Significance
High pressure has emerged as a powerful tool for exploring the energy landscape of proteins, but structural origins of the pressure response remain controversial. The results of this study on a cavity mutant of T4 lysozyme (L99A) provide direct evidence for a structure-relaxation mechanism wherein pressure shifts conformational equilibria toward states with alternative packing arrangements that fill cavities or voids in the core. Both structure relaxation and cavity hydration can occur in response to pressure, and which dominates is found to depend on details of the energy landscape. The results also address conflicting views regarding the pressure response of L99A that have recently been published.
Keywords: DEER, EPR, conformational exchange, protein structural dynamics
Abstract
Application of hydrostatic pressure shifts protein conformational equilibria in a direction to reduce the volume of the system. A current view is that the volume reduction is dominated by elimination of voids or cavities in the protein interior via cavity hydration, although an alternative mechanism wherein cavities are filled with protein side chains resulting from a structure relaxation has been suggested [López CJ, Yang Z, Altenbach C, Hubbell WL (2013) Proc Natl Acad Sci USA 110(46):E4306–E4315]. In the present study, mechanisms for elimination of cavities under high pressure are investigated in the L99A cavity mutant of T4 lysozyme and derivatives thereof using site-directed spin labeling, pressure-resolved double electron–electron resonance, and high-pressure circular dichroism spectroscopy. In the L99A mutant, the ground state is in equilibrium with an excited state of only ∼3% of the population in which the cavity is filled by a protein side chain [Bouvignies et al. (2011) Nature 477(7362):111–114]. The results of the present study show that in L99A the native ground state is the dominant conformation to pressures of 3 kbar, with cavity hydration apparently taking place in the range of 2–3 kbar. However, in the presence of additional mutations that lower the free energy of the excited state, pressure strongly populates the excited state, thereby eliminating the cavity with a native side chain rather than solvent. Thus, both cavity hydration and structure relaxation are mechanisms for cavity elimination under pressure, and which is dominant is determined by details of the energy landscape.
Proteins in solution exist in conformational equilibria that cannot be appreciated from structures observed in crystal lattices (1–5). The members of a folded conformational ensemble may have distinct functions and hence are of interest in elucidating mechanisms of protein action (5–7). The free-energy differences between the conformations can range from zero to a few kilocalories per mole; the higher free-energy states are referred to as “invisible” or “excited” (E) states owing to their low equilibrium populations. The structural transition between the native ground state (G) and the E state may involve rigid body motion of the peptide backbone (5, 8) or local unfolding (9).
For a complete understanding of molecular mechanisms underlying function, characterization of functionally relevant conformational substates is required. However, in the case of E states, low populations and short lifetimes present a challenge for biophysical characterization. The elegant high-resolution NMR studies from Akasaka (10, 11) and coworkers suggest that application of hydrostatic pressure on the order of a few kilobars may solve this problem by reversibly populating functional E states, making them amenable for study by spectroscopic methods. For example, high-pressure NMR has been used to identify and characterize E states crucial to ligand binding in ubiquitin (12) and dihydrofolate reductase (13). As a result, high-pressure biophysics is currently of mainstream interest in protein science.
Application of sufficiently high pressure leads to population of a “pressure-denatured” state. For many proteins this state retains a relatively compact fold, at least near neutral pH and in the absence of denaturants, and is thus distinct from the unfolded states produced thermally or by chemical denaturants wherein both secondary and tertiary structures are largely lost (14–20). The pressure-denatured state is often labeled as the “unfolded state” (10), but here we reserve the term “unfolded” to describe a state with little tertiary or secondary structure. In this paper attention is focused on moderate pressures (<4 kbar) that shift conformational equilibria rather than leading to a pressure-denatured state.
Pressure shifts equilibria in a direction to reduce the total volume of the system. The current view is that the volume reduction that accompanies pressure-modulated transitions in proteins, including formation of the denatured state, is dominated by the elimination of voids or cavities in the protein’s interior (14, 21–24) via hydration, although other factors contribute (15, 25–28). In the equilibrium between two folded conformations, G ↔ E for example, an alternative “structure-relaxation” mechanism may play a role in the pressure response. In this model, voids are eliminated by pressure owing to an increase in the population of an alternative packing arrangement of the core in which cavities are filled with native side chains rather than solvent. This model has been suggested to play a role in certain proteins at high pressure (1, 29–31), although to our knowledge direct observation of structure relaxation under pressure has not been reported.
That a structure-relaxation mechanism may play a role in the pressure response is suggested by recent studies of cavity-creating mutants in T4 lysozyme (T4L) at atmospheric pressure (1). Although the crystal structures of the cavity mutants are nearly identical to those of the WT protein (32–35), in solution there are multiple conformations in equilibrium (1, 4). For example, in the T4L cavity mutants L121A/L133A, L133G, and W138A, two conformations of similar free energy were identified in solution (1). Perhaps the most extensively investigated cavity-forming mutation is T4L L99A that enlarges a preexisting cavity in the rigid four-helix bundle of the protein. In this mutant, a minor conformation (E) in equilibrium with the ground state (G) was detected that accounted for ∼3% of the population (4, 36). Remarkably, in each of the above cavity-creating mutants, one member of the conformational ensemble corresponds to a structural rearrangement that fills the cavity with a side chain; for L99A it is Phe114 that occupies the engineered cavity (5). Such conformations in which cavities are absent or reduced are expected to have a lower molar volume, and thus may be populated by pressure.
The pressure dependence of the L99A mutant was recently studied with NMR methods by Nucci et al. (27) and independently by Maeno et al. (31). In the study of Maeno et al. (31), the disappearance of cross-peaks in a 1H-13C heteronuclear single-quantum coherence (HSQC) NMR experiment at pressures up to 3 kbar was interpreted to reflect an increase in population of the E conformation with Phe114 occupying the cavity, consistent with a structure-relaxation mechanism rather than cavity hydration or unfolding. However, based on 1H-15N HSQC NMR studies on the same protein, Nucci et al. (27) concluded that cavity hydration and unfolding occurred at pressures less than 2.5 kbar rather than populating the E conformation. Commentaries on these conflicting interpretations were recently published (37, 38).
In the present study, we investigate the pressure dependence of T4L WT*, L99A, and L99A containing the additional mutations G113A and G113A/R119P, both of which have been shown to lower the free energy of the E conformation and hence increase the equilibrium population (5). The experimental approach is based on site-directed spin labeling EPR (SDSL-EPR) and the recently developed technologies of high-pressure continuous wave (CW) EPR spectroscopy for SDSL (30, 39), pressure-resolved double electron–electron resonance (PR DEER) spectroscopy (40), and high-pressure circular dichroism (HP-CD) (30). HP-CD reveals the global secondary structure of the protein at pressures up to 2.4 kbar. CW EPR line shapes of spin-labeled proteins are sensitive to backbone fluctuations on the nanosecond time scale (41, 42) and can unambiguously identify site-specific unfolding under pressure to 4 kbar, as well as identify sequences in slow conformational exchange on the microsecond–millisecond time scale (30, 39). The intrinsic time scale of SDSL-EPR is much shorter than NMR, such that spectral averaging of microsecond–millisecond protein conformational exchange does not occur. Hence, EPR spectra of a spin-labeled protein provide a “snapshot” of conformational equilibria frozen in time. Although the CW spectra can reveal conformational equilibria with exquisite sensitivity, they do not provide quantitative information on the structures involved or resolve the true heterogeneity of a conformational manifold. This information is uniquely provided by distance mapping using pairs of spin labels and DEER spectroscopy. At the present state of development, PR DEER allows direct structure mapping of the states populated by pressure to 6 kbar.
The results reported below indicate that at pressures up to 2.4 kbar the secondary structure content of T4L L99A is unchanged, eliminating the possibility of global or subglobal unfolding of the helical C domain, in agreement with the results of Maeno et al. (31). Collectively, the data do not support a large-scale shift to the E conformation of T4L L99A under high pressure, but rather cavity hydration and finally transition to a partially disordered state at pressures of 4 kbar. However, pressure strongly shifts the G ↔ E equilibrium toward E in the presence of the additional G113A and R119P mutations that lower the energy of the E conformation, thus demonstrating a structure-relaxation mechanism for the pressure response. Moreover, ligand binding to the engineered cavity strongly stabilizes the G conformation. Taken together, the results show that both cavity hydration and structure relaxation are valid models for the pressure dependence in proteins, and which prevails depends on the details of the energy landscape.
Results
Experimental Strategy.
