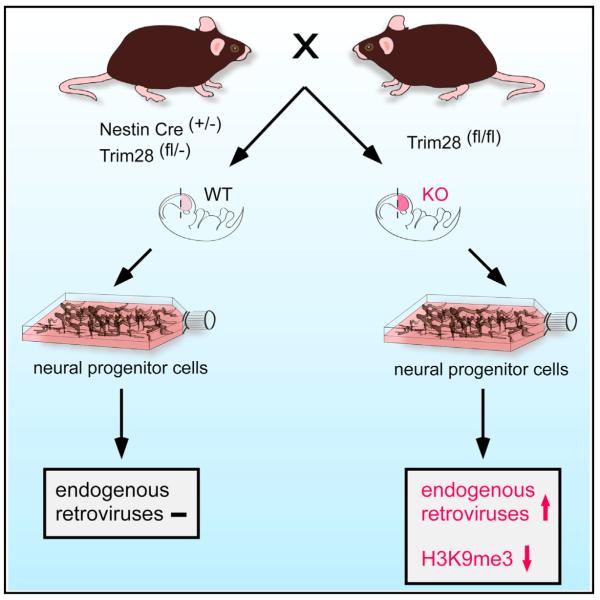
SUMMARY
TRIM28 is a corepressor that mediates transcriptional silencing by establishing local heterochromatin. Here, we show that deletion of TRIM28 in neural progenitor cells (NPCs) results in high-level expression of two groups of endogenous retroviruses (ERVs): IAP1 and MMERVK10C. We find that NPCs use TRIM28-mediated histone modifications to dynamically regulate transcription and silencing of ERVs, which is in contrast to other somatic cell types using DNA methylation. We also show that derepression of ERVs influences transcriptional dynamics in NPCs through the activation of nearby genes and the expression of long noncoding RNAs. These findings demonstrate a unique dynamic transcriptional regulation of ERVs in NPCs. Our results warrant future studies on the role of ERVs in the healthy and diseased brain.
Graphical Abstract
INTRODUCTION
The mammalian brain is an extremely complex organ harboring more than a thousand different types of neurons that serve a wide variety of functions. How this complexity is achieved remains largely unknown. However, epigenetic mechanisms such as DNA methylation, histone modification, and noncoding RNAs are thought to be important in establishing a high diversity of gene expression from the same template, leading to a spatial pattern of transcription. How distinct transcriptional programs are established in different neuronal populations remains poorly understood, but one interesting recently proposed hypothesis suggests transposable elements (TEs) to be involved in this process (Muotri et al., 2007; Reilly et al., 2013). TEs are repetitive mobile genetic elements that were originally considered to be parasitic DNA without any function, popularly termed “junk DNA.” Today, it is becoming increasingly clear that TEs can act as gene regulatory elements by serving as hubs for chromatin modifications and by acting as transcriptional start sites for noncoding RNAs. Consequently, TEs are very well suited to influence gene expression and may play an important role in controlling and fine-tuning gene networks in the brain (Jern and Coffin, 2008; Cowley and Oakey, 2013).
Retroviruses are found in most vertebrates and can transform their genetic material and integrate into the host genome as proviruses to produce new viruses. Occasionally, retroviruses infect germline cells allowing the integrated proviruses to be passed on to the offspring as an endogenous retrovirus (ERV). Around 8%–10% of the human and mouse genome are composed of this type of TE, and, despite up to millions of years since their integration in host germline, many ERVs contain sequences that can serve as transcriptional start sites or as cis-acting regulatory elements in the host genomes (Jern and Coffin, 2008). The large amount of ERVs in mammalian genomes suggest that they play important roles in the host organisms, for instance, by influencing gene regulatory networks (Kunarso et al., 2010; Feschotte, 2008), but ERVs have also been linked to diseases. In humans, aberrant expression of ERVs has been found in both cancer and brain disorders, although causality remains to be established (Jern and Coffin, 2008; Douville et al., 2011). Thus, ERVs may contribute both beneficial and detrimental effects, which have been balanced throughout evolution, to the host organism.
ERVs are silenced during the first few days of embryogenesis by TRIM28 (tripartite motif-containing protein 28, also known as KAP1 or TIF1beta), a transcriptional corepressor essential for early mouse development (Cammas et al., 2000; Rowe et al., 2010). During the extensive genome reprogramming that takes place at this period, TRIM28 is recruited to ERVs via sequence-specific Krüppel-associated box zinc-finger proteins (KRAB-ZFPs), a family of transcription factors that has undergone a rapid expansion in mammalian genomes in parallel with the expansion of ERVs (Wolf and Goff, 2009; Thomas and Schneider, 2011). TRIM28 then induces repressive histone modifications by recruiting multiprotein complexes including the H3K9 methyltransferase SETDB1 (also known as ESET), the histone deacetylase-containing NuRD complex, and heterochromatin protein 1 (HP1) (Schultz et al., 2002; Sripathy et al., 2006). Deletion of Trim28 or Setdb1 in ESCs leads to loss of the H3K9me3-mark at ERVs, resulting in transcriptional activation of these elements (Matsui et al., 2010; Rowe et al., 2010).
