Abstract
Rosacea is a common, chronic skin disease that is currently incurable. Although environmental factors influence rosacea, the genetic basis of rosacea is not established. In this genome-wide association study, a discovery group of 22,952 individuals (2,618 rosacea cases and 20,334 controls) was analyzed, leading to identification of two significant single-nucleotide polymorphisms (SNPs) associated with rosacea, one of which replicated in a new group of 29,481 individuals (3,205 rosacea cases and 26,262 controls). The confirmed SNP, rs763035 (P=8.0 × 10−11 discovery group; P=0.00031 replication group), is intergenic between HLA-DRA and BTNL2. Exploratory immunohistochemical analysis of HLA-DRA and BTNL2 expression in papulopustular rosacea lesions from six individuals, including one with the rs763035 variant, revealed staining in the perifollicular inflammatory infiltrate of rosacea for both proteins. In addition, three HLA alleles, all MHC class II proteins, were significantly associated with rosacea in the discovery group and confirmed in the replication group: HLA-DRB1*03:01 (P=1.0 × 10−8 discovery group; P=4.4 × 10−6 replication group), HLA-DQB1*02:01 (P=1.3 × 10−8 discovery group; P=7.2 × 10−6 replication group), and HLA-DQA1*05:01 (P=1.4 × 10−8 discovery group; P=7.6 × 10−6 replication group). Collectively, the gene variants identified in this study support the concept of a genetic component for rosacea, and provide candidate targets for future studies to better understand and treat rosacea.
Introduction
Many causes of rosacea have been proposed over the years. In the lay population, rosacea has been commonly attributed to multiple external factors such as alcohol excess or sun exposure, which can exacerbate, but are unlikely, to cause rosacea (reviewed by Cribier (2013). Other reported associations have included small intestinal bacterial overgrowth and skin surface microbes (Weinstock and Steinhoff, 2013). The important role of antimicrobial peptides (Yamasaki et al., 2007) and the toll-like receptors2 pathway (Yamasaki et al., 2011) has been demonstrated in rosacea pathophysiology.
Nevertheless, the potential genetic basis of this common, disfiguring yet incurable condition is not known. Evidence for a genetic component to rosacea has been hypothesized, with a retrospective study showing that rosacea patients have a greater than fourfold increased odds of having a family member with rosacea (Abram et al., 2010; Steinhoff et al., 2013), but the genes leading to this association are not known. This current study explores genes that associate with rosacea in a large population of individuals of European descent by genome-wide association study.
Results
A genome-wide association study was conducted in 22,952 individuals whose genomes showed >97% European ancestry. Because of the sample size needed for this study, cases and controls were identified by an online questionnaire in which participants responded to a survey item on whether a healthcare professional had ever diagnosed them with rosacea. Participants who answered “yes” were defined as “cases” (n=2,618), and those who answered “no” (n=20,334) were defined as “controls”. With this sample size, we would have 80% power to detect an association with an odds ratio of 1.25 and minor allele frequency of 20%, with P<5 × 10−8 (Freidlin et al., 2002).
Basic demographics of the case and control groups are reported in Table 1a for the discovery group and in Table 1b for the replication group. As this study was conducted in individuals with European ancestry, the proportion of cases versus controls (11 vs. 89%) was consistent with population data from European countries, in which the prevalence of rosacea has been reported at up to 10% of the population (Elewski et al., 2011).
Table 1. Gender and age characteristics of rosacea cases and controls (a) from the discovery group and (b) from the replication group.
| Characteristic | Rosacea cases | Controls |
|---|---|---|
| (a)Discovery group, n=22,952 | ||
| n (%) | 2,618 (11%) | 20,334 (89%) |
| Female (%) | 1,848 (65%) | 8,655 (43%) |
| Age (years) | ||
| <30 | 3 (0.001%) | 3 (0.00015%) |
| 30–45 | 528 (20%) | 7,190 (35%) |
| >45–60 | 968 (37%) | 6,554 (32%) |
| >60 | 1,119 (43%) | 6,587 (32%) |
| (b) Replication group, n=29,481 | ||
| n (%) | 3,207(11%) | 26,274 (89%) |
| Female (%) | 2,305 (72%) | 13,216 (50%) |
| Age, (years) | ||
| <30 | 0 (0%) | 0 (0%) |
| 30–45 | 527 (16%) | 7,554 (29%) |
| >45–60 | 1,172 (37%) | 9,052 (35%) |
| >60 | 1,508 (47%) | 9,668 (37%) |
The questionnaire was targeted to participants who had self-reported their age as ⩾30 years. A small number of participants in the discovery group may have subsequently changed their age to be <30 years. Their data was included, as the small numbers do not significantly alter the analysis.
Our study's proportion of individuals with rosacea over the age of 60 years is 43% in the discovery group (1,119/2,618) and 47% in the replication group (1,508/3,207), which is consistent with known demographics of rosacea. Rosacea prevalence is reported to increase with age in the medical literature with about 43% diagnosed over the age of 50 years (Elewski et al., 2011). However, this study is not a population cohort as individuals choose whether to participate in this study and participants may not necessarily match a natural population.
