Abstract
Asexual reproduction via thelytokous parthenogenesis is widespread in the Hymenoptera, but its genetic underpinnings have been described only twice. In the wasp Lysiphlebus fabarum and the Cape honey bee Apis mellifera capensis the origin of thelytoky have each been traced to a single recessive locus. In the Cape honey bee it has been argued that thelytoky (th) controls the thelytoky phenotype and that a deletion of 9 bp in the flanking intron downstream of exon 5 (tae) of the gemini gene switches parthenogenesis from arrhenotoky to thelytoky. To further explore the mode of inheritance of thelytoky, we generated reciprocal backcrosses between thelytokous A. m. capensis and the arrhenotokous A. m. scutellata. Ten genetic markers were used to identify 108 thelytokously produced offspring and 225 arrhenotokously produced offspring from 14 colonies. Patterns of appearance of thelytokous parthenogenesis were inconsistent with a single locus, either th or tae, controlling thelytoky. We further show that the 9 bp deletion is present in the arrhenotokous A. m. scutellata population in South Africa, in A. m. intermissa in Morocco and in Africanized bees from Brazil and Texas, USA, where thelytoky has not been reported. Thus the 9 bp deletion cannot be the cause of thelytoky. Further, we found two novel tae alleles. One contains the previously described 9 bp deletion and an additional deletion of 7 bp nearby. The second carries a single base insertion with respect to the wild type. Our data are consistent with the putative th locus increasing reproductive capacity.
Introduction
There are few exceptions to sexual reproduction in the animal kingdom. Sexual reproduction may confer advantages over asexual reproduction by facilitating the purging of deleterious mutations (Muller, 1964) and by generating genetic variability among offspring, so that at least some of them will be resistant to the parasites and pathogens that they may encounter (Hamilton, 1980; Hamilton and Axelrod, 1990). Curiously, asexual reproduction via thelytokous parthenogenesis (the production of diploid females without fertilisation) is widespread in one important group of animals, the Hymenoptera (ants, wasps and bees), and particularly the social Hymenoptera. Up to 50 social Hymenopterans have been associated with thelytokous parthenogenesis (Crozier and Pamilo, 1996; Wenseleers and Oystaeyen, 2011; Rabeling and Kronauer, 2013). In Hymenopterans thelytokous populations are often derived from sexual populations (Wenseleers and Oystaeyen, 2011). This suggests a simple molecular switch from arrhenotoky, the normal mode of male production in haplodiploid insects, to thelytoky. Indeed, genetic control of thelytokous parthenogenesis via a single locus has been reported in the parasitoid wasp Lysiphlebus fabarum (Sandrock and Vorburger, 2011) and the honey bee Apis mellifera capensis (Lattorff et al., 2005; Lattorff et al., 2007; Jarosch et al., 2011). In L. fabarum, all thelytokously reproducing individuals are homozygous for allele 183 at microsatellite locus Lysi07, whereas arrhenotokously reproducing individuals are never homozygous for this allele at this locus and it is found at low frequency (<5%) in arrhenotokously reproducing individuals (Sandrock and Vorburger, 2011). In A. m. capensis, thelytokous individuals are thought to be homozygous for a particular deletion near the gene gemini (see below; Lattorff et al., 2005; Lattorff et al., 2007; Jarosch et al., 2011). It is unknown if the loci that apparently affect thelytoky are homologous in bee and wasp.
Apis mellifera capensis (hereafter ‘Capensis') is native to the southern portion of South Africa (Ruttner, 1988; Hepburn and Crewe, 1990; Beekman et al., 2008; Goudie and Oldroyd, 2014). Like many other social insects and all other honey bee species, when a Capensis colony loses its queen a proportion of workers begin to lay eggs. Although a minority of workers produce males via arrhenotoky (Hepburn and Radloff, 2002; Goudie et al., 2012), most of the workers' unfertilised eggs develop as females via thelytokous parthenogenesis (Verma and Ruttner, 1983; Oldroyd et al., 2008). These diploid eggs may be reared either as a worker (Beekman et al., 2009) or a queen (Jordan et al., 2008b; Allsopp et al., 2010; Holmes et al., 2010). In the form of thelytoky found in Capensis, the central pair of the four haploid products of meiosis fuse to produce a diploid zygote (Verma and Ruttner, 1983).
In the rest of South Africa, and in countries to its north, a second subspecies is extant, A. m. scutellata (hereafter ‘Scutellata') (Ruttner, 1988; Hepburn and Crewe, 1990). This subspecies, like most other honey bee (sub)species is arrhenotokous (for some rare exceptions see Mackensen, 1943; DeGrandi-Hoffman et al., 1991; Holmes et al., 2015). Thus, when queenless Scutellata workers lay eggs they develop as haploid males (Hepburn and Crewe, 1990). A tension zone exists between the Scutellata and Capensis populations that reduces gene flow between the two subspecies (Beekman et al., 2008). However, in 1990 an anthropogenic introduction of Capensis colonies to an area near Pretoria, in the north, gave rise to a remarkable phenomenon: a clonal lineage originating from a single Capensis worker (Neumann et al., 2010; Oldroyd et al., 2011). Workers of this lineage enter Scutellata colonies and activate their ovaries to produce new generations of parasites (Martin et al., 2002). This lineage (hereafter ‘the Clone') infests and kills hundreds of commercial Scutellata colonies each year as a kind of ‘social cancer' (Oldroyd, 2002). Previous Capensis invasions due to anthropomorphic movements have also been recorded, but these were rapidly contained (Onions, 1912; Lundie, 1954; Johannsmeier, 1983).
The genetic basis of thelytoky in Capensis appears to be under the control of a single recessive locus (th) mapped to chromosome 13 (Lattorff et al., 2007; Supplementary Information 1). This putative locus was additionally shown to control two queen-like reproductive traits pleiotropically: early onset of oviposition and excess production of the queen mandibular pheromone component 9-oxy-decanoic acid involved in reproductive dominance (Lattorff et al., 2007). Following up on Lattorff et al.'s (2007) study, Jarosch et al. (2011) screened two candidate genes, both transcription factors, from within the region associated with th in Capensis workers with and without activated ovaries and in arrhenotokous A. m. carnica workers. Of these candidates, they found an association between worker fertility and the splice forms of one of the genes in the region of th: a homologue of the Drosophila transcription factor gemini (Figure 1; Supplementary Information 1). Jarosch et al. (2011) further showed that in the Clone there is a 9 base pair deletion in the intron flanking the alternatively spliced exon 5 (Figure 1) compared with the arrhenotokously reproducing A. m. carnica individuals they examined. They suggested that this deletion, thelytoky associated element 1 (tae1), could be the putative molecular switch that controls whether an unmated worker will reproduce thelytokously or arrhenotokously (Jarosch et al., 2011). Individuals subjected to RNAi knockdown of this region were significantly more likely to have activated ovaries than the individuals treated with scrambled RNA, indicating an effect of tae1 on reproductive behaviour. However, the mode of parthenogenesis (arrhenotoky or thelytoky) was not determined in these workers (Jarosch et al., 2011). Thus the 9 bp deletion may be specific to the Clone, or be involved in reproductive dominance or capacity, without necessarily causing thelytoky.
Figure 1.
