Significance
An increased rate of mutation, or “mutator phenotype,” generates genetic diversity that can accelerate cancer progression or confer resistance to chemotherapy drugs. New therapeutic strategies are needed that target mutator phenotypes directly. Mutator phenotypes due to defects in DNA polymerase ε have been implicated in colorectal and endometrial cancers and may emerge in other cancers during treatment. Here, we show in budding yeast that such mutator phenotypes are influenced by the levels of dNTPs, the building blocks of DNA. Lowering dNTP pool levels lessens the mutator phenotypes, whereas increasing dNTP pools accentuates the mutator phenotypes. These findings suggest that mutator phenotypes due to error-prone polymerases may be modulated by treatments that target dNTP pools.
Keywords: DNA replication and repair, polymerase fidelity, cancer, lethal mutagenesis
Abstract
Mutator phenotypes create genetic diversity that fuels tumor evolution. DNA polymerase (Pol) ε mediates leading strand DNA replication. Proofreading defects in this enzyme drive a number of human malignancies. Here, using budding yeast, we show that mutator variants of Pol ε depend on damage uninducible (Dun)1, an S-phase checkpoint kinase that maintains dNTP levels during a normal cell cycle and up-regulates dNTP synthesis upon checkpoint activation. Deletion of DUN1 (dun1Δ) suppresses the mutator phenotype of pol2-4 (encoding Pol ε proofreading deficiency) and is synthetically lethal with pol2-M644G (encoding altered Pol ε base selectivity). Although pol2-4 cells cycle normally, pol2-M644G cells progress slowly through S-phase. The pol2-M644G cells tolerate deletions of mediator of the replication checkpoint (MRC) 1 (mrc1Δ) and radiation sensitive (Rad) 9 (rad9Δ), which encode mediators of checkpoint responses to replication stress and DNA damage, respectively. The pol2-M644G mutator phenotype is partially suppressed by mrc1Δ but not rad9Δ; neither deletion suppresses the pol2-4 mutator phenotype. Thus, checkpoint activation augments the Dun1 effect on replication fidelity but is not required for it. Deletions of genes encoding key Dun1 targets that negatively regulate dNTP synthesis, suppress the dun1Δ pol2-M644G synthetic lethality and restore the mutator phenotype of pol2-4 in dun1Δ cells. DUN1 pol2-M644G cells have constitutively high dNTP levels, consistent with checkpoint activation. In contrast, pol2-4 and POL2 cells have similar dNTP levels, which decline in the absence of Dun1 and rise in the absence of the negative regulators of dNTP synthesis. Thus, dNTP pool levels correlate with Pol ε mutator severity, suggesting that treatments targeting dNTP pools could modulate mutator phenotypes for therapy.
Many cancers defy treatment despite substantial investments in the development of anticancer drugs. Those cancers that do respond to chemotherapy often evolve resistance, necessitating a steady supply of new therapies. An alternative strategy is needed that takes into account evolutionary theory. The recalcitrant nature of cancer lies in its origin and treatment. Sustained selective pressure during neoplasia and chemotherapy favors cells with an elevated mutation rate (“mutator phenotype”) that acquire adaptive mutations more readily (1, 2). Mutator phenotypes result in reservoirs of genetically diverse cells from which resistance arises. The unifying feature of many of these cells is a mutator allele. Thus, therapies that target mutator phenotypes represent a rational way forward. Attenuation of mutator phenotypes may slow tumor progression or improve conventional chemotherapy by slowing the evolution of drug resistance. Alternatively, synthetic–lethal interactions between mutator phenotypes and other pathways may be used to kill tumor cells selectively. Increasing mutation rate beyond a threshold may compromise replicative fitness directly or enhance the overall immunogenicity of the tumor clone.
The best-characterized mutator phenotype in cancer derives from mismatch repair (MMR) defects, which increase point mutation rate and microsatellite instability. MMR defects lead to colorectal cancer (CRC) and endometrial cancer (EC) (3), among others. MMR cooperates with DNA polymerase proofreading to correct polymerase errors, which are the most abundant known source of potential mutations in dividing cells (4). Recently, germline and somatic mutations have been described affecting human POLE, encoding the catalytic subunit of polymerase (Pol) ε. Evidence from yeast and human studies indicates that Pol ε performs the bulk of leading strand DNA replication (5–9). POLE mutations affecting the proofreading exonuclease are found in 3% of CRC and 7% of EC (6, 10–14). Mutations affecting the proofreading function of Pol δ, the main lagging strand DNA polymerase, were also observed, although less frequently (6, 10–14). These observations support the hypothesis that maintenance of DNA replication fidelity restrains neoplasia in humans, as first observed in mice (15–17), and advance mutator polymerases as important targets to consider for therapeutic intervention.
Given the high conservation of DNA replication machinery, the yeast Saccharomyces cerevisiae represents an ideal system with which to identify genetic pathways that influence mutator phenotypes (18–20). The pol2-4 allele, encoding proofreading-deficient Pol ε, is lethal in strains lacking all MMR activity [e.g., deletion of MutS homologue (MSH) 2 (msh2Δ)] (19). This “error-induced extinction” is surmounted by “antimutator” mutations affecting the Pol ε catalytic subunit, as well as by unidentified mutations elsewhere in the genome (19). Mutants harboring these unidentified mutations constitute the vast majority (96%) of strains that escape error-induced extinction of pol2-4 msh2Δ cells, providing tantalizing evidence that other factors besides Pol ε and MMR influence leading strand replication fidelity.
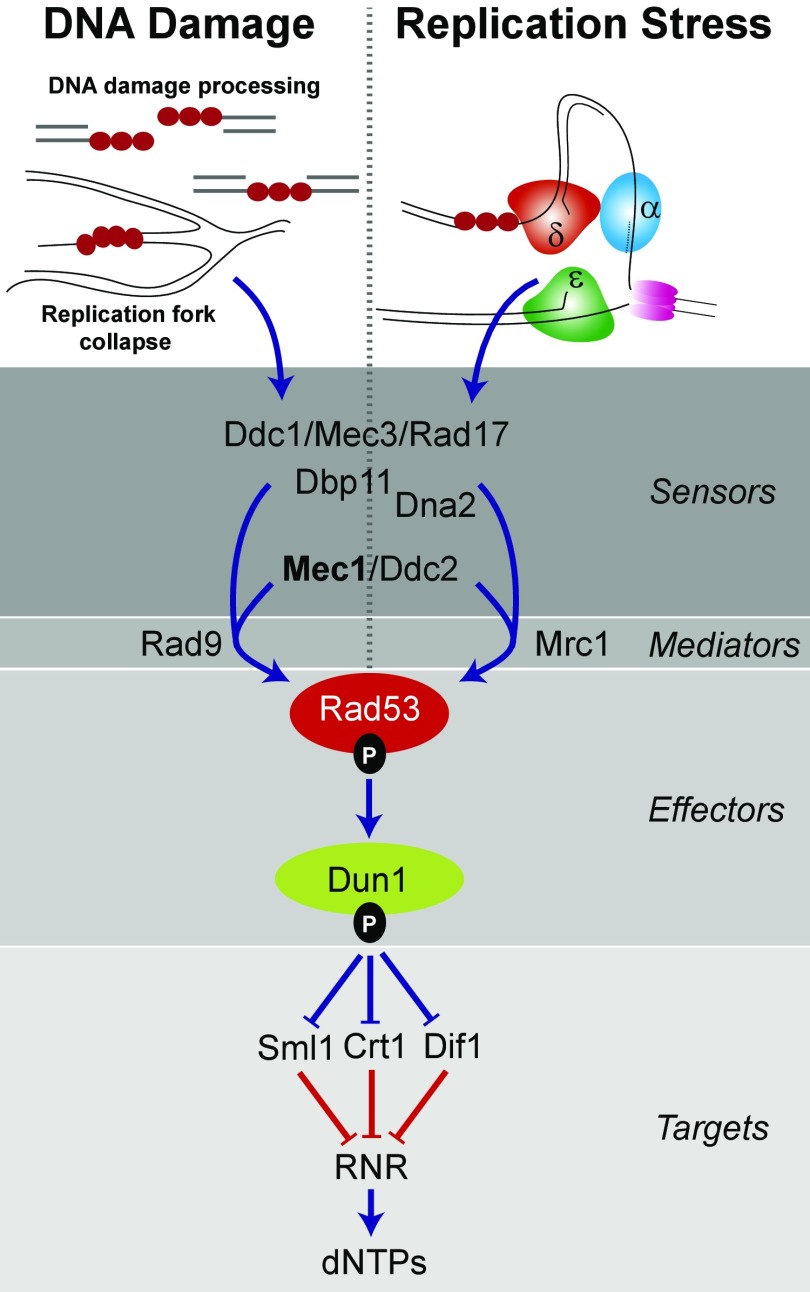
Previous studies observed that the mutator phenotype of proofreading-deficient Pol δ (encoded by the pol3-01 allele) partially depends on the Dun1 effector kinase (20, 21) (Fig. 1), which lies directly downstream of the mitosis entry checkpoint 1 (Mec1) [mammalian ataxia telangiectasia and Rad3-related protein (ATR)] and Rad53 [mammalian checkpoint kinase (Chk)1] kinases in the S-phase checkpoint pathway (22, 23) (Fig. 1). One function of Dun1 is to modulate dNTP pools by controlling negative regulators of ribonucleotide reductase (RNR), a central enzyme in the dNTP biosynthetic pathway that reduces NDPs to dNDPs (24–26). The RNR holoenzyme contains a large Rnr1 homodimeric subunit and a small dimeric subunit of Rnr2 and Rnr4 (27–30). A minor isoform contains Rnr3 instead of Rnr1 in the large subunit (28). Dun1 targets three proteins that repress expression, assembly, or activity of RNR: constitutive RNR transcription (Crt)1 represses transcription of RNR2, RNR3, and RNR4 (28, 30, 31); damage-regulated import facilitator (Dif)1 prevents RNR assembly by mediating import of Rnr2–Rnr4 into the nucleus, where it is sequestered from Rnr1 (32–34); and suppressor of Mec1 lethality (Sml)1 binds and inhibits RNR activity (35, 36). Phosphorylation of Sml1 and Dif1 by Dun1 results in their proteolysis, releasing the negative regulation of RNR catalysis and allowing cytoplasmic localization of Rnr2 and Rnr4, followed by RNR holoenzyme assembly (32–34). At the same time, phosphorylation of Crt1 increases transcription of RNR2, RNR3, and RNR4. A separate damage-inducible system controls RNR1 transcription (37). Overall, dNTP pools increase as much as eightfold during the DNA damage response (38). Mutants of the checkpoint proteins Mec1 and Rad53 are inviable but are rescued by deletions of SML1, DIF1, or CRT1 or by overexpression of RNR subunits. Thus, regulation of dNTP pools is a key function of the Mec1-Rad53-Dun1 pathway (31, 33, 35, 39).
