Abstract
Background
Thromboxane A synthase (TXAS) metabolizes the cyclooxygenase product prostaglandin (PG) H2 into thromboxane A2 (TXA2), a potent inducer of blood vessel constriction and platelet aggregation. Non-synonymous polymorphisms in the TBXAS gene have the potential to alter TXAS activity and affect TXA2 generation.
Objectives
Assess the functional effects of genetic variants in the TXAS protein, including K258E, L357V, Q417E, E450K and T451N.
Methods
Wild-type TXAS and the variant proteins were expressed in a bacterial system and purified by affinity and hydroxyapatite chromatography. The two characteristic catalytic activities of TXAS were assayed in each of the purified recombinant proteins: isomerization of PGH2 to TXA2 and fragmentation of PGH2 to 12-hydroxyheptadecatrienoic acid and malondialdehyde.
Results
All of the variants exhibited both isomerization and fragmentation activities. The KM values of the variants ranged from 27–52 µM PGH2 (wild-type value: 32 µM PGH2); Vmax values of the variants ranged from 18–40 units/mg (wild-type value: 41 units/mg). The kinetic differences were largest for the L357V variant, whose Vmax / KM ratio was just 27% of the wild-type value.
Conclusion
The increased KM and decreased Vmax values observed with L357V suggest this variant may generate less TXA2 at the low levels of PGH2 expected in vivo, raising the possibility of attenuated signaling via the thromboxane pathway.
Keywords: cytochrome p450, thromboxane synthase, genetic polymorphism, enzyme kinetics
INTRODUCTION
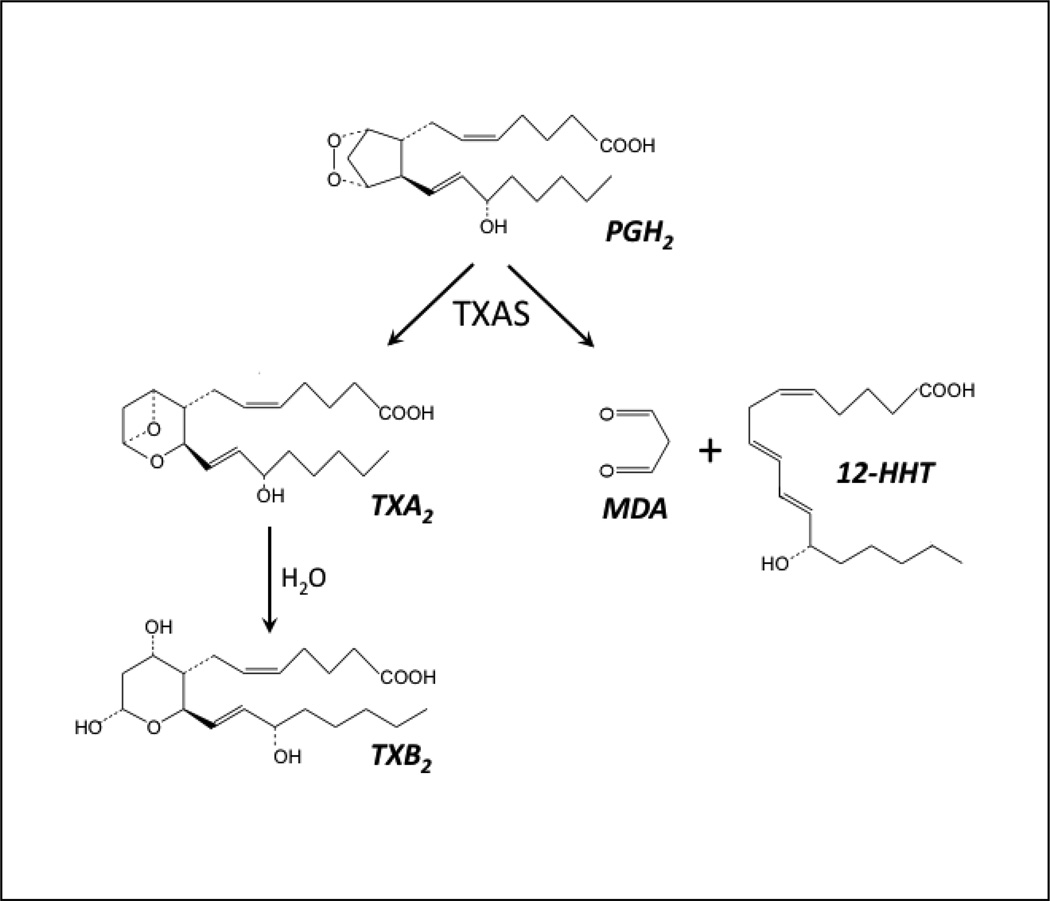
Thromboxane A2 (TXA2) is a potent stimulator of platelet secretion and aggregation as well as a vasoconstrictor with an important role in major cardiovascular diseases, such as atherosclerosis and myocardial infarction [1,2,3]. TXA2 is biosynthesized by the sequential actions of prostaglandin H synthase (also known as cyclooxygenase), which converts arachidonic acid to prostaglandin H2 (PGH2), and thromboxane synthase (TXAS) which subsequently converts PGH2 to TXA2 (Fig. 1). TXAS is a cytochrome P450 enzyme (also known as CYP5A1[4]) that is associated with the endoplasmic reticulum membrane and is primarily expressed in hematopoietic cells, such as platelets, monocytes, leukocytes and macrophages [5]. Unlike other microsomal P450s, which catalyze a mono-oxygenation reaction and require P450 reductase as electron donor, TXAS catalyzes an isomerizaton reaction without an external electron donor or molecular oxygen [6]. Notably, besides forming TXA2, TXAS fragments PGH2 to 12-hydroxy-5,8,10-heptadecatrienoic acid (12-HHT) and malondialdehyde (MDA) in a ratio of 1:1:1 [6] (Fig. 1). The biological functions of MDA and HHT are unclear, although MDA can form adducts with amino groups of proteins or phospholipids and such adducts have been detected in atherosclerotic lesions of human aorta [7]. MDA also participates in formation of an important endogenous DNA adduct that may contribute to human genetic disease and cancer [8,9].
