Significance
Monovalent ions, such as Na+, play important roles in biology, yet few sensors that image intracellular Na+ have been reported. Although deoxyribozymes (DNAzymes) have been shown to be a promising platform for detection of metal ions, most reported DNAzymes require multivalent metal ions for catalytic activity. Existing monovalent ion-responsive DNAzymes have poor selectivity for Na+, low catalytic rate, and require high ion concentrations for function. Here, we report in vitro selection of the first (to our knowledge) highly selective, sensitive, and efficient Na+-specific, RNA-cleaving DNAzyme and its conversion into a catalytic beacon sensor for imaging Na+ in living cells, using an efficient cationic polypeptide delivery method, together with a photocaging strategy, to allow controllable activation of the DNAzyme probe inside cells.
Keywords: deoxyribozyme, in vitro selection, sodium, fluorescent sensor, intracellular imaging
Abstract
Over the past two decades, enormous progress has been made in designing fluorescent sensors or probes for divalent metal ions. In contrast, the development of fluorescent sensors for monovalent metal ions, such as sodium (Na+), has remained underdeveloped, even though Na+ is one the most abundant metal ions in biological systems and plays a critical role in many biological processes. Here, we report the in vitro selection of the first (to our knowledge) Na+-specific, RNA-cleaving deoxyribozyme (DNAzyme) with a fast catalytic rate [observed rate constant (kobs) ∼0.1 min−1], and the transformation of this DNAzyme into a fluorescent sensor for Na+ by labeling the enzyme strand with a quencher at the 3′ end, and the DNA substrate strand with a fluorophore and a quencher at the 5′ and 3′ ends, respectively. The presence of Na+ catalyzed cleavage of the substrate strand at an internal ribonucleotide adenosine (rA) site, resulting in release of the fluorophore from its quenchers and thus a significant increase in fluorescence signal. The sensor displays a remarkable selectivity (>10,000-fold) for Na+ over competing metal ions and has a detection limit of 135 µM (3.1 ppm). Furthermore, we demonstrate that this DNAzyme-based sensor can readily enter cells with the aid of α-helical cationic polypeptides. Finally, by protecting the cleavage site of the Na+-specific DNAzyme with a photolabile o-nitrobenzyl group, we achieved controlled activation of the sensor after DNAzyme delivery into cells. Together, these results demonstrate that such a DNAzyme-based sensor provides a promising platform for detection and quantification of Na+ in living cells.
Metal ions play crucial roles in a variety of biochemical processes. As a result, the concentrations of cellular metal ions have to be highly regulated in different parts of cells, as both deficiency and surplus of metal ions can disrupt normal functions (1–4). To better understand the functions of metal ions in biology, it is important to detect metal ions selectively in living cells; such an endeavor will not only result in better understanding of cellular processes but also novel ways to reprogram these processes to achieve novel functions for biotechnological applications.
Among the metal ions in cells, sodium (Na+) serves particularly important functions, as changes in its concentrations influence the cellular processes of numerous living organisms and cells (5–8), such as epithelial and other excitable cells (9). As one of the most abundant metal ions in intracellular fluid (10), Na+ affects cellular processes by triggering the activation of many signal transduction pathways, as well as influencing the actions of hormones (11). Therefore, it is important to carefully monitor the concentrations of Na+ in cells. Toward this goal, instrumental analyses by atomic absorption spectroscopy (12), X-ray fluorescence microscopy (13), and 23Na NMR (14) have been used to detect the concentration of intracellular Na+. However, it is difficult to use these methods to obtain real-time dynamics of Na+ distribution in living cells. Fluorescent sensors provide an excellent choice to overcome this difficulty, as they can provide sensitive detection with high spatial and temporal resolution. However, despite significant efforts in developing fluorescent metal ion sensors, such as those based on either genetically encoded probes or small molecular sensors, most fluorescent sensors reported so far can detect divalent metal ions such as Ca2+, Zn2+, Cu2+, and Fe2+ (15–21). Among the limited number of Na+ sensors, such as sodium-binding benzofuran isophthalate (22), Sodium Green (23), CoroNa Green/Red (24, 25), and Asante NaTRIUM Green-1/2 (26), most of them are not selective for Na+ over K+ (22–25, 27, 28) or have a low binding affinity for Na+ (with a Kd higher than 100 mM) (25, 27–31). Furthermore, the presence of organic solvents is frequently required to achieve the desired sensitivity and selectivity for many of the Na+ probes (32–34), making it difficult to study Na+ under physiological conditions. Therefore, it is still a major challenge to design fluorescent sensors with strong affinity for Na+ and high selectivity over other monovalent and multivalent metal ions that work under physiological conditions.
To meet this challenge, our group and others have taken advantage of an emerging class of metalloenzymes called DNAzymes (deoxyribozymes or catalytic DNA) and turned them into metal ion probes. DNAzymes were first discovered in 1994 through a combinatorial process called in vitro selection (35). Since then, many DNAzymes have been isolated via this selection process. Among them, RNA-cleaving DNAzymes are of particular interest for metal ion sensing, due to their fast reaction rate and because the cleavage, which is catalyzed by a metal ion cofactor, can easily be converted into a detectable signal (36–38). Unlike the rational design of either small-molecule or genetically encoded protein sensors, DNAzymes with desired sensitivity and specificity for a metal ion of interest can be selected from a large library of DNA molecules, containing up to 1015 different sequences (35, 39). A major advantage of DNAzymes as metal ion sensors is that metal-selective DNAzymes can be obtained without prior knowledge of necessary metal ion binding sites or specific metal–DNA interaction (40, 41). In addition, through the in vitro selection process, metal ion binding affinity and selectivity can be improved by tuning the stringency of selection pressure and introducing negative selection against competing metal ions (39, 40). Finally, DNA is easily synthesized with a variety of useful modifications and its biocompatibility makes DNAzyme-based sensors excellent tools for live-cell imaging of metal ions. As a result, several metal-specific DNAzymes have been isolated and converted into sensors for their respective metal ion cofactors, including Pb2+ (35, 42, 43), Cu2+ (44, 45), Zn2+ (46), UO22+ (47), and Hg2+ (48). They have recently been delivered into cells for monitoring UO22+ (41, 49), Pb2+ (50), Zn2+ (51), and histidine (52) in living cells.