Fig. 1.
Two S-phase checkpoint responses in S. cerevisiae. The S-phase checkpoint is triggered in the DNA damage signaling pathway by replication protein A (RPA)-coated DNA (red circles) resulting from ssDNA repair intermediates, resected ends of double-stranded breaks, or collapsed replication forks. In the replication stress signaling pathway, the S-phase checkpoint is triggered by stalled replication forks (α, δ, ε; replicative DNA polymerases). The two signaling pathways converge at Mec1/Ddc2, which phosphorylates Rad53 with the help of Rad9 (DNA damage) or Mrc1 (replication stress). Phospho-Rad53 then activates Dun1, which, in turn, inactivates three repressors (Sml1, Crt1, and Dif1) of RNR. RNR generates dNDPs from NDPs, which are then converted to dNTPs by nucleoside diphosphate kinases. Higher dNTP concentrations facilitate DNA repair and replication fork restart. Blue lines are signals that increase dNTP pools. Red lines are signals that repress dNTP pools. Adapted from ref. 63.
Datta et al. (21) observed that pol3-01 induced an S-phase progression defect, suggesting that the mutator phenotype may be checkpoint-dependent. They hypothesized that dun1Δ suppresses the pol3-01 mutator phenotype by altering the balance between error-prone and error-free repair pathways. The same study found that the mutator phenotype of the pol2-4 allele was unaffected by the absence of Dun1. This result is surprising, given the role of Pol ε in helping to mediate the S-phase checkpoint (40–44). However, the weak pol2-4 mutator phenotype or the acquisition of a suppressor mutation may have limited the ability to detect a decrease in mutation rate in dun1Δ strains.
Here, we present evidence that elevated mutation rates conferred by pol2 mutator alleles require Dun1. Our findings suggest that mutator polymerases or their errors are sensed as replication stressors that trigger Dun1 activation. We provide evidence that Dun1-dependent modulation of dNTP pools, either in response to DNA damage or as part of normal dNTP homeostasis, influences mispair extension by mutator DNA polymerases. Our findings suggest that perturbation of dNTP pools may provide the means to modulate human POLE mutator phenotypes in cancers.
Results
Dun1 Is Required for the Mutator Phenotype or Viability of pol2 Mutator Cells.
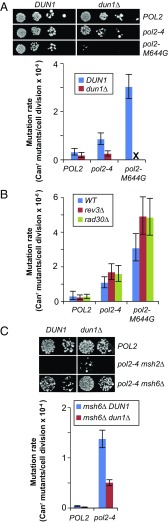
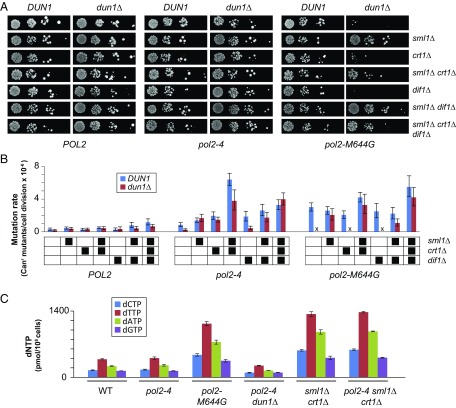
To determine the effect of dun1Δ on Pol ε mutator phenotypes, we introduced POL2 (WT), pol2-4 (proofreading-defective), or pol2-M644G (reduced polymerase accuracy) alleles into DUN1 and dun1Δ strains by plasmid shuffling (a detailed description is provided in Materials and Methods) (19). The pol2-4 and pol2-M644G alleles increase mutation rates threefold and 10-fold, respectively (Fig. 2A and Table 1) in Dun1-proficient cells, similar to what has been reported (5, 19, 21, 45). Mutation rates of pol2-4 dun1Δ cells were not statistically different from mutation rates of WT or dun1Δ cells, whereas pol2-M644G dun1Δ cells failed to form visible colonies upon shuffling (Fig. 2A). Thus, Dun1 is required for the mutator phenotype of pol2-4 and the viability of pol2-M644G cells.
Fig. 2.
Dun1 promotes Pol ε error-prone replication. (A) Dun1 dependence of pol2 mutator alleles. (Top) Viability measurements. Dun1 and dun1Δ strains were transformed with LEU2 plasmids carrying POL2, pol2-4, or pol2-4,M644G. Ten-fold serial dilutions of the resulting transformants were then plated onto FOA media to select for loss of the complementing POL2-URA3 plasmid and were photographed after 3 d at 30 °C. (Bottom) Spontaneous mutation rates determined from multiple independent fluctuation analyses of each strain. Error bars represent 95% confidence intervals. “X” indicates synthetic lethality. (B) pol2-4 and pol2-M644G mutator phenotypes do not require the translesion polymerase Pol ζ (rev3Δ) or Pol η (rad30Δ). Mutation rates were performed as described above. (C) Error-induced extinction in pol2-4 msh2Δ cells requires Dun1. (Top) Viability measurements. DUN1 and dun1Δ cells proficient for MMR, deficient in all MMR (msh2Δ), or deficient in only base–base MMR (msh6Δ) were transformed with POL2 or pol2-4 plasmids and plated on FOA using 10-fold serial dilutions. (Bottom) Mutation rates were determined as above.
Table 1.
Mutation rates of pol2 shuffling strains
| POL2† | pol2-4† | pol2-M644G† | |||||
| Strain* | Relevant genotype | MR | 95% CI | MR | 95% CI | MR | 95% CI |
| Figs. 2A and 5B‡ | |||||||
| LW14 | WT | 3.0 | (1.8–4.5) | 8.4 | (6.2–11.0) | 30.1 | (25.2–35.3) |
| AH7808 | + sml1Δ | 4.4 | (3.2–5.8) | 13.8 | (11.2–16.8) | 25.9 | (21.4–30.7) |
| AH8009 | + crt1Δ | 2.7 | (1.6–4.3) | 19.4 | (15.1–24.2) | 20.5 | (16.1–25.5) |
| AH8209 | + sml1Δ crt1Δ | 4.8 | (3.7–6.0) | 63.7 | (56.4–71.4) | 42.0 | (36.3–48.2) |
| AH8403 | + dif1Δ | 2.5 | (1.2–4.6) | 18.4 | (13.0–25.0) | 24.9 | (17.0–34.5) |
| AH8604 | + sml1Δ dif1Δ | 7.9 | (5.1–11.5) | 26.0 | (19.5–33.5) | 22.0 | (15.6–29.8) |
| AH9704 | + sml1Δ crt1Δ dif1Δ | 11.6 | (8.2–15.7) | 32.7 | (26.9–39.0) | 54.8 | (42.7–68.4) |
| LW15 | dun1Δ | 1.6 | (0.8–2.9) | 2.3 | (1.4–3.6) | — | |
| AH7905 | + sml1Δ | 3.6 | (2.4–5.2) | 16.6 | (12.4–21.4) | 20.5 | (14.2–28.1) |
| AH8110 | + crt1Δ | 3.1 | (2.0–4.4) | 14.6 | (11.8–17.9) | — | |
| AH8306 | + sml1Δcrt1Δ | 4.3 | (2.8–6.2) | 37.7 | (26.5–51.1) | 32.7 | (22.2–45.9) |
| AH8506 | + dif1Δ | 3.3 | (1.6–5.9) | 4.3 | (2.6–6.8) | — | |
| AH8702 | + sml1Δ dif1Δ | 4.0 | (2.3–6.4) | 17.1 | (11.9–23.6) | 10.7 | (6.9–15.6) |
| AH9806 | + sml1Δ crt1Δ dif1Δ | 6.6 | (4.7–8.9) | 39.7 | (32.6–47.5) | 42.0 | (31.5–54.0) |
| Figs. 2B and 4B‡ | |||||||
| LW14 | WT | 2.9 | (1.0–6.2) | 11.5 | (8.4–15.3) | 33.2 | (25.2–42.4) |
| LW17 | + rev3Δ | 2.2 | (1.1–3.9) | 18.1 | (13.5–23.5) | 53.2 | (42.0–65.5) |
| LW18 | +rad30Δ | 2.8 | (1.7–4.3) | 17.0 | (12.4–22.4) | 52.4 | (41.4–64.6) |
| LW19 | +mrc1Δ | 2.6 | (1.2–4.6) | 12.4 | (8.6–17.2) | 17.1 | (11.9–23.4) |
| LW21 | +rad9Δ | 2.8 | (1.7–4.3) | 18.1 | (13.9–22.8) | 41.2 | (31.6–52.3) |
| Fig. 2C‡ | |||||||
| LW3 | msh6Δ | 38.4 | (30.3–47.4) | 1,370 | (1,200–1,550) | ||
| LW16 | +dun1Δ | 16.1 | (11.6–21.6) | 504 | (445–563) | ||
Mutation rate (MR) and 95% confidence interval (CI) were calculated using Salvador 2.3 with Mathematica 8.0 from the fluctuation of Canr mutants arising in at least three isolates of each strain–plasmid combination (eight replica cultures per experiment), derived from independent plasmid shuffling experiments. These values are multiplied by 1 × 10−7 Canr mutants per cell division. A dash (—) indicates the strain was inviable.