Figure 1.
Reactions of PGH2 catalyzed by TXAS. Left branch: isomerization to TXA2. Right branch: fragmentation to MDA and 12-HHT. Hydrolysis of TXA2 to TXB2 is nonenzymatic.
TXAS is a single copy gene, located on chromosome 7q33-35 [10]; the gene is 193 kb long and has 13 exons [11]. Knockout of the corresponding mouse gene resulted in mild bleeding disorders and altered vascular responses to arachidonic acid [12], and knockdown of the zebrafish gene for TXAS resulted in defective heart development [13]. Further, human TXAS mutations have been linked to a bone density disorder [14]. Fourteen non-synonymous polymorphisms in the TXAS coding region were identified in the first three reports on variations in the gene [15,16,17], with evidence for selective evolutionary pressure against genetic mutations in the gene [17]. In view of the roles of TXAS in physiology and pathology, it is important to understand the effects of human TXAS protein variants on catalytic activity. We have developed a prokaryotic expression system that provides sufficient recombinant TXAS with ~90 % purity for enzymatic characterization [18,19]. Structural analysis based on homology modeling suggested that four of the TXAS variants (K258E, Q417E, E450K and T451N) were likely to have altered enzymatic activities [17]. In addition, the L357V variant, which has a minor allele frequency of almost 11% in African-Americans [17] was predicted to have altered catalytic activity by the PolyPhen algorithm [20]. The present report describes the results of our enzymatic characterization of the five targeted TXAS variants as purified recombinant proteins.
MATERIALS AND METHODS
Materials
Arachidonic acid was from NuChek Preps (Elysian, MN). IPTG, δ-aminolevulinic acid, Igepal CA630 and Lubrol PX were acquired from Sigma. Nickel nitrilotriacetate agarose and hydroxyapatite were obtained from Qiagen and Bio-Rad, respectively. PGH2 was synthesized from arachidonic acid using detergent-solubilized ovine prostaglandin H synthase-1 and was purified by normal phase chromatography [21].
Protein expression and purification
The recombinant TXAS proteins were prepared following procedures described previously [18,19]. Briefly, the wild-type TXAS cDNA was modified by replacement of the first 28 amino acid codons with a sequence coding for a hydrophilic segment (MAKKTSS) and by addition of four histidine codons for a his-tag at the carboxyl terminus. The cDNA was subcloned into the pCW expression plasmid to produce the wild-type vector, pCW-TXAS. Site-directed mutagenesis to produce expression vectors for the five variants was performed using the Quick-Change kit (Stratagene, La Jolla, CA, USA) with pCW-TXAS as the template. The mutations were confirmed by DNA sequencing. Each construct was transformed into E. coli BL21(DE3)pLys and expression of recombinant protein was induced by IPTG in the presence of the heme precursor, δ-aminolevulinic acid. Each recombinant TXAS protein was solubilized with 2% Igepal CA630 and was purified by sequential chromatography on nickel affinity resin and hydroxyapatite, and stored at −70°C. The concentration of each purified recombinant TXAS was determined using a Soret extinction coefficient of 100 mM−1 cm−1 [18]. Protein concentrations were determined using a Bio-Rad Protein Assay Kit with bovine serum albumin as standard.
