Significance
Cancer cell proliferation is highly dependent on cap-dependent protein synthesis, which is generally assumed to be inhibited during mitosis. Using a viral oncoprotein that enforces mitosis, we show that CDK1 substitutes for mTOR interphase functions to phosphorylate eukaryotic initiation factor 4E-binding protein (4E-BP1) to a mitosis-specific δ isoform. Flow cytometric assays reveal that mitotic cells have high levels of inactivated 4E-BP1 and do not generally show specific loss of cap-dependent translation compared with interphase cells. This appears to be due to cyclin-dependent kinase 1 (CDK1) activity during mitosis. Mitotic cells typically represent less than 1% of all cells in bulk culture, and mitosis-arresting drugs, such as nocodazole, can directly inhibit mitotic protein translation, potentially explaining differences between our findings and previous studies showing reduced cap-dependent translation during mitosis.
Keywords: Merkel cell, small T, 4E-BP1, cyclin-dependent kinase 1, mitosis
Abstract
Mitosis is commonly thought to be associated with reduced cap-dependent protein translation. Here we show an alternative control mechanism for maintaining cap-dependent translation during mitosis revealed by a viral oncoprotein, Merkel cell polyomavirus small T (MCV sT). We find MCV sT to be a promiscuous E3 ligase inhibitor targeting the anaphase-promoting complex, which increases cell mitogenesis. MCV sT binds through its Large T stabilization domain region to cell division cycle protein 20 (Cdc20) and, possibly, cdc20 homolog 1 (Cdh1) E3 ligase adapters. This activates cyclin-dependent kinase 1/cyclin B1 (CDK1/CYCB1) to directly hyperphosphorylate eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP1) at authentic sites, generating a mitosis-specific, mechanistic target of rapamycin (mTOR) inhibitor-resistant δ phospho-isoform not present in G1-arrested cells. Recombinant 4E-BP1 inhibits capped mRNA reticulocyte translation, which is partially reversed by CDK1/CYCB1 phosphorylation of 4E-BP1. eIF4G binding to the eIF4E–m7GTP cap complex is resistant to mTOR inhibition during mitosis but sensitive during interphase. Flow cytometry, with and without sT, reveals an orthogonal pH3S10+ mitotic cell population having higher inactive p4E-BP1T37/T46+ saturation levels than pH3S10– interphase cells. Using a Click-iT flow cytometric assay to directly measure mitotic protein synthesis, we find that most new protein synthesis during mitosis is cap-dependent, a result confirmed using the eIF4E/4G inhibitor drug 4E1RCat. For most cell lines tested, cap-dependent translation levels were generally similar between mitotic and interphase cells, and the majority of new mitotic protein synthesis was cap-dependent. These findings suggest that mitotic cap-dependent translation is generally sustained during mitosis by CDK1 phosphorylation of 4E-BP1 even under conditions of reduced mTOR signaling.
Eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP1) is a principal target for mechanistic target of rapamycin complex 1 (mTORC1) (1–3). mTOR regulates a variety of metabolic signaling pathways related to ribosomal biosynthesis and autophagy that contribute to cancer cell survival (1, 3–6). Increasing evidence indicates that direct mTORC1 phosphorylation of 4E-BP1 may be the key event in mTOR-associated tumorigenesis (2). In the absence of activated mTOR, hypophosphorylated 4E-BP1 sequesters eIF4E to prevent assembly of eIF4F complex components onto capped mRNA, inhibiting cap-dependent translation. When 4E-BP1 is phosphorylated by mTOR (7), first at critical priming threonine (T) 37 and T46 residues and then at other sites, 4E-BP1 is inactivated and releases eIF4E to allow initiation of cap-dependent translation (8). Other non-mTOR kinases, including cyclin-dependent kinase 1 (CDK1), have been shown to be able to phosphorylate 4E-BP1 (9–12) but have not been extensively examined in vivo for their effects on 4E-BP1–regulated cap-dependent translation.
Protein synthesis has been described to decrease during mitosis relative to interphase in reports dating back to the 1960s (13, 14). There are two issues, however, with this conclusion: (i) Mitotic cells represent less than 1% of the total cell population in bulk culture, and even under stringent conditions, high levels of interphase cell contamination can occur. (ii) Many studies of mitotic cap-dependent translation rely on cell cycle synchronization studies with microtubule inhibitors (e.g., nocodazole), which are also mitotic translation inhibitors (15). Under these conditions, comparisons of interphase and mitotic translation can be imprecise. Single-cell measurements, such as flow cytometry, can potentially overcome these concerns. Additionally, a highly hyperphosphorylated 4E-BP1 isoform called δ-4E-BP1 is present in mitotic cells (10, 16). This hyperphosphorylated isoform is predicted to promote rather than inhibit cap-dependent protein translation and is therefore inconsistent with the standard model.