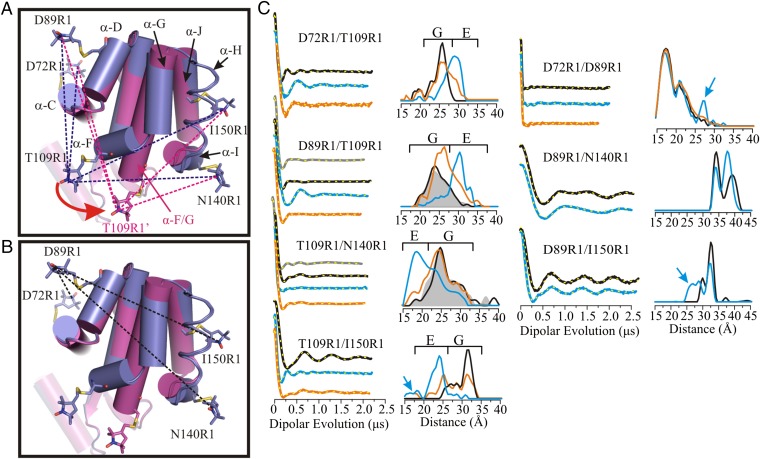
An objective of this study is to monitor the structure and dynamics of T4L cavity mutants under pressure using SDSL-EPR methods, and to determine whether the E conformation is populated as opposed to local or global unfolding. The structure of the E state of T4L L99A at atmospheric pressure and pH 5.5 determined by NMR and Rosetta modeling is shown in Fig. 1A, where it is compared with the WT* or G state (5). In the context of these models, the major structural rearrangement in the G-to-E transition involves dramatic rigid body motions of helices F and G to form a single helix, with only minor changes elsewhere in the structure. The motion of helices F and G place Phe114 in the cavity created by the L99A mutation. An R1 nitroxide side chain (43, 44) placed at position 109 in helix F (T109R1) is well-suited to monitor changes in the position of the F helix, and hence to identify the E conformation. For example, the change in environment that accompanies helix F movement would in general lead to changes in the mobility of T109R1, and hence the EPR spectral line shape. More importantly, T109R1 would move toward helices I and J, but further from helices C and D. These changes can be monitored experimentally using DEER spectroscopy to measure the distance between T109R1 and a second R1 residue placed at a reference site in a helix that shows comparably little or no movement. For this purpose, residues D72R1 (helix C), D89R1 (helix D), N140R1 (helix I), and I150R1 (helix J) were selected (Fig. 1 A and B). Residues D72R1, D89R1, and I150R1 are in helices for which NMR showed essentially no change in structure between G and E; residue N140R1 in helix I is displaced in E relative to G, but the magnitude is relatively small (∼2 Å) compared with that involving helix F. Thus, these sites are suitable for monitoring the position of T109R1 in the structure and consequently for identifying the E conformation. This simple strategy assumes the veracity of the NMR-based model of E, an assumption that can be checked by a quantitative comparison between measured and modeled distances, including distances between the references themselves, which should remain relatively constant independent of the position of T109R1.
Fig. 1.
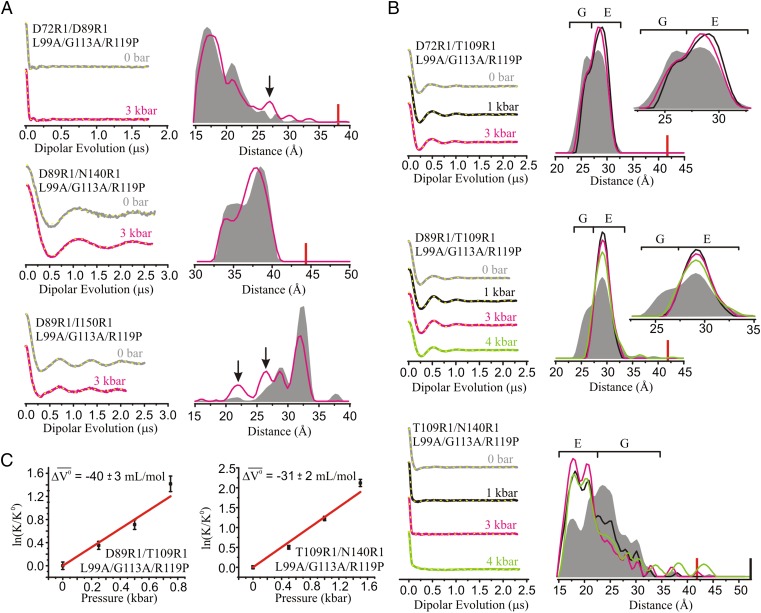
Distance mapping of the G and E conformations at atmospheric pressure and pH 5.5 with DEER spectroscopy. (A) An overlay in cylinder representation of the G (PDB ID code 3DMV) and E (PDB ID code 2LC9) (5) conformations of L99A in blue and magenta, respectively. Models of the R1 side chain are shown in stick representation; helix H is rendered in ribbon form to show the 150R1 side chain and its parent helix J. The direction of movement of helix F in the G→E structural transition is indicated by a red arrow. The dashed lines show the distances measured involving residue T109R1 with respect to an R1 reference for the G state (blue) and E state (magenta). (B) Ribbon diagram showing the interspin distances measured between the indicated reference sites in the G and E conformations. (C) DEFs, model-free fits of the DEFs (dashed yellow traces), and corresponding distance distributions for the indicated spin-labeled mutants in the WT* (black), L99A (gray), and L99A/G113A/R119P (blue) backgrounds in buffer consisting of 50 mM phosphate, 25 mM NaCl, and 20% (vol/vol) glycerol at pH 5.5. The DEFs and distance distributions after addition of benzene to mutants in the L99A/G113A/R119P background are shown in orange. The blue arrows identify distances only observed in the E conformation. The range of distances corresponding to the G and E conformations are indicated by brackets above the distributions.
To evaluate these ideas, and confirm the structure of the E conformation, we use additional mutants G113A and G113A/R119P in the L99A background. These mutations were shown to strongly shift the G ↔ E equilibrium toward E, resulting in large equilibrium populations of E (34% and 96%, respectively, at 274 K) (5). Thus, these additional mutations enable a direct characterization of the E conformation at atmospheric pressure using SDSL technology. Armed with SDSL-based metrics for identifying the E conformation, it is then possible to ask whether or not this conformation is populated by pressure in L99A, and to study the pressure dependence of the G ↔ E equilibrium in the G113A and G113A/R119P variants. The results, described below, provide new insight into mechanisms regarding the response of proteins to pressure.
Characterization of the G and E States of T4 Lysozyme with SDSL.
To quantitatively characterize the G and E conformations at atmospheric pressure, interspin distance distributions were measured between seven pairs of R1 residues in the WT*, L99A, and/or L99A/G113A/R119P backgrounds using DEER spectroscopy. Additionally, one of the pairs was also engineered in the L99A/G113A background. Four of the R1 pairs measure interspin distances between residue T109R1 in helix F and a reference at D72R1, D89R1, N140R1, or I150R1 (Fig. 1A), and three pairs measure distances between the reference sites themselves (Fig. 1B).
Fig. 1C, Left, shows the dipolar evolution functions (DEFs), the model free fits to the DEFs, and the corresponding distance distributions for T109R1 paired with each of the reference R1 residues in the WT*, L99A, and L99A/G113A/R119P backgrounds. The distance measurements in the WT* background provide a means for assigning distances corresponding to the pure G state for each spin pair. The distance distributions in the WT* and L99A backgrounds are nearly identical for the sites investigated, as expected owing to the low population of the E state in the L99A background. Addition of the G113A/R119P mutations drives the G ↔ E equilibrium strongly toward the E state; therefore, the distances corresponding to the E state for each spin pair may be easily identified in the L99A/G113A/R119P background. Indeed, changes in the most probable distance are observed for all pairs involving residue T109R1 in L99A/G113A/R119P relative to WT* and/or L99A (Fig. 1C). Distributions for these pairs reveal that in the E state, residue T109R1 moves away from residues D72R1 and D89R1, and closer to N140R1 and I150R1. The direction and magnitude of changes are in reasonable agreement with the rotation and translation of helix F reported by NMR for the G-to-E transition (Table S1). The T109R1/I150R1 pair that measures the position of helix F relative to J was also engineered in the L99A/G113A background. Compared with the same pair in L99A/G113A/R119P, the population of the G conformation is substantially increased, further demonstrating qualitative consistency with NMR relaxation dispersion measurements (Fig. S1A) (5). Interestingly, for the T109R1/I150R1 pair in the L99A/G113A/R119P and L99A/G113A backgrounds, there is a minor population at ∼18 Å that does not correspond to the G or E states (Fig. 1C and Fig. S1A). The possible origins of this minor population will be discussed below.
Previous DEER studies have shown that analysis of complex distance distributions using a multiple-Gaussian model provides a means to estimate equilibrium constants and relative energies between states in equilibrium (45). To obtain the fractional populations of the G and E states from the distance distributions, the DEFs were fit to a multiple-Gaussian model for the distance distribution, which was found to be equivalent to the model-free analysis in reproducing the details of the distance distributions (Figs. S2 and S3). Based on this metric, the energy difference at 295 K between the G and E (ΔGG↔E) for D89R1/T109R1 and T109R1/I150R1 in the L99A/G113A/R119P background is ∼1 kcal/mol. For D72R1/T109R1 and T109R1/N140R1 the states are nearly isoenergetic (Fig. S2, Left). The differences in the equilibrium constant and derived free energy among the four doubles investigated likely arises from a population bias due to site-specific attractive or repulsive interactions of the spin label with the local protein environment. This inevitable consequence of labeling methodology has been previously discussed in detail and does not influence the overall conclusions regarding shifts in the equilibrium due to other factors (1).
