Abstract
HMGA2, a high-mobility-group AT-hook protein, is an oncogene involved in tumorigenesis of many malignant neoplasms. HMGA2 overexpression is common in both early and later stage of high grade ovarian serous papillary carcinoma. To test whether HMGA2 participates in the initiation of ovarian cancer and promotion of aggressive tumor growth, we examined the oncogenic properties of HMGA2 in ovarian surface epithelial (OSE) cell lines. We found that introduction of HMGA2 overexpression was sufficient to induce OSE transformation in vitro. HMGA2-mediated OSE transformation resulted in tumor formation in xenografts of nude mice. By silencing HMGA2 in HMGA2 overexpressing OSE and ovarian cancer cell lines, the aggressiveness of tumor cell growth behaviors was partially suppressed. Global gene profiling analyses revealed that HMGA2-mediated tumorigenesis was associated with expression changes of target genes and microRNAs that are involved in epithelial-to-mesenchymal transition (EMT). Lumican (LUM), a tumor suppressor that inhibits EMT, was found to be transcriptionally repressed by HMGA2 and was frequently lost in human high-grade serous papillary carcinoma.
Keywords: HMGA2, ovarian cancer, miRNA, oncogene, EMT
INTRODUCTION
High grade ovarian serous carcinoma (HG-PSC) is the most lethal type of gynecologic malignancy. The cause of this disease remains poorly understood. P53 is the most commonly detected mutation in this tumor (1–2). In addition, mutation or loss of expression of BRCA1 and or BRCA2 are frequently detected in high grade serous carcinoma (3). In mice, conditional inactivation of P53 and BRCA1, the most common genetic changes in HG-PSC, in ovaries results in the development of leiomyosarcomas but not of ovarian surface epithelial (OSE) carcinomas (4–5). These findings indicate that in addition to loss of these tumor suppressors, the early tumorigenesis of HG-PSC may require the activation of oncogenes. HMGA2, the high-mobility group AT-hook 2 gene, is one of a few oncogenes found to be overexpressed in human HG-PSC (6–7). The rate of HMGA2 overexpression was found to be significantly higher in type II ovarian cancer than in type I ovarian cancer (7). Most importantly, about 75% of serous carcinomas in situ in fallopian tube have HMGA2 overexpression along with P53 mutations (8).
HMGA2 is a nonhistone nuclear binding protein and an important regulator of cell growth and differentiation (8–9). It is an oncofetal protein that is overexpressed in embryonic tissue and many malignant neoplasms, including ovarian cancer (6–7), but rarely in normal adult tissues. However, the roles of HMGA2 in tumorigenesis and tumor progression remain to be fully understood. The oncogenic properties of HMGA2 are demonstrated to be involved in aggressive tumor growth (10–11), stem cell self-renewal (12–13), the DNA damage response (14), and tumor cell differentiation (6, 15). HMGA2 has also been found to participate in the epithelial-to-mesenchymal transition (EMT) (16–17). Silencing of HMGA2 expression in ovarian cancer cells has also been found to have a therapeutic effect on ovarian cancer (18).
To test the role of HMGA2 in tumorigenesis of OSE cells, we used the immortalized OSE cells as a model system to investigate the oncogenic properties of HMGA2. We found that HMGA2 overexpression was sufficient to induce OSE transformation in vitro and in vivo and resulted in aggressive tumor cell growth. HMGA2-induced neoplastic transformation is at least partially attributable to a group of target genes that are involved in EMT. Our findings suggest that HMGA2 overexpression can be a critical molecular event responsible for the early tumorigenesis of human HG-PSC.
MATERIALS AND METHODS
Patients and Tissue Samples
This study included 30 cases of high-grade serous carcinoma (HG-PSC) and matched fallopian tube tissues. In addition, 30 ovaries from non-cancer patients were included as normal controls. All cases were collected from Northwestern Memorial Hospital between 2007 and 2010. The study was approved by the Northwestern University institutional review boards. The tissue sections were collected and prepared for tissue microarray (TMA) as described previously (8). Fresh-frozen tissues from 12 HG-PSC with HMGA2 overexpression and matched normal ovarian tissue were also collected for the study.
Cell lines and Cell Cultures
OSE cell lines T29 and T80 were immortalized using SV40 and hTERT. The cell lines maintained cell biomarkers of normal OSE and detail information of these two cell lines have been described elsewhere (19). Cell lines were maintained in cell culture medium consisting of 1:1 Medium199 (Sigma-Aldrich, St. Louis, MO) and MCDB105 medium (Sigma-Aldrich) with 10% heat-inactivated fetal bovine serum (USA Scientific, Ocala, FL) and 10 ng/ml epidermal growth factor (Sigma-Aldrich) in a humidified atmosphere of 95% air and 5% CO2. Additional ovarian cancer cell lines used in this study included ovarian cancer cell lines Skov3, Hey, and Ovcar-3 (from American Type Culture Collection) and T29H (T29 with stable mutant H-RASH12 expression) (19).
T29 and T80 with Stable Overexpression of HMGA2
pBabe retroviral constructs containing HMGA2 with and without the 3’ untranslated region (UTR) were prepared as described previously (S Figure 1) (10). The constructs were transfected into the Phoenix amphotropic packaging cell line (from American Type Culture Collection) per manufacturer’s protocol with the help of 25 µM of chloroquine (Sigma-Aldrich). The infected cells were subjected to 1.5 µg/ml puromycin for 10–30 days (Detailed information is provided in S Figure 1). Single colonies with HMGA2 overexpression were isolated and confirmed by reverse transcription–polymerase chain reaction (RT-PCR) and Western blot analysis (Figure 1A).
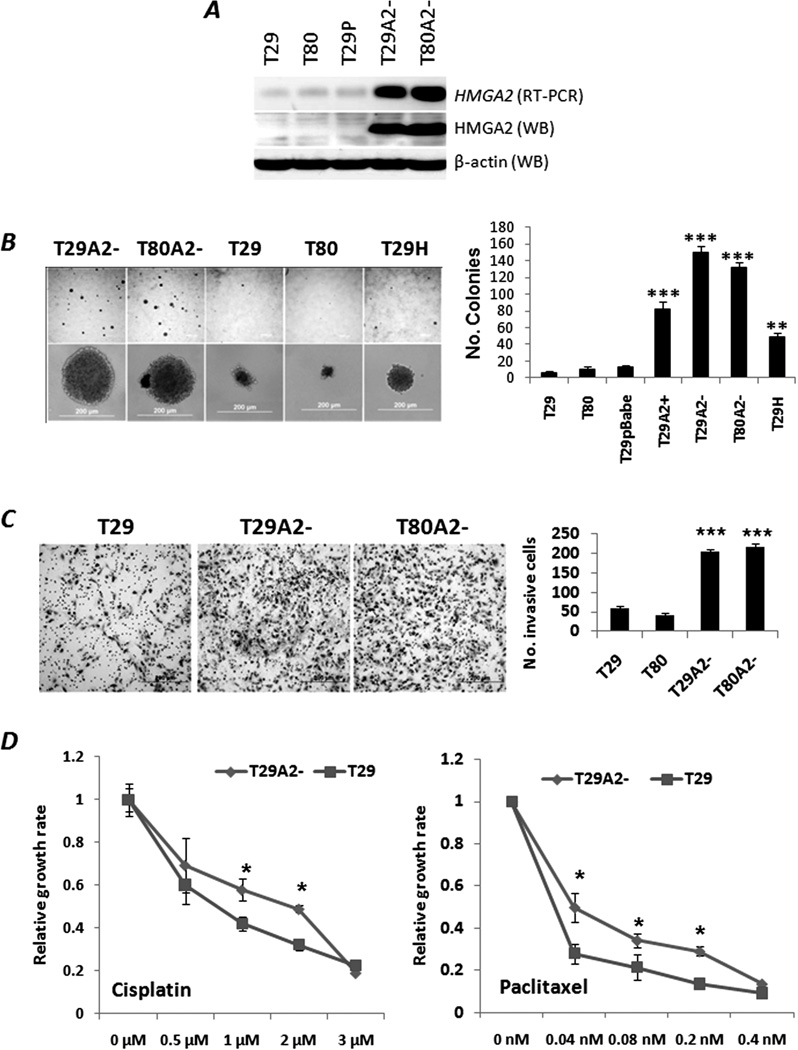
Figure 1.