TXAS enzymatic assays
To determine the TXA2-synthesizing activity, TXAS (30 nM) in 200 µl of 20 mM NaPi, pH 7.4, and 0.2 % Lubrol PX at room temperature (23 °C) was reacted with the desired level of PGH2. The reaction was terminated after 15 s by acidification with 21 µl of 2 M citric acid. Formation of TXB2, the stable hydrolysis product of TXA2, was measured by radioimmunoassay [22]. To monitor continuously the initial rate for steady-state kinetic analysis, the assay was carried out at room temperature (23 °C) in a cuvette containing 400 µl of 20 mM NaPi, pH 7.4, 0.2 % Lubrol PX and 5 nM of recombinant TXAS. The reaction was started by an addition of the desired level of PGH2 and the A268 monitored for the first 30 s. The initial rate was calculated from the increase in A268 resulting from formation of MDA (ε = 31.5 mM−1 cm−1). One unit is the amount of TXAS forming 1 µmol of MDA per min. The TXAS rate data for a series of PGH2 concentrations were fitted to the Michaelis-Menten equation by non-linear regression to estimate values for Km and Vmax. Each assay was performed in triplicate.
Statistical analysis
The p-values were generated using the t-test to determine any significant differences in kinetic parameters between the means of wild-type and mutants.
RESULTS AND DISCUSSION
Production of variant TXAS proteins
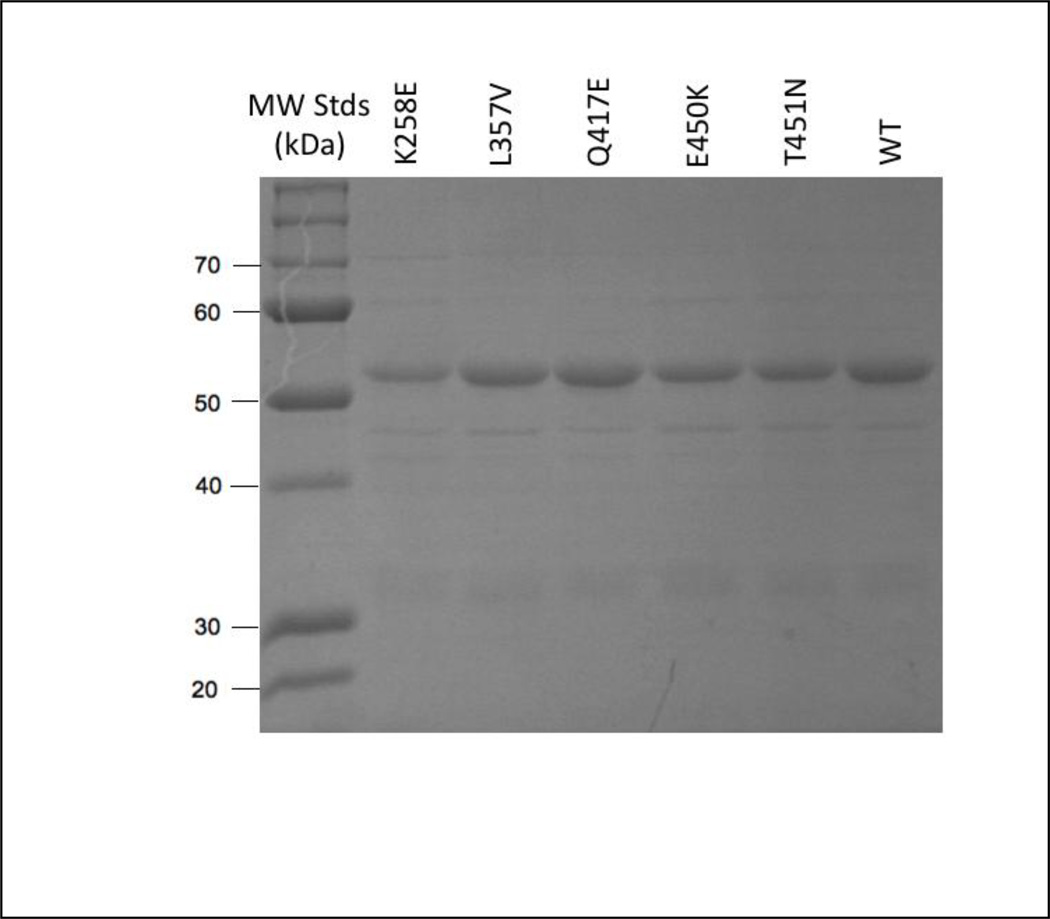
The expression levels of the K258E, L357V and E450K variants were similar to that of a wild-type TXAS (~0.27 µmol/liter of culture), whereas the Q417E and T451N variants were expressed at lower levels (0.07–0.09 µmol/liter of culture). Note that differences in expression level among TXAS variants in the present bacterial system are unlikely to reliably predict the expression levels of the variants in native human tissues. Each of the recombinant proteins was purified to ~90% homogeneity, as judged by SDS-PAGE analysis (Fig. 2). The optical absorbance spectrum of resting wild-type TXAS had a Soret peak at 418 nm, as previously observed [18]. Each of the TXAS variants also exhibited a Soret peak near 418 nm, except for K258E, whose Soret peak was at 422 nm (data not shown). These results indicate that the heme in the wild-type and each of the TXAS variants is predominantly in the expected six-coordinate, low-spin ferric state.
Figure 2.
Electrophoretic analysis of purified recombinant TXAS wild-type (WT) and variant proteins. Aliquots of purified wild-type and variant TXAS (~ one µg protein) were separated on a 10 % polyacrylamide gel and stained with Coomassie blue.
