Abstract
New neurons are continuously generated from neural stem cells with astrocyte properties, which reside in close proximity to the ventricle in the postnatal and adult brain. In this study we found that microRNA-124 (miR-124) dictates postnatal neurogenesis in the mouse subventricular zone. Using a transgenic reporter mouse we show that miR-124 expression is initiated in the rapid amplifying progenitors and remains expressed in the resulting neurons. When we stably inhibited miR-124 in vivo, neurogenesis was blocked, leading to the appearance of ectopic cells with astrocyte characteristics in the olfactory bulb. Conversely, when we overexpressed miR-124, neural stem cells were not maintained in the subventricular zone and neurogenesis was lost. In summary, our results demonstrate that miR-124 is a neuronal fate determinant in the subventricular zone.
Introduction
In the postnatal mammalian brain, neural stem cells (NSCs) reside in the subventricular zone (SVZ) (Doetsch et al., 1999). These cells generate new neurons that migrate via the rostral migratory stream (RMS) to the olfactory bulb (OB). The NSCs, often termed type B cells, are of astrocytic lineage expressing glial fibrillary acidic protein (GFAP) and have the morphology and ultrastructural properties of astrocytes (Doetsch et al., 1999; Laywell et al., 2000; Skogh et al., 2001; Imura et al., 2003; Merkle et al., 2004). Type B astrocytes undergo asymmetric cell division to generate rapid amplifying progenitors (type C cells) that subsequently differentiate into neurons. This cellular transformation is accompanied by transcriptional changes for which the underlying molecular mechanisms remain largely unknown.
MicroRNAs (miRNAs) are small, noncoding, endogenous RNA that bind to and regulate mRNA targets, making them central players of gene regulation (Bartel, 2009). In neurons, miR-124 is one of the most abundant miRNAs (Lagos-Quintana et al., 2002). The expression of microRNA-124 (miR-124) is initiated upon neural differentiation and reaches high levels in mature neurons (Cao et al., 2007; Makeyev et al., 2007; Visvanathan et al., 2007; Cheng et al., 2009; Sanuki et al., 2011). Several in vitro studies suggest that miR-124 plays a role in neuronal fate determination. For example, delivery of miR-124 duplexes to HeLa cells induces a neuronal gene profile (Lim et al., 2005), and forced expression of miR-124 in progenitor cells causes neuronal differentiation (Visvanathan et al., 2007; Yu et al., 2008).
Nevertheless, despite the observed upregulation of miR-124 during NSC neuronal differentiation (Cheng et al., 2009), in vivo blockade of this miRNA by antisense oligonucleotides only resulted in subtle phenotypes, with mild delays in differentiation and no detectable malformation (Cao et al., 2007; Visvanathan et al., 2007; Cheng et al., 2009). Based on these findings, it was concluded, that miR-124 regulates proliferation but does not affect the glia/neuronal fate choice (Cheng et al., 2009). However, these results were obtained using a technique, in vivo infusions of antisense molecules, which only induced a transient inhibition. In addition, targeting of specific cell types, such as type B astrocytes, could not be ascertained since the antisense molecules were not coupled to a reporter.
Here, to study the role of miR-124 in the postnatal SVZ, we generated a transgenic reporter mouse that allows visualization of miR-124 activity in the brain in vivo. We demonstrate that in the SVZ stem cell niche, miR-124 activity is initiated already in transient amplifying progenitors, type C cells, suggesting that it is not linked to cell cycle exit. This finding was substantiated with loss of function studies of miR-124 in the SVZ, demonstrating that stable inhibition of this miRNA does not force progenitors to remain cycling. miR-124 loss of function, rather, blocks adult neurogenesis and induces the formation of ectopic astrocytes in the OB derived from the SVZ. Furthermore, when we overexpressed miR-124 in the SVZ, we found that NSCs lost their stem cell characteristics, resulting in a loss of continuous supply of new neurons to the OB.
Materials and Methods
Viral vectors.
The lentiviral vectors used in this study were third-generation self-inactivating vectors. miRNA target sequences were cloned into the 3′ untranslated region (UTR) of the transgene expression cassette as follows: four tandem repeats of a target sequence perfectly complementary to miR-124 (TTAAGGCACGCGGTGAATGCCA) were synthesized starting from the following 5′ phosphorylated, SDS-PAGE purified DNA oligonucleotides: sense1: CTAGATAATGGCATTCACCGCGTGCCTTAATTCGAATGGCATTCACCGCGTGCCTTAAACGCGT; sense2: TGGCATTCACCGCGTGCCTTAAATGCATTGGCATTCACCGCGTGCCTTAAC; antisense1: TTAAGGCACGCGGTGAATGCCATTCGAATTAAGGCACGCGGTGAATGCCATTAT; antisense2: CCGGGTTAAGGCACGCGGTGAATGCCAATGCATTTAAGGCACGCGGTGAATGCCAACGCGT. Sense1 and antisense1, as well as sense2 and antisense2, were hybridized by slow cooling from 95°C down to 37°C, respectively. The resulting double-stranded DNA oligonucleotides could anneal to each other by an ACGCGT sticky end, producing the 119 bp long fragment containing four miR-124 targets with a 5′ CTAG and a 3′ GGCC overhang allowing insertion into the XbaI-XmaI site, contained immediately downstream the woodchuck hepatitis virus posttranscriptional response element (WPRE) of a third-generation lentiviral vector, expressing green fluorescent protein (GFP) from the phosphoglycerate kinase (PGK) promoter derived from the phosphoglycerol kinase housekeeping gene. This promoter results in robust, ubiquitous, moderate-level transgene expression, with very little tendency to transgene silencing/variegation.
Similarly, eight tandem repeats of an imperfectly complementary sequence forming a central bulge when binding to miR-124 (hereafter referred to as “sponge”) were synthesized and cloned into a third-generation lentiviral vector containing a strong promoter derived from cytomegalovirus (CMV), which drives overexpression of the miR-124 sponge, a requirement for successful miR-124 knockdown. The miR-124 sponge sequence is as follows: TTAAGGCACG-TA-TGAATGCCA. The miR-125b sponge sequence is as follows: TCACAAGTTTAGTCTCAGGGA. A detailed description of how to design and use lentiviral miRNA reporter and sponge vectors has been described previously (Brown et al., 2007; Gentner et al., 2009).
A miR-124–1 overexpression vector (referred to as miR-124OE vector) was obtained by PCR amplification of a 484 bp (miR sequence 84 bp, 200 bp flanking on either side) genomic fragment from mouse DNA. Primers were as follows: forward, ATGAATTCTCGCCAGCTTTTTCTTTCTC; reverse, ATGAATTCATTTGCATCTGCACAAACCC. This fragment was subsequently cloned between the GFP and the WPRE sequence in pFUGW (Lois et al., 2002). The fragment encoding a human miR-125b was amplified from human DNA. Primers were as follows: forward, ATTCTAGAGTTGCGCTCCCCTCAGTC; reverse, ATGGTACCGCAGCTCCCAAGAGCCTAAC. All cloning was performed using standard techniques.
Lentiviral vectors were produced as previously described (Zufferey et al., 1997). Lentiviral vectors were titrated using flow cytometry and quantitative PCR analysis as previously described (Georgievska et al., 2004). The titers of the vectors in this study were in the range of 2 × 108 – 1 × 109 TU/ml.
To validate the miR-124OE-vector, 50,000 HeLa cells were plated in 12 well plates. Two hours after plating, cells were transduced with volumes of 0.3, 0.5, 1, 2, or 5 μl of concentrated miR-124OE-vector. RNA was extracted 72 h after transduction, followed by locked nucleic acid (LNA) qRT-PCR. The titer of this vector batch was 2 × 108.