HMGA2-induced OSE cell transformation in vitro. A. Stable HMGA2 overexpression in OSE T29 and T80 (designated as T29A2− and T80A2−), established by pBabe viral infection, was confirmed by RT-PCR and Western blot (WB) analysis. T29P indicates pBabe viral vector only. B. Photomicrographs illustrating examples of soft agar colonies (left) and histobars indicating the statistical significance of the numbers of colonies (right) in OSE cell lines with HMGA2 overexpression (T29A2−, T29A2+ and T80A2−), H-Ras overexpression (T29H), and controls (T29, T80 and T29P). Stable HMGA2 OSE cell lines with (T29A2+) and without (T29A2−, T80A2−) the HMGA2 3’UTR were also analyzed. C. Photomicrographs illustrating examples of Matrigel invasion (left) and histobars indicating the statistical significance for the numbers of invasive cells (right) in OSE cell lines with and without HMGA2 overexpression. D. Different growth rates of OSE cell lines with (T29) and without (T29A2−) HMGA2 overexpression in cisplatin (left) and paclitaxel (right) treatments. *: p<.05; **: p<.01; ***: p<.001.
Soft Agar Assay
The test cells (3×104) were suspended in 5 ml of culture medium containing 0.4% agar (USB Corporation, OH) and seeded onto a base layer of 5 ml of 0.7% agar bed in 10-cm tissue-culture dishes. Colonies >0.1 mm in diameter were counted after 2 weeks.
Matrigel Invasion Assay
2×105 test cells were placed in Matrigel-coated inserts (8-µm pores) (BD Biosciences, MA) in accordance with the manufacturer’s protocol. After 24 hrs, cells in the upper chamber were removed by cotton swab. Cells on the lower surface of the membrane were fixed by 10% formalin, stained by Giemsa and Eosin, and counted under a light microscope of 100× magnification.
Cellular Proliferation Assay
Cells were seeded in 24-well plates in triplicate at densities of 1×104 cells per well. Cell proliferation was monitored at 24, 48, 72, and 96 hrs using the colorimetric MTS assay (CellTiter 96® Aqueous Assay, Promega). In brief, the cells were rinsed with PBS and then incubated with 20% MTS reagent in serum-free medium for 3 hrs. Aliquots were then pipetted into 96-well plates, and the samples were read in a spectrophotometric plate reader at 490 nm (FLUOstar OPTIMA, BMG Lab Technologies, Germany).
siRNA and microRNA Transient Transfection
Cells were placed in standard medium without antibiotics for 24 hrs. Individual microRNA, siRNA, and control small RNA (Block-iT fluorescent double-stranded random 22mer RNA from Invitrogen), were used at a concentration of 60 pmol/well in a 6-well plate with Lipofectamine 2000 (Invitrogen). Cells receiving only the tagged random sequence double-strand 22mer were used as nonspecific references at all data points. After transfection, cells were harvested and analyzed at the indicated times. HMGA2 siRNAs were purchased from Invitrogen, and the efficacy of HMGA2 inhibition was tested by RT-PCR and Western blot analysis (Figure 2A).
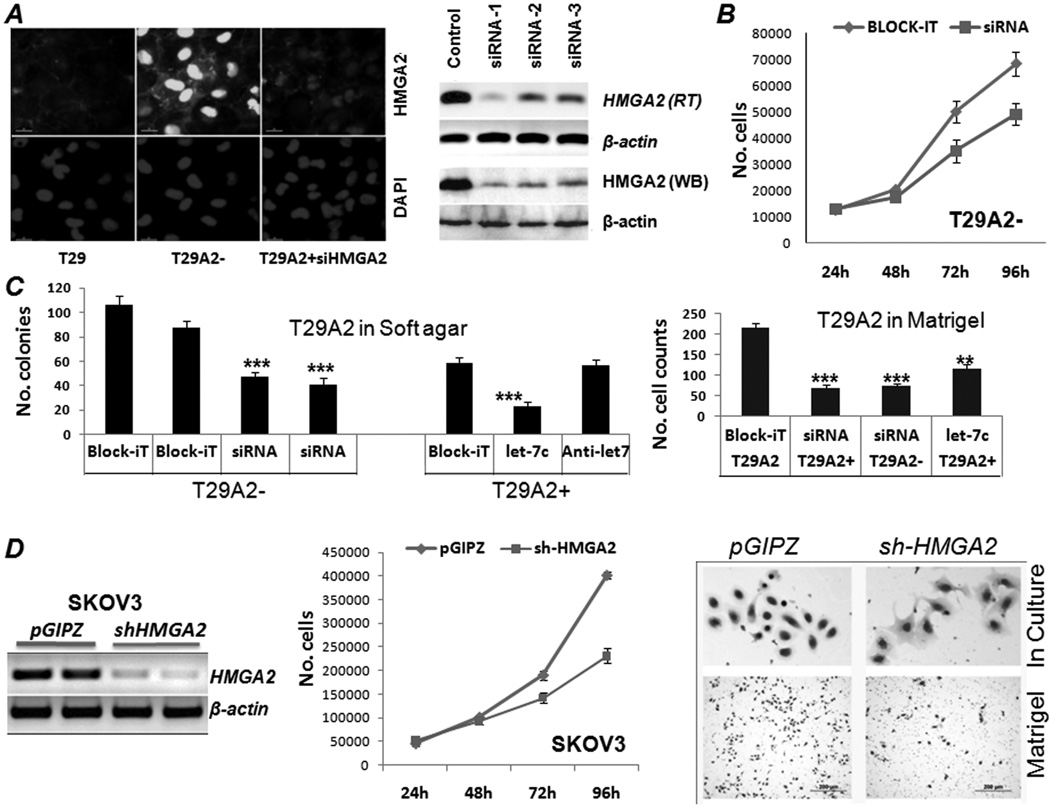
Figure 2.
Repression of HMGA2 expression reduced aggressive OSE tumor cell growth in vitro. A. Efficacy of HMGA2 repression by HMGA2 siRNAs in OSE cells illustrated by immunofluorescence staining (left) and RT-PCR and Western blot (right) analyses. B and C. Transient transfection of HMGA2 siRNA or let-7c significantly reduced T29A2 cell growth rate (B), soft agar colonies (C, left), and the numbers of invasive cells in Matrigel (C, right). ***: p<0.001. D. Stable HMGA2 repression by lentiviral shRNA infection in Skov-3 cell lines, demonstrated by RT-PCR analysis (left), significantly reduced Skov-3 cell proliferation (middle) and restored the epithelioid-like cell morphology (right upper) and reduced Matrigel invasion (right lower).