However, KRAB-ZFP/TRIM28 histone-based repression of ERVs rapidly gives place to a more permanent silencing mechanism, as the TRIM28-mediated recruitment of de novo DNA methyltransferases leads to cytosine methylation at CpG dinucleotides (Ellis et al., 2007; Wiznerowicz et al., 2007; Rowe and Trono, 2011). The maintenance DNA methyltransferase complex then ensures that DNA methylation is maintained, alleviating the need for sequence-specific KRAB-ZFPs and TRIM28. In mouse embryonic fibroblasts as well as in all adult tissues examined so far, TRIM28 depletion has no impact on ERV silencing, which is instead released by drugs such as 5-azacytidine or by deletion of DNA methyltransferases (Jackson-Grusby et al., 2001; Hutnick et al., 2010).
DNA methylation has long been considered as a stable epigenetic mark resulting in maintenance of DNA-methylation patterns throughout the lifespan of an organism. However, several recent studies demonstrate a unique dynamic regulation of DNA-methylation patterns in the brain (Sweatt, 2013). There is also evidence that retroelements and transposons are highly active during brain development and in neural progenitor cells (NPCs) (Muotri et al., 2005, 2010; Baillie et al., 2011; Evrony et al., 2012; Li et al., 2013; Perrat et al., 2013). For example, LINE-1 elements have been found to be transcriptionally active and to retrotranspose in NPCs (Muotri et al., 2005, 2010; Coufal et al., 2009). In addition, we have previously found that deletion of TRIM28 in postmitotic forebrain neurons results in complex behavioral alterations, including vulnerability to stress (Jakobsson et al., 2008). In the present work, we demonstrate that NPCs use TRIM28-mediated histone modifications to dynamically regulate the transcription and silencing of ERVs, rather than the DNA methylation at play in other somatic tissues. We also unveil that derepression of ERVs influences transcriptional dynamics in NPCs, by activating nearby genes and the expression of long noncoding RNAs (lncRNAs).
RESULTS
TRIM28-Deficient NPCs Express High Levels of ERVs
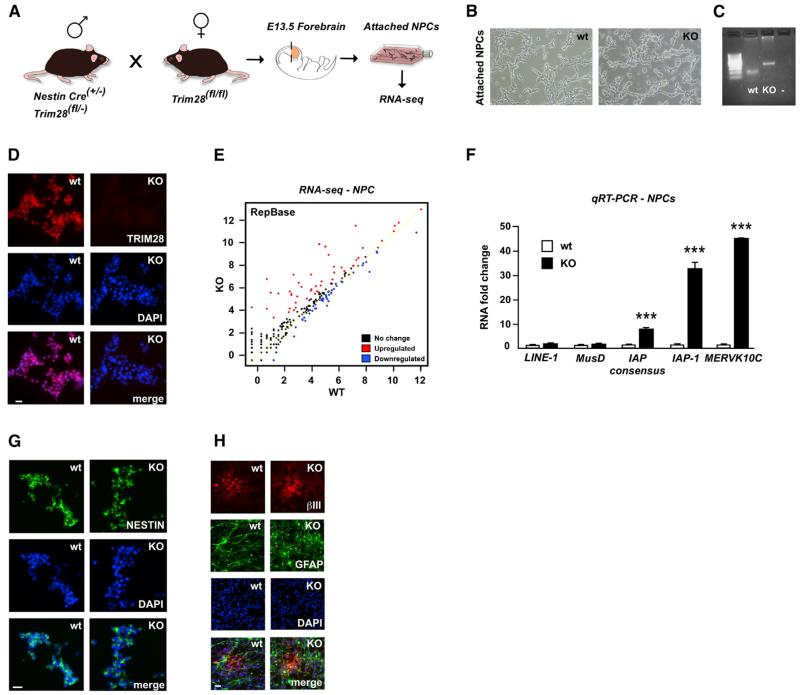
To investigate if TRIM28 contributes to ERV silencing in NPCs we established Trim28-deficient NPC cultures. We crossed transgenic NestinCre mice (Tronche et al., 1999) with mice carrying floxed Trim28-alleles (Trim28fl/fl) (Weber et al., 2002), resulting in excision of Trim28 in neural progenitors at the time when Nestin-expression is initiated, starting around embryonic day 10 (E10). At E13.5, we collected embryos, dissected the forebrain, and established NPC cultures from individual embryos (Figures 1A and 1B). We confirmed the deletion of Trim28 by genotyping for the excised allele and by verifying the absence of TRIM28 protein (Figures 1C and 1D). We collected RNA from Trim28−/− NPCs and wild-type controls and performed RNA extraction followed by deep sequencing (RNA-seq). The resulting reads were mapped against reference sequences from Repbase, a database containing consensus sequences for known repetitive elements (Jurka et al., 2005). We found that several ERVs were highly upregulated in Trim28−/− NPCs, including, e.g., Mus musculus ERV using tRNALys type 10C (MMERVK10C) and intracisternal A-particles class 1 (IAP1) (Figure 1E; Tables S1 and S2). Other retroelements such as MusD and LINE-1 were modestly upregulated, whereas several other types of common repetitive elements were unaffected (Figure 1E;Tables S1 and S2).
