Abstract
Non-viral gene delivery systems are important transport vehicles that can be safe and effective alternatives to currently available viral systems. A new family of multifunctional spider silk-based gene carriers was bioengineered and found capable of targeting human mesenchymal stem cells (hMSCs). These carriers successfully delivered DNA to the nucleus of these mammalian cells. The presence of specific functional sequences in the recombinant proteins, such as a nuclear localization sequence (NLS) of the large tumor (T) antigen of the Simian virus 40 (SV40), an hMSC high affinity binding peptide (HAB), and a translocation motif (TLM) of the hepatitis-B virus surface protein (PreS2), and their roles in mitigation and enhancement of gene transfection efficiency towards hMSCs were characterized. The results demonstrate that these bioengineered spider silk proteins serve as effective carriers, without the well-known complications associated with viral delivery systems.
Keywords: spider silks, non-viral gene delivery, nuclear targeting, recombinant DNA technology, human mesenchymal stem cells
Introduction
Gene therapy is an attractive approach for treating many complicated inherited (e.g. cystic fibrosis, hemophilia) and acquired (e.g., cancer, Paroxysmal nocturnal hemoglobinuria) diseases [1]. For success, therapeutic genes (i.e., pDNA, ODNs, siRNA, miRNA) must be safely delivered to a targeted site of action (i.e., cytosol, nucleus, mitochondria). These genes must pass through cell and nuclear membranes to efficiently reach target sites, and this transit is complicated due to the high-molecular weights and negative charges of the genes to be delivered. Therefore, safe and efficient gene delivery systems are needed. Viral and non-viral systems have been pursued, with viral systems (e.g., lentivirus, adenovirus, herpes simplex virus) widely used to deliver exogenous genes to different cell types, due to ease of use and their high efficiency [2].
Viruses have evolved successful machinery to deliver their load to a cell, possess intrinsic mechanisms for endosomal escape, and have developed mechanisms for nuclear import [3–6]. However, viral vectors can also exhibit significant immunogenicity, oncogenicity and acute toxicity [7]. In particular, the use of viral vectors has led to the occurrence of vector-related leukemia in a significant number of treated children [4]. As a result, research has focused on safe and effective non-viral gene vectors. Current non-viral vector systems have relatively low risk of immunogenicity and oncogenicity, and can carry large sized genes. However, these systems suffer from low transfection rates, lack intrinsic mechanisms for endosomal escape and nuclear import and can be toxic when used at high doses. Therefore, there is a need to develop more effective non-viral gene delivery systems.
Human bone marrow-derived mesenchymal stromal cells (hMSCs) are a useful source of cells for specialized tissue outcomes due to their native ability to differentiate into a variety of cell types including adipocytes, osteoblasts, tenocytes, chondrocytes and smooth muscles cells [8, 9]. hMSCS are harvested from adults and do not induce immune responses upon local transplantation or systemic administration. The introduction of exogenous genes in this cell type is desirable in order to influence cell differentiation and/or reprogram cells to perform desired or lost functions. Thus, it might be possible to reprogram these cells and, as a result, treat a variety of serum protein deficiencies and genetic or acquired diseases (such as bone, cartilage or bone marrow disorders) with gene therapy. Current non-viral gene delivery systems have had limited success when used to transfect exogenous genes in primary cell lines, such as hMSCs. Hence, it is important to develop safe and effective non-viral gene delivery systems for the transfection of MSCs [6].
Spider silks provide a unique avenue to produce well-defined gene carriers that exhibit low toxicity and high transfection efficiency [10–12], as well as being accessible to bioengineering for precise tailoring of chemistry and sequence. Previously, spider silk gene delivery systems have been demonstrated to be safe and efficient gene carriers for targeting metastatic human breast tumor cells [11]. Furthermore, silks are biocompatible and biodegradable and can be modified to target various cell types and specialized subcellular subunits [13–15]. Silks generate robust materials, thus generating carriers that can survive handling, injection and in vivo function [16, 17].
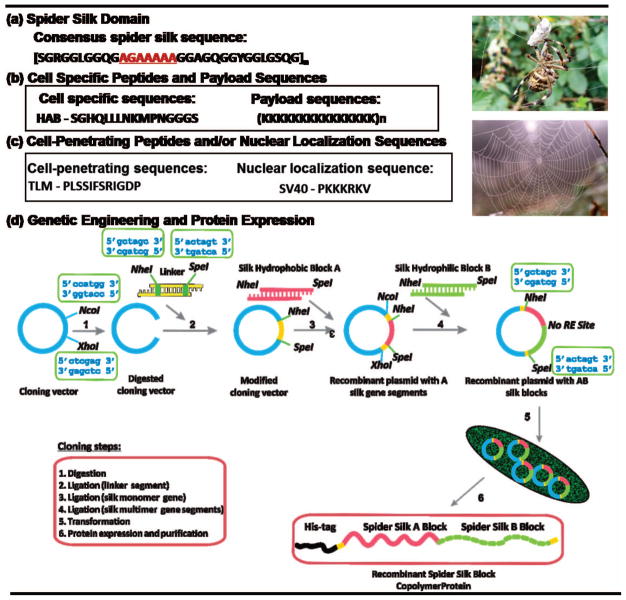
The goal of the present study was to bioengineer silk-based gene delivery systems with enhanced capabilities to deliver a therapeutic gene to hMSCs. Our carrier design has included a penta-block polymer composed of five distinct elements: (1) a spider silk block that is based on the monomeric sequence of the major ampullate spidroin I (MaSp I) of a golden orb-weaver spider, Nephila clavipes, to define carrier (e.g., nanoparticle) size, (2) a payload sequence (poly-L-lysine) to define gene loading capacity, (3) a nuclear localization sequence (NLS) of the large tumor (T) antigen of the Simian virus 40 (SV40) to govern intracellular delivery, (4) an MSC binding peptide known as high affinity binding peptide (HAB) for cell specific targeting, and (5) a histidine-rich peptide to facilitate the purification of the recombinant proteins and to serve as an endosomal escape sequence (Figure 1). Here, we report the synthesis, characterization and evaluation of these non-viral gene delivery vectors based on spider silk sequences linked with functional peptide domains. We demonstrated that by fusing an HAB peptide recognized by receptors at the surface of hMSC membranes and a nuclear localization sequence SV40 to the spider silk sequences transfection efficiency was improved. The presence of these cell-specific and organelle-tailored elements in our non-viral spider-silk systems provide advantages over gene delivery systems with synthetic polymers that cannot be easily decorated with these specific elements. The unique aspect of this system is that it can be easily tailored to accommodate precise delivery needs and has potential to be used as a versatile platform for both intracellular and cell specific targeting.
Figure 1.