ShRNA HMGA2 Transfection
Human HMGA2 shRNA in pGIPZ was purchased from Open Biosystems (RHS4430-99160621). Lentivirus express HMGA2 shRNA were produced in HEK293T cells packaged by pMD2G and psPAX2. For stable infection, 8×104 cells were plated in each well of 6-well plates along with 2 ml of medium without antibiotics. After overnight incubation, medium was removed and replaced with 1 ml per well of Opti-MEM® I Reduced-Serum Medium containing 12 µg/mL polybrene. Next, 50 µL of concentrated lentiviral particles were added to each well. 48 h later, fresh medium containing 2 µg/ml puromycin was added to each well. Fresh medium containing puromycin was replaced every 3–4 days. Single colonies were obtained after 2 weeks of puromycin selection.
Luciferase Reporter Assay
The constructs were described in S Materials and Methods. Transfection was done in 293T and 293A cells with the aid of Lipofectamine 2000: 7×104 cells were seeded in each well of 24-well plates 1 day prior to transfection. Next, 0.1 µg of pRL-TK, 0.5 µg of pGL3-LUM, and indicated amounts of pcDNA3.1-HMGA2, together with varied amounts of blank pcDNA3.1 plasmid to keep constant the amounts of total DNA, were transfected into cells. Luciferase activity was measured 48 h after transfection using a dual-luciferase reporter assay system (Promega, WI), and results were normalized by Renilla luciferase activity. All experiments were repeated at least in triplicate for each sample.
Western Blotting
Cells were lysed at 4°C in NP40 cell lysis buffer (Invitrogen) supplemented with 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail. Equal amounts of total proteins from each sample were loaded onto a 12.5% SDS-PAGE gel and transferred to PVDF membranes. Membranes were incubated with primary antibody at 4°C overnight. The dilution rates of the primary antibodies were 1:100 to 1:1000 for LUM (goat anti-human IgG for LUM, R & D Systems, MN), HMGA2 (rabbit anti-human IgG (20)), Vimentin (mouse anti-human IgG, Santa Cruz), N-Cadherin (rabbit anti-human IgG, Santa Cruz), E-Cadherin (mouse anti-human IgG, Abcam) and β-actin (mouse anti-human IgG for β-actin, Sigma-Aldrich, MO). The secondary antibody was detected by Western ECL-enhanced luminol reagent (Perkinelmer Inc. MA).
Xenografts in Nude Mice
For xenograft experiments, 6-week old female BALB/c nude mice (NCI-Frederick, MD) were injected with 5×106 cells in bilateral flanks. Cells were resuspended in 0.1 ml of PBS for subcutaneous injection. Five types of cell lines were prepared for xenografts: T29 (10 implants); T80 (10 implants), T29H (7 implants), T29A2− (16 implants); T80A2− (18 implants). Tumor sizes were measured weekly, and mice were euthanized when tumors reached 1.5 cm in diameter.
MicroRNA Array
Five micrograms of total RNA were annealed with oligo mix from the miRNA profiling assay kit (Signosis, CA). The miRNA/oligo hybrids were purified by beads, and a ligation of miRNA-directed oligos was created to form a single molecule. The ligated molecule was incorporated with biotin-UTP. The labeled RNAs were hybridized with cancer miRNA array membrane (Signosis, CA) and detected using a chemiluminescence imaging system (Signosis). MiRNA expression data sets have been deposited into the NCBI with accession number of GSE23634.
mRNA Expression Array
Total RNA was isolated from 3 control cell lines (T29-1, T29-2, and T80) and testing cell lines (T29A2-8, T29A2-15, and T80A2-11) using an miRNA isolation kit (Ambion, TX). RNAs were labeled with Cy5/Cy3 and hybridized to the Human Whole Genome OneArray (Phalanx Biotech Group, Taiwan). The hybridized chips were scanned by an Axon 4000 scanner (Molecular Devices, CA). Spot quantification was done with the use of genepix 4.1 software. Hierarchical clustering of the expression profiles was analyzed by Cluster 3.0 (Molecular Devices). Differentially expressed genes with fold changes >2.0 were further analyzed by Pathway-Express (Onto-Tools, Wayne State University, MI). Gene expression data sets have been deposited into the NCBI with accession number of GSE23599.
Histological and Immunohistochemical Analyses
Formalin-fixed and paraffin-embedded tissues were sectioned at 4 µm. Antigen retrieval was performed by either heat-induced epitope retrieval or by proteolytic enzyme digestion as previously described (7). All immunohistochemical staining procedures were performed on a Ventana Nexus automated system (Tucson, Arizona, USA).
Immunofluorescence analysis
After being fixed with 4% paraformaldehyde, cells were permeabilized with 0.2% Triton X-100 and blocked with 10% goat serum. Then cells were incubated with specific primary antibody overnight at 4 °C. After cells were incubated with appropriate FITC-conjugated secondary antibodies (Abcam, MA) and stained by 4,6-diamidino-2-phenylindole (DAPI), the fluorescence images were examined with microscope (model IX70, Olympus).
Chemoresistance Assay
cis-Diamineplatinum (II) dichloride (cisplatin) and paclitaxel were purchased from Sigma (Sigma-Aldrich). Cultured cells were plated in 96-well plates (1×104 cells/well) overnight. Then, cells were treated with indicated dosages of cisplatin or paclitaxel for 48 hrs. Cell proliferation rates were evaluated by MTS assay (Promega, CA).
Statistical Analysis
All data are presented as means and standard errors of at least three independent experiments. Student’s t test was used for comparisons between two groups of experiments, and one-way ANOVA analysis was used for comparisons among three or more groups. P < 0.05 was considered significant.
RESULTS
HMGA2 Overexpression Results in OSE Cell Transformation In Vitro
Immortalized OSE cell lines T29 and T80 showed an epithelioid morphology in culture, and they maintained OSE cell markers, including cytokeratin 7 (CK-7), WT-1 and CA-125, as well as low levels of estrogen and progesterone receptor (ER and PR) expression in immunohistochemistry (19). T29 and T80 cells had very low levels of HMGA2 expression (Figure 1A). After pBabe viral constructs of HMGA2 were transfected into T29 and T80 cell lines (S Figure 1A), cell lines with stable HMGA2 overexpression were established and designated as T29A2 and T80A2. Five individual colonies were prepared from each of three cell lines with stable HMGA2 overexpression with (T29A2+) and without (T29A2− and T80A2−) 3’ untranslated region (3’UTR) (S Figure 1B). Levels of HMGA2 expression were evaluated by RT-PCR and Western blot analysis (Figure 1A, Supplementary Figure 1C). T29 cell line with pBabe virus vector only (designated as T29P) and stable overexpression of mutant H-RAS gene (defined as T29H) was used as controls (19) (S Figure 1B).
To test whether HMGA2 overexpression leads to OSE transformation, we examined anchorage-independent cell growth. Numbers of colonies formed by T29A2− (150±8.33) and T80A2− (132±5.69) were significantly higher than those by T29 (7±1.53), T80 (11±2.52), and T29P (12±8.33) (Figure 1B) (p<0.001). The average number of colonies in T29A2+ was half of that in T29A2− and T80A2−, indicating the functional role of endogenous microRNAs in regulation of HMGA2 expression. There was significant increase of colonies in T29H (49±4.21) in comparison to T29, T80 and T29P (Figure 1B).
To evaluate the mitogenic role of HMGA2, we compared cell proliferation rates in OSE cell lines with and without HMGA2 overexpression. We did not see a significant difference of cell proliferation rate between T29/T80 and T29A2/T80A2, due to fast growth rate of T29/T80. However, the growth rates of T29A2− were significantly higher (p<0.05) than those of T29 when cells were treated with chemotherapy reagents cisplatin and paclitaxel (Figure 1D), indicating an increase of chemoresistance in the presence of HMGA2 overexpression. This effect was dosage dependent.