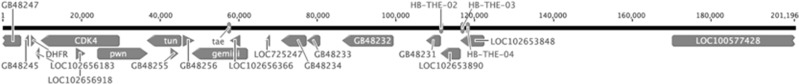
Genetic map of the thelytoky region showing the three HB-THE microsatellite loci, the gene gemini thought to regulate worker reproductive dominance and thelytoky and the tae region within gemini postulated to cause thelytoky. The region in its full genomic context is given in Supplementary Information 1 SI1.
Despite the above findings, our ad hoc observations of worker reproduction in crosses between thelytokous Capensis and arrhenotokous Scutellata were inconsistent with a simple Mendelian pattern of inheritance of thelytoky. For example, we have seen thelytokous reproduction in F1 worker hybrids of Capensis and Scutellata, a scenario that is incompatible with a single recessive locus. Moreover, the production of males by arrhenotoky in the Clone workers (Goudie et al., 2012) cannot be explained by the current model. We therefore performed reciprocal backcrosses to further clarify the inheritance of thelytoky in Capensis and to reconcile our observations with the studies of Lattorff et al. (2005, 2007) and Jarosch et al. (2011). If we could confirm the presence of a single-locus control of the thelytoky/arrhenotoky switch in honey bees, then this would be only the second example of such a molecular switch in Hymenoptera. Most other examples of thelytoky are thought to be mediated by bacterial endosymbionts such as Wolbachia (Huiens et al., 2000; Rabeling and Kronauer, 2013), or more commonly, the mechanism is unknown (Wenseleers and Oystaeyen, 2011; Rabeling and Kronauer, 2013).
Materials and methods
Crosses
Five sister Capensis and two sister Scutellata queens were inseminated with the semen of a single Scutellata or Capensis drone, respectively. A different drone was used for each queen and within subspecies the drones were brothers. The mother of the Scutellata queens was acquired from Douglas, (26°01'S, 29°22′E), well inside the zone where there are no Capensis (except for the rare Clone, which is easily identified both visually and genetically; Neumann et al., 2010; Oldroyd et al., 2011), and no thelytokous reproduction by workers (Hepburn and Crewe, 1990; Hepburn et al., 1994, 1998). The Capensis mother came from Stellenbosch, (33°56′S, 18°51′E), where there are no Scutellata and thelytokous reproduction by workers is common (Hepburn and Crewe, 1990; Hepburn et al., 1994, 1998), and was unrelated to the Clone. F1 queens were then reared from one Capensis x Scutellata and one Scutellata x Capensis colony and inseminated with the semen of either a single Scutellata or Capensis drone, creating four parent-specific backcross colony types (Figure 2, Table 1).
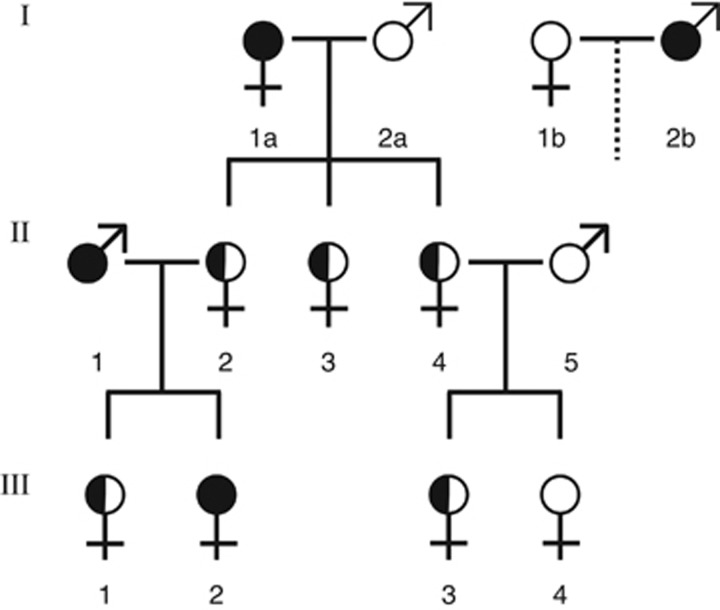
Figure 2.
Pedigree showing backcross crosses performed using artificial insemination. Solid black fill indicates individuals homozygous for the putatively recessive thelytoky-determining allele of the thelytoky locus. Half-filled circles indicate heterozygous individuals and white fill indicates individuals homozygous for the putatively dominant arrhenotoky-determining allele. For the first generation, both Capensis queen (I:1a) x Scutellata male (I:2a) and Scutellata queen (I:1b) x Capensis male (I:2b) crosses were created. Queen offspring of these queens were reciprocally backcrossed to single Scutellata and Capensis males. Offspring of workers were collected in all colonies after dequeening. Under a recessive mode of inheritance, as proposed by Lattorff et al. (2005), only workers with solid fill (i.e.III-2) should reproduce thelytokously. Colonies without workers of this genotype will therefore produce only males.
Table 1. Theoretical expectation of th locus genotypes carried by the queens and drones and their worker offspring in each F1 or BC colony.
| Colony | Cross type | Cross direction (queen x drone) | Expected th genotype queen | Expected th genotype drone | Expected th genotype workers | Thelytoky expected | Arrhenotoky expected | Thelytokous offspring | Arrhenotokous offspring | Origin of HB-THE alleles carried by thelytokous offspring |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | F1 | SxC | Th,Th | th | Th,th | No | Yes | 0 | 47 | — |
| 4 | F1 | SxC | Th,Th | th | Th,th | No | Yes | 0 | 31 | — |
| 7 | F1 | CxS | th,th | Th | Th,th | No | Yes | 0 | 20 | — |
| 9 | F1 | CxS | th,th | Th | Th,th | No | Yes | 1 | 16 | 1 C |
| 11 | F1 | CxS | th,th | Th | Th,th | No | Yes | 1 | 15 | 1 S |
| 14 | F1 | CxS | th,th | Th | Th,th | No | Yes | 0 | 58 | — |
| 10 | BC | (SxC)xS | Th,th | Th | Th,Th or Th,th | No | Yes | 0 | 8 | — |
| 15 | BC | (SxC)xC | Th,th | th | Th,th or th,th | Yes | Yes | 20 | 8 | 1 S,C and 18 C,C |
| 16 | BC | (SxC)xC | Th,th | th | Th,th or th,th | Yes | Yes | 26 | 1 | 6 S,C and 19 C,C |
| 21 | BC | (SxC)xC | Th,th | th | Th,th or th,th | Yes | Yes | 2 | 0 | 2 C,C |
| 19 | BC | (CxS)xS | Th,th | Th | Th,Th or Th,th | No | Yes | 0 | 6 | — |
| 20 | BC | (CxS)xS | Th,th | Th | Th,Th or Th,th | No | Yes | 21 | 0 | 11 S,S and 9 S,C |
| 22 | BC | (CxS)xC | Th,th | th | Th,th or th,th | Yes | Yes | 23 | 0 | 9 S,C and 14 C,C |
| 26 | BC | (CxS)xC | Th,th | th | Th,th or th,th | Yes | Yes | 14 | 21 | 14 C,C |
Abbreviation: BC, backcross.