The initial enzymatic characterization of the purified TXAS variants involved measuring their ability to convert PGH2 to TXA2, which was quantified as its stable hydrolysis product, TXB2. These screening reactions used fixed concentration of PGH2 (290 µM) and enzyme (30 nM). The results, shown in Table 1, confirm that each of the variants was able to synthesize TXA2 at initial rates that were 40–90% of the rate for the wild-type enzyme. Thus, none of the structural changes in the variants blocked the proper active site positioning of the PGH2 substrate that is thought to be crucial for the TXAS isomerization reaction (left branch in Fig. 1) [23]. The rank order of the rate of thromboxane formation was: wild-type > T451N > Q417E > E450K > K258E > L357V (Table 1). This ranking gives a qualitative indication of the relative effects of the variants on TXAS catalysis. However, the discontinuous nature of the assay which has only one time-point, 15 s in the current assay, and possible variations in the extent of conversion of TXA2 to TXB2 limit quantitative interpretation of the results.
Table 1.
TXB2 synthesis by purified recombinant wild-type and variant TXAS proteins.
| TXAS construct |
Specific activity (µmol TXB2/min/mg)a |
|---|---|
| wild-type | 9.9 ± 0.5 |
| K258E | 5.7 ± 0.1 |
| L357V | 4.2 ± 0.1 |
| Q417E | 7.1 ± 0.4 |
| E450K | 6.3 ± 1.2 |
| T451N | 8.8 ± 0.9 |
In reactions with 290 µM PGH2 and 30 nM enzyme. Mean ± SEM are shown for three measurements.
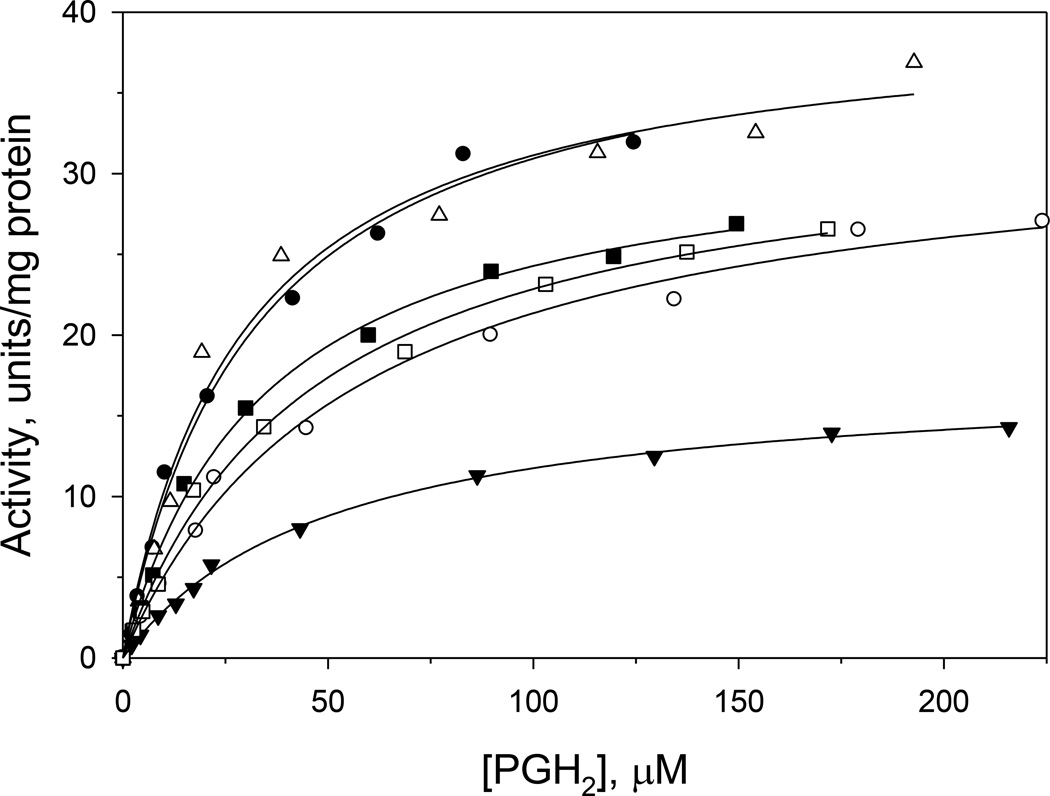
For a more detailed analysis of kinetic parameters in the variants, we monitored the second catalytic activity of TXAS, namely the fragmentation of PGH2 into 12-HHT and MDA (right branch in Fig. 1). Formation of MDA gives an increase in A268, allowing the initial, steady-state rate in each reaction to be calculated from the ΔA268/Δt over the first 30 s. The initial rate was found to be a saturable function of the PGH2 level for wild-type TXAS and each of the variants (Fig. 3). Accordingly, each set of rate vs. [PGH2] data was fitted to the Michaelis-Menten equation to estimate values for Vmax and KM, which are shown in Table 2. Wild-type TXAS had a KM of 32 µM PGH2 and Vmax of 41 units /mg protein, values comparable to those previously reported [19,23]. The KM values of Q417E and E450K were essentially indistinguishable from the wild-type value, but those of the K258E, L357V and T451N variants were about 1.5-fold higher (Table 2), suggesting that the interaction with substrate was modestly weaker in the latter three variants. The L357V variant exhibited a marked decrease in Vmax (only 44% of the wild-type value), whereas modestly lower Vmax values were seen with K258E, E450K and T451N (81–83% of wild-type value). The Vmax was not affected in Q417E (Table 2).
