Significance
Mutations affecting replicative DNA polymerases δ (Polδ) and ε (Polε) are linked to sporadic and hereditary colorectal cancer and sporadic endometrial cancer in humans. How these mutations promote the genome instability and tumorigenesis is unclear. Here, we deciphered the mechanism of mutagenesis caused by the colon cancer-associated variant Polδ-R696W in a yeast model. It is a previously unappreciated pathway in which erroneous DNA synthesis by Polδ-R696W induces checkpoint-dependent expansion of dNTP pools. The increase in dNTP levels, in turn, causes further dramatic reduction in the fidelity of Polδ-R696W, resulting in more errors and continuous activation of the signaling cascade that keeps the dNTP pools expanded, thus forming a “vicious circle.” This phenomenon may provide insight into the processes shaping the genomes of hypermutated human cancers.
Keywords: DNA polymerase δ, colon cancer, dNTP pools, mutagenesis, DNA replication fidelity
Abstract
Defects in DNA polymerases δ (Polδ) and ε (Polε) cause hereditary colorectal cancer and have been implicated in the etiology of some sporadic colorectal and endometrial tumors. We previously reported that the yeast pol3-R696W allele mimicking a human cancer-associated variant, POLD1-R689W, causes a catastrophic increase in spontaneous mutagenesis. Here, we describe the mechanism of this extraordinary mutator effect. We found that the mutation rate increased synergistically when the R696W mutation was combined with defects in Polδ proofreading or mismatch repair, indicating that pathways correcting DNA replication errors are not compromised in pol3-R696W mutants. DNA synthesis by purified Polδ-R696W was error-prone, but not to the extent that could account for the unprecedented mutator phenotype of pol3-R696W strains. In a search for cellular factors that augment the mutagenic potential of Polδ-R696W, we discovered that pol3-R696W causes S-phase checkpoint-dependent elevation of dNTP pools. Abrogating this elevation by strategic mutations in dNTP metabolism genes eliminated the mutator effect of pol3-R696W, whereas restoration of high intracellular dNTP levels restored the mutator phenotype. Further, the use of dNTP concentrations present in pol3-R696W cells for in vitro DNA synthesis greatly decreased the fidelity of Polδ-R696W and produced a mutation spectrum strikingly similar to the spectrum observed in vivo. The results support a model in which (i) faulty synthesis by Polδ-R696W leads to a checkpoint-dependent increase in dNTP levels and (ii) this increase mediates the hypermutator effect of Polδ-R696W by facilitating the extension of mismatched primer termini it creates and by promoting further errors that continue to fuel the mutagenic pathway.
When functioning properly, DNA replication is phenomenally accurate, making ∼10−10 mutations per base pair during each replication cycle (1). In respect to human biology, it means that, on average, less than one mutation occurs each time the human genome is replicated. This amazing exactitude is contingent upon the serial action of DNA polymerase selectivity, exonucleolytic proofreading, and DNA mismatch repair (MMR). MMR defects increase spontaneous mutagenesis in numerous model systems (2) and give rise to cancer in mice (3). In humans, inherited mutations in MMR genes predispose to colorectal cancer (CRC) in Lynch syndrome (4, 5). Additionally, MMR genes are inactivated via hypermethylation in ∼15% of sporadic CRC, endometrial cancer (EC), and gastric cancer (6).
Like defects in MMR, mutations that decrease the base selectivity or proofreading activity of replicative DNA polymerases elevate spontaneous mutagenesis in eukaryotic cells (7–14) and cancer incidence in mice (15–18). Germline mutations affecting the exonuclease domains of DNA polymerases δ (Polδ) and ε (Polε) cause hereditary CRC (19), and somatic changes in the exonuclease domain of Polε were found in sporadic hypermutated CRC and EC (20–23). Two of these exonuclease domain mutations were modeled in yeast and found to increase mutation rate, supporting their role in the development of hypermutated tumors (19, 24). Because of these discoveries, recent publications emphasized proofreading deficiency as the initiating cause of some human tumors (25–28). The potential role of base selectivity defects in human cancers received much less attention. At the same time, evidence is accumulating that base selectivity defects do occur in tumors and can have dramatic consequences for genome stability. Modeling in yeast of the Polε exonuclease domain mutation most prevalent in CRC and EC produced an increase in the mutation rate far exceeding the increase expected from loss of proofreading, suggesting additional fidelity defects (24). Several studies of colon cancer cell lines and primary tumors reported amino acid changes in Polδ and Polε outside of the exonuclease domain, including some in the conserved DNA polymerase motifs (20, 23, 29). We have previously shown that the yeast analog of one such variant, Polδ-R696W, is an extremely mutagenic DNA polymerase that, notably, retains full exonuclease activity (30). Expression of the pol3-R696W allele was estimated to result in a more than 10,000-fold increase in the mutation rate, which is incompatible with life in both haploid and diploid cells and represents the strongest mutator effect described to date in eukaryotic cells. The corresponding human mutation, POLD1-R689W, was found in two colon cancer cell lines, DLD-1 (29) and HCT15 (31), which were independently derived from the same primary tumor (32), indicating that the R689W change existed in the tumor before the establishment of these cell lines.
To understand how a single amino acid change in Polδ could cause such a phenomenal increase in mutagenesis, we first set out to characterize the fidelity of purified Polδ-R696W during in vitro DNA synthesis. Surprisingly, Polδ-R696W was not dramatically less accurate than previously studied Polδ variants that are only moderately mutagenic in vivo. We found, however, that yeast cells producing Polδ-R696W undergo S-phase checkpoint-dependent expansion of dNTP pools. Simulating this dNTP increase in vitro reduced the fidelity of Polδ-R696W to a level that explains its extraordinary mutator effect in vivo. We present evidence that the mutagenesis in pol3-R696W strains results from a combination of a Polδ nucleotide selectivity defect with activation of the signaling pathway that elevates dNTP pools and further increases the rate of Polδ errors. This vicious circle represents a new mechanism through which DNA polymerase alterations can promote the genome instability and which should be taken into consideration when trying to understand the development of hypermutated human cancers.
Results
Effect of pol3-R696W on the Mutation Rate in Diploid Yeast.
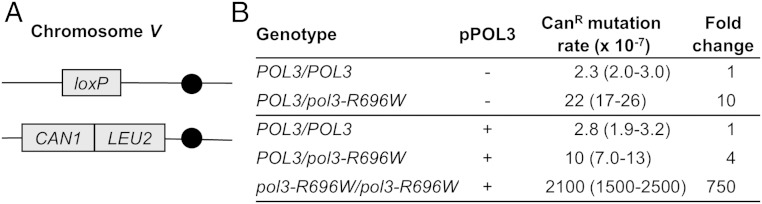
We first set out to quantify the mutator effect of the pol3-R696W allele in diploid cells accurately. Prior studies indicated that measurements in haploid yeast strains grossly underestimate the mutagenic potential of the pol3-R696W, because the majority of mutants also acquire mutations in essential genes and escape detection (30). Accurate evaluation of the mutator effect in diploids, however, was hampered by the fact that the commonly used forward mutation assays involve selection for recessive mutations. These mutations are not detectable in diploids unless the second allele of the reporter gene is also inactivated. To overcome this limitation, we developed an assay to measure the rate of forward mutation at a single-copy CAN1 gene in diploid yeast (Fig. 1A). The CAN1 reporter scores a variety of base substitutions, frameshifts, deletions, and complex events that inactivate the gene as canavanine-resistant (CanR) mutants. In our assay, one CAN1 locus has the LEU2 gene from Kluyveromyces lactis inserted immediately downstream of the CAN1 ORF and the second CAN1 locus is deleted. In nonselective conditions, diploids with a single copy of the CAN1::LEU2 cassette accumulate CanR clones predominantly due to homologous recombination between the CAN1 and the centromere, resulting in loss of the entire locus. Maintenance of cells on media lacking Leu selects against the recombination events and allows us to detect mutations within the CAN1 gene. A similar diploid mutagenesis assay has been independently developed and recently published by Herr et al. (33). Using this assay, we found that the rate of CanR mutation in wild-type diploids, 2.3 × 10−7, was not different from the rate of CanR mutation in haploid wild-type yeast (24) (Tables S1 and S2). The pol3-R696W/pol3-R696W diploids, which expressed a small amount of the wild-type POL3 to suppress the lethality conferred by the pol3-R696W mutation, showed a 750-fold increase in the CanR mutation rate compared with the wild-type strains (Fig. 1B). This analysis confirms that the pol3-R696W is the strongest mutator DNA polymerase variant known in eukaryotes and provides an accurate estimate of its mutator effect.
Fig. 1.
Mutator effects of the pol3-R696W allele in diploid yeast strains. (A) Structure of chromosome V in diploid strains used in the measurement of CanR mutation rate. The filled circles (●) designate centromeres. The strains have a deletion of the CAN1 ORF and 100 bp of flanking DNA in one copy of chromosome V and an insertion of K. lactis LEU2 immediately downstream of CAN1 in the homologous chromosome. Propagation of can1Δ/CAN1::LEU2 strains on media lacking Leu selects against cells that lose CAN1 via mitotic recombination and allows for selection of cells with intragenic CAN1 mutations. (B) Rate of CanR mutation in diploids with the chromosome V configuration shown in A. Full genotypes of yeast strains are described in Table S4. Mutation rates are medians for at least 18 cultures, with the 95% confidence limits shown in parentheses. The fold change in the mutation rate is relative to the wild-type diploid strain in each section.