The interspin distance distributions between reference pairs D72R1/D89R1, D89R1/I150R1, and D89R1/N140R1 in the WT* and L99A/G113A/R119P backgrounds are shown in Fig. 1C, Right. For D72R1/D89R1and D89R1/I150R1 the distance distributions characteristic of the G conformation are preserved in E, but with the appearance of small new populations at ∼27 Å that are not observed in G (arrows, Fig. 1C, Right), suggesting the presence of a conformational substate in the cavity mutant that is not represented in the structural model of Fig. 1. The potential significance of this population will be discussed below. As anticipated from the models, there are small differences in the interspin distance distributions between the G and E conformations for reference pair D89R1/N140R1, corresponding to the small displacement of helix I (N140R1) in the structural models of Fig. 1. Under any condition, the differences in the distance distributions between the reference residues are small compared with those involving T109R1 and a reference residue, validating their use as reference sites. The origin of the strongly bimodal distribution in D89R1/N140R1 could arise from positional heterogeneity of the flexible helix I or rotamers of R1, as discussed previously (1).
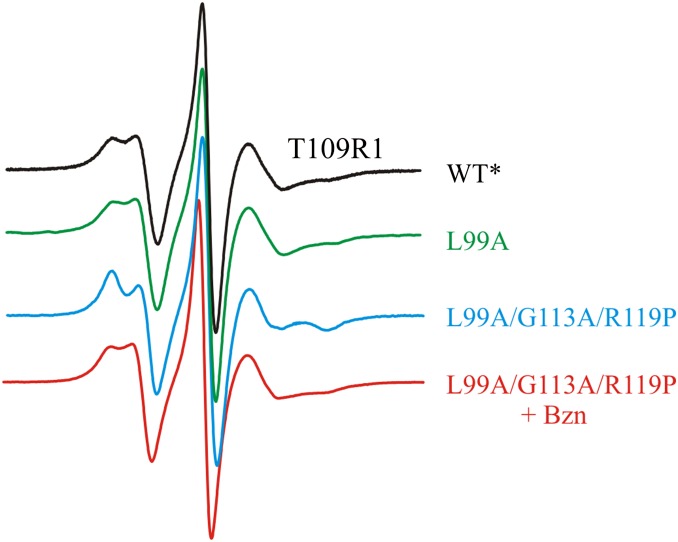
CW EPR spectra of residue T109R1 in the WT*, L99A, and L99A/G113A/R119P backgrounds (Fig. 2) support the changes of helix F position documented by PR DEER. For example, the CW spectra are essentially identical for WT* and L99A and show two relatively mobile components, the possible origins of which have been previously discussed (46). However, in the L99A/G113A/R119P background in which the E state is stabilized, the spectrum becomes highly ordered, showing restricted mobility of the R1 side chain, which confirms a change in the environment around R1 when helix F rotates and translates to a new position. Modeling suggests that an interaction of the nitroxide with a nearby carboxylate (residue E108) may be responsible for the ordering (Fig. S1B). Interaction of nitroxides with carboxylates has been previously identified (47, 48).
Fig. 2.
CW EPR spectra of T109R1 in the indicated genetic background. Spectra were recorded in 30% (wt/wt) sucrose at pH 6.8.
Shifting the G ↔ E Equilibrium at Atmospheric Pressure with Ligand Binding and pH Change.
The L99A cavity mutant binds benzene with submillimolar affinity (32, 49–51). Structurally, the ligand-bound state is similar to the G state (4, 32, 49), and thus addition of benzene to the E-stabilized mutant (L99A/G113A/R119P) should shift the equilibrium population toward the G state. Indeed, CW spectra of T109R1/L99A/G113A/R119P and DEER distance distributions for all doubles involving T109R1 show shifts in population toward the G state upon addition of benzene (Figs. 1C and 2 and Figs. S1A and S2), demonstrating that the distances assigned as G and E states represent states in equilibrium. Interestingly, benzene binding to the reference pair D72R1/D89R1 in the L99A/G113A/R119P background essentially eliminates the population of the new minor state (27 Å) observed in the apo-protein (Fig. 1C), suggesting a possible role of this minor state in ligand binding (Discussion).
The equilibrium populations of G and E are not only shifted by ligand binding, but by pH as well. The data in Fig. 1 were collected at pH 5.5, the same as in the NMR experiments that modeled the G and E states (5). At this pH, the population is strongly biased toward E in the L99A/G113A/R119P background (Fig. 1C). As will be shown below, pressure further populates the E state in this mutant, and to quantitatively investigate the pressure dependence of the G ↔ E equilibrium, experimental conditions are desired where the populations of G and E are similar so that shifts in equilibrium are readily detectable. The equilibrium population of the G state can be conveniently increased in the L99A/G113A/R119P background by an increase in the pH from 5.5 to 6.8, as assayed by the changes in distance distributions for all doubly labeled mutants involving T109R1 (Fig. S2). For D72R1/T109R1, D89R1/T109R1, and T109R1/N140R1 the G population is increased by 16 ± 4% at pH 6.8 relative to pH 5.5. For T109R1/I150R1, the equilibrium is still biased toward E (E 61%, G 21%) even at pH 6.8. The population of G is increased to 48% in the L99A/G113A background (without the R119P mutation), as qualitatively expected from Bouvignies et al. (5).
In the following sections, experiments designed to monitor pressure-dependent shifts in the G ↔ E equilibrium for L99A/G113A/R119P or L99A/G113A were carried out at pH 6.8 for the reason outlined above. Experiments designed to monitor the pressure dependence of WT* or L99A alone were done under the same conditions used in the NMR studies that led to the models of Fig. 1 (i.e., pH 5.5).
Far-UV CD Measurements of Global Secondary Structure at High Pressure.
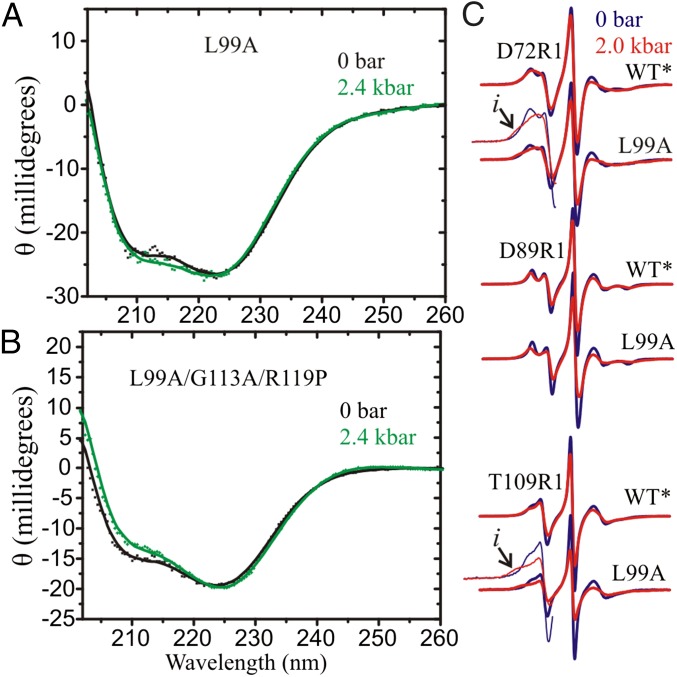
Recent development of a modified high-pressure optical cell suitable for use in far-UV CD allowed for monitoring global secondary structure in the range of 0–2.4 kbar (gauge pressure; 0 bar is equal to atmospheric pressure) (30). The pressure dependence for L99A and L99A/G113A/R119P was investigated to assess whether pressure affects the global secondary structure. CD spectra and secondary structure composition are shown in Fig. 3 A and B and Table S2, respectively. Both mutants exhibited expected levels of helical content (∼60%) and β-sheet content (∼6%) at atmospheric pressure, corresponding to the WT* protein. The HP-CD data for both indicate little change in the secondary structure content up to 2.4 kbar (∼1%), showing that for L99A at pH 5.5 the protein does not unfold in the pressure range investigated. The same conclusion applies to L99A/G113A/R119P at pH 6.8, with the addition that there is also little difference in secondary structure between the G and E conformations, in agreement with the models of Fig. 1.
Fig. 3.