Th is the putative arrhenotoky allele derived from the Scutellata parent (S), while th is the recessive thelytoky-determining allele derived from the Capensis (C) parent. Based on the expected genotype the expected mode of reproduction in workers is presented, followed by the observed number of thelytokous and arrhenotokous offspring (eggs, larvae and pupae) of workers. The empirically determined origin of the linked HB-THE alleles in thelytokously produced offspring is given in the last column. Queens heading colonies 10, 15, 16 and 21 were offspring of colony 3. Queens heading colonies 19, 20, 22 and 26 were offspring of colony 6. All the worker-produced offspring collected from colony 6 at the time of sampling were non-natal.
Inseminated queens were introduced into colonies and maintained for at least six weeks to ensure the replacement of all workers with daughters of the queen. Queens were then removed from the F1 and backcross colonies to stimulate worker reproduction. Eggs, larvae and pupae—offspring of the workers—were collected and genotyped to verify that they were the offspring of the resident workers, and not the offspring of parasitic workers from other colonies—a common phenomenon in queenless Capensis colonies (Jordan et al., 2008b; Holmes et al., 2010) and to determine their tae alleles and infer their th alleles by linkage to HB-THE microsatellites (Shaibi et al., 2008; Figure 1).
DNA extraction and genotyping
We genotyped ~24 egg, larval or pupal offspring of workers from each colony, the sire of those workers and the mother queen of the workers that we had removed to stimulate worker reproduction. Genotypes had to be inferred for two drones and one queen that were not available. DNA was extracted as described in Aljanabi and Martinez (1997). We genotyped the bees using ten genetic markers, both to verify their parentage and to determine whether they had been produced arrhenotokously (that is, were haploid) or thelytokously (that is, were diploid). The loci were: (1) five unlinked microsatellites—A79, B124, A29, A14 and A107 (Estoup et al., 1993; Solignac et al., 2003); (2) three tightly linked loci within the th locus—HB-THE-02, HB-THE-03 and HB-THE-04 (Shaibi et al., 2008; Figure 1); (3) the thelytoky associated element (tae) found in the gemini homologue (Jarosch et al., 2011; Figure 1); and (4) complementary sex determiner (csd) (Oldroyd et al., 2011).
Analysis
To exclude offspring of non-natal workers, we discarded all individuals that carried alleles absent from the queen and the drone parents of the egg-laying workers that produced the sampled offspring. In colonies where we determined that a large number of offsprings were not offsprings of resident workers, we genotyped additional offsprings when these were available.
In honey bees sex is determined by zygosity at a single sex-determining locus complementary sex determiner or csd (Beye et al., 2003). Individuals that are heterozygous at csd are female, individuals that are hemizygous (that is, haploid) are male. Individuals that are homozygous at csd and diploid at other loci are diploid males, but these individuals are normally inviable beyond the larval stage (Woyke, 1963). Oldroyd et al. (2011) developed a polymerase chain reaction-based test of zygosity at csd that relies on the extreme sexual phenotype-determining length polymorphism of exon 7 (Beye et al., 2003). By using this marker of zygosity at csd, we declared a worker-produced offspring to be thelytokously produced if it was heterozygous at csd and an arrhenotokously produced haploid male when only one allele could be identified at csd and each of the nine other loci studied. Individuals that were homozygous at csd but heterozygous at any other locus were most likely diploid males (Goudie et al., 2012) but, because the csd polymerase chain reaction product does not encompass the entire csd region, we cannot completely exclude the possibility that they were females. In either case, diploidy at any locus is unequivocal evidence of thelytokous reproduction.
Each backcross colony (Figure 2) was comprised of workers of two different genotypes with respect to parental or grand-parental origin. Th is the putative arrhenotoky-causing allele derived from the Scutellata parent and th is the putative thelytoky-causing allele derived from the Capensis parent. We expected that thelytokously produced offspring would only occur in colonies where workers were homozygous th,th (Lattorff et al., 2007). To uncover the relationship between genotype at the putative th locus and the thelytoky/arrhenotoky phenotype, we assigned the offspring of workers based on their genotype at the linked HB-THE loci from within the th locus (Figure 1) as being offspring of workers carrying two Scutellata-derived alleles, two Capensis-derived alleles, or one Capensis-derived allele and one Scutellata-derived allele. We expected that thelytokously produced offspring would only derive from the workers carrying two Capensis-derived HB-THE alleles (Lattorff et al., 2005, 2007; Table 1) at each HB-THE locus. We further expected that the tae1 allele would only be present in Capensis, and that thelytoky would only occur in colonies in which the workers were homozygous for this allele (Jarosch et al., 2011; Table 2).
Table 2. The putative thelytoky associated element, tae, (Jarosch et al., 2011) allele carried by the queens and drones and their worker offspring in each F1 or BC colony, expected presence of thelytokously and arrhenotokously produced offspring if (a) both the tae1 and tae2 alleles result in thelytoky, or (b) only the tae1 allele results in thelytoky.
| Colony | Cross type | Cross direction (queen x drone) | tae queen | tae drone | tae workers | A: Thelytoky expected tae1 & tae2 cause thelytoky | A: Arrhenotoky expected tae1 & tae2 cause thelytoky | B: Thelytoky expected tae1 causes thelytoky | B: Arrhenotoky expected tae1 causes thelytoky | Thelytokous offspring | Arrhenotokous offspring | tae allele carried by thelytokous offspring |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | F1 | SxC | tae1,tae1 | tae2 | tae1,tae2 | Yes | No | No | Yes | 0 | 47 | — |
| 4 | F1 | SxC | tae1,tae1 | tae2 | tae1, tae2 | Yes | No | No | Yes | 0 | 31 | — |
| 7 | F1 | CxS | tae1,tae2 | tae4 | tae1,tae4 or tae2,tae4 | No | Yes | No | Yes | 0 | 20 | — |
| 9 | F1 | CxS | tae1,tae1 | tae2 | tae1,tae2 | Yes | No | No | Yes | 1 | 16 | 1 tae1,tae1 |
| 11 | F1 | CxS | tae1,tae1 | tae2 | tae1,tae2 | Yes | No | No | Yes | 1 | 15 | 1 tae2,tae2 |
| 14 | F1 | CxS | tae2,tae2 | tae1 | tae1,tae2 | Yes | No | No | Yes | 0 | 58 | - |
| 10 | BC | (SxC)xS | tae1,tae1 | tae3 | tae1,tae3 | No | Yes | No | Yes | 0 | 8 | - |
| 15 | BC | (SxC)xC | tae1,tae2 | tae1 | tae1,tae1 or tae1,tae2 | Yes | No | Yes | Yes | 20 | 8 | 1 tae1,tae1 and 14 tae1,tae2 |
| 16 | BC | (SxC)xC | tae1,tae2 | tae1 | tae1,tae1 or tae1,tae2 | Yes | No | Yes | Yes | 26 | 1 | 6 tae1,tae1 and 17 tae1,tae2 |
| 21 | BC | (SxC)xC | tae1,tae2 | tae1 | tae1,tae1 or tae1,tae2 | Yes | No | Yes | Yes | 2 | 0 | 2 tae1,tae2 |
| 19 | BC | (CxS)xS | tae1,tae2 | tae3 | tae1,tae3 or tae2,tae3 | No | Yes | No | Yes | 0 | 6 | — |
| 20 | BC | (CxS)xS | tae1,tae2 | tae1 | tae1,tae1 or tae1,tae2 | Yes | No | Yes | Yes | 21 | 0 | 9 tae1,tae1 and 10 tae1,tae2 |
| 22 | BC | (CxS)xC | tae1,tae2 | tae1 | tae1,tae1 or tae1,tae2 | Yes | No | Yes | Yes | 23 | 0 | 13 tae1,tae1 and 8 tae1,tae2 |
| 26 | BC | (CxS)xC | tae1,tae2 | tae1 | tae1,tae1 or tae1,tae2 | Yes | No | Yes | Yes | 14 | 21 | 12 tae1,tae1 |
Abbreviation: BC, backcross.