The single-copy CAN1 assay also allowed us to determine the effect of heterozygosity for the pol3-R696W allele on the mutation rate. Sequencing of the DLD-1 and HCT15 cell lines indicated that the tumor from which they were derived was heterozygous for the POLD1-R689W (29, 31). Further, no evidence of loss of heterozygosity was reported for any other POLD1 or POLE mutation found in sporadic human tumors (discussed in ref. 25). To understand the potential contribution of these polymerase variants to tumor development, it is important to determine if the heterozygosity is sufficient to elevate the mutation rate. In the single-copy CAN1 assay, the CanR mutation rate was elevated 10-fold in POL3/pol3-R696W heterozygotes (Fig. 1B), consistent with the proposed pathogenic role of the analogous human mutation. As observed previously (30), the mutator effect was highly sensitive to the relative amount of the normal and mutant polymerases: the addition of pPOL3 plasmid expressing the wild-type POL3 significantly reduced the mutation rate (Fig. 1B).
Errors Made by Polδ-R696W Are Corrected by Exonucleolytic Proofreading and MMR.
The base selectivity of replicative DNA polymerases, proofreading, and MMR act in series to avoid replication errors and reduce the likelihood of mutation (34). Defects in any of these individual steps elevate the mutation rate but have not been previously reported to cause inviability. However, error-induced extinction could readily be achieved if two of the replication fidelity mechanisms are simultaneously inactivated (33–37). Therefore, we asked whether the pol3-R696W cells were incapable of correcting Polδ-R696W errors by the proofreading activity of Polδ and/or MMR. We reasoned that if the correction mechanisms are functional in the pol3-R696W mutants, combining the R696W substitution with a mutation inactivating the exonuclease activity of Polδ or MMR would result in a synergistic increase in the mutation rate. In contrast, if the single pol3-R696W mutants were deficient in proofreading or MMR, no further increase in mutagenesis would occur when additional mutations inactivating the Polδ exonuclease or MMR are introduced. It is also formally possible that the mutator effect of pol3-R696W results from mismatches not susceptible to MMR, such as mismatches generated outside of the replication fork. In this case, an additive interaction of the mutator effects of pol3-R696W and MMR deficiency would be expected.
We first combined pol3-R696W with a deletion of the MSH6 gene in haploid yeast. The msh6 mutation disrupts the MutSα-dependent branch of MMR that is responsible for the repair of base/base mismatches (38). Consistent with previously reported results (30, 39), single pol3-R696W and msh6Δ mutations increased the CanR mutation rate 23-fold and 13-fold, respectively. Combining msh6Δ and pol3-R696W led to a synergistic (580-fold) increase in the CanR mutation rate (Fig. 2A). The same synergistic interaction was observed when heterozygosity for the pol3-R696W was combined with MSH6 deficiency in diploid strains (200-fold increase in mutagenesis in the double mutant vs. 9.6-fold and 18-fold increases in the POL3/pol3-R696W and msh6Δ/msh6Δ diploids, respectively; Fig. 2B). These results indicate that (i) MMR is not impaired by the pol3-R696W mutation and (ii) mispairs created by Polδ-R696W are subject to correction by MMR, and therefore represent DNA replication errors. Importantly, the human R689W Polδ variant was found as a heterozygous mutation in cell lines lacking functional MSH6 (31, 40). The synergy we observed between heterozygosity for pol3-R696W and the loss of MSH6 in diploids suggests that the two defects likely synergized in a similar manner to promote genome instability in the human tumor. Indeed, the HCT15 cell line is highly hypermutated, with the number of genetic changes far exceeding the mutation load of MMR-deficient cancer cell lines without DNA polymerase mutations (31).
Fig. 2.
Mistakes made by Polδ-R696W are corrected by MMR and the proofreading function of Polδ-R696W in vivo. (A) Combining the pol3-R696W allele with a defect in MMR leads to a synergistic increase in mutagenesis in haploid yeast. The mutation rates were determined for haploid strains carrying chromosomal pol3-R696W and/or msh6Δ mutations and plasmid pPOL3. (B) Combining the pol3-R696W mutation with a defect in MMR leads to a synergistic increase in mutagenesis in diploid yeast. The mutation rates were measured in diploid strains with a single copy of CAN1 (Fig. 1A) and no pPOL3. (C) Combining the R696W with an amino acid substitution in the exonuclease domain of Polδ (the pol3-5DV mutation) leads to a synergistic increase in mutagenesis in haploid yeast. The mutation rates were determined in wild-type haploids expressing wild-type or mutant POL3 variants from plasmid pPOL3-GST. The fold change in the mutation rate relative to the wild-type strain is displayed above the bars. Mutation rates are medians for at least 18 cultures. Error bars denote the 95% confidence intervals. All data are from Tables S1 and S2. Strain genotypes are described in Table S4.
Next, we tested a possibility that the extreme mutator effect of pol3-R696W results from defects in both base selectivity and proofreading conferred by this single mutation. It has been shown that a combination of two Polδ mutations, one affecting selectivity and another affecting proofreading, could produce high levels of mutagenesis incompatible with survival (33). It was also suggested, based on crystallographic studies of yeast Polδ, that the R696W change may affect proofreading (41). We previously observed that Polδ-R696W and wild-type Polδ have comparable 3′→5′ exonuclease activity in vitro (30), but the ability of Polδ-R696W to proofread its errors in vivo has never been tested. To address this issue, we created a double-mutant version of Polδ that combined the R696W change with the 5DV mutation eliminating the exonuclease activity (12). The mutator effect of the pol3-5DV,R696W allele was compared with the mutator effects of single pol3-R696W and pol3-5DV mutations when the variants were expressed from plasmid pPOL3.GST in wild-type strains. Under these conditions, pol3-R696W and pol3-5DV expression elevated the CanR mutation rate 79-fold and 5.2-fold, respectively. Expression of the pol3-5DV,R696W allele resulted in a synergistic (410-fold) increase in mutagenesis (Fig. 2C). This finding demonstrated that the vast majority of mispairs created by Polδ-R696W in vivo are corrected by the intrinsic proofreading activity of Polδ-R696W.
Spectrum of Mutations Generated by Polδ-R696W in Vivo Is Dominated by GC→AT Transitions.
To understand the mutational specificity of Polδ-R696W–driven DNA synthesis in vivo, we sequenced the CAN1 gene from 189 independent CanR mutants of msh6Δ pol3-R696W haploid strains containing pPOL3. Because the majority of Polδ-R696W errors are corrected by MMR, mutation spectrum analysis in the msh6Δ background allowed us to eliminate potential bias resulting from more efficient repair of certain mismatches. In addition, because the human POLD1-R689W mutation was found in an MSH6-deficient tumor, the yeast msh6Δ pol3-R696W strains provide a model to understand the mutational processes acting in tumors. Of the 189 CanR mutants analyzed, 188 contained base pair substitutions and one contained a frameshift mutation in the CAN1 gene (Table 1). Fourteen of the 189 mutants had two different base pair substitutions in CAN1 separated by distances from 4 to 1,345 nt, further indicating that the msh6Δ pol3-R696W strains experience extraordinary levels of mutagenesis. The vast majority of mutations (73%) were GC→AT transitions (Table 1). As a percentage of total mutations, GC→AT transitions were twofold to threefold more common in the msh6Δ pol3-R696W than in the previously published mutational spectra of wild-type and msh6Δ yeast strains (35, 39, 42). The vast majority of the 139 GC→AT transitions appear as G→A changes in the nontranscribed strand, and most occur in ∼20 hotspots spread throughout the CAN1 ORF (Fig. S1).
Table 1.
Spontaneous can1 mutations in msh6Δ pol3-R696W yeast strains
| Mutation | No. of detectable mutations | Mutation rate (×10−7)* | Percentage of total |
| Base substitutions | |||
| GC to AT | 139 | 1,100 | 73 |
| GC to TA | 24 | 190 | 12 |
| GC to CG | 1 | 8.0 | 0.5 |
| AT to GC | 14 | 110 | 8 |
| AT to CG | 4 | 32 | 2 |
| AT to TA | 7 | 56 | 4 |
| −1 frameshift | 1 | 8.0 | 0.5 |
| Total | 190 | 1,500 | 100 |
The data are based on DNA sequence analysis of 189 independent CanR mutants of msh6Δ pol3-R696W strains containing pPOL3. Fourteen mutants contained two different point mutations in the CAN1 gene. For these mutants, only mutations previously reported to produce a detectable CanR phenotype are shown; the others were identified as likely hitchhiker mutations and were excluded from the mutation rate calculation. The location of mutations in the CAN1 sequence is shown in Fig. S1.
The rate for each type of mutation was calculated as follows: MRi = (Mi/MT) × MR, where Mi is the number of mutations of the particular type, MT is the total number of mutations, and MR is the rate of CanR mutation in msh6Δ pol3-R696W strains containing pPOL3 as determined by fluctuation analysis (1,500 × 10−7; Table S1).
