Significance
Mitochondrial dysfunction contributes significantly to glucose toxicity in diabetes. Increased O-GlcNAcylation is emerging as a major molecular cause of glucose toxicity via many mechanisms. The studies herein provide a direct molecular link between hyperglycemia and mitochondrial dysfunction. We show that mitochondrial O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) expression levels and localizations are strikingly different between normal and diabetic rat hearts. We also discover how UDP-GlcNAc enters the mitochondrial space. Finally our data demonstrate that OGT and OGA play significant roles in ATP production, mitochondrial membrane potential, and oxygen consumption. These studies are of general interest not only with respect to nutrient regulation of mitochondrial function, but also are important to elucidate mechanisms of diabetic complications.
Keywords: diabetes, diabetic cardiomyopathy, mitochondria, O-GlcNAc, O-GlcNAc transferase
Abstract
Elevated mitochondrial O-GlcNAcylation caused by hyperglycemia, as occurs in diabetes, significantly contributes to mitochondrial dysfunction and to diabetic cardiomyopathy. However, little is known about the enzymology of mitochondrial O-GlcNAcylation. Herein, we investigated the enzymes responsible for cycling O-GlcNAc on mitochondrial proteins and studied the mitochondrial transport of UDP-GlcNAc. Analyses of purified rat heart mitochondria from normal and streptozocin-treated diabetic rats show increased mitochondrial O-GlcNAc transferase (OGT) and a concomitant decrease in the mito-specific O-GlcNAcase (OGA). Strikingly, OGT is mislocalized in cardiac mitochondria from diabetic rats. Interaction of OGT and complex IV observed in normal rat heart mitochondria is visibly reduced in diabetic samples, where OGT is mislocalized to the matrix. Live cell OGA activity assays establish the presence of O-GlcNAcase within the mitochondria. Furthermore, we establish that the inner mitochondrial membrane transporter, pyrimidine nucleotide carrier, transports UDP-GlcNAc from the cytosol to the inside of the mitochondria. Knockdown of this transporter substantially lowers mitochondrial O-GlcNAcylation. Inhibition of OGT or OGA activity within neonatal rat cardiomyocytes significantly affects energy production, mitochondrial membrane potential, and mitochondrial oxygen consumption. These data suggest that cardiac mitochondria not only have robust O-GlcNAc cycling, but also that dysregulation of O-GlcNAcylation likely plays a key role in mitochondrial dysfunction associated with diabetes.
Protein O-GlcNAcylation is a nutrient sensor occurring within nuclear, cytoplasmic, and mitochondrial cellular compartments (1–3). Using UDP-GlcNAc, O-GlcNAc transferase (OGT) modifies myriad proteins on serine and/or threonine residues (4). Cycling of O-GlcNAc is also maintained by an O-GlcNAcase (OGA) (5, 6). O-GlcNAcylation regulates protein functions in response to nutrient availability, metabolic state, and cellular stress levels (7, 8). The extent of protein O-GlcNAcylation at a site on a protein is determined collectively by the cycling enzyme activities, specificities, localizations, and by the concentration of UDP-GlcNAc. Thus, an understanding of protein O-GlcNAcylation’s biological roles must take into account the presence, expression, interactions, and levels of these important players in the O-GlcNAc cycle.
Increased protein O-GlcNAcylation within chronic high glucose-mediated diabetic tissues indicates a causal link between high O-GlcNAcylation and diabetes (9). Within the cardiovascular system, significant levels of O-GlcNAcylation have been observed on myofilament proteins, which severely affect contractile force generation (10). Alterations in cardiac O-GlcNAc levels significantly affect Ca2+ handling and Serca2a function (11). STIM1 a modulator of cardiomyocyte calcium entry is modified by O-GlcNAc, which affects cellular calcium translocation (12). Modification of CaMKII by O-GlcNAc during diabetes activates it to sensitize Ca2+ release from the sarcoplasmic reticulum causing cardiac mechanical dysfunction and arrhythmias (13, 14). Both ADP and Ca2+ signals control ATP production by the mitochondria. Mitochondria are also significantly regulated by nutrient levels and by calcium trafficking in the cell. Because O-GlcNAc is an end point of nutrient-sensitive pathways, and O-GlcNAcylation significantly affects calcium handling, it stands to reason that this Post translational modification (PTM) exerts influence on mitochondrial function. Indeed, PGC1α, the master regulator of mitochondrial biogenesis and survival is O-GlcNAc modified and serves as a cofactor targeting OGT to FOXO transcription factors (15, 16). O-GlcNAcylation of Milton affects mitochondrial motility in neurons (17). Hu et al. reported the modification of mitochondrial electron chain complexes by O-GlcNAc, which affected activities of complexes I, II, and IV (18). Altering O-GlcNAc levels by overexpression of cytoplasmic OGT or OGA had significant effects on mitochondrial function and mitochondrion-related protein expression (19). The existence of a mitochondrial isoform of OGT suggests that O-GlcNAcylation occurs within the mitochondria and on mito-specific proteins (20, 21). However, there have been no reports of an O-GlcNAc hydrolase activity within the mitochondria, as well as no reported mechanism of donor nucleotide sugar (UDP-GlcNAc) presence or import inside the mitochondria (22).
In this paper, we find that not only are the levels of mitochondrial OGT and OGA dramatically altered in mitochondria from diabetic hearts, but we also show that OGT is mislocalized in diabetic mitochondria. We use live cell imaging to show the presence of OGA within the mitochondria and report the presence of a specific mitochondrial transporter that imports UDP-GlcNAc. Together our data suggest that not only does O-GlcNAcylation appear to regulate mitochondrial functions in normal cells, but also that diabetes-associated changes in O-GlcNAcylation likely contribute to mitochondrial dysfunction associated with diabetic cardiomyopathy.