Schematic representation of the structure of spider silk penta-block polymer. (a) Spider silk core sequence; (b) cell specific peptides and payload sequences; (c) cell-penetrating peptide and nuclear localization sequences used in the study; (d) recombinant DNA strategy used to prepare the spider silk gene delivery systems for the study. NcoI, NheI, SpeI, XhoI are restriction enzymes (RE). A box above/below a specific RE indicates an oligonucleotide sequence recognized by this RE. Pet30a(+) plasmid is in blue, cloning linker is in yellow, spider silk monomer is in dark blue, poly-L-lysine sequence is in magenta, HAB peptide is in orange and NLS is in green.
Material and Methods
Construction of cloning vector pET30L
The construction of the cloning vector pET30L was performed in a similar fashion to the procedure we described previously [18–20]. The cloning cassette linker was generated with NcoI and XhoI sites and prepared by annealing two synthetic nucleotides, 30aNHistop and 30aNHisbottom (Figure 2). Annealing was accomplished by decreasing the temperature from 95 to 20°C at a gradient of 0.1°C per second. Mismatched double strands were denatured at 70°C followed by another temperature decrease to 20°C. This cycle was repeated three times. The resulting double stranded linker was ligated into pET30a(+) (Novagen, San Diego, CA) previously digested with NcoI and XhoI. Both restriction sites were preserved after ligation. The resulting cloning vector was referred to as pET30L.
Figure 2.
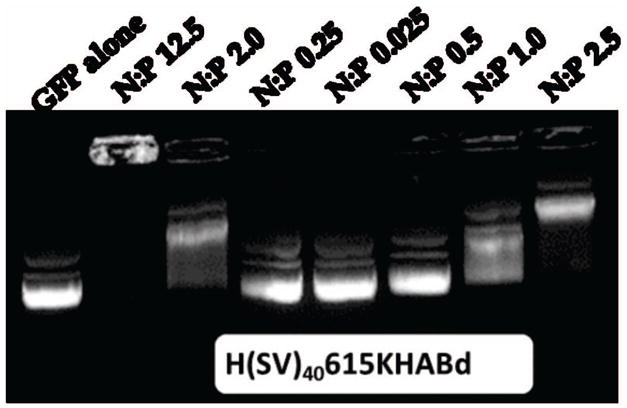
Agarose gel retardation assay results for N/P ratios ranging from 0 (pDNA-GFP only) to 62. pDNA-silk complexation reaction is shown by the inhibition of pDNA electrophoretic mobility. Two confirmations of pDNA are seen on a gel: relaxed (lower band) and supercoiled (upper band). HTLMM15KHABt was chosen as a representative example.
Construction of Spider Silk Penta-Block Polymers
A core amino acid module derived from N. clavipes MaSp I was back translated into DNA sequences and adopted for cloning in bacteria based on codon bias (Figure 1). The coding sequences of multiple DNA monomers were designed in such way that they were caring SpeI and NheI restriction sites at the ends of their sequences. A step-by-step directional ligation approach was used to generate spider silk polymers containing six repeats of MaSp I. All gene cassettes were digested with NheI and SpeI and ligated into the pET30L vector. Ligation reactions were carried out with T4 DNA ligase (Invitrogen, Carlsbad, CA) at 16°C. First, the silk block was ligated into the vector followed by stepwise ligations of remaining functional sequences. E. coli DH5α cells (Invitrogen, Carlsbad CA) were transformed with the ligation products and successful transformants were identified by plating and incubation on medium containing 30 μg/ml kanamycin. The presence of correct inserts in each construct was confirmed by PCR and DNA sequencing. Table 1 depicts constructs employed in the current study.
Table 1.
Amino acid sequences of the recombinant spider silk gene delivery constructs, their molecular weights (Mw), and isoelectric points (PI).
| Name | Sequence | Mw, Da | PI |
|---|---|---|---|
| H6mer |
|
21,005 | 9.45 |
| H615K |
|
23,115 | 10.37 |
| H(TLM)1615K(HAB)d |
|
27,618 | 10.41 |
| H(SV40)1615K(HAB)t |
|
28,734 | 10.59 |
| HM15K |
|
9962 | 10.23 |
| HM15K(HAB)t |
|
14527 | 10.33 |
| H(TLM)1M15K(HAB)t |
|
16447 | 10.16 |
| H(SV40)1M15K(HAB)t |
|
15581 | 10.51 |
Note: Spider silk sequence in blue, HAB sequence in red, PLL sequence in green, TLM/NLS in orange, poly-his sequence in black.
Expression of Spider Silk Penta-Block Polymers
The spider silk penta-block polymers genes were expressed in E. coli Bl21DE3 (Invitrogen, Carlsbad, CA). Cells were grown at 37°C in LB medium to an OD600 of 0.8 at which point protein expression was induced with 0.5 mM ITPG (isopropyl β-D-thiogalactoside) (Fisher Scientific, Hampton, NH). The cells were harvested 4 hours after induction by centrifugation at 8,000 g (Sorvall, Fisher Scientific, Hampton, NH). All culture media contained 30 μg/ml of kanamycin.
Purification of Spider Silk Penta-Block Polymers
Protein purification was performed under denaturing conditions on Ni-NTA resin (Qiagen, Valencia, CA) using the manufacturer’s guidelines. Briefly, cells were resuspended in denaturing buffer B (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl, pH 8.0) containing 10 mM imidazole. The cells were lysed by stirring for 30 min at 4°C followed by sonication for 10 seconds with 30 second time intervals until fully lysed. Insoluble cell fragments and soluble proteins were separated by centrifugation at 9,000 rpm at 4°C for 25 min. Cell lysate was mixed with Ni-NTA resin (Qiagen, Valencia, CA) in a 4 to 1 ratio and left overnight under constant agitation at 4°C. The supernatant-Ni-NTA mixture was loaded into a column and washed four times with denaturing buffer B (pH 8.0), twice with denaturing buffer C ((8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl, pH 6.7), and further two times using buffer D ((8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl, pH 5.3). The recombinant silk based penta-block polymers were eluted using the denaturing buffer E (pH 4.7). The proteins were dialyzed against 10 mM Tris buffer (pH = 7.5) followed by dialysis against water using Slide-A-Lyzer Cassette (Pierce, Rockford, IL) with MWCO of 2,000 Da. Dialyzed proteins were lyophilized. The purity of expressed proteins was verified by SDS-PAGE electrophoresis followed by Colloidal Blue and silver staining. Protein identity was confirmed by matrix-assisted laser desorption ionization mass spectrometry (MALDI-TOF) (Tufts Core Chemistry Facility, Boston, MA).
Polyplex Preparations
To prepare non-viral silk based gene delivery complexes, bioengineered spider silk-based pentapeptide solution and the pDNA solution were mixed together at different N:P ratios (where N is a number of primary amines in the protein; P is a number of phosphate groups in the pDNA backbone) and incubated for 30 min at room temperature. 10 mM tris-HCl (pH 7.5) was used to prepare these solutions unless otherwise stated. Hexamer silk (H6mer) and naked pDNA were used as negative controls. Lipofectamine™2000 (Invitrogen, Carlsbad, CA) was used as a reference in gene delivery experiments. Lipofectamine™2000 pDNA complexes were prepared according to the manufacturer’s instructions.