Hypermutator Effect of pol3-R696W Is Mediated by a Dun1-Dependent Elevation of dNTP Pools.
Because the Polδ proofreading and MMR are both functional in the pol3-R696W cells, we deduced that the strong mutator effect of the R696W substitution likely results from severely reduced nucleotide selectivity of the enzyme. Using primer extension assays with oligonucleotide templates, we have previously shown (30) that Polδ-R696W does indeed have a profound ability to misinsert nucleotides, at least in one particular DNA sequence context used in that study. To understand better the extent to which the nucleotide selectivity of Polδ is impaired by the R696W substitution, we set out to characterize the fidelity of DNA synthesis by Polδ-R696W in vitro using the M13mp2 lacZ forward mutation assay (43). The assay involves filling a 407-nt single-stranded gap in a double-stranded M13mp2 DNA molecule by Polδ-R696W in vitro and detection of nucleotide changes introduced during copying of the lacZ sequence by genetic selection in Escherichia coli. The assay can detect all types of base substitutions in a variety of DNA sequence contexts, as well as frameshift mutations, large deletions, and more complex changes. Surprisingly, at protein and nucleotide concentrations typically used in this assay, synthesis by Polδ-R696W did not show a higher error rate than what was previously observed for Polδ-5DV, a much weaker mutator in vivo (44) (Table 2). To explain this unexpected result, we hypothesized that there must be factor(s) in vivo that contribute to mutagenesis in pol3-R696W strains and that are not present in DNA synthesis reactions with purified proteins. For example, increased incorporation of noncanonical nucleotides by Polδ-R696W might contribute to the high levels of mutagenesis observed in vivo. We tested the ability of Polδ-R696W to incorporate several nucleotide analogs in primer extension assays. These analogs included 8-oxo-dGTP and 6-Me-dGTP, which are common mutagenic products of oxidation and methylation damage, respectively; natural contaminants of intracellular dNTP pools 2′-deoxyinosine 5′-triphosphate (dITP) and 2′-deoxyxanthosine 5′-triphosphate; and model mutagenic nucleotide analogs 2′-deoxy-6-hydroxylaminopurine 5′-triphosphate and 2′-deoxy-2-amino-6-hydroxylaminopurine 5′-triphosphate (dAHAPTP). Although Polδ-R696W was slightly more efficient at incorporating dAHAPTP and dITP than wild-type Polδ, no major differences between the two enzymes were found (Fig. S3).
Table 2.
Fidelity of in vitro DNA synthesis by wild-type Polδ and Polδ-R696W
| WT Polδ (100 μM dNTPs) | Polδ-R696W (100 μM dNTPs) | WT Polδ (S-phase dNTPs) | Polδ-R696W (S-phase dNTPs) | |||||
| No. | ER (×10−5) | No. | ER (×10−5) | No. | ER (×10−5) | No. | ER (×10−5) | |
| Base substitutions (mispair) | 32 | 1.4 | 77 | 5.1 | 34 | 1.3 | 118 | 15 |
| Transitions | 25 | 1.5 | 39 | 3.5 | 22 | 1.2 | 79 | 13 |
| A→G (A-dCTP) | 0 | <0.24 | 3 | 1.1 | 1 | 0.21 | 1 | 0.67 |
| G→A (G-dTTP) | 3 | 0.92 | 4 | 1.8 | 9 | 2.5 | 45 | 39 |
| C→T (C-dATP) | 18 | 4.0 | 20 | 6.8 | 8 | 1.6 | 15 | 9.4 |
| T→C (T-dGTP) | 4 | 0.82 | 12 | 3.7 | 4 | 0.73 | 18 | 10 |
| Transversions | 7 | 0.26 | 38 | 2.2 | 12 | 0.40 | 39 | 4.1 |
| A→C (A-dGTP) | 0 | <0.34 | 2 | 1.0 | 2 | 0.60 | 1 | 0.94 |
| A→T (A-dATP | 3 | 0.67 | 12 | 4.1 | 2 | 0.40 | 6 | 3.8 |
| C→A (C-dTTP) | 0 | <0.29 | 4 | 1.8 | 0 | <0.26 | 9 | 7.4 |
| C→G (C-dCTP) | 0 | <0.45 | 0 | <0.68 | 0 | <0.40 | 0 | <1.3 |
| G→C (G-dGTP) | 3 | 0.84 | 4 | 1.7 | 5 | 1.2 | 5 | 3.9 |
| G→T (G-dATP) | 1 | 0.27 | 10 | 4.1 | 3 | 0.72 | 13 | 10 |
| T→A (T-dTTP) | 0 | <0.40 | 3 | 1.8 | 0 | <0.35 | 3 | 3.3 |
| T→G (T-dCTP) | 0 | <0.27 | 3 | 1.2 | 0 | <0.24 | 2 | 1.5 |
| Frameshifts | 15 | 0.50 | 41 | 2.1 | 8 | 0.24 | 38 | 3.5 |
| Minus 1 | 15 | 0.50 | 40 | 2.0 | 6 | 0.18 | 38 | 3.5 |
| Plus 1 | 0 | <0.03 | 0 | <0.05 | 2 | 0.06 | 0 | <0.09 |
| Large deletions | 1 | 0 | 1 | 3 | ||||
| Total | 48 | 118 | 43 | 159 | ||||
| lacZ mutant frequency | 0.0019 | 0.0071 | 0.0015 | 0.017 | ||||
The S-phase dNTP concentrations used for wild-type Polδ and Polδ-R696W are the same as calculated for wild-type and pol3-R696W + pPOL3 yeast strains (Table 3). All lacZ mutants recovered from reactions with wild-type Polδ contained single mutations. Sixteen percent of mutants obtained from reactions with Polδ-R696W contained more than one mutation. Only phenotypically detectable mutations are listed. Error rates (ERs) for individual mutation types were calculated as described in Materials and Methods. Silent mutations were excluded from the error rate calculation. The location of mutations in the lacZ sequence is shown in Fig. S2, along with the description of large deletions and other rare mutations. The background mutation frequency for unfilled M13mp2 gapped substrate was 0.0016.
We also used a candidate approach to determine what proteins and pathways might be required for mutagenesis caused by pol3-R696W. We transformed selected yeast mutants defective in DNA repair or cell cycle checkpoint pathways with plasmid pPOL3.GST-R696W and performed a semiquantitative assessment of mutation rates by patch tests as described by Daee et al. (30). In this candidate screen, we found that inactivation of the checkpoint kinase Dun1 largely suppressed mutagenesis caused by pol3-R696W expression. Quantitative analysis of the mutation rate showed that the mutator effect of pPOL3.GST-R696W was reduced from 79-fold to only 2.5-fold in dun1Δ strains (Table S1). To verify the suppressive effect of DUN1 deletion, we measured the rate of CanR mutation in DUN1+ and dun1Δ strains expressing pol3-R696W from the natural chromosomal POL3 locus. Similar to the results obtained with ectopic pol3-R696W expression, the mutator effect of pol3-R696W was completely eliminated in the dun1Δ strain (Fig. 3A). In contrast, deletion of DUN1 led to a minor 1.5-fold increase in the CanR mutation rate in the wild-type strain, indicating that the suppressive effect was specific to the Polδ mutator.
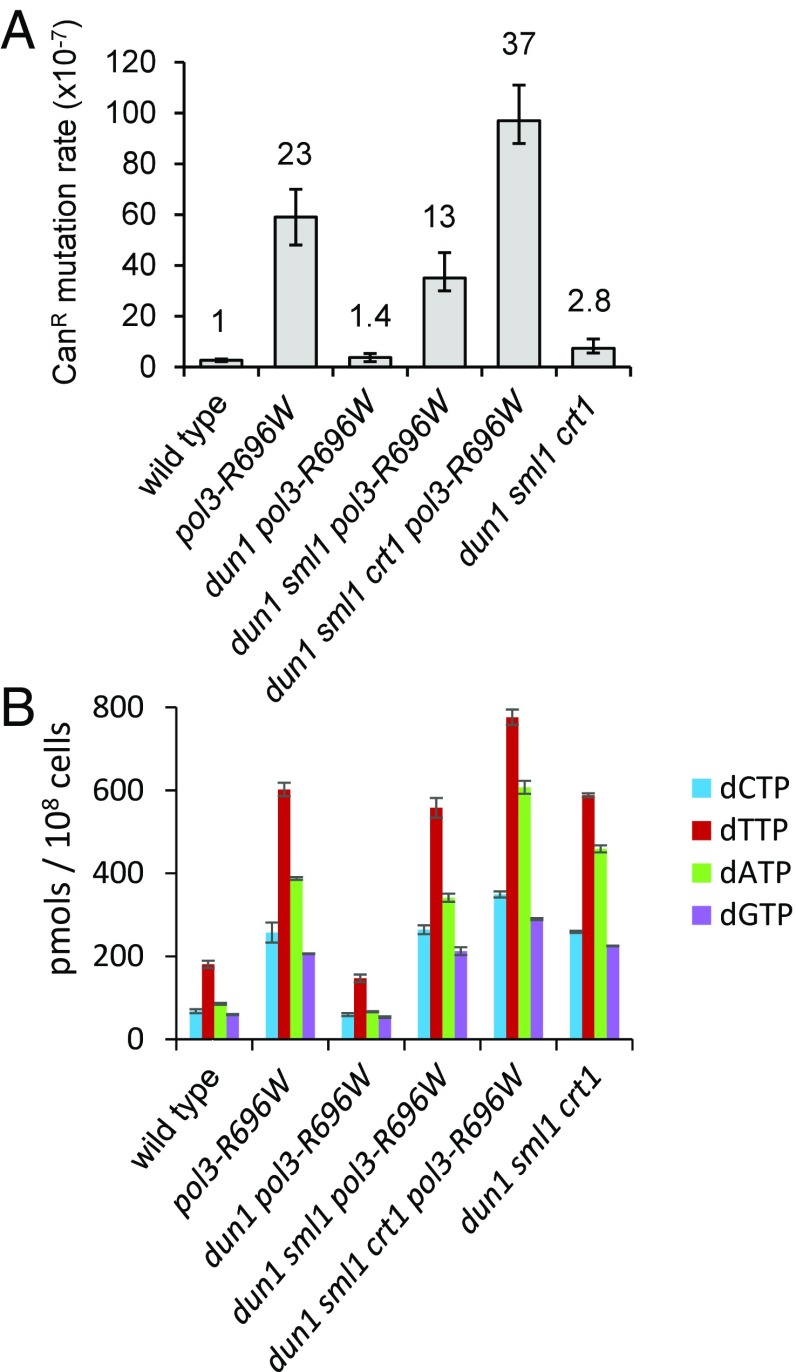
Fig. 3.