The number of thelytokously and arrhenotokously produced offspring of workers (eggs, larvae and pupae) observed in F1 and backcross colonies of Capensis (C) and Scutellata (S), and the tae alleles carried by thelytokously produced offspring. Queens heading colonies 10, 15, 16 and 21 were offspring of colony 3. Queens heading colonies 19, 20, 22 and 26 were offspring of colony 6. All the worker-produced offspring collected from colony 6 at the time of sampling were non-natal.
DNA sequencing of tae
During our analysis we found length polymorphisms that were incompatible with a simple presence or absence of the 9 bp deletion at tae. We therefore directly sequenced examples of each observed allele from drones using the primers gem_taeI (Jarosch et al., 2011) and gem_taeIV (5′-GACATTTAACACAGAAAGATACCG-3′) in the reverse direction only (Macrogen, Seoul, South Korea). DNA for sequencing was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD, USA). DNA sequences were manually edited and aligned using Sequencher (v5.2, Gene Codes Corporation, Ann Arbor, MI, USA) and compared with the annotated A. mellifera genome (Honey Bee Genome Sequencing Honey Bee Genome Sequencing Consortium, 2014; Amel_4.5). A map of the linked HB-THE and tae markers (Figure 1) and a map of genes occurring within the th region on both sides of the LOD peak (Lattorff et al., 2007; Supplementary Information 1) were created using Geneious (v7.1.4, Biomatters, Auckland, New Zealand).
tae length polymorphisms in populations
As we located the length polymorphisms associated with the tae1 allele in four Scutellata individuals within our crosses, we characterized tae length polymorphisms using the primers gem_taeI and gem_taeII (Jarosch et al., 2011) to determine the frequency and distribution of these polymorphisms at a population level based on one individual from each of 80 Scutellata colonies collected in 1984, 1993, 2006, 2008 and 2012, 56 Capensis colonies collected in 1984, 2004, 2009, 2011; 2012 and 2013; 25 colonies of A. m. ligustica from Italy; 8 colonies of A. m. intermissa from Morocco; 6 colonies of A. m. carnica from former Yugoslavia; 5 colonies of A. m. mellifera from France; 17 colonies of Africanized bees from Brazil (for details of collections see Clarke et al., 2001, 2002; Supplementary Information 3); 23 colonies of Africanized bees we collected from Texas, USA in 2013 and 30 colonies of European-derived bees from Ohio and California, USA collected in 2013 (Supplementary Information 3). DNA was extracted using phenol/chloroform, and the lengths of the polymerase chain reaction products were determined using an ABI 3130 DNA analyser (Applied Biosystems, Life Technologies, Carlsbad, CA, USA). We confirmed the sequence of the tae allele in an Africanized (hybrid of Scutellata and the previously introduced bees of European origin in Americas) worker that was homozygous for the tae1 deletion based on the length polymorphism.
Results
The th locus is not associated with thelytoky
The Capensis parent, but not the Scutellata parent, was expected to carry the putative-recessive thelytoky-causing th allele. Therefore we expected an absence of thelytoky in workers of our F1 colonies and in our backcross to Scutellata colonies (Table 1). Contrary to this expectation, we observed thelytokous offspring in three such colonies (Table 1; colony 9, 11 and 20; Supplementary Information 2). First, in a colony in which a hybrid queen had been backcrossed to a Scutellata male (colony 20), the 21 offspring were produced solely by thelytoky but were heterozygous at the linked HB-THE loci (Table 1). Second, we observed a single thelytokously produced individual in each of two F1 colonies. One individual carried allele combinations unique to its colony and we note that this individual carried only a single Capensis parent-derived allele at each of the linked HB-THE markers, while it was heterozygous at all the unlinked markers (Table 1; colony 9; Supplementary Information 2). The second individual from an F1 colony carried a single Scutellata allele apparently derived from her Scutellata grandfather at each of the linked HB-THE loci (Table 1; colony 11; Supplementary Information 2). Furthermore, in three backcross to Capensis colonies where both thelytoky and arrhenotoky were expected, there were 16 thelytokously produced individuals carrying HB-THE alleles derived from their Scutellata great-grandparent (Table 1; colony 15, 16 and 22; Supplementary Information 2). Thus, based on the appearance of thelytoky at high frequency in a backcross to Scutellata colony and the production of offspring that are heterozygous and carry HB-THE alleles derived from Scutellata, we cannot support the assertion that a recessive locus, th, tightly linked to the HB-THE loci is the sole cause of thelytoky in Capensis.
In the five backcross to Capensis colonies, and assuming the absence of reproductive dominance (which would lead to only a subset of the workers laying eggs), we expected an equal frequency of thelytokous and arrhenotokous parthenogenesis as workers are expected to be either Th,th or th,th in equal frequency (Table 1) (Lattorff et al., 2005; Lattorff et al., 2007). However, two colonies did not produce any arrhenotokous offspring (23 thelytokous individuals in colony 22 and 2 in colony 21). There was a higher than expected frequency of thelytoky in a further two colonies (colonies 15 and 16), and in the remaining backcross to Capensis colony (colony 26) the frequency of arrhenotokous offspring was slightly greater than the number of thelytokous offspring (14 vs 21 individuals; Table 1; Supplementary Information 2).
Reproductive dominance and the th locus
To determine whether the putative th locus is associated with reproductive dominance we computed expected allelic frequencies among offspring assuming the absence of reproductive dominance. As both th,th and Th,th backcross workers reproduced thelytokously in our experiment, under the assumption that they do so at equal frequency we can calculate the expected frequency of Th,th and th,th offspring in backcross to Capensis colonies. Half of the mother workers are expected to be th,th and all their offspring will be th,th. The remaining half of backcross workers will be Th,th. Because the th locus is distal to the centromere (Lattorff et al., 2005; Lattorff et al., 2007), multiple recombination events are expected between th and the centromere at each meiosis. Thus 1/3 of the offspring of Th,th workers will lose heterozygosity via recombination (Goudie et al., 2012). In all, we expect (0.583 offspring workers to be th,th (that is, 1/2+1/6 × 1/2 due to crossovers); 0.33 offspring to be Th,th (1/2 × 2/3 where there was no crossover with respect to the centromere) and 0.083 to be Th,Th (1/6 × 1/2 due to crossovers). Contrary to the prediction of 58.3% th,th offspring (~48), we found that 80.7% (67) of diploid offspring produced in backcross to Capensis colonies were homozygous for the HB-THE markers putatively linked to th. The remaining 19.3% (16) individuals were heterozygous and carried an allele at each HB-THE marker originating from their Scutellata great-grandfather. No Th,Th individuals were observed even though ~7 were expected (Table 1; Supplementary Information 2). Overall, the observed and expected offspring worker genotypes differed significantly from expectation (χ22=18.783, P<0.001) in backcross to Capensis colonies because of an excess of th,th individuals. This may suggest that even though th is clearly not associated with thelytoky itself, th is associated with reproductive dominance as shown by Lattorf et al. (2005, 2007).