The genome-wide association study was conducting by adjusting for age, sex, and the first five principal components. Q-Q plot of the P-values for this discovery group are shown in Supplementary Figure 1 online.
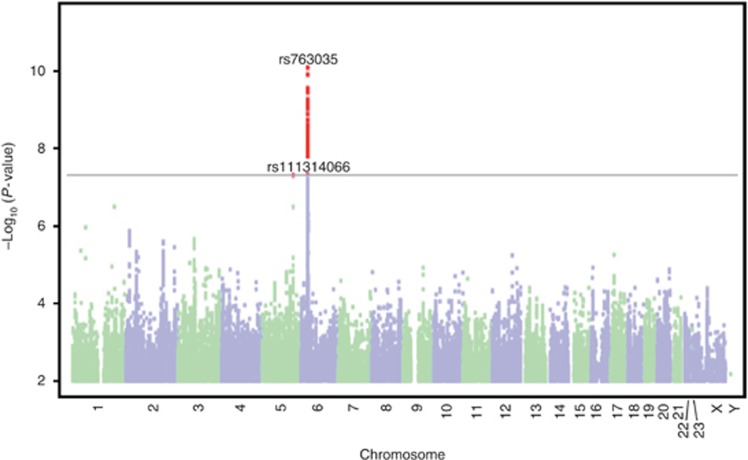
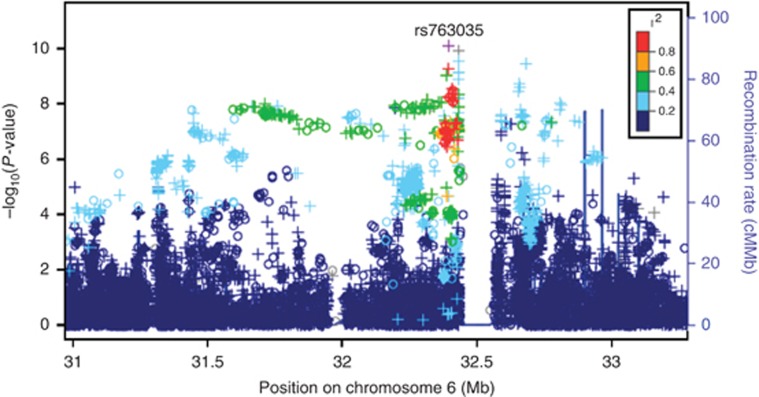
Two single-nucleotide polymorphisms (SNPs) met the Bonferroni threshold for significance (P<5 × 10−8) in the discovery cohort: rs763035 (P=8.0 × 10–11) and rs111314066 (P=4.9 × 10–8). A Manhattan plot for these results is shown in Figure 1. Targeted interrogation of the two significant SNPs from the discovery group in a replication group (n=29,481) consisting of new cases (n=3,205) and new controls (n=26,262; and using a significance threshold of P<0.05 and correction for the same covariates as the discovery group) revealed that only rs763035 was associated with rosacea (P=0.00031; Table 2). Regional association plot of the confirmed SNP rs763035 showed linkage disequilibrium with increasing P-values of the SNPs adjacent to rs763035 (Figure 2). Quality statistics for index SNPs are shown in Supplementary Table 1 online.
Figure 1.
Manhattan plot of single-nucleotide polymorphisms in the rosacea genome-wide association study of the discovery group. Single-nucleotide polymorphisms meeting a Bonferroni threshold of significance at P<5 × 10−8 (gray line) were rs763035 and rs111314066; however, only rs763035 was confirmed in the replication group (Table 2). Results are plotted as −log10-transformed P-values from the association test.
Table 2. Results of the discovery group GWAS for rosacea and replication group.
| (a) | ||||||||
|---|---|---|---|---|---|---|---|---|
|
P-value
OR (95% CI) |
Gene context | |||||||
| rsSNP | Chromosome | Position | Allele | Imputed SNP or genotyped SNP | MAF | Discovery group | Replication group | |
| 763035 | 6 | 32,394,845 | A/G | Imputed | 0.1387 | 8.0 × 10−11 1.36 (1.24–1.50) | 0.00031 1.18 (1.08–1.30) | Intergenic: 12,774 bp upstream of HLA-DRA 19,945 bp downstream of BTNL2 |
| 111314066 | 5 | 144,469,907 | A/T | Imputed | 0.0167 | 4.9 × 10−8 1.98 (1.57–2.50) | 0.326 1.15 (0.87–1.53) | Intergenic: 666,000 bp upstream of PRELID2 612,963 bp downstream of KCTD16 |
| (b) | ||||
|---|---|---|---|---|
|
P-value
OR (95% CI) |
||||
| HLA allele | Chromosome | MAF | Discovery group | Replication group |
| DRB1*03:01 | 6 | 0.1149 | 1.0 × 10−8 0.75 (0.68–0.83) | 4.4 × 10−6 0.81 (0.74–0.89) |
| DQB1*02:01 | 6 | 0.1140 | 1.3 × 10−8 0.75 (0.68–0.83) | 7.2 × 10−6 0.82 (0.75–0.89 |
| DQA1*05:01 | 6 | 0.1142 | 1.4 × 10−8 0.75 (0.68–0.83) | 7.6 × 10−6 0.82 (0.75–0.89) |
Abbreviations: CI, confidence interval; GWAS, genome-wide association study; HLA, human leukocyte antigen; MAF, minor allele frequency; OR, odds ratio; SNP, single-nucleotide polymorphism.