Elevated spontaneous mutagenesis in yeast strains expressing pol3-R696W requires a DUN1-dependent increase in dNTP levels. (A) Mutations in the RNR regulation pathway modulate the mutator effect of pol3-R696W. Mutation rates are medians for at least 18 cultures. The fold change in the mutation rate relative to the wild-type strain is displayed above the bars. Error bars denote the 95% confidence intervals. The data are from Table S1. (B) Intracellular dNTP levels in strains assayed in A. The dNTP concentrations were measured in asynchronous logarithmic phase cultures, as described in Materials and Methods. The values are the mean and SE of three measurements, each performed with an independently created isolate. All strains assayed in A and B are haploids containing pPOL3.
The Dun1 kinase is activated upon DNA damage or replication stress as part of a signaling cascade that includes Mec1 and Rad53 (45), the homologs of human ATR and Chk1, respectively. During the damage response, activated Dun1 increases ribonucleotide reductase (RNR) activity (46), which results in an elevation of dNTP levels (47). This dNTP pool increase is important for survival and mutagenesis after exposure to DNA damaging agents (47, 48). Dun1 has also been reported to promote expansion of dNTP pools in response to mutations that impair DNA replication (49). Considering these findings and the suppression of mutagenesis in pol3-R696W strains by the DUN1 deletion, we hypothesized that the mutator effect of pol3-R696W requires a Dun1-dependent increase in dNTP pools. To test this hypothesis, we first determined if dNTP pools are elevated in the pol3-R696W strains in a Dun1-dependent manner. Indeed, we found that the dNTP levels measured in logarithmically growing haploid pol3-R696W mutants were ∼3.8-fold higher compared with wild-type strains, with the increases ranging from 3.3- to 4.5-fold for individual dNTPs (Fig. 3B). In contrast, the dNTP levels in dun1Δ pol3-R696W haploids were indistinguishable from the dNTP levels in the wild-type strains (Fig. 3B).
Regulation of RNR activity by Dun1 occurs by at least three separate mechanisms (50–52). One of these mechanisms involves Sml1, a small protein that binds to and inhibits RNR (50). Upon DNA damage, activated Dun1 phosphorylates Sml1, triggering its degradation (53). Consistent with a Dun1-dependent elevation of dNTP pools, we found that the level of Sml1 was decreased in pol3-R696W strains, albeit to a smaller extent than in cells arrested by exposure to the DNA-damaging agent methyl methanesulfonate (Fig. S4A). There was, however, no detectable Rad53 phosphorylation in pol3-R696W strains (Fig. S4B). It is likely that only a very small amount of Rad53 and/or a few of the many sites within Rad53 (54) are phosphorylated, consistent with the absence of growth arrest in these strains and a moderate elevation of the dNTP pools. In comparison, the full activation of the checkpoint by DNA damaging agents leads to a sixfold to eightfold increase in dNTP levels (47). Other studies similarly reported that Rad53 phosphorylation is often not detectable in cells with chronic or low checkpoint activation even though effects of downstream signaling are apparent (49, 55, 56). Due to the signal amplification by the kinase cascade, the levels of the downstream targets, such as Sml1 or the RNR subunits (46, 57), are much more sensitive indicators of the checkpoint activation.
Next, because both dNTP pools and mutagenesis are elevated in pol3-R696W strains in a Dun1-dependent manner, we investigated whether mutagenesis in pol3-R696W strains was, in fact, modulated by dNTP levels. Although dun1Δ mutants are not able to expand dNTP pools in response to the pol3-R696W mutation (Fig. 3B), the dNTP levels can be increased in dun1Δ strains by deleting the SML1 gene (58). RNR activity is also modulated by Crt1, a transcriptional repressor that binds to and inhibits transcription from RNR2, RNR3, and RNR4 promoters (52). Deletion of CRT1 further increases dNTP pools in sml1Δ strains (58). We found that deletion of SML1 in dun1Δ pol3-R696W mutants restored dNTP pools to the levels observed in pol3-R696W strains (Fig. 3B) and, concomitantly, brought back the mutator phenotype (Fig. 3A). In quadruple crt1Δ dun1Δ sml1Δ pol3-R696W mutants, both the dNTP pools and the mutation rate were further increased and exceeded the levels observed in single pol3-R696W mutants (Fig. 3). These observations provide strong support for the idea that the mutator effect of pol3-R696W depends on dNTP pool elevation. Furthermore, although dNTP levels in crt1Δ dun1Δ sml1Δ strains with wild-type POL3 and in single pol3-R696W mutants were similar (Fig. 3B), the mutation rate was increased only 2.8-fold in crt1Δ dun1Δ sml1Δ strains (Fig. 3A). Thus, synthesis by Polδ-R696W at the high dNTP concentrations is responsible for the dramatic increase in mutagenesis in pol3-R696W strains, and not the high dNTP levels per se. This finding is consistent with earlier observations that in contrast to the strong mutagenic effect of dNTP pool imbalances (59), proportional elevation of all four dNTPs only mildly affects spontaneous mutagenesis in yeast (47).
Fidelity and Error Specificity of Polδ-R696W in Vitro.
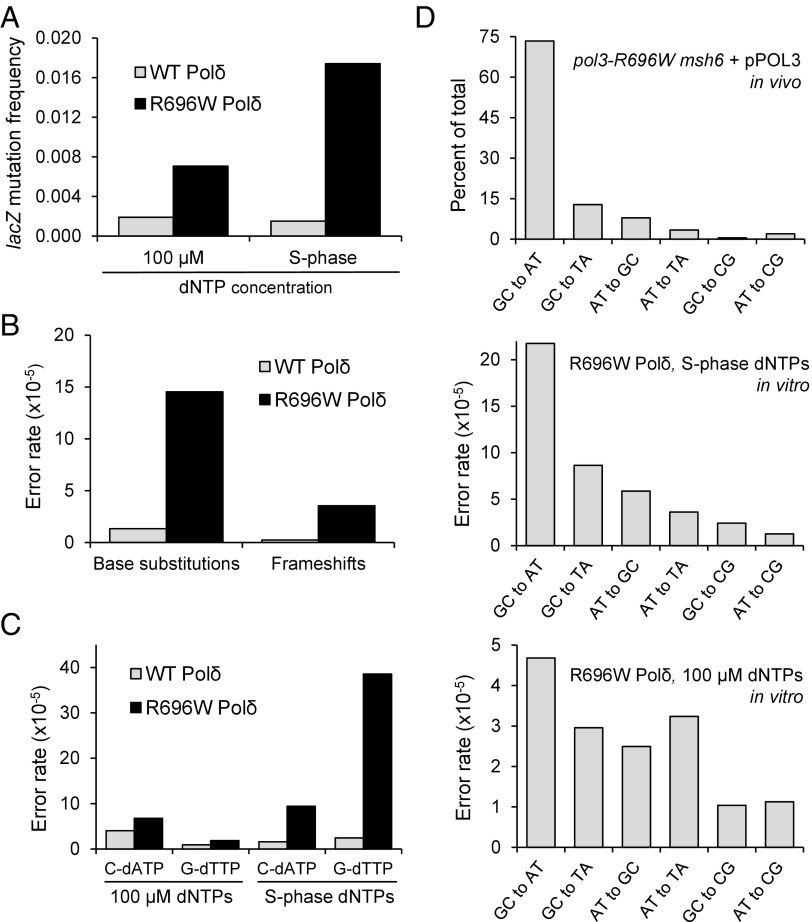
The experiments described in the previous section provided convincing evidence that the strong mutator effect of pol3-R696W requires elevated intracellular dNTP levels. Next, we sought to determine if the low fidelity and error specificity of Polδ-R696W could be recapitulated in vitro by adjusting dNTP concentrations to match the dNTP levels present during DNA replication in pol3-R696W cells. For comparison, we also estimated the fidelity of the wild-type Polδ at the dNTP concentrations present in wild-type yeast. The S-phase dNTP concentrations were calculated using measurements of dNTP levels in wild-type and pol3-R696W haploids containing pPOL3, as described in Table 3. Similar to other studies (8, 44), the frequency of mutations resulting from synthesis by wild-type Polδ was not significantly different from the background mutation frequency for unfilled M13mp2 gapped substrate, regardless of whether the S-phase or standard 100 μM dNTP concentration was used (Table 2). In contrast, DNA synthesis by Polδ-R696W was highly error-prone. The fold increase in the lacZ mutation frequency over the wild-type Polδ reactions grew from 3.7-fold to 11-fold when dNTP concentrations were changed from 100 μM to the concentrations estimated to be present in the corresponding yeast strains (Fig. 4A and Table 2). Rates of base substitution and frameshift errors at the S-phase dNTP concentrations were at least 12-fold and 15-fold higher for Polδ-R696W compared with the wild-type Polδ (Fig. 4B and Table 2). These numbers likely underestimate the difference in the fidelity of the two enzymes, because the error rate for the wild-type Polδ could be well below the background level of mutation in the lacZ M13mp2 assay. The R696W substitution increased the Polδ error rate for every type of base substitution with the possible exception of C to G, which was not present in either mutation spectrum (Table 2). As observed for other replicative polymerase variants, Polδ-R696W generated base substitutions far more frequently than frameshifts, in agreement with the predominance of base substitutions in the in vivo mutation spectrum of msh6Δ pol3-R696W strains (Table 1).