High-pressure CD and CW EPR of T4L mutants. (A) High-pressure far-UV CD of L99A and (B) L99A/G113A/R119P. Both proteins contained spin labels D89R1/T109R1. CD Spectra were recorded in 5 mM MES and 2.5 mM NaCl at pH 5.5 for L99A and 10 mM MES and 25 mM NaCl at pH 6.8 for L99A/G113A/R119P at the indicated pressure. The typical protein concentration was ∼15 μM. (C) CW EPR spectra of the indicated protein at 0 and 2 kbar are shown in blue and red traces, respectively. For clarity, the low field lines in the spectra of D72R1/L99A and T109R1/L99A are amplified. Arrows identify a new component observed at 2 kbar. Spectra were recorded in 25% (wt/wt) Ficoll-70 at pH 5.5.
Pressure Effects on Conformational Equilibria in WT*, L99A, L99A/G113A, and L99A/G113A/R119P.
CW EPR spectra were collected at 0 and 2 kbar at pH 5.5 for the single R1-containing mutants D72R1, D89R1, and T109R1 in the WT* and L99A backgrounds (Fig. 3C). Spectra of R1 at the selected sites serve to sample local backbone dynamics and conformational substates involving the interdomain helix C and helices D and F in the C-terminal domain, respectively. This pressure range corresponds to that where NMR studies reported either enhancement of fluctuations within a G state ensemble and an increase in the population of E (31) or unfolding of the C domain of T4L L99A (27). In the WT* background, application of pressure to 2 kbar results in a slight reduction in nanosecond nitroxide motion as evidenced by a minor line broadening in the spectra of R1 at each site. Such effects have been previously reported and interpreted to reflect a limited compressibility of the protein in the region of the label site (30, 39). In the L99A background, the pressure-dependent changes are of substantially larger magnitude. Indeed, for D72R1 and T109R1, pressure produces an increase in spectral intensity corresponding to partially immobilized states of the nitroxide (arrows, Fig. 3C), clearly evident at 2 kbar, that signals a corresponding increase in the population of conformational substates not detected in the WT*. Consistent with HP-CD data, the CW spectra in L99A unequivocally show that the C-terminal domain is not globally unfolded at 2 kbar; in that case the CW resonance line shape would consist of sharp resonance lines rather than the reversible appearance of components corresponding to immobile states of R1 (Fig. S1C) (30, 39). As shown below, the new conformational substates sensed by R1 in L99A at 2 kbar are not related to the E conformation, but likely reflect low-amplitude structural fluctuations within the ground state ensemble of L99A (but see Discussion). Within the context of this model, the enhanced pressure dependence of L99A relative to WT* is interpreted to arise from an increased compressibility owing to the presence of the cavity, in agreement with Maeno et al. (31).
Although the CW spectra qualitatively identify protein compression, they do not directly reveal the magnitude of structural changes with pressure. For this purpose, PR DEER is required. In this method, high-pressure states of a protein are trapped by rapid freezing in dry ice/ethanol (200 K) (Materials and Methods) and the distance and distance distributions between pairs of R1 residues introduced into the protein are measured with DEER after depressurization to atmospheric pressure. In agreement with previous work (40), freezing in dry ice/ethanol and the more conventional liquid nitrogen results in similar distance distributions for T4L doubly labeled mutants in the WT*, L99A, and L99A/G113A/R119P backgrounds at atmospheric pressure (Fig. S4A).
For reference, the effect of pressure on the WT* protein was investigated to 3 or 4 kbar with PR DEER and four pairs of R1 residues that sampled distances between a reference pair (D89R1/N140R1) and distances that monitored the position of T109R1 in helix F (D72R1/T109R1, T109R1/N140R1, and D89R1/T109R1). Remarkably, the distance distributions at 3 or 4 kbar were essentially identical to those at atmospheric pressure (Fig. S5), showing that the tertiary fold of the WT* protein throughout the broad domain sampled by the above sites (Fig. 1 A and B) is unchanged. Therefore, any pressure dependence observed in the L99A, L99A/G113A, and L99A/G113A/R119P backgrounds can be attributed to conformational changes in the protein due to the presence of the cavity.
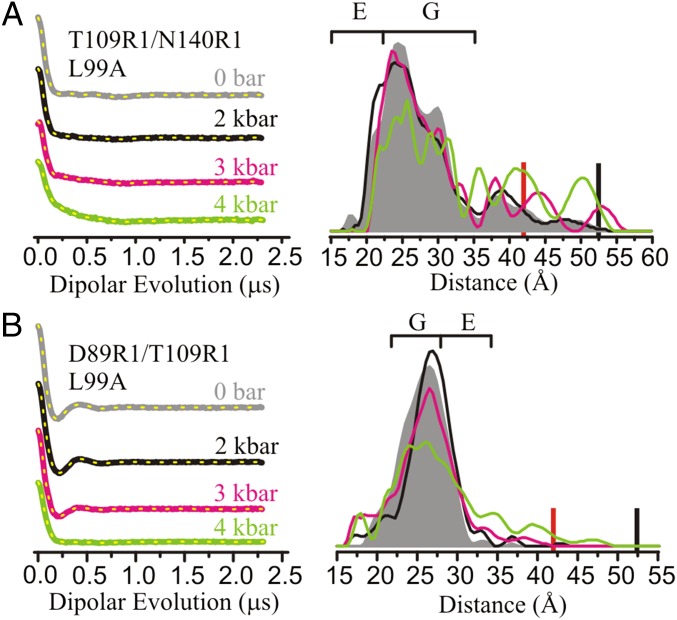
The effect of pressure on L99A was explored with DEER using R1 pairs T109R1/N140R1 and D89R1/T109R1 (Fig. 4 and Fig. S6A). As shown above (Fig. 1C) distances between R1 residues in these pairs monitor the position of helix F and are diagnostic for the formation of the E state. In the range of 0–3 kbar (Fig. 4), only subtle shifts in the relative population of individual distances are observed for the two mutants. Although this sparse dataset is insufficient to draw global conclusions about the protein conformation, it serves to demonstrate that the E conformation characterized by NMR is not substantially populated with pressure up to 3 kbar. At 4 kbar, the distance distributions begin to broaden in L99A (full width >30 Å), but not in WT*, suggesting a transition to a nonnative state for L99A in the regions sampled by the R1 sites. The 4-kbar conformation is not fully unfolded, because in this case the distance distribution would be far broader, as illustrated by that for the doubly labeled T4 lysozyme mutants in the guanidine hydrochloride-unfolded state (Fig. S1C). Additional PR DEER data will be needed to draw conclusions regarding the global fold, but for the spin pairs investigated, discrete distances corresponding to a highly populated E conformation (as illustrated in Fig. 1C) are not observed in the L99A background at any pressure, in agreement with Nucci et al. (27). The pressure response for the two mutants in the L99A was fully reversible (Fig. S4B).
Fig. 4.
Effect of pressure on L99A monitored with DEER using spin pairs (A) T109R1/N140R1 and (B) D89R1/T109R1. DEFs and model-free fits (dashed yellow traces) (Left) and corresponding distance distributions (Right) are shown from 0 to 4 kbar. The DEFs and distance distributions are color-coded as indicated. The approximated distances corresponding to the G and E states are indicated with brackets above the distributions. PR DEER experiments were conducted in buffer consisting of 50 mM MES, 25 mM NaCl, and 20% (vol/vol) glycerol at pH 5.5. The red and black bars indicate the upper limit of reliable shape and distance of the distribution, respectively (Materials and Methods).
Of particular interest is the effect of pressure on L99A/G113A/R119P, the mutant that lowers the free energy of the E state. In this mutant, the position of helix F is again monitored using spin pairs involving T109R1 and one of the reference sites D72R1, D89R1, N140R1, or I150R1 at pH 6.8. Distances between reference pairs D72R1/D89R1 and D89R1/N140R1 are rather weakly dependent on pressure to 3 kbar, although there are small shifts in populations (Fig. 5A) in the latter; in D72R1/D89R1 a minor population at ∼27 Å, also observed at atmospheric pressure and pH 5.5 (Fig. 1C), is increased by pressure (arrows, Fig. 5A). For D89R1/I150R1 populations at short distances (∼22 and 27 Å), also observed at atmospheric pressure and pH 5.5 (Fig. 1C), are substantially enhanced (Fig. 5A, arrows); the significance of states identified by these distances will be considered below.
Fig. 5.
The effect of pressure on the G ↔ E equilibrium in L99A/G113A/R119P. PR DEER was used to map distance changes (A) between indicated reference sites and (B) between T109R1 and selected references to monitor the position of helix F. DEFs, model-free fits of the DEFs (dashed yellow traces), and corresponding distance distributions are shown at various pressures between 0 and 4 kbar. DEFs and distance distributions are color-coded as indicated. The black arrows identify populations in reference pairs increased by pressure. The approximate distance ranges corresponding to G and E are indicated by brackets above the distributions. PR DEER data were collected for the protein in buffer consisting of 50 mM MOPS, 25 mM NaCl, and 20% (vol/vol) glycerol at pH 6.8. The red and black bars indicate the upper limit of reliable shape and distance of the distribution, respectively (Materials and Methods). (C) Plots of ln(K/Ko) vs. pressure for the indicated mutants, and fits (red trace) using a two-state model to measure Δ for the G → E transition.
