Abstract
Genome sequence comparison across the Drosophila genus revealed that some fixed inversion breakpoints had been multiply reused at this long timescale. Cytological studies of Drosophila inversion polymorphism had previously shown that, also at this shorter timescale, some breakpoints had been multiply reused. The paucity of molecularly characterized polymorphic inversion breakpoints has so far precluded contrasting whether cytologically shared breakpoints of these relatively young inversions are actually reused at the molecular level. The E chromosome of Drosophila subobscura stands out because it presents several inversion complexes. This is the case of the E1+2+9+3 arrangement that originated from the ancestral Est arrangement through the sequential accumulation of four inversions (E1, E2, E9 and E3) sharing some breakpoints. We recently identified the breakpoints of inversions E1 and E2, which allowed establishing reuse at the molecular level of the cytologically shared breakpoint of these inversions. Here, we identified and sequenced the breakpoints of inversions E9 and E3, because they share breakpoints at sections 58D and 64C with those of inversions E1 and E2. This has allowed establishing that E9 and E3 originated through the staggered-break mechanism. Most importantly, sequence comparison has revealed the multiple reuse at the molecular level of the proximal breakpoint (section 58D), which would have been used at least by inversions E2, E9 and E3. In contrast, the distal breakpoint (section 64C) might have been only reused once by inversions E1 and E2, because the distal E3 breakpoint is displaced >70 kb from the other breakpoint limits.
Introduction
Cytological studies as well as comparative studies of the genome sequences from related species revealed that genome reorganization is widespread among taxa (for example, Krimbas and Powell, 1992; Newman et al., 2005; Clark et al., 2007; Ferguson-Smith and Trifonov, 2007; Schaeffer et al., 2008). Structural variation ranging from chromosome fusions to paracentric inversions underlies the detected reorganization. These studies also detected an uneven distribution of rearrangement breakpoints across the genome of both mammals and dipters, with some regions having suffered multiple disruptions (for example, Coluzzi et al., 2002; Pevzner and Tesler, 2003). This coincidental occurrence of a rearrangement breakpoint in the same region in two or more species is referred to as breakpoint reuse. In Drosophila, there is both cytological and molecular evidence that the breakpoints of some fixed inversions have been used more than once through evolutionary time (for example, Tonzetich et al., 1988; Ruiz and Wasserman, 1993; Ranz et al., 2007; Bhutkar et al., 2008; von Grotthuss et al., 2010). Even at the much shorter intraspecific timescale, a similar observation of some reused breakpoints has emerged from cytological studies of chromosomal inversion polymorphism in different Drosophila species (Kunze-Mühl and Müller, 1958; Aulard et al., 2002).
The identification and molecular characterization of inversion breakpoints constitute a starting point not only to address questions about the mechanisms for the origin of inversions but also to contrast whether breakpoints reused at the cytological level are or not reused at the molecular level. Polymorphic inversions offer an advantage over fixed inversions due to the shorter time elapsed since their origin. Indeed, extant sequences of polymorphic inversion breakpoints are expected to better reflect the sequences in the original inverted chromosome. Despite the extensive knowledge accumulated on inversion polymorphism in Drosophila, there are relatively few polymorphic inversion breakpoints molecularly characterized both in any particular species and across species (but see Wesley and Eanes, 1994; Andolfatto et al., 1999; Cáceres et al., 1999; Casals et al., 2003; Matzkin et al., 2005; Richards et al., 2005; Delprat et al., 2009; Corbett-Detig et al., 2012; Papaceit et al., 2013; Puerma et al., 2014). The number of molecularly characterized breakpoints drops even more when inversions with cytological evidence of reused breakpoints are considered (Corbett-Detig et al., 2012; Puerma et al., 2014). This paucity of molecularly characterized polymorphic inversion breakpoints has so far precluded to extensively contrast whether cytologically shared breakpoints of the relatively young polymorphic inversions are actually reused at the molecular level. It has also precluded ascertaining why disruptions occur repeatedly at multiply reused breakpoints.
Drosophila subobscura has a rich chromosomal inversion polymorphism in its five major chromosomes. Its E chromosome (Muller's C element) stands out because it presents several inversion complexes. One such case is the E1+2+9+3 arrangement that originated from the ancestral Est arrangement through the sequential accumulation of four inversions sharing some of their breakpoints (E1, E2, E9 and E3; Figure 1). These characteristics render this chromosomal element particularly suitable not only to study the mechanisms underlying the origin of inversions through the molecular characterization of their breakpoints, but also to study breakpoint reuse, and more specifically to test whether breakpoints of polymorphic inversions are actually reused, or multiply reused, at the molecular level. Moreover, this characterization may provide new insights into the role played by asynapsis in inversion heterokaryotype breakpoints in the sequential accumulation of polymorphic inversions with shared breakpoints.
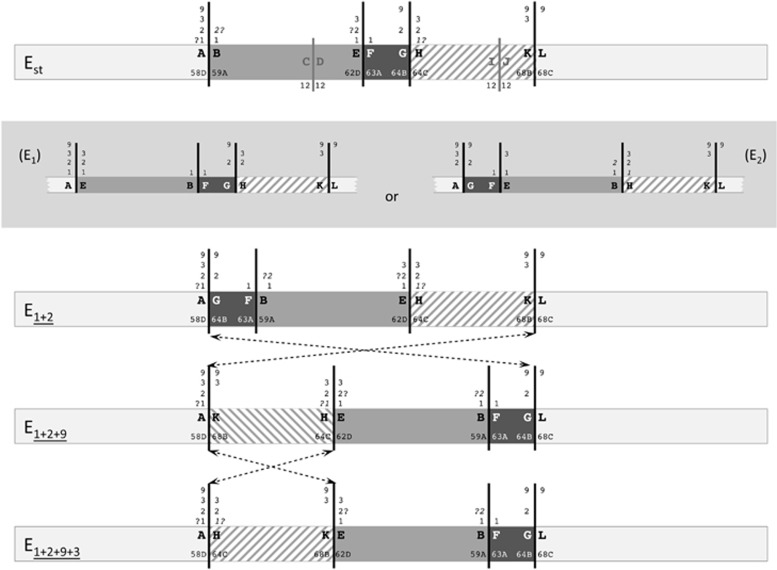
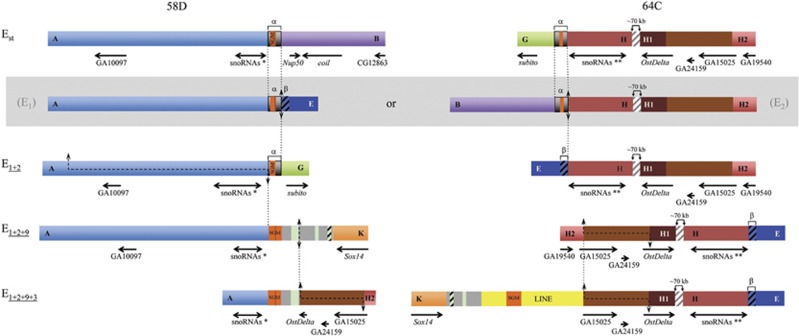
Figure 1.