However, in contrast to the previously reported DNAzymes with divalent metal ion selectivity, no DNAzymes have been reported to have high selectivity toward a specific monovalent metal ion. Although DNAzymes that are independent of divalent metal ions have been obtained (53–55), including those using modified nucleosides with protein-like functionalities (i.e., guanidinium and imidazole) (56–58), no DNAzyme has been found to be selective for a specific monovalent metal ion over other monovalent metal ions. For example, the DNAzyme with the highest reported selectivity for Na+ still binds Na+ over K+ with only 1.3-fold selectivity (54). More importantly, those DNAzymes require very high concentrations of monovalent ions (molar ranges) to function and display very slow catalytic rates (e.g., 10−3 min−1) (53–55). The poor selectivity, sensitivity, and slow catalytic rate render these DNAzymes unsuitable for cellular detection of Na+, due to interference from other monovalent ions such as K+ (which is present in concentrations about 10-fold higher than Na+), and the need to image the Na+ rapidly.
In this study, we report the in vitro selection and characterization of an RNA-cleaving DNAzyme with exceptionally high selectivity (>10,000-fold) for Na+ over other competing metal ions, with a dynamic range covering the physiological Na+ concentration range (0.135–50 mM) and a fast catalytic rate (kobs, ∼0.1 min−1). This Na+-specific DNAzyme was transformed into a DNAzyme-based fluorescent sensor for imaging intracellular Na+ in living cells, by adopting an efficient DNAzyme delivery method using a cationic polypeptide, together with a photocaging strategy to allow controllable activation of the probe inside cells.
Results and Discussion
In Vitro Selection of Na+-Specific DNAzyme.
With the goal of selecting Na+-specific DNAzymes, we carried out two parallel experiments using a combination of column-based and gel-based in vitro selection methods reported previously (35, 46, 47), one in the presence of 135 mM total Na+ (selection A) and another in the presence of 400 mM total Na+ (selection B). Each selection used a 110-mer oligonucleotide containing a 50-nt random sequence, flanked by conserved sequences at both sides as primer-binding regions for PCR amplification. The intended cleavage site, a single adenosine ribonucleotide (rA), was contained in the 5′-conserved region of the library sequence. Two distinct pairing regions were designed in the library to confine the folded random region to be in proximity of the rA cleavage site (SI Appendix, Fig. S1A). For both selections A and B, the positive selection buffers contained 10 mM citrate and 1 mM EDTA to minimize the likelihood of selecting DNAzymes that could exploit trace multivalent metal ions to catalyze cleavage of the rA in the cleavage site.
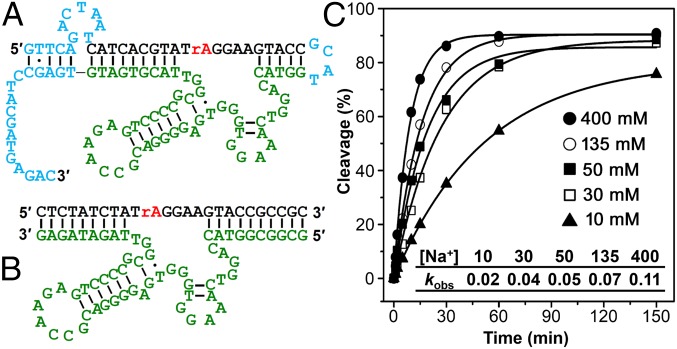
Initially, the selection pools were immobilized on NeutrAvidin columns through the 5′-biotin moiety on the DNA molecules (SI Appendix, Fig. S1B). Functional sequences that could catalyze cleavage of the RNA bond in the presence of Na+ were eluted off the columns. Before using PCR to amplify the cleaved DNA for the next round of selection, we performed an additional isolation step using denaturing PAGE purification to collect the cleaved DNA product. We added this step to prevent collecting nonspecific detachment of uncleaved DNA sequences from the column to seed the next-round selection. This additional purification step was necessary due to the fact that a monovalent metal ion was used as the cleavage cofactor and the fraction of active sequences in the initial rounds of the in vitro selection was much lower than nonspecific detachment of uncleaved sequences (SI Appendix, Fig. S2). Selective-amplification cycles were repeated until the DNA pools were enriched with Na+-specific DNAzymes. To increase the stringency of the selection and thus search for more efficient DNAzymes, the reaction time was gradually decreased from 2 h in round 1, to 45 s in round 14. Selection progress and enrichment of the DNA pools with Na+-specific DNAzymes were monitored using several methods (SI Appendix). The most active pools from selection A (round 13) and B (round 15) were chosen for cloning and sequencing to find the best DNAzymes based on the percentage of the pool cleaved, the signal-to-background ratio, and the apparent kobs of different pools (SI Appendix, Figs. S3–S6). Analysis of 95 resulting sequences revealed the existence of two and three major classes of similar sequences in selection A and B, respectively (SI Appendix, Tables S1 and S2). The class A-II and B-I shared a common consensus sequence in the 50-nt random region (SI Appendix, Table S3). The catalytic activity of the obtained sequences was then screened to find the most active DNAzyme. One such clone, named NaA43, displayed a kobs of 0.11 ± 0.01 min−1 in the presence of 400 mM Na+ at 20 °C, which is about two to three orders of magnitude faster than those of other divalent-independent DNAzymes reported previously (53–55). It is also known that naturally occurring ribozymes such as the hammerhead, hairpin, and hepatitis delta virus ribozymes can catalyze the RNA cleavage reaction in the presence of very high concentrations of monovalent metal ions (as high as 4 M) with low catalytic activity (10−3 min−1 in 4 M Na+) (59, 60). Therefore, in contrast to the previously reported DNAzymes and ribozymes, NaA43 is the first (to our knowledge) monovalent-specific RNA-cleaving nucleic acid catalyst with substantial activity at relatively low concentrations of Na+. The NaA43 DNAzyme achieved a rate enhancement of ∼109-fold over uncatalyzed cleavage of a single RNA phosphodiester bond under similar conditions (61, 62). No noticeable difference was observed in the activity of the Na+-dependent NaA43 DNAzyme in the presence or absence of 50 mM EDTA (SI Appendix, Fig. S7), suggesting that the DNAzyme activity was not caused by any trace multivalent metal ions in the buffer. In addition, we performed a series of control experiments by following strict RNase-free handling procedures, including decontaminating bench surfaces and pipettes with RNase AWAY reagent, preparing all of the buffers freshly with diethylpyrocarbonate-treated water, and using NaCl either baked at 260 °C for 36 h or treated with proteinase K. The results from these experiments showed that RNA degradation caused by RNase did not contribute to the cleavage activity of the NaA43 DNAzyme (SI Appendix, Fig. S8).