(a) rs76035 was significantly associated with rosacea in both the discovery and replication groups. (b) Three HLA alleles were significantly associated with rosacea in the discovery and replication groups. Bold font indicate the statistically significant results.
Figure 2.
Regional association plot for rs763035, the single-nucleotide polymorphism confirmed in the replication group. The y-axis shows negative transformed −log10 of association test P-values, and x-axis shows the adjacent region on the chromosome. Colors indicate the strength of linkage disequilibrium with the index single-nucleotide polymorphism, and plot symbols indicate imputed (+) and genotyped (o) single-nucleotide polymorphisms. The human leukocyte antigen association is supported by numerous genotyped and imputed variants spanning an interval of about 1.5 Mb.
The confirmed SNP, rs763035, is intergenic and located 12,774 bp upstream of HLA-DRA (human leukocyte antigen (HLA) class II histocompatibility antigen, DR alpha chain) and 19,945 bp downstream of BTNL2 (butyrophilin-like 2, major histocompatibility complex class I associated).
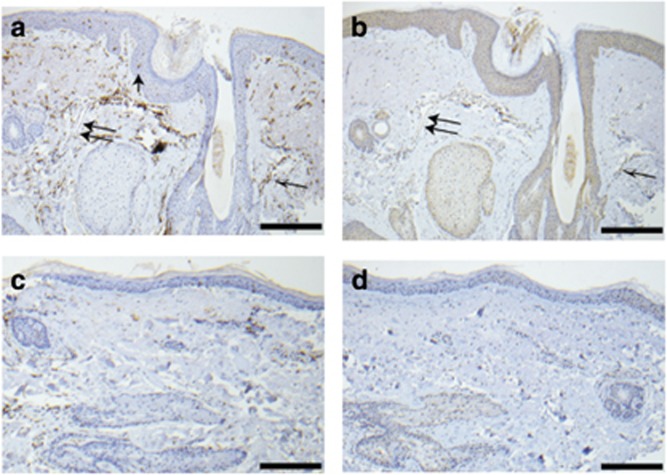
Immunohistochemistry using antibodies against HLA-DRA and BTNL2 both showed staining in skin biopsies of papulopustular rosacea lesions from the lower face of six individuals of European descent whose rosacea phenotype was confirmed on clinical history and physical examination by a board-certified dermatologist experienced in the treatment of rosacea. Out of the six individuals, one was heterozygous at rs763035 for the rosacea-associated allele and five did not possess this allele. As this was an exploratory study, it was not powered to enable comparison of genotype at rs763035 and semiquantitative differences in immunohistochemical staining level for either HLA-DRA or BTNL2. Nevertheless, the presence of HLA-DRA and BTNL2 in rosacea skin suggests potential biologic relevance.
In all six rosacea skin samples, the HLA-DRA antibody strongly stained the perifollicular inflammatory infiltrate characteristic of rosacea, as well as epidermal Langerhans cells and endothelial cells (example shown in Figure 3a). In adjacent nonaffected skin from the same individuals, there was no heavy inflammatory infiltrate, and hence much fewer cells were stained. However, visible staining of Langerhans cells and endothelial cells could be discerned (example shown in Figure 3b). Endothelial cell staining is very interesting, as a cardinal feature of rosacea is vascular dilation and proliferation (Fowler et al., 2012). Negative controls in which the primary antibody was omitted showed no staining.
Figure 3.
Genes associated with rs763035 are present in rosacea skin. (a) Rosacea skin from an individual of European descent and heterozygous for the minor allele at rs763035 immunostained with human leukocyte antigen-DRA antibody with a strong signal in the perifollicular inflammatory infiltrate (black and white arrow), epidermal Langerhans cells (single black arrow), and endothelial cells (double arrows). (b) Rosacea skin from the same individual as a and b immunostained with BTNL2 antibody showed mild–moderate staining in keratinocytes, perifollicular infiltrate (black and white arrow), and endothelial cells (double arrow). (c) Normal skin from the same individual stains with human leukocyte antigen-DRA antibody but lacks perifollicular infiltrate, numerous epidermal Langerhans cells, and dilated blood vessels seen in rosacea skin. (d) Normal skin from the same individual stained with BTNL2 antibody in keratinocytes but does not contain inflammatory infiltrate or dilated blood vessels. Negative controls were performed for all the tissues above by omitting either human leukocyte antigen-DRA or BTNL2 primary antibodies, and these displayed no staining (data not shown). Scale bar=100 μm.