For the sponge validation experiments, HeLa cells were transduced at a multiplicity of infection (MOI) of 10 for the miR-124 sponge vector and an MOI of 5 for the miR-124OE vector. RNA was extracted 72 h after transduction followed by qRT-PCR. Extraction of RNA, generation of cDNA, and qRT-PCR were performed as previously described (Sachdeva et al., 2010). Primer sequences were as follows: Evovl1: forward, CCCCGGGAGGAATGGGCTCTT, reverse, TAGGGTTGTGCCACAGGGCCA; Jag1: forward, AGGCCACGTGCAACAACGGT, reverse, AGGGGTTGGGCAGGCAGCTA. An unpaired t test was performed to test for statistical significance. Data are presented as mean ± SEM.
Luciferase reporter assay.
Sequences incorporating the putative miR-124 binding sites of Jag1 3′UTR were amplified from mouse genomic DNA and cloned in the dual luciferase reporter vector pSICHECK-2 (Promega). Primers were as follows: Jag1utr forward, CTCGAGCGCTCTCACAGCTATGCAAAA; Jag1utr reverse, GCGGCCGCTGGCTTACAGGCAACACGTTA. The luciferase reporter construct was cotransfected with the lentiviral miR-124 overexpression construct and the miR-124 sponge construct into HeLa cells using Turbofect (Fermentas). Forty-eight hours after transfection, cells were assayed for Luciferase activity using a dual-luciferase assay (Promega). An unpaired t test was performed to test for statistical significance. Data are presented as mean ± SEM.
Animals.
All animal-related procedures were approved by and conducted in accordance with the committee for the use of laboratory animals at Lund University and École Polytechnique Fédérale de Lausanne.
Lentiviral transgenesis was performed as previously described (Sauvain et al., 2008). Offspring were genotyped, and number of integrated transgenes was estimated using quantitative real-time PCR. Primers were as follows: WPRE: forward, GGCACTGACAATTCCGTGGT; reverse, AGGGACGTAGCAGAAGGACG; probe WPRE: ACGTCCTTTCCATGGCTGCTCGC; Titine forward, AAAACGAGCAGTGACGTGAGC; reverse, TTCAGTCATGCTGCTAGCGC; probe Titine: TCGACGGAAGCGTCTCGTCTCAGTC. The data were quantified using the ΔΔCt-method. The transgene data (WPRE primer) was normalized with the Titine data to give the number of transgenes in the genome of each animal.
For vector injections, 1 μl of concentrated lentiviral vector was unilaterally injected into the right ventricles of NMRI mice at postnatal day (P) 3 (males and females; Charles River). The pups were placed on ice for 5 min before positioning them in an ice-cold stereotaxic frame. Coordinates from bregma were as follows: AP + 0.8, ML − 0.8, DV − 1.9. Mice were perfused 1, 2, 3, 4, or 8 weeks postinjection. A proportion of the animals were given an intraperitoneal pulse of 50 mg/kg BrdU (Sigma) 2 h before the 4 week time point. We used similar titers for all experiments. We found similar results when analyzing either the ipsilateral or the contralateral hemisphere after vector injection.
Immunofluorescence.
Mice were transcardially perfused with 4% paraformaldehyde (PFA) (Sigma), and the brains were postfixed for 2 h and transferred to PBS with 25% sucrose. Brains were coronally sectioned on a microtome (35 μm) and put in potassium PBS. Embryonic brains were fixed in 4% PFA overnight followed by PBS with 15% sucrose and sectioned coronally on a cryostat (14 μm).
Similar procedures was used for both fixed sections and cells, as published in detail previously (Thompson et al., 2005; Sachdeva et al., 2010). Primary antibodies were diluted as follows: chicken anti-GFP 1:1000 (Abcam), rabbit anti-GFAP 1:1000 (DAKO), goat anti-DCX 1:400 (Santa Cruz Biotechnology), mouse anti-NeuN 1:1000 (Millipore), mouse anti-S100β 1:500 (Sigma), rabbit anti-IBA1 1:1000 (Wako Chemicals), mouse anti-MASH1 1:200 (BD Biosciences), mouse anti-O4 1:100 (Millipore), and rabbit anti-PERIPHERIN 1:200 (Covance). The dilution factor of the secondary antibodies was 1:500 (Invitrogen) or 1:200 (Jackson Laboratories).
For BrdU staining, the slices were fixed for 20 min in 4% PFA followed by incubation at 65°C in 1 M HCl before addition of the primary antibody (1:500 rat anti-BrdU; Serotec).
Quantification of GFP expressing cells in the OB.
Three representative OB sections (35 μm) from each of three control injected, three sponge injected, and three overexpressing injected animals were selected for quantification. Fluorescent images of GFP expression were taken with the same exposure, and all GFP-expressing cells of the sections were counted. The OB was divided into different cell layers, including the granule cell layer (GCL), mitral cell layer, internal plexiform layer, external plexiform layer, and glomerular layer based on DAPI staining. An unpaired t test was performed to test for statistical significance. Data are presented as mean ± SEM.
Neurosphere cultures.
Generation and culturing of embryonic day (E) 13.5 and adult neurospheres were performed as previously described with minor modifications (Ahlenius and Kokaia, 2010). In brief, half the forebrain from each miR-124.T or GFP control E13.5 fetus was dissected and placed in DMEM/F12 basic medium with trypsin and DNase. The tissue was mechanically suspended by pipetting followed by incubation in 37°C for 30 min. Cells were plated out in DMEM/F12 supplemented with 20 ng/ml EGF and 10 ng/ml bFGF at a density of 100,000 cells/ml and were cultured in a humidified incubator at 37°C with 5% CO2. Neurosphere cultures were passaged using mechanical dissociation approximately every 7 d.
For adult neurospheres, 8 week old wt NMRI mice or 4.3 week old NMRI mice injected with CMV.1000 in the ventricle at P3 were used. The mice were lethally anesthetized, and each brain was removed from the skull. The brain was put in ice-cold L15 media and transferred to a coronal brain matrix, and 1 mm thick sections were cut. From those, the SVZ region was dissected out under a dissection light microscope. SVZ from four different animals was pooled in L15 medium. The tissue was dissociated in HBSS with HEPES, glucose, trypsin, DNase, hyaluronidase, and kynurenic acid in 37°C for 30 min, followed by titration by pipetting. Adult neurospheres were cultured in Neurobasal basic medium supplemented with EGF, bFGF, and heparin. Cultures were passaged every 7 d using Accutase (Sigma) and mechanical dissociation. FACS analysis of dissociated neurospheres was performed at passages 1–2, as previously described (Jonsson et al., 2009).
Fluorescence in situ hybridization.
LNA-FISH was performed as previously described with minor modifications (Silahtaroglu, 2010). In brief, fresh frozen brain tissue was cut at 14 μm on a cryostat on charged slides and kept at −20°C until use. Upon staining, slides were dried and fixed with 4% PFA for 10 min. The slides were hybridized with a DIG-labeled miR-124 LNA-probe (Exiqon) diluted 1:200 for 1 h at 67°C. Following hybridization, slides were washed in 0.1× SSC at 75°C followed by treatment with 3% H2O2. The slides were then incubated with an anti-DIG-antibody (1:100; Roche) overnight and developed using Cy3-tyramide (PerkinElmer) for 2 h. Slides were counterstained with DAPI and coverslipped with polyvinyl alcohol mounting medium with DABCO (Sigma).