Figure 3.
Effect of PGH2 level on reaction rate in TXAS wild-type and variants. Continuous assays contained 5 nM of recombinant TXAS. Lines represent fits of the data to the Michaelis- Menten equation. Filled circles, wild-type TXAS; open circles, K258E; filled triangles, L357V; open triangles, Q417E; filled squares, E450K, and open squares, T451N. Data shown is from a representative set of experiments
Table 2.
TXAS kinetic parameters for purified wild-type and variant proteinsa.
| wild-type | K258E | L357V | Q417E | E450K | T451N | |
|---|---|---|---|---|---|---|
| KM, µM PGH2 | 32.0 ± 2.7 | 51.9 ± 4.3 (p= 0.017)b |
51.0 ± 2.5 (p= 0.007) |
27.1 ± 3.6 (p> 0.05) |
33.8 ± 2.4 (p> 0.05) |
46.7 ± 3.9 (p= 0.036) |
| Vmax, units /mg protein |
40.8 ± 1.0 | 32.9 ± 1.0 (p= 0.005) |
18.1 ± 1.0 (p= 0.0001) |
39.7 ± 1.0 (p> 0.05) |
32.9 ± 1.0 (p= 0.005) |
34.0 ± 1.0 (p= 0.009) |
| Vmax /KM, units/mg/µM PGH2 |
1.28 ± 0.14 | 0.64 ± 0.07 (p= 0.015) |
0.35 ± 0.04 (p=0.0031) |
1.46 ± 0.23 (p> 0.05) |
0.97 ± 0.10 (p> 0.05) |
0.72 ± 0.08 (p= 0.025) |
Activity was assayed by following the absorbance changes at 268 nm for MDA formation in the presence of 5 nM TXAS. Values represent mean ± SEM of three experiments.
p-values are calculated as compared to the wild-type, and values larger than 0.05 are considered to be statistically in-significant.
The Vmax/KM ratio, or specificity constant, is an index of catalytic rate at substrate concentrations well below the KM value [24]. Given that cellular PGH2 levels are likely to be well below the KM values of all the TXAS proteins, comparison of the specificity constant values in Table 2 predicts the possible in vivo functional impacts of the TXAS polymorphisms. The specificity constant for Q417E is not significantly different from the wild-type value, suggesting that the variant has little functional effect (p value > 0.05). Moderate decreases in TXAS catalytic efficiency might be anticipated for the K258E, E450K and T451N variants (Vmax/KM 50–56% of wild-type value; 0.05 > p values > 0.001). On the other hand, the Vmax/KM value for L357V is only 27% of the wild-type value (Table 2; p value= 0.0004), predicting a large decrease in catalytic function for this variant in vivo.
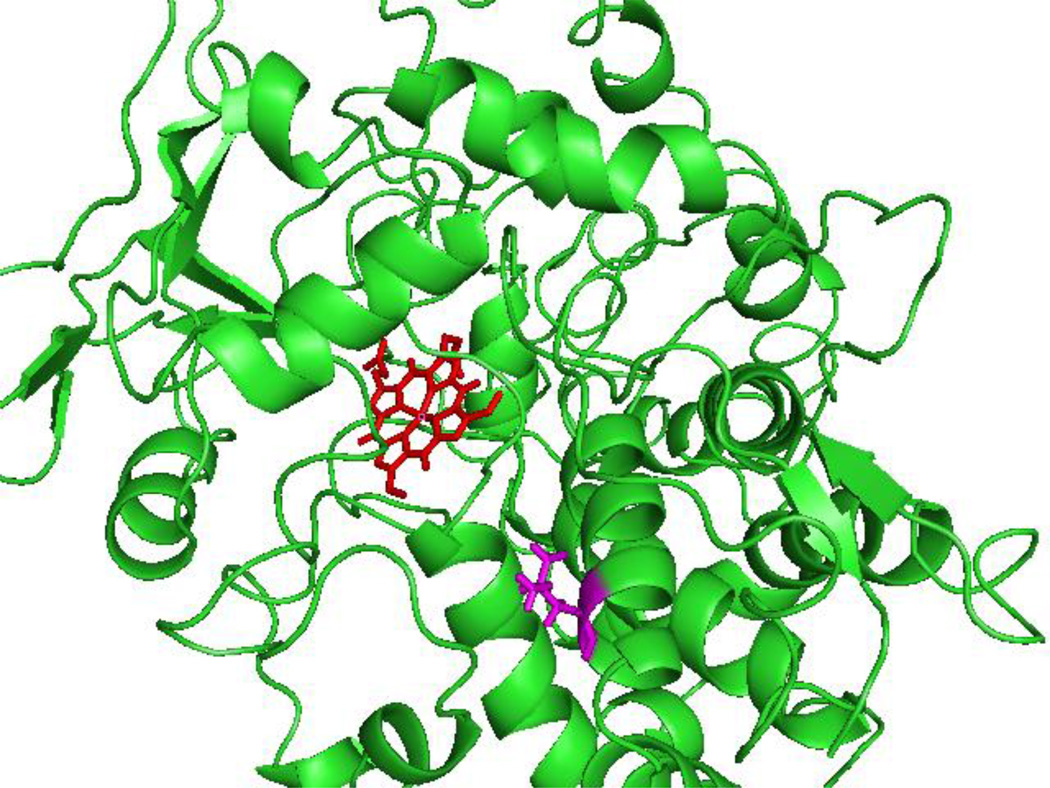
Taken together, L357V is the most affected variant for TXA2 synthesis. It should be noted that of the variants examined here, Leu357 is the only amino acid residue that is conserved in the wild-type for human, chimpanzee, dog, cow, mouse, rat, chicken and zebrafish TXAS. We have postulated previously that Leu357 is located at the end of the Helix I and suggested that a mutation at Leu357 would affect catalytic activity [17,25]. This prediction from our computational model of the TXAS structure (Fig. 4) remains to be tested by a direct structure determination. The prevalence of L357V is an important factor in evaluating its potential impact. The dbSNP data base lists a minimal allele