Results
Mitochondrial OGT Is Increased and Mislocalized in Diabetic Rat Hearts.
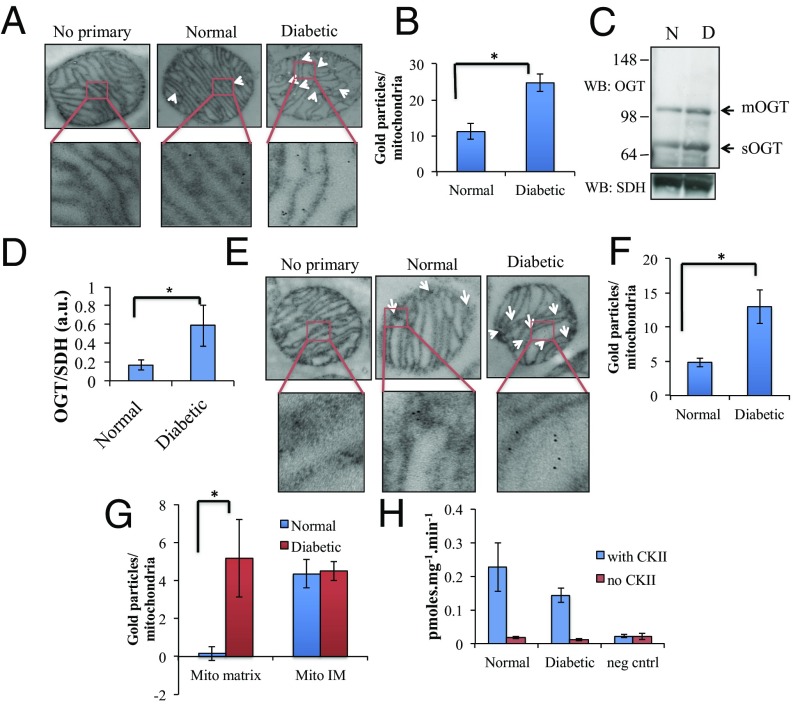
Mitochondria purified (Fig. S1) from normal and streptozocin (STZ)-treated rat hearts were analyzed by Transmission Electron Microscopy (TEM) for levels of overall protein O-GlcNAc. Using an anti-O-GlcNAc antibody (RL2), we observed significant increases in the total O-GlcNAc levels in the STZ-treated rat mitochondria compared with control rats (Fig. 1A). Quantifying gold particles indicated more than twofold increases in mitochondrial protein O-GlcNAcylation in diabetic heart samples (Fig. 1B). Western blot analysis of purified mitochondrial lysates also showed a twofold increase in the levels of mitochondrial OGT (observed around 105 kDa) in diabetic samples (Fig. 1 C and D). Western blots showed a band at 78 kDa, indicating that the short isoform of OGT is also present in the mitochondria (Fig. 1C). TEM analysis on mito sections validates the twofold increase in OGT levels as observed by immunoblotting (Fig. 1 E and F). Closer examination of OGT-labeled mitochondria revealed a distinct difference in the localization of gold particles between normal and diabetic samples. Localization of total mitochondrial OGT for control rats was mostly on the inner membrane of the purified mitochondria. In contrast, for diabetic rats, there is a significant amount of matrix localization of OGT (Fig. 1 E and G). The overall in vitro activity of mitochondrial OGT from purified mitochondrial lysate was observed to be similar for the normal and diabetic samples (Fig. 1E) (23). Thus, more OGT protein is required to maintain activity levels similar to normal, indicating a potential reduction in specific activity of mito-specific OGT in diabetic samples.
Fig. 1.
TEM analysis of mitochondrial O-GlcNAc and OGT from normal and diabetic rat hearts. (A) TEM image of rat heart mitochondria labeled with anti–O-GlcNAc antibody, RL2. Control samples were treated with only secondary antibody. (B) Quantification of immunogold labeling in normal and diabetic mitochondria from A above (n = 3). (C) Western blotting of purified mitochondrial lysate obtained from normal and diabetic rat hearts immunoblotted with an anti-OGT and a succinate dehydrogenase (SDH) antibody (N, normal and D, diabetic). (D) Quantification of OGT intensity upon SDH intensity from purified mitochondria lysate (n = 3). (E) TEM image of rat heart mitochondria labeled with anti-OGT antibody. Control samples were treated with only secondary antibody. (F) Quantification of immunogold labeling in normal and diabetic mitochondria from E above (n = 3). (G) Quantification of immunogold labeling on anti–OGT-treated mitochondria at inner membrane (mito IM) and within the matrix (mito matrix) regions of the sample. (H) Total activity of mitochondrial OGT from normal and diabetic samples as assessed by radioactive transferase assay on purified mitochondrial lysate with an external substrate peptide from CKII. (*P < 0.05).
Interaction of OGT with Mitochondrial Electron Transport Chain Complex IV in Normal Rats Is Greatly Reduced in Diabetic Animals.