LNA-qRT-PCR.
Total RNA, including miRNA, was extracted from dissociated neurospheres using the miRNeasy kit (Qiagen) followed by Universal cDNA synthesis kit (Exiqon). LNA–PCR primer sets, hsa-miR-124 and hsa-miR-103 (normalization miRNA), were purchased from Exiqon and were used together with LightCycler 480 SYBR Green I Master (Roche). Standard procedures of qRT-PCR were used, and data were quantified using the ΔΔCt-method.
Results
Generation of miR-124 sensor transgenic mice
Studies of miRNA expression patterns in the brain in histological preparations have up to now mostly relied on ISH techniques (Pena et al., 2009). However, ISH requires high stringency, and it is often incompatible with other labeling techniques such as immunohistochemistry. In addition, discriminating between the pre-miRNA and mature miRNA is difficult through ISH, making it complicated to detect potential posttranscriptional regulation, which is thought to play a prominent role in controlling the activity of miRNA (Obernosterer et al., 2006). To circumvent these issues, we decided to generate a transgenic sensor mouse that can report the activity of miR-124.
Lentiviral vectors expressing a reporter gene regulated by four copies of a perfectly matched miRNA target sequence can be exploited to monitor miRNA activity (Brown et al., 2007; Sachdeva et al., 2010). For this study we used a lentiviral vector containing a GFP reporter gene followed by four copies of perfect matching miR-124-target sequences (miR-124.T) (Fig. 1A). When miR-124 is present in the cell, it binds the miR target sequence in the vector-derived mRNA and efficiently inhibits GFP expression. In cells in which miR-124 is not expressed, the mRNA remains active, and GFP expression from the ubiquitous PGK promoter is easily detected (Fig. 1B). We used the miR-124.T-vector to generate transgenic mice through lentiviral transgenesis (Lois et al., 2002; Sauvain et al., 2008). High-concentration lentiviral vectors were injected into the perivitelline space of fertilized single-cell embryos followed by transfer to surrogate mothers (Fig. 1A). We generated mice with either the miR-124.T-containing or control GFP vectors (without miRNA regulatory sites). Noteworthy, the simplicity and high efficiency of lentiviral transgenesis allowed us to generate several founders that each carried multiple copies of the transgenes. We then analyzed offspring from six founders for each construct, all carrying 5–15 copies of the vector, and found no difference in GFP expression patterns among lines. Furthermore, we performed intercrosses between founders that, in theory, further reduce the risk for position-dependent effects, and when analyzing such animals we found the same expression pattern as with F0 generation mice.
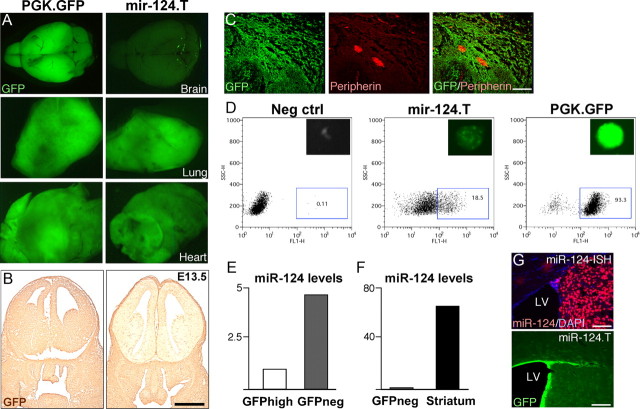
Figure 1.
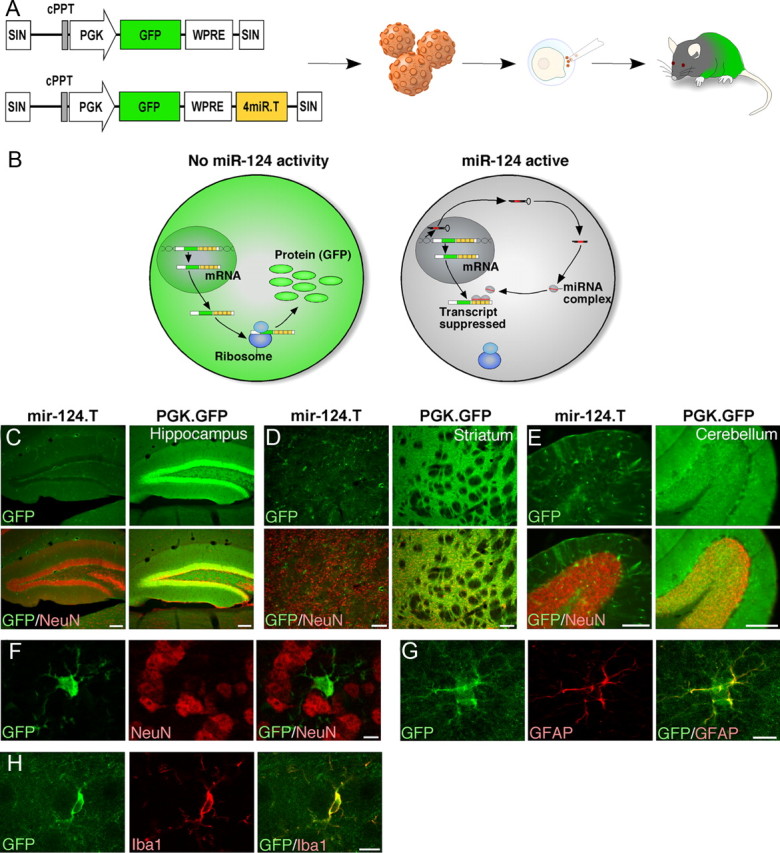
miR-124 is a neuron-specific miRNA. A, Illustration of the vectors and experimental approach to generate miR-124.T reporter mice using lentiviral transgenesis. Lentiviral vectors were injected into the perivitelline space of fertilized embryos, which were transplanted into a pseudopregnant mouse. B, The vector-encoded transcript is expressed in both miR-124 expressing cells and nonexpressing cells. In the miR-124-expressing cells, the target site containing transcripts is suppressed, resulting in no GFP expression. C–E, GFP expression in miR-124.T reporter mice was found exclusively in nonneuronal cells throughout the brain. Control mice (PGK.GFP) displayed GFP activity in all cells of the brain. Scale bars, 100 μm. F–H, Confocal analysis of GFP cells in miR-124.T mice showing that neurons (NeuN) do not express GFP while astrocytes (GFAP) and microglia (IBA1) do. Scale bars, 10 μm. cPPT, Central polypurine tract; SIN, self-inactivating.
miR-124 activity is detected in all neuronal populations in the adult brain but not in glia
Histological examination of adult brains from miR-124.T mice revealed that all neuronal populations throughout the anterior/posterior axis lack GFP expression (exemplified in Fig. 1C–E by low-magnification images from hippocampus, striatum, and cerebellum). Using double-labeled immunohistochemistry for GFP and the neuronal marker NeuN, we found no evidence for GFP-expressing neurons in any brain region examined (Fig. 1C–F). In contrast, GFP control mice showed abundant GFP expression in NeuN-positive cells (Fig. 1C–E). The GFP-expressing cells found in the brains of the miR-124.T mice had a nonneuronal morphology resembling different glial cell types. Colabeling with GFAP and IBA1 confirmed that astrocytes and microglia do not display miR-124 activity (Fig. 1G,H).