Complete genotypes of all strains can be found in Table S1.
All plasmids were derived from pRS415 (LEU2).
The figures shown in italics were constructed from the set of mutation rate measurements that immediately follow.
Error-prone synthesis by Pol ζ and Pol η allows bypass of replication-blocking DNA lesions (46), and several mutator polymerases are known to create mutations in a Pol ζ-dependent manner (47–52). To test whether Pol ζ or Pol η lies downstream of Dun1 and contributes to the pol2 mutator phenotypes, we engineered pol2-4 and pol2-M644G strains lacking Pol ζ (rev3Δ) or Pol η (rad30Δ). We found that the pol2-4 and pol2-M644G mutator phenotypes remained robust in rev3Δ and rad30Δ strains (Fig. 2B and Table 1), suggesting that neither Pol ζ nor Pol η is responsible for the Dun1 effect on mutation rates or viability.
Dun1 Contributes to pol2-4 msh2Δ Synthetic Lethality.
Haploid yeast cells with combined defects in Pol proofreading and MMR succumb to error-induced extinction (18, 19, 53–56). The majority of spontaneous mutants that escape pol2-4 msh2Δ synthetic lethality harbor unknown mutations that are extragenic to pol2 (19). We observed that Dun1 deficiency suppressed pol2-4 msh2Δ error-induced extinction, although the pol2-4 msh2Δ dun1Δ cells exhibited poor colony-forming capacity compared with WT (Fig. 2B). A twofold to threefold decrease in the mutation rate is known to rescue haploid yeast from error-induced extinction (18, 19). To quantitate the influence of Dun1 on the synergy between MMR and polymerase proofreading defects, we examined the contribution of Dun1 to the pol2-4 mutator phenotype in the absence of msh6Δ, which is not synthetically lethal with pol2-4. We found the dun1Δ mutation rescued the synthetic sick phenotype of pol2-4 msh6Δ and reduced the mutation rate almost threefold (Fig. 2C and Table 1), indicating that more than half of pol2-4–dependent mutagenesis also depends on Dun1 in an MMR-deficient background. Thus, mutations in DUN1 or genes encoding upstream signaling components may account for some of the weak extragenic antimutator alleles that suppress the lethality of pol2-4 msh2Δ (19).
pol2-4 and pol2-M644G Exhibit Divergent Effects on Cell Cycle Progression.
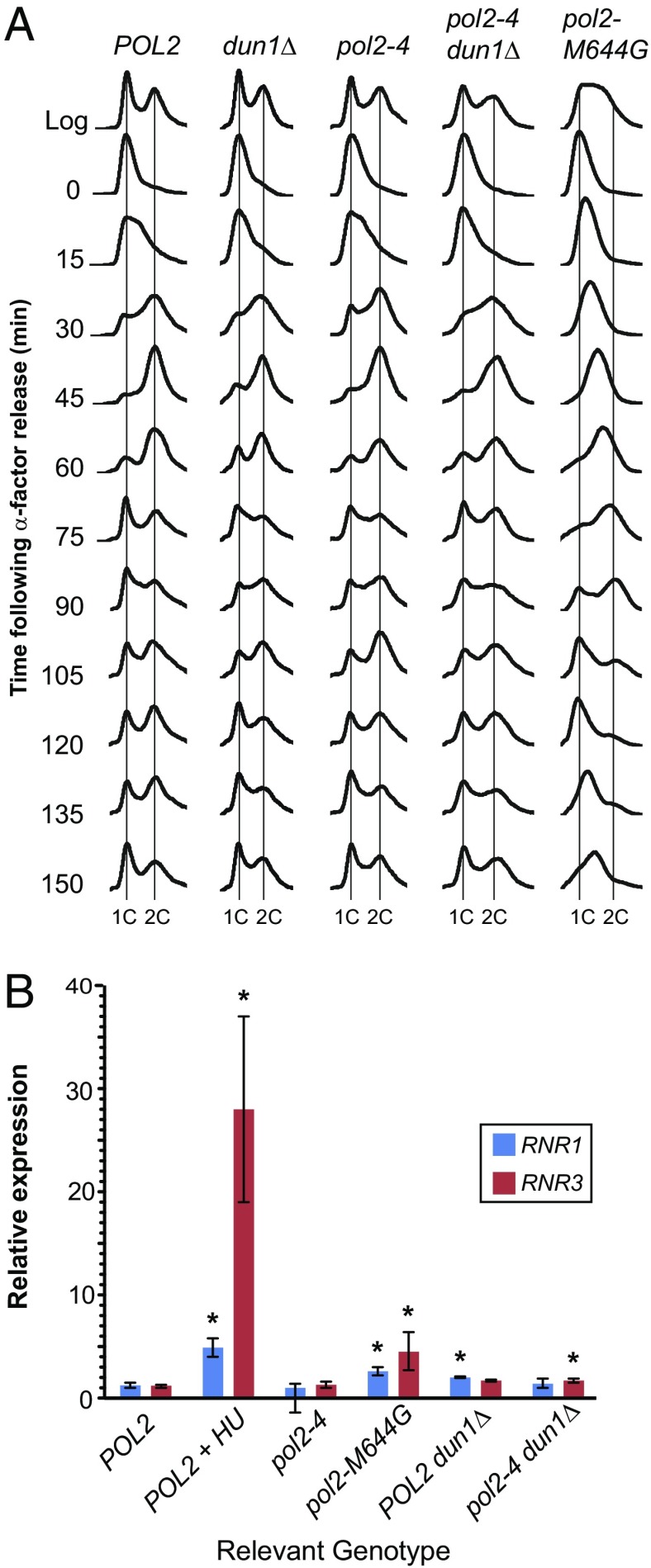
The pol3-01 allele confers a delayed progression through S-phase that is rescued by dun1Δ (21). To determine whether Pol ε mutator alleles elicit similar S-phase defects, we monitored the cell cycle progressions of pol2-4 and pol2-M644G strains by fluorescence-activated cell sorting (FACS). The cell cycle kinetics of the pol2-4 strain were indistinguishable from WT (Fig. 3A). The dun1Δ and pol2-4 dun1Δ strains were delayed in the initiation of S-phase (Fig. 3A) but appeared to divide at the same rate as WT. The delay in replication initiation in dun1Δ strains has been previously observed (32). The pol2-M644G strain progressed more slowly through S-phase, finally completing division 25 min later than WT. The majority of asynchronously dividing cells collected in the log-phase samples of pol2-M644G strains were in S-phase, further demonstrating the cell cycle defect of these mutants. These data suggest the pol2-M644G allele, but not pol2-4, confers an S-phase progression defect consistent with a requirement for Dun1 activity.
Fig. 3.
Cell cycle progression and RNR transcript expression in pol2-4 and pol2-M644G strains. (A) Cell cycle progression of pol2 mutator strains. Cells were synchronized with α-factor, released into rich medium, and monitored for DNA content by FACS. The x axis represents relative DNA content, and the y axis represents cell number at the indicated time points. Vertical lines indicate ploidy (1C and 2C). (B) Checkpoint induction in pol2 mutator strains. Transcript levels of RNR1 and RNR3 were measured by quantitative PCR assay, normalized to actin expression, and then normalized to the levels found in POL2 control cells. RNA isolated from POL2 cells treated with 200 mM hydroxyurea for 2 h was used as a positive control for checkpoint activation. Error bars show 95% confidence intervals for transcript expression based on three independent biological replicas. Asterisks denote expression levels that are significantly different (P ≤ 0.05) from POL2 control cells.
To complement the cell cycle studies, we measured the log-phase doubling times of each strain. The dun1Δ strain had the same growth rate as WT, with a doubling time of 90 min. The pol2-4 and pol2-M644G strains both had longer doubling times of 100 min and 115 min, respectively, whereas the dun1Δ mutation restored the doubling time of the pol2-4 strain to 90 min. Thus, Dun1 activity correlates with increased doubling time of cells expressing pol2 mutator phenotypes.
pol2-M644G, but Not pol2-4, Induces the Expression of RNR1 and RNR3.