Schematic representation of E chromosomal arrangements of D. subobscura. The scheme includes four extant E chromosomal arrangements of D. subobscura as well as the two possible intermediate, and now extinct, arrangements connecting Est and E1+2 (within a gray box). On the Est chromosome scheme, black and gray vertical lines represent the cytological location of the breakpoints of five inversions, with the black lines referring to those of inversions included in the present study (E1, E2, E9 and E3). The fragments separating these inversion breakpoints are differentiated either by color (gray, dark gray) or texture (striped) to facilitate their identification in the different arrangements. Inversion breakpoints are labeled consecutively with pairs of capital letters (for example, AB, CD and EF) from the most proximal to the most distal breakpoint, as in Puerma et al. (2014), with numbers on both sides of each continuous vertical line referring to the inversions delimited by each breakpoint and map sections indicating their location on the Kunze-Mühl and Müller (1958) map. Discontinuous lines connecting two arrangements refer to the region inverted in each case.
We recently characterized through chromosome walking the breakpoints of the two inversions leading from the ancestral arrangement of the E chromosome in the D. subobscura subgroup (Est) to the E1+2 arrangement (Puerma et al., 2014). At the cytological level, inversions E1 and E2 share one breakpoint. Comparison of the three breakpoint regions in the two arrangements—AB, EF and GH in Est, and AG, FB and EH in E1+2 (Figure 1)—pointed to each inversion having been generated by a different mechanism: E1 through staggered double-strand breaks, and E2 through ectopic recombination (Puerma et al., 2014). This study also revealed that the breakpoint reused at the cytological level had also been reused at the molecular level, even though we could not establish the order of occurrence of both inversions and, therefore, which breakpoint had been reused. The breakpoint reused would be the most proximal one corresponding to chromosomal section 58D if E1 had occurred before E2, whereas in the other case (E2 before E1) it would be the most distal one corresponding to chromosomal section 64C (Figure 1).
Here, we have identified and characterized the breakpoints of inversions E9 and E3 that occurred sequentially and generated the E1+2+9 and E1+2+9+3 arrangements (Figure 1). As previously indicated, some breakpoints of inversions E1, E2, E9 and E3 are multiply shared at the cytological level (Figure 1). Indeed, if inversion E1 had occurred first, the most proximal breakpoint at section 58D would have been shared by all four inversions, whereas the most distal breakpoint at section 64C would have been shared by inversions E2 and E3. Alternatively, if inversion E2 had occurred first, the proximal breakpoint would have been shared by inversions E2, E9 and E3, and the distal breakpoint by inversions E1, E2 and E3. The molecular characterization of the breakpoints of inversions E9 and E3 in the corresponding ancestral and derived arrangements will not only allow establishing each inversion generating mechanism but also contrasting breakpoint reuse at the molecular level. Most importantly, it will provide novel molecular information concerning breakpoints that have been multiply reused at least at the cytological level.
Materials and Methods
Drosophila strains
Four D. subobscura strains were used to identify the breakpoints of inversions E9 and E3 and to sequence the breakpoint regions: ch cu, OF21, OF82 and FO_12b, which are homokaryotypic for the Est, E1+2, E1+2+9 and E1+2+9+3 chromosomal arrangements, respectively. OF strains were obtained through over 13 generations of sib mating from isofemale lines established upon collection in Observatori Fabra (Barcelona, Catalonia, Spain), as reported in Puerma et al. (2014). Strain FO_12b was made homokaryotypic for the E1+2+9+3 chromosomal arrangement upon 11 generations of sib mating from the segregating F1 of a cross between a wild-caught male from Observatori Fabra and a strain homokaryotypic for the Est arrangement (ch cu).
Identification of breakpoint regions: chromosome walk, in situ hybridization and sequencing
The starting point to identify breakpoint KL in Est and E1+2 chromosomes (Figure 1) was a molecular marker previously located at section 68B of the E chromosome (Laayouni et al., 2007). Our walk was based on both the D. pseudoobscura and D. melanogaster genome sequences, with oligonucleotides for PCR amplification designed using D. subobscura sequences whenever available (Barcelona Subobscura Initiative [BSI]). Labeled probes were in situ hybridized on polytene chromosomes of D. subobscura (see below), which allowed walking toward the breakpoint and to eventually cross it.
For breakpoint regions AK and GL in E1+2+9 chromosomes, and breakpoint regions AH and KE in E1+2+9+3 chromosomes (see Results section for the renaming of these breakpoints), the fragment spanning each breakpoint was PCR amplified using oligonucleotides anchored at its flanking regions (Supplementary Figure S1). Different Taq polymerases (GoTaq DNA polymerase from Promega Corporation, Fitchburg, WI, USA and TaKaRa DNA polymerase from Takara Bio Inc, Otsu, Japan) were used for PCR amplification according to the expected length of the fragment to be amplified.
All steps of the in situ hybridization procedure were performed as described in Montgomery et al. (1987). Hybridization signals were located on the cytological map of D. subobscura (Kunze-Mühl and Müller, 1958) with the standard arrangement for all chromosomes.
Only fragments spanning inversion breakpoints were sequenced upon their amplification by PCR (Supplementary Figure S1), using primer walking whenever necessary. MultiScreen PCR (Merck Millipore, Darmstadt, Germany) was used to purify amplicons before sequencing them with the ABI PRISM version 3.2 cycle sequencing kit (Thermo Fisher Scientific Inc, Waltham, MA, USA), with sequencing products separated on an ABI PRISM 3730 sequencer (Thermo Fisher Scientific Inc). All sequences were obtained on both strands and assembled using the DNASTAR package (Burland, 2000). When sequences could not be obtained directly from PCR products, we used the cloning and sequencing strategy described in Puerma et al. (2014).
Sequence analysis
All breakpoint regions were annotated with genes by comparison with the D. pseudoobscura genome of FlyBase (http://flybase.org/) using BLAST tools and analyzed to detect repeated motifs using RepeatMasker. The newly sequenced breakpoint regions as well as those of the extended E1+2 breakpoint regions were compared using the Align Sequences Nucleotide BLAST utility at NCBI webpage to accurately map each breakpoint and to determine putative duplications resulting from the inversion process.