Agarose Gel Electrophoresis Retardation Assay
Gel electrophoresis in agarose gels was carried out at 80 V. Agarose gel (0.8% w/v) containing ethidium bromide (0.05 μL/mL) was prepared in Tris-acetate-EDTA buffer. Polyplex solutions were prepared at different N:P ratios using 1 μg of pDNA (g-Wiz-GFP, Aldevron, Madison, WI). Before subjecting the samples to gel electrophoresis, 2 μL of 6X Blue/Orange Loading Dye (Promega, Madison, WI) were added. Recombinant spider silks-pDNA interaction is shown by a lack of migration of the pDNA in the electrophoretic field.
Dynamic Light Scattering
Dynamic Light Scattering (DLS) was performed using a DynaPro Titan instrument (Wyatt Technology, Santa Barbara, CA). Analysis was performed using the DYNAMICS V6 software. The pDNA – protein complex solution at N/P ratio of 2.5 was added to 450 μL of ultra-pure water (Invitrogen, Carlsbad, CA) and, then used as a sample for DLS measurement. All samples were filtered through a 0.2 μm filter before measurements (n=10). All measurements were performed with n=10.
Scanning Electron Microscopy
Scanning Electron Microscopy (SEM) was used to assess morphological features of the spider silk penta-block polymers. The experiments were performed using a 55 Ultra System (Harvard University Center for Nanoscale Systems, Cambridge, MA) from Zeiss (Jena, Germany). Lyophilized peptides were dissolved in water or 10 mM Tris (pH = 7.5) to a final concentration of 1 μm/mL or 5 μm/mL and dried on a silicon chip in a closed container at room temperature. Images were taken using InLense and SE2 detectors at 1.00 kV.
Cytotoxicity Studies
The cytotoxicity of the gene delivery vectors with pDNA was evaluated by determining cell viability with respect to unexposed cells using the CellTiter96 AQueous assay (Promega, Madison, WI). The assay is composed of solutions of tetrazonium compound (MTS) and electron coupling reagent (PMS) and establishes a correlation between the cellular metabolic activity and the number of viable cells in culture. The conversion of MTS into aqueous, soluble formazan is accomplished by dehydrogenase enzymes found in metabolically active cells. Cell viability was studied as a function of the gene delivery vector type at N:P ratio 2.5. 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO) was used as a positive control. Lipofectamine 2000 (Invitrogen, Casablanca, CA) was used as a reference. Cells were seeded in 96-well tissue culture plates at a density of 3,200 cells per well. After 24 h, medium was replaced with fresh DMEM medium and 10 μL of each silk-pDNA complex at N/P ratio of 2.5 was added and incubated 24 h. After 24 h, the medium was exchanged and 20 μL the combined MTS/PMS solution was added into each well. Plates were incubated for 3 h at 37°C in humidified, 5% CO2 atmosphere. The amount of soluble formazan was measured at 490 nm using a microplate reader SpectraMax M2 (Molecular Devices, Sunnyvale, CA). The relative cell viability (%) related to the negative control was calculated by test sample/negative control × 100. The experiment was run in triplicate three times.
Cell Culture
hMSCs were extracted according to common procedures [21] from commercially obtained fresh human bone marrow aspirates (Lonza, Basel, Switzerland). Aspirate donors were male, under 25 years of age and free of HIV, hepatitis B and hepatitis C, and had a cell count of at least 15×106 leukocytes/mL (as measured by a Coulter counter). Briefly, an aspirate volume of 25mL was diluted 10-fold with MSC expansion medium consisting of DMEM:F12 basal medium (DMEM:F12) supplemented with 10% fetal bovine serum (FBS), antibiotics-antimycotics (100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL fungizone), 0.1 mM nonessential amino acids and 1 ng/mL basic fibroblast growth factor (bFGF) (Invitrogen). The diluted bone marrow was plated on tissue culture treated plastic (TCP) T-175 flasks at an average seeding density of 350,000 leukocytes/cm2. After 10 days the non-adherent cells were removed and the adherent cells were kept in expansion medium to reach confluence. Upon reaching confluence, the expanded cells from each flask were passaged using 0.25% trypsin-EDTA (Invitrogen) and expanded without bFGF at a passaging seeding density of 6,750 cells/cm2.
Gene Delivery Studies
Gene delivery was studied based on reporter gene expression. A reporter gene that codes for the green fluorescence protein (GFP) was used in three independent experiments. Each experiment was performed three times in triplicates.
Expression of the GFP Gene
Cells (hMSCs) were seeded at 1.25 × 104 cell/cm2, in 12-well plates, 24 h prior to transfection using a basic culture medium consisting of DMEM, 10% FBS, and 1% PSF (penicillin, streptomycin, and fungizone). Cells were transfected after reaching 70% confluence. Before contact with spider silk gene delivery complexes, medium was exchanged for 1 mL media. 100 μL of polyplex solution was added to the cells and, then after 6h the culture media was replaced with fresh basic media. Twenty-four hours after transfection, cells were observed with a fluorescence microscope (Leica Microsystems, Wetzlar, Germany).
Cellular Uptake Studies by Fluorescence-Activated Cell Sorting (FACS)
For flow cytometric analysis, growth medium was aspirated from confluent wells. After washing with PBS, 0.25% trypsin/EDTA (Invitrogen) was added to each flask and kept in an incubator for 10 min at 37°C. Trypsinization was halted by adding 10 mL serum containing medium and the cell suspension was collected into a conical tube. The conical tube was centrifuged for 10 min at 450 g and 4°C. After removing the supernatant, the pellet was re-suspended in FACS-buffer, consisting of PBS supplemented with 0.5% FBS (Invitrogen) and 5 mM EDTA (Sigma). The cell suspension was split into samples in 2 mL Eppendorf tubes. 1 mL of FACS-buffer was added to each sample and centrifuged for 10 min at 450 g and 4°C. The supernatant was removed and the pellets were re-suspended in 1 mL FACS-buffer. After a second centrifugation at the same settings, the samples were re-suspended in 250 μL FACS-buffer, transferred into their respective FACS-tubes and kept on ice until measurement. Using forward and side scatter parameters, dead cells and debris were eliminated from the analysis. GFP was excited by an argon laser and fluorescence was detected using a 530/30 nm band pass filter in the FL1 channel. Flow cytometric analysis was performed on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) with a 488nm argon laser for optimal GFP excitation. Analysis was performed using FlowJo (TreeStar, Ashland, OR) on dot plots to obtain a more accurate quantification as compared to histogram analysis. Fluorescence analysis was performed on a FL1 vs. FL3 dot plot, with the FL1 detector having a 530/30 nm and the FL3 detector having a 670/LP band-pass filter respectively. GFP was predominantly detected by the FL1 detector and the FL3 detector allowed for increased spatial separation and identification of weakly fluorescent cells.