frequency (MAF) of 0.109 for the L357V polymorphism [26]. Thus, most individuals with this TXAS variant will be heterozygotes. Assuming that the wild type and L357V variant proteins act independently, TXAS activity in a heterozygous individual might be expected to be an average of wild type and L357V variant values. Notably, we found no evidence for linkage disequilibrium between any two polymorphisms in the study (r2< 0.005). Furthermore, these polymorphisms are relatively rare (0.6 %–11 %) and have different minor allele frequencies in different ethnic groups [17], so that any two SNPs are rarely, if ever, in combination with one another, making it very unlikely that these polymorphisms would form common haplotypes.
Figure 4.
Computation model of TXAS structure [38] showing the position of Leu357 (magenta) and heme (red). The figure was generated using PyMol (The PyMol Molecular Graphic System, Version 1.3, Schrödinger, LLC).
In general, the type and quantity of prostanoids produced from PGH2 in a given cell type are governed by the specific secondary prostanoid-synthesizing enzymes present in that particular cell type [27,28]. The many variables in the prostanoid synthetic pathway in vivo make it difficult to model accurately in a cultured cell model. However, it is reasonable to expect that any altered function in a TXAS variant would change the proportion of TXA2 in the prostanoid profile, with the potential to affect cellular signaling via TXA2. TXA2 is a critical signal in platelet stimulation, aggregation, and vasoconstriction, and increases in TXAS expression have been associated with a variety of cancers (reviewed in [29]). Thus, TXAS has become a suggested target for chemotherapy, with the development of TXAS-specific inhibitors such as ozagrel and furegrelate yielding promising early results in vitro [30,31,32]. TXAS variants with decreased activity could affect targeted drug intervention, as individuals with lower baseline TXAS activity may get less benefit from selective TXAS inhibitors. Recent findings indicate that genetic variation in TXAS is associated with several pathologies, including non-fatal myocardial infarction [33], cerebral infarction [34], and breast cancer [35]. In addition, pharmacogenetic consequences of TXAS polymorphisms on aspirin sensitivity have been investigated in stroke [36] and asthma patients [37]. Given the several pathophysiological functions of TXAS and the potential clinical utility of TXAS pathway inhibitors, it will be of interest to evaluate phenotypic and pharmacogenetic effects in individuals with the L357V polymorphism in TXAS.
ACKNOWLEDGEMENTS
This work was supported by NIH grants CA114467 (C.M.U.) and HL60625 (L.H.W.) from the United States Public Health Service, and by American Heart Association 10GRNT3610014 (L.H.W.). The authors thank Ms. Laura M. Heath, Fred Hutchinson Cancer Research Center, for helpful discussion on the health consequences and genetics.