To test whether HMGA2 overexpression induced a more aggressive growth of OSE cells, we examined the rate of OSE cell migration and invasion through Matrigel. T29A2− and T80A2− cells had significantly higher numbers of cells migrating through Matrigel than T29 and T80 had (p<0.01) (Figure 1C). The number of cells migrating through Matrigel in T29A2− and T80A2− was also significantly higher than that in T29H (data not shown), indicating more aggressive tumor invasion in HMGA2-mediated tumor transformation.
Repression of HMGA2 Overexpression Can Reduce the Oncogenic Activities of OSE and Ovarian Cancer Cell Lines In Vitro
To determine whether inhibition of HMGA2 expression can reduce the oncogenic activities in OSE and ovarian cancer cell lines, we prepared and tested three HMGA2-interfering RNA targeting molecules, including let-7s, HMGA2 siRNA, and shRNA (see Materials and Methods). As shown in Figure 2A, transient transfection of HMGA2 siRNAs significantly inhibited HMGA2 expression at both transcription and translational levels. Repression of HMGA2 expression by let-7s was also reproducible in T29A2+ (data not shown).
Repression of HMGA2 expression by siRNA in T29A2− significantly (p<0.01) reduced the rate of cell proliferation at 72–96 hours (Figure 2B). The rate of cell proliferation can also be reduced in T29A2+ cells by let-7c (data not shown). Similar results were found in SKOV-3 cells, in which complete repression of HMGA2 expression by shRNA expression revealed a more than 2-fold reduction of tumor cell proliferation (Figure 2D).
To test whether inhibition of HMGA2 overexpression interrupts anchorage-independent cell growth of OSE cell lines, we examined the cell growth in soft agar. Repression of HMGA2 expression by HMGA2 siRNA in T29A2− (Figure 2C) significantly decreased the numbers of colonies compared with controls (Block-iT) (p<0.001) (Figure 2C). The same results were obtained in T29A2+ treated with let-7 (Figure 2C). Matrigel invasion was partially blocked by repression of HMGA2 expression with either siRNA or let-7 mimics (Figure 2C). Notably, efficient repression of HMGA2 expression by shRNA in SKOV-3 cells significantly reduced cell proliferation (Figure 2D) and changed cell morphology in culture from a spindle shape to a polygonal-type differentiation (Figure 2D). These findings suggested a role for HMGA2 in the EMT and in promoting cell migration through Matrigel.
HMGA2 Overexpression in OSE Cell Lines Enhances Tumor Formation in Xenografts in Nude Mice
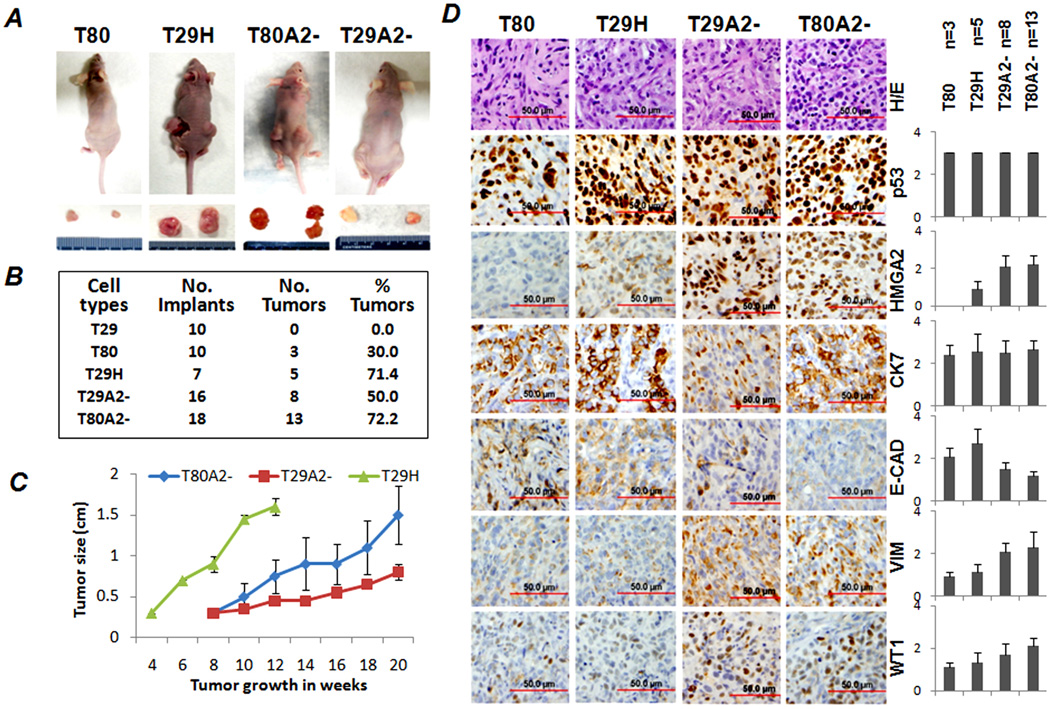
To explore whether HMGA2-mediated tumor transformation in OSE cell lines can promote tumor formation in vivo, we inoculated OSE cell lines with and without HMGA2 overexpression as xenografts in nude mice. We found that T29H, the positive control, required about 3 months to reach a tumor size of 1.5 cm in diameter and that 71.4% (5/7) of mice developed tumors (Figure 3A and 3B). T29H tumors showed mostly a solid growth pattern with extensive tumor necrosis. T29A2− and T80A2− grew visible tumors in about 8 weeks and reached 1.5 cm in 20 weeks (Figure 3C). Of the injection sites, 50% (8/16) of T29A2− and 72.2% (13/18) of T80A2− had tumor formation (Figure 3B). The tumor growth rates are summarized in Figure 3C. By the end of 20 weeks, no tumor was identified in mice receiving T29 (0/10), and three tumors of <0.5 cm were found in mice receiving T80 (3/10).
Figure 3.