The tae 9 bp deletion is not associated with thelytoky
Our sequencing revealed two novel tae alleles, one containing an additional 7 bp deletion close to the tae1 9 bp deletion (tae2; Figure 3) and another that had a single base insertion (tae4; Figure 3) with respect to the Scutellata sequence reported by Jarosch et al. (2011); tae3; Figure 3. If homozygosity for the 9 bp deletion is both necessary and sufficient for thelytokous worker reproduction, then individuals carrying any combination of the tae1 or tae2 alleles should reproduce thelytokously. On this hypothesis, one expects thelytoky in 11 of the 14 colonies (Table 2). However, we found that thelytoky occurred in only 8 of these colonies, and that offspring were produced solely by thelytoky in only three of them (colony 20, 21 and 22), even though all workers were homozygous for the 9 bp deletion. Our data are thus inconsistent with the hypothesis that homozygosity at the 9 bp deletion is necessary and sufficient for thelytoky.
Figure 3.
Alignment of the tae region sequenced with the gem_taeIV primer in the reverse direction. Wild type represents the annotated sequence (A mel 4.5). The tae1 allele was associated with thelytoky.
Alternatively, if homozygosity for the tae1 allele alone is required for thelytoky then we would expect both thelytoky and arrhenotoky in six of our colonies, (half of the workers in these colonies were expected to be homozygous for tae1 and half were expected to be heterozygous tae1,tae2). In the six colonies (colonies 15, 16, 20, 21, 22 and 26) where both thelytokous and arrhenotokous parthenogenesis is expected, 106 individuals were produced thelytokously and 30 were produced arrhenotokously (Table 2; Supplementary Information 2). Both thelytokous and arrhenotokous offspring were produced in three of these colonies (colony 15, 16 and 26), but in the other three colonies (20, 21 and 22) of this type only thelytokously produced offspring were identified (Table 2; Supplementary Information 2). In two of the three backcross to Scutellata colonies and all F1 colonies, all workers were either heterozygous for tae1 or did not carry tae1 (Table 2), and thus only arrhenotoky should have been observed. As expected under this hypothesis, we found 201 arrhenotokously produced individuals in these colonies, but also two thelytokously produced individuals (see above; Table 2). However, of the 94 thelytokously produced offspring we genotyped at tae, only 45.2% were homozygous for tae1 (Table 2, Supplementary Information 2), while the remainder were heterozygous. Therefore both homozygous tae1 and heterozygous tae1,tae2 individuals were reproducing thelytokously. Thus our data do not support the hypothesis that homozygosity of the 9 bp deletion, tae1, is responsible for thelytokous reproduction.
The tae deletion is widespread in bees of African descent
Genotyping and sequencing of tae revealed that the tae1 allele was present in the Scutellata parent from Douglas (Table 2; colony 3, 4, 14 and 20; Supplementary Information 2). Thelytokous parthenogenesis has not been observed in Scutellata workers (Hepburn and Crewe, 1990; Hepburn et al., 1994, 1998). Following this unexpected finding, we determined the frequency of tae1 in various honey bee populations to see if there is any association between the presence of the tae1 allele and thelytokous reproduction. We found that the tae1 and tae2 alleles are present throughout South Africa, including the Scutellata population we had sampled from four locations north of Douglas in 1984, prior to the inception of the Clone (Table 3; Supplementary Information 3). Moreover, the 9 bp deletion at tae occurs in the Africanized honey bee population in the Americas (Table 3; Supplementary Information 3). Africanized honey bees from the Americas are hybrids between the previously introduced bees of European origin and the more recently introduced Scutellata (Winston, 1992). Yet to our knowledge thelytoky has never been reported from Africanized bees.
Table 3. Presence (X) or absence (—) of tae alleles in global honey bee populations.
| Population | tae1 | tae2 | tae3 | tae4 | Sample Size | Year collected |
|---|---|---|---|---|---|---|
| Africanized, Texas, USA | X | X | X | X | 23 | 2013 |
| Africanized, Brazil | X | X | X | X | 17 | 1993 |
| A. m. capensis | X | X | X | X | 56 | 1984, 2004, 2009, 2011, 2012, 2013 |
| A. m. carnica | — | — | — | X | 6 | 1990 |
| A. m. intermissa | X | X | X | — | 8 | 1989 |
| A. m. ligustica | — | — | X | X | 25 | 1992 |
| A. m. mellifera | — | — | X | X | 5 | 1991 |
| A. m. scutellata | X | X | X | X | 80 | 1984, 1993, 2006, 2008, 2012 |
| Euro-American | — | — | X | X | 30 | 2013 |
The tae1 and tae2 alleles contain a 9 bp deletion. The tae1 allele was previously found to be associated with thelytokous parthenogenesis. The location of each collection point is in Supplementary Information 3 SI3.
Scutellata was introduced into Brazil in 1956, and has since spread as far south as Argentina and north into the south-western states of the United States (Winston, 1992). As we did not find the tae1 or tae2 alleles in a sample of 36 bees from Europe (A. m. mellifera, A. m. carnica and A. m. ligustica), nor in the sample of 30 European-derived bees from the United States, it seems likely that the 1956 introduction is the source of the tae1 and tae2 alleles in the Africanized bees (Table 3; Supplementary Information 3). The tae1 and tae2 alleles are also present in A. m. intermissa from Morocco, a subspecies that, like Capensis and Scutellata, is from the African evolutionary lineage (Ruttner, 1988; Franck et al., 2001; Whitfield et al., 2006; Harpur et al., 2014). It thus appears that the tae1 and tae2 alleles are associated with the geographic origin of bees—from Africa or hybridised with bees from Africa. The fact that thelytokous parthenogenesis has not been reported in the other populations where the 9 bp deletion occurs is further evidence that the tae locus is not the genetic switch between thelytoky and arrhenotoky.
Reproductive dominance and the tae locus
Jarosch et al. (2011) showed that differential splicing of gemini is associated with the different reproductive states of queens and workers, and knockdown of exon 5 resulted in workers with more active ovaries. Unfortunately, we have too few data to determine whether individuals homozygous for alleles with the 9 bp deletion (tae1 and tae2) have reproductive dominance over the individuals carrying alleles without the deletion (tae3 and tae4). However, there were six colonies (colony 15, 16, 20, 21, 22 and 26) where half of the workers are expected to be homozygous tae1,tae1 and the other half to be heterozygous tae1,tae2. Following the logic (above) for the th locus, their diploid offspring should be in the following proportions: 0.583 tae1,tae1, 0.333 tae1,tae2 and 0.083 tae2,tae2. However, we found an excess of thelytokously produced individuals that were heterozygous tae1,tae2 (51) and a deficit of homozygous tae1,tae1 (41) and tae2,tae2 (0) individuals (χ22=24.138, P<0.001; Table 3; Supplementary Information 2) suggesting that workers heterozygous tae1,tae2 are reproductively dominant over the workers homozygous tae1,tae1.