Immunocytochemistry
Following transfection, cells were washed with PBS, fixed with 4% paraformaldehyde, and then washed in PBS. Cells were then permeablized with 0.2% Triton-X 100 in PBS and blocked with 1% Bovine serum albumin (BSA) prior to incubation with primary antibodies. Penta·His Antibody (primary) (EMD Millipore, Billerica, MA) and Cy5-conjugated goat anti-mouse IgG antibody (secondary) (Abcam, Cambridge, MA) were used in 3% BSA for 3 h at room temperature or 16 h at 4°C on a shaker. Cells were washed with PBST (0.05% Tween 20 in PBS), then mounted in Vectashield medium and the nuclei counterstained with DAPI (4,6-diamidino-2-phenylindole) (1.5 mg/mL) for conventional immunofluoresence. For spider silk-pDNA complexes staining, Penta·His Antibody followed by Cy5-conjugated goat anti-mouse IgG antibody were used. After the staining positive GFP expression and silk gene carriers were observed with a fluorescence microscope (Leica Microsystems, Wetzlar, Germany).
Statistical Analysis
Student’s t-test was used to calculate the significance of the observed results. One-way analysis of variance (ANOVA) was used to determine significant differences between different experimental groups. Differences were considered significant when p<0.05. SEM and DLS results are reported as the mean ± standard deviation of the mean.
Results
Design and Purification of Spider Silk Block Copolymers
Our previously developed cloning strategy [18, 22, 23] for the biosynthesis of recombinant spider silk genes was employed to engineer a new family of spider silk penta-block polymers with enhanced capabilities to specifically target hMSCs. Figure 1 provides the overall strategy to generate these new proteins. By using a step-by-step directional ligation approach, we gained direct control over the assembly of monomeric genes into complex sequences.
Two sets of non-viral gene delivery contracts were prepared (Table 1). MaSp I monomers as well as MaSp I hexamers were used as the core sequences. The monomeric spider silk block consisted of one poly-alanine/poly-glycine repeat (GAGAAAAAGGAG) that is responsible for β-sheet formation and of eight GGX repeats, separated by the GSQGSGR sequence [24]. The GGX repeats are thought to adopt a helical conformation and serve as a more hydrophilic link between crystalline β-sheet regions and neighboring GGX helices in adjacent protein molecules that can help reinforce fiber alignment [25, 26]. By increasing the number of spider silk blocks in the recombinant silk penta-block constructs, we intended to provoke the formation of zipper-like β-sheet structures leading to a higher degree of crystallinity for the overall structure, and thus formation of stable nanoparticles. The nanoparticle diameter was also controlled by the length of the silk block. The presence of glutamine (Q) and arginine (R) residues were considered for the hydrophilic properties of the spider silk block; while serine (S) and tyrosine (Y) were responsible for hydrogen bonding between the silk-based peptides. SDS-PAGE analysis indicated successful expression and purification of the spider silk block polymers.
Agarose Gel Electrophoresis Retardation Assay
Recombinant spider silk gene delivery systems were investigated for their ability to bind, neutralize and compact plasmid DNA (pDNA). Plasmid DNA encoding for Green Fluorescent Protein (5.7 kb) was used. Prior to all studies, silk polymers were mixed with pDNA at different N:P ratios in 10 mM Tris Buffer or water for polyplex formation. Agarose gel retardation assays revealed that binding to and charge neutralization of pDNA occurred at N:P ratios of 1.25 and higher (Figure 2). By using pDNA migration patterns, which when not bound to a protein the DNA migrates according to molecular weight (i.e. linearized plasmid), complete pDNA packaging was achieved at an N:P ratio of 2. Based on these results, an N:P ratio of 2.5 was selected for further experiments to assure silk-pDNA complex formation.
Morphological characterizations
Dynamic light scattering (DLS) measurements were performed to examine the size of the formed polyplexes at an N:P ratio of 2.5. The diameter of pDNA complexes of the recombinant silk with PLL and HAB sequences enlarged with increase in the length of the silk domain. In the case of HM15HABt, HTLMM15HABt, H(SV40)M15HABt, H(SV40)615HABd and HTLM615HABd, diameters were bimodal, indicating the presence of both small and large complexes (Table 2). The formation of large complexes indicates that silk cationic gene delivery proteins have a tendency to aggregate when pDNA is added. HTLM615HABd had the largest particle size of 783 ± 13 nm, whereas HM15K peptide had the smallest size of 5 ± 3 nm.
Table 2.
Sizes of spider silk nanoparticles obtained by DLS.
| Silk Constructs | R1 (ST), nm | R2 (ST), nm |
|---|---|---|
| HM15K | 5 (3) | 56 (3) |
| H(SV40)M15KKHABt | 72 (21) | 292 (32) |
| HM15HABt | 121 (24) | 257 (24) |
| HTLMM15HABT | 32 (12) | 65 (20) |
| H6mer* | 68 (14) | - |
| H615K | 17 (3) | 63 (5) |
| H(SV40)615KHABd | 53 (8) | 225 (12) |
| HTLM615KHABd | 69 (5) | 783 (13) |
Results are expressed as mean ± standard deviation (ST) obtained from three independent experiments.
indicates that a single peak was observed.
SEM was used to visualize the morphologies of the spider silk penta-block polymers prepared at N:P ratio 2.5. Figure 3 depicts the morphologies that were observed for spider silk polymers in water. Recombinant spider silk polyplexes assembled into spherical aggregates upon complexation with pDNA. The diameter of the aggregates was in agreement with the DLS measurements.
Figure 3.
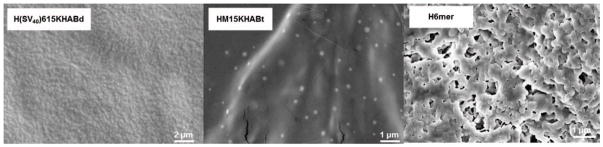
SEM images of spider silk complexes with pDNA. The complexation reaction was performed at N/P ratio of 2.5. Silk-pDNA complexes formed spherical aggregates; H6mer showed no formation of spherical morphologies due to the inability to complex pDNA.