To examine the associations of mitochondrial OGT, we analyzed the mitochondrial samples using Blue Native PAGE. With an anti-OGT antibody, we observed a band at 78 kDa for both normal and diabetic samples (Fig. 2A). Whereas the normal samples showed a high association of OGT with complex IV proteins, in the diabetic samples more OGT is associated with complex III (Fig. 2A). To confirm this observation, we performed immunoprecipitation (IP) experiments of these complexes and immunoblotting for OGT. For complex IV immunoprecipitations, we observed a similar band at 78 kDa for the normal samples, which was reduced in the diabetic samples (Fig. 2 B and C). The O-GlcNAcylation pattern of complex IV subunits in the immunoprecipitated samples show changes in O-GlcNAc levels in a number of bands (Fig. 2D). We also observed a reduction in complex IV activity within diabetic rat heart mitochondria (Fig. 2 F and G). It has previously been reported that high-glucose–treated cardiomyocytes show significant loss in complex IV activity (11, 18). Our results directly implicate O-GlcNAcylation in the mechanism of this loss of complex IV activity. We also confirmed that the 78-kDa band is indeed OGT by assaying complex IV immunoprecipitated samples for OGT enzymatic activity. Higher OGT transferase activity was observed within IPs of complex IV from normal samples (Fig. 2E).
Fig. 2.
Interaction of mitochondrial complex IV with the putative short OGT isoform within normal and diabetic cardiac mitochondria and the corresponding complex IV activity assay. (A) BN PAGE analysis of purified heart mitochondria samples followed by second dimensional SDS gel immunoblotted with anti-OGT antibody. (B) Immunoprecipitation of complex IV from normal and diabetic mitochondria with anti-complex IV antibody labeled beads (Abcam) and immunoblotted against OGT and complex IV antibody. (C) Quantification of immunoprecipitated OGT/complex IV levels (n = 3). (D) CTD blot on complex IV immunoprecipitated sample showing changes in O-GlcNAcylation pattern of complex IV subunits between normal and diabetic samples. Arrows indicate bands that show altered O-GlcNAc labeling. (E) OGT activity on immunoprecipitated complex IV beads with and without external substrate peptide (NH, normal and DH, diabetic samples). (F) Complex IV activity assay as analyzed by following oxidation of cytochrome c resulting in loss of absorbance at 550 nm. (G) Rate of cytochrome c oxidation from normal and diabetic samples obtained by the initial slope from F above (n = 3). (*P < 0.05).
OGA Levels Are Reduced in Diabetic Heart and OGA Activity Occurs Within the Mitochondria.
Using an antibody against OGA, thin sections containing purified mitochondria were analyzed by TEM for OGA levels with secondary immune-gold labeling. The data (Fig. 3A) suggest the presence of mitochondria-specific OGA. The levels of OGA decrease in diabetic rat heart mitochondria. These findings partially explain why O-GlcNAc levels in mitochondria are up-regulated in diabetic samples. To further visualize the presence of mitochondrial OGA, we immunostained for OGA and mitochondria (using mito-tracker RED). Immunofluoresence imaging of labeled cells show significant colocalization between OGA antibody and mito-tracker (Fig. S2).
Fig. 3.
Mitochondria OGA and mito-specific live cell OGA activity. (A) TEM image of rat heart mitochondria labeled with anti-OGA antibody. Control samples were treated with only secondary antibody. (B) Quantification of immunogold labeling in normal and diabetic mitochondria from A above (background subtracted, n = 3). (C) Live cell OGA activity assay on neonatal rat cardiomyocytes treated with M-31850 and TMG to inhibit lysosomal hydrolases and OGA, respectively. Nuclear labeling is seen with Hoest, hydrolase activity with fluorescein (green), mitochondria with TMRM (red), and merged image. White circles represent regions used for mitochondrial segmentation using TMRM signals. (D) Total mean fluorescence intensity obtained from green channel from specific region of interest between TMG- and M-31850–treated samples (n = 3). (E) Mean fluorescence intensity obtained from green channel using specific segmented regions as specified by white circles in C between TMG- and M-31850–treated samples (n = 3), representing mito-specific regions within the cell. (*P < 0.05).
Because hydrolase activity from purified mitochondria is inherently problematic due to the low levels of lysosomal impurities (24), we used live cell imaging analysis to ascertain mito-specific OGA activity. Neonatal rat cardiomyocytes were plated and allowed to grow in 5 mM glucose media (25), which were then treated with two inhibitors, Thiamet G (TMG), which is highly specific for OGA (26), and M-31850 for lysosomal hexosaminidases (IC50 for Hex A and Hex B are 6 and 3.1 µM, respectively). Two hours after treatment, the cells were fed the OGA substrate, fluorescein-GlcNAc. The cells were also treated with Tetramethyl Rhodamine Methyl ester (TMRM) and Hoesht for mito-tracking and nuclear localization. Live cell imaging showed significantly enhanced fluorescence signal from hydrolyzed fluorescein-GlcNAc within cells treated with M-31850 compared with TMG-treated cardiomyocytes (Fig. 3 C and D, green filter), indicating that the observed activity differences were OGA specific. For mitochondria-specific activity, TMRM signals were used to segment areas within cells and the fluorescein intensity within these areas were measured. Total segmented fluorescein intensity is also significantly higher within M-31850–treated cells (Fig. 3D), indicating that cardiomyocyte mitochondria retain substantial OGA hydrolysis activity. This is the first demonstration to our knowledge that OGA is indeed present within the mitochondria.
Radioactive 3H UDP-GlcNAc Is Efficiently Transported into Purified Rat Heart Mitochondria.