Figure 1. Establishment of Trim28-Deficient Neural Progenitor Cultures.
(A) Illustration of the experimental approach.
(B) Representative images of early passage Trim28−/− NPCs.
(C) PCR analysis of genomic DNA from wild-type and Trim28−/− NPCs demonstrates the presence of the 152 and 290 bp products corresponding to loxP-flanked or excised Trim28, respectively.
(D) Verification of a complete lack of TRIM28 protein via immunocytochemistry.
(E) RNA-seq analysis. The graph shows KO samples plotted versus wild-type samples, where each dot represents a Repbase sequence.
(F) qRT-PCR of RNA isolated from wild-type and Trim28−/− NPCs.
(G) Trim28-deficient NPCs display a homogenous expression of NESTIN.
(H) Immunofluorescent analysis of differentiated NPCs.
Data are presented as mean of relative values ± SEM. **p < 0.01, ***p < 0.001, Student’s t test. Scale bars, 200 (A) and 50 (B) μm. See also Tables S1 and S2.
We confirmed increased transcription of MMERVK10C and IAP1 elements using quantitative RT-PCR (qRT-PCR) (Figure 1F). In contrast, when we used primer pairs designed to recognize the consensus sequence of the entire IAP-family, including more ancient IAP elements, we detected only a modest upregulation (Figure 1F). This finding is in line with the results of the RNA-seq, which indicated that only certain types of IAP elements were upregulated in Trim28−/− NPCs. Also in agreement with the RNA-seq, qRT-PCR analyses indicated that deletion of Trim28 in NPCs only modestly increased the expression of other retroelements such as LINE-1 or MusD (Figure 1F). We confirmed these results in cultures derived from two separate embryos (data not shown).
Trim28−/− NPCs proliferated at a similar rate compared to cells generated from wild-type and heterozygous siblings and could be expanded for more than 60 passages. However, we observed that Trim28−/− NPCs were growing in dense cluster-like formations, which seemed to attach less to the flask surface compared to the wild-type control. Trim28−/− NPCs could also be differentiated to both neurons and astrocytes suggesting that TRIM28 has no major influence on the self-renewal and differentiation of NPCs (Figures 1G and 1H).
MMERVK10C Elements Are Controlled by TRIM28
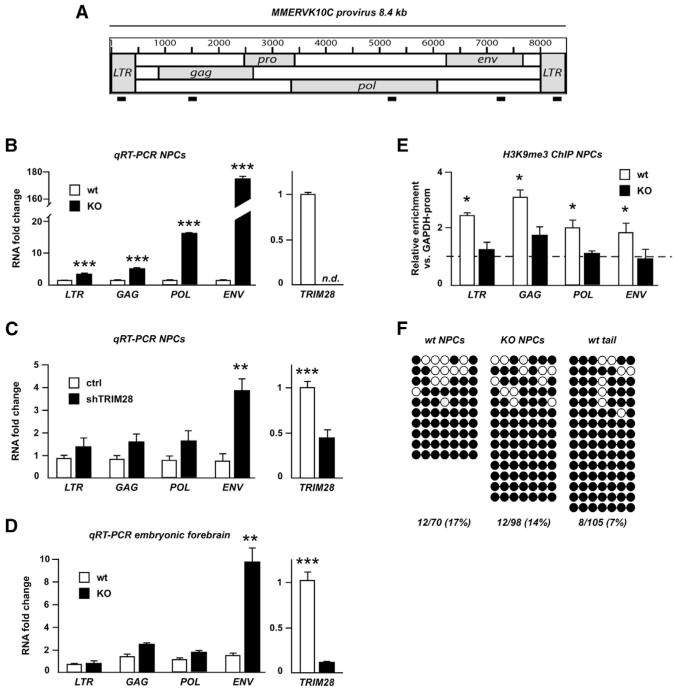
The RNA-seq analysis indicated that MMERVK10C elements were among the most upregulated ERVs following Trim28-deletion in NPCs. MMERVK10C is a beta-like ERV similar to HERVK (HML2), one of the most recent ERVs to invade the human genome (Reichmann et al., 2012) (Belshaw et al., 2005). MMERVK10C sequences flanked by RLTR10C make up putative proviral sequences of around 8.4 kb. In the mouse genome, MMERVK10C is present in a few complete provirus loci (~20) and more than 1,000 incomplete loci (Reichmann et al., 2012). We performed sequence analysis of the MMERVK10C provirus for the presence of retroviral features using the RetroTector software (Sperber et al., 2007). Based on this analysis, we designed primers recognizing the LTRs, gag, pol, and env of the MMERVK10C provirus and investigated expression levels in Trim28−/− NPCs (schematics in Figure 2A). We found that transcripts over the entire region of the provirus were increased, including a massive expression of env sequences when compared to wild-type controls (170-fold; Figure 2B).