In all six rosacea skin samples, BTNL2 antibody led to diffuse staining of the keratinocytes, as well as perifollicular inflammatory infiltrate and endothelial cells (example shown in Figure 3c). In adjacent nonaffected skin from the same individuals, there was no perifollicular inflammatory infiltrate, and hence fewer cells were stained. This staining was visible diffusely in the keratinocytes, in the scant lymphocytes, Langerhans cells, and endothelial cells.
Of note, HLA-DRA is invariant at its peptide-binding site, but differences in HLA-DRA gene expression levels or differences outside of the peptide-binding site could still occur. Indeed, variation in the HLA-DRA promoter region has been associated with multiple sclerosis (International Multiple Sclerosis Genetics Consortium, 2007). Both multiple sclerosis and rosacea are conditions with pathologic inflammation.
BTNL2 is a member of the immunoglobulin gene superfamily and is associated with a number of human autoimmune diseases including inflammatory bowel disease and sarcoidosis (Valentonyte et al., 2005; Sirota et al., 2009; Anderson et al., 2011), the latter being a granulomatous condition involving skin. Rosacea pathology is characterized by inflammation, and a subtype can display granulomas as well (Rallis and Korfitis, 2012).
To explore whether rs763035 is transcribed or found near known regulatory elements, the Encyclopedia of DNA Elements database was searched. However, rs763035 was not located in H3K27Ac or H3K4Me1 marks often found near active regulatory elements in seven cell lines (University of California Santa Cruz genome browser, accessed on 3 February 2014), nor was rs763035 in DNAseI hypersensitivity clusters, indicating open chromatin sites in 125 cell types from Encyclopedia of DNA Elements. Although the SNP was not found to be within known regulatory elements, Encyclopedia of DNA Elements includes only some cell types, such as basal keratinocytes in skin. Although the basal keratinocytes in rosacea skin did stain with BTNL2 antibody, they did not stain with HLA-DRA antibody. Notably, cutaneous Langerhans cells and endothelial cells, which stained in rosacea skin with both HLA-DRA and BTNL2 antibodies, were not represented in the Encyclopedia of DNA Elements database.
As the genotyping platform covered a maximum of 1,008,948 SNPs and an estimated 10 million SNPs exist in the human genome, we searched for SNPs in high linkage disequilibrium with rs763035. Three SNPs on chromosome 6 were found to be in linkage disequilibrium with the rs763035 polymorphism using the 1000GenomesProject database: rs1894552 (r2=0.953), rs2105903 (r2=0.953), and rs2105902 (r2=0.857). These three SNPs in linkage disequilibrium with rs763035 were not intragenic or in regulatory regions.
Rs763035 was not reported as an expression quantitative trait locus (eQTL) for skin in a Genotype-Tissue Expression search of skin (http://commonfund.nih.gov/GTex/index, accessed on 25 November 2013). However, in monocytes, rs763035 was associated with an eQTL at a distance of 418,831 bp, associated with the gene HLA-DOB (P=1.9 × 10−19).
A gene network analysis to assess gene relationships of the genes closest to the validated significant SNP was performed by GeneMANIA (accessed on 13 September 2014). HLA-DRA and BTNL2 are connected by coexpression via HLA-DRB5 and CIITA (class II, major histocompatibility complex, trans-activator).
Because the HLA system is highly polymorphic, has a key role in presentation of antigen, and has been associated with multiple human diseases, HLA allelic association was analyzed by imputation using attribute bagging with haplotype inference from unphased SNPs and HLA types (HIBAG; Zheng et al, 2013). Regression using the same set of covariates as in the SNP-based genome-wide association study (GWAS) for rosacea phenotype resulted in three significant associations: HLA-DRB1*03:01 (P=1.0 × 10−8), HLA-DQB1*02:01 (P=1.3 × 10−8), and HLA-DQA1*05:01 (P=1.4 × 10−8; Table 2). All of these associations were confirmed through the replication group described above, using a significance threshold of P<0.05 and were as follows: HLA-DRB1*03:01 (P=4.4 × 10−6), HLA-DQB1*02:01 (P=7.2 × 10−6), and HLA-DQA1*05:01 (P=7.6 × 10−6; Table 2).
Discussion
All of these HLA allele associations have links to other autoimmune diseases. The DRB1*03:01-DQB1*02:01-DQA1*05:01 haplotype has been associated with type I diabetes (Erlich et al., 2008), and HLA-DRB1*03:01 is associated with retinopathy in type I diabetics (Kastelan et al., 2013), a vascular proliferative disorder of the eye. Rosacea also exhibits abnormal proliferation of blood vessels including on the ocular conjunctiva. HLA-DQB1*02:01 has been associated with celiac disease in humans (Krini et al., 2012), a condition that can have skin manifestations. Celiac disease involves the small intestine, and rosacea has been associated with conditions located in the small intestine, namely small intestinal bacterial overgrowth (Weinstock and Steinhoff, 2013). Together, these data strongly suggest a role for antigen presentation by class II HLA in the etiology of rosacea.