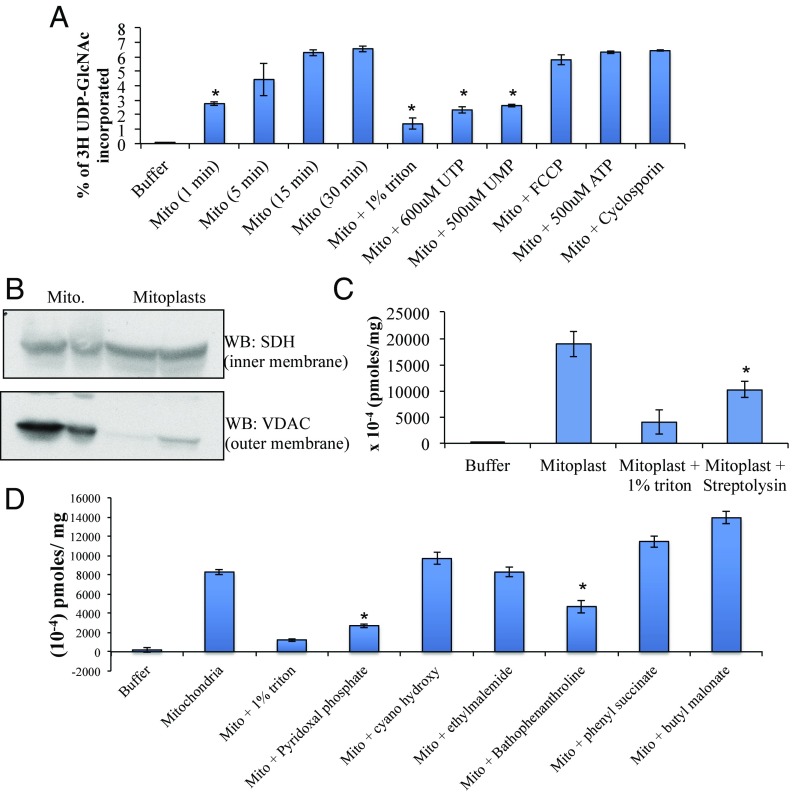
The transport of nucleotide sugars into the mitochondrial matrix has not been investigated. To ascertain whether UDP-GlcNAc could be transported through the mitochondrial membranes, we used radioactive UDP-GlcNAc to query whether purified mitochondria from rat hearts would efficiently incorporate the nucleotide sugar. As shown in Fig. 4A, we observed effective uptake of the nucleotide sugar by purified rat heart mitochondria. The transport was affected by uridine nucleotides, UTP and UMP, but remained unaffected by ATP, membrane decouplers [Carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone, FCCP] and mitochondrial permeability transition pore inhibitors. Control mitochondria treated with 1% triton lost a significant amount of the transported UDP-GlcNAc. We analyzed the ability of the inner mitochondrial membrane to control transport function. Mitoplast preparations (Fig. 4B) used in the transport assay (Fig. 4C) show that mitoplasts are also capable of transporting UDP-GlcNAc, which is affected by pore-forming toxins (27), hinting that inner membrane transporter proteins may be responsible for UDP-GlcNAc transport. We confirmed this hypothesis by using known mitochondrial inner membrane transporter inhibitors in our radioactive uptake assay. The data showed that only two inhibitors produced a significant loss in transport of radioactive UDP-GlcNAc. Pyridoxal phosphate, a universal mitochondrial transporter inhibitor and bathophenanthroline, reportedly affecting one of the pyrimidine nucleotide carriers (pnc1) or deoxy nucleotide carrier (dnc) (28, 29). Our data suggest that either of these two transporters might be responsible for uptake of UDP-GlcNAc into the mitochondrial matrix (Fig. 4D).
Fig. 4.
Transport assay showing uptake of radioactive UDP-GlcNAc into purified rat heart mitochondria and mitoplasts in the presence of different substrates and inhibitors. (A) Incorporation of time-dependent UDP-[3H]GlcNAc into purified mitochondria and its effect on presence of detergent, UTP, UMP, FCCP, ATP, and cyclosporine A (n = 3). (B) Immunoblot on mitoplast preparations showing intact inner mitochondrial membrane and ruptured outer membrane. (C) Uptake assay on mitoplasts showing efficient uptake on only mitoplast prep but reduced uptake on mitoplasts treated with either detergent or streptolysin O (n = 3). (D) Effect of major mitochondrial transport inhibitors on UDP-GlcNAc transport (n = 3). (t test was carried out for mito samples vs. different treatments with P < 0.05 considered significant.)
Pnc1 Transporter Protein (SLC25A33) Is Responsible for Transport of Mitochondrial UDP-GlcNAc.