Results
Identification and characterization of inversion E9 breakpoints
According to available cytological information (Kunze-Mühl and Müller, 1958), inversions E2 and E9 (and possibly E1; see Introduction and Figure 1) share their proximal breakpoint at section 58D. The previously sequenced ~7.1-kb fragment that spans this breakpoint in E1+2 (AG in Figure 2 in Puerma et al., 2014) was PCR amplified using DNA from strain OF21 (E1+2) and used as a probe for in situ hybridization on E1+2+9 chromosomes (strain OF82). This probe gave two strong signals at sections 58D next to 68B (breakpoint AK) and 64B next to 68C (breakpoint GL) (Figure 1 and Supplementary Figure S2), confirming that it included the proximal breakpoint of inversion E9.
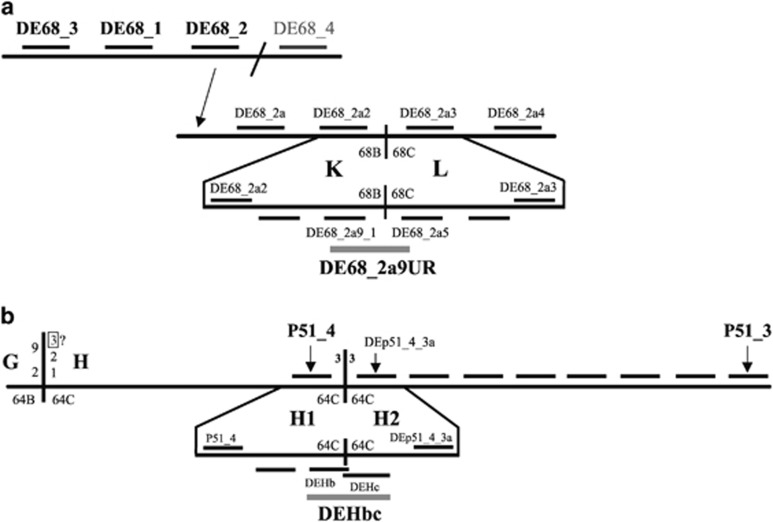
Figure 2.
Chromosome walks. Schematic representation of chromosome walks performed on Est (ch cu) chromosomes to identify the KL (a) and H1H2 (b) breakpoint regions (not at scale). Horizontal lines above the long horizontal lines that represent D. subobscura chromosomes indicate probes used in the first steps, whereas horizontal lines below the corresponding chromosome indicate those used in the final steps. Only the names of the most informative probes are shown. Probes spanning the breakpoints are highlighted in gray. In each breakpoint region, a thick vertical line represents the breakpoint itself. In the case of the KL breakpoint region (a), the inclined line at the upper scheme separates two noncontiguous regions in D. subobscura, whereas the arrow indicates the location of the corresponding probe in the lower scheme. In the case of the H1H2 breakpoint region (b), the vertical line on the left side of the D. subobscura chromosome corresponds to the GH breakpoint that according to cytological information would not only be shared by inversions E1, E2 and E9 but also by inversion E3.
To identify the distal breakpoint of inversion E9 (KL in Est and E1+2; Figure 1), a chromosome walk was started from a molecular marker previously located at section 68B of the E chromosome (Laayouni et al., 2007). Four probes were designed, two at each side of the marker, on the D. pseudoobscura genome sequence: probes DE68_1, DE68_2, DE68_3 and DE68_4 (Figure 2 and Supplementary Figure S3). Probes were amplified using DNA from the ch cu (Est) strain and subsequently hybridized on E1+2+9 chromosomes. Only the first three probes hybridized close to the inversion breakpoint, probes DE68_3 and DE68_1 at section 68A/B and probe DE68_2 at section 68B (Supplementary Table S2), indicating the direction in which to proceed with our walk. Moreover, a break of collinearity between the D. subobscura and D. pseudoobscura genomes was detected between fragments DE68_2 and DE68_4 (Supplementary Figure S3 and Supplementary Table S1). We designed four new probes based on the D. melanogaster genome sequence: DE68_2a, DE68_2a2, DE68_2a3 and DE68_2a4 (Figure 2 and Supplementary Figure S3). When in situ hybridized on E1+2+9 chromosomes, the first two probes gave a single strong signal at section 68B next to 58D, whereas the latter two probes did at section 68C next to 64B (that is, at a certain distance), indicating that probes DE68_2a2 and DE68_2a3 flanked the KL breakpoint (Figures 1 and 2; Supplementary Figures S3 and Supplementary Table S1). Four additional probes were designed in this ~30-kb-long interval and subsequently hybridized on E1+2+9 chromosomes, which allowed narrowing down the KL breakpoint region to an ~8-kb-long fragment flanked by probes DE68_2a9_1 and DE68_2a5 (Figure 2 and Supplementary Figure S3). A new probe (DE68_2a9UR) was designed anchored at genes Sox14 and Phm (Figure 2 and Supplementary Figure S3) that gave a single signal when in situ hybridized on E1+2 chromosomes, and two distant signals (at sections 68B and 68C) on E1+2+9 chromosomes both when amplified using DNA from strains ch cu (Est) and OF21 (E1+2) (Supplementary Figure S2).
Upon identification of inversion breakpoint regions on E1+2 chromosomes (AG and KL in Figure 1) and sequencing of the KL fragment in both Est (ch cu) and E1+2 (OF21) chromosomes, fragments spanning the E9 breakpoint regions in the E1+2+9 arrangement (AK and GL in Figure 1) were amplified with the corresponding combination of oligonucleotides (Supplementary Figure S1) and subsequently in situ hybridized on both E1+2 and E1+2+9 chromosomes. In both cases, the amplified fragments gave one strong signal on E1+2+9 chromosomes and two strong distant signals on E1+2 chromosomes at the expected locations, confirming that the breakpoints were included in the amplified fragments (Figure 1 and Supplementary Table S1). Each of these probes gave an additional weak signal at the alternative breakpoint on E1+2+9 chromosomes (see next paragraph and Supplementary Figure S2). The fragments spanning the breakpoints were completely sequenced in the OF82 strain (E1+2+9).
Pairwise comparison of the AG, AK and GL breakpoint regions allowed detecting the presence of the A part not only in the AG and AK regions but also in the GL region (Figure 3). Actually, the A fragment present in the GL region extended further upstream than that previously sequenced in the Est and E1+2 arrangements (Puerma et al., 2014), which prompted us to also sequence the extended stretch in the Est, E1+2 and E1+2+9 arrangements. In all three cases (Figure 3), the duplicated A fragment (~7.8-kb long) does not only contain the orthologs of the snoRNA genes GA29824 to GA29818 that are encoded in introns of the Uhg5 gene but also that of the GA10097 gene. The presence of this duplicated region in the AK and GL breakpoints accounts for the weak additional signal detected when probes containing these breakpoints are hybridized on E1+2+9 chromosomes (Supplementary Figure S2).