Because the viability of pol2-M644G cells and the mutator phenotype of pol2-4 are Dun1-dependent, we expected both alleles to activate the S-phase checkpoint. We first assessed whether Rad53 was phosphorylated by looking for a shift in Rad53 gel mobility. Although WT cells treated with hydroxyurea displayed decreased mobility of Rad53, no evidence for a gel shift was observed in pol2-4 or pol2-M644G cells (Fig. S1). Increased expression of RNR-encoding genes provides a more sensitive indicator of checkpoint activation (28, 31, 37). Others have observed that RNR1 expression is independent of Dun1 activity and induced up to fivefold during checkpoint activation (28, 37), whereas RNR3 induction is induced up to 100-fold (28). Although initially thought to be solely Dun1-dependent (31), RNR3 induction can also occur by a Dun1-independent pathway (30, 57, 58). To investigate the checkpoint response to Pol ε mutations, we measured RNR1 and RNR3 transcript levels in POL2, pol2-4, and pol2-M644G strains. Increased levels (P ≤ 0.05) of RNR1 and RNR3 were detected in pol2-M644G cells (Fig. 3B). However, RNR3 induction in pol2-M644G cells was significantly lower than in hydroxyurea-treated cells, suggesting that the nature of the checkpoint signal is quantitatively or qualitatively different from the checkpoint signal found in cells experiencing chronic replication stress. Neither transcript was increased in pol2-4 cells (Fig. 3B), suggesting that Dun1 may influence the pol2-4 mutator phenotype independent of the S-phase checkpoint.
MRC1 Contributes to the Mutator Phenotype of pol2-M644G, but Not pol2-4.
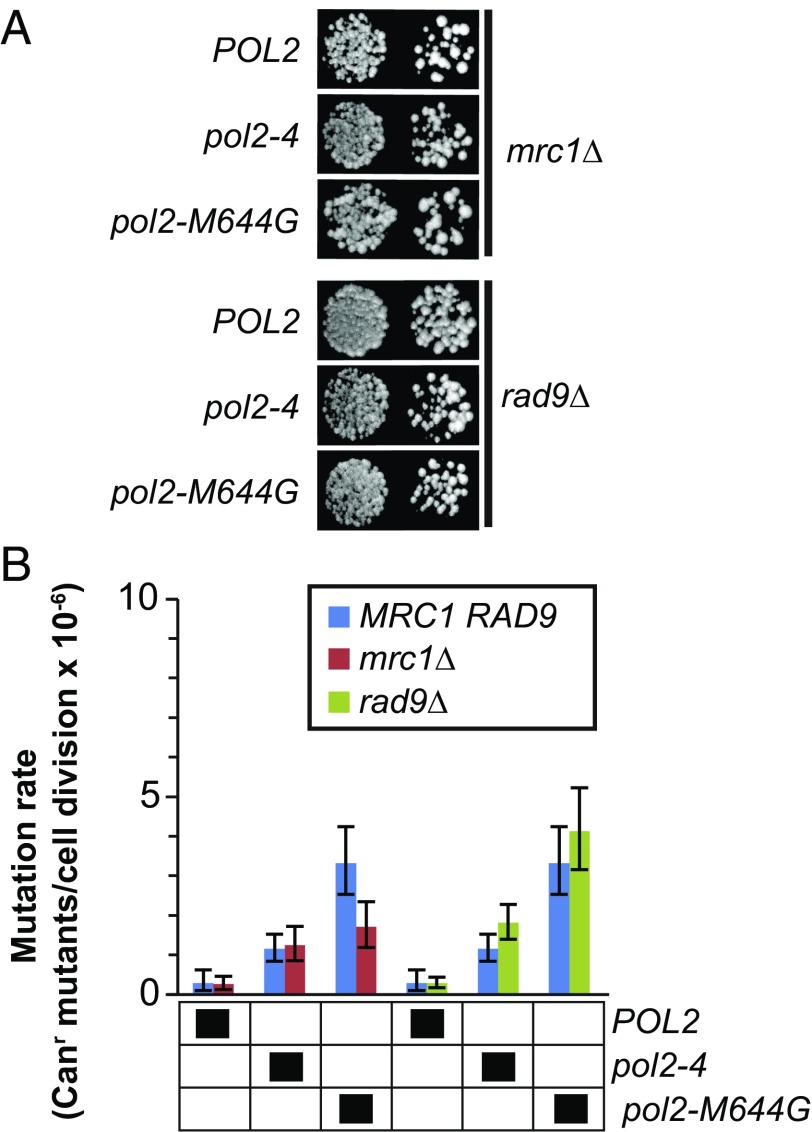
Mrc1 and Rad9 define two pathways by which Dun1 may be activated by the Mec1-Rad53 signaling pathway during the S-phase checkpoint (59, 60) (Fig. 1). Mutator Pol ε variants have the potential to activate Mrc1, which associates with Pol ε (43, 61, 62), through mispair-induced pausing of the replication machinery. Mutator Pol ε variants may also induce Rad9-dependent signaling through ssDNA produced by MMR processing of polymerase errors (63) or through replication fork collapse (64). We introduced pol2-4 and pol2-M644G into strains lacking Mrc1 or Rad9. The mrc1Δ and rad9Δ strains expressing either pol2 allele were viable, although mrc1Δ strains grew slowly. If Mrc1 or Rad9 participates in the activation of Dun1, then deletion of the corresponding genes should suppress the pol2-4 and pol2-M644G mutator phenotypes. The rad9Δ allele had no effect on the mutator phenotypes of either pol2-4 or pol2-M644G (Fig. 4B and Table 1). The mrc1Δ allele also had no effect on the pol2-4 mutator phenotype but did dampen the pol2-M644G mutator phenotype by 50% (Fig. 4B). These results agree with the lack of checkpoint activation in pol2-4 cells and suggest that pol2-M644G (Fig. 2) activates the checkpoint through Mrc1. Moreover, the viability of pol2-M644G mrc1Δ cells, in contrast to the synthetic lethality observed in pol2-M644G dun1Δ cells, indicates that Dun1 performs an essential function for pol2-M644G cells that does not require Mrc1-dependent checkpoint signaling.
Fig. 4.
Effects of Mrc1 and Rad9 on pol2-4 and pol2-M644G phenotypes. (A) Viability measurements. The mrc1Δ or rad9Δ strain was transformed with LEU2 plasmids carrying POL2, pol2-4, or pol2-M644G. Transformants were plated onto FOA media using 10-fold serial dilutions to select for loss of the complementing POL2-URA3 plasmid and photographed after 3 d at 30 °C. (B) Spontaneous mutation rates. Canr mutants per cell division were determined from multiple independent fluctuation analyses of each strain. The 95% confidence intervals for each mutation rate are shown as error bars. In the grid below the graphs, black boxes designate strain genotypes.
Modulation of dNTP Pools Accounts for the Dun1 Effect on pol2-4 and pol2-M644G.
The influence of Dun1 on the viability and mutator phenotypes of mutator cells could be mediated by a basal function of Dun1 that is amplified by checkpoint signaling. Because one of the main roles of Dun1 during checkpoint signaling is to regulate RNR, we hypothesized that Dun1 influences the Pol ε mutator polymerases through the modulation of dNTP pools. To test this hypothesis, we engineered DUN1- and dun1Δ-deficient strains with defects in Sml1, Crt1, and/or Dif1, the three main Dun1 targets that negatively regulate RNR. We found that deletion of SML1, but not of CRT1 or DIF1, suppressed the synthetic lethality between pol2-M644G and dun1Δ (Fig. 5A). Of the three proteins encoded by these genes, Sml1 plays the most direct role in regulating dNTP synthesis by binding RNR1 and inhibiting RNR activity. Thus, increased dNTP pools appear to be essential for survival of pol2-M644G cells.
Fig. 5.
dNTP levels correlate with mutator phenotypes. (A) Viability measurements. Strains with the indicated genotypes were transformed with LEU2 plasmids carrying Pol ε alleles (POL2, pol2-4, or pol2-M644G), plated onto FOA media to select for loss of the complementing POL2-URA3 plasmid, and photographed after 3 d at 30 °C. (B) Spontaneous mutation rates. Canr mutants per cell division were determined from multiple independent fluctuation analyses of each strain. The 95% confidence intervals for each mutation rate are shown as error bars. In the grid below the graphs, black boxes designate strain genotypes. (C) dNTP measurements. dNTPs were harvested from duplicate midlog-phase cultures of cells with the indicated genotypes, normalized to NTP levels in each sample, and then divided by total number of cells used for the preparation. Error bars reflect SE measurement based on the variance between the duplicate samples.
We measured mutation rates of the viable WT, pol2-4, and pol2-M644G strains (Fig. 5B) to determine if factors that influence dNTP pools also influence pol2 mutator phenotypes. The sml1Δ or crt1Δ mutation, but not the dif1Δ mutation, restored the pol2-4 mutator phenotype in dun1Δ cells (Fig. 5B). The pol2-4 sml1Δ crt1Δ cells had mutation rates eightfold higher than pol2-4 SML1 CRT1 cells. In contrast, the pol2-M644G mutator phenotype observed in DUN1 cells increased only modestly in sml1Δ crt1Δ cells, suggesting that the Sml1 and Crt1 proteins are already inactivated in pol2-M644G cells as part of the checkpoint response. No further increases in mutation rates were induced by elimination of all three downstream targets of Dun1 (sml1Δ, crt1Δ, and dif1Δ). Thus, mutations known to increase the levels of dNTP synthesis markedly enhance the pol2-4 mutator phenotype, but not the mutator phenotype of pol2-M644G.