The PR DEER data for D89R1/T109R1, T109R1/N140R1, and D72R1/T109R1 that define the position of helix F in the L99A/G113A/R119P background are shown in Fig. 5B and Fig. S6B. The primary effect of pressure on the distance distributions of these mutants is to shift the two-state G ↔ E equilibrium toward the E conformation, as can be seen by comparison with the data in Fig. 1C (note that the data in Fig. 1 were obtained at pH 5.5, whereas those in Fig. 5 are for pH 6.8 at which the population of the G conformation is increased). For the three R1 pairs, the population of the E state is already substantially increased at 1 kbar. In all cases, pressures up to 3 kbar shift the equilibrium without populating new states, and the widths of individual populations in the distributions do not increase. A similar result was observed for T109R1/I150R1 in the L99A/G113A background, where the distance distribution shows a nearly complete shift to the E conformation at 2 kbar (Fig. S6C).
For D72R1/T109R1, D89R1/T109R1, and T109R1/N140R1, the distance distributions remain narrow up to 4 kbar, showing that introduction of the G113A/R119P mutations in L99A dramatically stabilizes the protein against the pressure-dependent structure changes seen in L99A alone. Remarkably, distributions for D72R1/T109R1 and D89R1/T109R1 remain narrow even to 6 kbar, with only T109R1/N140R1 showing an increased breadth, likely due to local structural changes in the short helix I in which N140R1 resides (Fig. S6) (Discussion).
The pressure dependence of the G ↔ E equilibrium constant (K) was determined using fits of the DEF to multiple-Gaussian distance distributions as described in Supporting Information. Fig. S7 A and B contains a subset of the fits and associated multiple-Gaussian distance distributions along with their model-free counterparts, highlighting the similar goodness of fit for the two methods. Plots of ln(K/Ko) vs. pressure (p) for T109R1/N140R1 and D89R1/T109R1 in the L99A/G113A/R119P background (Fig. 5C) and T109R1/I150R1 in the L99A/G113A background (Fig. S7C) were fit to determine the change in partial molar volume (Δ) for the G-to-E transition as described in Materials and Methods. Values of Δ were −31, −40, and −29 mL/mol, respectively. It is noted that for this analysis the dataset was fit in the low pressure range (≤1.5 kbar) to eliminate contributions from compressibility of the individual states.
Discussion
The present study was undertaken to elucidate the response of T4L cavity mutants to hydrostatic pressure, with the expectation that the results will have general applicability to proteins with native cavities or packing imperfections. In particular, we are interested in the structural origins of the volume changes that underlie the shift in equilibrium between folded conformations at moderate pressures, rather than those that lead to a pressure-denatured state (14–20), or to an unfolded state formed at high pressure in the presence of a chemical denaturant (21, 52). In the present study unfolding will refer to a process in which loss of tertiary and secondary structure occurs.
The interpretations of the pressure-dependent EPR data rely on the NMR/Rosetta models of the G and E conformations of T4L L99A. Collectively, all SDSL and CD data for WT*, L99A, and L99A with G113A or G113A/R119P at atmospheric pressure are in good agreement with these models (Figs. 1–3, Fig. S1, and Tables S1 and S2). In particular, the essentially identical CD spectra as a function of pressure for the L99A and L99A/G113A/R119P mutants, which differ greatly in the populations of the G and E states, indicate that the global secondary structure of the two states is the same, as expected from the models, and any changes in intramolecular distances measured by DEER must arise from changes in tertiary structure. The distinctive distance changes involving movement in the F helix detected by DEER distance mapping are in good agreement with the models. Thus, we assume that the models of Fig. 1 reliably reflect the salient features of the G and E conformations, and that the SDSL-EPR data can be interpreted in the context of these models. The structural change involving the F helix, readily identified by DEER distance mapping, is used as a “fingerprint” to identify the E state population upon pressure application. In addition, the overall breadth of distance distributions reflects structural heterogeneity, whereas CW EPR spectra of single R1 residues monitor backbone dynamics and identify regions in conformational exchange on a time scale long compared with nanoseconds (41, 42). The main conclusions using this strategy are discussed in relationship to the earlier NMR results of Maeno et al. (31) and Nucci et al. (27) in the sections to follow. It is noted that the conclusions refer only to the C-terminal domain of T4L that contains the engineered L99A cavity.
Pressurization of L99A Drives Cavity Hydration and an Increase in Structural Heterogeneity.
Distance distributions in L99A that monitor the position of helix F show only subtle changes between 0 bar and 3 kbar (Fig. 4) and HP-CD reveals no secondary structure changes up to 2.4 kbar (Fig. 3A and Table S2). These data unambiguously show that the folded conformation of T4L L99A is maintained to 2.4 kbar, without loss of global secondary structure or tertiary structure in the regions sampled. This conclusion is strongly supported by the CW EPR data, which reveal no detectable dynamic disorder in the form of sharp resonance line shapes (Fig. S1C), but instead indicate structural fluctuations of the tertiary fold within the ground state ensemble of L99A at pressures in the range of 0–2 kbar (Fig. 3C). These fluctuations correspond to an increased compressibility in L99A relative to WT*, possibly resulting from hydration of the L99A cavity (discussed below). Interestingly, the structural fluctuations inferred from the line shapes of T109R1 and D72R1 must be of small amplitude because the DEER distance distributions involving these sites show only small changes in the same pressure range. This reinforces a previous conclusion that the line shapes of single R1 residues with multiple components are exquisitely sensitive to very small changes in structure owing to the r−6/r−12 dependence of attractive/repulsive interactions of the nitroxide with the local environment (41).
The above results are in general agreement with Maeno et al. (31) but apparently at odds with the conclusions of Nucci et al. (27), who interpret the responses observed for the C domain of L99A under pressure in the range of 1–2.5 kbar in terms of local unfolding of secondary structural elements. The HP-CD data obtained in the present study show no significant loss of secondary structure up to 2.4 kbar, and the PR DEER data show the tertiary fold to be intact.
Interestingly, large changes in tryptophan fluorescence were reported in the pressure range of 1–3 kbar at neutral pH with a midpoint at 2.4 kbar. As discussed by Ando et al. (14), these changes apparently correspond to cavity hydration. The fact that PR DEER detects no rearrangements in the tertiary fold in this pressure range shows that the putative hydrated cavity ground state (GH) has essentially the same conformation as that for the empty cavity (G), at least in the domains sampled by the spin labels. This is in accord with the previous high-pressure crystallography of L99A (22) and NMR studies of the same mutant contained in inverse micelles (27), although this result might not be anticipated considering that the available conformational space in a crystal lattice and trapped in an inverse micelle is highly limited. Overall, the fluorescence, HP-CD, CW EPR, and PR DEER data are consistent with a model wherein the L99A cavity becomes hydrated in the range of 2–3 kbar, concomitant with the onset of conformational flexibility involving small-amplitude tertiary structure fluctuations without unfolding.
The PR DEER data do not show discrete distances corresponding to the E conformation in L99A within the detection limit of the experiment (∼10%) in the pressure range of 0–3 kbar, apparently at odds with the conclusions of Maeno et al. (31). However, the E state is an established member of the conformational ensemble (5) and is surely present at some level, so the differences may be quantitative rather than qualitative. Maeno et al. (31) note that, in addition to an increase in the E state population, pressure dependence of the G ↔ E exchange rates could also account for their data and this would be consistent with the PR DEER data presented here. At pressures above ca. 3 kbar, the distance distributions for D89R1/T109R1 and T109R1/N140R1 in the L99A background become markedly broadened, with a full distribution width of ∼30–35 Å at 4 kbar, reflecting the onset of significant structural heterogeneity. The relatively broad distance distributions may arise from local/global unfolding driven by internal hydration and the formation of a “wet” molten globule state, as was recently observed for apomyoglobin by PR DEER (40).
Distance Mapping in L99A/G113A and L99A/G113A/R119P Supports a Structure-Relaxation Mechanism Rather Than Cavity Hydration for the Pressure Response.
In the G113A and G113A/R119P mutations in L99A, the energy of the E state is lowered relative to G (5) and the G ↔ E equilibrium is readily resolved at atmospheric pressure using DEER distance mapping (Fig. 1). Remarkably, the equilibrium is shifted toward the E conformation with increasing pressure in the range of 0–3 kbar (Fig. 5B), with no new states appearing as judged by the shift in position of helix F, the constancy of the CD spectra (to 2.4 kbar) (Fig. 3B), and the lack of substantial distance change between reference sites (Fig. 5A) (but see below). Thus, we tentatively conclude that the G and E structures are essentially pressure-independent at low pressures, and that the primary effect of pressure is to shift the G ↔ E equilibrium. In the context of this model, the data show that the E conformation has a lower partial molar volume compared with G and they provide direct experimental evidence for a structure-relaxation mechanism as the basis for the volume reduction.