The abbreviations used are
- TXA2 and TXB2
thromboxane A2 and B2
- TXAS
thromboxane synthase
- PGH2
prostaglandin H2
- MDA
malondialdehyde
- HHT
12-hydroxy-5,8,10-heptadecatrienoic acid
REFERENCES
- 1.Dogne JM, Hanson J, de Leval X, Pratico D, Pace-Asciak CR, Drion P, et al. From the design to the clinical application of thromboxane modulators. Curr Pharm Des. 2006;12:903–923. doi: 10.2174/138161206776055921. [DOI] [PubMed] [Google Scholar]
- 2.Nakahata N. Thromboxane A2: physiology/pathophysiology, cellular signal transduction and pharmacology. Pharmacol Ther. 2008;118:18–35. doi: 10.1016/j.pharmthera.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Yuhki K, Kojima F, Kashiwagi H, Kawabe J, Fujino T, Narumiya S, et al. Roles of prostanoids in the pathogenesis of cardiovascular diseases: Novel insights from knockout mouse studies. Pharmacol Ther. 2011;129:195–205. doi: 10.1016/j.pharmthera.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, et al. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- 5.Ullrich V, Nusing R. Thromboxane synthase. From isolation to function. Stroke. 1990;21:IV134–IV138. [PubMed] [Google Scholar]
- 6.Haurand M, Ullrich V. Isolation and characterization of thromboxane synthase from human platelets as a cytochrome P-450 enzyme. J Biol Chem. 1985;260:15059–15067. [PubMed] [Google Scholar]
- 7.Uchida K. Current status of acrolein as a lipid peroxidation product. Trends Cardiovasc Med. 1999;9:109–113. doi: 10.1016/s1050-1738(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhary AK, Nokubo M, Reddy GR, Yeola SN, Morrow JD, Blair IA, et al. Detection of endogenous malondialdehyde-deoxyguanosine adducts in human liver. Science. 1994;265:1580–1582. doi: 10.1126/science.8079172. [DOI] [PubMed] [Google Scholar]
- 9.Otteneder MB, Knutson CG, Daniels JS, Hashim M, Crews BC, Remmel RP, et al. In vivo oxidative metabolism of a major peroxidation-derived DNA adduct, M1dG. Proc Natl Acad Sci USA. 2006;103:6665–6669. doi: 10.1073/pnas.0602017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chase MB, Baek SJ, Purtell DC, Schwartz S, Shen RF. Mapping of the human thromboxane synthase gene (TBXAS1) to chromosome 7q34-q35 by two-color fluorescence in situ hybridization. Genomics. 1993;16:771–773. doi: 10.1006/geno.1993.1264. [DOI] [PubMed] [Google Scholar]
- 11.Tazawa R, Green ED, Ohashi K, Wu KK, Wang LH. Characterization of the complete genomic structure of human thromboxane synthase gene and functional analysis of its promoter. Arch Biochem Biophys. 1996;334:349–356. doi: 10.1006/abbi.1996.0464. [DOI] [PubMed] [Google Scholar]
- 12.Yu IS, Lin SR, Huang CC, Tseng HY, Huang PH, Shi GY, et al. TXAS-deleted mice exhibit normal thrombopoiesis, defective hemostasis, and resistance to arachidonate-induced death. Blood. 2004;104:135–142. doi: 10.1182/blood-2003-10-3661. [DOI] [PubMed] [Google Scholar]
- 13.Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135:1179–1187. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- 14.Genevieve D, Proulle V, Isidor B, Bellais S, Serre V, Djouadi F, et al. Thromboxane synthase mutations in an increased bone density disorder (Ghosal syndrome) Nat Genet. 2008;40:284–286. doi: 10.1038/ng.2007.66. [DOI] [PubMed] [Google Scholar]
- 15.Chevalier D, Lo-Guidice JM, Sergent E, Allorge D, Debuysere H, Ferrari N, et al. Identification of genetic variants in the human thromboxane synthase gene (CYP5A1) . Mutat Res. 2001;432:61–67. doi: 10.1016/s1383-5726(00)00004-2. [DOI] [PubMed] [Google Scholar]
- 16.Saito S, Iida A, Sekine A, Kawauchi S, Higuchi S, Ogawa C, et al. Catalog of 680 variations among eight cytochrome p450 ( CYP) genes, nine esterase genes, and two other genes in the Japanese population. J Hum Genet. 2003;48:249–270. doi: 10.1007/s10038-003-0021-7. [DOI] [PubMed] [Google Scholar]
- 17.Ulrich CM, Carlson CS, Sibert J, Poole EM, Yu JH, Wang LH, et al. Thromboxane synthase (TBXAS1) polymorphisms in African-American and Caucasian populations: evidence for selective pressure. Hum Mutat. 2005;26:394–395. doi: 10.1002/humu.9371. [DOI] [PubMed] [Google Scholar]
- 18.Hsu PY, Tsai AL, Kulmacz RJ, Wang LH. Expression, purification, and spectroscopic characterization of human thromboxane synthase. J Biol Chem. 1999;274:762–769. doi: 10.1074/jbc.274.2.762. [DOI] [PubMed] [Google Scholar]