We analyzed grossly dissected organs from the miR-124.T mice and found that the brain was mostly devoid of GFP expression when compared with control GFP mice (Fig. 2A). In other organs such as lung and heart there was no major difference in GFP expression (Fig. 2A). FACS analysis of white blood cells from miR-124.T mice showed similar GFP expression levels as GFP control mice (data not shown). In addition, we analyzed GFP expression in embryos at E13.5 and found that miR.124.T embryos displayed a lack of GFP expression exclusively in the developing brain (Fig. 2B). We also used miR.124.T embryos to show that miR-124 is not active in the peripheral nervous system, as demonstrated by GFP expression in PERIPHERIN-expressing ganglion (Fig. 2C). Together, these data provide evidence that miR-124 is a CNS neuron-specific miRNA.
Figure 2.
Characterization of miR.124.T mice. A, Adult miR.124.T mice display low GFP expression in the brain, while other tissues, here exemplified by lung and heart, display similar GFP levels as control animals. B, The developing brain is almost entirely depleted of miR-124 activity at E13.5, as shown by diaminobenzidine-staining for GFP. Scale bar, 1 mm. C, Confocal analysis of E13.5 peripheral ganglion reveals GFP expression in peripheral nervous system neurons labeled with PERIPHERIN. Scale bars, 100 μm. D, Generation of neurospheres from forebrain of E.13.5 miR.124.T embryos. FACS plots showing that in miR.124.T cultures, ∼20% of the cells express GFP at early passages. E, F, FACS sorting of the GFP-expressing cells followed by LNA-qRT-PCR for mature miR-124 reveals that mature miR-124 levels are higher in the GFP negative fraction. G, LNA-ISH for miR-124 reveals high-level expression in neurons (top). There is also an absence of miR-124 expression in the ependymal layer that mirrors the expression pattern in miR-124.T mice (bottom). LV, lateral ventricle. Scale bars, 100 μm. Neg, Negative.
To validate that the miR-124.T mice actually report miR-124 activity, we first generated neurosphere cultures from the forebrains of E13.5 embryos. These neurosphere cultures displayed high-level GFP expression only in a proportion of the cells, corresponding to approximately 20% of the total number of cells in the sphere (Fig. 2D). We FACS sorted GFP-expressing and GFP-negative cells from the neurospheres and performed LNA-qRT-PCR for miR-124 levels on RNA extracted from the sorted populations. As expected, we found miR-124 expression only in the GFP-negative population (Fig. 2E). It is worth noting that the levels of miR-124 in the GFP-negative population of neurospheres were approximately 60-fold lower than in adult mouse brain tissue, suggesting that the miR-124.T-reporter system is very sensitive (Fig. 2F).
We compared the GFP staining of miR-124.T mice with fluorescent LNA-ISH for miR-124. miR-124-LNA-ISH results in a high signal that appears to be present primarily in neurons, thus supporting that the miR-124 reporter mice display an inverse picture of miR-124 activity (Fig. 2G). However, with LNA-ISH it was difficult to determine whether cell populations expressing lower levels of miR-124 exist. A faint miR-124 signal appeared in the SVZ that could correspond to progenitors (Fig. 2F). Careful phenotypic identification of cells in the SVZ was again impossible due to technical difficulties in performing colabeling for several markers, including, for example, BrdU-staining (data not shown).
With this in mind, lentiviral reporter mice expressing GFP incorporating miRNA target sites provide a valuable tool to monitor miRNA activity in the brain.
miR-124 activity is initiated in neurogenic niches in the adult brain
We then focused our analysis on the SVZ stem cell niche in the adult brain. In the SVZ, we detected GFP-expressing cells (i.e., not expressing miR-124) lining the lateral ventricle (Fig. 3A), and colabeling with S100β confirmed that ependymal cells express GFP, thus not exhibiting miR-124 activity (Fig. 3B). To detect proliferating cells, we injected BrdU 2 h before killing the mice. Only rare GFP/BrdU-positive cells could be found, which were in close contact with the ventricular wall, while the majority (estimated to be >95%) of BrdU-positive cells did not express GFP (Fig. 3C). BrdU primarily labels the rapid amplifying progenitors type C, while only a small proportion of BrdU-labeled cells are type B.
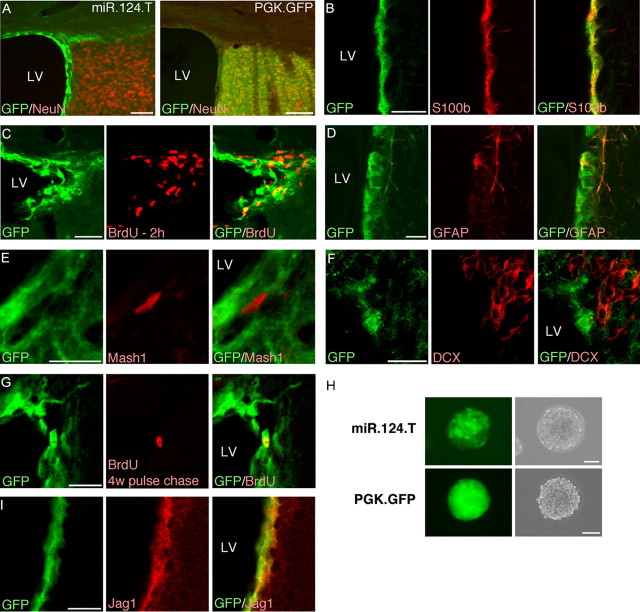
Figure 3.
miR-124 activity is absent from stem cells but initiated in rapid-amplifying progenitors in the adult SVZ. A, GFP/NeuN expression in the adult SVZ in miR-124 reporter mice demonstrates that miR-124 is absent from parts of the niche. Scale bars, 100 μm. B–D, Cells that express GFP, thus lacking miR-124, colabel with S100β and GFAP but only rarely incorporate BrdU in the SVZ. Scale bars, 20 μm. E, F, GFP-expressing cells do not colabel with markers for rapid-amplifying progenitors (MASH1) and migrating neuroblasts (DCX) in miR-124 reporter mice. Scale bars, 20 μm. G, BrdU pulse chase experiment in which the thymidine analog was injected 4 weeks before killing of the animals demonstrates that slowly dividing cells in the SVZ express GFP in miR-124.T mice. Scale bars, 20 μm. H, GFP expression in neurosphere cultures derived from the adult SVZ revealing a small population of green cells in spheres. Scale bar, 100 μm. I, GFP-expressing cells in the miR-124.T mice express the miR-124 target JAG1. Scale bars, 20 μm. LV, Lateral ventricle; w, weeks.
This concept was further confirmed by the finding that GFAP-expressing cells in the SVZ coexpressed GFP while both MASH1-expressing cells, which identifies type C cells, and doublecortin (DCX)-expressing cells, which correspond to early migrating neuroblasts, type A cells, were GFP negative (Fig. 3D–F). Moreover, we performed a pulse-chase experiment in which BrdU was injected 4 weeks before the animals were sacrificed. In these animals we found that the majority of the rare remaining BrdU-cells in the SVZ, corresponding mostly to slowly proliferating stem cell-like cells, were GFP positive (Fig. 3G). In contrast, the BrdU-positive cells in the OB that correspond to adult generated neurons were GFP negative (data not shown). We also derived neurosphere cultures from the adult SVZ of miR-124.T mice. These neurospheres had a spotty appearance, with GFP-expressing cells intermingled with non-GFP-expressing cells (Fig. 3H). One percent to 2% of the neurosphere in early passage culture consisted of GFP-expressing cells, resembling the number of expected self-renewing cells in neurosphere cultures (data not shown). Finally, we investigated the expression pattern of JAGGED1 (JAG1), which is a target gene of miR-124 (Cheng et al., 2009; Liu et al., 2011). In our miR-124 reporter mice, we detected JAG1 expression in cells lining the ventricle and in the SVZ. These cells colabeled with GFP, confirming that miR-124 activity and JAG1 expression do not overlap, providing in vivo evidence of miR-124 suppression of Jag1 in the SVZ niche (Fig. 3I).