Interestingly, the Δ estimated for the G-to-E transition is ∼−36 mL/mol, close to the molar volume of the Phe114 side chain that occupies the cavity in the E state (42 mL/mol). The Δ for the G-to-E transition is similar in the presence and absence of R119P (Fig. 5C and Fig. S7C), further reinforcing that the G113A and R119P mutations shift the relative energies of G and E without significantly altering their structures. Moreover, the widths of the distance distributions for peaks corresponding to the G and E conformations are essentially pressure-independent (Fig. 5B), suggesting that the two states have similar flexibility in the structural elements sampled by the distance measurements. Viewed in combination with the results from HP-CD, this indicates that, in the case of equilibria involving nearly isoenergetic states, pressure may populate a conformational state with backbone configurational entropy similar to the G state. For truly excited states with substantially higher energy than the ground state, a large body of evidence suggests that the excited state has higher configurational entropy [the “volume rule” (53)].
A remaining question regarding ligand binding to L99A is the ingress/egress pathway for ligand exchange (5). The structure of the T4L L99A G state shows the cavity to be completely solvent-inaccessible, and in the E state the ligand binding site is occupied by Phe114. Access to the empty cavity must then be via an as-yet-unresolved conformation, distinct from both the G and E states, as suggested by Bouvignies et al. (5). Interestingly, three of the R1 pairs (D72R1/D89R1, D89R1/I150R1, and T109R1/I150R1) show populations at distances not corresponding to the E or G states in the L99A/G113A/R119P background (Fig. 1C), and the size of those populations increases with pressure (Fig. 5A and Fig. S6C). This suggests that both pressure and the additional G113A and R119P mutations populate not only the E state, but also the putative ligand binding state to a small extent. For the one case investigated involving reference sites (D72R1/D89R1), the new additional distance disappears upon ligand binding. Although the data are too sparse draw conclusions regarding possible structures, the third state observed may correspond to the ligand accessible conformation that must exist to enable rapid (approximately millisecond) binding of ligands to the otherwise buried cavity (51). Because D89R1 is common to the two reference pairs that reveal the new populations and it is located at a helix (helix D) closest to the engineered cavity, we tentatively suggest that it is helix D that moves in the new state, closer to I150R1 and further from D72R1. Such a motion could open a direct path to the cavity. Interestingly, structural analysis using Caver (54, 55) reveals putative tunnels in the interface between helices D and G that could enable ligand entry (Fig. S1D). This pathway is different from that identified by MD simulations for water penetration and escape from the L99A cavity (22).
The Thermodynamic Basis for Hydration vs. Structure Relaxation.
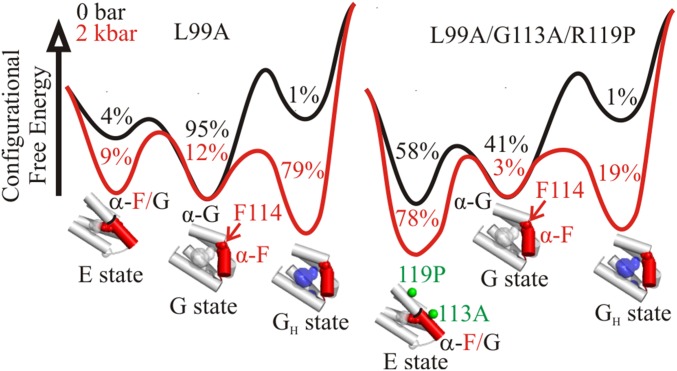
Collectively, the above data suggest that pressure hydrates the enlarged cavity in the L99A mutant but shifts the conformation from G to E in L99A/G113A/R119P and L99A/G113A. Why is the cavity not hydrated by pressure in these latter mutants? A likely explanation for this difference lies in details of the energy landscape. The combination of atmospheric and high-pressure data from NMR, fluorescence, X-ray crystallography, and EPR suggest an equilibrium model for L99A and L99A/G113A/R119P with at least four states in the pressure range of 0–3 kbar, namely E, G, GH, and L, where L is a proposed conformation where the ligand has access to the cavity, and GH is a state wherein the L99A cavity is hydrated. At higher pressures, the putative molten globule state would be included. To illustrate a simple model that can account qualitatively for structure relaxation vs. cavity hydration, consider a three-state equilibrium excluding a minor L state:
Although thermodynamically distinct, the protein is structurally similar in GH and G based on high-pressure crystallography (22), HP-CD (Fig. 3 A and B), and PR DEER at 2 kbar (Fig. 4B). To compute pressure-dependent free-energy changes, values for free-energy differences between states at atmospheric pressure (ΔGo) and corresponding volume changes (Δ) are needed. ΔGo can be estimated from NMR (4, 5), fluorescence (14), and the DEER data presented above. Δ values corresponding to the transitions are obtained from the pressure dependence of the corresponding equilibria. For the G → GH transition, a value of −75 mL/mol was taken because it generates landscapes that illustrate the main features of the experimental results presented above. This value is intermediate between that for complete cavity elimination (−100 mL/mol) and a value of −56 mL/mol measured from fluorescence at pH 7. The Δ for the G → E transition identified by PR DEER is ∼−36 mL/mol, and because the G and E states are expected to be structurally similar in L99A, L99A/G113A/R119P, and L99A/G113A based on the evidence presented above, we assume this Δ to be the same for all three genetic backgrounds. With these values, free-energy profiles as a function of pressure can be generated and are shown in Fig. 6 for 0–2 kbar. These landscapes are not intended to reproduce the experimental data in detail because of the neglect of other conformations and compressibility effects, but only to illustrate a concept with realistic values of ΔGo and Δ.
Fig. 6.
Effect of pressure on the equilibrium between the GH, G, and E states of T4L mutants L99A (Left) and L99A/G113A/R119P (Right). Relative configurational free energies (ΔGo) are shown for 0 and 2 kbar; values for the GH and E states are relative to G. Values for Δ and ΔGo are −36 mL/mol and −0.2 kcal/mol, respectively, for the G-to-E transition (based on PR DEER of L99A/G113A/R119P), and −75 mL/mol and 2.5 kcal/mol, respectively, for the G-to-GH transition [based on high-pressure tryptophan fluorescence of L99A (14) and the total L99A cavity volume]. Populations of each state are indicated in the local minima of the landscape. Structural models for the C domain are shown below the corresponding energy minima. Helix F is shown in red. The location of Phe114 (red spheres) in the G and E states are indicated. The empty cavity is shown in gray surface representation. A blue surface is used to represent pressure-populated hydration of the cavity in the L99A mutant. Green spheres at the Cα are used to indicate the position of G113A and R119P mutants in the E state structure (PDB ID code 2LC9) (5).
The volumetric contribution to stabilization of the E state under pressure is less than that for hydration, and pressure will drive hydration instead of repacking unless the ΔGo for repacking is markedly smaller than ΔGo for hydration. This is not the case for the L99A mutant, so pressure populates the GH state. The addition of G113A and R119P mutations lowers ΔGo for formation of the E conformation (5) sufficiently that pressure drives the equilibrium to the E state in the L99A/G113A and L99A/G113A/R119P mutants.
The preference for repacking in L99A/G113A/R119P rather than hydration has important implications regarding the stability of proteins under pressure. For example, the spin pairs T109R1/N140R1 and D89R1/T109R1 have narrower distance distributions in L99A/G113A/R119P backgrounds (Fig. 5B) compared with the L99A background at 4 kbar (Fig. 4), suggesting increased disorder in L99A but not in L99A/G113A/R119P. Thus, hydration of the cavity in L99A at intermediate pressures leads to destabilization compared with L99A/G113A/R119P, because in the E conformation that dominates L99A/G113A/R119P, the side chain of Phe114 occupies the cavity and must be displaced before water can penetrate the protein. Similarly, both cavity-filling mutations and ligand binding have been found to stabilize proteins against denaturation at high pressure (24, 56, 57). In the context of this model, cavities in flexible proteins that can repack are expected to be more stable to pressure than a corresponding rigid protein in which the cavity will hydrate with pressure. Indeed, repacking has been proposed to explain the stabilizing effect of proline-to-glycine mutations against pressure denaturation observed in staphylococcal nuclease (58). In that study, the authors speculated that the increased flexibility caused by the mutations could have allowed the protein to sample alternative, more stable, packing modes. Evolutionarily, mutations such as G113A and R119P that modify the energy landscape to stabilize or expand the number of accessible conformations that the protein can sample within the native state ensemble may prove advantageous for adaptation to various environmental stresses such as pressure and temperature and give rise to new functionality.