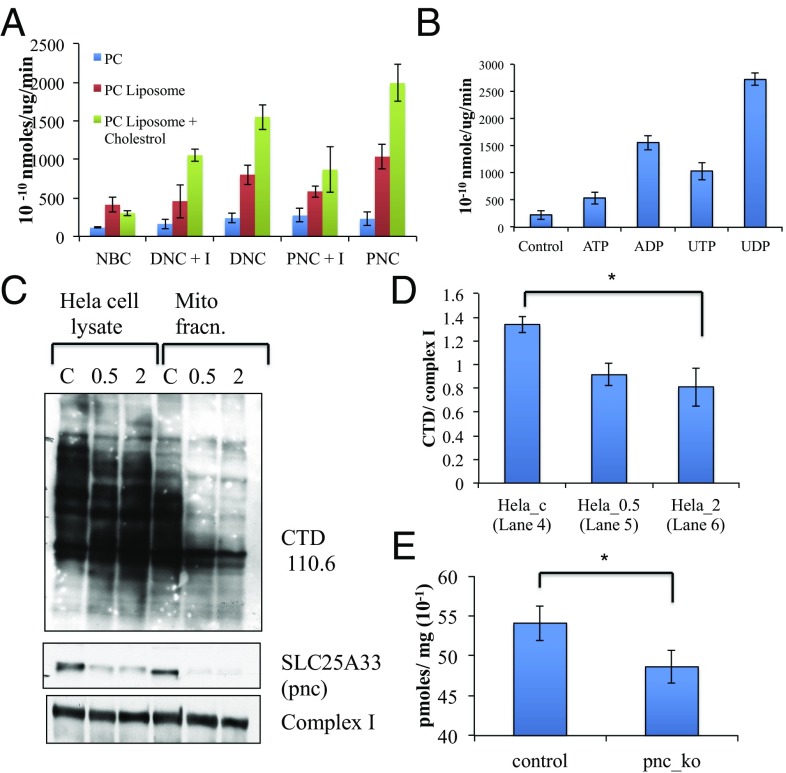
We reconstituted transport activity in vitro using liposome–protein preparations. Toward this aim, we obtained the expression plasmid for the dnc (29) and pnc (30) genes. Proteins were expressed in Escherichia coli, and overexpressed proteins were purified (31) and used for preparation of proteoliposomes for analysis of transport activity. Phosphatidylcholine (PC) liposomes mixed with protein and 10 mM nucleotide sugar in buffer were detergent exchanged to generate proteoliposomes. Dynamic light scattering spectroscopy was used to determine the size and polydispersity (Fig. S3A) of the liposomes. Subsequently, the liposomes were used to determine transport of radioactive UDP-GlcNAc. Control experiments were set up using liposomes that were preincubated with 20 mM bathophenanthroline and liposomes containing the Sodium Bicarbonate Co-transporter (NBC). Transport data suggested that the pyrimidine nucleotide carrier, pnc1, may be a major transport protein responsible for uptake of UDP-GlcNAc (Fig. 5A), although the deoxy nucleotide carrier also showed significant uptake. We also analyzed a number of nucleotide substrates that may act as exchange substrates for UDP-GlcNAc and found UDP to give the highest transport activity (Fig. 5B), indicating that pnc1 may act as an antiporter. Using a stealth siRNA, we were successful in knocking down pnc1 protein partially from HeLa cells (Fig. 5 C and D). The mitochondria-enriched fractions of these cells showed significant reduction in protein O-GlcNAcylation compared with controls. Finally, we used enriched mitochondria from control and pnc knockdown cells to analyze uptake of radioactive UDP-GlcNAc. The data (Fig. 5E) show a significant reduction in transport activity in the pnc knockdown-enriched mitos, supporting the role of pnc as a transporter for UDP-GlcNAc.
Fig. 5.
SLC25A33 gene encoding for mitochondrial pyrimidine nucleotide carrier is majorly responsible for UDP-GlcNAc incorporation to mitochondrial matrix. (A) Transport assay on phosphatidylcholine (PC)-based preformed proteoliposome made without PC extrusion (blue), with PC extrusion and protein (red), and with 1% cholesterol + PC with extrusion and protein (green). DNC samples contain deoxyribonucleotide carrier and PNC samples contain pyrimidine nucleotide carrier. +I samples contain 20 mM bathophenanthroline. NBC samples contain the cell membrane sodium bicarbonate transporter as a negative control. (B) Transport assay with internal substrate to assess antiporter activity of pnc protein. Control sample contains no protein and others contain ATP, ADP, UDP, and UTP inside preformed proteoliposome. (C) Knockdown of SLC25A33 protein from HeLa cells using 500 pmol (0.5) and 2 nmol (2) of stealth siRNA. Control samples contain 500 pmol control siRNA. Both whole cell lysate and mitochondrial fraction are run on gel to analyze total O-GlcNAc, pnc1, and mito complex I levels. (D) Quantification of mito-enriched fractions of O-GlcNAc blot from C above against complex I (n = 3). (E) Transport assay of radioactive UDP-GlcNAc uptake in enriched mitochondria prepared from HeLa cells after treatment with control and pnc siRNA (n = 3). (*P < 0.05).
TMG and Ac4SGlcNAc Affect ATP Levels and Mitochondrial Membrane Potential in Neonatal Rat Cardiomyocyte Cells.
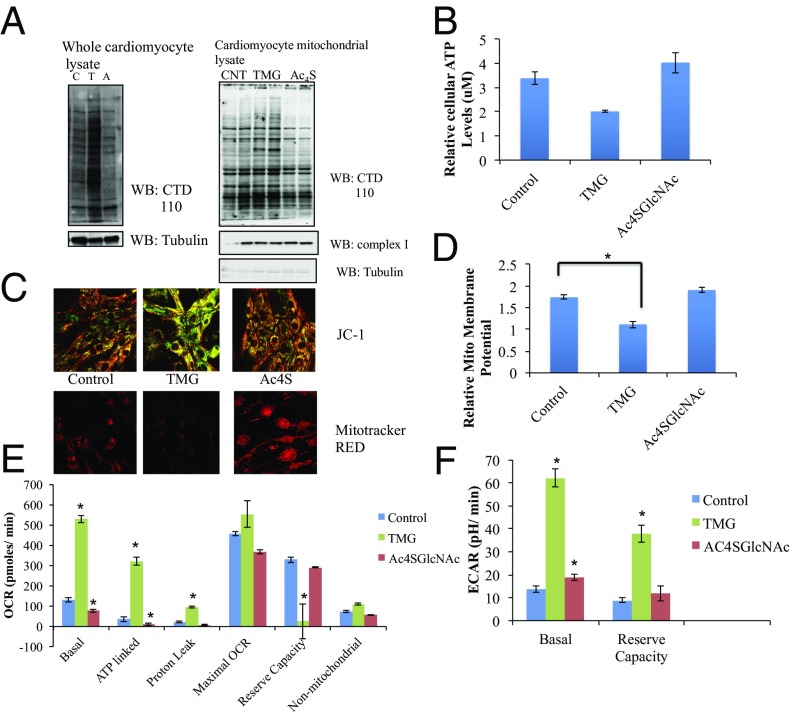
Our data show that rat heart mitochondria contain both OGT and OGA, whose levels and localization are altered in diabetes. To interrogate effects of inhibiting these key enzymes on cardiac tissue, we used neonatal rat cardiomyocyte (NRCM) culture to access the functional effects of altering O-GlcNAcylation on NRCM. For this analysis we used a short-term treatment of TMG and peracetyl thio-GlcNAc (Ac4SGlcNAc) as inhibitors for OGA and OGT, respectively (6 h for TMG and 18 h for Ac4SGlcNAc). Western blotting of whole cell lysates and mitochondrial fraction of cardiomyocytes confirm effective inhibition of both these enzymes (Fig. 6A). Inhibitor-treated cells were also used to determine total cellular ATP levels. OGA inhibition using TMG lowers the total cellular ATP levels by 40% compared with normal glucose, whereas inhibition of OGT did not significantly alter the overall ATP levels (Fig. 6B). Reduced mitochondrial membrane potential attenuates total ATP generated by the mitochondria and can cause dysfunction in cardiac mitochondria. To investigate how O-GlcNAc affects membrane potential, we tested mitochondrial potential in NRCMs after inhibition of OGT and OGA. Using live NRCMs, relative membrane potentials were determined by using a membrane potential activated dye, JC-1. We also used a mitotracker RED dye to look at fixed cells that were pretreated with the inhibitors. Both live and fixed cell analysis showed that cells treated with TMG had significantly reduced membrane potential compared with control cells (Fig. 6 C and D), whereas upon Ac4SGlcNAc treatment, a slight strengthening of the potential was observed (Fig. 6 C and D).