Based on the secondary structure of the cis-cleaving NaA43 DNAzyme (Fig. 1A), predicted by the UNAFold web package (63), the NaA43 DNAzyme was truncated into the minimal catalytic sequence by removing the sequences in blue and converted into a true enzyme with catalytic turnover (trans-form), by removing the connecting loop between the substrate strand (NaA43S) and the enzyme strand (NaA43E) and then extending the double-stranded arm to ensure efficient hybridization (Fig. 1B). Activity assays (Fig. 1C and SI Appendix, Fig. S9) indicate that the truncation has little effect on the activity of the NaA43 DNAzyme. It is known that the substrate-binding arms of different DNAzymes, such as those of 8–17 and 10–23, are interchangeable and that their sequences do not play a significant role in the DNAzymes activity (42, 64). Similarly, the NaA43 DNAzyme showed no significant change in activity when tested with four different substrate-binding arms (SI Appendix, Fig. S10).
Fig. 1.
The sequence and activity of a Na+-specific DNAzyme. (A) Secondary structure of an in vitro selected Na+-specific DNAzyme (A43 clone) in its cis-cleaving form. Regions that are colored in blue were removed by truncation. (B) Secondary structure of the trans-cleaving NaA43 DNAzyme. (C) Plot of the fraction of cleavage product (percentage) versus the incubation time for the cis-cleaving NaA43 DNAzyme at different concentrations of Na+. (Inset) kobs (per minute) values for the NaA43 DNAzyme from C. The calculated SD was less than 10% for all samples.
Converting the DNAzyme into a Fluorescent Sensor for Na+ Detection.
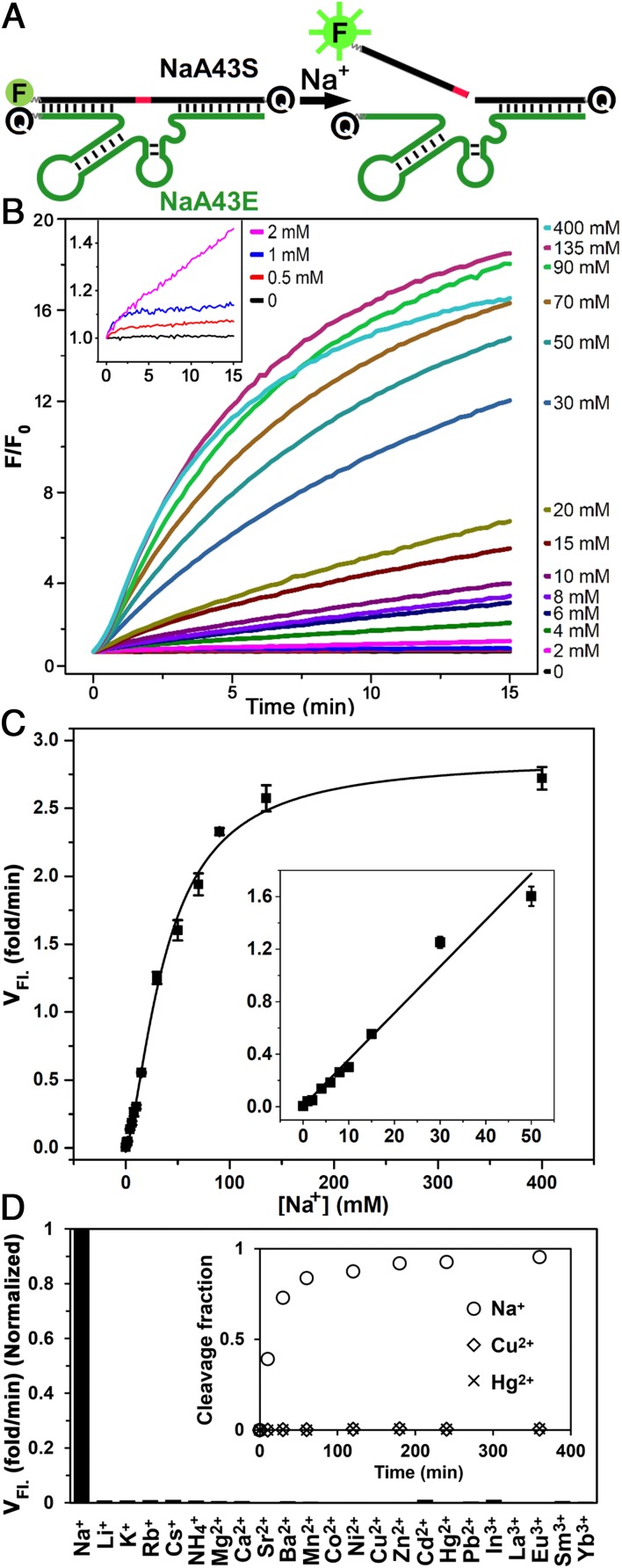
To convert the Na+-dependent catalytic activity of the NaA43 DNAzyme into a turn-on fluorescence response, we next designed a catalytic beacon by labeling the NaA43S with a 6-carboxyfluorescein fluorophore (FAM) at its 5′ end and the NaA43E with an Iowa Black FQ quencher at its 3′ end (65). In addition, a second quencher was added at the 3′ end of the NaA43S to minimize background fluorescence (Fig. 2A) (66). To ensure stable duplex formation at room temperature, the 3′ arm of NaA43S was extended by 5 nt and designed to have a high (∼80%) GC content (Fig. 1B). To ensure the release of the product fragment containing the fluorophore after cleavage, the 5′ end of NaA43S was designed with a much lower (33%) GC content (Fig. 1B). In the presence of sufficient Li+, the NaA43ES complex was formed because it has a melting temperature higher than room temperature (>44 °C). As a result, the fluorescence signal was quenched due to the close proximity of the fluorophore and quencher. Upon addition of Na+, NaA43S was cleaved at the rA. Because the melting temperature of the fluorophore-containing arm after cleavage was below room temperature (∼10 °C), dehybridization caused the fluorophore to release from its quenchers, resulting in fluorescence increase (47, 65). Indeed, as shown in Fig. 2C, the observed rate of fluorescence increase was accelerated with additional Na+, until saturation at ∼135 mM Na+, with an apparent dissociation constant (Kd) of 39.1 ± 2.3 mM (SI Appendix, Fig. S11). The limit of detection was determined to be 135 µM or 3.1 ppm (3σ/slope), with the dynamic range up to 50 mM (Fig. 2C, Inset). This range covers the likely cellular concentrations of Na+ very well (67, 68).