Cytotoxicity Studies
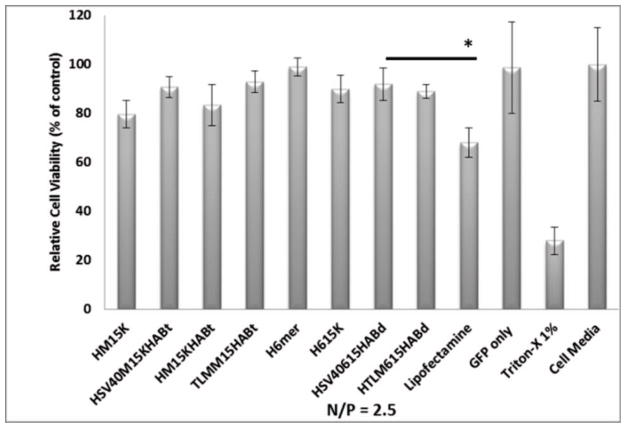
The dehydrogenase enzyme activity, and by extension, relative cell viability was measured by the CellTiter96 AQueous assay. hMSCs were maintained in DMEM supplemented with different concentrations of spider silk pDNA complexes for 24 h. Relative cell viability (%) was calculated by test sample/negative control × 100. After 24 h of incubation, silk gene delivery pDNA complexes did not reduce cell viability over the range of constructs (Figure 4). Triton X-100 showed 70% reduction in cell viability. The silk hexamer (H6mer) showed the highest cell viability (99%). The other silk construct demonstrated a slight reduction in cell viability (~10%). The reduction in cell viability in other constructs when compared to H6mer can be attributed to the poly-L-lysine domain which is known to cause toxicity when present. When the length of PLL increases, cell viability decreases due to cytotoxicity [27]. Relative cell viability was reduced to 68% when Lipofectamine 2000 was used as the gene carrier. Hence, spider silk gene carriers demonstrate significantly higher levels of cell viability when compared to Lipofectamine 2000.
Figure 4.
The percentage of relative cell viability of spider silk penta-block polymers. The relative percentage was calculated as described in Materials and Methods section. *indicates that significant difference was observed between Lipofectamine and a silk construct (H(SV40)615KHABd) based on Student’s t-test (*p<0.05).
Gene Delivery Studies
To evaluate the feasibility of the pDNA complexes with recombinant spider silk proteins possessing an HAB peptide and SV40 sequences for gene delivery, in vitro transfection experiments were performed. Two sets of constructs, spider silk monomers and hexamers, were employed in the study to assess the length of the silk domain and the amino acid composition on transfection. An N:P ratio of 2.5 was selected for transfection experiments based on gel retardation results.
Quantitative expression of GFP by Fluorescence-Activated Cell Sorting (FACS)
To quantify the expression of GFP in the transfected cells, FACS was employed after 24 h of transfection. All constructs were prepared at the N/P ratio of 2.5. Non-transfected cells were used to quantify background fluorescence, purified GFP was used as a negative control, and Lipofectamine 2000 (Invitrogen, Casablanca, CA) was used as a reference.
Only silk carriers possessing both (SV40) sequence and HAB peptide showed significant GFP transfection (Figure 6). H(SV40)M15KHABt and H(SV40)615KHABt transfected 3.52% ± 2.32% and 4.11% ± 0.47% of the hMSCs. The Lipofectamine reference samples showed a transfection rate of 9.08% ± 3.34%. When the HAB peptide was present without a SV40 or TLM sequences, the transfection efficiency did not change when compared to silk domains alone. This indicates that the HAB peptide is important for targeting hMSCS, but probably does not play a central role in the uptake process. Table 3 summarizes the relative percentage of the GFP signal as detected by FACS.
Figure 6.
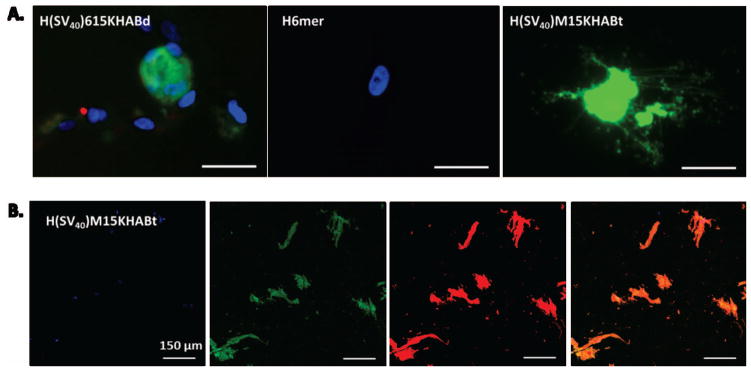
(A) Fluorescent microscopy images of live hMSCs transfected with H(SV40)615KHABd, H(SV40)M15HABt and silk 6mer showing Green Fluorescent Protein (GFP) expression 24 hours after transfection. DAPI (for nuclei staining) in blue, GFP in green. Scale bar 20 μm. (B) Confocal microscopy images of fixed hMSCs transfected with H(SV40)M15KHABt. Scale bar 150 μm. DAPI (for nuclei staining) in blue, GFP in green, Phalloidin (for cytoskeleton staining) in red, and overlay in orange.
Table 3.
Green Fluorescent Protein expression achieved with silk-based gene delivery systems. H(SV40)M15HABt and H(SV40)615KHABd showed the highest transfection levels when compare to other silk constructs. Lipofectamine 2000 was used as a positive control, while untransfected hMSCs served as a negative control.
| Silk Constructs | Relative Percentage, % | Standard Deviation,% |
|---|---|---|
| HM15K | 0.02 | 0.02 |
| H(SV40)M15KKHABt | 3.52 | 2.32 |
| HM15HABt | 0.02 | 0.02 |
| HTLMM15HABT | 0.06 | 0.02 |
| H6mer | 0.02 | 0.02 |
| H615K | 0.08 | 0.02 |
| H(SV40)615KHABd | 4.11 | 0.47 |
| HTLM615KHABd* | 0.24 | 0.23 |
| Lipofectamine 2000* | 9.08 | 3.34 |
| GFP only | 0.05 | 0.04 |
| Negative Control | 0.02 | 0.01 |
Results are expressed as the relative percentage of the mean ± standard deviation. All experiments were performed in triplicates.
Qualitative expression of GFP as detected by Fluorescence and Confocal Microscopy
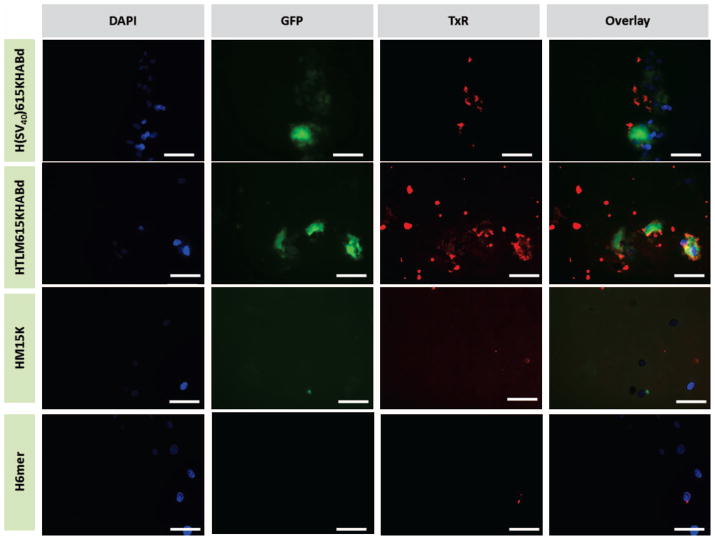
pDNA delivery by spider silk-based cationic complexes was qualitatively studied by visualization of the expressed GFP protein using fluorescent and confocal microscopy. As demonstrated by green fluorescence, the pDNA/silk-based protein complexes containing both the cell recognition domain and nuclear localization sequence (H(SV40)M15HABt and H(SV40)615KHABd) delivered a target gene to the nucleus, confirmed by subsequent expression of GFP (Figure 6). Low levels of GFP expression were observed when hMSCs were transfected with a silk carrier without NLS (HTLM615KHABd). Additionally, pDNA complexes with silk proteins only, demonstrated no nuclear import. The H(SV40)M15KHABt construct exhibited the highest transfection efficiency as indicated by both fluorescence imaging and FACS. Figure 6 depicts the observed results.