Figure 2. Analysis of the Putative MMERVK10C Provirus.
(A) Schematic drawing of the MMERVK10C provirus and approximate primer positions.
(B) Quantitative analysis of transcript levels of different regions of the MMERVK10C provirus in Trim28−/− and wild-type NPCs.
(C) qRT-PCR analysis of MMERVK10C following TRIM28-shRNA knockdown.
(D) qRT-PCR analysis of E13.5 forebrain dissected from intercrosses of NestinCre Trim28floxed mice.
(E) ChIP for H3K9me3 in Trim28−/− and wild-type NPCs.
(F) Bisulfite sequencing analysis of the 3′ end region of MMERVK10C. Empty and full circles represent unmethylated and methylated CpGs, respectively.
Data are presented as mean of relative values ± SEM. *p < 0.05, Student’s t test.
Ascertaining that the ERV induction observed in NPCs isolated from Trim28−/− animals was not secondary to more general developmental anomalies, knocking down TRIM28 in wild-type NPCs by lentivector-mediated RNA interference led to a marked upregulation of these retroelements (Figure 2C). Furthermore, increased ERV expression was detected in forebrain tissue from Trim28−/− embryos (Figure 2D).
In ESCs, TRIM28 controls ERV expression via histone modifications including H3K9 trimethylation (Rowe et al., 2010), whereas it is DNA methylation that instead prevails in somatic tissues. In NPCs, we found that the MMERVK10C provirus was enriched in H3K9me3, and that this repressive mark was markedly reduced in Trim28−/− NPCs (Figure 2E).
Because MMERVK10C appeared to be under TRIM28 control in NPCs, we hypothesized that at least a proportion of these retroelements escaped DNA methylation in these cells. To probe this issue, we examined the DNA methylation status of full-length MMERVK10C, which were among the most highly upregulated retroelements in Trim28−/− NPCs. Bisulfite sequencing of a CpG-island located in the 3′ region of MMERVK10C revealed several clones with some unmethylated CpGs (17% unmethylated CpGs, Figure 2F) in NPCs, whereas this region was almost fully methylated in DNA extracted from mouse tail (7% unmethylated CpGs, Figure 2F, Fisher’s exact test one-sided p < 0.05). Moreover, we found no difference in the level of CpG methylation between wild-type and Trim28−/− NPCs. In summary, these data suggest that a proportion of the MMERVK10C elements are spared from undergoing DNA methylation specifically in NPCs during early development.
Increased Expression of IAP1 Results in ERV-Derived Protein Expression
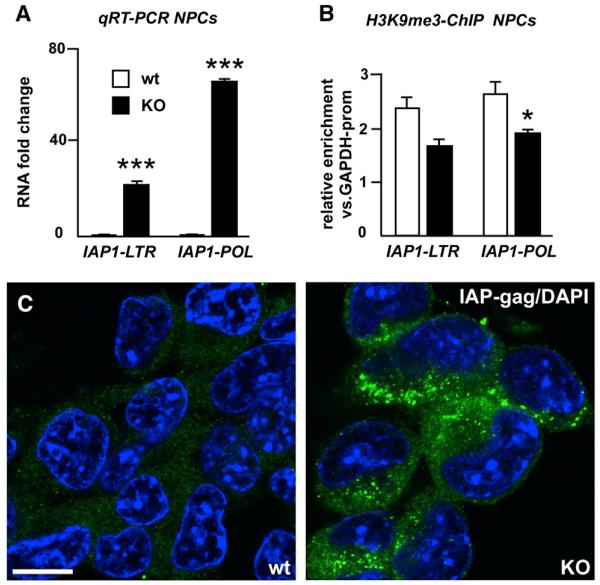
IAP1 elements, which lose H3K9me3 marks and were also highly upregulated in Trim28−/− NPCs (Figures 3A and 3B), are internalized env-lacking mouse ERVs that demonstrate a large degree of polymorphism among different mouse strains and maintain the capacity to retrotranspose. Using immunocytochemistry with an IAP-specific antibody, we found a uniform, high-level IAP-gag expression located to the cytoplasm in Trim28−/− NPCs (Figure 3C).
Figure 3. Analysis of IAP1 Expression.
(A) Quantitative analysis of transcript levels of different regions of IAP1 provirus in Trim28−/− and wild-type NPCs.
(B) ChIP for H3K9me3 in Trim28−/− and wild-type NPCs.