Fig. 2.
Design, sensitivity, and selectivity of the NaA43 DNAzyme-based Na+ fluorescent sensor. (A) Design of the DNAzyme beacon. (B) Fluorescence increase of the sensor over time at different Na+ concentrations. (Inset) Sensor response to Na+ concentrations below 2 mM. (C) Initial rate of fluorescence enhancement (VFl.). (Inset) Linear response at Na+ concentrations lower than 50 mM. The error bars represent the SD calculated from three independent experiments. (D) Response of the sensor to different competing metal ions. The rate of fluorescence enhancement was measured in the presence of 100, 2, and 0.2 mM of monovalent, divalent, and trivalent metal ions, respectively. (Inset) Fraction of cleavage product in a gel-based assay in the presence of Na+, Cu2+, and Hg2+ (SI Appendix, Fig. S13).
To determine the selectivity of the sensor for Na+ over other metal ions, we monitored sensor response to 22 different metal ions (SI Appendix, Fig. S12). None of the tested ions showed a significant change in fluorescence signal (Fig. 2D), suggesting that the NaA43 DNAzyme-based sensor has excellent selectivity for Na+, with at least 10,000-fold better activity versus the next best competing metal ion (Li+) (SI Appendix, Fig. S14). The sensor remained selective in the presence of a mixture of 100 mM Na+ and other monovalent or divalent metal ions (SI Appendix, Fig. S15).
Intracellular Na+ Imaging Using the NaA43 DNAzyme.
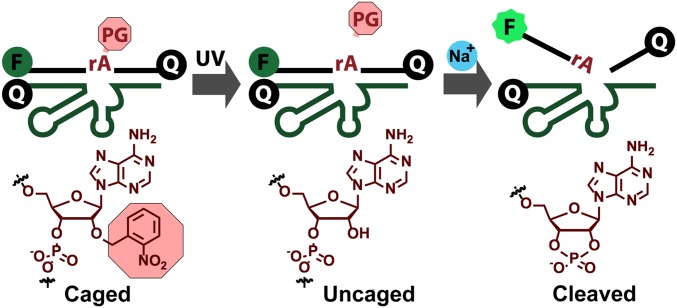
Having demonstrated detection of Na+ in a buffer, we then explored application of the Na+-specific DNAzyme for imaging Na+ in living cells. First, the stability of either NaA43S alone or the NaA43ES complex was tested in the presence of cell lysate and various RNases over the period of 2 h (SI Appendix, Fig. S16). No obvious cleavage was observed unless we spiked the solutions with NaCl, which suggests that the NaA43 DNAzyme remains intact in the presence of RNases and other components from cells. To further prevent cleavage of NaA43S during the delivery of the DNAzyme into cells and to allow for controlled activation of the sensor, we used a photocaging strategy, in which the 2′-hydroxyl (2′-OH) group at the rA cleavage site in the substrate strand (NaA43S) was modified with a photolabile o-nitrobenzyl group (Fig. 3) (69). The caging of the 2′-OH group prevented cleavage of NaA43S by blocking activity of the 2′-OH as a nucleophile in the transesterification reaction (64). The caging group was readily removed upon brief irradiation at 365 nm, which switched the substrate from being noncleavable to cleavable, in a controllable manner.
Fig. 3.
Scheme of the decaging process for the photolabile Na+-specific DNAzyme.
To convert the caged DNAzyme into a fluorescent sensor, fluorophore and quenchers were attached to the DNAzyme, as shown in Fig. 2A. The caged DNAzyme showed no activity even in the presence of a high concentration of Na+ (300 mM) (SI Appendix, Fig. S17). After 365-nm irradiation for 30 min, ∼80% of the DNAzymes were decaged, estimated based on HPLC, using a protocol reported previously (51). Two saline solutions, commonly used for in vivo calibration of Na+ probes (22), were made for testing the performance of the decaged DNAzyme. One solution contains 12.5 mM Hepes (pH 7.4), 140 mM NaCl, 10 mM glucose, 1.2 mM MgCl2, and 1 mM CaCl2. The other buffer has the same components, except 140 mM NaCl was replaced by 140 mM KCl. A mixture of these two solutions was used to generate buffers with a range of different concentrations of Na+. Increased fluorescent signal with increasing concentrations of Na+ was observed, indicating that the activity of the DNAzyme can be restored after reactivation by 365-nm irradiation (SI Appendix, Fig. S17).