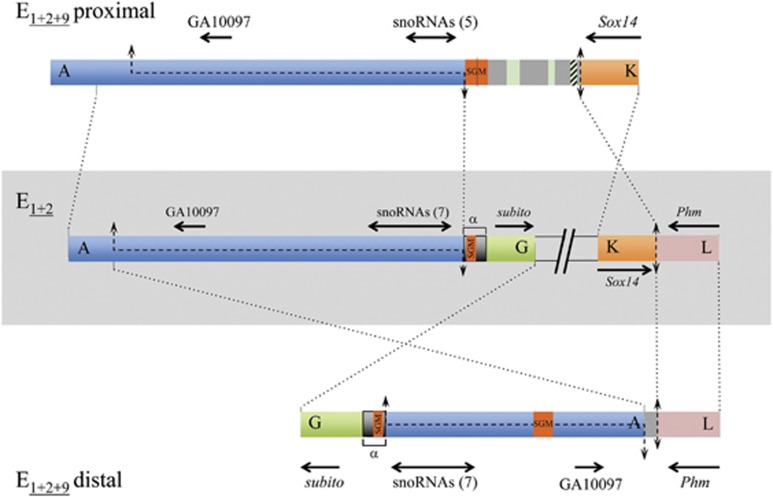
Figure 3.
E9 inversion breakpoints. Schematic representation of the E9 inversion breakpoint regions on E1+2 and E1+2+9 arrangements with breakpoints labeled as in Figure 1. Thick colored bars represent the different breakpoint regions. The central part shows, within a light gray rectangle, the scheme of both breakpoints of the E9 inversion in gene arrangement E1+2 (AG and KL), separated by two inclined lines. Schemes of the proximal and distal breakpoints in gene arrangement E1+2+9 (AK and GL) are represented above and below, respectively, of the E1+2 scheme. The lengths of the four sequenced breakpoint regions are AG, ~14.4-kb; KL, ~3.8-kb; AK, ~17.0-kb; and GL, ~13.0-kb. Black discontinuous lines along a chromosomal region represent staggered breaks with their limits indicated by arrows. Vertical double-headed arrows indicate cut-and-paste breakpoints. Dotted lines between arrangements indicate the limits and orientation of homologous regions. On each flanking region, the names of the orthologous coding regions in either D. melanogaster or D. pseudoobscura are given, with black arrows indicating their sense and approximate size. Double-headed horizontal arrows refer to the multiple snoRNAs generated from the corresponding Uhg gene introns, with their number given in parenthesis. Orange boxes labeled SGM refer to either near canonical copies of the SGM transposable element, or remnants thereof within a larger repeat sequence named α-motif in Puerma et al. (2014) and marked with an α. Gray regions present in the E9 breakpoints (AK and GL) in the E1+2+9 arrangement are intervening regions not present in any of the E1+2 breakpoint regions that in the proximal breakpoint present different-sized fragments of the gypsy (green) and Pao (striped green) transposable elements.
Pairwise comparison of the KL (~3.8-kb long), AK and GL breakpoint regions revealed the precise nucleotide at which the KL region had been broken in the inversion process (Figure 3). The K (~1.9-kb long) and L (~2.0-kb long) sequenced fragments contain the Sox14 gene (partially) and the Phm gene (partially), respectively.
In the AK (~18.2-kb long) breakpoint region, there is an intervening stretch (3.6-kb long) between the flanking parts present in the non-inverted breakpoints—AG (~14.4-kb long) and KL; Figure 3—This stretch includes a rather conserved SubobscuraGuancheMadeirensis (SGM) transposable element (Miller et al., 2000) and fragments of other transposable elements (Figure 3). It should be noted that the ~700 bp α-motif present in the G part of the AG breakpoint region—and also in that of the GL breakpoint region—constitutes the E2 proximal breakpoint itself (Puerma et al., 2014). The E2 breakpoint motif (Figure 2 in Puerma et al., 2014) exhibits only two fragments with some similarity to the SGM element, unlike the nearly canonical SGM element detected both at the AK breakpoint region and within the A part of the GL breakpoint region (Figure 3).
Identification and characterization of inversion E3 breakpoints
According to available cytological information (Kunze-Mühl and Müller, 1958), inversion E3 shares its proximal breakpoint at section 58D with those of inversions E2 (or E1 and E2; see Introduction and Figure 1) and E9, whereas it shares its distal breakpoint at section 64C with those of inversions E2 (or E1 and E2; see Introduction and Figure 1). Since inversion E3 occurred on an E1+2+9 chromosome, the fragment spanning the proximal breakpoint in this arrangement (AK in Figure 1) was PCR amplified using DNA from strain OF82 (E1+2+9) and subsequently in situ hybridized on E1+2+9+3 chromosomes (strain FO_12b), where it gave two strong signals at section 58D next to 64C (breakpoint AH) and 68B next to 62D (breakpoint KE) on E1+2+9+3 chromosomes (Figure 1 and Supplementary Figure S4), confirming that it included the proximal breakpoint of inversion E3. It should be noted that in both gene arrangements probe AK gave a fainter signal at section 64B next to 68C due to the presence of part A in the GL breakpoint as a result of its duplication during the E9 inversion process (Figure 3 and Supplementary Figure S4).
For the distal breakpoint, the fragment spanning it in E1+2 and E1+2+9 chromosomes (EH and HE, respectively, in Figure 1) was PCR amplified using DNA from strain OF21 (E1+2) and subsequently in situ hybridized on E1+2+9+3 chromosomes (strain FO_12b), where it gave only one signal at section 64C close to section 62D (Figure 1 and Supplementary Table S1), indicating that it did not span the distal breakpoint of inversion E3. To identify this breakpoint region, we took advantage of the previous chromosome walk that we had performed to identify the distal breakpoint (GH) of inversions E1 and E2 (Supplementary Figure S7 in Puerma et al., 2014), a walk that extended over 700 kb outside the inversion (that is, on its H part that is the one that at the cytological level is reused by inversion E3; Figure 1). To identify the new breakpoint region that was renamed H1H2, two probes from our previous walk that are at an appreciable distance from the GH breakpoint—probes P51_4 and P51_3 approximately 74-kb and 168-kb apart of that breakpoint (Supplementary Figure S7 in Puerma et al., 2014)—were in situ hybridized on E1+2+9+3 chromosomes. Probe P51_4 mapped at section 64C in the proximity of section 62D (that is, in the non-inverted H1 region; Supplementary Figure S1), whereas probe P51_3 did at section 64C in the proximity of section 58D (that is, in the inverted H2 region) (Supplementary Figure S1 and Table S1). The seven new probes designed in that ~94-kb-long interval mapped, like probe P51_3, at section 64C in the proximity of section 58D, like probe P51_3 (Figure 2, and Supplementary Figure S5 and Supplementary Table S1), indicating that the breakpoint was located between probes P51_4 and DEp51_4_3a. Upon narrowing down the breakpoint region with three new probes, a final probe (DEHbc) was designed to span the breakpoint (Figure 2 and Supplementary Figure S5). When this probe was in situ hybridized on E1+2+9+3 and E1+2+9 chromosomes, it gave two signals on the former chromosomes—at section 64C next to 62D (KH1) and at section 64C next to 58D (AH2), respectively; Supplementary Figures S1 and S4—while it gave a single signal at section 64C close to 62D (H2H1) on E1+2+9 chromosomes (Supplementary Figures S1 and S4), confirming that it contained the distal breakpoint of the E3 inversion.