Table 3.
Estimated S-phase dNTP concentrations in the wild-type and pol3-R696W yeast strains
| Genotype | dCTP, μM | dTTP, μM | dATP, μM | dGTP, μM |
| POL3 + pPOL3 | 30 | 80 | 38 | 26 |
| pol3-R696W + pPOL3 | 114 | 266 | 171 | 91 |
S-phase dNTP concentrations were calculated from experimentally determined dNTP levels (picomoles per 108 cells) as in the study by Nick McElhinny et al. (91) using a cell volume estimate of 45 μm3 and assuming that dNTP levels in the S phase are approximately twice as high as the average levels measured in asynchronous logarithmically growing cells (47).
Fig. 4.
Use of in vivo dNTP concentrations for in vitro DNA synthesis reveals the infidelity, error rate asymmetry, and error specificity of Polδ-R696W. (A) lacZ mutation frequencies resulting from in vitro DNA synthesis by wild-type Polδ and Polδ-R696W. (B) Rates of single-base substitutions and single-base frameshifts generated by wild-type Polδ and Polδ-R696W at estimated S-phase dNTP concentrations. (C) Error rates for reciprocal mispairs that result in GC→AT transitions at 100 μM dNTPs and the S-phase dNTP concentrations. (D) Comparison of base substitution spectrum of the msh6Δ pol3-R696W yeast strain and the spectra of errors generated by Polδ-R696W in vitro. (Top) Proportions of individual base substitutions in the CAN1 gene of the msh6Δ pol3-R696W yeast strain containing pPOL3. (Middle and Bottom) Rates of base substitutions generated by Polδ-R696W in vitro at the S-phase dNTP concentrations and 100 μM dNTPs, respectively.
Remarkably, in addition to increasing the overall error rate of Polδ-R696W, the use of physiologically relevant dNTP concentrations dramatically affected the spectrum of Polδ-R696W errors and made it resemble the in vivo mutation spectrum of msh6Δ pol3-R696W strains very closely (Fig. 4D, Top and Middle). In contrast, the error specificity of Polδ-R696W derived from reactions with equimolar (100 μM) dNTPs bears much less similarity to the in vivo spectrum (Fig. 4D, Top and Bottom). A particularly striking effect of dNTP concentrations was observed for the two reciprocal mispairs G-dTTP and C-dATP, which lead to GC→AT transitions (Fig. 4C and Table 2). These transitions constitute the majority of mutations in the msh6Δ pol3-R696W strains (73%). At 100 μM dNTPs, neither of the two mispairs was generated by Polδ-R696W at a significantly higher rate than the rate observed with the wild-type Polδ. The use of S-phase dNTPs increased the rate of G-dTTP mispair 22-fold, making it by far the most frequent error in the Polδ-R696W spectrum, consistent with the in vivo mutational specificity of pol3-R696W. In contrast, the rate of C-dATP errors changed only slightly when 100 μM dNTPs were replaced with S-phase dNTPs, although Polδ-R696W still generated this mispair sixfold more frequently than the wild-type Polδ at its respective S-phase dNTP concentrations. The greatly higher rate of G-dTTP mispairs suggested that incorrect T incorporation opposite G is the main source of Polδ-R696W–induced mutations in vivo. We also attempted to increase dNTP levels in the gap-filling reactions threefold over the S-phase concentrations shown in Table 3 while preserving the correct ratio of individual dNTPs. Although this change further increased both the rate of Polδ-R696W errors and the proportion of lacZ clones with multiple mutations (Table S3), the similarity to the in vivo msh6Δ pol3-R696W spectrum became less pronounced. The deviations from the in vivo spectrum included a decreased proportion of GC→AT transitions and a high proportion of AT→TA transversions. Thus, adequate dNTP levels were critical for recapitulating the properties of the mutator Polδ variant in vitro, and the concentrations shown in Table 3 must mimic the in vivo dNTP levels rather closely.
Therefore, the base substitution error rate of 1.5 × 10−4 derived from Polδ-R696W reactions with S-phase dNTP concentrations provides a good estimate of Polδ fidelity in pol3-R696W mutant cells. We used this value to examine whether the Polδ-R696W error rate is sufficient to explain the lethal levels of mutagenesis observed in pol3-R696W strains. Data presented in Fig. 2B imply that MMR corrects ∼95% of Polδ-R696W errors, bringing the accuracy of DNA synthesis in pol3-R696W cells to 7.5 × 10−6 errors per nucleotide. When multiplied by the average size of the yeast ORF (1,385 nucleotides) and by the estimated average fraction of mutations (0.3) that produce a detectable phenotype (60), this number translates into 3.1 × 10−3 phenotypically detectable mutations per gene in each replication cycle. With ∼1,200 yeast genes being essential for growth under standard laboratory conditions, pol3-R696W haploids are expected to accumulate 3.7 lethal mutations per cell division. Diploid cells can tolerate a 10-fold higher mutational load than haploids (33), so only every third division of the homozygous pol3-R696W/pol3-R696W diploid cells would be accompanied by a lethal mutation according to these calculations. It is reasonable to suggest, however, that such a high rate of lethal mutations would be incompatible with long-term strain survival. The in vitro error rate of Polδ-R696W at the S-phase dNTP concentrations is thus in good agreement with the theoretically calculated rate of mutation needed to extinguish a cell population. This finding provides strong support for the idea that the infidelity of Polδ-R696W promoted by the expansion of the dNTP pools is the primary cause of lethality in pol3-R696W mutants.
Discussion
Polδ-R696W Promotes Its Own Infidelity by Expansion of dNTP Pools.
Recent reports implicating POLE and POLD1 mutations in the etiology of human cancers (19–24, 31, 61) revived the interest in the mechanisms through which replicative DNA polymerase variants elevate genome instability. A widely held belief is that the hypermutability of tumors with POLE or POLD1 alterations merely results from an increased rate of nucleotide misincorporation by the mutant polymerases. In dissonance with this simplistic view, we show here that mutagenesis induced by the cancer-associated variant Polδ-R696W in yeast occurs by a multifaceted process. We found that (i) the mutator effect of pol3-R696W requires checkpoint-dependent elevation of intracellular dNTP pools, (ii) restoration of the high dNTP levels in checkpoint-deficient strains is sufficient to restore the mutator effect of pol3-R696W, (iii) the increase in dNTP levels greatly reduces the fidelity of DNA synthesis by Polδ-R696W, and (iv) in vitro DNA synthesis by Polδ-R696W at dNTP concentrations present in pol3-R696W strains shows an error rate and specificity that fully explains the mutator properties of pol3-R696W in vivo.