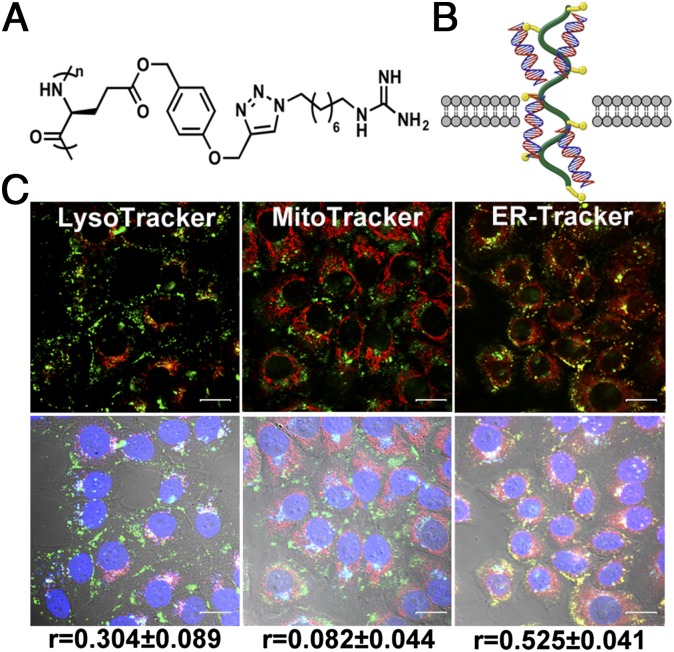
To use the photocaged Na+-specific DNAzyme to image Na+ in cells, a delivery method that can transport the DNAzyme into the cytoplasm of cells without accumulation in specific subcellular organelles is required. It has been shown previously that a class of α-helical cationic polypeptide was able to deliver siRNA and DNA plasmid into various types of mammalian cells with high efficacy and capability of escaping the endocytic pathway (70–72). These studies suggest that this class of polypeptides can be electrostatically attracted to anionic cell membranes and facilitate transient cell membrane disruption, which allows the peptide–nucleic acid complex to diffuse into the cell cytosol. To deliver the NaA43ES complex into cell cytosol, we chose one polypeptide, G8, from this polypeptide family (Fig. 4A), which contains guanidine side chains that are known to play a crucial role in cell penetration efficiency (73). It has been demonstrated that G8 forms an ultrastable helical structure within a pH range of 1–9 and has sufficient water solubility. By maintaining its helical structure at both neutral and acidic pH, G8 was able to penetrate cell membranes as well as escape from endosomes and lysosomes, resulting in highly efficient gene delivery (Fig. 4B) (73). Using G8 polypeptide, we achieved high delivery efficiency of the NaA43ES DNAzymes into the cytoplasm of living HeLa cells after a 4-h incubation (Fig. 4C and SI Appendix, Fig. S18). To further investigate localization of the DNAzymes inside cells, FAM-labeled noncleavable NaA43S, in which the rA at the cleavage site was substituted by deoxyribonucleic adenosine (dA), was used to form complex with NaA43E and was delivered using G8. Staining using organelle-specific trackers was carried out subsequently. The degree of colocalization of the two fluorophores was quantified by Pearson correlation coefficient as a standard technique (74). As suggested by both this calculation and microscopic images (Fig. 4C and SI Appendix, Fig. S19), NaA43ES was mainly located inside the cytosol of the cell, without showing organelle localization in early endosomes, lysosomes, mitochondria, or the endoplasmic reticulum.
Fig. 4.
Intracellular delivery of NaA43ES into HeLa cells by G8 polypeptide. (A) Chemical structure of G8. (B) Scheme of the proposed delivery mechanism. (C) Internalization of FAM-labeled NaA43ES and its colocalization with LysoTracker (Left), MitoTracker (Middle), and ER-Tracker (Right). (r refers to the Pearson correlation coefficients.) (Scale bar: 20 μm.)
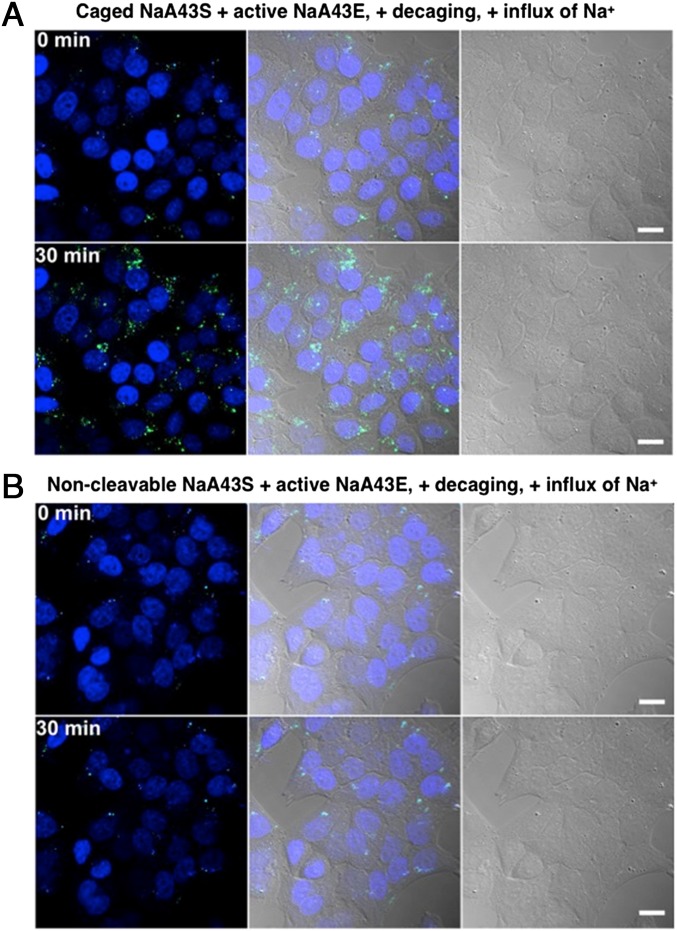
Given the fact that the G8 polypeptide is a very efficient carrier for NaA43ES, we used it to deliver the caged NaA43ES complex into living HeLa cells (Fig. 5 A and B) for the detection of Na+. The sensor showed minimal background fluorescence after its delivery, indicating that most of the caged NaA43S remained intact during the delivery process. After washing the cells to remove excess probes and G8 in the culture medium, the cells were incubated in Dulbecco’s PBS solution and irradiated with light at 365 nm for 30 min to uncage the DNAzyme complex. Immediately after uncaging, the intracellular Na+ level was elevated by adding gramicidin, monensin, and ouabain. The combination of these three ionophores is known to equilibrate the intracellular Na+ concentration with extracellular concentration within several minutes (22, 30). The influx of Na+ from extracellular medium caused the fluorescence inside cells to gradually increase over a time course of 30 min (Fig. 5A). In comparison, with the same treatment, but using a noncleavable NaA43S, the turn-on fluorescence was not observed inside cells (Fig. 5B).
Fig. 5.
Confocal microscopy images of HeLa cells transfected with (A) caged NaA43ES complex and (B) noncleavable NaA43S in complex with NaA43E. After transfection, both groups were irradiated with light at 365 nm for 30 min, followed by induced Na+ influx. The images show the fluorescence of the probe inside cells before and 30 min after Na+ influx, respectively. (Scale bar: 20 μm.)