Our studies on Merkel cell polyomavirus (MCV) provide insights into these issues. MCV is a small double-stranded DNA virus discovered in 2008 by our laboratory that causes most cases of the human skin cancer Merkel cell carcinoma (MCC) (17) (for review, see refs. 18–20). The 19-kDa MCV small T (sT) antigen is a transforming oncoprotein required for MCC cell growth (21, 22). A region of the sT protein spanning amino acid residues 91–95, called the Large T stabilization domain (LSD), promotes δ-4E-BP1 hyperphosphorylation (23), rodent cell transformation (24), and fibroblast proliferation in a mouse transgenic model (21, 25). Expression of the phosphorylation-defective, dominant-positive 4E-BP1 (26) with alanine substitution mutations at priming T37/T46 (4E-BP1T37A/T46A) reverses sT-induced rodent fibroblast transformation, suggesting a direct link between 4E-BP1 phosphorylation status and sT-induced transformation (21). Surprisingly, sT-induced δ-4E-BP1 hyperphosphorylation is not dependent on mTOR activity (21). The sT LSD region is known to bind the Fbw7 E3 ligase to promote cell proliferation, but Fbw7 targeting is not sufficient to explain either cell transformation or 4E-BP1 hyperphosphorylation (24).
We show here that MCV sT, through its LSD domain, also promotes mitogenesis and 4E-BP1 hyperphosphorylation by functioning as a promiscuous E3 ligase inhibitor that targets cellular anaphase-promoting complex/cyclosome (APC/C) E3 ligase activity. During sT-induced mitosis, sT-induced CDK1/CYCB1 rather than mTOR directly phosphorylates 4E-BP1 to the mitosis-specific δ isoform. Using a flow cytometry-based method to directly measure mitotic cap-dependent protein synthesis for the first time, to our knowledge, we do not detect a general shift from cap-dependent to cap-independent protein translation in mitotic cells compared with interphase cells. Mitotic cells actually show higher saturation levels of p4E-BP1T37/T46+, consistent with 4E-BP1 inactivation, than interphase cells. Consistent with this, and in contrast to previous studies, we find that δ-4E-BP1–positive mitotic cells show high levels of cap-dependent protein translation that is reduced by the cap translation inhibitor 4E1RCat. When accentuated or sustained, high levels of mitotic cap-dependent protein translation may play a role in cancer cell transformation and contribute to mTOR inhibitor resistance in subsets of cancers.
