN, Payne RP, Sacha JB, Rasaiyaah JT, Chen F, Takiguchi M, Yang OO, Towers GJ, Goulder P, Prado JG. Early Antigen Presentation of Protective HIV-1 KF11Gag and KK10Gag Epitopes from Incoming Viral Particles Facilitates Rapid Recognition of Infected Cells by Specific CD8+ T Cells. J Virol. 2013;87:2628–2638. doi: 10.1128/JVI.02131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chao A, Jost L. Coverage-based rarefaction and extrapolation: standardizing samples by completeness rather than size. Ecology. 2012;93:2533–2547. doi: 10.1890/11-1952.1. [DOI] [PubMed] [Google Scholar]
- 59.Costa AI, Koning D, Ladell K, McLaren JE, Grady BP, Schellens IM, van Ham P, Nijhuis M, Borghans JA, Kesmir C, Price DA, van Baarle D. Complex T-Cell Receptor Repertoire Dynamics Underlie the CD8+ T-Cell Response to HIV-1. J Virol. 2015;89:110–119. doi: 10.1128/JVI.01765-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gold MC, McLaren JE, Reistetter JA, Smyk-Pearson S, Ladell K, Swarbrick GM, Yu YY, Hansen TH, Lund O, Nielsen M, Gerritsen B, Kesmir C, Miles JJ, Lewinsohn DA, Price DA, Lewinsohn DM. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. 2014;211:1601–1610. doi: 10.1084/jem.20140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Draenert R, Altfeld M, Brander C, Basgoz N, Corcoran C, Wurcel AG, Stone DR, Kalams SA, Trocha A, Addo MM, Goulder PJ, Walker BD. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J Immunol Methods. 2003;275:19–29. doi: 10.1016/s0022-1759(02)00541-0. [DOI] [PubMed] [Google Scholar]
- 62.Llano A, Frahm N, Brander C. How to Optimally Define Optimal Cytotoxic T Lymphocyte Epitopes in HIV Infection? HIV Molecular immunology database 2009 [Google Scholar]
- 63.Leisner C, Loeth N, Lamberth K, Justesen S, Sylvester-Hvid C, Schmidt EG, Claesson M, Buus S, Stryhn A. One-pot, mix-and-read peptide-MHC tetramers. PLoS One. 2008;3:e1678. doi: 10.1371/journal.pone.0001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venturi V, Chin HY, Asher TE, Ladell K, Scheinberg P, Bornstein E, van Bockel D, Kelleher AD, Douek DC, Price DA, Davenport MP. TCR beta-chain sharing in human CD8+ T cell responses to cytomegalovirus and EBV. J Immunol. 2008;181:7853–7862. doi: 10.4049/jimmunol.181.11.7853. [DOI] [PubMed] [Google Scholar]
- 65.Turner SJ, La Gruta NL, Kedzierska K, Thomas PG, Doherty PC. Functional implications of T cell receptor diversity. Curr Opin Immunol. 2009;21:286–290. doi: 10.1016/j.coi.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kloverpris HN, Adland E, Koyanagi M, Stryhn A, Harndahl M, Matthews PC, Shapiro R, Walker BD, Ndung'u T, Brander C, Takiguchi M, Buus S, Goulder P. HIV Subtype Influences HLA-B*07:02-Associated HIV Disease Outcome. AIDS Res Hum Retroviruses. 2013 doi: 10.1089/aid.2013.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Draenert R, Verrill CL, Tang Y, Allen TM, Wurcel AG, Boczanowski M, Lechner A, Kim AY, Suscovich T, Brown NV, Addo MM, Walker BD. Persistent recognition of autologous virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection. J Virol. 2004;78:630–641. doi: 10.1128/JVI.78.2.630-641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen DY, Balamurugan A, Ng HL, Cumberland WG, Yang OO. Epitope targeting and viral inoculum are determinants of Nef-mediated immune evasion of HIV-1 from cytotoxic T lymphocytes. Blood. 2012;120:100–111. doi: 10.1182/blood-2012-02-409870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sacha JB, Chung C, Rakasz EG, Spencer SP, Jonas AK, Bean AT, Lee W, Burwitz BJ, Stephany JJ, Loffredo JT, Allison DB, Adnan S, Hoji A, Wilson NA, Friedrich TC, Lifson JD, Yang OO, Watkins DI. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J Immunol. 2007;178:2746–2754. doi: 10.4049/jimmunol.178.5.