To further confirm that the turn-on fluorescence was a result from the cleavage of uncaged NaA43S by active NaA43E, we also used the combination of caged NaA43S with an inactive variant of NaA43E (forming catalytically inactive NaA43ES complex) as a negative control. The inactive NaA43E contains a single point mutation, which completely abolishes the DNAzyme activity. In this case, fluorescence from the probes was maintained at a constant background level over 30 min (SI Appendix, Fig. S20), which strongly suggests that the turn-on fluorescence we observed from the active NaA43ES resulted from successful decaging and subsequent Na+-specific DNAzyme cleavage activity.
Conclusions
In conclusion, we have obtained the first (to our knowledge) Na+-specific, RNA-cleaving DNAzyme (NaA43ES) with fast catalytic rate and exceptionally high selectivity over other metal ions, and demonstrated the use of this DNAzyme for sensing and imaging intracellular Na+ in living cells, by adopting an efficient DNAzyme delivery method using a cationic polypeptide, together with a photocaging strategy to allow controllable activation of the probe inside cells. In the field of sensing monovalent metal ions, in particular Na+, obtaining highly selective sensors with proper sensitivity and selectivity has been a major challenge. Most previously developed Na+ fluorescent sensors suffer from poor sensitivity or are also responsive to other metal ions such as K+. The in vitro selection of DNAzymes selective for Na+ has allowed for the identification of a fluorescent sensor with exceptional selectivity and sensitivity, further validating this as a method for the simple identification of sensors for many other metal ions, even where existing design strategies may be lacking. Finally, delivery of the DNAzyme sensor into living cells to detect Na+ has been demonstrated, setting the stage for future work capable of offering deeper insight into the mechanisms and importance of sodium homeostasis in biology.
It is remarkable that the NaA43 DNAzyme has such a high selectivity for Na+ against Li+, K+, and other monovalent, divalent, and trivalent metal ions. To our knowledge, no Na+-specific nucleic acid, whether naturally occurring or in vitro selected has been previously reported. In the protein world, although K+ channels are quite selective for K+ over other metal ions, the Na+ channels are much less selective (75, 76). Being able to obtain the NaA43 DNAzyme with such a high selectivity for Na+ will not only provide a highly selective sensor for Na+, as demonstrated here, it will also give us an opportunity to elucidate the origin of such a high selectivity. Previous studies of a Pb2+-DNAzyme have shown the high selectivity is mainly due to the DNAzyme forming a specific binding pocket for the metal ion (77). It would be interesting to find out if the NaA43 DNAzyme uses the same mechanism for selectivity. We are performing this and other biochemical and biophysical studies of this DNAzyme and will report the results in a future publication.
Materials and Methods
Chemicals and DNA sequences are presented in SI Appendix, section S1, Materials. Detailed experimental procedures (in vitro selection, cloning, activity assays, design and analysis of the fluorescent sensor, cell culture, and microscopy) are available in SI Appendix, section S2, Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful for the financial support of the National Institutes of Health (Grant ES016865).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420361112/-/DCSupplemental.
References
- 1.Guengerich FP. Thematic series: Metals in biology. J Biol Chem. 2009;284(2):709. doi: 10.1074/jbc.R800060200. [DOI] [PubMed] [Google Scholar]
- 2.Domaille DW, Que EL, Chang CJ. Synthetic fluorescent sensors for studying the cell biology of metals. Nat Chem Biol. 2008;4(3):168–175. doi: 10.1038/nchembio.69. [DOI] [PubMed] [Google Scholar]
- 3.Fu D, Finney L. Metalloproteomics: Challenges and prospective for clinical research applications. Expert Rev Proteomics. 2014;11(1):13–19. doi: 10.1586/14789450.2014.876365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray HB. Biological inorganic chemistry at the beginning of the 21st century. Proc Natl Acad Sci USA. 2003;100(7):3563–3568. doi: 10.1073/pnas.0730378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant FD, et al. Low-renin hypertension, altered sodium homeostasis, and an alpha-adducin polymorphism. Hypertension. 2002;39(2):191–196. doi: 10.1161/hy0202.104273. [DOI] [PubMed] [Google Scholar]
- 6.Baartscheer A, et al. Chronic inhibition of Na+/H+-exchanger attenuates cardiac hypertrophy and prevents cellular remodeling in heart failure. Cardiovasc Res. 2005;65(1):83–92. doi: 10.1016/j.cardiores.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Moritz ML, Ayus JC. Disorders of water and sodium homeostasis. In: Warrell D, Cox T, Firth J, editors. Oxford Textbook of Medicine. Oxford Univ Press; Oxford: 2010. pp. 3818–3831. [Google Scholar]
- 8.Hollenberg NK. The influence of dietary sodium on blood pressure. J Am Coll Nutr. 2006;25(3) Suppl:240S–246S. doi: 10.1080/07315724.2006.10719573. [DOI] [PubMed] [Google Scholar]
- 9.Aronson PS, Boron WF, Boulpaep EL. Transport of solutes and water. In: Boron WF, Boulpaep EL, editors. Medical Physiology: A Cellular and Molecular Approach. Elsevier; Philadelphia: 2010. pp. 106–146. [Google Scholar]
- 10.Ferré-D’Amaré AR, Winkler WC. The roles of metal ions in regulation by riboswitches. Met Ions Life Sci. 2011;9:141–173. doi: 10.1039/9781849732512-00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaitovich A, Bertorello AM. Intracellular sodium sensing: SIK1 network, hormone action and high blood pressure. Biochim Biophys Acta. 2010;1802(12):1140–1149. doi: 10.1016/j.bbadis.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Malloy CR, et al. Influence of global ischemia on intracellular sodium in the perfused rat heart. Magn Reson Med. 1990;15(1):33–44. doi: 10.1002/mrm.1910150105. [DOI] [PubMed] [Google Scholar]