Upon identification of inversion E3 breakpoint regions on E1+2+9 chromosomes—AK and H2H1 (~6.3-kb long) in Figure 4—and sequencing of the H2H1 fragment in E1+2+9 (OF82) chromosomes, fragments spanning the E1+2+9+3 breakpoint regions (AH2 and KH1) were amplified with the corresponding combination of oligonucleotides (Supplementary Figure S1) and subsequently in situ hybridized on both E1+2+9 and E1+2+9+3 chromosomes. In both cases, the amplified fragments gave two signals on E1+2+9 chromosomes, confirming that the breakpoints were included in the amplified fragments (Supplementary Figure S4). Moreover, these fragments (AH2 and KH1) gave also two signals on E1+2+9+3 chromosomes, suggesting the presence of a duplicated region in the E3 breakpoints (see below), and, similarly to fragment AK (see above), fragment AH2 gave an additional weak signal at section 64B next to 68C of the E1+2+9+3 arrangement (Supplementary Figure S4). Fragments AH2 (~8.3-kb long) and KH1 (~14.3-kb long) were completely sequenced in the FO_12b (E1+2+9+3) strain.
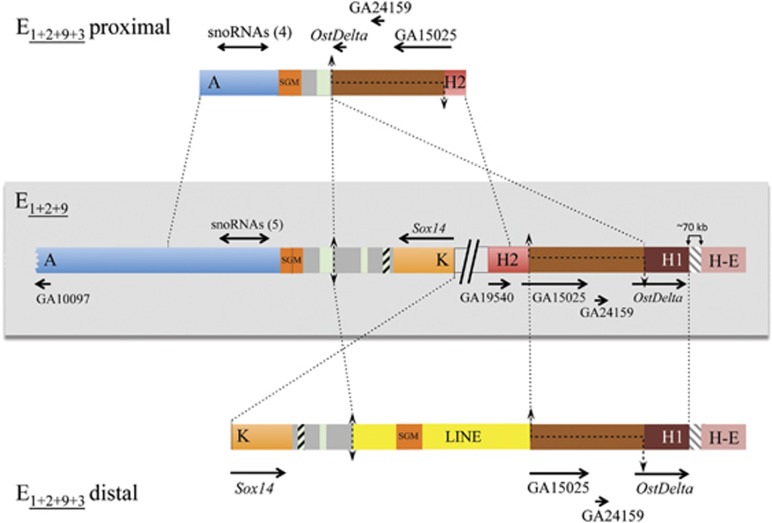
Figure 4.
E3 inversion breakpoints. Schematic representation of the E3 inversion breakpoint regions on E1+2+9 and E1+2+9+3 arrangements with the proximal breakpoint labeled as in Figure 1 (AK) and the distal breakpoint relocated to over 70 kb from the HE breakpoint and, therefore, renamed H2H1 (see text and Figure 2). Thick colored bars represent the different breakpoint regions. The central part shows, within a light gray rectangle, the scheme of both breakpoints of the E3 inversion in gene arrangement E1+2+9 (AK and H2H1), separated by two inclined lines. Schemes of the proximal and distal breakpoints in gene arrangement E1+2+9+3 (AH2 and KH1) are represented above and below, respectively, of the E1+2+9 scheme. The gray striped region present in the distal breakpoint (in both E1+2+9 and E1+2+9+3 arrangements) indicates the ~70-kb-long fragment separating the H part of the HE region from the H1 part of the H2H1 region. Black discontinuous lines along a chromosomal region represent staggered breaks with their limits indicated by arrows. Vertical double-headed arrows indicate cut-and-paste breakpoints. Dotted lines between arrangements indicate the limits and orientation of homologous regions. On each flanking region, the names of the orthologous coding regions in either D. melanogaster or D. pseudoobscura are given, with black arrows indicating their sense and approximate size. Double-headed horizontal arrows refer to the multiple snoRNAs generated from the corresponding Uhg gene introns, with their number given in parenthesis. Orange boxes labeled SGM refer to near canonical and canonical copies of the SGM transposable element, whereas green and striped green boxes refer to different-sized fragments of the gypsy and Pao transposable elements, respectively, and the two yellow boxes correspond to the two parts of a canonical LINE element. See Figure 3 legend for the gray boxes content.
The H2H1 breakpoint region contains the orthologs of genes GA19540, GA15025, GA24519 and OstDelta (Figure 4). Pairwise comparison of the H2H1, AH2 and KH1 breakpoint regions allowed detecting the presence of an ~3.6-kb fragment in the central part of the H2H1 region in both breakpoints of the E1+2+9+3 arrangement (Figure 4). Pairwise comparison of the AK, AH2 and KH1 regions revealed the precise nucleotide at which the AK region had been broken in the inversion process (Figure 4). Moreover, a LINE element was detected between the K and H1 parts of this breakpoint, with a canonical SGM element inserted within the LINE element (Figure 4).
Discussion
Origin of inversions in D. subobscura
An important question concerning the origin of chromosomal inversions, and more specifically of those that increase in frequency and either become polymorphic or attain fixation, refers to the relative importance of ectopic recombination between transposable elements (or other repetitive sequences) and of staggered double-strand breaks (Ranz et al., 2007) in their generation. A large number of breakpoints need to be molecularly characterized to reliably quantify the contribution of the different mechanisms generating inversions in any particular species or species group. In Drosophila, the currently largest data set corresponds to the 29 fixed inversions that differentiate D. melanogaster and its close relatives D. simulans and D. yakuba (Ranz et al., 2007). The presence of duplicated inverted non-repetitive fragments in the breakpoints of 17 inverted chromosomes led the authors to propose the staggered-break mechanism as the prevalent mechanism originating inversions in the melanogaster species group. The recent detection, also through genome sequence comparison, of inverted duplications at the breakpoints of 5 out of the 8 studied inversions segregating in natural populations of D. melanogaster (Corbett-Detig et al., 2012) would support the Ranz et al. (2007) proposal for the melanogaster species group. This proportion does not seem, however, to hold for the repleta group of species, neither for fixed inversions—with inverted duplications in 3 out of 10 fixed inversions (Calvete et al., 2012; Guillén and Ruiz, 2012)—nor for polymorphic inversions in D. buzzatii, where the generating mechanism of the three characterized inversions was ectopic recombination between transposable elements (Cáceres et al., 1999; Casals et al., 2003; Delprat et al., 2009).