(C) Confocal analysis of immunofluorescence staining for IAP-gag on Trim28−/− and wild-type NPCs. Scale bar, 10 μm.
Data are presented as mean of relative values ± SEM. *p < 0.05, Student’s t test.
Taken together, these data demonstrate that deletion of TRIM28 in NPCs results in a massive transcriptional increase of ERVs, including MMERVK10C and IAP1. NPCs thus appear to constitute a cellular environment distinct from that of other somatic cells studied so far, with the TRIM28-induced histone-based repressive mechanism playing a role in ERV control.
Activation of ERVs Correlates with Increased Transcription of Nearby Genes
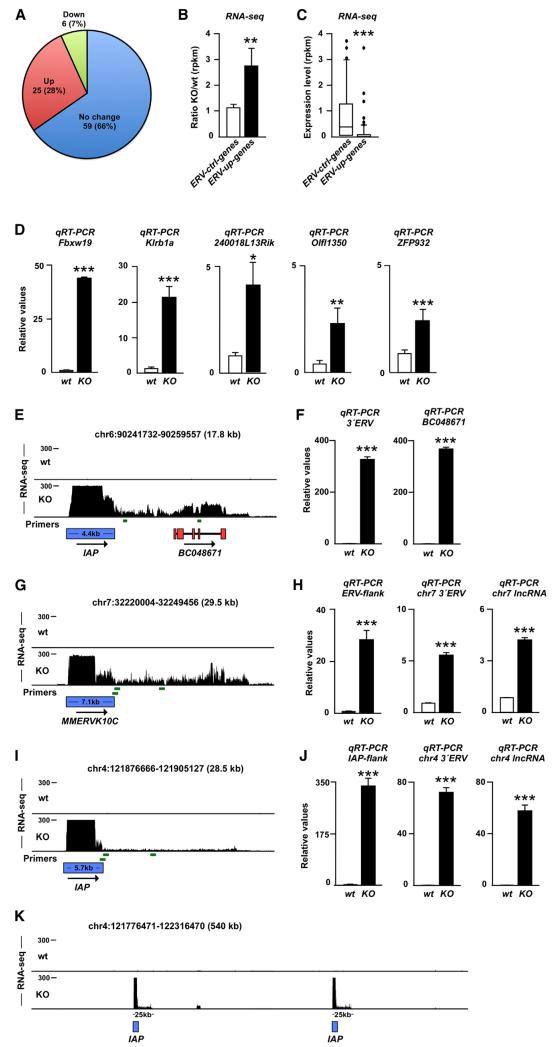
The ability of ERVs to attract transcription factors and silencing complexes has led to a reassessment of their role in the host genome. ERVs are now considered to be important transcriptional regulatory elements that shape and influence gene expression during early development (Isbel and Whitelaw, 2012). For example, we have recently found that TRIM28 controls the expression of developmental genes by repressing ERV-associated enhancers in pluripotent cells (Rowe et al., 2013). Twenty-six MMERVK10C proviruses and 361 IAP proviruses that were upregulated in Trim28−/− NPCs were mapped to precise genomic locations (Figure S1). Out of these 387 proviruses, 90 were situated close to genes (<50 kb). We found that 25 of those genes (28%) demonstrated significantly increased expression, whereas expression of only six of them was decreased (7%) (Figure 4A). We also found that those 90 genes located close to upregulated ERVs (ERV-up genes) were on average 3-fold upregulated in Trim28−/− cells (Figure 4B). In contrast, a random selection of ERVs that was not upregulated in Trim28−/− cells (n = 129, MMERVK10C and IAP1 elements) did not affect nearby genes (ERVs-ctrl genes, n = 50, Figure 4B). Interestingly, we also found that ERV-up genes were expressed at low levels in wild-type cells (Figure 4C), which is in agreement with a model where ERVs mediate repressive regulation of nearby genes caused by the attraction of the TRIM28 silencing complex to ERV sequences. We validated the increased expression of five ERV-up genes in Trim28−/− cells using qRT-PCR (Figure 4D).
Figure 4. Activation of ERVs Influences Expression of Nearby Genes and Results in the Expression of lncRNAs.
(A) Transcriptional change of genes located close (<50 kb) to ERVs in Trim28−/− NPCs.
(B) Mean transcriptional change of genes located to ERVs with increased transcription (ERV-up genes) and genes located close to unchanged ERVs (ERV-ctrl genes) in Trim28−/− NPCs.
(C) Absolute expression level of ERV-up genes and ERV-ctrl genes in wild-type NPCs.
(D) qRT-PCR of RNA isolated from wild-type and Trim28−/− NPCs.
(E) Screen shot from the USCS genome browser (mm9) showing induced transcription of BC048671 in Trim28−/− NPCs.
(G, I, and K) Activation of ERVs results in the expression of lncRNAs. Screen shot from the USCS genome browser (mm9).