- 13.Grubman A, et al. X-ray fluorescence imaging reveals subcellular biometal disturbances in a childhood neurodegenerative disorder. Chem Sci. 2014;5(6):2503–2516. doi: 10.1039/C4SC00316K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanics T, Blum H, Wroblewski K, Wang DJ, Osbakken M. Intracellular sodium in cardiomyocytes using 23Na nuclear magnetic resonance. Biochim Biophys Acta. 1994;1221(2):133–144. doi: 10.1016/0167-4889(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 15.Kikuchi K. Design, synthesis and biological application of chemical probes for bio-imaging. Chem Soc Rev. 2010;39(6):2048–2053. doi: 10.1039/b819316a. [DOI] [PubMed] [Google Scholar]
- 16.Fahrni CJ, O'Halloran TV. Aqueous coordination chemistry of quinoline-based fluorescence probes for the biological chemistry of zinc. J Am Chem Soc. 1999;121(49):11448–11458. [Google Scholar]
- 17.McRae R, Bagchi P, Sumalekshmy S, Fahrni CJ. In situ imaging of metals in cells and tissues. Chem Rev. 2009;109(10):4780–4827. doi: 10.1021/cr900223a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CJ, Jaworski J, Nolan EM, Sheng M, Lippard SJ. A tautomeric zinc sensor for ratiometric fluorescence imaging: Application to nitric oxide-induced release of intracellular zinc. Proc Natl Acad Sci USA. 2004;101(5):1129–1134. doi: 10.1073/pnas.0308079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu L, et al. A turn-on fluorescent Fe3+ sensor derived from an anthracene-bearing bisdiene macrocycle and its intracellular imaging application. Chem Commun (Camb) 2014;50(35):4631–4634. doi: 10.1039/c3cc49482a. [DOI] [PubMed] [Google Scholar]
- 20.Wegner SV, Arslan H, Sunbul M, Yin J, He C. Dynamic copper(I) imaging in mammalian cells with a genetically encoded fluorescent copper(I) sensor. J Am Chem Soc. 2010;132(8):2567–2569. doi: 10.1021/ja9097324. [DOI] [PubMed] [Google Scholar]
- 21.Carter KP, Young AM, Palmer AE. Fluorescent sensors for measuring metal ions in living systems. Chem Rev. 2014;114(8):4564–4601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donoso P, Mill JG, O’Neill SC, Eisner DA. Fluorescence measurements of cytoplasmic and mitochondrial sodium concentration in rat ventricular myocytes. J Physiol. 1992;448:493–509. doi: 10.1113/jphysiol.1992.sp019053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amorino GP, Fox MH. Intracellular Na+ measurements using sodium green tetraacetate with flow cytometry. Cytometry. 1995;21(3):248–256. doi: 10.1002/cyto.990210305. [DOI] [PubMed] [Google Scholar]
- 24.Meier SD, Kovalchuk Y, Rose CR. Properties of the new fluorescent Na+ indicator CoroNa Green: Comparison with SBFI and confocal Na+ imaging. J Neurosci Methods. 2006;155(2):251–259. doi: 10.1016/j.jneumeth.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Jayaraman S, Song Y, Vetrivel L, Shankar L, Verkman AS. Noninvasive in vivo fluorescence measurement of airway-surface liquid depth, salt concentration, and pH. J Clin Invest. 2001;107(3):317–324. doi: 10.1172/JCI11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim MK, et al. Sodium-ion-selective two-photon fluorescent probe for in vivo imaging. Angew Chem Int Ed Engl. 2010;49(2):364–367. doi: 10.1002/anie.200904835. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar AR, Heo CH, Park MY, Lee HW, Kim HM. A small molecule two-photon fluorescent probe for intracellular sodium ions. Chem Commun (Camb) 2014;50(11):1309–1312. doi: 10.1039/c3cc48342h. [DOI] [PubMed] [Google Scholar]
- 28.Martin VV, Rothe A, Gee KR. Fluorescent metal ion indicators based on benzoannelated crown systems: A green fluorescent indicator for intracellular sodium ions. Bioorg Med Chem Lett. 2005;15(7):1851–1855. doi: 10.1016/j.bmcl.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Dubach JM, Lim E, Zhang N, Francis KP, Clark H. In vivo sodium concentration continuously monitored with fluorescent sensors. Integr Biol (Camb) 2011;3(2):142–148. doi: 10.1039/c0ib00020e. [DOI] [PubMed] [Google Scholar]
- 30.Bernardinelli Y, Azarias G, Chatton JY. In situ fluorescence imaging of glutamate-evoked mitochondrial Na+ responses in astrocytes. Glia. 2006;54(5):460–470. doi: 10.1002/glia.20387. [DOI] [PubMed] [Google Scholar]
- 31.Kenmoku S, et al. Rational design of novel photoinduced electron transfer type fluorescent probes for sodium cation. Tetrahedron. 2004;60(49):11067–11073. [Google Scholar]
- 32.Gunnlaugsson T, Nieuwenhuyzen M, Richard L, Thoss V. Novel sodium-selective fluorescent PET and optically based chemosensors: Towards Na+ determination in serum. J Chem Soc Perkin Trans 2. 2002;(1):141–150. [Google Scholar]
- 33.Leray I, Valeur B, O’Reilly F, Jiwan JLH, Soumillion JP. A new calix[4]arene-based fluorescent sensor for sodium ion. Chem Commun (Camb) 1999;9:795–796. [Google Scholar]
- 34.Leray I, Lefevre JP, Delouis JF, Delaire J, Valeur B. Synthesis and photophysical and cation-binding properties of mono- and tetranaphthylcalix[4]arenes as highly sensitive and selective fluorescent sensors for sodium. Chemistry. 2001;7(21):4590–4598. doi: 10.1002/1521-3765(20011105)7:21<4590::aid-chem4590>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 35.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem Biol. 1994;1(4):223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 36.Xiang Y, Lu Y. DNA as sensors and imaging agents for metal ions. Inorg Chem. 2014;53(4):1925–1942. doi: 10.1021/ic4019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiuman W, Li Y. Simple fluorescent sensors engineered with catalytic DNA “MgZ” based on a non-classic allosteric design. PLoS One. 2007;2(11):e1224. doi: 10.1371/journal.pone.0001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlosser K, Li Y. A versatile endoribonuclease mimic made of DNA: Characteristics and applications of the 8-17 RNA-cleaving DNAzyme. ChemBioChem. 2010;11(7):866–879. doi: 10.1002/cbic.200900786. [DOI] [PubMed] [Google Scholar]