In the species here studied, D. subobscura, the number of polymorphic inversions with breakpoints molecularly characterized in both inverted and non-inverted chromosomes is still relatively low for any formal quantitation of the generating mechanisms: one inversion—O3—in Muller's E element (Papaceit et al., 2013) and four inversions—E1, E2, E3 and E9—in Muller's C element (Puerma et al., 2014; present work). This number allows, however, a qualitative evaluation of their relative importance in this species. In the three previously characterized inversions—O3, E1 and E2—, only in the E2 case was ectopic recombination between inverted repeated elements—α-motifs—considered the generating mechanism. In contrast, inverted duplicates of rather small fragments were detected in O3 and E1, suggesting the staggered-break mechanism of origin (Papaceit et al., 2013; Puerma et al., 2014). In the case of inversions E9 and E3 here studied, we have detected inverted duplicates in the corresponding derived chromosomal arrangement in both cases, that is, in E1+2+9 and E1+2+9+3 chromosomal arrangements, respectively (Figures 3 and 4). Our results from five inversions are similar to those obtained from eight polymorphic inversions in D. melanogaster (Corbett-Detig et al., 2012), suggesting that also in D. subobscura staggered double-strand breaks and subsequent repair might be the prevalent mechanism generating inversions. Although the number of polymorphic inversions with breakpoints molecularly characterized is rather similar in both species, it should be noted that in contrast to the 29 fixed inversions with breakpoints molecularly characterized since the D. melanogaster – D. yakuba split (Ranz et al., 2007), no fixed inversion breakpoint has been yet characterized in the D. subobscura lineage. Indeed, even if we previously characterized the breakpoint regions of an inversion fixed since the split of the D. melanogaster and D. subobscura lineages (Cirera et al., 1995), the phylogenetic comparative analysis of these regions has allowed us to infer that it did occur in the melanogaster group and more specifically in the ancestor of the melanogaster subgroup (results not shown).
The extent and content of the fragments duplicated in the inversion process differs between the two previously characterized D. subobscura inversions—O3 and E1—and those here characterized—E9 and E3. In both O3 and E1, a rather small fragment was duplicated: an ~300-bp-long intergenic fragment in the O3 case (Papaceit et al., 2013), and an ~400-bp-long repeat named β-motif in the E1 case (Puerma et al., 2014). During the E9 inversion process, an ~8-kb-long fragment that included one protein-coding gene (GA10097/CG10131) and one snoRNAs generating gene (Uhg5) became duplicated and is now present in the two breakpoints of the derived E1+2+9 arrangement (Figure 3). As the duplication includes the complete GA10097/CG10131 coding region and its 5′ regulatory region, the presence of two copies in the derived arrangement might have some phenotypic effect in case that the encoded protein (likely a 3-hydroxyacyl-CoA dehydrogenase involved in fatty acid metabolism) had a dose-dependent effect. Although the Uhg5 gene is also duplicated in the derived E1+2+9 arrangement, the number of snoRNAs encoded in each copy (five and seven in the AK and GL breakpoint regions, respectively) is either smaller or equal than in the E1+2 arrangement (seven in the AG breakpoint region; Figure 3). Thus, only five of the seven Uhg5 encoded snoRNAs are present twice in the inverted arrangement (results not shown). During the E3 inversion process, an ~3.5-kb-long fragment became duplicated and is now present in the two breakpoints of the derived E1+2+9+3 arrangement (Figure 4). In this case, the duplicated fragment includes three coding regions, with the OstDelta and the GA15025 genes being truncated in the proximal AH2 breakpoint and the distal KH1 breakpoint (Figure 4), respectively, which would result in a single complete and functional copy of these genes in the E1+2+9+3 arrangements and two complete and functional copies of the GA24519/CG14488 gene.
In all four D. subobscura inversions generated by staggered double-strand breaks, the fragment duplicated in the inverted arrangement corresponds to only one of the breakpoints in the non-inverted arrangement. The other breakpoint is clearly delimited between two adjacent nucleotides in some cases—for example, in the distal KL breakpoint of inversion E9 (Figure 3) and in the proximal AK breakpoint of inversion E3 (Figure 4)—, whereas in other cases a small fragment of the single-strand template might have been lost during the repair stage (for example, in the distal breakpoint of inversion O3; Papaceit et al., 2013). This observation of an asymmetric stagger in the two breakpoints of an inversion is not unique to D. subobscura inversions but extensive to most molecularly characterized inversions. Among the detected exceptions in inverted chromosomes with inverted duplicates corresponding to both breakpoints in non-inverted chromosomes are the five D. melanogaster polymorphic inversions—In(1)A, In(1)Be, In(2R)NS, In(3R)K and In(3R)Payne (Matzkin et al., 2005; Corbett-Detig et al., 2012)—, six of the 18 inversions with inverted duplicates fixed between D. melanogaster and D. yakuba (Ranz et al., 2007), and the 2q inversion fixed in D. mojavensis (Guillén and Ruiz, 2012).
Transposable elements and inversions
Although not directly involved in the origin of the here studied inversions, transposable elements—mostly defective—have been detected in the breakpoint regions of inversions E9 and E3. Although transposable elements can transpose to any region of the genome, their rate of excision varies with recombination rate (Charlesworth and Langley, 1989). In inversion heterokaryotypes, recombination in regions affected by chromosomal inversions is reduced, with the highest reduction near the breakpoints (Navarro et al., 1997). Transposable elements detected at breakpoint regions will not only include those copies present in the unique chromosome involved in the inversion process but also younger copies accumulated due to the highly suppressed recombination in inversion heterokaryotypes (Charlesworth et al., 1997). As both point and length mutations will lead to transposon degeneration through time, sequence comparison of transposable elements, or remnants thereof, detected at or near inversion breakpoints would thus allow detecting the periods of active transposition of particular TEs, and it might also be indicative of the relative age of inversions.
In D. subobscura, the characterization of the E2 breakpoints revealed that the breakpoints were within a repeated motif (α-motif) that exhibited two regions with low similarity to the SGM (Miller et al., 2000) family of transposable elements, indicating that they contained remnants of old copies of this element. Although the α-motif was present at each proximal and distal breakpoints of the E2 inversion (Figure 2 in Puerma et al., 2014), it was only present at the proximal breakpoint in the E1+2 arrangement (Figure 3). Moreover, it was also present in the GL, but not at the AK breakpoint of inversion E9, which allowed us to place this motif outside of the fragment duplicated during the inversion process (Figure 3). Surprisingly, at the same location of the AK breakpoint region, we detected a nearly complete canonical copy of the SGM element, which would be the result of a novel transposition to this location that might have been fostered by the staggered-break mechanism (Onozawa et al., 2014). We also detected two even more conserved copies of the SGM element, one in the distal (GL) breakpoint of inversion E9—within the A part duplicated fragment—and a second one in the distal (KH1) breakpoint of inversion E3—within a canonical and complete copy of the LINE transposable element. Although there is no evidence from our data for SGM transposition in the period immediately preceding the E2 inversion, and soon thereafter, this family seems to have been active at least in the period between the occurrence of inversion E9 and some time after that of the E3 inversion. We can also infer from our data that LINE elements were active most possibly after the occurrence of inversion E3.