Fig. 6.
Effect of inhibition of OGT (with Ac4SGlcNAc) and OGA (with TMG) on neonatal rat cardiomyocyte mitochondrial function. (A) Western analysis of whole cell and mitochondrial preps of cardiomyocytes treated with Ac4SGlcNAc and TMG (A, Ac4SGlcNAc treated; C, control; T, TMG treated). (B) Relative ATP levels of whole NRCM treated with OGT and OGA inhibitors, analyzed by luciferase ATP determination kit (n = 2). Standard curve with known ATP levels were used to determine concentration. (C) Mitochondrial membrane potential of NRCMs treated with Ac4SGlcNAc and TMG visualized by confocal microscopy with two different mito-tracker dyes, JC-1 for live cells and Mito-tracker Red for fixed cardiomyocyte cells. (D) Quantification of relative membrane potential obtained from JC-1–labeled cells, value calculated by dividing red fluorescence (for higher potential) by green fluorescence (for lower potential) (n = 3). (E) Mitochondrial oxygen consumption of inhibitor-treated NRCM cells under basal level, ATP linked (from oligomycin treatment), proton leak (basal – ATP linked), maximal OCR (FCCP treated – rotenone, antimycin treated) reserve capacity (FCCP treated – basal) and nonmitochondrial (rotenone, antimycin treated – iodoacetate treated) (n = 5). (F) Cellular glycolysis levels as measured by ECAR rates of NRCMs treated with TMG and OGT inhibitor under basal level and maximal ECAR (oligomycin treated – basal level) (n = 5). (t test was carried out for control vs. TMG- or Ac4SGlcNAc-treated samples; with P < 0.05 considered significant.)
Effects of TMG and Ac4SGlcNAc on Mitochondrial Oxygen Consumption.
Inhibition of neonatal rat cardiomyocyte OGT and OGA using Ac4SGlcNAc and TMG, respectively, showed that short-term treatment of these enzymes significantly affect total cellular ATP levels and mitochondrial membrane potential. We decided to query whether OGT and OGA inhibition had an effect on mitochondrial oxygen consumption. For these measurements, we used XF96 seahorse analysis. Cardiomyocytes plated in 96-well plates were treated with Ac4SGlcNAc and TMG as described. The cells were then put in the seahorse analyzer and basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were obtained, after which the cells were subsequently treated with oligomycin, FCCP, rotenone and antimycin, and both OCR and ECAR were measured. Our data (Fig. 6E and Fig. S4) suggest that short-term TMG treatment significantly increases basal oxygen consumption. A significant portion of this increase seems to be ATP linked, which points to the loss in total cellular ATP levels as observed in Fig. 6C and the corresponding lowering of membrane potential. Conversely, OGT inhibition slightly lowers basal mitochondrial oxygen consumption. Maximal OCR levels remain unchanged between untreated and TMG-treated cells, whereas inhibition of OGT seems to slightly lower maximal OCR levels. This may be due to the slight increase in mitochondrial membrane potential as observed in Fig. 6 C and D. Also, probably as a result of increased basal OCR, mitochondrial reserve capacity is lowered significantly upon TMG treatment. A look at the cellular glycolytic rate (Fig. 6F) reveals that both TMG and Ac4SGlcNAc increases the glycolytic rate, although the increase due to TMG is much higher. TMG also elevates the maximal glycolytic capacity, which OGT inhibition does not seem to affect. In this regard, lowering of the basal OCR levels and maximal OCR levels upon Ac4SGlcNAc treatment might indicate a mito-specific effect of OGT inhibition in the neonatal rat cardiomyocytes, underscoring the important role of mitochondrial OGT in its function.