These results suggest that dNTP levels directly modulate pol2 mutator phenotypes. To test this hypothesis, we measured dNTP pool levels of several key strains. The dNTP pool levels of pol2-4 cells were indistinguishable from WT, consistent with the normal cell cycle progression of pol2-4 cells. In contrast, dNTP pool levels in pol2-M644G cells were threefold higher than WT (Fig. 5C and Table 2), similar to what was observed previously by Kunkel and coworkers (65) and consistent with the checkpoint dependence of pol2-M644G. In pol2-4 dun1Δ cells, dNTP pool levels decreased by twofold. A similar twofold decrease in dNTP pool levels was previously observed in dun1Δ cells (66). Thus, a subtle decrease in dNTP pools may account for the antimutator effect of dun1Δ on the pol2-4 mutator phenotype. Strengthening the correlation between mutator phenotype severity and dNTP pool levels, pol2-4 sml1Δ crt1Δ cells, which displayed an eightfold increase in mutation rate, had a fourfold increase in dNTP pool levels (Fig. 5C).
Table 2.
dNTP concentrations
| Genotype | dCTP | dTTP | dATP | dGTP |
| WT | 139 ± 6 | 354 ± 13 | 219 ± 8 | 127 ± 1 |
| pol2-4 | 151 ± 9 | 376 ± 23 | 237 ± 18 | 126 ± 4 |
| pol2-M644G | 439 ± 24 | 1,051 ± 40 | 689 ± 39 | 325 ± 26 |
| pol2-4 dun1Δ | 91 ± 8 | 229 ± 5 | 132 ± 2 | 93 ± 4 |
| sml1Δ crt1Δ | 527 ± 15 | 1,238 ± 46 | 888 ± 43 | 381 ± 34 |
| pol2-4 sml1Δ crt1Δ | 541 ± 15 | 1,276 ± 16 | 898 ± 8 | 384 ± 8 |
dNTP concentrations (pmol per 108 cells) are the average of two biological replicates followed by the SE measurement.
Discussion
We have presented evidence that dNTP pools can modulate the severity of Pol ε fidelity defects in yeast. In what follows, we discuss the interplay between checkpoint activation, dNTP pools, and mutator polymerases, as well as the implications of our findings for the treatment of select human malignancies driven by POLE mutator alleles.
Mutator Polymerases and Checkpoint Signaling.
Dun1 was first identified as a regulator of damage-inducible RNR3 transcription (24). Since then, it has become one of the most well-characterized targets of the S-phase checkpoint in budding yeast (67–71). A previous study found that Dun1 contributed to the mutator phenotype of Pol δ proofreading deficiency but failed to find a role for Dun1 in the Pol ε proofreading deficiency encoded by the classic pol2-4 allele (21). Here, using an independently derived set of pol2-4 strains, we found that elimination of Dun1 suppresses the pol2-4 mutator phenotype fourfold. We speculate that the pol2-4 dun1Δ strain from the earlier study may have acquired a suppressor mutation like sml1Δ, which suppressed the dun1Δ effect on pol2-4 in our study (Fig. 5). Dun1 appears to influence the majority of Pol ε errors. Deletion of DUN1 rescues cells from lethal mutagenesis caused by combined defects in Pol ε proofreading and MMR (pol2-4 msh2Δ) (Fig. 2C) and suppresses the mutation rates of pol2-4 msh6Δ cells twofold to threefold (Fig. 2C). The sheer magnitude of Dun1-dependent mutations in these pol2-4 MMR-deficient strains suggests that Dun1 directly influences Pol ε fidelity. In support of this hypothesis, the pol2-4 mutator phenotype does not depend on the translesion polymerase Pol ζ or Pol η (Fig. 2B).
Despite the Dun1 dependence of the pol2-4 mutator phenotype, pol2-4 cells do not display overt signs of checkpoint activation. The pol2-4 cells traverse S-phase with the same kinetics as WT cells (Fig. 3A), lack increased Rad53 phosphorylation (Fig. S1), and do not display elevated RNR1 or RNR3 transcripts (Fig. 3B). Moreover, the pol2-4 mutator phenotype does not depend on the checkpoint mediator Rad9 or Mrc1 (Fig. 4). Finally, pol2-4 cells have similar dNTP pool levels as WT cells, which decrease twofold in the absence of Dun1 (66) (Fig. 5C). However, others have found that cells bearing hypomorphic alleles affecting the central checkpoint signaling component Mec1 or Rad53 also show a twofold reduction in dNTP pools in the absence of DNA damage (72). Thus, a low level of checkpoint signaling may contribute to normal dNTP homeostasis, explaining the influence of Dun1 on pol2-4. The Mec1-Rad53-Dun1 signaling axis can be regarded as a rheostat for dNTP pool regulation, which is always on but turned down to the lowest point in WT and pol2-4 cells. As dNTP pools rise, the pol2-4 mutator phenotype increases linearly. An approximately sixfold increase in dNTP pool levels from pol2-4 dun1Δ cells to pol2-4 sml1Δ crt1Δ cells produces a 27-fold increase in mutation rate. These findings are consistent with numerous biochemical studies indicating that the concentration of dNTPs influences the efficiency at which mismatched primer termini are extended (73–77).
Unlike pol2-4, pol2-M644G clearly activates the checkpoint. Although a clear Rad53 gel shift is also not apparent (Fig. S1), pol2-M644G cells transit S-phase more slowly than POL2 cells, display increased expression of RNR3, and have higher dNTP pools (65) (Fig. 5C). The elevated dNTP pools appear to feed back to enhance the pol2-M644G mutator phenotype, as indicated by the antimutator effect of mrc1Δ on pol2-M644G (Fig. 4). The viability of pol2-M644G mrc1Δ cells suggests that the role of Dun1 in maintaining normal dNTP pools, rather than its other checkpoint functions, is essential for survival of pol2-M644G cells. Consistent with this conclusion, pol2-M644G dun1Δ synthetic lethality is suppressed by sml1Δ. The pol2-M644G allele introduces a substitution in the polymerase active site that causes characteristic changes in the mutation spectrum generated by Pol ε, as well as a reduction in catalytic activity (5). Thus, pol2-M644G cells may require higher dNTP pools to compensate for low Pol ε catalytic efficiency or to extend misinsertion errors created by the mutant polymerase, which include elevated numbers of ribonucleotides (65). The characteristic mutation spectrum of pol2-M644G has been used to map the extent of Pol ε DNA synthesis in the yeast genome (5, 78, 79). The dependence of Pol ε-M644G on checkpoint activation suggests some caution should be used in inferring the full contribution of Pol ε to genome-wide synthesis. Pol ε-M644G replication complexes in checkpoint-activated cells may differ in processivity and/or composition from the Pol ε complexes found in normal cells.
Implications for Treatment of Mutator-Driven Cancer.
The findings presented here provide a potential avenue for treating mutator-driven cancers. It has long been appreciated that mutator clones emerge from cell populations subjected to multiple rounds of selection (80–86). This fact is highly relevant to tumorigenesis, where cancer cells overcome numerous endogenous and chemotherapeutic barriers during disease progression (2, 87).
Tumors with POLE or MMR mutations may be amenable to treatments that exploit the synergistic relationship between polymerase accuracy, proofreading, and MMR. Although theory and experimental evidence indicate that haploid and diploid mutator cells evolve more rapidly than nonmutators (2, 80–86, 88), prolonged expression of a strong mutator phenotype can also drive extinction (18, 19, 89, 90). We recently empirically defined the maximum mutation rate of diploid yeast (20) using combinations of mutator alleles. Once mutation rates are within an order of magnitude of the maximum, extinction appears inexorable. Mammalian cells may also be susceptible to error-induced extinction. Mice deficient in MMR and either Pol δ or Pol ε proofreading die by embryonic days 9.5 and 14.5, respectively (17). Recently, individuals with biallelic MMR deficiency were found to acquire early driver mutations in POLE that increased the mutation rate in brain tumors to an estimated 600 mutations per exome per cell division (91). Analysis of sequential tumor biopsies in that study found that mutation burden rapidly increased until a maximum mutation threshold was reached (mean = 249 × 10−6 mutations per base pair) (91). A similar average mutation frequency is observed in the exomes of EC POLE tumors (235 × 10−6 mutations per base pair), which do not appear to be MMR-deficient (10, 12, 14). Thus, we hypothesize that the POLE mutator phenotype drives these cancer cells to the verge of error-induced extinction. Our findings, as well as the findings in a companion paper in PNAS by Mertz et al. (92) and previous work in bacteria (93–95), suggest a treatment strategy for these patients may be to increase dNTP pools, thereby elevating polymerase errors. In normal cells that retain proofreading and MMR, such an approach would induce only a modest increase in mutation rates. In contrast, increasing the number of polymerase errors in cells with proofreading deficiency may have a multiplicative effect on mutation rates, pushing cells over the error threshold for extinction. The elevated dNTP pools may initially increase the rate of tumor cell replication, but we posit that the higher mutation rate will cause a rapid decline in replicative fitness.