Summary and Conclusions
Collectively, the data presented here show that a protein can respond to pressure by structure relaxation to fill a cavity or by the established mechanism of cavity solvation. In principle, any cavity-containing protein can have conformational substates in equilibrium in which the core is rearranged to fill or partially fill the cavity with protein atoms, thus reducing the partial molar volume. If the strain energy involved in forming such a conformation is high, cavity hydration will be of lower energy. This seems to be the case for L99A in solution. If the strain energy is lower than hydration, structure relaxation will be favored over hydration. The strain energy will be low in proteins that have nearly isoenergetic conformational substates, at least one of which has an alternative packing mode that fills the cavity. This is the case with L99A containing the additional mutations G113A and G113A/R119P that relieve steric hindrance for forming the cavity-filled conformation (the E conformation) (5). Although the principles were elucidated with cavity-creating mutants, they are expected to apply to proteins with naturally occurring packing imperfections.
Materials and Methods
Construction, Expression, and Purification of T4L Mutants.
All of the mutations designed for this study were introduced to the T4L gene (pET11a vector) using the QuikChange site-directed mutagenesis method. All of the mutants contain the pseudo-WT (WT*) mutations C54T and C97A. Mutations were verified by sequencing. Expression, purification, and spin labeling of cysteine mutants in the WT* background was done as previously described (44). All of the cysteine mutants in the L99A background were purified from inclusion bodies as described in López et al. (1).
EPR Spectroscopy.
CW EPR spectra of spin-labeled proteins were recorded at X-band in a Bruker ELEXSYS 580 spectrometer at room temperature in buffer at pH 5.5 (50 mM MES and 25 mM NaCl) or pH 6.8 (50 mM MOPS and 25 mM NaCl) containing 30% (wt/wt) sucrose or 25% (wt/wt) Ficoll-70 (Sigma). These concentrations of sucrose and Ficoll-70 cause an equivalent increase in the effective viscosity of the solution, and are thus equally effective in reducing the contribution of rotational diffusion of the protein to the spectral lineshape (39, 59, 60). Protein concentrations were in the range of 100–500 μM. Atmospheric pressure CW EPR samples were loaded into glass capillaries (0.60 i.d. × 0.84 o.d.; VitroCom Inc.). High-pressure CW EPR spectra were recorded at room temperature using ceramic sample cells and a computer-controlled pressure intensifier (HUB440-Cer and HUB440; Pressure BioSciences, Inc.) as described previously (30). Four-pulse DEER data at 80 K were obtained on a Bruker ELEXSYS 580 operated at Q-band. The protein concentration was at or below 200 μM. For DEER measurements at atmospheric pressure, samples of 12–20 μL in 50 mM phosphate and 25 mM NaCl at pH 5.5 or buffer at pH 6.8 containing 20% (vol/vol) glycerol were loaded in a glass capillary (1.4 i.d. × 1.7 o.d.; VitroCom Inc.) and flash-frozen in liquid nitrogen. A glycerol solution in the appropriate buffer was used as a stock in all cases. A π–pump pulse was set at the maximum peak of the absorption spectrum and the π/2 (16 ns) and π (32 ns) observe pulses were positioned 50 MHz (17.8 Gauss) upfield, which corresponds to the absorption maxima of the center-field line. Model-free and model-based (SI Materials and Methods) distance distributions were obtained from the raw dipolar evolution data using the program LongDistances available at http://www.chemistry.ucla.edu/directory/hubbell-wayne-l. The materials and detailed protocol for PR DEER experiments up to 3 kbar are described in Lerch et al. (40). For PR DEER above 3 kbar, a Barocycler HUB880 pressure intensifier (Pressure BioSciences, Inc.) was used with 100,000 psi-rated pressure tubing and connectors (Maxpro Technologies) according to the same protocol. Pressures specified in this article are gauge pressure, i.e., 0 bar is equal to atmospheric pressure. All pressure-resolved DEER experiments were done in buffer at pH 6.8 containing 20% (vol/vol) glycerol, except for mutants in the L99A background, for which the buffer used consists of 50 mM MES, 25 mM NaCl, and 20% glycerol at pH 5.5. For ligand binding studies, benzene was added via vapor diffusion as described in López et al. (1). The upper limit of reliable distance (r) and width determination (σ) for each mutant in nanometers was calculated using the following equations (61):
where tmax is the maximum time domain recorded for each sample.
High-Pressure Far-UV CD Spectroscopy.
High-pressure far-UV CD was done as previously described (30); details can be found in SI Materials and Methods.
Thermodynamic Analysis of the G ↔ E Equilibrium.
The use of multiple Gaussians to represent DEER distance distributions and relative populations of states has been previously described (45). Fits presented here were performed using LongDistances (SI Materials and Methods) for D89R1/T109R1 and T109R1/N140R1 in the L99A/G113A/R119P background and for T109R1/I150R1 in the L99A/G113A background in the range of 0–1.5 kbar. The data were fit as a function of pressure according to a two-state equilibrium, G ↔ E, where one or two individual Gaussians were used to account for each state at each pressure. The fractional populations of G and E (fG and fE) were calculated using the sum of the integrated areas for the individual Gaussians corresponding to the given state, normalized to the total area for all Gaussians. Errors reported by LongDistances for the areas of individual Gaussians were propagated throughout the analysis. The equilibrium constant (K) is given by Eq. 1:
| [1] |
A first-order approximation for the pressure dependence of the Gibbs free energy (ΔG), Eqs. 2 and 3, was used due to the relatively low pressures used in this analysis:
| [2] |
| [3] |
Combining Eqs. 2 and 3 yields Eq. 4, where R is the gas constant and T is temperature. The temperature was set to 298 K, which was the holding temperature for all samples before rapid freezing for DEER.
| [4] |
Plots of ln(K/Ko) vs. pressure (p) were fit to Eq. 4 using OriginPro 8.1 (OriginLab) to solve for the change in partial molar volume (Δ) associated with the pressure-populated transition from G to E.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01EY05216 (to W.L.H.), NIH Grant R01EY023588 (to J.H.), the Jules Stein Professor Endowment (W.L.H.), National Eye Institute Core Grant P30EY00331, and a Research to Prevent Blindness Unrestricted Grant (to the Jules Stein Eye Institute).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506505112/-/DCSupplemental.
References
- 1.López CJ, Yang Z, Altenbach C, Hubbell WL. Conformational selection and adaptation to ligand binding in T4 lysozyme cavity mutants. Proc Natl Acad Sci USA. 2013;110(46):E4306–E4315. doi: 10.1073/pnas.1318754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fanucci GE, Lee JY, Cafiso DS. Spectroscopic evidence that osmolytes used in crystallization buffers inhibit a conformation change in a membrane protein. Biochemistry. 2003;42(45):13106–13112. doi: 10.1021/bi035439t. [DOI] [PubMed] [Google Scholar]
- 3.Cafiso DS. Identifying and quantitating conformational exchange in membrane proteins using site-directed spin labeling. Acc Chem Res. 2014;47(10):3102–3109. doi: 10.1021/ar500228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulder FA, Hon B, Muhandiram DR, Dahlquist FW, Kay LE. Flexibility and ligand exchange in a buried cavity mutant of T4 lysozyme studied by multinuclear NMR. Biochemistry. 2000;39(41):12614–12622. doi: 10.1021/bi001351t. [DOI] [PubMed] [Google Scholar]
- 5.Bouvignies G, et al. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature. 2011;477(7362):111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5(11):808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 7.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450(7172):964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 8.Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A transient and low-populated protein-folding intermediate at atomic resolution. Science. 2010;329(5997):1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- 9.Neudecker P, et al. Structure of an intermediate state in protein folding and aggregation. Science. 2012;336(6079):362–366. doi: 10.1126/science.1214203. [DOI] [PubMed] [Google Scholar]
- 10.Akasaka K. Probing conformational fluctuation of proteins by pressure perturbation. Chem Rev. 2006;106(5):1814–1835. doi: 10.1021/cr040440z. [DOI] [PubMed] [Google Scholar]
- 11.Akasaka K. Pressure and protein dynamism. High Press Res. 2014;34(2):222–235. [Google Scholar]
- 12.Kitahara R, Yokoyama S, Akasaka K. NMR snapshots of a fluctuating protein structure: Ubiquitin at 30 bar-3 kbar. J Mol Biol. 2005;347(2):277–285. doi: 10.1016/j.jmb.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 13.Kitahara R, et al. High pressure NMR reveals active-site hinge motion of folate-bound Escherichia coli dihydrofolate reductase. Biochemistry. 2000;39(42):12789–12795. doi: 10.1021/bi0009993. [DOI] [PubMed] [Google Scholar]