Discussion
A significant volume of literature suggests that O-GlcNAcylation is a nutrient and stress sensor. Mitochondria—the cells’ own energy generator—is significantly affected by alterations in nutrient types and levels. Recent research in the field has attempted to understand how mitochondrial function is affected by alterations in cellular O-GlcNAc without focusing on the raw materials that alter O-GlcNAc cycling within the mitochondria. This paper attempts to understand how the enzymes responsible for O-GlcNAc cycling affect mitochondrial O-GlcNAc and its functioning. The observation that OGT levels are increased in diabetic mitochondria, without significantly affecting its in vitro specific activity, hints at the aberrant nature of OGT functioning within diabetic organelles. Interestingly, EM data suggest a loss of membrane association for OGT in diabetic mito samples; this alteration in localization likely changes the subset of proteins that get O-GlcNAc modified within diabetic tissues, thereby implicating altered O-GlcNAc status of mitoproteins in certain mitochondrial dysfunction observed during diabetes. This loss of membrane-associated protein–OGT interaction is observed in complex IV–OGT interaction that falls off in diabetic samples. The OGT isoform observed in this case seems to be the short 78-kDa form, although there is a possibility that this observed band is just a proteolytic fragment of the mitochondrial OGT. Complex IV activity is significantly reduced in diabetic mitochondria and its OGT association may be a reason for its loss of functionality.
In our study, we observe OGA levels in normal mito by EM, which is reduced in diabetic samples. We also show by Immuno Fluoresence (IF) analysis that OGA colocalizes with mito-tracker RED in both NRCMs and NIH 3T3 cells. Our live cell assay shows that a significant portion of the cellular OGA activity occurs within the mitochondria. Taken together with the presence, localization, and interaction of mito-specific OGT, this observation for the first time to our knowledge puts both of the two cycling enzymes within the mitochondria, thus indicating real-time O-GlcNAc cycling occurs inside the mitochondrion.
We also show here that purified mitochondria are indeed capable of transporting UDP-GlcNAc, and this transport is affected specifically by UTP nucleotides. Using mitoplast preparations, inhibitor-based transport assays and in vitro liposome assay, we identified the pyrimidine nucleotide carrier as a mitochondrial transporter responsible for transport of UDP-donor sugar. Finally using stealth siRNA-based knockdown assay, we observed significant loss of O-GlcNAc levels in mitochondrial O-GlcNAc and a lowering of UDP-GlcNAc transport capacity. This finding implicates pnc as the significant contributor to mitochondrial UDP-GlcNAc levels. It must be acknowledged that even though pnc seems to be the predominant transporter for UDP-GlcNAc, the deoxyribonucleotide carrier (dnc) also transported a certain amount of the external radioactivity. Given the structural similarities of these transporters as well as the substrates they reportedly transport, both of these proteins could be partially responsible for uptake of nucleotide sugars.
Finally, our results indicate a real effect of OGT/OGA inhibition on mitochondrial function. Short-term inhibition of OGA lowers ATP levels and mitochondrial membrane potential. Concurrently at the basal level, we observe a significant increase in mitochondrial oxygen consumption upon TMG treatment. There is also an increase in basal glycolytic rate upon TMG treatment. These observations can be understood as an increase in overall cellular metabolic rate, which effects glycolysis, oxygen consumption, and membrane potential. These data indicate short-term increase in O-GlcNAc levels allows the cell to become more metabolically active and possibly assists in cardioprotection. Conversely, targeting OGT slightly strengthens mitochondrial membrane potential and cellular ATP levels. The ECAR data show a slight increase in glycolytic rates upon OGT inhibition, whereas a decrease in OCR is observed. This finding indicates that OGT’s effects may be more mitoprotein specific and that lowering of O-GlcNAc possibly lowers mitochondrial metabolism rates.
In conclusion, through this work we characterize the players in O-GlcNAc cycling within the mitochondria. We delineate how the enzyme levels, their location, and their interactions change in going from normal to diabetic heart samples. We show for the first time to our knowledge mito-specific OGA activity within live neonatal rat cardiomyocytes. And we report the presence of a mitochondrial transporter that uptakes UDP-GlcNAc. A knockout of one of these transporters also lowers the O-GlcNAc levels in the mitochondria. Because O-GlcNAcylation is highly dynamic, the presence of the full repertoire of responsible elements in the mitochondria indicates effective machinery that possibly alters the mitochondria’s functioning in response to not just nutrient and other cellular stresses but also to alteration in overall cellular O-GlcNAcylation conditions. Overall, current data suggest that O-GlcNAcylation not only plays a role in nutrient regulation of mitochondrial functions, but also its alterations in diabetes, both in terms of extent and types of proteins modified, plays a role in mitochondrial dysfunction in diabetic heart tissue.
Materials and Methods
Animals and Mitochondrial Purification.
All rats were treated in accordance with animal safety regulations according to The Johns Hopkins University School of Medicine. Rat hearts were taken from both diabetic and control animals and used for mitochondrial purification. See SI Materials and Methods for further details.
NRCM Mitochondrial Oxygen Consumption.
Live cell oxygen consumption was measured by using a XF96 flux analyzer. The oxygen consumption rate was measured over a period of 100 min over which time oligomycin, FCCP, antimycin A, and rotenone and iodoacetate were sequentially added to each well. See SI Materials and Methods for further details.
Live Cell OGA Activity Assay.
Cardiomyocytes were treated with OGA and lysosomal hexosaminidase inhibitors and used to determine live cell mito-specific OGA activity. See SI Materials and Methods for further details.
Generation of Proteoliposomes for UDP-GlcNAc Uptake Assay.
Liposomes were generated and transport of radioactive UDP-[3H]GlcNAc was carried out as described (31, 32). See SI Materials and Methods for details.
Statistics.
Data from Western blots and IF samples were processed by ImageJ quantification software. All data are expressed as mean ± SD. Each ‘n” represents the number of rats used or the number of biological repeats as applicable. Comparisons between different groups were performed with the use of a two-tailed, unpaired Student t test, with P < 0.05 considered significant.