Two bouts of SGM activity are now documented in the subobscura subgroup lineage (Miller et al., 2000; present work). The first one would have occurred before the split of D. guanche from the D. subobscura-D. madeirensis lineage as previously inferred from its detection in the P-neogene cluster present in all three species (Miller et al., 2000), inference that is supported by the presence of old degenerated copies at both breakpoints of the E2 inversion (Figure 5; Puerma et al., 2014). The presence of near canonical and canonical copies of the SGM element at the breakpoints of both the E9 and E3 inversions, points to a second more recent bout of SGM (as well as of LINE) activity possibly after the D. subobscura-D. madeirensis split.
Figure 5.
Comparison of two reused breakpoints. The regions spanning the two breakpoints (at sections 58D and 64C, respectively) that have been multiply reused, at least at the cytological level (see Figure 1), in extant chromosomal arrangements Est, E1+2, E1+2+9 and E1+2+9+3 as well as in the two possible intermediate, and now extinct, arrangements connecting Est and E1+2 (within a gray box). Given that inversions occurred sequentially, vertical discontinuous lines connect a particular breakpoint in the corresponding non-inverted and inverted chromosomes. The blue striped fragments refer to a repeat motif, named β-motif in Puerma et al. (2014) and, therefore, marked with a β. The asterisk (*) and double asterisks (**) are meant to differentiate the snoRNAs generated from the Uhg5 and Uhg1 gene introns, respectively. The sequenced AB fragment is ~18.3-kb long. See legends of Figures 3 and 4 for all other notations.
Contrasting breakpoint reuse at the cytological and molecular levels
In Drosophila, paracentric inversions constitute the main source of gene reorganization within each chromosomal element. Classical cytological studies of chromosomal polymorphism (that is, of inversions segregating in natural populations) revealed that some breakpoints are shared by two or more inversions (for example, Kunze-Mühl and Müller, 1958; Aulard et al., 2002), which could also be ascertained from comparisons of species of the same group or subgroup. Comparative genomic analysis across the complete, or partial, Drosophila phylogeny also revealed that some genes had flanked two or more chromosomal breaks (Bhutkar et al., 2008; von Grotthuss et al., 2010). To assess reuse at a fine molecular level (that is, more precisely than in either of the above-mentioned types of studies), detailed sequence comparison of the breakpoint regions in non-inverted and inverted chromosomes is needed, and preferably of polymorphic inversions because, as evolutionary change accumulates with time, breakpoint sequences better reflect the inversion process the shorter the time elapsed since their origin.
Our previous characterization of the breakpoints of inversions E1 and E2 in D. subobscura was one of the first studies that molecularly characterized a polymorphic inversion breakpoint cytologically shared by two inversions (Corbett-Detig and Hartl, 2012; Puerma et al., 2014). Ours was a difficult endeavor because both inversions occurred sequentially and only the Est and E1+2 arrangements segregate in extant populations. Although we could not establish which breakpoint had been reused—that is, whether it was the proximal AB or the distal GH breakpoint (Figure 1)—, we could, however, infer that the E2 break occurred somewhere within an ~700-bp-long repeat motif—α-motif; Figure 2 in Puerma et al. (2014)—and the E1 break most possibly at this motif distal limit. These results indicate that the shared breakpoint (be it the proximal or the distal breakpoint) was reused not only at the cytological level but also at the molecular level.
In the present study, we contrast at the molecular level the multiple cytological and sequential reuse of the breakpoints at sections 58D and 64C by inversions E1, E2, E9 and E3 (Figures 1 and 5). The breakpoint shared by inversions E1 and E2 (be it the proximal or the distal breakpoint) was previously considered an example of strict reuse (even if not at the strictest 1-bp level) as opposed to broad sense reuse adopted in interspecific genome comparisons (breakpoint flanked by the same gene; von Grotthuss et al., 2010). Indeed, breakpoints of inversions E1 and E2 were narrowed down to repeats α and β that ranged in size between 400 and 700 bp (Figures 1 and 5; Puerma et al., 2014). In the case of the proximal breakpoint at section 58D, the distal limit of the region duplicated in the E9 inversion process is actually the proximal limit of the α-motif present in the E1+2 arrangement (Figures 3 and 5), whereas the proximal E3 breakpoint is displaced ~1.5 kb from that limit (Figures 4 and 5). In the case of the distal breakpoint at section 64C, both limits of the E3 breakpoint are displaced >70 kb from the other breakpoint limits (Figures 4 and 5).
It should be noted that even if the four inversions considered segregate in natural populations, they are not extremely young (Puerma et al., 2014) raising the possibility of length mutations (insertions and deletions of different size and kind) having blurred the actual limits of breakpoints (see above for transposable elements and other intervening sequences; Figures 3, 4, 5). If possible indel events occurring during or after the inversion process are considered, our data would support the strict reuse of the proximal breakpoint at section 58D by inversions either E1, E2, E9 and E3, or E2, E9 and E3 (Figure 5). In the case of the distal breakpoint at section 64C, our data would support its strict reuse by inversions E1 and E2—but not by inversion E3—if E2 had occurred before E1, whereas in the alternative case (E1 before E2) our data would similarly not support the cytological observation that indicated its reuse by inversions E2 and E3.
Our inference of strict multiple reuse of the proximal breakpoint at section 58D is supported at the broad sense by the presence of gene Uhg5 flanking this breakpoint (Figure 5). Similarly, our inference of strict reuse of the distal breakpoint at section 64C only by inversions E1 and E2, if E2 had preceded E1, is supported at the broad sense by gene Uhg1 flanking the breakpoint of only these inversions. Uhg genes would seem prone to breakage maybe as a result of the secondary structures that might be adopted by their snoRNAs encoding introns. It is also important to consider that the inversions generating the different arrangements of this complex system occurred sequentially. The presence in inversion heterokaryotypes of asynapsed regions near breakpoints might add some tension to those regions and render them more breakage prone than completely synapsed regions. The added fragility associated with Uhg genes and with asynapsed regions might not, however, fully explain the strict reuse, detected one or multiple times, of the breakpoints here considered.
In summary, we have characterized the breakpoints of four inversions involved in a complex inversion system and shown that these inversions originated more frequently by the staggered-break mechanism than by repeat-mediated ectopic recombination. Moreover, we have shown that the transposable elements that accumulate at the breakpoint regions as a result of reduced recombination in heterokaryotypes can be indicative of the periods of their active transposition. Finally, we have shown that one of the multiply shared breakpoints at the cytological level—at section 58D—has also been multiply reused at the molecular level, whereas the other one—at section 64C—might have been reused only once at this level.