The data presented from this large discovery and replication group provide evidence for a genetic component of rosacea. The association of rosacea with HLA-DRA and other HLA alleles is consistent with the inflammatory nature of the disease. In fact, HLA-DRA, HLA-DRB, HLA-DQB, and HLA-DQA all belong to major histocompatibility complex class II, which presents antigen from extracellular sources, potentially explaining the connection of various microbes with rosacea suggested in the medical literature (Weinstock and Steinhoff, 2013).
One of the most interesting findings of this study is the potential connection between rosacea and other human diseases such as diabetes and celiac disease. As previously mentioned, HLA-DRB1*03:01 is associated with diabetic retinopathy in a Croatian population of type I diabetics (Kastelan et al., 2013). The connection between rosacea and diabetes merits further study, as rosacea has been reported to associate negatively with diabetic individuals on insulin or oral antidiabetic drugs in a retrospective study (Spoendlin et al., 2013). This reduction in rosacea could be owing to treatment for diabetes with insulin or antidiabetic drugs.
As rosacea is a highly visible disease, it may be a cutaneous sign that cues healthcare providers to consider screening for diabetes. Future study may even identify individuals at risk for both rosacea and diabetes based on allele profiles, enabling these individuals to avoid triggers such as sun exposure (for rosacea), seek regular blood glucose screening (for diabetes), and/or treat the diseases early to avoid long-term adverse health sequelae (such as scarring or permanent disfigurement in rosacea, or cardiovascular damage and neuropathy in diabetes).
Within the study, the rosacea phenotype was based on self-report of diagnosis by a healthcare provider. We cannot exclude the possibility that some cases and controls are incorrectly classified. Misclassification would likely reduce study power and not lead to false associations. Traditional clinical phenotyping would not have been feasible for the large sample size used in this study. However, the use of a self-reported phenotype using an unvalidated instrument remains a limitation of the work. Forthcoming studies will use dermatologists' assessments of rosacea phenotype to validate the gene variants identified in this study in a targeted manner.
Future studies will also be powered to investigate the effect of the significant SNP on gene expression and/or protein levels in rosacea skin, with the goal of establishing a more direct mechanistic link between the gene variants identified here and the mechanisms by which these changes lead to the rosacea phenotype. Additional study will also involve extension to individuals of non-European descent.
These data could also be used to identify individual risk for rosacea based on genotype, and explore differences in response to various rosacea treatments based on genotype, so that treatment choices could be more “personalized”. Additional research based on the data in this study could lead to the identification of targets that could be tested and harnessed to better treat this chronic and disfiguring disease.
Materials and Methods
Study design and population
The genome-wide association study was conducted in accordance with the Declaration of Helsinki Principles, including approval by an external Association for the Accreditation of Human Research Protection Programs-accredited Institutional Review Board, E & I Review Services. Participants in the GWAS were customers of the genetics company 23andMe (Mountain View, CA), with the vast majority residing in the United States who voluntarily consented to release their aggregated genetic information in an anonymous manner, as well as answer research questions via online questionnaires. Individual results were not relayed back to the participants. Therefore, the risks of psychological harm or loss of confidentiality from participating in this study are extremely low. The discovery group was accrued before the replication group. Accrual methods and criteria for inclusion and exclusion in the study were the same for both groups, as described above. None of the participants in the discovery were in the replication group.
Skin samples from individuals with rosacea were obtained as part of a gene expression study for rosacea, a protocol approved by the Stanford Human Subjects Review panel (#22419). Written informed consent in accord with the Declaration of Helsinki Principles was obtained before all study procedures.
Statistical adjustment for covariates
Age, sex, and population stratification in the cases versus controls were adjusted for by multivariate regression in the GWAS. This was performed by assuming an additive model for allelic effects with logistic regression using the model: rosacea diagnosis ~ age+sex+pc.0+pc.1+pc.2+pc.3+pc.4+genotype. The results in this study were adjusted for a genomic control inflation factor of 1.032.
Determination of rosacea phenotype
Rosacea phenotype of participants in both the discovery and replication groups was determined using the same methods. To identify cases for the purposes of GWAS, participants who self-identified as age 30 years or older voluntarily answered the online question “Have you ever been diagnosed with rosacea (sometimes called adult acne or acne rosacea) by a medical professional?” Answer choices consisted of “yes,” “no,” and “I'm not sure.” To ensure that facial redness was not due to concomitant medical condition unrelated to rosacea, individuals who answered “yes” to the prior question were asked “Was the onset of your rosacea associated with any of the following conditions? Please check all that apply.” Choices included “lupus,” “pregnancy,” “rheumatoid arthritis,” “scleroderma,” “other arthritis,” “other autoimmune disease,” “I'm not sure,” and “None of the above”. Individuals who selected one of the disease conditions were excluded from the GWAS. Individuals who answered “No” to the first question were assigned to be controls. Owing to the large sample size needed for this study, clinical examination for rosacea phenotype for both the discovery group and the replication group was not feasible.