- 39.Joyce GF. Directed evolution of nucleic acid enzymes. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 40.Ihms HE, Lu Y. In vitro selection of metal ion-selective DNAzymes. Methods Mol Biol. 2012;848:297–316. doi: 10.1007/978-1-61779-545-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu P, Hwang K, Lan T, Lu Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J Am Chem Soc. 2013;135(14):5254–5257. doi: 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA. 1997;94(9):4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deibler K, Basu P. Continuing issues with lead: Recent advances in detection. Eur J Inorg Chem. 2013;2013(7):1086–1096. doi: 10.1002/ejic.201200997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmi N, Shultz LA, Breaker RR. In vitro selection of self-cleaving DNAs. Chem Biol. 1996;3(12):1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- 45.Carmi N, Balkhi SR, Breaker RR. Cleaving DNA with DNA. Proc Natl Acad Sci USA. 1998;95(5):2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Zheng W, Kwon AH, Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28(2):481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, et al. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc Natl Acad Sci USA. 2007;104(7):2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM. A highly selective DNAzyme sensor for mercuric ions. Angew Chem Int Ed Engl. 2008;47(23):4346–4350. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]
- 49.Torabi SF, Lu Y. Functional DNA nanomaterials for sensing and imaging in living cells. Curr Opin Biotechnol. 2014;28:88–95. doi: 10.1016/j.copbio.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Huang H, Xu N, Yin Q. Functionalization of cationic poly(p-phenylene ethynylene) with dendritic polyethylene enables efficient DNAzyme delivery for imaging Pb2+ in living cells. J Mater Chem B. 2014;2(30):4935–4942. doi: 10.1039/c4tb00680a. [DOI] [PubMed] [Google Scholar]
- 51.Hwang K, et al. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew Chem Int Ed Engl. 2014;53(50):13798–13802. doi: 10.1002/anie.201408333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng H-M, et al. DNA dendrimer: An efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014;8(6):6171–6181. doi: 10.1021/nn5015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faulhammer D, Famulok M. Characterization and divalent metal-ion dependence of in vitro selected deoxyribozymes which cleave DNA/RNA chimeric oligonucleotides. J Mol Biol. 1997;269(2):188–202. doi: 10.1006/jmbi.1997.1036. [DOI] [PubMed] [Google Scholar]
- 54.Geyer CR, Sen D. Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem Biol. 1997;4(8):579–593. doi: 10.1016/s1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- 55.Carrigan MA, Ricardo A, Ang DN, Benner SA. Quantitative analysis of a RNA-cleaving DNA catalyst obtained via in vitro selection. Biochemistry. 2004;43(36):11446–11459. doi: 10.1021/bi049898l. [DOI] [PubMed] [Google Scholar]
- 56.Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M2+) Nucleic Acids Res. 2009;37(5):1638–1649. doi: 10.1093/nar/gkn1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smuga D, Majchrzak K, Sochacka E, Nawrot B. RNA-cleaving 10–23 deoxyribozyme with a single amino acid-like functionality operates without metal ion cofactors. New J Chem. 2010;34(5):934–948. [Google Scholar]
- 58.Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates. ACS Comb Sci. 2013;15(4):174–182. doi: 10.1021/co3001378. [DOI] [PubMed] [Google Scholar]
- 59.Perrotta AT, Been MD. HDV ribozyme activity in monovalent cations. Biochemistry. 2006;45(38):11357–11365. doi: 10.1021/bi061215+. [DOI] [PubMed] [Google Scholar]
- 60.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5(10):587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 61.Weinstein LB, Earnshaw DJ, Cosstick R, Cech TR. Synthesis and characterization of an RNA dinucleotide containing a 3′-S-phosphorothiolate linkage. J Am Chem Soc. 1996;118(43):10341–10350. [Google Scholar]
- 62.Matsumoto Y, Komiyama M. Efficient cleavage of adenylyl(3ʹ-5ʹ)adenosine by triethylenetetraminecobalt(III) J Chem Soc Chem Commun. 1990;15:1050–1051. [Google Scholar]
- 63.Markham NR, Zuker M. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 64.Brown AK, Li J, Pavot CM, Lu Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry. 2003;42(23):7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]
- 65.Li J, Lu Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J Am Chem Soc. 2000;122(42):10466–10467. [Google Scholar]
- 66.Liu J, Lu Y. Improving fluorescent DNAzyme biosensors by combining inter- and intramolecular quenchers. Anal Chem. 2003;75(23):6666–6672. doi: 10.1021/ac034924r. [DOI] [PubMed] [Google Scholar]
- 67.Murphy E, Eisner DA. Regulation of intracellular and mitochondrial sodium in health and disease. Circ Res. 2009;104(3):292–303. doi: 10.1161/CIRCRESAHA.108.189050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cinelli AR, Efendiev R, Pedemonte CH. Trafficking of Na-K-ATPase and dopamine receptor molecules induced by changes in intracellular sodium concentration of renal epithelial cells. Am J Physiol Renal Physiol. 2008;295(4):F1117–F1125. doi: 10.1152/ajprenal.90317.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaulk SG, MacMillan AM. Synthesis of oligo-RNAs with photocaged adenosine 2′-hydroxyls. Nat Protoc. 2007;2(5):1052–1058. doi: 10.1038/nprot.2007.154. [DOI] [PubMed] [Google Scholar]
- 70.Lu H, et al. Ionic polypeptides with unusual helical stability. Nat Commun. 2011;2:206. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]
- 71.Gabrielson NP, Lu H, Yin L, Kim KH, Cheng J. A cell-penetrating helical polymer for siRNA delivery to mammalian cells. Mol Ther. 2012;20(8):1599–1609. doi: 10.1038/mt.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gabrielson NP, et al. Reactive and bioactive cationic α-helical polypeptide template for nonviral gene delivery. Angew Chem Int Ed Engl. 2012;51(5):1143–1147. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang R, Zheng N, Song Z, Yin L, Cheng J. The effect of side-chain functionality and hydrophobicity on the gene delivery capabilities of cationic helical polypeptides. Biomaterials. 2014;35(10):3443–3454. doi: 10.1016/j.biomaterials.2013.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008;3(4):619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- 75.Sheng S, McNulty KA, Harvey JM, Kleyman TR. Second transmembrane domains of ENaC subunits contribute to ion permeation and selectivity. J Biol Chem. 2001;276(47):44091–44098. doi: 10.1074/jbc.M108522200. [DOI] [PubMed] [Google Scholar]
- 76.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 77.Kim HK, Rasnik I, Liu J, Ha T, Lu Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat Chem Biol. 2007;3(12):763–768. doi: 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.