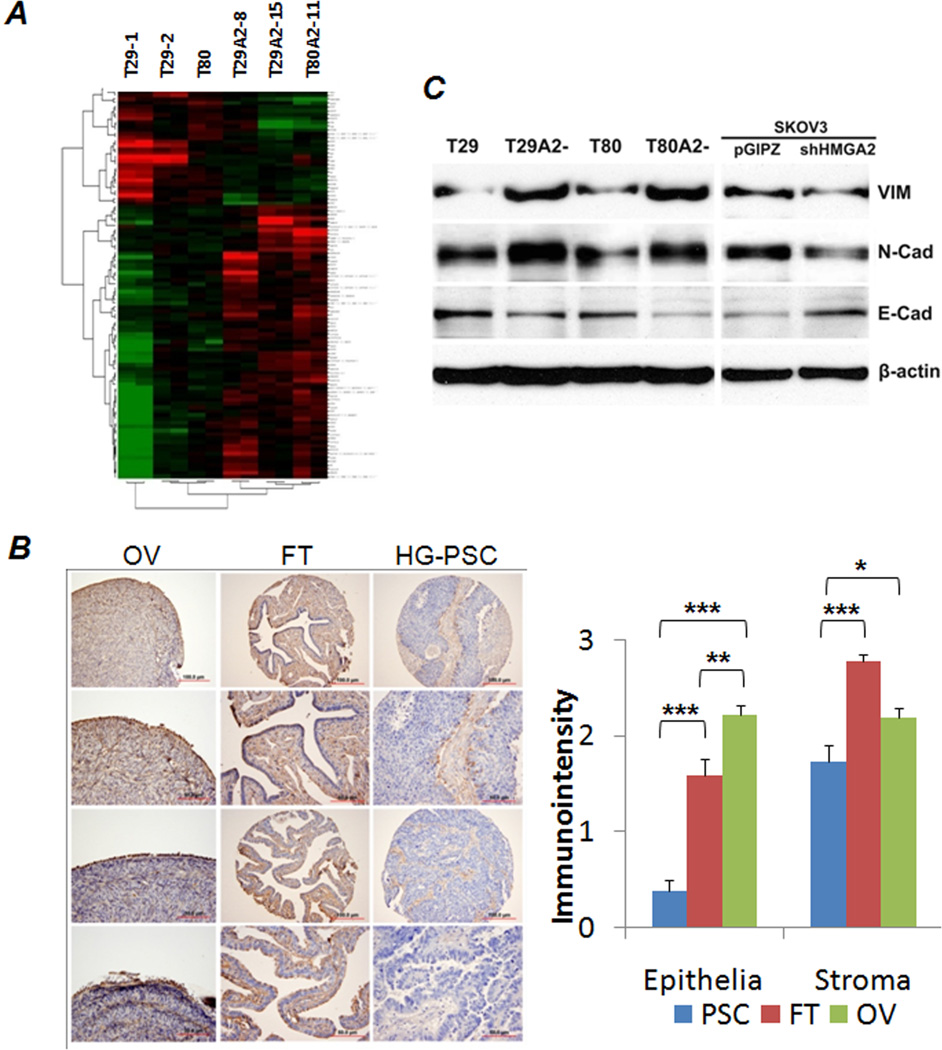
Tumor formations in xenografts of OSE cell lines with HMGA2 overexpression in nude mice. A. Photomacrographs illustrated examples of tumor formation in flanks of nude mice. Dissected tumors are listed below. B. Numbers of mice and the rate of tumor formation in each OSE cell type are summarized in this table. C. Growth curves of xenograft tumors in OSE with H-Ras (T29H) and HMGA2 overexpression (T29A2− and T80A2−) were measured biweekly (x-axis) in cm (y-axis). D. Immunophenotypes of xenograft tumors in OSE cell lines without (T80) and with (T29A2− and T80A2−) HMGA2 overexpression. Left panel: examples of immunostains in selected markers; right panel: statistical results. N represents the number of xenograft tumors collected for analysis. VIM: Vimentin; E-CAD: E-cadherin; CK7: Cytokeratin 7; WT1: Wilm’s tumor 1.
To characterize the pathological features of HMGA2-induced OSE tumors, we collected all xenograft tumors (29 tumors) and prepared a TMA from formalin-fixed and paraffin-embedded tumor tissues. T29A2− and T80A2− tumors had solid growth patterns with areas of nesting and papillary features (S Figure 2). Tumor cells had moderate to high-grade nuclei, brisk mitosis, and areas of tumor necrosis. Immunoprofiling showed that T29A2− and T80A2− tumor cells were strongly immunoreactive for HMGA2 and P53. Tumor cells maintained CK-7 and WT-1, characteristics of Mullerian epithelial origin. T29A2− and T80A2− showed increased immunoreactivity for vimentin and decreased immunoreactivity for E-cadherin compared with Ras-induced tumor (T29H) and T80 (Figure 3D). These findings supported a functional role of HMGA2 in the EMT in OSE tumors.
HMGA2-Induced Tumor Transformation Is Associated with Dysregulation of a Group of Target Genes and microRNAs in OSE Cell Lines
HMGA2 can induce OSE transformation in vitro (Figure 1) and in vivo (Figure 3). Identification of HMGA2-regulated target genes may increase the understanding of the oncogenic properties of HMGA2 in ovarian tumorigenesis. To test whether HMGA2-mediated tumor transformation acts through regulation of its target genes and microRNAs, we examined and compared global gene expression of OSE cell lines with and without HMGA2 overexpression (T29/T80 vs. T29A2−/T80A2−).
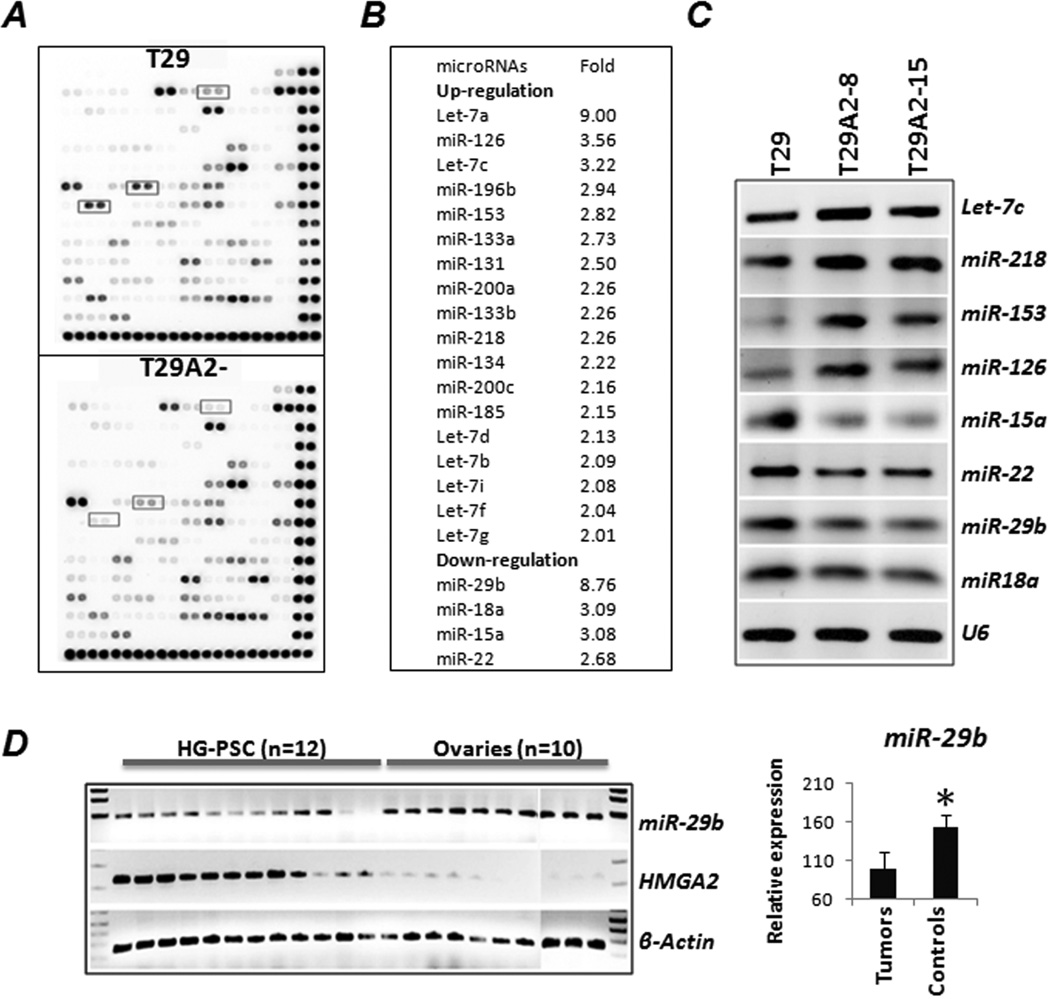
Of 124 cancer-related microRNAs, we found that 18% (22/124) were expressed differently between T29 and T29A2− by at least a 2-fold change (Figure 4A and 4B). Among these, four microRNAs were down-regulated (miR-15a, miR-18a, miR-22, and miR-29b). Differential expression of the selected microRNAs was validated by end-product (Figure 4C) and real-time (data not shown) RT-PCR. MiR-29b showed an almost 9-fold reduction in HMGA2-transformed OSE cell lines. MiR-29b is an important tumor suppressor in regulating the P53 pathway (21), apoptosis, and tumor growth, and was down-regulated in HG-PSC (22). In the selected 12 HG-PSC tissues with HMGA2 overexpression, significant down-regulation of miR-29b expression was evident compared with expression in matched normal ovarian tissues (Figure 4D).