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, Tang J, Farmer P, Ndung'u T, Lakhi S, Gilmour J, Goepfert P, Walker BD, Kaslow R, Mulenga J, Allen S, Goulder PJ, Hunter E. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206:909–921. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneidewind A, Brockman MA, Sidney J, Wang YE, Chen H, Suscovich TJ, Li B, Adam RI, Allgaier RL, Mothe BR, Kuntzen T, Oniangue-Ndza C, Trocha A, Yu XG, Brander C, Sette A, Walker BD, Allen TM. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J Virol. 2008;82:5594–5605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prince JL, Claiborne DT, Carlson JM, Schaefer M, Yu T, Lahki S, Prentice HA, Yue L, Vishwanathan SA, Kilembe W, Goepfert P, Price MA, Gilmour J, Mulenga J, Farmer P, Derdeyn CA, Tang J, Heckerman D, Kaslow RA, Allen SA, Hunter E. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog. 2012;8:e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koning D, Costa AI, Hoof I, Miles JJ, Nanlohy NM, Ladell K, Matthews KK, Venturi V, Schellens IM, Borghans JA, Kesmir C, Price DA, van Baarle D. CD8+ TCR repertoire formation is guided primarily by the peptide component of the antigenic complex. J Immunol. 2013;190:931–939. doi: 10.4049/jimmunol.1202466. [DOI] [PubMed] [Google Scholar]
- 74.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 75.Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 77.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cole DK, Yuan F, Rizkallah PJ, Miles JJ, Gostick E, Price DA, Gao GF, Jakobsen BK, Sewell AK. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. J Biol Chem. 2009;284:27281–27289. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hislop AD, Annels NE, Gudgeon NH, Leese AM, Rickinson AB. Epitope-specific evolution of human CD8(+) T cell responses from primary to persistent phases of Epstein-Barr virus infection. J Exp Med. 2002;195:893–905. doi: 10.1084/jem.20011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishizuka J, Stewart-Jones GB, van der Merwe A, Bell JI, McMichael AJ, Jones EY. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vbeta domain. Immunity. 2008;28:171–182. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 81.Zhang SC, Martin E, Shimada M, Godfrey SB, Fricke J, Locastro S, Lai NY, Liebesny P, Carlson JM, Brumme CJ, Ogbechie OA, Chen H, Walker BD, Brumme ZL, Kavanagh DG, Le Gall S. Aminopeptidase substrate preference affects HIV epitope presentation and predicts immune escape patterns in HIV-infected individuals. J Immunol. 2012;188:5924–5934. doi: 10.4049/jimmunol.1200219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kloverpris HN, Stryhn A, Harndahl M, Payne R, Towers GJ, Chen F, Riddell L, Walker BD, Ndung'u T, Leslie A, Buus S, Goulder P. HLA-specific intracellular epitope processing shapes an immunodominance pattern for HLA-B*57 that is distinct from HLA-B*58:01. J Virol. 2013 doi: 10.1128/JVI.01122-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmidt J, Iversen AK, Tenzer S, Gostick E, Price DA, Lohmann V, Distler U, Bowness P, Schild H, Blum HE, Klenerman P, Neumann-Haefelin C, Thimme R. Rapid antigen processing and presentation of a protective and immunodominant HLA-B*27-restricted hepatitis C virus-specific CD8+ T-cell epitope. PLoS Pathog. 2012;8:e1003042. doi: 10.1371/journal.ppat.1003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 85.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 86.Harndahl M, Rasmussen M, Roder G, Dalgaard Pedersen I, Sorensen M, Nielsen M, Buus S. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur J Immunol. 2012;42:1405–1416. doi: 10.1002/eji.201141774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.