In mammals, dNTP pool size and composition during S-phase are largely regulated by transcriptional, allosteric, and proteolytic control of RNR, which is composed of large and small subunits encoded by RRM1 and RRM2, respectively. Transcription of these genes markedly increases at the G1/S transition, peaking in the middle of S-phase (96). Due to a long t1/2, RRM1 protein remains relatively constant in proliferating cells. In contrast, RRM2 levels fall during G1 and G2 due to ubiquitin-mediated degradation, ensuring dNTP pools decline to levels that limit unscheduled DNA replication (97, 98). Unlike the case in yeast, dNTP pools in mammals appear unperturbed by DNA damage signaling (99). The core checkpoint signaling components ATR (MEC1) and CHK1/CHK2 (RAD53) are conserved, but mammalian cells do not encode orthologs of Dun1 or Sml1. Activation of mammalian p53 by checkpoint signaling does induce the production of the RRM2 paralog, p53R2 (100, 101), but its role is limited to the maintenance of mtDNA and DNA repair in nondividing differentiated cells (102). Checkpoint signaling also leads to translocation of the RRM1/RRM2 complex into the nucleus and recruitment to the sites of DNA damage through an interaction with the histone acetylase, Tip60 (103), where it presumably increases the local concentration of dNTPs. In principle, it is possible that RNR is also recruited to replication forks slowed by mutator variants of Pol ε. Such RNR localization could elevate dNTP pools just in the vicinity of the polymerase, thereby increasing mutagenesis. Because there currently is no evidence for this mechanism, we favor the hypothesis that global dNTP pool changes do not normally enhance the POLE mutator phenotype during bulk DNA synthesis. Therefore, implementation of an error-induced extinction strategy for POLE-driven tumors should ideally focus on ways to enhance the activity of the main RNR complex during S-phase. Targeting allosteric control of RNR would be a logical first step. Two allosteric sites regulate RNR activity (26). The allosteric specificity site controls NDP selection in the catalytic site, ensuring that dNTP pools remain balanced. The allosteric activity site controls overall RNR activity: Binding of dATP inactivates the enzyme, whereas ATP stimulates catalysis (104). Substitution of a highly conserved amino acid (D57N) in the allosteric activity site of yeast and mammalian RNR leads to elimination of dATP feedback inhibition and an elevated nucleotide pool (38, 104). These studies provide proof of concept that an ATP mimic with higher affinity for the activity site could boost dNTP pools during S-phase. The challenge will be to design an ATP analogue that is specific to RNR to minimize off-target effects. An alternative strategy would be to use drugs that cause nucleotide pool imbalances by perturbing the allosteric specificity sites of RNR or dCMP deaminase (26). The resulting increase in nucleotide misinsertion events would increase mutation rates synergistically with proofreading deficiency in POLE tumor cells but would be largely suppressed in normal cells.
In the age of precision medicine, it is worth noting that this treatment strategy will not work for patients with rare germline POLE mutations (13). Their normal cells would suffer the same dramatic increase in mutation rate as the cancer cells. However, the work presented here and the study by Mertz et al. (92) suggest that before the appearance of any cancer, such patients may lower their cancer risk through prophylactic treatment with drugs that diminish overall dNTP pools. Many drugs already in use for chemotherapy suppress dNTP pools but create mutagenic dNTP pool imbalances. Because POLE patients will need dNTPs for somatic maintenance, the goal should be to find treatments that lead to a balanced reduction in dNTP pools in a manner similar to Sml1.
Materials and Methods
Media and Growth Conditions.
Standard media and growth conditions were used to culture yeast strains (105). Cells were propagated nonselectively on YPD (10 g/L yeast extract, 20 g/L bacto peptone, 20 g/L dextrose) or synthetic complete (SC) media [1.7 g/L yeast nitrogen base (Difco), 37.8 mM ammonium sulfate, 20 g/L dextrose, 2 g/L SC amino acid supplement (Bufferad)]. Prototrophic clones were selected on SC media lacking the appropriate amino acid(s). Preformulated SC amino acid supplement minus Ura and Leu was purchased from Bufferad. All other amino acid-limited supplements were mixed from individual amino acids as described (105). Cells lacking URA3 were selected with media containing 5-fluorootic acid (FOA; 1 mg/mL; Zymo Research) (106). The can1 mutants were selected on SC media lacking Arg and containing 50 μg/mL canavanine. Unless otherwise specified, reagents were purchased from Sigma–Aldrich or Fischer Scientific.
Yeast Plasmids and Strains.
POL2 plasmids.
We previously described POL2 or pol2-4 CEN6/ARS4 plasmids containing URA3 (pRS416POL2) or LEU2 (pRS415POL2 and pRS415pol2-4) (19). For this study, we engineered a pol2-M644G-LEU2 plasmid (pRS415pol2-M644G) using mutagenic primers (Table S2) and the QuikChange protocol (107), with the following PCR conditions: Phusion polymerase (New England Biolabs) at 95 °C for 1 min and 16 cycles of 95 °C for 40 s; 53 °C for 1 min; 68 °C for 7 min. Following cloning in bacteria, the entire pol2 gene was sequenced to confirm the presence of the pol2-M644G mutation and the absence of any additional mutations.
Strains.
All strains used in this study (Table S1) were derived from the BY4733 strain (108) and carry a pol2Δ::KANMX insertion complemented by the pRS416POL2 plasmid, which provides the essential activity of Pol ε. All other chromosomal gene disruptions were made by transforming yeast (109) with PCR products generated with Phusion polymerase and the primers and PCR conditions indicated in Table S2. Correct clones were identified by PCR genotyping of zymolyase (MP Biomedicals)-treated cells using primers that amplify unique junctions between the transgenes and endogenous loci.
Plasmid Shuffling.
We used plasmid shuffling to freshly derive all WT and mutator strains used in our studies, thereby avoiding artifacts due to genetic drift. Cells were transformed with pRS415POL2, pRS415pol2-4, or pRS415pol2-M644G and plated on SC media lacking Leu and Ura, which maintains selection for pRS416POL2 as well as the incoming LEU2 plasmid. Selection for both WT and mutator alleles at this stage masks any lethal phenotypes due to the mutator allele. To examine the effect of the mutator pol2 alleles in the absence of pRS416POL2, the transformants were picked, suspended in water, and plated in serial dilutions onto media containing FOA, which selects for cells that have lost the URA3-containing plasmid. In the resulting colonies, the sole source of Pol ε is the LEU2 plasmid, which allows the influence of the mutator allele on growth and mutation rates to be assessed. Please note that the WT reference strain for our experiments was LW14 (Table S1), shuffled with the pRS415POL2 plasmid.
Mutation Rates.
We measured canavanine-resistant (Canr) mutation rates as described (18). Briefly, 2 d after plating POL2 plasmid shuffling strains onto FOA media, at least 24 FOA-resistant colonies representing at least three independent plasmid shuffling experiments were suspended separately in 100 μL of water. We used 10 μL of each cell suspension for 10-fold serial dilutions to determine the total number of cells per colony (Nt). The remaining 90 μL was plated onto canavanine selection plates to determine the number of Canr mutants. Plates were incubated at 30 °C, and colonies were counted after 3 d. Mutation rates were calculated from the mutant counts in each replica culture by estimating the probable number of mutations (m) by maximum likelihood using newtonLDPlating in Salvador 2.3 (110) with Mathematica 8.0 and then dividing by the number of cell divisions inferred from the Nt. Confidence intervals (95%) were calculated with CILDplating in Salvador 2.3.
Cell Cycle Analysis.
Cell synchronization.
Cells were synchronized using α-factor (Zymo Research) (111). Strains were grown overnight in 50 mL of YPD at 30 °C to an OD600 of 0.2–0.3. The α-factor was diluted in methanol and added at a final concentration of 5 μM. Cells were checked for synchrony after 90 min (10 μL of culture was sonicated for 10 s and examined on the microscope for unbudded cells). If unbudded cells remained, more α-factor was added and cells were reexamined every 30 min. When only unbudded cells were present, cultures were collected in 50-mL Falcon tubes and centrifuged to pellet cells. Cells were rinsed once in water and resuspended into 25 mL of warm YPD containing Pronase E (10 μg/mL) to release them from α-factor arrest.
Cell collection and ethanol fixation.
One milliliter of cells was collected before α-factor arrest (log-phase sample) and before release from arrest (0-min sample). Following release, 1 mL of cells was collected every 15 min for 210 min. Immediately following collection, cells were spun down in 1.5-mL microcentrifuge tubes. YPD was aspirated, and cells were rinsed once in water. The cell pellet was then resuspended in 300 μL of water and vigorously vortexed. Seven hundred microliters of 95% ethanol was immediately added, and cells were vortexed again. Cells were fixed overnight at 4 °C.
SYTOX green staining.
Fixed cells were spun down and washed in 500 μL of water, and then resuspended in 500 μL of sodium citrate (50 mM) solution containing RNase (10 μg/mL; Thermo Scientific). Samples were heated to 95 °C for 15 min and then incubated at 37 °C for at least 2 h. Samples were spun down and resuspended in 500 μL of 50 mM sodium citrate without RNase and probe-sonicated for 20 s. Finally, samples were spun down and stained with 500 μL of 50 mM sodium citrate containing SYTOX green (2 μM; Invitrogen) and immediately analyzed using FACS. FACS data were analyzed using WinCycle (Phoenix Flow Systems) and FCS Express (De Novo Software).
Doubling time.