Figure 4.
Differential expression of microRNAs in OSE cells with (T29A2−) and without (T29) HMGA2 overexpression. A. Dot-blot analysis of cancer-related microRNA expression (a total of 128 microRNAs) in T29 and T29A2−. Boxed dots illustrate significantly down-regulated miRNAs in T29A2−. B. List of microRNAs significantly up- or down-regulated (>2-fold) in OSE cell lines with HMGA2 overexpression. C. Differential expression of the selected miRNAs was validated by RT-PCR. D. Differential expression of miR-29b was examined in 12 human HG-PSC samples with HMGA2 overexpression and in 10 normal ovaries. A significant inverse correlation of HMGA2 and miR-29b expression was noted (right panel).
Seven of 11 let-7 family members were up-regulated in T29A2− and T80A2−. HMGA2-mediated up-regulation of let-7 family members in OSE cell lines could be related to a cellular response to HMGA2 overexpression. MiR-196 and miR29, the direct targets of HMGA1 (23), were dysregulated in HMGA2-transformed OSE cell lines. Whether HMGA2 can directly regulate miR-196 and miR-29 deserves further investigation. Among 22 miRNAs dysregulated by HMGA2 transduction, 73% (16/22) were found to be significantly dysregulated in ovarian carcinomas (24). The findings suggest that HMGA2-induced tumor transformation in vitro might share many of the molecular changes seen in human ovarian cancer.
HMGA2, a nuclear binding protein, directly regulates or interferes with the expression of many genes (25). We further examined the global gene expression of OSE cell lines with and without HMGA2 overexpression. Three OSE cell lines without HMGA2 overexpression (T29-1, T29-2, and T80) were used as controls, and three OSE cell lines with HMGA2 overexpression (T29A2-8, T29A2-15, and T80A2-11) were used as the testing group. Among a total of 32,000 genes, 621 were significantly dysregulated in OSE cell lines with HMGA2 overexpression (p<0.05). Among these, 36 genes showed at least 2-fold changes (Figure 6A, S Table 2), of which 25 genes were up-regulated and 11 genes were down-regulated. The genes most dysregulated in OSE cell lines with HMGA2 overexpression were confirmed by RT-PCR (data not shown).
Figure 6.
A. Dendrogram and cluster analysis revealed that 36 genes were differentially expressed (>2-fold) between in OSE cell lines with (right) and without (left) HMGA2 overexpression. Green indicates down-regulation and red up-regulation. B. Photomicrographs (left) illustrate examples of immunoreactivity for LUM in human high grade serous papillary carcinoma (HG-PSC), matched normal fallopian tubes and ovaries. Histobars (right) show the relative expression level of LUM scaled by immunointensity as mean values of LUM (y-axis) in HG-PSC (PSC, blue), fallopian tube (FT, red) and ovary (OV, green). *p<0.05; **p<0.01; ***p<0.001. C. Western blot analysis of Vimentin (VIM, upper), N-cadherin (N-Cad, mid) and E-cadherin (E-Cad, lower) in cells with (T29A2−, T80A2−, SKOV3) and without (T29, T80, SKOV3+shHMGA2) HMGA2 overexpression. β-actin is protein loading control.
Of the 11 down-regulated genes, 4 were functionally related to the EMT, including ID1, LUM, POSTN, and WNT2. Of the 25 up-regulated genes, 1 was EMT genes (STC2), 2 were mitogenic factors (CDKN1a and IFI27L2), and 4 were ribosomal protein complex genes (SNORD58B, RPS15A, RPS27L, and RPS14). In the list of HMGA2 target genes, 5 (16%) were associated with the EMT, suggesting the strong role of HMGA2 in EMT. This was compatible with our observation of frequent morphological changes in cultured ovarian cancer cell lines from a more spindle type (high HMGA2) to an epithelial type (low HMGA2) (Figure 2D). Our experiment identified a list of possible HMGA2 target genes in association with ovarian carcinogenesis. We selected LUM for further validation as shown below.
LUM, an EMT-Associated Gene, Is a Target of HMGA2
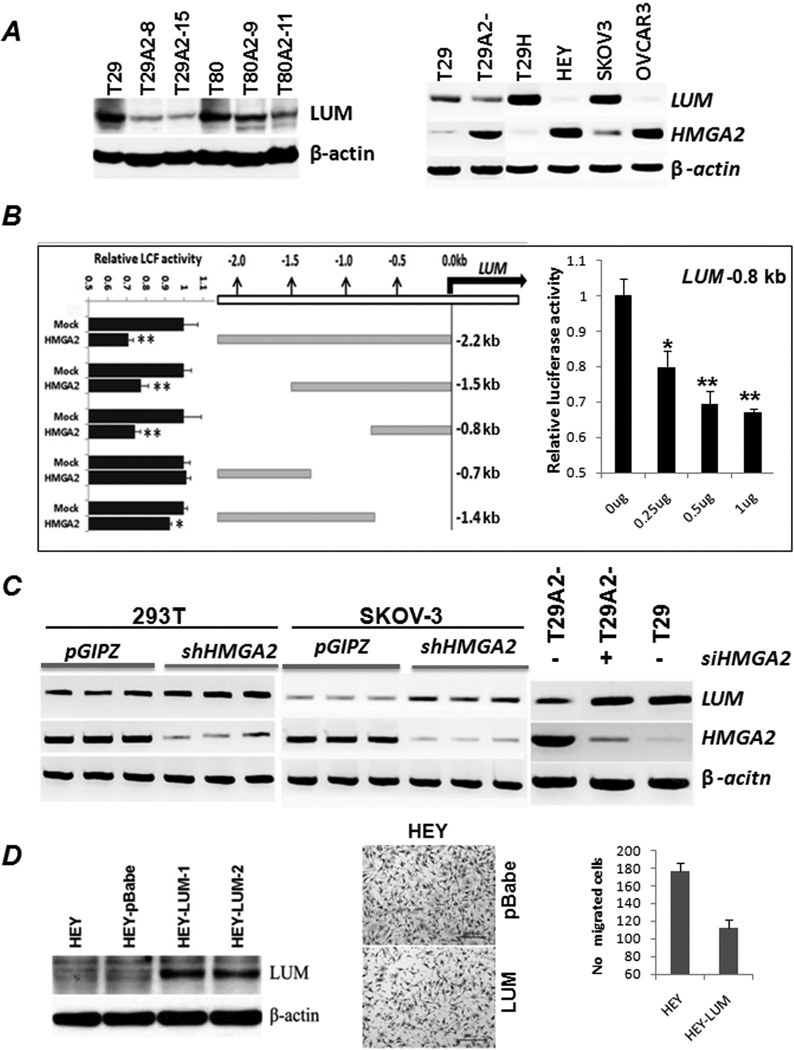
A near 3-fold reduction in LUM expression was found in all three HMGA2-induced OSE cell lines (S Table 2). Western blot analysis confirmed that HMGA2 induced a 2–5-fold reduction in LUM expression in OSE cell lines (Figure 5A). Interestingly, in the six OSE and ovarian cancer cell lines used in this study, HMGA2 expression was inversely correlated with LUM expression (Figure 5A). To determine whether this HMGA2-mediated LUM reduction was through transcription regulation, we examined LUM promoter regions.