Discussion
Our results show that the 9 bp deletion tae1 at the tae locus that has been associated with thelytoky (Jarosch et al., 2011) is also found in non-thelytokously reproducing honey bees of African origin in South Africa (Scutellata), Morocco (A. m. intermissa) and Africanized bees in Brazil and USA. This polymorphism was not found in European (A. m. mellifera, A. m. ligustica and A. m. carnica) and European-derived populations from the United States. It is therefore highly unlikely that the 9 bp deletion in the tae locus is responsible for thelytokous parthenogenesis in Capensis, but rather is associated with bees with African lineage ancestry. Our study has further revealed two novel ‘tae' alleles, one of which carries the 9 bp deletion hypothesised to be associated with thelytoky with a second deletion nearby (tae2). The tae2 polymorphism was also only found in African and African-derived populations. The remaining polymorphism (tae4) had a single-base insertion compared with wild type (tae3). The existence of multiple polymorphisms at the tae locus casts further doubt on the hypothesis that thelytoky is under the control of a simple short deletion in this locus.
If any polymorphism containing the 9 bp deletion resulted in thelytoky then we would expect thelytoky to have occurred in the majority of colonies (11 of 14). Instead we observed thelytoky in only 8 colonies and solely thelytokous reproduction in only 3. Thus our results are inconsistent with either of the 9 bp deletion alleles causing thelytoky. If homozygosity for the tae1 allele alone is required for thelytoky then all thelytokously produced individuals would be homozygous for this allele. However approximately half of the thelytokously produced offspring were heterozygous for tae1.
Our data are also inconsistent with the th locus controlling thelytoky. Unlike tae1, th is a hypothetical locus linked to the HB-THE microsatellite loci (Figure 1). If the recessive th allele from Capensis results in thelytoky then thelytokous reproduction should have only been observed in the offspring of F1 queens backcrossed to Capensis drones. However we observed thelytokously produced offspring in a colony headed by a F1 queen backcrossed to a Scutellata drone and we also observed a single thelytokously produced individual in two F1 colonies. Furthermore some thelytokously produced individuals in backcross colonies, where thelytoky was expected, carried HB-THE alleles derived from their Scutellata grandparent.
In agreement with Lattorff et al. (2005), we can also rule out maternally transmitted, microbially induced thelytoky in Capensis, because we found paternal transmission of thelytoky in the offspring of three colonies headed by F1 queens who had a Scutellata mother. Currently there are no examples of microbial induced thelytoky in eusocial Hymenoptera (Wenseleers and Billen, 2000). Wolbachia is common in ants (Wenseleers et al., 1998) and is present both in Capensis and Scutellata (Hoy et al., 2003). Furthermore, microbial induction of thelytoky in Hymenoptera is only rarely associated with automixis (Leach et al., 2009), the mode of thelytoky in Capensis (Verma and Ruttner, 1983).
Previous studies have also found variable patterns of expression of the thelytoky phenotype depending on the genetic background of the sire and queen. When Capensis queens were crossed with A. m. carnica males, thelytoky was expressed in offspring workers, but when crossed with A. m. ligustica males, offspring workers reproduced arrhenotokously (Ruttner, 1988), indicating an effect of sire independent of the genetic background of the queen. Similarly, aspects of honey bee defensiveness (Guzman-Novoa et al., 2005) and reproductive physiology (Jordan et al., 2008a; Linksvayer et al., 2009; Beekman et al., 2012; Oldroyd et al., 2014) are more strongly transmitted via males than via females. Honey bees have a fully functional DNA methylation system (Wang et al., 2006; Foret et al., 2009) and differential methylation depending on parent-of-origin could explain the observed effects of sire (Drewell et al., 2012; Drewell et al., 2014; Oldroyd et al., 2014). We found that two colonies headed by F1 queens backcrossed to Scutellata drones produced a similar proportion of offspring thelytokously (21 thelytokous vs 6 arrhenotokous) as two colonies headed by F1 queens backcrossed to Capensis drones (37 thelytokous vs 21 arrhenotokous; χ21=1.662, P=0.197). Although we did not find a paternal effect on the likelihood of thelytokous reproduction, our sample sizes were small and it remains possible that epigenetics effects play a role in the expression of thelytoky.
In colonies where both thelytokous and arrhenotokous reproduction were expected we found a higher frequency of thelytokously produced individuals than we expected on the basis of simple Mendelian ratios of a putative th locus. However thelytoky in Capensis and the Clone is associated with a suite of characteristics associated with reproductive dominance, including increased frequency of ovary activation, faster activation of ovaries, production of queen-like pheromones and inhibition of ovary activation in other workers (for example, Neumann et al., 2000; Dietemann et al., 2007; Lattorff et al., 2007; Goudie and Oldroyd, 2014). Thus in colonies where both types of parthenogenesis occur there will be more thelytokously produced offspring than arrhenotokously produced offspring as a result of reproductive dominant thelytokously reproducing individuals. Our data are consistent with this hypothesis. The th locus may be associated with reproductive dominance, and this may have led to the belief that it also controlled thelytoky as most individuals would be produced by workers carrying the reproductive dominance trait (Lattorff et al., 2005; Lattorff et al., 2007; Jarosch et al., 2011). Individuals heterozygous tae1,tae2 were reproductively dominant over individuals homozygous tae1,tae1. We were unable to determine if individuals carrying either of these alleles were dominant over individuals carrying alleles that did not have the 9 bp deletion.
The absence of thelytokously or arrhenotokously produced individuals in a colony does not prove the inability of the workers in that colony to use either form of parthenogenesis. It may be that ecological factors play a role in determining the type of parthenogenesis used by queenless workers. Some colonies in our study were heavily parasitised by workers from other colonies and this is likely to have inhibited the reproduction of the host colony workers.
In summary, we found that (1) a recessive allele at the thelytoky (th) locus (Lattorff et al., 2005, 2007) is unlikely to be the sole cause of thelytokous parthenogenesis in Capensis workers. (2) A recessive gene linked to the three tightly linked markers HB-THE-02, HB-THE-03 and HB-THE-04 within th may be associated with reproductive dominance. (3) The tae length polymorphism within gemini is unrelated to thelytoky. (4) The 9 bp deletion suggested as being causative of thelytoky is common in arrhenotokous populations of African origin, including Scutellata and Africanized bees. (5) We identified two previously unreported tae polymorphisms. (6) Individuals heterozygous for the two 9 bp deletion alleles tae1 and tae2 are reproductively dominant over individuals homozygous tae1. (7) There is no evidence for maternally transmitted microbial induced thelytoky. Thus while the cause of thelytoky in Capensis remains unknown, a number of postulated mechanisms can now been ruled out. The wasp Lysiphlebus fabarum remains the only hymenopteran in which thelytoky is determined by a single locus.
Data accessibility
All data have been deposited at the University of Sydney: http://ses.library.usyd.edu.au/handle/2123/12446.
Acknowledgments
BPO and MB are supported by the Australian Research Council. NC and BPO are supported by the honey bee program of the Rural Industries Research and Development Corporation. We thank members of the Behaviour and Genetics of Social Insects Lab and four anonymous reviewers for constructive criticism that improved the manuscript.