Strain doubling times were measured using a Bioscreen C MBR precision incubator and growth-monitoring instrument (Growth Curves Inc.). Five colonies of each genotype were inoculated into standard 2-mL cultures and grown overnight to saturation. Cultures were then diluted 1:200 and incubated in the Bioscreen, which monitored growth by measuring by OD. Doubling times were calculated as previously described (112).
Quantitative Reverse Transcriptase PCR.
cDNA preparation.
Cells from three biological replicates were grown at 30 °C in 25 mL of YPD until log-phase. One hundred-microliter cell pellets were recovered by centrifugation and stored at −80 °C. RNA was extracted from each pellet with 1.0 mL of Tri-Reagent (Sigma), 300 μL of acid-washed glass beads, and 14 min of vigorous vortexing with a Disrupter-Genie (Scientific Industries). Chloroform (0.2 mL) was added. The tubes were mixed by inversion, incubated for 3 min, and then spun for 15 min (12,000 × g) at 4 °C. Roughly 1 mL of supernatant was transferred to a fresh microfuge tube, precipitated with 0.5 mL of isopropanol, and resuspended in 100 μL H2O. After assessing RNA quality and quantitation, 10 μg of RNA was DNase-treated with DNase I (New England Biolabs), phenol-chloroform extracted, precipitated, and resuspended in 20 μL H2O. cDNA was synthesized from 1.25 μg of RNA using a Transcriptor First Strand cDNA Synthesis Kit (Roche) and oligo-dT.
Quantitative PCR Analysis.
The cDNA levels were measured using quantitative PCR analysis. RNR1 and RNR3 transcripts were measured using ACT1 as a reference control. cDNA was amplified using the following primers: Rnr1 forward (F) 5′-TGCCCCAACTATGGGTAAACTAACA-3′ and Rnr1 reverse (R) 5′-CACTGGAAGGCATATATGA-3′; Rnr3F 5′-GATCGTCCAGTTTATGTTCCAAAGGGTA-3′ and Rnr3R 5′-TATTGTTTCCGTTGGAACTGCT-3′; and Act1F, 5′-GAAATGCAAACCGCTGCTCA-3′ and Act1R 5′-TACCGGCAGATTCCAAACCC-3′. Quantitative PCR assays were performed using Brilliant III Ultra-Fast QPCR Master Mix (Agilent Technologies).
Immunoblot Analysis.
Cells from each genotype were grown to saturation in 3 mL of synthetic media lacking Leu to maintain selection for the POL2, pol2-4, or pol2-M644G plasmids. Cells for positive control of checkpoint activation were diluted to OD0.3 and treated with 200 mM hydroxyurea for 2 h. All samples were then resuspended in 300 μL of 0.1 NaOH and incubated at room temperature for 10 min. After centrifugation, cells were resuspended in 50 μL of Western sample buffer and boiled for 5 min. Cellular debris was pelleted, and 10 μL of supernatant for each sample was run on a 10% SDS polyacrylamide gel and electroblotted to a Hybond ECL nitrocellulose blotting membrane (Amersham). The primary antibody was goat yC-19 anti-Rad53 (Santa Cruz Biotechnology) diluted 1:1,000. The secondary antibody was an HRP-conjugated donkey anti-goat IgG antibody (Jackson ImmunoResearch) diluted 1:5,000. Antibody binding was visualized using ECL Prime Western Blotting Detection Reagent (GE Healthcare) and visualized with an AlphaImager (ProteinSimple).
dNTP Measurements.
Two independent isolates of select plasmid shuffling strains used for mutation rate measurements were patched onto SC plates. These plates were used to inoculate overnight cultures, which were then diluted the following morning. Strains were grown in YPAD media (1% yeast extract, 2% bactopeptone, 0.002% adenine, 2% dextrose) at 30 °C. Approximately 3.7 × 108 cells (determined by OD600) were harvested by filtration through 25-mm AAWP nitrocellulose filters (0.8 μm; Millipore AB). The filters were immersed in 700 μL of ice-cold extraction solution [12% (wt/vol) trichloroacetic acid, 15 mM MgCl2] and frozen in liquid nitrogen. The following steps were carried out at 4 °C. The tubes were vortexed for 30 s and vortex-mixed on an Intelli-Mixer (ELMI Ltd.) for 15 min in a cold room. The filters were removed, and the 700-μL supernatants were collected after centrifugation at 20,000 × g for 1 min and added to 800 μL of ice-cold Freon-trioctylamine mixture [10 mL of Freon (1,1,2-trichloro-1,2,2-trifluoroethane); Sigma–Aldrich Sweden AB (>99%) and 2.8 mL of trioctylamine; Sigma–Aldrich Sweden AB (98%)]. The samples were vortexed and centrifuged for 1 min at 20,000 × g. The aqueous phase was collected and added to 700 μL of ice-cold Freon-trioctylamine mixture and vortexed and centrifuged for 1 min at 20,000 × g. Volumes of 475 μL and 47.5 μL of the aqueous phase were collected. The 475-μL aliquots of the aqueous phase were pH-adjusted with 1 M ammonium carbonate [(NH4)2CO3] at pH 8.9, loaded on boronate columns (Affi-Gel Boronate Gel; Bio-Rad), and eluted with 50 mM ammonium carbonate [(NH4)2CO3] at pH 8.9 and 15 mM MgCl2 to separate dNTPs and NTPs. The eluates (purified dNTPs) were adjusted to pH 3.4 with 6 M HCl, separated on a Partisphere SAX HPLC column (125 mm × 4.6 mm; Hichrome) under isocratic elusion with 0.35 M potassium phosphate buffer [pH 3.4; containing 2.5% (vol/vol) acetonitrile] and quantified using a LaChrom Elite HPLC system (Hitachi). The 47.5-μL aliquots of the aqueous phase were adjusted to pH 3.4 and used to quantify NTPs by HPLC in the same way (38, 66).
The raw NTP values (height, milli-absorbance units (mAU)) were highly similar between the different strains and preparations (Fig. S2) and were used as a normalization factor/internal standard. To correct for subtle differences due to sample processing and to normalize the levels of dNTPs, we used several steps. First, to normalize the relative differences in NTPs between samples, we divided the values of each NTP in each sample by the value of each NTP found in one of the WT samples. The resulting four normalized values were averaged to obtain a single NTP normalization factor for each sample. We then normalized the raw dNTP values (height, mAU) between preparations by dividing the dCTP, dTTP, dATP, and dGTP values from each sample by the aforementioned NTP normalization factors. These normalized dNTP data were then multiplied by known amounts of dNTP standards recovered from the same chromatography columns. Finally, the values were corrected for the number of cells used in the preparations to obtain picomoles of dNTPs per 108 cells.
Supplementary Material
Acknowledgments
We thank Brad Preston and the laboratory of Larry Loeb for valuable advice and discussions. This project was supported by Grant R21 ES021544 from the National Institute of Environmental Health Sciences. Additional funding for A.J.H. was provided by a Hitchings–Elion Fellowship from the Burroughs Wellcome Fund and a University of Washington Nathan Shock Junior Faculty Fellowship. L.N.W. was supported by Public Health Service National Research Service Award T32 GM07270. L.M. and A.C. were supported by the Knut and Alice Wallenberg Foundation and the Swedish Cancer Society. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Sciences, the National Institute on Aging, the National Institutes of Health, or the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.A.L. is a guest editor invited by the Editorial Board.