- 14.Ando N, et al. Structural and thermodynamic characterization of T4 lysozyme mutants and the contribution of internal cavities to pressure denaturation. Biochemistry. 2008;47(42):11097–11109. doi: 10.1021/bi801287m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalikian TV, Macgregor RB., Jr Origins of pressure-induced protein transitions. J Mol Biol. 2009;394(5):834–842. doi: 10.1016/j.jmb.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Peng X, Jonas A, Jonas J. NMR study of the cold, heat, and pressure unfolding of ribonuclease A. Biochemistry. 1995;34(27):8631–8641. doi: 10.1021/bi00027a012. [DOI] [PubMed] [Google Scholar]
- 17.Smeller L. Pressure-temperature phase diagrams of biomolecules. Biochim Biophys Acta. 2002;1595(1-2):11–29. doi: 10.1016/s0167-4838(01)00332-6. [DOI] [PubMed] [Google Scholar]
- 18.Chryssomallis GS, Torgerson PM, Drickamer HG, Weber G. Effect of hydrostatic pressure on lysozyme and chymotrypsinogen detected by fluorescence polarization. Biochemistry. 1981;20(14):3955–3959. doi: 10.1021/bi00517a002. [DOI] [PubMed] [Google Scholar]
- 19.Meersman F, et al. High-pressure biochemistry and biophysics. Rev Mineral Geochem. 2013;75(1):607–648. [Google Scholar]
- 20.Meersman F, Smeller L, Heremans K. Comparative Fourier transform infrared spectroscopy study of cold-, pressure-, and heat-induced unfolding and aggregation of myoglobin. Biophys J. 2002;82(5):2635–2644. doi: 10.1016/S0006-3495(02)75605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roche J, et al. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci USA. 2012;109(18):6945–6950. doi: 10.1073/pnas.1200915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins MD, Hummer G, Quillin ML, Matthews BW, Gruner SM. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc Natl Acad Sci USA. 2005;102(46):16668–16671. doi: 10.1073/pnas.0508224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamatari YO, Smith LJ, Dobson CM, Akasaka K. Cavity hydration as a gateway to unfolding: An NMR study of hen lysozyme at high pressure and low temperature. Biophys Chem. 2011;156(1):24–30. doi: 10.1016/j.bpc.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Lassalle MW, et al. Filling a cavity dramatically increases pressure stability of the c-Myb R2 subdomain. Proteins. 2001;45(1):96–101. doi: 10.1002/prot.1128. [DOI] [PubMed] [Google Scholar]
- 25.Hummer G, Garde S, García AE, Paulaitis ME, Pratt LR. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci USA. 1998;95(4):1552–1555. doi: 10.1073/pnas.95.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouget J-B, et al. Size and sequence and the volume change of protein folding. J Am Chem Soc. 2011;133(15):6020–6027. doi: 10.1021/ja200228w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nucci NV, Fuglestad B, Athanasoula EA, Wand AJ. Role of cavities and hydration in the pressure unfolding of T4 lysozyme. Proc Natl Acad Sci USA. 2014;111(38):13846–13851. doi: 10.1073/pnas.1410655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voloshin VP, Medvedev NN, Smolin N, Geiger A, Winter R. Exploring volume, compressibility and hydration changes of folded proteins upon compression. Phys Chem Chem Phys. 2015;17(13):8499–8508. doi: 10.1039/c5cp00251f. [DOI] [PubMed] [Google Scholar]
- 29.Vidugiris GJA, Truckses DM, Markley JL, Royer CA. High-pressure denaturation of staphylococcal nuclease proline-to-glycine substitution mutants. Biochemistry. 1996;35(12):3857–3864. doi: 10.1021/bi952012g. [DOI] [PubMed] [Google Scholar]
- 30.Lerch MT, Horwitz J, McCoy J, Hubbell WL. Circular dichroism and site-directed spin labeling reveal structural and dynamical features of high-pressure states of myoglobin. Proc Natl Acad Sci USA. 2013;110(49):E4714–E4722. doi: 10.1073/pnas.1320124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeno A, et al. Cavity as a source of conformational fluctuation and high-energy state: High-pressure NMR study of a cavity-enlarged mutant of T4 lysozyme. Biophys J. 2015;108(1):133–145. doi: 10.1016/j.bpj.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morton A, Matthews BW. Specificity of ligand binding in a buried nonpolar cavity of T4 lysozyme: Linkage of dynamics and structural plasticity. Biochemistry. 1995;34(27):8576–8588. doi: 10.1021/bi00027a007. [DOI] [PubMed] [Google Scholar]
- 33.Eriksson AE, et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 34.Baldwin E, Baase WA, Zhang XJ, Feher V, Matthews BW. Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions. J Mol Biol. 1998;277(2):467–485. doi: 10.1006/jmbi.1997.1606. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Baase WA, Baldwin E, Matthews BW. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7(1):158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat Struct Biol. 2001;8(11):932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 37.Kitahara R, Mulder FA. Is pressure-induced signal loss in NMR spectra for the Leu99Ala cavity mutant of T4 lysozyme due to unfolding? Proc Natl Acad Sci USA. 2015;112(9):E923. doi: 10.1073/pnas.1423279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wand AJ, Nucci NV. Reply to Kitahara and Mulder: An ensemble view of protein stability best explains pressure effects in a T4 lysozyme cavity mutant. Proc Natl Acad Sci USA. 2015;112(9):E924. doi: 10.1073/pnas.1424002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy J, Hubbell WL. High-pressure EPR reveals conformational equilibria and volumetric properties of spin-labeled proteins. Proc Natl Acad Sci USA. 2011;108(4):1331–1336. doi: 10.1073/pnas.1017877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lerch MT, Yang Z, Brooks EK, Hubbell WL. Mapping protein conformational heterogeneity under pressure with site-directed spin labeling and double electron-electron resonance. Proc Natl Acad Sci USA. 2014;111(13):E1201–E1210. doi: 10.1073/pnas.1403179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: A case study of myoglobin. Biochemistry. 2012;51(33):6568–6583. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4-58 bZip free and bound to DNA. Biochemistry. 2004;43(23):7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 43.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18(5):893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35(24):7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 45.Georgieva ER, Borbat PP, Ginter C, Freed JH, Boudker O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat Struct Mol Biol. 2013;20(2):215–221. doi: 10.1038/nsmb.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bridges MD, Hideg K, Hubbell WL. Resolving conformational and rotameric exchange in spin-labeled proteins using saturation recovery EPR. Appl Magn Reson. 2010;37(1-4):363. doi: 10.1007/s00723-009-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lietzow MA, Hubbell WL. Motion of spin label side chains in cellular retinol-binding protein: Correlation with structure and nearest-neighbor interactions in an antiparallel beta-sheet. Biochemistry. 2004;43(11):3137–3151. doi: 10.1021/bi0360962. [DOI] [PubMed] [Google Scholar]
- 48.Guo Z, Cascio D, Hideg K, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17(2):228–239. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eriksson AE, Baase WA, Wozniak JA, Matthews BW. A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature. 1992;355(6358):371–373. doi: 10.1038/355371a0. [DOI] [PubMed] [Google Scholar]
- 50.Morton A, Baase WA, Matthews BW. Energetic origins of specificity of ligand binding in an interior nonpolar cavity of T4 lysozyme. Biochemistry. 1995;34(27):8564–8575. doi: 10.1021/bi00027a006. [DOI] [PubMed] [Google Scholar]
- 51.Feher VA, Baldwin EP, Dahlquist FW. Access of ligands to cavities within the core of a protein is rapid. Nat Struct Biol. 1996;3(6):516–521. doi: 10.1038/nsb0696-516. [DOI] [PubMed] [Google Scholar]
- 52.Roche J, et al. Remodeling of the folding free energy landscape of staphylococcal nuclease by cavity-creating mutations. Biochemistry. 2012;51(47):9535–9546. doi: 10.1021/bi301071z. [DOI] [PubMed] [Google Scholar]
- 53.Fourme R, Girard E, Akasaka K. High-pressure macromolecular crystallography and NMR: Status, achievements and prospects. Curr Opin Struct Biol. 2012;22(5):636–642. doi: 10.1016/j.sbi.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Kozlikova B, et al. CAVER Analyst 1.0: Graphic tool for interactive visualization and analysis of tunnels and channels in protein structures. Bioinformatics. 2014;30(18):2684–2685. doi: 10.1093/bioinformatics/btu364. [DOI] [PubMed] [Google Scholar]
- 55.Chovancova E, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput Biol. 2012;8(10):e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toleikis Z, Cimmperman P, Petrauskas V, Matulis D. Determination of the volume changes induced by ligand binding to heat shock protein 90 using high-pressure denaturation. Anal Biochem. 2011;413(2):171–178. doi: 10.1016/j.ab.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 57.Toleikis Z, Cimmperman P, Petrauskas V, Matulis D. Serum albumin ligand binding volumes using high pressure denaturation. J Chem Thermodyn. 2012;52(0):24–29. [Google Scholar]
- 58.Royer CA, et al. Effects of amino acid substitutions on the pressure denaturation of staphylococcal nuclease as monitored by fluorescence and nuclear magnetic resonance spectroscopy. Biochemistry. 1993;32(19):5222–5232. doi: 10.1021/bi00070a034. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Z, et al. Multifrequency electron spin resonance study of the dynamics of spin labeled T4 lysozyme. J Phys Chem B. 2010;114(16):5503–5521. doi: 10.1021/jp910606h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.López CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18(8):1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.