Author Contributions
BPO, MB and MHA designed the experiment and performed field work. NCC, JL, MB and PRO performed genotyping. NCC and PRO analysed data. NCC, MB, PRO and BPO wrote the paper. MHA and TER provided samples.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp MH, Beekman M, Gloag RS, Oldroyd BP. Maternity of replacement queens in the thelytokous Cape honey bee Apis mellifera capensis. Behav Ecol Sociobiol. 2010;64:567–574. [Google Scholar]
- Beekman M, Allsopp MH, Holmes MJ, Lim J, Noach-Pienaar LA, Wossler TC, et al. Racial mixing in South African honeybees: the effects of genotype mixing on reproductive traits of workers. Behav Ecol Sociobiol. 2012;66:897–904. [Google Scholar]
- Beekman M, Allsopp MH, Jordan LA, Lim J, Oldroyd BP. A quantitative study of worker reproduction in queenright colonies of the Cape honey bee Apis mellifera capensis. Mol Ecol. 2009;18:2722–2727. doi: 10.1111/j.1365-294X.2009.04224.x. [DOI] [PubMed] [Google Scholar]
- Beekman M, Allsopp MH, Wossler TC, Oldroyd BP. Factors affecting the dynamics of the honeybee (Apis mellifera hybrid zone of South Africa. Heredity. 2008;100:13–18. doi: 10.1038/sj.hdy.6801058. [DOI] [PubMed] [Google Scholar]
- Beye M, Hasselmann M, Fondrk MK, Page RE, Omholt SW. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114:419–429. doi: 10.1016/s0092-8674(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Clarke KE, Oldroyd BP, Javier J, Quezada-Euan G, Rinderer TE. Origin of honeybees (Apis mellifera L.) from the Yucatan peninsula inferred from mitochondrial DNA analysis. Mol Ecol. 2001;10:1347–1355. doi: 10.1046/j.1365-294x.2001.01274.x. [DOI] [PubMed] [Google Scholar]
- Clarke KE, Rinderer TE, Franck P, Quezada-Euan JG, Oldroyd BP. The Africanization of honeybees (Apis mellifera L.) of the Yucatan: a study of a massive hybridization event across time. Evolution. 2002;56:1462–1474. doi: 10.1111/j.0014-3820.2002.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Crozier RH, Pamilo P. Evolution of Social Insect Colonies. Oxford University Press: Oxford, UK; 1996. [Google Scholar]
- DeGrandi-Hoffman G, Erickson EHJ, Lusby D, Lusby E. Thelytoky in a strain of U.S. honey bees (Apis mellifera L.) Bee Sci. 1991;1:166–171. [Google Scholar]
- Dietemann V, Neumann P, Härtel S, Pirk CWW, Crewe RM. Pheromonal dominance and the selection of a socially parasitic honeybee worker lineage (Apis mellifera capensis Esch.) J Evol Biol. 2007;20:997–1007. doi: 10.1111/j.1420-9101.2007.01303.x. [DOI] [PubMed] [Google Scholar]
- Drewell RA, Bush EC, Remnant EJ, Wong GT, Beeler SM, Stringham JL, et al. The dynamic DNA methylation cycle from egg to sperm in the honey bee Apis mellifera. Development. 2014;141:2702–2011. doi: 10.1242/dev.110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewell RA, Lo N, Oxley PR, Oldroyd BP. Kin conflict in insect societies: a new epigenetic perspective. Trends Ecol Evol. 2012;27:367–373. doi: 10.1016/j.tree.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Estoup A, Solignac M, Harry M, Cornuet JM. Characterization of (GT)n and (CT)n microsatellites in two insect species: Apis mellifera and Bombus terrestris. Nucleic Acids Res. 1993;21:1427–1431. doi: 10.1093/nar/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret S, Kucharski R, Pittelkow Y, Lockett GA, Maleszka J. Epigenetic regulation of the honey bee transcriptome: unravelling the nature of methylated genes. BMC Genomics. 2009;10:472. doi: 10.1186/1471-2164-10-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck P, Garnery L, Loiseau A, Oldroyd BP, Hepburn HR, Solignac M, et al. Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data. Heredity. 2001;86:420–430. doi: 10.1046/j.1365-2540.2001.00842.x. [DOI] [PubMed] [Google Scholar]
- Goudie F, Allsopp MH, Beekman M, Oxley PR, Lim J, Oldroyd BP. Maintenance and loss of heterozygosity in a thelytokous lineage of honey bees (Apis mellifera capensis. Evolution. 2012;66:1897–1906. doi: 10.1111/j.1558-5646.2011.01543.x. [DOI] [PubMed] [Google Scholar]
- Goudie F, Oldroyd BP. Theltyoky in the honey bee. Apidologie. 2014;45:306–326. [Google Scholar]
- Guzman-Novoa E, Hunt GJ, Page RE, Uribe-Rubio JL, Prieto-Merlos D, Becerra-Guzman F. Paternal effects on the defensive behavior of honeybees. J Heredity. 2005;96:376–380. doi: 10.1093/jhered/esi038. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. Sex versus non-sex versus parasite. Oikos. 1980;35:282–290. [Google Scholar]
- Hamilton WD, Axelrod R. Sexual reproduction as an adaptation to resist parasites. Proc Natl Acad Sci USA. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpur BA, Kent CF, Molodtsova D, Lebon JM, Alqarni AS, Owayss AA, et al. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc Natl Acad Sci USA. 2014;111:2614–2619. doi: 10.1073/pnas.1315506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn HR, Crewe RM. Defining the Cape honeybee: reproductive traits of queenless workers. South African J Sci. 1990;86:524–527. [Google Scholar]
- Hepburn HR, Jones GE, Kirby R. Introgression between Apis mellifera capensis Escholtz and Apis mellifera scutellata Lepeletier: the sting pheromones. Apidologie. 1994;25:557–565. [Google Scholar]
- Hepburn HR, Radloff SE, Fuchs S. Population structure and the interface between Apis mellifera capensis and Apis mellifera scutellata. Apidologie. 1998;29:333–346. [Google Scholar]
- Hepburn R, Radloff S. Apis mellifera capensis: an essary on the subspecific classification of honeybees. Apidologie. 2002;33:105–127. [Google Scholar]
- Holmes MJ, Oldroyd BP, Allsopp MH, Lim J, Wossler TC, Beekman M. Maternity of emergency queens in the Cape honey bee Apis mellifera capensis. Mol Ecol. 2010;19:2792–2799. doi: 10.1111/j.1365-294X.2010.04683.x. [DOI] [PubMed] [Google Scholar]
- Holmes MJ, Tan K, Wang Z, Oldroyd BP, Beekman M. Genetic reincarnation of workers as queens in the Eastern honeybee Apis cerana. Heredity. 2015;144:65–68. doi: 10.1038/hdy.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey Bee Genome Sequencing Consortium Finding the missing honey bee genes: lessons from a genome upgrade. BMC Genomics. 2014;15:86. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy MA, Jeyaprakash A, Alvareza JM, Allsopp MH. Wolbachia is present in Apis mellifera capensisA. m. scutellata, and their hybrid in Southern Africa. Apidologie. 2003;34:53–60. [Google Scholar]
- Huiens ME, Luck RF, Klaassen RHG, Maas MF, Timmermans MJ, Stouthamer R. Infectious parthenogenesis. Nature. 2000;4005:178–179. doi: 10.1038/35012066. [DOI] [PubMed] [Google Scholar]