When we analyzed miR-124 activity in the dentate gyrus (DG) of the hippocampus, which is the second neurogenic niche in the adult brain, we found that GFP-expressing cells had an elaborate morphology with a prominent radial process resembling what has previously been described for neural progenitor cells in the DG. A proportion of BrdU cells were GFP positive, while neuroblasts expressing DCX did not express GFP. This suggests also that in the DG, miR-124 activity is rapidly initiated once the cell exits the self-renewing state (Fig. 4A,B).
Figure 4.
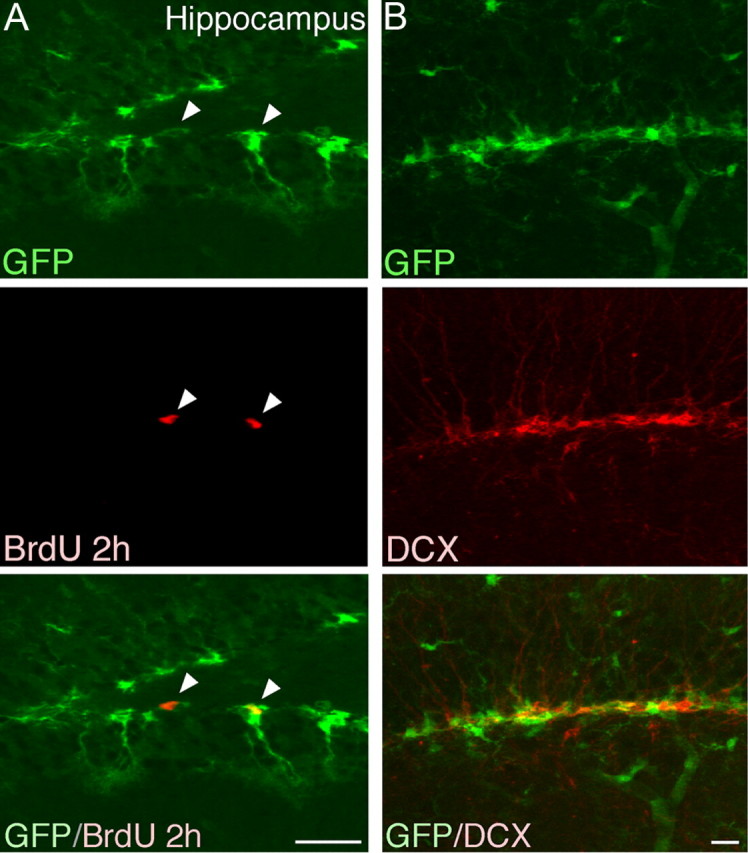
GFP expression in hippocampus in adult brain. A, In the DG we found GFP-expressing cells with morphologies resembling what have previously been attributed to progenitor cells. Confocal analysis revealed that a proportion of the GFP-expressing cells incorporated BrdU. It is worth noting that the proportion of BrdU-incorporating cells that expressed GFP was higher in the DG when compared with the SVZ, which reflects differences in the proliferation of progenitors in the two niches. B, GFP-expressing cells did not colabel with DCX. Scale bars, 20 μm.
Together, these findings demonstrate that miR-124 activity is absent from the actual NSC populations in the adult brain but is initiated as soon as the cell is determined to become a neuron. An interesting aspect is that in the SVZ this corresponds to type C cells that rapidly undergo several cell cycles. Thus, miR-124 initiation does not correlate with cell cycle exit.
Inhibition of miR-124 activity impairs neurogenesis
Having found that miR-124 is absent from the GFAP-expressing and BrdU-label retaining stem cells but is upregulated in rapid amplifying progenitors, we hypothesized that miR-124 plays a critical role in fate determination during SVZ neurogenesis. We investigated this by stably inhibiting miR-124 in the SVZ stem cell niche using a lentiviral sponge strategy.
To achieve efficient targeting of the SVZ niche, including all relevant cell types, we injected lentiviral vectors into the lateral ventricles of mice at P3. At this time point, the ependymal layer is not fully developed and the vector can therefore efficiently transduce all cell types in the niche. Using a control CMV.GFP vector, without miRNA sponge sequences, we found that injection of lentiviral vectors into the lateral ventricles of P3 mice allows efficient targeting of the SVZ stem cell niche. At 1 week postinjection, cells lining the ventricle expressed a high level of GFP (Fig. 5A). At 4 and 8 weeks after injection, we found that there were still numerous GFP-expressing cells in the ventricular zone, and colabeling demonstrated that ependymal cells (s100β) and astrocyte-like cells (GFAP) were expressing GFP (Fig. 5B–D). Eight weeks after vector injection, there were GFP-expressing cells in the RMS coexpressing DCX, demonstrating a continuous supply of cells from the SVZ (Fig. 5E). In the OB there was an increasing number of GFP-expressing interneurons that with time colabeled with NeuN (Fig. 5F). To further confirm stable genetic modification of SVZ stem cells, we performed a pulse chase experiment in which animals were injected with BrdU 4 weeks after vector injection and then sacrificed 4 weeks later, at 8 weeks of age. GFP-expressing neurons in the OB colabeled with BrdU (Fig. 5G), and to estimate the proportion of transduced type B cells, we calculated the number of GFP/BrdU-expressing cells in the OB to be 21 ± 6% (n = 4). Together, this demonstrates that injection of lentiviral vectors into the lateral ventricle of P3 mice allows efficient targeting of the SVZ stem cell niche, including the slowly dividing GFAP-expressing type B stem cells.
Figure 5.
Injection of lentiviral vectors into the ventricles of p3 mice allows efficient targeting of SVZ stem cells. A, B, We found abundant GFP expression in the ventricular zone at both 1 week and 8 weeks after vector injection. Scale bars, 100 μm. C, D, Eight weeks after injection we found GFP-expressing cells colabeling with either GFAP or S100β in the SVZ. Scale bars, 50 μm. E, In the RMS we found GFP-expressing migrating neuroblasts (DCX) 8 weeks after vector injection demonstrating an ongoing migration of GFP-expressing progenitors. Scale bars, 100 μm. F, In the OB we found numerous GFP-expressing neurons that colabeled with the neuronal marker NeuN 8 weeks after vector injection. Scale bars, 100 μm. G, We also performed a pulse chase experiment in which animals were injected with BrdU 4 weeks after vector injection and then killed 4 weeks later, at 8 weeks of age. We found GFP-expressing neurons in the OB that colabeled with BrdU. This demonstrates that cells dividing 4 weeks after vector injection still retain the GFP transgene, showing that slowly dividing stem cells are targeted. Scale bar, 10 μm. w, Weeks.