To visualize the presence of silk particles inside the cells, immunocytochemistry was employed. Since spider silk gene carriers possess an N-terminal histidine tag, they were visualized through non-covalent interactions of a histidine block with a penta-His antibody followed by Cy5-conjugated goat anti-mouse IgG antibody. The results revealed the presence of silk particles for those cells that demonstrated high transfection levels (Figure 7).
Figure 7.
Fluorescent microscopy images of fixed hMSCs cells transfected with spider silk-pDNA complexes at N/P ratio of 2.5. DAPI (for nuclei staining) in blue, GFP in green, silk-pDNA complexes in red. Scale bars are 20 μm.
Spider silk-based gene carriers (in red) were observed in close proximity to the nuclei (blue) indicating importance of NLS for gene delivery. Moreover, spider silk micro-particles were present at a higher number (empirical observations) in the cells expressing GFP (green) when compared to non-transfected cells indicating the role of the particle stability in gene delivery. Based on the current results, spider silk gene delivery carriers exhibited low toxicity and moderate levels of transfection efficiency towards the difficult to transfect hMSCs. Furthermore, recombinant carriers can be easily modified to target cell types of interest and desired specialized subcellular subunits.
Discussion
Naturally occurring polymers are an emerging class of non-viral gene delivery systems that can provide safe and efficient vehicles to transfer a therapeutic gene. These non-viral systems have a unique ability for active cell specific targeting via receptor-mediated endocytosis due to ease of modification through the recombinant DNA approach. The polymers can be further functionalized with target-specific sequences for cytosolic or nuclear delivery. Moreover, in contrast to viral systems, non-viral polymers are capable of delivering different types of nucleic acids such as pDNA, siRNA, and miRNA [28].
In our design, we took advantage of the cationic nature of the lysine groups adjacent to the spider silk domain together with the hMSCs receptor-specific targeting capabilities of the HAB peptide to improve gene delivery towards hMSCs. It was recently demonstrated by Santos and coworkers that the HAB peptide can be specifically recognized by hMSCs [29]. Poly-L-Lysine (PLL), a repeat of the amino acid lysine, was the first cationic polymer-based peptide to be employed as a gene delivery vehicle [30] due to its plasmid complexation properties. However, PLL peptides when used alone possess cytotoxicity due to a tendency to precipitate and aggregate and demonstrated poor delivery of pDNA [27]. However, after the PLL peptide was conjugated with a naturally occurring lipid (i.e. palmitic acid (PA)) via amide linkages transfection efficiency improved significantly [31, 32]. PLL-PA conjugates resulted in an approximately 10-fold increase in cellular uptake of genes due to better cell binding and internalization of the conjugates, thus improving delivery of pDNA (EGFP) to hMSCs. The conjugates achieved a maximum transfection efficiency of ~22%, which was significantly higher than Lipofectamine™2000 mediated transfection (11%) [32].
The large tumor antigen of the Simian virus 40 (SV40) was chosen as it is the most studied and used NLS [33, 34]. This peptide is comprised of basic amino acids (PKKKRKV) and is capable of increasing the rate of DNA uptake by 100-fold when compared with its reverse sequence [35]. Finally, to avoid vesicular entrapment due to an endolytic uptake, a histidine-rich peptide containing six histidines (HHHHHH) was added as an N-terminal sequence. Histidine-rich peptides have been used to promote an endosmolitic function of non-viral cationic gene carriers [36, 37]. The mechanism of action of histidine-rich peptides is unknown, however, the imidazole group of histidine is key with a pKa about 6.0 indicating that it protonates in the acidic environment. This response would turn on membrane disruption properties when entrapped by endolytic vesicles. At physiological pH of the cytosol, the imidazole ring remains neutral in terms of charge [27, 38].
In the present study, we designed, produced, and evaluated spider silk penta-block polymers as potential gene carriers capable of targeting hMSCs among other cell types. hMSCs were chosen due to their ability to differentiate into various tissue outcomes as well as current difficulties associated with hMSC transfection. We demonstrated that by combining the HAB peptide, the nuclear localization sequence SV40, and poly-lysine and poly-histidine units with the spider silk sequences, transfection efficiency can be improved and hMSCs targeting achieved. Moreover, the recombinant spider silk gene carriers have higher cell viability than compared to Lipofectamine 2000, even for those constructs with PLL sequences. The high pKa value of 10 enables PLL to package pDNA into nanoparticulate complexes. However, it is also known that PLL is inefficient in terms of endosomal escape and transfection efficiency when used alone [39]. By adding a histidine-rich peptide together with NLS, endosomal escape was achieved resulting in the expression of GFP (e.g. H(SV40)615KHABd and H(SV40)M15KHABt). Silk gene delivery constructs without a NLS sequence (e.g. HTLM615KHABd, H615K, HM15K) displayed low levels of transfection. Thus, a short PKKKRKV peptide is important if nuclear delivery is the goal. This was confirmed when the translocation motif (TLM) of the hepatitis-B virus surface protein (PreS2) known for its cell permeability properties was used without NLS. TLM sequence (PLSSFSRIGDP) is known for its cell penetrating properties [40]. Silk constructs bearing TLM sequences demonstrated poor nuclear import resulting in negligible levels of the expressed GFP as indicated by FACS and fluorescent microscopy studies. When using HTLM615KHABd and HTLMM15KHABt constructs, 0.4% and 0.07% of cells expressed GFP as detected by FACS. H6mer and H615K constructs have demonstrated even lower transfection levels, 0.04% and 0.06%, respectively, indicating the importance of NLS. However, these constructs are desirable when targeting the cell cytosol to achieve gene silencing by RNAi or antisense technology.
Intracellular organelle-targeted delivery of genes is essential for therapeutic action. Therapeutic DNA molecules must be transcribed to mRNA in the nucleus to perform their programmed functions, however, the nuclear envelope represent a physical barrier for the gene entry [41]. The lipid bilayer is perforated by large nuclear pore complexes that control nuclear import. Only small molecules and ions have access to the nuclear compartment via passive diffusion, whereas molecules larger than 40 kDa required active transport [42]. Two class of receptors, β-importin and β-karyopherin, are key mediators of active transport. The receptors bind macromolecules possessing specific basic peptide sequences (NLS) and transport them across the nuclear envelop [42]. This active transport is how pDNA enters the nuclear compartment in non-dividing cells. The silk penta-block polymers possessing NLS (e.g. H(SV40)615KHABd and H(SV40)M15KHABt) described here, demonstrated expression of GFP as evidence of successful nuclear import.