Data archiving
Sequences newly obtained have been deposited in the EMBL/GenBank Data Libraries under accession numbers LM999978 to LM999984, and updated accession numbers LK022764.2 and LK022779.2.
Acknowledgments
We thank David Salguero for his excellent technical assistance, and Servei de Genòmica, Serveis Cientifico-Tècnics, Universitat de Barcelona, for automated sequencing facilities. We also thank reviewers for their critical comments. This paper was prepared with full knowledge and support of the Barcelona Subobscura Initiative (BSI). This work was supported by grants BFU2012-35168 from Ministerio de Economía y Competitividad, Spain, and 2009SGR-1287 from Comissió Interdepartamental de Recerca i Innovació Tecnològica, Generalitat de Catalunya, Spain to MA.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Andolfatto P, Wall JD, Kreitman M. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics. 1999;153:1297–1311. doi: 10.1093/genetics/153.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulard S, David JR, Lemeunier F. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet Res. 2002;79:49–63. doi: 10.1017/s0016672301005407. [DOI] [PubMed] [Google Scholar]
- Bhutkar A, Schaeffer SW, Russo SM, Xu M, Smith TF, Gelbart WM, et al. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- Calvete O, González J, Betrán E, Ruiz A. Segmental duplication, microinversion and gene loss associated with a complex inversion breakpoint region in Drosophila. Mol Biol Evol. 2012;29:1875–1889. doi: 10.1093/molbev/mss067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals F, Cáceres M, Ruiz A. The foldback-like transposon Galileo is involved in the generation of two different natural chromosomal inversions of Drosophila buzzatii. Mol Biol Evol. 2003;20:674–685. doi: 10.1093/molbev/msg070. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH. The population genetics of Drosophila transposable elements. Annu. Rev Genet. 1989;23:251–287. doi: 10.1146/annurev.ge.23.120189.001343. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH, Sniegowski PD. Transposable element distributions in Drosophila. Genetics. 1997;147:1993–1995. doi: 10.1093/genetics/147.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera S, Martín-Campos JM, Segarra C, Aguadé M. Molecular characterization of the breakpoints of an inversion fixed between Drosophila melanogaster and D. subobscura. Genetics. 1995;139:321–326. doi: 10.1093/genetics/139.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Corbett-Detig RB, Cardeno C, Langley CH. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics. 2012;192:131–137. doi: 10.1534/genetics.112.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 2012;8:e1003056. doi: 10.1371/journal.pgen.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprat A, Negre B, Puig M, Ruiz A. The transposon Galileo generates natural chromosomal inversions in Drosophila by ectopic recombination. PLoS One. 2009;4:e7883. doi: 10.1371/journal.pone.0007883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith MA, Trifonov V. Mammalian karyotype evolution. Nat Rev Gen. 2007;8:950–962. doi: 10.1038/nrg2199. [DOI] [PubMed] [Google Scholar]
- Guillén Y, Ruiz A. Gene alterations at Drosophila inversion breakpoints provide prima facie evidence for natural selection as an explanation for rapid chromosomal evolution. BMC Genomics. 2012;13:53. doi: 10.1186/1471-2164-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas CB, Powell J. Drosophila inversion polymorphism. CRC: Boca Raton, FL, USA; 1992. [Google Scholar]
- Kunze-Mühl E, Müller E. Weitere Untersuchungen über die chromosomale Struktur und die natürlichen Strukturtypen von Drosophila subobscura Coll. Chromosoma. 1958;9:559–570. [PubMed] [Google Scholar]
- Laayouni H, García-Franco F, Chávez-Sandoval BE, Trotta V, Beltran S, Corominas M, et al. Thermal evolution of gene expression profiles in Drosophila subobscura. BMC Evol Biol. 2007;7:42. doi: 10.1186/1471-2148-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Merritt TJ, Zhu CT, Eanes WF. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics. 2005;170:1143–1152. doi: 10.1534/genetics.104.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WJ, Nagel A, Bachmann J, Bachmann L. Evolutionary dynamics of the SGM transposon family in the Drosophila obscura species group. Mol Biol Evol. 2000;17:1597–1609. doi: 10.1093/oxfordjournals.molbev.a026259. [DOI] [PubMed] [Google Scholar]
- Montgomery EB, Charlesworth B, Langley CH. A test for the role of natural selection in the stabilization of transposable element copy number in a population of D. melanogaster. Genet Res. 1987;49:31–41. doi: 10.1017/s0016672300026707. [DOI] [PubMed] [Google Scholar]
- Navarro A, Betrán E, Barbadilla A, Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics. 1997;146:695–709. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TL, Tuzun E, Morrison VA, Hayden KE, Ventura M, McGrath SD, et al. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 2005;15:1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozawa M, Zhang Z, Kim YJ, Goldberg L, Varga T, Bergsagel PL, et al. Repair of DNA double-strand breaks by templated nucleotide sequence insertions derived from distant regions of the genome. Proc Natl Acad Sci USA. 2014;111:7729–7734. doi: 10.1073/pnas.1321889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaceit M, Segarra C, Aguadé M. Structure and population genetics of the breakpoints of a polymorphic inversion in Drosophila subobscura. Evolution. 2013;67:66–79. doi: 10.1111/j.1558-5646.2012.01731.x. [DOI] [PubMed] [Google Scholar]
- Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci USA. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerma E, Orengo DJ, Salguero D, Papaceit M, Segarra C, Aguadé M, et al. Characterization of the breakpoints of a polymorphic inversion complex detects strict and broad breakpoint reuse at the molecular level. Mol Biol Evol. 2014;31:2331–2341. doi: 10.1093/molbev/msu177. [DOI] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, von Grotthuss M, Hillier LW, Roote J, et al. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:1366–1381. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Wasserman M. Evolutionary cytogenetics of the Drosophila buzzatii species complex. Heredity. 1993;70:582–596. doi: 10.1038/hdy.1993.85. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW, Bhutkar A, McAllister BF, Matsuda M, Matzkin LM, O'Grady PM, et al. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonzetich J, Lyttle TW, Carson HL. Induced and natural break sites in the chromosomes of Hawaiian Drosophila. Proc Natl Acad Sci USA. 1988;85:1717–1721. doi: 10.1073/pnas.85.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grotthuss M, Ashburner M, Ranz JM. Fragile regions and not functional constraints predominate in shaping gene organization in the genus Drosophila. Genome Res. 2010;20:1084–1096. doi: 10.1101/gr.103713.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley CS, Eanes W. Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proc Natl Acad Sci USA. 1994;91:3132–3136. doi: 10.1073/pnas.91.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.