Figure 5.
HMGA2 regulates target gene LUM expression. A. Western blot analysis confirmed the down-regulation of LUM in OSE cell lines T29 and T80 with HMGA2 overexpression (left). RT-PCR analysis of HMGA2 and LUM expression in several ovarian cancer cell lines (T29H, Hey, Skov-3, and Ovcar-3) revealed an inverse correlation of HMGA2 and LUM expression (right). B. Five constructs containing up to 2,200 nt of genomic DNA upstream of the LUM promoter were prepared for the luciferase assay. Significant reduction of luciferase expression was noted when the region from 0 to −800 nt was included in the construct and cotransfected with HMGA2 expression (left). Reduced luciferase expression rendered by the −800 nt DNA sequence was HMGA2 dosage dependent (right). C. Repression of HMGA2 expression by transfection of HMGA2 siRNA (right) or shRNA (left) enhanced LUM expression in T29A2−, 293T and Skov-3 cell lines. D. Ovarian cancer cell line Hey with stable LUM overexpression by western blot (left panel) significantly reduces the cell migration in Matrigel (middle and right panel) in comparison to the controls.
Computer analysis revealed that there were multiple HMGA2 AT binding domains (ATATT) along a 2,200 nt upstream region from the 5’ transcription start site. In several constructs representing different parts of 5’ LUM promoter regions for a luciferase assay (S Table 1), the first 800 nt, immediately adjacent to the LUM transcription start site, showed a significant reduction in luciferase expression when cotransfected with HMGA2 expression (Figure 5B). Reduction in luciferase activity was dose dependent (Figure 5B). In contrast, the upstream region starting from −1,000 nt showed minimal activity in driving luciferase expression, indicating that HMGA2 regulatory region is mostly confined to the 0 to −1,000 nt promoter region.
To examine whether HMGA2 interferes with LUM expression, we examined LUM expression in different cell types. LUM expression was up-regulated in OSE cells with transient transfection of HMGA2 siRNA or with shRNA (Figure 5C). These findings further support that HMGA2 can directly regulate LUM expression.
To test whether LUM is associated with invasion and migration in ovarian cancer, we selected HEY cell line (with very low level of endogenous LUM expression, Figure 5A) and established stable LUM overexpression (Figure 5D). We found that induction of LUM overexpression in ovarian cancer cell line HEY significantly reduced tumor cell migration through Mitragel (Figure 5D).
We examined LUM expression in 30 HG-PSC samples with matched fallopian tubes and 30 normal ovaries by immunohistochemistry. Overall, LUM expression was found to be significantly higher in stroma than in epithelial cells (Figure 6B). Furthermore, HG-PSC tumor cells showed significant down-regulation of LUM compared with normal ovarian surface and fallopian tube epithelia (Figure 6B). When we compared whether HMGA2 overexpression was associated with LUM expression in the 30 cases of HG-PSC, a weak correlation (r=0.29) was noted. The weak correlation was largely owing to the very low level of LUM immunoreactivity in almost all HG-PSC samples.
DISCUSSION
HMGA2 was shown to enhance tumor transformation in different cell types (26–27). It has been found that HMGA2 overexpression is associated with aggressive tumor growth, early metastasis, and a poor prognosis, typically seen in papillary thyroid carcinoma (28), pancreatic cancer (11), breast cancer (29), and HG-PSC (18). In this study, we found that HMGA2 enhances OSE cell transformation (Figure 1 and 3), and that the oncogenic properties of HMGA2 in ovarian cancer may be mediated by its regulation of genes involved in EMT.
EMT, a complex process involving multiple extracellular signal pathways (30), confers tumor plasticity by converting adherent epithelial cells to motile mesenchymal cells through the functional loss of E-cadherin, which is required to maintain epithelial cell-cell adhesion (30). EMT plays a key role in embryonic development and is important in the pathogenesis of cancer. Several EMT-associated genes have been identified and their functions have been characterized in breast and ovarian cancers (30). Until recently, HMGA2 has not been linked to EMT regulation. Shell et al. found that HMGA2 is one of a few gene markers that can distinguish most type I (mesenchymal gene signature) from type II (epithelial gene signature) cancer cell lines (6, 15). In addition, Thauault et al identified HMGA2 to be a transcriptional regulator of SNAIL1 by directly binding to the promoter (16). SNAIL1, a key EMT molecule, is associated with aggressive tumor growth in pancreatic cancer (17).
In this study, we found that HMGA2-induced OSE transformation displays many cellular and molecular features common to the EMT process. First, we observed that OSE cells with high HMGA2 expression had a spindle-like cell morphology in culture and that repression of HMGA2 restored their epithelioid phenotype (Figure 2D). Second, HMGA2 expressing OSE xenograft tumors had a significant loss of E-cadherin and increase of vimentin (Figure 3D). Third, miRNAs miR-200s (31) and miR-29b (32), critical microRNAs in the EMT, were significantly dysregulated in OSE cells with HMGA2 overexpression (Figure 4B). Fourth, we found that induction of HMGA2 overexpression in OSE cell lines (T29 and T80) and inhibition of HMGA2 expression in ovarian cancer cell line SKOV3 had an impact on EMT gene expression, including vimentin, N-cadherin and E-Cadherin (Figure 6C).
One of most important findings in this study was that about 16% of the HMGA2-regulated genes were associated with EMT (S Table 2). Among them is LUM which is down regulated by HMGA2 overexpression. LUM, a member of a small leucine-rich proteoglycan family, constitutes a fraction of noncollagenous extracellular matrix proteins (33). LUM can bind type I collagen and regulate the assembly of collagen fibers. LUM functions in the EMT in the wound healing process (34) and affects the bioavailability of SMAD 2 activators (TGF-β) (35). Nikitovic et al (35) reported that a human osteosarcoma cell line with a higher level of LUM expression displays low metastatic capability. In breast cancer, low LUM expression was shown to be correlated with rapid progression and poor survival (36). Although the functional role of LUM in cancer has not been fully characterized, the accumulating evidence suggests that it may serve as a tumor suppressor by inhibiting EMT. Thus, the identification of LUM as target of HMGA2 in ovarian cancer provides further evidence that HMGA2 promotes ovarian tumorigenesis through regulation of EMT.
Several other EMT-associated genes indentified in this study may also be directly regulated by HMGA2. For example, T29A2− and T80A2− cell lines have STC2 overexpression (Supplementary Table 2). Recent study demonstrates that STC2 overexpression promotes epithelial and stromal transition in ovarian cancer cell lines in the hypoxic condition (37). Further characterization of how HMGA2 regulates these gene expressions and their EMT function will provide new insight for our understanding of HG-PSC.
In summary, we have, to our knowledge for the first time, achieved HMGA2 overexpression in human OSE cell lines, and we found that HMGA2 overexpression was sufficient to induce OSE cell transformation and tumor formation. HMGA2-induced ovarian carcinogenesis occurs at least in part through controlling tumor-associated EMT gene expression. We postulate that induction of HMGA2 overexpression in early ovarian serous carcinoma may be responsible for the short latency of noninvasive carcinoma to the rapid progression of ovarian serous carcinoma. Further characterization of the functional relationship between HMGA2 and HMGA2-mediated EMT target gene regulation will help us understand the tumorigenesis of ovarian serous carcinoma.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Miguel Segura and Guangyu Yang, Ximing Yang and Chunyan Luan for technical support.