It is important to note that the mechanism by which silk-pDNA complexes interact with hMSCs and other cells are not fully understood [43]. To improve transfection efficiency of the above described constructs, it is necessary to capture atomistic interactions between the protein and gene components at specific cellular and intracellular environments as well as to understand the pathways by which silk-pDNA complexes enter cells. To achieve this goal, extensive experimental and simulation studies are needed.
In the present study, non-viral silk gene delivery carriers demonstrated moderate levels of transfection efficiency when compared with their viral counter-parts. However, high cell viability, low toxicity and the lack of immunogenicity suggest that these bioengineered silk-based gene delivery systems are attractive candidates for further refinement as nuclear delivery vehicles.
Conclusions
Spider silk-based gene delivery systems are promising candidates as gene carriers with a unique capability to be easily designed, tailored, and modified via genetic engineering to enhance specificity and targeting. In the current study, hMSCs were chosen as the target due to their unique ability to differentiate into various cell types, as well as challenges associated with their transfection. We concluded that in addition to previously studied cell types (e.g. human embryonic kidney cells, MDA-MB-435 melanoma cells, fibroblast-like cell lines), the multifunctional silk-based complexes can be engineered to transfect hMSCs. The complexes are non-toxic and have potential to be used as vehicles in stem cell engineering. pDNA trafficking mechanisms from the cytoplasm into the nucleus of the cells, along with in vivo studies are needed to fully evaluate the potential of these non-viral gene delivery systems.
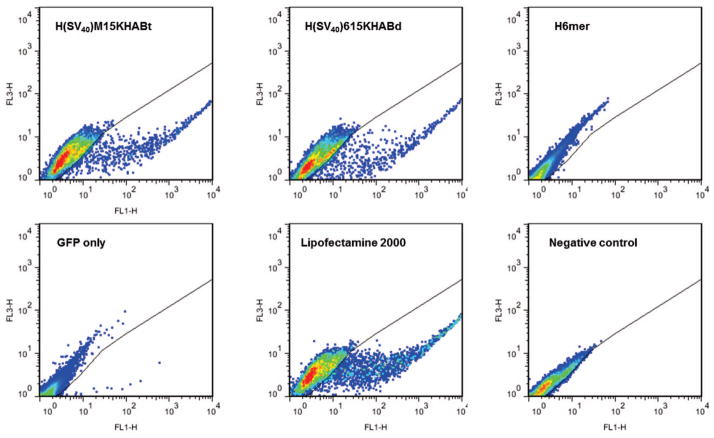
Figure 5.
Flow cytometry analysis of Green Fluorescent Protein expression achieved with H(SV40)M15HABt, H(SV40)615KHABd, H6mer, naked pDNA encoded GFP (GFP only), and Lipofectamine 2000 24 hours after transfection. Untransfected population of hMSCs served as a negative control. FL1-H vs.FL3-H dot plot was used for analysis.
Acknowledgments
We thank the NIH (NIH 1F32EB018138-01) for support of this research.
References
- 1.Giatsidis G, Dalla Venezia E, Bassetto F. The role of gene therapy in regenerative surgery: updated insights. Plast Reconstr Surg. 2013;131:1425–35. doi: 10.1097/PRS.0b013e31828bd153. [DOI] [PubMed] [Google Scholar]
- 2.Bergen J, Schaffer D. Engineering viruses for gene therapy. In: Ducheyne P, editor. Comprehensive Biomaterials. Oxford: Elsevier; 2011. pp. 17–33. [Google Scholar]
- 3.Wang T, Upponi JR, Torchilin VP. Design of multifunctional non-viral gene vectors to overcome physiological barriers: Dilemmas and strategies. International Journal of Pharmaceutics. 2012;427:3–20. doi: 10.1016/j.ijpharm.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Escors D, Breckpot K. Lentiviral vectors in gene therapy: their current status and future potential. Arch Immunol Ther Exp (Warsz) 2010;58:107–19. doi: 10.1007/s00005-010-0063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pluta K, Kacprzak MM. Use of HIV as a gene transfer vector. Acta Biochim Pol. 2009;56:531–95. [PubMed] [Google Scholar]
- 6.Picanco-Castro V, de Sousa Russo-Carbolante EM, Tadeu Covas D. Advances in lentiviral vectors: a patent review. Recent Pat DNA Gene Seq. 2012;6:82–90. doi: 10.2174/187221512801327433. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Q-Y, Lai L-H, Shen J, Wang Q-Q, Xu F-J, Tang G-P. Gene delivery to tumor cells by cationic polymeric nanovectors coupled to folic acid and the cell-penetrating peptide octaarginine. Biomaterials. 2011;32:7253–62. doi: 10.1016/j.biomaterials.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 8.Deans RJ, Moseley AB. Mesenchymal stem cells: Biology and potential clinical uses. Experimental hematology. 2000;28:875–84. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 9.Glettig D, Kaplan D. Long-term phenotypic characterization of human bone marrow and adipose tissue derived mesenchymal stromal cells. Stem Cell Discovery. 2013;3:99–116. [Google Scholar]
- 10.Numata K, Kaplan DL. Silk-based delivery systems of bioactive molecules. Advanced Drug Delivery Reviews. 2010;62:1497–508. doi: 10.1016/j.addr.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Numata K, Mieszawska-Czajkowska AJ, Kvenvold LA, Kaplan DL. Silk-based nanocomplexes with tumor-homing peptides for tumor-specific gene delivery. Macromolecular Bioscience. 2012;12:75–82. doi: 10.1002/mabi.201100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Numata K, Reagan MR, Goldstein RH, Rosenblatt M, Kaplan DL. Spider silk-based gene carriers for tumor cell-specific delivery. Bioconjugate Chemistry. 2011;22:1605–10. doi: 10.1021/bc200170u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kluge JA, Rabotyagova O, Leisk GG, Kaplan DL. Spider silks and their applications. Trends in Biotechnology. 2008;26:244–51. doi: 10.1016/j.tibtech.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Rabotyagova OS, Cebe P, Kaplan DL. Protein-based block copolymers. Biomacromolecules. 2011;12:269–89. doi: 10.1021/bm100928x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dams-Kozlowska H, Majer A, Tomasiewicz P, Lozinska J, Kaplan DL, Mackiewicz A. Purification and cytotoxicity of tag-free bioengineered spider silk proteins. Journal of Biomedical Materials Research Part A. 2013;101A:456–64. doi: 10.1002/jbm.a.34353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Hu X, Daley A, Rabotyagova O, Cebe P, Kaplan DL. Nanolayer biomaterial coatings of silk fibroin for controlled release. Journal of Controlled Release. 2007;121:190–9. doi: 10.1016/j.jconrel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Pritchard E, Hu X, Valentin T, Panilaitis B, Omenetto FG, et al. Stabilization of vaccines and antibiotics in silk and eliminating the cold chain. Proc Natl Acad Sci U S A. 2012;109:11981–6. doi: 10.1073/pnas.1206210109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Rabotyagova OS, Cebe P, Kaplan DL. Self-assembly of genetically engineered spider silk block copolymers. Biomacromolecules. 2009;10:229–36. doi: 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- 19.Rabotyagova OS, Cebe P, Kaplan DL. Role of polyalanine domains in β-sheet formation in spider silk block copolymers. Macromolecular Bioscience. 2010;10:49–59. doi: 10.1002/mabi.200900203. [DOI] [PubMed] [Google Scholar]