(F, H, and J) qRT-PCR of RNA isolated from wild-type and Trim28−/− NPCs. Primers are indicated as green bars and include primers over the ERV junction as well as close and more distant from the 3′ end of the ERVs.
Data are presented as mean of relative values ± SEM. *p < 0.05, Student’s t test. See also Figures S1 and S2.
ERVs Produce Long Noncoding RNAs
We looked in detail at BC048671, which is a protein-coding transcript that is induced in Trim28−/− NPCs but completely absent in wild-type NPCs. BC048671 is located 5 kb downstream of an IAP element, which is also highly upregulated in Trim28−/− NPCs. The RNA-seq data show that transcriptional initiation at the IAP element results in the formation of a long transcript (>10 kb) that extends into the coding sequence of BC048671 (Figure 4E). The presence of high levels of this long transcript was verified using qRT-PCR primers located both upstream and within the coding sequence of BC048671 (Figure 4F). Thus, readthrough of an ERV-derived transcript into another locus is likely to be one of several mechanisms by which nearby gene expression can be affected (see also Figure S2). This finding supports the notion that a general feature of ERVs might be to act as transcriptional start sites for long noncoding RNAs (lncRNAs). Indeed, when we scrutinized ERV elements located in gene free regions, we found that both IAP and MMERVK10C elements serve as start sites for lncRNAs (Figures 4G and 4I). Using qRT-PCR, we confirmed high-level expression of two ERV-derived lncRNAs in Trim28−/− NPCs (Figures 4H and 4J). The length of the ERV-derived lncRNAs did in many cases extend 25 kb (Figure 4K). These data demonstrate that derepression of ERVs in NPCs can result in the expression of multiple lncRNAs. The functional role of lncRNAs in NPCs remains largely unexplored, but they are thought to play important regulatory roles and have been implicated as scaffolds for nuclear protein complexes and as antisense transcripts in the control of epigenetic pathways (Guttman and Rinn, 2012).
DISCUSSION
In pluripotent stem cells, TRIM28 is a master corepressor of retroelements including ERVs (Matsui et al., 2010; Rowe et al., 2010). When these cells differentiate into various somatic cell types, DNA methylation is instated on ERV sequences, which ultimately results in stable silencing that is no longer dependent on TRIM28 (Wiznerowicz et al., 2007; Rowe et al., 2013). Thus, when TRIM28 is deleted from various somatic cell types such as fibroblasts, hepatocytes, and white blood cells, no increased ERV expression is detected (Rowe et al., 2010; Bojkowska et al., 2012; Santoni de Sio et al., 2012a, 2012b). Here, we describe an exception to this rule. When TRIM28 is deleted in NPCs, several ERVs become highly expressed. This finding unravels a unique transcriptional regulation of ERVs in NPCs.
ERVs regulated by TRIM28 in NPCs are recent invaders of the mouse genome. IAP1 is the most recent member of the well-studied IAP ERVs (Qin et al., 2010). IAPs are ERVs that have lost the env gene and adopted an intracellular life cycle (Ribet et al., 2008). IAP1 has been shown to retrotranspose and has distinct integration patterns in different strains of laboratory mice (Li et al., 2012). MMERVK10C, another ERV massively upregulated in Trim28−/− NPCs, is poorly characterized, and it is unclear if it is still endowed with retrotransposition potential, whether on its own or with the support of factors provided in trans. A previous study that analyzed the structure of MMERVK10C elements in the mouse genome found that the majority of these elements have 3′ deletions removing the start of the gag open reading frame as well as the major part of env (Reichmann et al., 2012). Our data demonstrate that, in NPCs, TRIM28 controls the rare copies of env-containing MMERVK10C elements, which are most likely to be the youngest ones, raising the possibility that these recent invaders of the mouse genome contain cis-acting genomic elements that allow them to escape DNA methylation in NPCs.
The classic view of repetitive mobile genetic elements as parasitic DNA without beneficial function to the host is challenged in many ways. There are a number of recent studies indicating that transposable elements (TEs) play important roles in establishing and rewiring gene networks (Kunarso et al., 2010; Chuong et al., 2013). TEs have been shown to act as enhancers, repressors, and alternative promoters. In addition, TEs can affect splicing patterns and produce peptides with important functional roles (Jern and Coffin, 2008). In this study, we demonstrate that activated ERVs can influence gene expression of nearby genes, such as BC048671, and serve as start sites for lncRNAs. Taken together, our findings indicate that ERVs participate in the control of gene networks in the brain.