To inhibit miR-124 we used a lentiviral sponge vector expressing eight copies of an imperfect miR-124 target sequence driven by a strong CMV promoter (Fig. 6A), a design that has been optimized to inhibit miRNA activity in vivo (Gentner et al., 2009). The vector contains a GFP reporter gene that is only detectably expressed when miR-124 is inhibited or in cells lacking miR-124, allowing for simple detection of sponged cells (Gentner et al., 2009). We validated the functionality of the miR-124 sponge construct by demonstrating its ability to de-repress transcripts controlled by miR-124 using a luciferase reporter assay as well as by performing qRT-PCR for endogenous target genes (Fig. 6B,C). When we inhibited miR-124 expression in the SVZ using miR-124 sponge vectors, we found that transduced GFP sponge-expressing cells displayed reduced OB neurogenesis 4 weeks after vector injection (n = 7). In the OB there were very few GFP sponge-expressing neurons when compared with CMV.GFP-injected animals, and all these neurons expressed a low level of GFP (Fig. 6D,E). These low GFP-expressing neurons most likely correspond to cells in which the sponge is expressed at low levels due to low copy number, resulting in insufficient miRNA inhibition. However, even after amplification of the GFP signal using immunohistochemistry, these cells corresponded to <10% of GFP cells when compared with control inject animals. In the SVZ we found numerous GFP-expressing cells lining the ventricle (Fig. 6F). In the RMS we could not find any DCX-GFP-expressing cells (Fig. 6G). Careful examination of transduced cells in the SVZ and RMS revealed that they were expressing either S100β or GFAP but never DCX (Fig. 6H,I). When we pulsed with BrdU 2 h before sacrifice, we only detected very few GFP-expressing cells that were BrdU positive in the SVZ (estimated to be <1%; Fig. 5J). These data suggest that miR-124 inhibition affects the transition from type B stem cells into rapid amplifying progenitors, type C cells, and subsequent neuronal differentiation. It is worth noting that ependymal cells (not expressing miR-124) expressed high levels of the sponge (GFP) at all time points investigated (1, 4, and 8 weeks) and displayed a normal phenotype throughout, indicating that there is no toxicity associated with the miR-124 sponge per se.
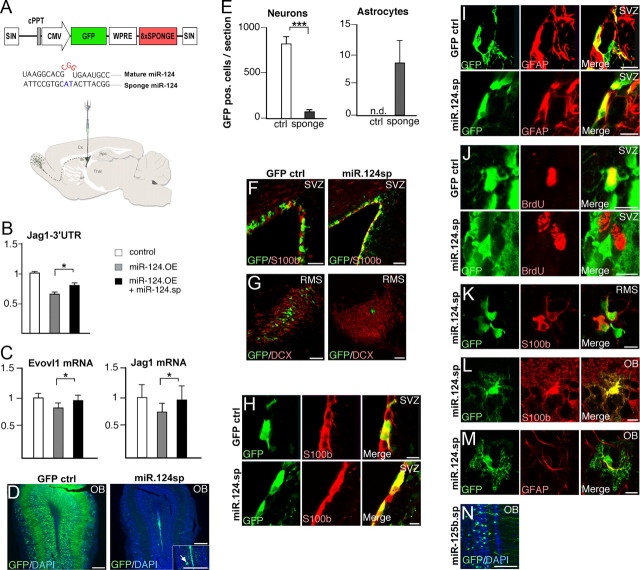
Figure 6.
Stable inhibition of miR-124 in the SVZ blocks neurogenesis, leading to the formation of ectopic glial cells in the OB. A, Illustration of the vectors and experimental approach to inhibit miR-124 in the postnatal SVZ. Lentiviral sponge vectors were injected at P3. Illustration shows the migratory route of neuroblasts from the SVZ via the RMS to the OB. B, Luciferase assay in HeLa cells for the miR-124 target gene Jag1 demonstrating the efficiency of the lentiviral constructs. Graph display ratio of Renilla to firefly luciferase activity normalized to cells transfected with the Jag1–3′UTR construct only. Data represent a mean of four independent experiments. Data are presented as mean ± SEM. *p < 0.05, Student's t test. C, Examples of two miR-124 target genes (Evovl1 and Jag1) that display reduced expression in HeLa cells following miR-124 overexpression (black bars). Coexpression of miR-124 sponge rescues the downregulation. Data are presented as mean ± SEM; n = 6. *p < 0.05, Student's t test. Evovl1 is taken from Conaco et al. (2006) and Jag1 from Cheng et al. (2009). D, GFP expression in the OB 4 weeks after control and sponge vector injection. In animals injected with miR-124 sponge vectors, only very few GFP-expressing cells can be detected. OB is counterstained with DAPI. Scale bars, 250 μm. E, Quantification of GFP-expressing neurons and astrocytes in the OB after amplification of the GFP signal using immunohistochemistry. Data are presented as mean ± SEM; n = 3. ***p < 0.001. n.d., not detected. F, G, GFP expression in the SVZ and RMS 4 weeks after vector injection. SVZ is counterstained with s100β and the RMS with DCX. Scale bars: 50 μm (F); 75 μm (G). H–J, GFP-expressing cells in the SVZ 4 weeks after injection of ctrl or miR-124 sponge vector. In animals injected with sponge vector, it was possible to detect GFP cells expressing s100β and GFAP, but these GFP cells did not incorporate BrdU. BrdU was injected 2 h before kill. Scale bars, 10 μm. K, GFP-expressing cells in the RMS 4 weeks after injection of sponge vector colabeled with s100β. Scale bars, 10 μm. L, M, GFP-expressing cells in the OB 4 weeks after sponge injection colabeled with S100β and GFAP. Scale bars, 10 μm. N, GFP-expressing neurons in the OB 4 weeks after injection of a control lentiviral vector expressing a miR-125b sponge sequence. Scale bars, 100 μm. cPPT, Central polypurine tract; SIN, self-inactivating; w, weeks.
Interestingly, we found GFP-expressing cells in the RMS with a glial-like morphology that colabeled with S100β 4 weeks after miR-124 sponge injection (Fig. 6K). Adding to this, we detected cells with an astrocyte-like morphology in the OB expressing GFP. These cells colabeled with both GFAP and S100β (Fig. 6L,M). The number of GFP-expressing astrocytes corresponded to ∼10 cells/section and could be found in all cell layers of the OB (Fig. 6F). In animals injected with the CMV.GFP vector or a sponge vector targeting the unrelated miR-125b, we never detected GFP/S100β- or GFP/GFAP-expressing cells in the RMS or the OB (Fig. 6D,E,N).
Together, these results show that miR-124 inhibition reduces neurogenesis and increases gliogenesis, leading to the formation of ectopic glial cells in the OB. This demonstrates that miR-124 plays a critical role in regulating the molecular machinery that governs the cellular transformation from an SVZ astrocyte-like stem cell into an OB neuron.
Overexpression of miR-124 in the SVZ leads to loss of self-renewing NSCs
Since miR-124 regulates postnatal neurogenesis, we furthermore decided to investigate the consequence of miR-124 overexpression in the SVZ niche. To this end, we generated a lentiviral vector containing a 484 bp genomic fragment containing miR-124–1 (miR-124OE) that allowed high-level expression of mature miR-124 and a GFP marker (Fig. 7A).
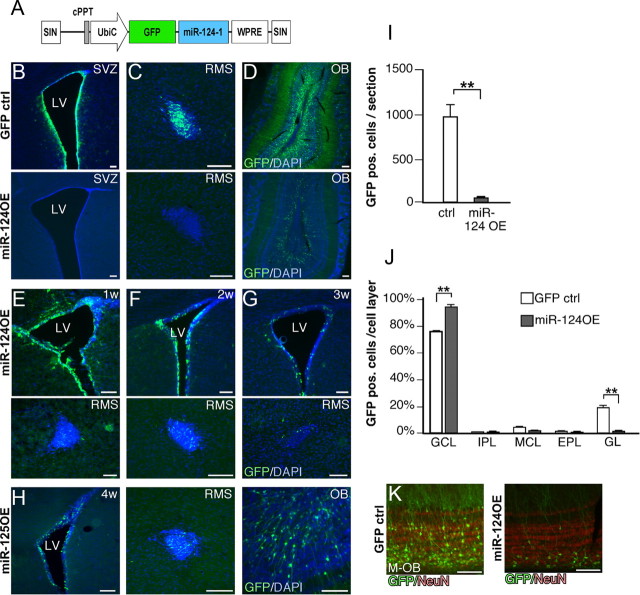
Figure 7.