- 20.Tokareva O, Michalczechen-Lacerda VA, Rech EL, Kaplan DL. Recombinant DNA production of spider silk proteins. Microbial Biotechnology. 2013;6:651–63. doi: 10.1111/1751-7915.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glettig DL, Kaplan DL. Extending human hematopoietic stem cell survival in vitro with adipocytes. Biores Open Access. 2013;2:179–85. doi: 10.1089/biores.2013.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Numata K, Subramanian B, Currie HA, Kaplan DL. Bioengineered silk protein-based gene delivery systems. Biomaterials. 2009;30:5775–84. doi: 10.1016/j.biomaterials.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Numata K, Hamasaki J, Subramanian B, Kaplan DL. Gene delivery mediated by recombinant silk proteins containing cationic and cell binding motifs. Journal of Controlled Release. 2010;146:136–43. doi: 10.1016/j.jconrel.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL. Construction, cloning, and expression of synthetic genes encoding spider dragline silk. Biochemistry. 1995;34:10879–85. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- 25.Breslauer DN, Kaplan DL. 9.04 - Silks. In: Krzysztof M, Martin M, editors. Polymer Science: A Comprehensive Reference. Amsterdam: Elsevier; 2012. pp. 57–69. [Google Scholar]
- 26.Tokareva O, Jacobsen M, Buehler M, Wong J, Kaplan DL. Structure–function–property–design interplay in biopolymers: Spider silk. Acta Biomaterialia. 2013 doi: 10.1016/j.actbio.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin ME, Rice KG. Peptide-guided gene delivery. Aaps J. 2007;9:E18–29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akita H, Hatakeyama H, Khalil IA, Yamada Y, Harashima H. Delivery of nucleic acids and gene delivery. In: Ducheyne P, editor. Comprehensive Biomaterials. Oxford: Elsevier; 2011. pp. 411–44. [Google Scholar]
- 29.Santos JL, Pandita D, Rodrigues J, Pêgo AP, Ganja PL, Balkan G, et al. Receptor-mediated gene delivery using PAMAM dendrites conjugated with peptides recognized by mesenchymal stem cells. Molecular Pharmaceutics. 2010;7:763–74. doi: 10.1021/mp9002877. [DOI] [PubMed] [Google Scholar]
- 30.Waxhaw MS, Collard WT, Adam RC, McKenzie DL, Rice KG. Peptide-mediated gene delivery: influence of peptide structure on gene expression. Bioconjug Chem. 1997;8:81–8. doi: 10.1021/bc960079q. [DOI] [PubMed] [Google Scholar]
- 31.Incani V, Tunis E, Clements BA, Olson C, Kucharski C, Lavasanifar A, et al. Palmitic acid substitution on cationic polymers for effective delivery of plasmid DNA to bone marrow stromal cells. J Biomed Mater Res A. 2007;81:493–504. doi: 10.1002/jbm.a.31249. [DOI] [PubMed] [Google Scholar]
- 32.Abbasi M, Uludag H, Incani V, Olson C, Lin X, Clements BA, et al. Palmitic acid-modified poly-L-lysine for non-viral delivery of plasmid DNA to skin fibroblasts. Biomacromolecules. 2007;8:1059–63. doi: 10.1021/bm060940x. [DOI] [PubMed] [Google Scholar]
- 33.Collas P, Alestrom P. Rapid targeting of plasmid DNA to zebrafish embryo nuclei by the nuclear localization signal of SV40 T antigen. Mol Mar Biol Biotechnol. 1997;6:48–58. [PubMed] [Google Scholar]
- 34.Collas P, Alestrom P. Nuclear localization signal of SV40 T antigen directs import of plasmid DNA into sea urchin male pronuclei in vitro. Mol Reprod Dev. 1996;45:431–8. doi: 10.1002/(SICI)1098-2795(199612)45:4<431::AID-MRD4>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 35.Collas P, Husebye H, Aleström P. The nuclear localization sequence of the SV40 T antigen promotes transgene uptake and expression in zebrafish embryo nuclei. Transgenic Research. 1996;5:451–8. doi: 10.1007/BF01980210. [DOI] [PubMed] [Google Scholar]
- 36.Bure C, Maget R, Delmas AF, Pichon C, Midoux P. Histidine-rich peptide: evidence for a single zinc-binding site on H5WYG peptide that promotes membrane fusion at neutral pH. J Mass Spectrom. 2009;44:81–9. doi: 10.1002/jms.1473. [DOI] [PubMed] [Google Scholar]
- 37.Kichler A, Leborgne C, Marz J, Danos O, Bechinger B. Histidine-rich amphipathic peptide antibiotics promote efficient delivery of DNA into mammalian cells. Proc Natl Acad Sci U S A. 2003;100:1564–8. doi: 10.1073/pnas.0337677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrer-Miralles N, Corchero JL, Kumar P, Cedano JA, Gupta KC, Villaverde A, et al. Biological activities of histidine-rich peptides; merging biotechnology and nanomedicine. Microb Cell Fact. 2011;10:101. doi: 10.1186/1475-2859-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elouahabi A, Ruysschaert JM. Formation and intracellular trafficking of lipoplexes and polyplexes. Molecular therapy: the journal of the American Society of Gene Therapy. 2005;11:336–47. doi: 10.1016/j.ymthe.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Oess S, Hildt E. Novel cell permeable motif derived from the PreS2-domain of hepatitis-B virus surface antigens. Gene Ther. 2000;7:750–8. doi: 10.1038/sj.gt.3301154. [DOI] [PubMed] [Google Scholar]
- 41.Rajendran L, Knolker HJ, Simons K. Subcellular targeting strategies for drug design and delivery. Nature reviews Drug discovery. 2010;9:29–42. doi: 10.1038/nrd2897. [DOI] [PubMed] [Google Scholar]
- 42.Huang JG, Leshuk T, Gu FX. Emerging nanomaterials for targeting subcellular organelles. Nano Today. 2011;6:478–92. [Google Scholar]
- 43.Yigit S, Tokareva O, Varone A, Georgakoudi I, Kaplan DL. Bioengineered Silk Gene Delivery System for Nuclear Targeting. Macromolecular Bioscience. 2014 doi: 10.1002/mabi.201400113. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]