Supports: This study is supported by MFH Dixon translation research fund.
Footnotes
This study was presented in 99th United States and Canadian Academy of Pathology (2010) at Washington DC.
REFERENCES
- 1.Bartel F, Jung J, Bohnke A, et al. Both germ line and somatic genetics of the p53 pathway affect ovarian cancer incidence and survival. Clin Cancer Res. 2008;14:89–96. doi: 10.1158/1078-0432.CCR-07-1192. [DOI] [PubMed] [Google Scholar]
- 2.Salani R, Kurman RJ, Giuntoli R, 2nd, et al. Assessment of TP53 mutation using purified tissue samples of ovarian serous carcinomas reveals a higher mutation rate than previously reported and does not correlate with drug resistance. Int J Gynecol Cancer. 2008;18:487–491. doi: 10.1111/j.1525-1438.2007.01039.x. [DOI] [PubMed] [Google Scholar]
- 3.Ramus SJ, Gayther SA. The contribution of BRCA1 and BRCA2 to ovarian cancer. Mol Oncol. 2009;3:138–150. doi: 10.1016/j.molonc.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinn BA, Brake T, Hua X, et al. Induction of ovarian leiomyosarcomas in mice by conditional inactivation of Brca1 and p53. PLoS One. 2009;4:e8404. doi: 10.1371/journal.pone.0008404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark-Knowles KV, Senterman MK, Collins O, Vanderhyden BC. Conditional inactivation of Brca1, p53 and Rb in mouse ovaries results in the development of leiomyosarcomas. PLoS One. 2009;4:e8534. doi: 10.1371/journal.pone.0008534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park SM, Shell S, Radjabi AR, et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2090. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 7.Mahajan A, Liu Z, Gellert L, et al. HMGA2: a biomarker significantly overexpressed in high-grade ovarian serous carcinoma. Mod Pathol. 2010;23:673–681. doi: 10.1038/modpathol.2010.49. [DOI] [PubMed] [Google Scholar]
- 8.Wei JJ, Wu J, Luan C, et al. HMGA2: a potential biomarker complement to P53 for detection of early-stage high-grade papillary serous carcinoma in fallopian tubes. Am J Surg Pathol. 2010;34:18–26. doi: 10.1097/PAS.0b013e3181be5d72. [DOI] [PubMed] [Google Scholar]
- 9.Reeves R. Structure and function of the HMGI(Y) family of architectural transcription factors. Environ Health Perspect. 2000;108(Suppl 5):803–809. doi: 10.1289/ehp.00108s5803. [DOI] [PubMed] [Google Scholar]
- 10.Peng Y, Laser J, Shi G, et al. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6:663–673. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- 11.Hristov AC, Cope L, Reyes MD, et al. HMGA2 protein expression correlates with lymph node metastasis and increased tumor grade in pancreatic ductal adenocarcinoma. Mod Pathol. 2009;22:43–49. doi: 10.1038/modpathol.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzatsos A, Bardeesy N. Ink4a/Arf regulation by let-7b and Hmga2: a genetic pathway governing stem cell aging. Cell Stem Cell. 2008;3:469–470. doi: 10.1016/j.stem.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li AY, Boo LM, Wang SY, et al. Suppression of nonhomologous end joining repair by overexpression of HMGA2. Cancer Res. 2009;69:5699–5706. doi: 10.1158/0008-5472.CAN-08-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283:33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe S, Ueda Y, Akaboshi S, Hino Y, Sekita Y, Nakao M. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am J Pathol. 2009;174:854–868. doi: 10.2353/ajpath.2009.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malek A, Bakhidze E, Noske A, et al. HMGA2 gene is a promising target for ovarian cancer silencing therapy. Int J Cancer. 2008;123:348–356. doi: 10.1002/ijc.23491. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Yang G, Thompson-Lanza JA, et al. A genetically defined model for human ovarian cancer. Cancer Res. 2004;64:1655–1663. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 20.Narita M, Narita M, Krizhanovsky V, et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 21.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 22.Flavin R, Smyth P, Barrett C, et al. miR-29b expression is associated with disease-free survival in patients with ovarian serous carcinoma. Int J Gynecol Cancer. 2009;19:641–647. doi: 10.1111/IGC.0b013e3181a48cf9. [DOI] [PubMed] [Google Scholar]
- 23.De Martino I, Visone R, Fedele M, et al. Regulation of microRNA expression by HMGA1 proteins. Oncogene. 2009;28:1432–1442. doi: 10.1038/onc.2008.495. [DOI] [PubMed] [Google Scholar]
- 24.Dahiya N, Morin PJ. MicroRNAs in ovarian carcinomas. Endocr Relat Cancer. 2010;17:F77–F89. doi: 10.1677/ERC-09-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fedele M, Visone R, De Martino I, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 26.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Cello F, Hillion J, Hristov A, et al. HMGA2 participates in transformation in human lung cancer. Mol Cancer Res. 2008;6:743–750. doi: 10.1158/1541-7786.MCR-07-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiappetta G, Ferraro A, Vuttariello E, et al. HMGA2 mRNA expression correlates with the malignant phenotype in human thyroid neoplasias. Eur J Cancer. 2008;44:1015–1021. doi: 10.1016/j.ejca.2008.02.039. [DOI] [PubMed] [Google Scholar]
- 29.Fabjani G, Tong D, Wolf A, et al. HMGA2 is associated with invasiveness but not a suitable marker for the detection of circulating tumor cells in breast cancer. Oncol Rep. 2005;14:737–741. [PubMed] [Google Scholar]
- 30.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 31.Bendoraite A, Knouf EC, Garg KS, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 2010;116:117–125. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luna C, Li G, Qiu J, Epstein DL, Gonzalez P. Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol Vis. 2009;15:2488–2497. [PMC free article] [PubMed] [Google Scholar]
- 33.Blochberger TC, Cornuet PK, Hassell JR. Isolation and partial characterization of lumican and decorin from adult chicken corneas. A keratan sulfate-containing isoform of decorin is developmentally regulated. J Biol Chem. 1992;267:20613–20619. [PubMed] [Google Scholar]
- 34.Saika S, Miyamoto T, Tanaka S, et al. Response of lens epithelial cells to injury: role of lumican in epithelial-mesenchymal transition. Invest Ophthalmol Vis Sci. 2003;44:2094–2102. doi: 10.1167/iovs.02-1059. [DOI] [PubMed] [Google Scholar]
- 35.Nikitovic D, Berdiaki A, Zafiropoulos A, et al. Lumican expression is positively correlated with the differentiation and negatively with the growth of human osteosarcoma cells. FEBS J. 2008;275:350–361. doi: 10.1111/j.1742-4658.2007.06205.x. [DOI] [PubMed] [Google Scholar]
- 36.Troup S, Njue C, Kliewer EV, et al. Reduced expression of the small leucine-rich proteoglycans, lumican, and decorin is associated with poor outcome in node-negative invasive breast cancer. Clin Cancer Res. 2003;9:207–214. [PubMed] [Google Scholar]
- 37.Law AY, Wong CK. Stanniocalcin-2 promotes epithelial-mesenchymal transition and invasiveness in hypoxic human ovarian cancer cells. Exp Cell Res. 2010 doi: 10.1016/j.yexcr.2010.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.