Overexpression of miR-124 in the postnatal SVZ leads to loss of neural stem cells. A, Illustration of the vector to overexpress miR-124 in the postnatal SVZ. Lentiviral vectors were injected at P3. B, C, Four weeks after the injection of miR-124OE, no GFP-expressing cells can be detected in either SVZ or RMS, suggesting that all stem and progenitor cells overexpressing miR-124 have migrated away and differentiated to OB neurons. D, In animals injected with miR-124OE vectors, OB neurons are generated to a lesser extent when compared with control injections. E–G, GFP-expressing cells are gradually lost from the SVZ and RMS after injection of miR-124OE vector. H, Injection of a control lentiviral vector overexpressing the unrelated miR-125b does not cause loss of GFP expression in the SVZ and RMS. I, Quantification of GFP-expressing neurons in the OB. Data are presented as mean ± SEM; n = 3. **p < 0.01. J, Altered integration into the OB after miR-124 overexpression. Data are presented as mean ± SEM; n = 3. *p < 0.05. K, Newborn neurons overexpressing miR-124 integrate primarily in the inner parts of the GCL. LV, Lateral ventricle; M-OB, most medial part of the OB. All scale bars, 100 μm. cPPT, Central polypurine tract; EPL, external plexiform layer; GL, glomerular layer; IPL internal plexiform layer; MCL, mitral cell layer; SIN, self-inactivating; UbiC, ubiquitin C; w, weeks.
When overexpressing miR-124 in the SVZ, we were unable to detect GFP-expressing cells in the SVZ or the RMS 4 weeks after delivery of the miR-124 overexpression vector although we did detect GFP-neurons in the OB (n = 3; Fig. 7B–D). When using a control GFP vector, we detected high-level GFP expression in the SVZ, RMS, and OB (Fig. 7B–D). These data suggest that miR-124 overexpression is not compatible with the maintenance of type B stem cells, leading to a loss of continuous supply of new neurons to the OB via the RMS. To investigate this phenomenon in detail, we killed mice 1, 2, and 3 weeks after miR-124OE vector injection (n = 9). At 1 week postinjection, miR-124 overexpressing GFP cells were found in the SVZ and the RMS, and we also found cells with immature morphology that had entered the OB (Fig. 7E) (data not shown). Starting at 2 weeks and even more so at 3 weeks postinjection, there was a gradual loss of GFP-expressing cells in the SVZ and in the RMS (Fig. 7F,G). These data suggest that overexpression of miR-124 results in only a single wave of GFP-expressing cells reaching the OB, which is in contrast to the constant supply of new GFP neurons when using control vectors. In control experiments, in which we used a lentiviral vector with a similar design overexpressing the unrelated miR-125b, we found GFP-expressing cells in the SVZ, RMS, and OB 4 weeks postinjection demonstrating a continuous supply of new neurons (Fig. 7H).
The number of GFP-expressing neurons detected in the OB 4 weeks after miR-124OE injection were fewer and arranged differently when compared with control injected animals (Fig. 7D,I–K). When we analyzed the distribution of GFP cells in the different cell layers in the OB, we found that miR-124 overexpression led to an increased integration into the GCL, with fewer cells integrating into the glomerular layer compared with the control GFP cells (Fig. 7J). Furthermore, the miR-124-overexpressing cells were primarily found in the inner parts of the GCL (Fig. 7K). This indicates that the level of miR-124 expression is important for the timing of integration of newborn neurons and that forced expression of miR-124 leads to premature differentiation once the cell reaches the OB.
In summary, these data suggest that overexpression of miR-124 in NSCs leads to loss of cells with stem cell characteristics while it does not impair migration of progenitors from the SVZ to the OB. In addition, the precise level of miR-124 expression appears to regulate neuronal integration into the OB. Noteworthy, we only detected very few GFP-expressing ependymal cells after miR-124 overexpression. This suggests that ependymal cells overexpressing miR-124 either are lost due to cell death or also enter the migratory path to the OB to form interneurons.
Discussion
In this study we show that miR-124 is expressed and active in all neuronal populations of the adult brain. We also demonstrate that miR-124 activity is rapidly initiated once NSCs, type B cells, transit to rapid-amplifying progenitors, type C cells. When the upregulation of miR-124 expression is blocked, type C cells are not generated, leading to a subsequent inhibition of neurogenesis. On the other hand, if miR-124 expression is forced in type B cells they lose their stem cell characteristics.
Our strategy to detect miR-124 activity relies on the generation of a GFP reporter transgenic mouse. The detection of GFP expression only in glial cell types, including astrocytes and microglia, demonstrates that miR-124 is active only in neurons and cells destined to become neurons. This is in contrast to a recent study in which miR-124 was reported to be expressed in resting microglia (Ponomarev et al., 2011). This discrepancy is most likely explained by the ability of the reporter mouse to detect the activity of miR-124 at single cell resolution whereas the study by Ponomarev et al. (2011) solely relies on PCR-based techniques that are limited to measure expression levels. The possibility that we would detect GFP expression in cells that are also displaying miRNA activity is very unlikely. A false signal could be potentially caused by loss of the miR.T sequence, but this would occur in a random fashion and not specifically in astrocytes and oligodendrocytes, and the fact that we see similar expression patterns in different founder mice further rules out this possibility. Alternatively, a false signal could be caused by a lack of sensitivity of the miRNA reporter system in detecting low levels of miRNA. Still, such a low level of miRNA expression is most likely not relevant for endogenous miRNA targets either (Brown et al., 2007; Mukherji et al., 2011).
We found the reporter mouse particular useful when analyzing neurogenesis since it allows for straightforward colabeling with relevant markers such as BrdU, which otherwise would be complicated to combine with standard miRNA detection techniques such as LNA-ISH. The GFP reporter also permits morphological analysis, which is not possible when using ISH-staining to detect miRNA. In future studies, the miR.124.T mice generated here could be used in studies to monitor changes in miR-124 levels in models of stroke, neurodegenerative disorders, and brain cancer. Additionally, the simplicity of lentiviral transgenesis should allow for the generation of a large number of this type of miRNA reporter mice, which would greatly enhance our understanding of miRNA expression patterns in the brain.
Our loss-of-function and gain-of-function experiments suggest that miR-124 is an essential part of the neuronal network. This is in agreement with previous studies in which it has been shown that overexpression of miR-124 in HeLa cells induces a broad neuronal gene profile (Lim et al., 2005). Furthermore, two recent reports demonstrate that overexpression of miR-124 contributes to direct conversion of fibroblasts into functional neurons (Ambasudhan et al., 2011; Yoo et al., 2011). Together, these studies and ours indicate that miR-124 is a factor necessary for the induction of the neuronal gene program. However, our results contrast, to some extent, a previous study by Cheng et al. (2009)in which the authors concluded that miR-124 does not influence neuron/glia fate decisions in the SVZ. The difference between the studies lies primarily in our lentiviral strategy, which leads to stable targeting of the type B stem cells, whereas Cheng et al. (2009) used infusions of 2′OMe-RNA antisense molecules that are limited to a transient, nontargeted inhibition of miR-124 most likely occurring primarily in rapid-amplifying progenitors.
An interesting observation in our study is that at least a proportion of the cells in which miR-124 was inhibited continued to migrate to the OB without obtaining neuronal characteristics. These data suggest that intracellular gene programs other than those inducing a neuronal fate governs migration to the OB.