We have previously demonstrated that deletion of Trim28 in postmitotic forebrain neurons results in complex behavioral changes (Jakobsson et al., 2008). In addition, heterozygous germline deletion of Trim28 has been described to result in abnormal behavioral phenotypes (Whitelaw et al., 2010). In this study, we found that deletion of Trim28 during brain development is lethal (Figure S3). In addition, we also noted that heterozygous deletion of Trim28 during brain development resulted in behavioral changes characterized by hyperactivity (Figure S3). Together, these findings demonstrate that disruption of TRIM28 levels in the mouse brain results in behavioral changes that are similar to impairments found in humans with certain psychiatric disorders. With this in mind, it is noteworthy that increased levels of ERV transcripts have been detected in patients with several neurological and psychiatric disorders (Jeong et al., 2010; Douville et al., 2011; Li et al., 2012; Karlsson et al., 2001). The significance of these findings has been questioned because the human genome does not appear to harbor ERVs with known retrotransposing capacity (Jern and Coffin, 2008). However, the increasing evidence that derepression of ERVs influence gene networks, including the findings presented here, provides a potential mechanistic explanation for these observations.
In summary, our data suggest that ERVs may be involved in the regulation of gene expression in NPCs and may hereby offer a link between ERVs and brain disorders. It seems unlikely that behavioral phenotypes would arise from the derepression of a single ERV-induced gene. Instead, the presence of ERVs in multiple copies scattered throughout the genome allows for a powerful network-like control of gene expression, where dysregulation could result in widespread consequences. However, due to the large numbers of ERVs present in the mouse and human genome and their sequence variation, it is currently unfeasible to demonstrate a causal role for ERVs in controlling complex behavior or brain disorders using loss-of-function approaches, such as gene targeting and small hairpin RNA (shRNA) knockdown. Instead, improving our knowledge of critical host factors and networks controlling ERVs is essential to appreciate their impact on the genome and pathologies that may stem from their dysregulation. The demonstration that there is an ongoing dynamic TRIM28-mediated regulation of ERVs in NPCs is a step in this direction and warrants future studies of epigenetic and posttranscriptional regulation of ERVs in the healthy and diseased brain.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures can be found in the Supplemental Experimental Procedures.
Procedures
Transgenic Animals
All animal-related procedures were approved by and conducted in accordance with the committee for use of laboratory animals at Lund University. NestinCre and floxed Trim28 mice have been described previously (Weber et al., 2002; Tronche et al., 1999).
Cell Culture
NPC was established from embryonic day 13.5 (E13.5) forebrain and cultured as previously described (Conti et al., 2005).
Immunofluorescence
Immunofluorescence was performed as previously described (Thompson et al., 2005; Sachdeva et al., 2010).
RNA Studies
RNA-seq and qRT-PCR was performed as previously described (Rowe et al., 2010). The 50-base-paired end reads were mapped onto the RepBase version 16.08 (Jurka et al., 2005) and to the mouse genome (mm9) assembly. Mapping was done using the bowtie short read aligner (Langmead et al., 2009).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed with iDeal chromatin immunoprecipitation sequencing (ChIP-seq) kit (Diagenode) according to supplier’s recommendations.
DNA-Methylation Analysis
Bisulfite sequencing was performed with the EpiTect bisulfite kit (QIAGEN) according to the supplier’s recommendations. Sequence data were analyzed with the QUantification tool for Methylation Analysis (Kumaki et al., 2008).
Statistical Analysis
An unpaired t test was performed in order to test for statistical significance. Data are presented as mean ± SEM.
Supplementary Material
Highlights.
Deletion of TRIM28 in NPCs results in transcriptional activation of ERVs
ERVs are marked by H3K9me3 in NPCs, which is lost upon TRIM28 deletion
Activation of ERVs in NPCs influences expression levels of nearby genes
Activation of ERVs in NPCs results in the production of long noncoding RNAs
ACKNOWLEDGMENTS
We are grateful to A. Björklund, S. Quenneville, and all members of the J.J. and Parmar laboratories for stimulating discussions. We thank U. Jarl, A. Josefsson, C. Isaksson, I. Nilsson, A.-K. Oldén, E. Ling, S. Smiljanic, M. Sparrenius, and E. Tjon for technical assistance. This study was supported by grants from Swedish Research Council (J.J.), Formas (P.J.), the Swedish Cancer Foundation (J.J.), the Lundqvist, Jeansson, and Crafoord foundations (J.J.), the Swedish Government Initiative for Strategic Research Areas MultiPark (J.J.), the French government, CNRS and INSERM (F.C.), and the French Agence Nationale pour la Recherche (F.C.).
Footnotes
ACCESSION NUMBERS
The RNA-seq data were deposited in the NCBI Gene Expression Omnibus and are available under accession number GSE45930.
Supplemental Information includes Supplemental Experimental Procedures, three figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.12.004.