Supplementary Material
Acknowledgments
The authors thank Prof. Rosemary O’Conner (National University of Ireland, Cork) and Prof. Fernando Palmieri (University of Bari, Italy) for generously providing the plasmids for PNC and DNC, respectively; Junaid Afzal and Genaro Ramirez-Correra for assistance with the seahorse analysis and live cell OGA activity assay, respectively; and the Johns Hopkins Microscopy facility for assistance with TEM and immunofluoresence analysis. This work was supported by NIH Grants P01HL107153, R01DK61671, and N01-HV-00240.
Footnotes
Conflict of interest statement: G.W.H. receives a share of royalty received by the university on sales of the CTD 110.6 antibody, which are managed by Johns Hopkins University.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424017112/-/DCSupplemental.
References
- 1.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80(1):825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259(5):3308–3317. [PubMed] [Google Scholar]
- 3.Harwood KR, Hanover JA. Nutrient-driven O-GlcNAc cycling: Think globally but act locally. J Cell Sci. 2014;127(Pt 9):1857–1867. doi: 10.1242/jcs.113233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272(14):9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 5.Dong DL, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J Biol Chem. 1994;269(30):19321–19330. [PubMed] [Google Scholar]
- 6.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276(13):9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- 7.Zachara NE, et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem. 2004;279(29):30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 8.Zachara NE, Hart GW. O-GlcNAc modification: A nutritional sensor that modulates proteasome function. Trends Cell Biol. 2004;14(5):218–221. doi: 10.1016/j.tcb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Dassanayaka S, Jones SP. O-GlcNAc and the cardiovascular system. Pharmacol Ther. 2014;142(1):62–71. doi: 10.1016/j.pharmthera.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramirez-Correa GA, et al. O-linked GlcNAc modification of cardiac myofilament proteins: A novel regulator of myocardial contractile function. Circ Res. 2008;103(12):1354–1358. doi: 10.1161/CIRCRESAHA.108.184978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suarez J, et al. Alterations in mitochondrial function and cytosolic calcium induced by hyperglycemia are restored by mitochondrial transcription factor A in cardiomyocytes. Am J Physiol Cell Physiol. 2008;295(6):C1561–C1568. doi: 10.1152/ajpcell.00076.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu-Mauldin X, Marsh SA, Zou L, Marchase RB, Chatham JC. Modification of STIM1 by O-linked N-acetylglucosamine (O-GlcNAc) attenuates store-operated calcium entry in neonatal cardiomyocytes. J Biol Chem. 2012;287(46):39094–39106. doi: 10.1074/jbc.M112.383778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias WB, Cheung WD, Wang Z, Hart GW. Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification. J Biol Chem. 2009;284(32):21327–21337. doi: 10.1074/jbc.M109.007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson JR, et al. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013;502(7471):372–376. doi: 10.1038/nature12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Housley MP, et al. A PGC-1α-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem. 2009;284(8):5148–5157. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Housley MP, et al. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283(24):16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pekkurnaz G, Trinidad JC, Wang X, Kong D, Schwarz TL. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell. 2014;158(1):54–68. doi: 10.1016/j.cell.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, et al. Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J Biol Chem. 2009;284(1):547–555. doi: 10.1074/jbc.M808518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan EP, et al. Altering O-linked β-N-acetylglucosamine cycling disrupts mitochondrial function. J Biol Chem. 2014;289(21):14719–14730. doi: 10.1074/jbc.M113.525790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanover JA, et al. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys. 2003;409(2):287–297. doi: 10.1016/s0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- 21.Love DC, Kochan J, Cathey RL, Shin S-H, Hanover JA. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci. 2003;116(Pt 4):647–654. doi: 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- 22.Darley-Usmar VM, Ball LE, Chatham JC. Protein O-linked β-N-acetylglucosamine: A novel effector of cardiomyocyte metabolism and function. J Mol Cell Cardiol. 2012;52(3):538–549. doi: 10.1016/j.yjmcc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-linked β-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J Biol Chem. 2008;283(49):33935–33941. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4(11):1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 25.Takimoto E, et al. Chronic inhibition of cyclic GMP phosphodiesterase 5A prevents and reverses cardiac hypertrophy. Nat Med. 2005;11(2):214–222. doi: 10.1038/nm1175. [DOI] [PubMed] [Google Scholar]
- 26.Yuzwa SA, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008;4(8):483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 27.Bhakdi S, Tranum-Jensen J, Sziegoleit A. Mechanism of membrane damage by streptolysin-O. Infect Immun. 1985;47(1):52–60. doi: 10.1128/iai.47.1.52-60.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmieri F, et al. Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta. 2006;1757(9–10):1249–1262. doi: 10.1016/j.bbabio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Dolce V, Fiermonte G, Runswick MJ, Palmieri F, Walker JE. The human mitochondrial deoxynucleotide carrier and its role in the toxicity of nucleoside antivirals. Proc Natl Acad Sci USA. 2001;98(5):2284–2288. doi: 10.1073/pnas.031430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Floyd S, et al. The insulin-like growth factor-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol Biol Cell. 2007;18(9):3545–3555. doi: 10.1091/mbc.E06-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmieri F, Indiveri C, Bisaccia F, Iacobazzi V. Mitochondrial metabolite carrier proteins: Purification, reconstitution, and transport studies. Methods Enzymol. 1995;260:349–369. doi: 10.1016/0076-6879(95)60150-3. [DOI] [PubMed] [Google Scholar]
- 32.Fiermonte G, Dolce V, Palmieri F. Expression in Escherichia coli, functional characterization, and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J Biol Chem. 1998;273(35):22782–22787. doi: 10.1074/jbc.273.35.22782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.