Our data show that ectopic expression of miR-124 in the SVZ is not compatible with continuous neurogenesis. Rather, we observed a single wave of neurogenesis when miR-124 was overexpressed. This “washout” of GFP-expressing cells follows a similar temporal profile to what has been observed when injecting MLV retroviral vectors into the ventricles of newborn rodents (Rogelius et al., 2005). MLV vectors target primarily the rapidly proliferating type A and type C cells, suggesting that overexpression of miR-124 in type B cells caused forced differentiation into neural progenitors without self-renewing capacities. Alternatively, it is possible that the loss of self-renewing GFP cells after miR-124 is at least partly due to cell death. We have analyzed our material for cell death markers but have not been able to detect dying cells (data not shown). However, during the first postnatal weeks the SVZ is a dynamic and expanding region, and cell death in response to the overexpression may not be detectable. Still, the presence of GFP-expressing cells in the SVZ, RMS, and OB at different time points after the injection of miR-124OE demonstrates that miR-124 overexpression does not always cause cell death. Furthermore, our observations after miR-124 overexpression are in line with in vitro experiments in which miR-124 has been overexpressed in cultured cells with neural progenitor characteristics (Makeyev et al., 2007; Silber et al., 2008; Cheng et al., 2009; Liu et al., 2011). In these studies, overexpression of miR-124 led to reduced proliferation associated with forced neuronal differentiation. These studies and our findings indicate that miR-124 may in part act by the downregulation of genes that are associated with stemness.
Of particular interest is the identification of the Notch-ligand receptor JAG1 as a miR-124 target (Cheng et al., 2009; Liu et al., 2011). Recent studies demonstrate a critical role for the Notch pathway not only in regulating NSCs, but also in maintaining ependymal cells in a nonproliferating state (Carlen et al., 2009). The consequences of miR-124 overexpression in the SVZ are very similar to Notch inhibition, suggesting that miR-124 is a crucial player in this network. However, the molecular machinery controlled by miR-124 appears to be complex; an mRNA target prediction algorithm suggests >1000 potential targets, and several of those have been experimentally validated (targetscan.org). In the adult SVZ, miR-124 has, in addition to JAG1, been demonstrated to regulate SOX9, which is a transcription factor that controls adult neurogenesis, as well as DLX2, a transcription factor mediating neuronal subtype specification (Cheng et al., 2009). In light of this, it will be interesting to identify the complete set of miR-124 targets in neural progenitor cells. Recent studies demonstrate that a combination of computational approaches, loss-of-function experiments, and experimental determination of either mRNA-Ago2 complexes (PAR-CLIP) or miRNA-mRNA interaction is necessary to obtain a reliable list of true miRNA targets (Lal et al., 2011; Lipchina et al., 2011). The application of such approaches to a small cell population in vivo is challenging but will be a focus in our future work.
Footnotes
This work was supported by grants from Swedish Research Council and The Swedish Cancer Foundation. We thank A. Björklund, O. Lindvall, M. Parmar, and L. Naldini for providing reagents and for stimulating discussions, and we thank U. Jarl, A. Josefsson, C. Isaksson, I. Nilsson, S. Smiljanic, A.-K. Oldén, Z. Ma, E. Ling, M. Sparrenius, and B. Mattsson for technical assistance.
References
- Ahlenius H, Kokaia Z. Isolation and generation of neurosphere cultures from embryonic and adult mouse brain. Methods Mol Biol. 2010;633:241–252. doi: 10.1007/978-1-59745-019-5_18. [DOI] [PubMed] [Google Scholar]
- Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlén M, Meletis K, Göritz C, Darsalia V, Evergren E, Tanigaki K, Amendola M, Barnabé-Heider F, Yeung MS, Naldini L, Honjo T, Kokaia Z, Shupliakov O, Cassidy RM, Lindvall O, Frisén J. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat Neurosci. 2009;12:259–267. doi: 10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- Georgievska B, Jakobsson J, Persson E, Ericson C, Kirik D, Lundberg C. Regulated delivery of glial cell line-derived neurotrophic factor into rat striatum, using a tetracycline-dependent lentiviral vector. Hum Gene Ther. 2004;15:934–944. doi: 10.1089/hum.2004.15.934. [DOI] [PubMed] [Google Scholar]
- Imura T, Kornblum HI, Sofroniew MV. The predominant neural stem cell isolated from postnatal and adult forebrain but not early embryonic forebrain expresses GFAP. J Neurosci. 2003;23:2824–2832. doi: 10.1523/JNEUROSCI.23-07-02824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson ME, Ono Y, Björklund A, Thompson LH. Identification of transplantable dopamine neuron precursors at different stages of midbrain neurogenesis. Exp Neurol. 2009;219:341–354. doi: 10.1016/j.expneurol.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, Deutsch A, Hofmann O, Ventura A, Hide W, Lieberman J. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laywell ED, Rakic P, Kukekov VG, Holland EC, Steindler DA. Identification of a multipotent astrocytic stem cell in the immature and adult mouse brain. Proc Natl Acad Sci U S A. 2000;97:13883–13888. doi: 10.1073/pnas.250471697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lipchina I, Elkabetz Y, Hafner M, Sheridan R, Mihailovic A, Tuschl T, Sander C, Studer L, Betel D. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25:2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Chopp M, Zhang RL, Tao T, Wang XL, Kassis H, Hozeska-Solgot A, Zhang L, Chen C, Zhang ZG. MicroRNA profiling in subventricular zone after stroke: MiR-124a regulates proliferation of neural progenitor cells through Notch signaling pathway. PLoS One. 2011;6:e23461. doi: 10.1371/journal.pone.0023461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Tramontin AD, García-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci U S A. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogelius N, Ericson C, Lundberg C. In vivo labeling of neuroblasts in the subventricular zone of rats. J Neurosci Methods. 2005;142:285–293. doi: 10.1016/j.jneumeth.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Sachdeva R, Jönsson ME, Nelander J, Kirkeby A, Guibentif C, Gentner B, Naldini L, Björklund A, Parmar M, Jakobsson J. Tracking differentiating neural progenitors in pluripotent cultures using microRNA-regulated lentiviral vectors. Proc Natl Acad Sci U S A. 2010;107:11602–11607. doi: 10.1073/pnas.1006568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Chérasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- Sauvain MO, Dorr AP, Stevenson B, Quazzola A, Naef F, Wiznerowicz M, Schütz F, Jongeneel V, Duboule D, Spitz F, Trono D. Genotypic features of lentivirus transgenic mice. J Virol. 2008;82:7111–7119. doi: 10.1128/JVI.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silahtaroglu AN. LNA-FISH for detection of microRNAs in frozen sections. Methods Mol Biol. 2010;659:165–171. doi: 10.1007/978-1-60761-789-1_11. [DOI] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogh C, Eriksson C, Kokaia M, Meijer XC, Wahlberg LU, Wictorin K, Campbell K. Generation of regionally specified neurons in expanded glial cultures derived from the mouse and human lateral ganglionic eminence. Mol Cell Neurosci. 2001;17:811–820. doi: 10.1006/mcne.2001.0973. [DOI] [PubMed] [Google Scholar]
- Thompson L, Barraud P, Andersson E, Kirik D, Björklund A. Identification of dopaminergic neurons of nigral and ventral tegmental area subtypes in grafts of fetal ventral mesencephalon based on cell morphology, protein expression, and efferent projections. J Neurosci. 2005;25:6467–6477. doi: 10.1523/JNEUROSCI.1676-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]