- Jarosch A, Stolle E, Crewe RM, Moritz RFA. Alternative splicing of a single transcription factor drives selfish reproductive behavior in honeybee workers (Apis mellifera. Proc Natl Acad Sci USA. 2011;108:15282–15287. doi: 10.1073/pnas.1109343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsmeier MF. Experiences with the Cape bee in the Transvaal. South African Bee J. 1983;55:130–138. [Google Scholar]
- Jordan LA, Allsopp MH, Beekman M, Wossler TC, Oldroyd BP. Inheritance of traits associated with reproductive potential in Apis mellifera capensis and Apis mellifera scutellata workers. J Heredity. 2008;99:376–381. doi: 10.1093/jhered/esn023. [DOI] [PubMed] [Google Scholar]
- Jordan LA, Allsopp MH, Oldroyd BP, Wossler TC, Beekman M. Cheating honeybee workers produce royal offspring. Proc R Soc B-Biol Sci. 2008;275:345–351. doi: 10.1098/rspb.2007.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattorff HMG, Moritz RFA, Crewe RM, Solignac M. Control of reproductive dominance by the thelytoky gene in honeybees. Biol Lett. 2007;3:292–295. doi: 10.1098/rsbl.2007.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattorff HMG, Moritz RFA, Fuchs S. A single locus determines thelytokous parthenogenesis of laying honeybee workers (Apis mellifera capensis. Heredity. 2005;94:533–537. doi: 10.1038/sj.hdy.6800654. [DOI] [PubMed] [Google Scholar]
- Leach IM, Pannebakker BA, Schneider MV, Driessen G, Van de Zande L, Beukeboom LW.2009Thelytoky in Hymenoptera with Venturia canescens and Leptopilinaclavipes as Case StudiesIn: Schon I, Martens K, Dijk PJ, van Dijk P (eds)Lost Sex: The Evolutionary Biology of Parthenogenesis Springer: London, UK [Google Scholar]
- Linksvayer TA, Rueppell O, Siegel A, Kaftanoglu O, Page RE, Jr, Amdam GV. The genetic basis of transgressive ovary size in honeybee workers. Genetics. 2009;183:693–707. doi: 10.1534/genetics.109.105452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundie AE. Laying worker bees produce worker bees. South African Bee J. 1954;29:10–11. [Google Scholar]
- Mackensen O. The occurrence of parthenogenetic females in some strains of honeybees. J Econ Entomol. 1943;36:465–467. [Google Scholar]
- Martin S, Wossler T, Kryger P. Usurpation of African Apis mellifera scutellata colonies by parasitic Apis mellifera capensis workers. Apidologie. 2002;33:215–231. [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutation Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Neumann P, Härtel S, Kryger P, Crewe RM, Moritz RFA. Reproductive division of labour and thelytoky result in sympatric barriers to gene flow in honeybees (Apis mellifera L.) J Evol Biol. 2010;24:286–294. doi: 10.1111/j.1420-9101.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- Neumann P, Hepburn HR, Radloff SE. Modes of worker reproduction, reproductive dominance and brood cell construction in queenless honeybee (Apis mellifera L.) colonies. Apidologie. 2000;31:479–486. [Google Scholar]
- Oldroyd BP. The Cape honeybee: an example of a social cancer. Trends Ecol Evol. 2002;17:249–251. [Google Scholar]
- Oldroyd BP, Allsopp MH, Gloag RS, Lim J, Jordan LA, Beekman M. Thelytokous parthenogenesis in unmated queen honeybees (Apis mellifera capensis: central fusion and high recombination rates. Genetics. 2008;180:359–366. doi: 10.1534/genetics.108.090415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd BP, Allsopp MH, Lim J, Beekman M. A thelytokous lineage of socially parasitic honey bees has retained heterozygosity despite at least 10 year of inbreeding. Evolution. 2011;65:860–868. doi: 10.1111/j.1558-5646.2010.01164.x. [DOI] [PubMed] [Google Scholar]
- Oldroyd BP, Allsopp MH, Roth KM, Remnant EJ, Drewell RA, Beekman M. A parent-of-origin effect on honeybee worker ovary size. Proc R Soc B-Biol Sci. 2014;281:7. doi: 10.1098/rspb.2013.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onions GW. South African 'fertile worker bees'. South African Agric J. 1912;1:720–728. [Google Scholar]
- Rabeling C, Kronauer DJC. Thelytokous parthenogenesis in eusocial Hymenoptera. Annu Rev Entomol. 2013;58:273–292. doi: 10.1146/annurev-ento-120811-153710. [DOI] [PubMed] [Google Scholar]
- Ruttner F. Biogeography and Taxonomy of Honeybees. Springer-Verlag: Berlin, Germany; 1988. [Google Scholar]
- Sandrock C, Vorburger C. Single-locus recessive inheritance of asexual reproduction in a parasitoid wasp. Curr Biol. 2011;21:433–437. doi: 10.1016/j.cub.2011.01.070. [DOI] [PubMed] [Google Scholar]
- Shaibi T, Lattorff HMG, Moritz RFA. A microsatellite DNA toolkit for studying population structure in Apis mellifera. Mol Ecol Resour. 2008;8:1034–1036. doi: 10.1111/j.1755-0998.2008.02146.x. [DOI] [PubMed] [Google Scholar]
- Solignac M, Vautrin D, Loiseau A, Mougel F, Baudry E, Estoup A, et al. Five hundred and fifty microsatellite markers for the study of the honeybee (Apis mellifera L.) genome. Mol Ecol Notes. 2003;3:307–311. [Google Scholar]
- Verma S, Ruttner F. Cytological analysis of the thelytokous parthenogenesis in the Cape honeybee (Apis mellifera capensis Escholtz) Apidologie. 1983;14:41–57. [Google Scholar]
- Wang Y, Jorda M, Jones PL, Maleszka R, Ling X, Robertson HM, et al. Functional CpG methylation system in a social insect. Science. 2006;314:645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- Wenseleers T, Billen J. No evidence for Wolbachia-induced parthenogenesis in the social Hymenoptera. J Evol Biol. 2000;13:277–280. [Google Scholar]
- Wenseleers T, Ito F, van Borm S, Huybrechts R, Volckaert F, Billen J. Widespread occurence of the micro-organism Wolbachia in ants. Proc R Soc B Biol Sci. 1998;265:1447–1452. doi: 10.1098/rspb.1998.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenseleers T, Oystaeyen V. Unusual modes of reproduction in social insects: shedding light on the evolutionary paradox of sex. Bioessays. 2011;33:927–937. doi: 10.1002/bies.201100096. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Behura SK, Berlocher SH, Clark AG, Johnston JS, Sheppard WS, et al. Thrice out of Africa: ancient and recent expansions of the honey bee, Apis mellifera. Science. 2006;314:642–645. doi: 10.1126/science.1132772. [DOI] [PubMed] [Google Scholar]
- Winston ML. Killer Bees: The Africanized Honey Bee in the Americas. Harvard University Press: Harvard, USA; 1992. [Google Scholar]
- Woyke J. What happens to diploid drone larvae in a honeybee colony. J Apicult Res. 1963;2:73–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data have been deposited at the University of Sydney: http://ses.library.usyd.edu.au/handle/2123/12446.