Methods of genotyping and quality control
For both discovery and replication groups, DNA extraction and genotype analyses were performed on saliva samples by the National Genetics Institute, a Clinical Laboratory Improvement Amendments–certified clinical laboratory, and subsidiary of the Laboratory Corporation of America.
Samples for the discovery and replication groups were genotyped on one of two platforms. A portion of participants were genotyped on the Illumina HumanHap550+ BeadChip platform, which included SNPs from the standard HumanHap550 panel augmented with a custom set of ~25,000 SNPs selected by 23andMe. The remaining participants were genotyped on the Illumina Human OmniExpress+ BeadChip. This platform has a base set of 730,000 SNPs augmented with ~250,000 SNPs to obtain a superset of the HumanHap550+ content, as well as a custom set of about 30,000 SNPs. To confirm that results from both platforms were concordant, several thousand samples were processed with both arrays. SNPs with significant rates of discordant genotype calls were removed. Data from the two platforms were imputed against the same reference panel, and quality control filters were applied to the imputed data to remove variants with batch effects.
Every sample that did not reach a 98.5% call rate for SNPs on the platforms was reanalyzed. Individuals whose analyses repeatedly failed were contacted by 23andMe customer service to provide additional samples.
Participants in both the discovery and replication groups were restricted to individuals with >97% European ancestry, as determined through an analysis of local ancestry via comparison with the three HapMap 2 populations. A maximal set of unrelated individuals was chosen for the analysis using a segmental identity-by-descent estimation algorithm (Henn et al., 2013). Individuals were defined as related if they shared more than 700 cM identity-by-descent, including regions in which the two individuals share either one or both genomic segments identical-by-descent. This level of relatedness (roughly 20% of the genome) corresponds approximately to the minimal expected sharing between first cousins in an outbred population.
Participant genotype data were imputed against the August 2010 release of 1000Genomes reference haplotypes (Browning and Browning, 2007; Howie et al., 2012). First, Beagle (version 3.3.1) (University of Washington, Seattle, WA) was used to phase batches of 8000–9000 individuals across chromosomal segments of no more than 10,000 genotyped SNPs, with overlaps of 200 SNPs. We excluded SNPs with minor allele frequency <0.001, Hardy–Weinberg equilibrium P<10−20, call rate <95%, or with large allele frequency discrepancies compared with the 1000Genomes reference data. A total of 860,590 genotyped SNPs passed these filters.
Full-phased chromosomes were assembled by matching the phase of haplotypes across the overlapping segments. We imputed each batch against the European subset of 1000Genomes haplotypes using Minimac (2011-10-27) (University of Washington, Seattle, WA), using 5 rounds and 200 states for parameter estimation.
For the nonpseudoautosomal region of the X chromosome, men and women were phased together in segments, treating the men as already phased. The pseudoautosomal regions were phased separately. We assembled fully phased X chromosomes, representing men as homozygous pseudo-diploids for the nonpseudoautosomal region. We then imputed men and women together using Minimac as with the autosomes.
For imputed GWAS results, SNPs with average r2<0.5 or minimum r2<0.3 in any imputation batch, as well as SNPs that had strong evidence of an imputation batch effect, were flagged. To detect imputation batch effects, we performed an analysis of variance for the imputed genotype dosages versus imputation batch, and flagged SNPs with P<10−50 for the null hypothesis that dosages were independent of batch. A total of 7,381,496 imputed variants passed these filters.
In choosing between genotyped versus imputed GWAS results, the imputed result was chosen if it passed quality control or if a genotyped result was unavailable. Genotyped results were used for variants that either were not imputed (not in the 1000Genomes reference panel) or the imputed result failed a quality filter.
After merging genotyped and imputed association test results, logistic regression results that did not converge owing to complete separation were flagged. The final merged analysis included association results for 7,431,626 variants.
For case-control comparisons, we computed association test results by logistic regression assuming additive allelic effects. For tests using imputed data, we use the imputed dosages rather than best-guess genotypes. We included covariates for age, gender, and the top five principal components to account for residual population structure. The association test P-value was computed using a likelihood ratio test. Results for the X chromosome are computed similarly, with men coded as if they were homozygous diploid for the observed allele.
GWAS results were adjusted for genomic control to compensate for variance inflation owing to residual population stratification that has not been effectively controlled through principal components in the regression models. The genomic control inflation factor was computed from the median P-value for results that passed the quality control.
Gene associations, regulation, expression, and networks
A Manhattan plot was created to show the distribution of test statistics reported as P-values on the y-axis and chromosomal position on the x-axis. The regional association plots were generated with LocusZoom, using linkage disequilibrium data from the March 2012 release of 1000Genomes data (available from : www.ncbi.nlm.nih.gov/variation/tools//1000genomes/).