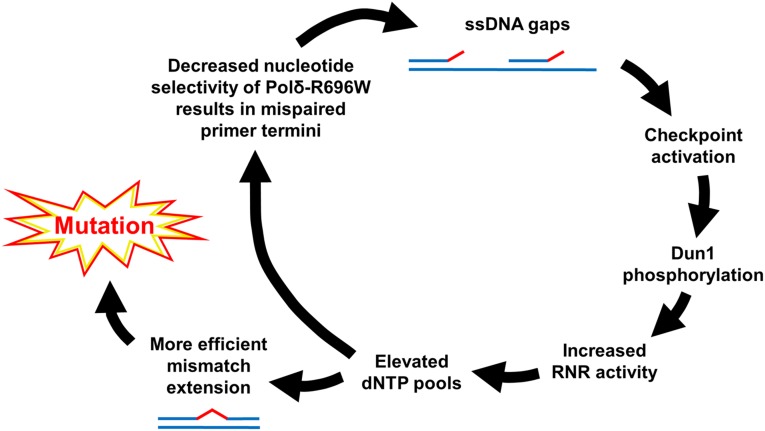
These observations led us to propose the following model (Fig. 5), in which Polδ-R696W increases its own infidelity by expansion of dNTP pools. We postulate that DNA synthesis by Polδ-R696W leads to an accumulation of incomplete replication intermediates. The incomplete replication intermediates could result from (i) pausing and/or dissociation of Polδ-R696W caused by mismatched primer termini, which it generates but does not extend efficiently at low dNTP levels, and (ii) suboptimal levels of Polδ activity. We previously reported that the R696W substitution moderately reduces the DNA polymerase activity of Polδ in primer extension assays (30), adding credence to the second idea. Slow DNA replication and incomplete replication intermediates, such as ssDNA gaps, are known triggers of the DNA damage checkpoint response, leading to sequential Dun1 activation, up-regulation of RNR activity, and elevated dNTP pools. We propose that at higher dNTP concentrations, Polδ-R696W more readily extends mismatched primer termini that it creates and is more likely to make further misinsertions. In addition to the fidelity measurements shown in Table 3 and Table S3, these last assumptions are supported by classic biochemical experiments showing that replicative DNA polymerases poorly extend mismatched primer termini, and that the efficiency of mismatch extension increases with dNTP concentrations (62).
Fig. 5.
Vicious circle model for mutagenesis caused by Polδ-R696W. Mismatched primer termini resulting from Polδ-R696W errors are inefficiently extended and beget an increased number of incomplete replication intermediates. These intermediates trigger a checkpoint response that involves Dun1-dependent up-regulation of RNR and subsequent elevation of intracellular dNTP pools. The increased dNTP levels facilitate extension of the mismatched primer termini created by Polδ-R696W, thereby stimulating mutagenesis. At the same time, expanded dNTP pools further increase the frequency of misincorporations that continue to fuel the mutagenic pathway.
Implications for Analysis of DNA Polymerase Signatures.
It is well documented that dNTP pool imbalances affect the rate and spectrum of spontaneous mutation, presumably by driving preferential incorporation of the nucleotide(s) in excess (59, 63). The potential effects of proportional increases or decreases in dNTP levels on DNA polymerase fidelity could also be predicted from kinetic considerations and have been demonstrated in a few biochemical studies (64–66). Significant stimulation of mutagenesis in E. coli by proportional dNTP increases has been reported (67, 68). Less recognized are the differences in the mutational specificity of DNA polymerases that could result from using a physiological concentration of each dNTP in the in vitro reactions as opposed to commonly used equimolar dNTPs. Two studies used physiological dNTP concentrations to explore the mechanism of DNA lesion bypass by replicative and TLS polymerases (69, 70). The effects of a proper dNTP ratio during processive copying of undamaged DNA, however, have never been investigated. The present work is illuminating in this respect. The S-phase dNTP concentrations estimated for the pol3-R696W strains and used for Polδ-R696W synthesis in vitro (114 μM, 266 μM, 171 μM, and 91 μM for dCTP, dTTP, dATP, and dGTP, respectively) may not seem radically different from standard 100 μM dNTPs. The use of the S-phase dNTP concentrations, however, dramatically changed both the rate and specificity of Polδ-R696W errors. Moreover, it made the spectrum of Polδ-R696W errors closely resemble the in vivo specificity of the pol3-R696W mutator (Fig. 4D). Analysis of the error rates for individual mispairs generated at physiological dNTP concentrations allowed us to determine that G-dTTP, and not C-dATP, mispairs are largely responsible for GC→AT transitions, which are the majority of spontaneous mutations in the pol3-R696W strains (Fig. 4C). This conclusion could not be derived from experiments with 100 μM dNTPs. In fact, the rate of neither G-dTTP nor C-dATP errors was significantly elevated in Polδ-R696W reactions with 100 μM dNTPs, in sharp disagreement with the abundance of GC→AT transitions in vivo. These results emphasize the importance of simulating relevant in vivo dNTP concentrations in in vitro studies of DNA polymerase signatures. These signatures are now used extensively and will likely be used even more in the future to understand the mutational processes operating in vivo in normal and pathological conditions, including cancer-associated hypermutation (71). As evident from the present study, it is imperative to take into account changes in dNTP levels that are induced by DNA damage, by the variant DNA polymerases themselves, or possibly by other factors acting during tumor development.
Implications for the Etiology of Human Cancers.
The POLD1-R689W mutation was found in the MSH6-deficient colon cancer cell lines DLD-1 and HCT15 in the heterozygous state (29, 31). The 10-fold increase in the mutation rate in yeast POL3/pol3-R696W heterozygotes (Fig. 1B) suggests that the heterozygosity for POLD1-R689W could have contributed to the accumulation of cancer-driving mutations in the human patient irrespective of MSH6 status. Moreover, a combination of POLD1-R689W with the biallelic MSH6 defect in the human tumor likely resulted in a synergistic increase in the rate of mutation accumulation, as we observed for the yeast POL3/pol3-R696W msh6Δ/msh6Δ diploids (Fig. 2B). Although the high conservation of Arg 689 and the strong mutator effect of the Trp substitution suggest that R689W is the pathogenic change, DLD-1 and HCT15 contain additional mutations in the POLD1 gene (29, 31). Two of these additional mutations are shared by the two cell lines, and therefore must have been present in the original tumor. Similarly, hypermutated sporadic tumors with functionally important POLE mutations often contain additional changes in POLE (20, 72). Although it is unclear from the published data if these mutations affect the same allele, it is possible that the additional changes were selected for during tumor evolution as suppressors of the strong mutator effect of R689W (or, in the case of POLE tumors, of the respective mutator POLE variant). A strong mutator phenotype could be disadvantageous for the tumor in the long term, and studies in yeast have shown that strains with mutator Polδ and Polε variants often accumulate secondary changes within the corresponding polymerase gene that reduce the mutation rate (35, 36).
As reviewed by Briggs and Tomlinson (25) and Seshagiri (28), mutator variants of Polδ and Polε are likely causative in 3% and 7% of CRC and EC, respectively. Our finding that such error-prone variants modeled in yeast can cause dNTP pool expansion, further reducing their own fidelity, may provide insight into the mechanism of mutation accumulation in these cancers. Future studies will help determine whether the “vicious circle” pathway depicted in Fig. 5 could be triggered by mutations other than pol3-R696W, and whether it can operate in human cells. In yeast, the requirement of Dun1 for the mutator effect has been observed previously for two other Polδ variants (33, 73). These findings suggest that high sensitivity of replicative DNA polymerase mutants to fluctuation of intracellular dNTP levels could be a general phenomenon. Regulation of dNTP metabolism in human cells experiencing DNA damage or other forms of replication stress is poorly understood. There is no clear functional homolog of Dun1 in humans. One study indicates that dNTP pools in human cells are weakly elevated in response to DNA damage, and this elevation is only observed during the G1 phase of the cell cycle, when dNTP pools are very low (74). Another study indicates that RNR in human cells localizes to sites of DNA damage, where it might locally expand dNTP pools (75). Tumor cells appear to have elevated dNTP pools in comparison to normal human cells (reviewed in ref. 76), although it is unclear if this elevation is due to the fact that tumors have an increased proportion of S-phase cells that have high dNTP pools. More interestingly, pathways that regulate dNTP pools are often mutated in human cancers and cancer cell lines (77–79). These observations suggest a possibility that the mutator effects of replicative DNA polymerase variants could change along with dNTP levels as the tumors evolve. Using the POLD1-R689W allele as a model, we are now conducting experiments to determine if, similar to our observations in yeast, cancer-associated DNA polymerase defects can lead to checkpoint activation and changes in dNTP metabolism in human cells.
Materials and Methods
Saccharomyces cerevisiae Strains and Plasmids.
All Saccharomyces cerevisiae strains used in this study (Table S4) were derived from the haploid strains 1B-D770 (MATa ade5-1 lys2-Tn5-13 trp1-289 his7-2 leu2-3,112 ura3-4) and E134 (MATα ade5-1 lys2::InsEA14 trp-289 his7-2 leu2-3,112 ura3-52) (80). The 1B-D770 strain was modified to create TM30 as follows. An integrative cassette containing the LEU2 gene from K. lactis was generated by PCR using plasmid pUG73 (81) as a template and primers with 5′ tail sequences homologous to the region downstream of CAN1. Sequences of all primers used in the study are available upon request. Following transformation of 1B-D770 with this cassette, integration of the K. lactis LEU2 gene immediately downstream of CAN1 was confirmed by PCR analysis. All haploids with the CAN1::LEU2 allele were derived from TM30. Haploids expressing pol3-R696W were constructed in a three-step process. In step 1, TM30 was transformed with BseRI-digested plasmid p170-R696W, a URA3-based integrative plasmid containing the C-terminal portion of the pol3-R696W allele (30). Integration of p170-R696W (BseRI) into the POL3 locus produced a strain containing a full-length wild-type POL3 gene, URA3, and a truncated copy of POL3 containing the R696W mutation. In step 2, this strain was transformed with pPOL3 (described below) creating strain TM34. In step 3, yeast cells that underwent recombination resulting in loss of URA3 and restoration of a single, full-length POL3 allele were selected on medium containing 5-fluoroorotic acid. The presence of the nucleotide change responsible for the R696W substitution was verified by PCR analysis and sequencing. TM44 was derived from E134 by deleting the CAN1 ORF and 100 bp of flanking DNA by transformation with a PCR-generated deletion cassette and CRE-mediated recombination as described (82). Strains with deletions of MSH6, DUN1, SML1, and CRT1 were created by transformation with PCR-generated deletion cassettes as described (82). To create strains with a combination of gene deletion(s) and pol3-R696W, the deletions were introduced between steps 2 and 3 described above. Diploids with a single copy of CAN1 (Fig. 1A) were constructed by crossing strains derived from TM30 with strains derived from TM44.