- 19.Yeh HC, Tsai AL, Wang LH. Reaction mechanisms of 15- hydroperoxyeicosatetraenoic acid catalyzed by human prostacyclin and thromboxane synthases. Arch Biochem Biophys. 2007;461:159–168. doi: 10.1016/j.abb.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LH, Tsai AL, Hsu PY. Substrate binding is the rate-limiting step in thromboxane synthase catalysis. J Biol Chem. 2001;276:14737–14743. doi: 10.1074/jbc.M009177200. [DOI] [PubMed] [Google Scholar]
- 22.Wu KK, Le Breton GC, Tai HH, Chen YC. Abnormal platelet response to thromboxane A2. J Clin Invest. 1981;67:1801–1804. doi: 10.1172/JCI110221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hecker M, Ullrich V. On the mechanism of prostacyclin and thromboxane A2 biosynthesis. J Biol Chem. 1989;264:141–150. [PubMed] [Google Scholar]
- 24.Cornish-Bowden A. Fundamentals of enzyme kinetics. 3rd ed. London: Portland Press; 2004. [Google Scholar]
- 25.Wang LH, Kulmacz RJ. Thromboxane synthase: structure and function of protein and gene. Prostaglandins Other Lipid Mediat. 2002;68–69:409–422. doi: 10.1016/s0090-6980(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 26.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgs EA, Moncada S, Vane JR. Prostaglandins and thromboxanes from fatty acids. Prog Lipid Res. 1986;25:5–11. doi: 10.1016/0163-7827(86)90005-6. [DOI] [PubMed] [Google Scholar]
- 28.Samuelsson B, Goldyne M, Granstrom E, Hamberg M, Hammarstrom S, Malmsten C. Prostaglandins and thromboxanes. Annu Rev Biochem. 1978;47:997–1029. doi: 10.1146/annurev.bi.47.070178.005025. [DOI] [PubMed] [Google Scholar]
- 29.Cathcart MC, Reynolds JV, O'Byrne KJ, Pidgeon GP. The role of prostacyclin synthase and thromboxane synthase signaling in the development and progression of cancer. Biochim Biophys Acta. 2010;1805:153–166. doi: 10.1016/j.bbcan.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Moussa O, Riker JM, Klein J, Fraig M, Halushka PV, Watson DK. Inhibition of thromboxane synthase activity modulates bladder cancer cell responses to chemotherapeutic agents. Oncogene. 2008;27:55–62. doi: 10.1038/sj.onc.1210629. [DOI] [PubMed] [Google Scholar]
- 31.Moussa O, Yordy JS, Abol-Enein H, Sinha D, Bissada NK, Halushka PV, et al. Prognostic and functional significance of thromboxane synthase gene overexpression in invasive bladder cancer. Cancer Res. 2005;65:11581–11587. doi: 10.1158/0008-5472.CAN-05-1622. [DOI] [PubMed] [Google Scholar]
- 32.Cathcart MC, Gately K, Cummins R, Kay E, O'Byrne KJ, Pidgeon GP. Examination of thromboxane synthase as a prognostic factor and therapeutic target in non-small cell lung cancer. Mol Cancer. 2011;10:25. doi: 10.1186/1476-4598-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemaitre RN, Rice K, Marciante K, Bis JC, Lumley TS, Wiggins KL, et al. Variation in eicosanoid genes, non-fatal myocardial infarction and ischemic stroke. Atherosclerosis. 2009;204:e58–e63. doi: 10.1016/j.atherosclerosis.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park SA, Park BL, Park JH, Lee TK, Sung KB, Lee YK, et al. Association of polymorphisms in thromboxane A2 receptor and thromboxane A synthase 1 with cerebral infarction in a Korean population. BMB Rep. 2009;42:200–205. doi: 10.5483/bmbrep.2009.42.4.200. [DOI] [PubMed] [Google Scholar]
- 35.Abraham JE, Harrington P, Driver KE, Tyrer J, Easton DF, Dunning AM, et al. Common polymorphisms in the prostaglandin pathway genes and their association with breast cancer susceptibility and survival. Clin Cancer Res. 2009;15:2181–2191. doi: 10.1158/1078-0432.CCR-08-0716. [DOI] [PubMed] [Google Scholar]
- 36.Kimouli M, Gourvas V, Konstantoudaki X, Basta M, Miyakis S, Spandidos DA. The effect of an exon 12 polymorphism of the human thromboxane synthase (CYP5A1) gene in stroke patients. Med Sci Monit. 2009;15:BR30–BR35. [PubMed] [Google Scholar]
- 37.Oh SH, Kim YH, Park SM, Cho SH, Park JS, Jang AS, et al. Association analysis of thromboxane A synthase 1 gene polymorphisms with aspirin intolerance in asthmatic patients. Pharmacogenomics. 2011;12:351–363. doi: 10.2217/pgs.10.181. [DOI] [PubMed] [Google Scholar]
- 38.Ruan KH, Milfeld K, Kulmacz RJ, Wu KK. Comparison of the construction of a 3-D model for human thromboxane synthase using P450cam and BM-3 as templates: implications for the substrate binding pocket. Protein Eng. 1994;7:1345–1351. doi: 10.1093/protein/7.11.1345. [DOI] [PubMed] [Google Scholar]