REFERENCES
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 2005;79:12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, Santoni de Sio F, Zangger N, Offner S, Cartoni C, Thomas C, Quenneville S, et al. Liver-specific ablation of Krüppel-associated box-associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology. 2012;56:1279–1290. doi: 10.1002/hep.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dollé P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, Smith A. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M, Oakey RJ. Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet. 2013;9:e1003234. doi: 10.1371/journal.pgen.1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Hotta A, Rastegar M. Retrovirus silencing by an epigenetic TRIM. Cell. 2007;131:13–14. doi: 10.1016/j.cell.2007.09.029. [DOI] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J. Biol. Chem. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbel L, Whitelaw E. Endogenous retroviruses in mammals: an emerging picture of how ERVs modify expression of adjacent genes. Bioessays. 2012;34:734–738. doi: 10.1002/bies.201200056. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, Bensadoun JC, Cammas F, Losson R, Mansuy IM, Sandi C, Trono D. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Jeong BH, Lee YJ, Carp RI, Kim YS. The prevalence of human endogenous retroviruses in cerebrospinal fluids from patients with sporadic Creutzfeldt-Jakob disease. J. Clin. Virol. 2010;47:136–142. doi: 10.1016/j.jcv.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Karlsson H, Bachmann S, Schröder J, McArthur J, Torrey EF, Yolken RH. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc. Natl. Acad. Sci. USA. 2001;98:4634–4639. doi: 10.1073/pnas.061021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–5. doi: 10.1093/nar/gkn294. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Akagi K, Hu Y, Trivett AL, Hlynialuk CJ, Swing DA, Volfovsky N, Morgan TC, Golubeva Y, Stephens RM, et al. Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance. Genome Res. 2012;22:870–884. doi: 10.1101/gr.130740.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Prazak L, Chatterjee N, Grüninger S, Krug L, Theodorou D, Dubnau J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013;16:529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Gage FH. The necessary junk: new functions for transposable elements. Hum. Mol. Genet. 2007;16(Spec No. 2):R159–R167. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrat PN, DasGupta S, Wang J, Theurkauf W, Weng Z, Rosbash M, Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science. 2013;340:91–95. doi: 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Wang Z, Shang J, Bekkari K, Liu R, Pacchione S, McNulty KA, Ng A, Barnum JE, Storer RD. Intracisternal A particle genes: Distribution in the mouse genome, active subtypes, and potential roles as species-specific mediators of susceptibility to cancer. Mol. Carcinog. 2010;49:54–67. doi: 10.1002/mc.20576. [DOI] [PubMed] [Google Scholar]
- Reichmann J, Crichton JH, Madej MJ, Taggart M, Gautier P, Garcia-Perez JL, Meehan RR, Adams IR. Microarray analysis of LTR retrotransposon silencing identifies Hdac1 as a regulator of retrotransposon expression in mouse embryonic stem cells. PLoS Comput. Biol. 2012;8:e1002486. doi: 10.1371/journal.pcbi.1002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 2013;33:17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T. An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res. 2008;18:597–609. doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva R, Jönsson ME, Nelander J, Kirkeby A, Guibentif C, Gentner B, Naldini L, Björklund A, Parmar M, Jakobsson J. Tracking differentiating neural progenitors in pluripotent cultures using microRNA-regulated lentiviral vectors. Proc. Natl. Acad. Sci. USA. 2010;107:11602–11607. doi: 10.1073/pnas.1006568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio FR, Barde I, Offner S, Kapopoulou A, Corsinotti A, Bojkowska K, Genolet R, Thomas JH, Luescher IF, Pinschewer D, et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J. 2012a;26:4561–4575. doi: 10.1096/fj.12-206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio FR, Massacand J, Barde I, Offner S, Corsinotti A, Kapopoulou A, Bojkowska K, Dagklis A, Fernandez M, Ghia P, et al. KAP1 regulates gene networks controlling mouse B-lymphoid cell differentiation and function. Blood. 2012b;119:4675–4685. doi: 10.1182/blood-2011-12-401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber GO, Airola T, Jern P, Blomberg J. Automated recognition of retroviral sequences in genomic data—RetroTector. Nucleic Acids Res. 2007;35:4964–4976. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L, Barraud P, Andersson E, Kirik D, Björklund A. Identification of dopaminergic neurons of nigral and ventral tegmental area subtypes in grafts of fetal ventral mesencephalon based on cell morphology, protein expression, and efferent projections. J. Neurosci. 2005;25:6467–6477. doi: 10.1523/JNEUROSCI.1676-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Weber P, Cammas F, Gerard C, Metzger D, Chambon P, Losson R, Mark M. Germ cell expression of the transcriptional co-repressor TIF1beta is required for the maintenance of spermatogenesis in the mouse. Development. 2002;129:2329–2337. doi: 10.1242/dev.129.10.2329. [DOI] [PubMed] [Google Scholar]
- Whitelaw NC, Chong S, Morgan DK, Nestor C, Bruxner TJ, Ashe A, Lambley E, Meehan R, Whitelaw E. Reduced levels of two modifiers of epigenetic gene silencing, Dnmt3a and Trim28, cause increased phenotypic noise. Genome Biol. 2010;11:R111. doi: 10.1186/gb-2010-11-11-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, Beermann F, Aebischer P, Trono D. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J. Biol. Chem. 2007;282:34535–34541. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.