The University of California Santa Cruz genome browser (http://genome.ucsc.edu; hg19) was used to search the candidate SNPs for gene associations and whether the SNPs were transcribed. GenePaint (www.genepaint.org) and GeneCards were used to search gene expression patterns in mice and humans. An association test for the most significant SNP and eQTLs within 500 kb was performed (Stranger et al., 2007; Montgomery et al., 2010), with cutoff r2>0.5. The analysis included data sets in lymphoblastoid tissue, liver (Schadt et al., 2008), fibroblast, B and T cell (Dimas et al., 2009), cerebellum, frontal cortex, pons, temporal cortex (Gibbs et al., 2010), and monocytes (Zeller et al., 2010). Genotype-Tissue Expression was used to query eQTLs in skin. To establish open chromatin signatures in and around the SNP sites, DNaseI hypersensitivity regions within the University of California Santa Cruz genome browser were searched. RegulomeDB was searched for potential transcription factor binding sites of significant SNPs. To query for SNPs in linkage disequilibrium with the significant SNPs, the NCBI short genetic variation (SNV) database (www.ncbi.nlm.nih.gov/SNP/) and 1000GenomesProject database were used. HapMap was used to interrogate allele frequencies in individuals of European ancestry. GeneMANIA (http://geneMANIA.org) was used to search for gene interactions in the significant SNP.
HLA imputation and association
HLA allele dosages were imputed from SNP genotype data using HIBAG (Zheng et al., 2013), an attribute bagging-based statistical method implemented in the freely available R package, including prefit classifier trained from a large database of individuals with known HLA alleles and SNP variation within the HLA region. Over 98% of the tagging SNPs used in HIBAG were genotyped and passed quality control on the 23andMe platform. Alleles for HLA-A, B, C, DPB1, DQA1, DQB1, and DRB1 loci were imputed at four-digit resolution. The default settings of HIBAG were used. To test associations between HLA allele dosages and phenotypes, logistic regression was performed using the same set of covariates used in the SNP-based GWAS for that phenotype. Separate association tests were performed for each imputed allele.
Immunohistochemistry
The institutional review board-approved consent form for the GWAS did not allow us to recontact participants in the discovery and replication groups for rosacea skin samples. However, we obtained separate institutional review board approval for an exploratory study of genotype and protein expression of six individuals of self-reported European ancestry with papulopustular rosacea diagnosed by a board-certified dermatologist recruited through Stanford Medical Dermatology Clinic. After written informed consent, peripheral blood from these individuals was obtained for genotyping by Illumina Human OmniExpress+ BeadChip (San Diego, CA) with imputation for rs763035 using the same methods as described for GWAS. In addition, rosacea-affected skin and adjacent normal skin were procured from the lower 1/3 of the face by 4mm skin punch biopsy device (Medichoice, Mechanicsville, VA) under local anesthesia with xylocaine. All rosacea-affected skin biopsies consisted of follicle-centered papules. Tissue was fixed in formalin solution and embedded in paraffin. Sections were stained with hematoxylin and eosin to confirm the rosacea diagnosis histologically. Additional sections were subjected to the immunohistochemistry with antibodies against HLA-DRA and BTNL2, to assess the protein distribution of HLA-DRA and BTNL2. The primary antibodies used were as follows: mouse anti-human monoclonal HLA-DRA antibody [clone TAL1B5, LifeSpan BioSciences (Seattle,WA)] at 1:200 dilution, and polyclonal rabbit anti-human primary BTNL2 antibody (Sigma-Aldrich, St. Louis, MO), product name HPA039844) at 1:100 dilution. Either horse or goat serum was used to block background; horseradish peroxidase-conjugated secondary antibody was used to visualize signal. Light microscopy was used to assess the immunohistochemical results. Negative controls were subjected to the same steps as above with omission of primary antibody, and showed no immunohistochemical staining. Owing to the small sample size, staining differences could not be quantified between the one individual with the rs763035 minor allele and the six individuals without the minor allele. MicroSuite Pathology version 5.0 software (Upper Sanon, PA) was used to capture images. Images were arranged with Adobe Illustrator (San Jose, CA).
Acknowledgments
We acknowledge the expertise of Jinah Kim and Kerri Rieger, faculty members in the department of pathology and dermatology at Stanford University School of Medicine, in the evaluation of the immunohistochemistry shown in this study. We thank Lisa Zaba for expert advice on immunohistochemistry; Jonathan Pritchard, professor of Genetics and Biology at Stanford University, and Timothee Flutre, Post-doctoral Fellow in Human Genetics at University of Chicago, for assistance in searching for the associated eQTLs; Anil Gupta, post-doctoral fellow in the Pritchard Laboratory, for imputation consultation; Marina Sirota, at the Stanford Center for Biomedical Informatics Research, for assistance in searching for SNPs in linkage disequilibrium; Shufeng Li, Biostatistician in Stanford Department of Dermatology, for statistical consultation; Howard Y Chang, professor of Epithelial Biology at Stanford University, for critical pre-review of the manuscript; and finally, the customers and employees of 23andMe for making this work possible. This study was funded by the National Human Genome Research Institute of the National Institutes of Health under grant number 2R44HG006981-02 and the National Rosacea Society.