Yeast 2μ-based plasmids expressing POL3 from the GAL1 promoter, pBL336 (83), pBL335, and pBL335-5DV (44), which are referred to as pPOL3, pPOL3.GST, and pPOL3.GST-5DV in this study, were provided by Peter Burgers (Washington University School of Medicine, St. Louis, MO). Plasmids pPOL3.GST-R696W and pPOL3.GST-5DV,R696W were constructed by gap repair in yeast strains, in which the effects of these plasmids were studied. The strains were cotransformed with two PCR-generated fragments: (i) the backbone of pPOL3.GST or pPOL3.GST-5DV and (ii) a part of the POL3 gene containing the R696W mutation amplified from p170-R696W. To construct plasmid pDUN1, the DUN1 gene from yeast strain 1B-D770 was amplified by PCR and cloned into the SmaI and EcoRI sites of pRS316 (84).
Proteins.
Yeast Polδ variants and replication protein A (RPA) were purified as described previously (30, 85). Yeast proliferating cell nuclear antigen (PCNA) was purified using DEAE-Sepharose, S-Sepharose, Mono-Q, and hydroxyapatite column chromatography using the methods described by Ayyagari et al. (86) and Fien and Stillman (87). Replication factor C (RFC) was kindly provided by Peter Burgers.
Mutation Rate Measurements.
The rate of CanR mutation was measured by fluctuation analysis as described previously (88). The cultures were grown in synthetic complete (SC) medium omitting amino acids as needed to maintain plasmids used in this study. Unless indicated otherwise, all strains contained pPOL3, which is necessary to suppress the lethality of pol3-R696W mutants. In this system, expression of the chromosomal POL3 (or pol3-R696W) allele from its natural promoter serves as the main source of the Pol3 protein, and a small amount of wild-type Pol3 is provided by a leaky expression of the plasmid-borne wild-type POL3 gene from the GAL1 promoter repressed by the presence of glucose (30). For CAN1::Kl.LEU2/can1∆::loxP diploid yeast strains, mutants were selected on SC medium lacking Leu to select against loss of CAN1 by homologous recombination.
In Vivo Mutation Spectrum.
Independent colonies of the msh6Δ pol3-R696W strain containing pPOL3 were streaked on SC medium lacking Trp, grown for 2 d at 30 °C, and replica-plated onto SC medium lacking Trp and Arg and containing 60 mg/L l-canavanine to select for can1 mutants. One Canr colony was randomly picked from each patch, and the CAN1 gene was amplified by PCR and sequenced.
Measurement of Intracellular dNTP Levels.
Three independent isolates of each strain were grown overnight at 30 °C in SC medium lacking Trp. Cultures were reinoculated in YPAD media [1% yeast extract, 2% (wt/vol) bactopeptone, 0.002% adenine, 2% (wt/vol) dextrose] and grown at 30 °C until midlog phase. Cells were harvested by filtration and processed as described previously (89). Briefly, NTPs and dNTPs were extracted in an ice-cold trichloroacetic acid-MgCl2 mixture, followed by two rounds of neutralization by Freon-trioctylamine mixture. A total of 475 μL of the aqueous phase was pH-adjusted with 1 M ammonium carbonate (pH 8.9), loaded on boronate columns (Affi-Gel Boronate Gel; Bio-Rad), and eluted with a 50 mM ammonium carbonate (pH 8.9) and 15 mM MgCl2 mixture to separate dNTPs from NTPs. The purified dNTP eluates were pH-adjusted to 3.4 and analyzed by HPLC on a LaChrom Elite HPLC system (Hitachi) with a Partisphere SAX HPLC column (Hichrom). NTPs were directly analyzed by HPLC similar to dNTPs by using 47.5-μL aliquots of the aqueous phase adjusted to pH 3.4.
Immunoblots.
Whole-cell protein extracts used for immunoblots were prepared as described by Northam et al. (90), except that a dual protease and phosphatase inhibitor mixture (catalog no. 88668; Pierce) was added to the lysis buffer. For Rad53, 30 μg of protein was separated by electrophoresis in a 6% (wt/vol) polyacrylamide Tris⋅glycine gel and transferred to a nitrocellulose membrane. After transfer, the membrane was cut horizontally at a position corresponding to a protein molecular mass of ∼60 kDa. The top and bottom halves were probed with goat polyclonal anti-Rad53 antibody (sc-6749; Santa Cruz Biotechnology) and rat monoclonal anti–α-tubulin antibody (sc-53030; Santa Cruz Biotechnology), respectively. For Sml1, 30 μg of protein was separated in a 10% (wt/vol) polyacrylamide Bis-Tris gel and transferred to a nitrocellulose membrane, which was subsequently cut horizontally at ∼30 kDa. The top and bottom halves were probed with the anti–α-tubulin antibody and a rabbit polyclonal anti-Sml1 antibody (AS10 847; Agrisera), respectively. The secondary antibodies were HRP-conjugated goat anti-rat (sc-2006; Santa Cruz Biotechnology), donkey anti-goat (sc-2020; Santa Cruz Biotechnology), and goat anti-rabbit (A00160; Genscript). Proteins were detected by using a chemiluminescent SuperSignal West Pico kit (Thermo Scientific) and autoradiography.
Measurement of in Vitro DNA Synthesis Fidelity.
M13mp2 gapped substrate was prepared as described by Bebenek and Kunkel (43) and gel-purified via electroelution using GelStar DNA stain (Lonza) and blue light illumination for visualization. DNA synthesis reactions (25 μL) contained 40 mM Tris (pH 7.5), 8 mM MgAc2, 125 mM NaAc, 0.5 mM ATP, 1 mM DTT, 200 mg/mL BSA, 500 fmol of PCNA, 200 fmol of RFC, 5 pmol of RPA, 150 fmol of Polδ or 300 fmol Polδ-R696W, and 25 fmol (1 nM) of gapped DNA. The dNTPs were included in the reactions at either 100 μM each or at the concentrations shown in Table 3. The reactions were incubated at 30 °C for 10 min. Synthesis by Polδ and Polδ-R696W appeared to fill the single-stranded gap completely without observable strand displacement, as determined by agarose gel electrophoresis. Transformation of E. coli with the gap-filling reactions, scoring of mutant plaques, and ssDNA isolation from purified plaques were as described by Bebenek and Kunkel (43). The lacZ gene from mutant M13mp2 plaques was sequenced, and error rates were calculated as previously described (66).
Supplementary Material
Acknowledgments
We thank Elizabeth Moore and Krista Brown for technical assistance, Peter Burgers for POL3 and pol3-5DV expression plasmids and the RFC protein, Brad Preston for discussions and communicating unpublished data, Tom Kunkel for sharing the database of phenotypically detectable mutations in the lacZ gene, and Youri Pavlov for the gift of nucleotide analogs and for valuable comments on the manuscript. This work was supported by National Institutes of Health Grant ES015869 and Nebraska Department of Health and Human Services Grant LB506 (to P.V.S.). T.M.M. was supported by a University of Nebraska Medical Center Graduate Studies Research Fellowship. S.S. and A.C. were supported by the Swedish Cancer Society and the Knut and Alice Wallenberg Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.A.L. is a guest editor invited by the Editorial Board.
See Commentary on page 5864.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422934112/-/DCSupplemental.