See Commentary on page 5864.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422948112/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA. Human cancers express mutator phenotypes: Origin, consequences and targeting. Nat Rev Cancer. 2011;11(6):450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 4.Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20(5):281–293. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317(5834):127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinbrot E, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24(11):1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C, et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56(4):551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clausen AR, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 2015;22(3):185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PMJ, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Church DN, et al. NSECG Collaborators DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22(14):2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida R, et al. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur J Hum Genet. 2011;19(3):320–325. doi: 10.1038/ejhg.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palles C, et al. CORGI Consortium WGS500 Consortium Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandoth C, et al. Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsby RE, et al. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc Natl Acad Sci USA. 2002;99(24):15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsby RE, et al. Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat Med. 2001;7(6):638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 17.Albertson TM, et al. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci USA. 2009;106(40):17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herr AJ, et al. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2011;7(10):e1002282. doi: 10.1371/journal.pgen.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams LN, Herr AJ, Preston BD. Emergence of DNA polymerase ε antimutators that escape error-induced extinction in yeast. Genetics. 2013;193(3):751–770. doi: 10.1534/genetics.112.146910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herr AJ, Kennedy SR, Knowels GM, Schultz EM, Preston BD. DNA replication error-induced extinction of diploid yeast. Genetics. 2014;196(3):677–691. doi: 10.1534/genetics.113.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datta A, et al. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3-01 mutants. Mol Cell. 2000;6(3):593–603. doi: 10.1016/s1097-2765(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 22.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297(5581):547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 23.Tourrière H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 2007;6(7):900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z, Elledge SJ. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75(6):1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- 25.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57(1):349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 26.Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20(9):1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 27.Elledge SJ, Davis RW. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol Cell Biol. 1987;7(8):2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elledge SJ, Davis RW. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4(5):740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 29.Chabes A, et al. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc Natl Acad Sci USA. 2000;97(6):2474–2479. doi: 10.1073/pnas.97.6.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang M, Elledge SJ. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17(10):6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94(5):595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA. 2002;99(6):3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell. 2008;32(1):70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Huang M. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol. 2008;28(23):7156–7167. doi: 10.1128/MCB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2(3):329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 36.Chabes A, Domkin V, Thelander L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J Biol Chem. 1999;274(51):36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- 37.Tsaponina O, Barsoum E, Aström SU, Chabes A. Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools. PLoS Genet. 2011;7(5):e1002061. doi: 10.1371/journal.pgen.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chabes A, et al. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112(3):391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 39.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12(18):2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80(1):29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 41.Navas TA, Sanchez Y, Elledge SJ. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 1996;10(20):2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- 42.Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274(32):22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 43.Lou H, et al. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell. 2008;32(1):106–117. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puddu F, Piergiovanni G, Plevani P, Muzi-Falconi M. Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase ε. PLoS Genet. 2011;7(3):e1002022. doi: 10.1371/journal.pgen.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′----5′ exonuclease activity. Proc Natl Acad Sci USA. 1991;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev. 2002;16(15):1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 47.Shcherbakova PV, Noskov VN, Pshenichnov MR, Pavlov YI. Base analog 6-N-hydroxylaminopurine mutagenesis in the yeast Saccharomyces cerevisiae is controlled by replicative DNA polymerases. Mutat Res. 1996;369(1-2):33–44. doi: 10.1016/s0165-1218(96)90045-2. [DOI] [PubMed] [Google Scholar]
- 48.Pavlov YI, Shcherbakova PV, Kunkel TA. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics. 2001;159(1):47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kai M, Wang TS-F. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17(1):64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 2006;25(18):4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Northam MR, Robinson HA, Kochenova OV, Shcherbakova PV. Participation of DNA polymerase ζ in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 2010;184(1):27–42. doi: 10.1534/genetics.109.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aksenova A, et al. Mismatch repair-independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase ε. PLoS Genet. 2010;6(11):e1001209. doi: 10.1371/journal.pgen.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12(4):1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greene CN, Jinks-Robertson S. Spontaneous frameshift mutations in Saccharomyces cerevisiae: Accumulation during DNA replication and removal by proofreading and mismatch repair activities. Genetics. 2001;159(1):65–75. doi: 10.1093/genetics/159.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran HT, Gordenin DA, Resnick MA. The 3′-->5′ exonucleases of DNA polymerases δ and ε and the 5′-->3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(3):2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sokolsky T, Alani E. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics. 2000;155(2):589–599. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zaim J, Speina E, Kierzek AM. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae. J Biol Chem. 2005;280(1):28–37. doi: 10.1074/jbc.M404669200. [DOI] [PubMed] [Google Scholar]
- 58.Davidson MB, et al. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012;31(4):895–907. doi: 10.1038/emboj.2011.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcasabas AA, et al. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3(11):958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- 60.Paulovich AG, Margulies RU, Garvik BM, Hartwell LH. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145(1):45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17(14):1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naylor ML, Li JM, Osborn AJ, Elledge SJ. Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc Natl Acad Sci USA. 2009;106(31):12765–12770. doi: 10.1073/pnas.0904623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reha-Krantz LJ, et al. Drug-sensitive DNA polymerase δ reveals a role for mismatch repair in checkpoint activation in yeast. Genetics. 2011;189(4):1211–1224. doi: 10.1534/genetics.111.131938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lopes M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412(6846):557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 65.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6(10):774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fasullo M, Tsaponina O, Sun M, Chabes A. Elevated dNTP levels suppress hyper-recombination in Saccharomyces cerevisiae S-phase checkpoint mutants. Nucleic Acids Res. 2010;38(4):1195–1203. doi: 10.1093/nar/gkp1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. BioEssays. 1993;15(5):333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 68.Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8(20):2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 69.Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104(3):397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 70.Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem. 2010;285(17):12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andreson BL, Gupta A, Georgieva BP, Rothstein R. The ribonucleotide reductase inhibitor, Sml1, is sequentially phosphorylated, ubiquitylated and degraded in response to DNA damage. Nucleic Acids Res. 2010;38(19):6490–6501. doi: 10.1093/nar/gkq552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20(13):3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petruska J, et al. Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci USA. 1988;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mendelman LV, Petruska J, Goodman MF. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J Biol Chem. 1990;265(4):2338–2346. [PubMed] [Google Scholar]
- 75.Perrino FW, Loeb LA. Differential extension of 3′ mispairs is a major contribution to the high fidelity of calf thymus DNA polymerase-alpha. J Biol Chem. 1989;264(5):2898–2905. [PubMed] [Google Scholar]
- 76.Perrino FW, Preston BD, Sandell LL, Loeb LA. Extension of mismatched 3′ termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1989;86(21):8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuchta RD, Benkovic P, Benkovic SJ. Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry. 1988;27(18):6716–6725. doi: 10.1021/bi00418a012. [DOI] [PubMed] [Google Scholar]
- 78.Larrea AA, et al. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci USA. 2010;107(41):17674–17679. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lujan SA, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24(11):1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chao L, Cox EC. Competition between high and low mutating strains of Escherichia coli. Evolution. 1983;37(1):125–134. doi: 10.1111/j.1558-5646.1983.tb05521.x. [DOI] [PubMed] [Google Scholar]
- 81.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387(6634):703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 82.Mao EF, Lane L, Lee J, Miller JH. Proliferation of mutators in A cell population. J Bacteriol. 1997;179(2):417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giraud A, et al. Costs and benefits of high mutation rates: Adaptive evolution of bacteria in the mouse gut. Science. 2001;291(5513):2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 84.Nilsson AI, Kugelberg E, Berg OG, Andersson DI. Experimental adaptation of Salmonella typhimurium to mice. Genetics. 2004;168(3):1119–1130. doi: 10.1534/genetics.104.030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Notley-McRobb L, Seeto S, Ferenci T. Enrichment and elimination of mutY mutators in Escherichia coli populations. Genetics. 2002;162(3):1055–1062. doi: 10.1093/genetics/162.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loh E, Salk JJ, Loeb LA. Optimization of DNA polymerase mutation rates during bacterial evolution. Proc Natl Acad Sci USA. 2010;107(3):1154–1159. doi: 10.1073/pnas.0912451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34(9):2311–2321. [PubMed] [Google Scholar]
- 88.Thompson DA, Desai MM, Murray AW. Ploidy controls the success of mutators and nature of mutations during budding yeast evolution. Curr Biol. 2006;16(16):1581–1590. doi: 10.1016/j.cub.2006.06.070. [DOI] [PubMed] [Google Scholar]
- 89.Zeyl C, Mizesko M, de Visser JAGM. Mutational meltdown in laboratory yeast populations. Evolution. 2001;55(5):909–917. doi: 10.1554/0014-3820(2001)055[0909:mmilyp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 90.Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of Escherichia coli dnaQ: Defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93(7):2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shlien A, et al. Biallelic Mismatch Repair Deficiency Consortium Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 92.Mertz TM, Sharma S, Chabes A, Shcherbakova PV. Colon cancer-associated mutator DNA polymerase δ variant causes expansion of dNTP pools increasing its own infidelity. Proc Natl Acad Sci USA. 2015;112:E2467–E2476. doi: 10.1073/pnas.1422934112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahluwalia D, Schaaper RM. Hypermutability and error catastrophe due to defects in ribonucleotide reductase. Proc Natl Acad Sci USA. 2013;110(46):18596–18601. doi: 10.1073/pnas.1310849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108(48):19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wheeler LJ, Rajagopal I, Mathews CK. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair (Amst) 2005;4(12):1450–1456. doi: 10.1016/j.dnarep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 96.Björklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29(23):5452–5458. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- 97.Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci USA. 2003;100(7):3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.D’Angiolella V, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149(5):1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281(12):7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 100.Nakano K, Bálint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19(37):4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 101.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404(6773):42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 102.Thelander L. Ribonucleotide reductase and mitochondrial DNA synthesis. Nat Genet. 2007;39(6):703–704. doi: 10.1038/ng0607-703. [DOI] [PubMed] [Google Scholar]
- 103.Niida H, et al. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 2010;24(4):333–338. doi: 10.1101/gad.1863810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reichard P, Eliasson R, Ingemarson R, Thelander L. Cross-talk between the allosteric effector-binding sites in mouse ribonucleotide reductase. J Biol Chem. 2000;275(42):33021–33026. doi: 10.1074/jbc.M005337200. [DOI] [PubMed] [Google Scholar]
- 105.Sherman F. 2002. Getting started with yeast. Part B: Guide to Yeast Genetics and Molecular and Cell Biology, Methods in Enzymology, eds Guthrie C, Fink GR (Academic, San Diego), Vol 350, pp 3–41.
- 106.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 107.Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26(4):680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 108.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 109.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Part B: Guide to Yeast Genetics and Molecular and Cell Biology, Methods in Enzymology, eds Guthrie C, Fink GR (Academic, San Diego), Vol 350, pp 87–96.
- 110.Zheng Q. A note on plating efficiency in fluctuation experiments. Math Biosci. 2008;216(2):150–153. doi: 10.1016/j.mbs.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 111.Breeden LL. 1997. α-Factor Synchronization of Budding Yeast. Methods in Enzymology, ed William GD (Academic, San Diego), Vol 283, pp 332–342.
- 112.Olsen B, Murakami CJ, Kaeberlein M. YODA: Software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC Bioinformatics. 2010;11:141. doi: 10.1186/1471-2105-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.