Author Contributions
ALSC, JYT, AKK, NKE, and DAH contributed to the design of the study. ALSC, JYT, RS, JX, RL, JC, IR, and AKK contributed data. RL provided technical support. CT, DAH, and NKE were responsible for biostatistical analysis. ALSC, JYT, and DAH helped to interpret results. ALSC drafted the manuscript. All authors contributed to the final paper, with ALSC and JYT having key roles.
Glossary
- eQTL
expression quantitative trait locus
- GWAS
genome-wide association study
- HIBAG
HLA Genotype Imputation with Attribute Bagging
- HLA
human leukocyte antigen
- SNP
single-nucleotide polymorphism
- UCSC
University of California Santa Cruz
JYT, CT, AKK, NKE, and DAH are current or former employees of 23andMe, Inc. The remaining authors state no conflict of interest.
Footnotes
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Supplementary Material
References
- Abram K, Silm H, Maaroos H-I, et al. Risk factors associated with rosacea. J Eur Acad Dermatol Venereol. 2010;24:565–571. doi: 10.1111/j.1468-3083.2009.03472.x. [DOI] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing data inference for whole genome association studies using localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier B. Medical history of the representation of rosacea in the 19th century. J Am Acad Dermatol. 2013;69:S2–13. doi: 10.1016/j.jaad.2013.04.046. [DOI] [PubMed] [Google Scholar]
- Dimas AS, Deutsch S, Stranger BE, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler J, Jarratt M, Moore A, et al. Once-daily topical brimonidine tartrate gel 0.5% is a novel treatment for moderate to severe facial erythema of rosacea: results of two multicenter, randomized and vehicle-controlled studies. Br J Dermatol. 2012;166:633–641. doi: 10.1111/j.1365-2133.2011.10716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elewski BE, Draelos Z, Dreno B, et al. Rosacea—global diversity and optimized outcome: proposed international consensus from the Rosacea International Expert Group. JEADV. 2011;25:188–200. doi: 10.1111/j.1468-3083.2010.03751.x. [DOI] [PubMed] [Google Scholar]
- Erlich H, Valdes AM, Noble J, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidlin B., Zheng G, Li Z, et al. Trend tests for case-control studies of genetic markers: power, sample size, and robustness. Hum Hered. 2002;53:146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Hon L, Macpherson JM, et al. Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS One. 2013;7:e34267. doi: 10.1371/journal.pone.0034267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium Risk alleles for multiple sclerosis identified by a genomewide study. New Eng J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Kastelan S, Tomic M, Salopek-Rabatic J, et al. The association between the HLA system and retinopathy development in patients with type 1 diabetes mellitus. Coll Antropol. 2013;37 (Suppl 1:65–70. [PubMed] [Google Scholar]
- Krini M, Chouliaras G, Kanariou M, et al. HLA class II high-resolution genotyping in Greek children with celiac disease and impact on disease susceptibility. Pediatr Res. 2012;72:625–630. doi: 10.1038/pr.2012.133. [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis E, Korfitis C. Isotretinoin for the treatment of granulomatous rosacea: a case report and review of the literature. J Cutan Med Surg. 2012;16:438–441. doi: 10.1177/120347541201600615. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota M, Schaub MA, Batzoglou S, et al. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009;5:e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoendlin J, Voegel JJ, Jick SS, et al. Risk of rosacea in patients with diabetes using insulin or oral antidiabetic drugs. J Invest Dermatol. 2013;133:2790–2793. doi: 10.1038/jid.2013.225. [DOI] [PubMed] [Google Scholar]
- Steinhoff M, Schauber J, Leyden JJ. New insights into rosacea pathophysiology: a review of recent findings. J Am Acad Dermatol. 2013;69:S15–S26. doi: 10.1016/j.jaad.2013.04.045. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentonyte R, Hampe J, Huse K, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- Weinstock L.B., Steinhoff M. Rosacea and small intestinal bacterial overgrowth: prevalence and response to rifaximin. J Am Acad Dermatol. 2013;68:875–876. doi: 10.1016/j.jaad.2012.11.038. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Di Nardo A, Bardan A, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat Med. 2007;13:975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kanada K, Macleod DT, et al. TLR2 expression is increased in rosacea and stimulates enhanced serine protease production by keratinocytes. J Invest Dermatol. 2011;131:688–697. doi: 10.1038/jid.2010.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, Wild P, Szymczak S, et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Shen J, Cox C, et al. HIBAG-HLA genotype imputation with attribute bagging. Pharmacogenom J. 2013;14:192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.