References
- 1.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148(4):1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34(1):359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 3.Wei K, Kucherlapati R, Edelmann W. Mouse models for human DNA mismatch-repair gene defects. Trends Mol Med. 2002;8(7):346–353. doi: 10.1016/s1471-4914(02)02359-6. [DOI] [PubMed] [Google Scholar]
- 4.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348(10):919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 5.Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition—Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20(4-5):269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: The interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29(4):673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 7.Agbor AA, Göksenin AY, LeCompte KG, Hans SH, Pursell ZF. Human Pol ε-dependent replication errors and the influence of mismatch repair on their correction. DNA Repair (Amst) 2013;12(11):954–963. doi: 10.1016/j.dnarep.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem. 2007;282(4):2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′ → 5′ exonuclease activity. Proc Natl Acad Sci USA. 1991;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon M, Giot L, Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10(8):2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase δ. J Biol Chem. 2006;281(7):4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 12.Jin YH, et al. The 3′-->5′ exonuclease of DNA polymerase δ can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci USA. 2001;98(9):5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Murphy KM, Kanevets U, Reha-Krantz LJ. Sensitivity to phosphonoacetic acid: A new phenotype to probe DNA polymerase δ in Saccharomyces cerevisiae. Genetics. 2005;170(2):569–580. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niimi A, et al. Palm mutants in DNA polymerases α and η alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24(7):2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albertson TM, et al. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci USA. 2009;106(40):17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsby RE, et al. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc Natl Acad Sci USA. 2002;99(24):15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkatesan RN, et al. Mutation at the polymerase active site of mouse DNA polymerase δ increases genomic instability and accelerates tumorigenesis. Mol Cell Biol. 2007;27(21):7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldsby RE, et al. Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat Med. 2001;7(6):638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 19.Palles C, et al. CORGI Consortium WGS500 Consortium Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandoth C, et al. Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Church DN, et al. NSECG Collaborators DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22(14):2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74(7):1895–1901. doi: 10.1158/0008-5472.CAN-13-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briggs S, Tomlinson I. Germline and somatic polymerase ε and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol. 2013;230(2):148–153. doi: 10.1002/path.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Church JM. Polymerase proofreading-associated polyposis: A new, dominantly inherited syndrome of hereditary colorectal cancer predisposition. Dis Colon Rectum. 2014;57(3):396–397. doi: 10.1097/DCR.0000000000000084. [DOI] [PubMed] [Google Scholar]
- 27.Heitzer E, Tomlinson I. Replicative DNA polymerase mutations in cancer. Curr Opin Genet Dev. 2014;24:107–113. doi: 10.1016/j.gde.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seshagiri S. The burden of faulty proofreading in colon cancer. Nat Genet. 2013;45(2):121–122. doi: 10.1038/ng.2540. [DOI] [PubMed] [Google Scholar]
- 29.Flohr T, et al. Detection of mutations in the DNA polymerase δ gene of human sporadic colorectal cancers and colon cancer cell lines. Int J Cancer. 1999;80(6):919–929. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 30.Daee DL, Mertz TM, Shcherbakova PV. A cancer-associated DNA polymerase δ variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci USA. 2010;107(1):157–162. doi: 10.1073/pnas.0907526106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abaan OD, et al. The exomes of the NCI-60 panel: A genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73(14):4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tibbetts LM, Chu MY, Hager JC, Dexter DL, Calabresi P. Chemotherapy of cell-line-derived human colon carcinomas in mice immunosuppressed with antithymocyte serum. Cancer. 1977;40(5) Suppl:2651–2659. doi: 10.1002/1097-0142(197711)40:5+<2651::aid-cncr2820400939>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 33.Herr AJ, Kennedy SR, Knowels GM, Schultz EM, Preston BD. DNA replication error-induced extinction of diploid yeast. Genetics. 2014;196(3):677–691. doi: 10.1534/genetics.113.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12(4):1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herr AJ, et al. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2011;7(10):e1002282. doi: 10.1371/journal.pgen.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams LN, Herr AJ, Preston BD. Emergence of DNA polymerase ε antimutators that escape error-induced extinction in yeast. Genetics. 2013;193(3):751–770. doi: 10.1534/genetics.112.146910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prindle MJ, Schmitt MW, Parmeggiani F, Loeb LA. A substitution in the fingers domain of DNA polymerase δ reduces fidelity by altering nucleotide discrimination in the catalytic site. J Biol Chem. 2013;288(8):5572–5580. doi: 10.1074/jbc.M112.436410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10(4):407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 39.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27(18):6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papadopoulos N, et al. Mutations of GTBP in genetically unstable cells. Science. 1995;268(5219):1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 41.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol. 2009;16(9):979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grabowska E, et al. Proper functioning of the GINS complex is important for the fidelity of DNA replication in yeast. Mol Microbiol. 2014;92(4):659–680. doi: 10.1111/mmi.12580. [DOI] [PubMed] [Google Scholar]
- 43.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 44.Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase δ. Nucleic Acids Res. 2006;34(16):4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bashkirov VI, Bashkirova EV, Haghnazari E, Heyer WD. Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol Cell Biol. 2003;23(4):1441–1452. doi: 10.1128/MCB.23.4.1441-1452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20(13):3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chabes A, et al. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112(3):391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 48.Lis ET, O’Neill BM, Gil-Lamaignere C, Chin JK, Romesberg FE. Identification of pathways controlling DNA damage induced mutation in Saccharomyces cerevisiae. DNA Repair (Amst) 2008;7(5):801–810. doi: 10.1016/j.dnarep.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davidson MB, et al. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012;31(4):895–907. doi: 10.1038/emboj.2011.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA. 2002;99(6):3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Huang M. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol. 2008;28(23):7156–7167. doi: 10.1128/MCB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94(5):595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 53.Andreson BL, Gupta A, Georgieva BP, Rothstein R. The ribonucleotide reductase inhibitor, Sml1, is sequentially phosphorylated, ubiquitylated and degraded in response to DNA damage. Nucleic Acids Res. 2010;38(19):6490–6501. doi: 10.1093/nar/gkq552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen ES-W, et al. Use of quantitative mass spectrometric analysis to elucidate the mechanisms of phospho-priming and auto-activation of the checkpoint kinase Rad53 in vivo. Mol Cell Proteomics. 2014;13(2):551–565. doi: 10.1074/mcp.M113.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30(1):73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsaponina O, Barsoum E, Aström SU, Chabes A. Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools. PLoS Genet. 2011;7(5):e1002061. doi: 10.1371/journal.pgen.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elledge SJ, Davis RW. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4(5):740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 58.Gupta A, et al. Telomere length homeostasis responds to changes in intracellular dNTP pools. Genetics. 2013;193(4):1095–1105. doi: 10.1534/genetics.112.149120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar D, et al. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011;39(4):1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88(16):7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valle L, et al. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum Mol Genet. 2014;23(13):3506–3512. doi: 10.1093/hmg/ddu058. [DOI] [PubMed] [Google Scholar]
- 62.Creighton S, Goodman MF. Gel kinetic analysis of DNA polymerase fidelity in the presence of proofreading using bacteriophage T4 DNA polymerase. J Biol Chem. 1995;270(9):4759–4774. doi: 10.1074/jbc.270.9.4759. [DOI] [PubMed] [Google Scholar]
- 63.Gawel D, Fijalkowska IJ, Jonczyk P, Schaaper RM. Effect of dNTP pool alterations on fidelity of leading and lagging strand DNA replication in E. coli. Mutat Res Fundam Mol Mech Mutagen. 2014;759:22–28. doi: 10.1016/j.mrfmmm.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eckert KA, Kunkel TA. DNA polymerase fidelity and the polymerase chain reaction. PCR Methods Appl. 1991;1(1):17–24. doi: 10.1101/gr.1.1.17. [DOI] [PubMed] [Google Scholar]
- 65.Pham PT, Olson MW, McHenry CS, Schaaper RM. The base substitution and frameshift fidelity of Escherichia coli DNA polymerase III holoenzyme in vitro. J Biol Chem. 1998;273(36):23575–23584. doi: 10.1074/jbc.273.36.23575. [DOI] [PubMed] [Google Scholar]
- 66.Shcherbakova PV, et al. Unique error signature of the four-subunit yeast DNA polymerase ε. J Biol Chem. 2003;278(44):43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 67.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108(48):19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wheeler LJ, Rajagopal I, Mathews CK. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair (Amst) 2005;4(12):1450–1456. doi: 10.1016/j.dnarep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 69.Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008;36(17):5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stone JE, et al. Lesion bypass by S. cerevisiae Pol ζ alone. DNA Repair (Amst) 2011;10(8):826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative ICGC Breast Cancer Consortium ICGC MMML-Seq Consortium ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shinbrot E, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24(11):1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Datta A, et al. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3-01 mutants. Mol Cell. 2000;6(3):593–603. doi: 10.1016/s1097-2765(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 74.Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281(12):7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 75.Niida H, et al. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 2010;24(4):333–338. doi: 10.1101/gad.1863810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140(1):1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 77.Angus SP, et al. Retinoblastoma tumor suppressor targets dNTP metabolism to regulate DNA replication. J Biol Chem. 2002;277(46):44376–44384. doi: 10.1074/jbc.M205911200. [DOI] [PubMed] [Google Scholar]
- 78.Onyenwoke RU, et al. AMPK directly inhibits NDPK through a phosphoserine switch to maintain cellular homeostasis. Mol Biol Cell. 2012;23(2):381–389. doi: 10.1091/mbc.E11-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aird KM, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Reports. 2013;3(4):1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shcherbakova PV, Kunkel TA. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 1999;19(4):3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30(6):e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15(14):1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 83.Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem. 1998;273(31):19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 84.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sibenaller ZA, Sorensen BR, Wold MS. The 32- and 14-kilodalton subunits of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry. 1998;37(36):12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- 86.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15(8):4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: A component of the leading-strand DNA replication complex. Mol Cell Biol. 1992;12(1):155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Northam MR, Robinson HA, Kochenova OV, Shcherbakova PV. Participation of DNA polymerase ζ in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 2010;184(1):27–42. doi: 10.1534/genetics.109.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38(12):3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase ζ regulated by PCNA. EMBO J. 2006;25(18):4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci USA. 2010;107(11):4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.