Results
MCV sT Increases Mitogenesis by Targeting the Cellular APC E3 Ligase.
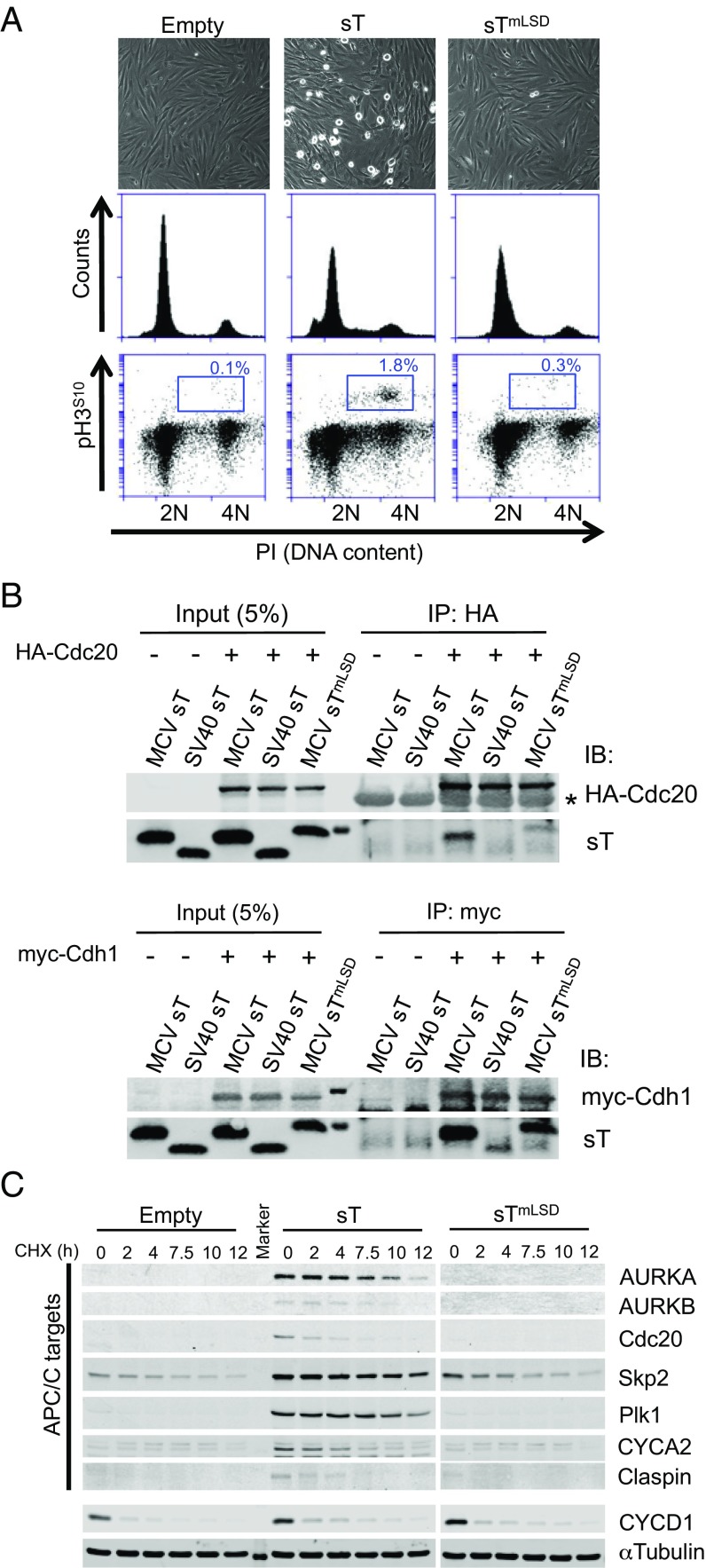
To search for factors contributing to MCV sT-induced transformation, the viral oncoprotein was expressed in hTERT-immortalized primary BJ-tert (BJ-T) human foreskin fibroblasts. These cells displayed a rounded phenotype in culture with increased phospho-histone H3 serine 10 (pH3S10) phosphorylation, characteristic for mitosis (Fig. 1A). Increased pH3S10 and increased expression of mitotic markers [including cyclin B1 (CYCB1) and phospho-aurora kinase B (pAURKB)] were also observed in 293 cells expressing MCV sT (Fig. S1). Immunoprecipitation of sT revealed an in vivo complex with the APC/C substrate recognition subunit cell division cycle protein 20 (Cdc20) that was dependent on an intact LSD (Fig. 1B). MCV sT also interacted with another APC/C substrate recognition subunit, cdc20 homolog 1 (Cdh1), but substantial Cdh1 binding occurred with sTmLSD having alanine substitutions at residues 91–95, suggesting that sT may bind Cdh1 at other sites in addition to the LSD. In line with these results, known APC/C E3 targets, including AURKA and AURKB, Skp2, polo-like kinase 1 (Plk1), and CYCA2, showed markedly reduced turnover in the presence of sT on cycloheximide (CHX) immunoblotting (Fig. 1C). These APC/C E3 targets retained rapid turnover in the presence of empty vector or sTmLSD expression. CYCD1, which is not directly regulated by APC/C (27), was unaffected by MCV sT or sTmLSD expression. Similarly, MCV sT expression stabilized FLAG-tagged AURKA and endogenous CYCB1, but not CYCD1, after nocodazole release of 293 cells, whereas MCV sTmLSD expression did not (Fig. S2).
Fig. 1.
MCV sT promotes mitosis by targeting APC/C E3-ubiquitin ligases. (A) MCV sT induces cellular mitogenesis. BJ-T cells stably transduced with MCV sT have increased mitotic rounding and a 6–18-fold increase in pH3S10+ mitotic cells compared with empty vector or sTmLSD transduced cells. (B) MCV sT interacts with APC/C E3 ligase substrate recognition subunit Cdc20 and Cdh1 proteins. HA-tagged Cdc20 or myc-tagged Cdh1 expression plasmids were cotransfected with MCV sT, MCV sTmLSD, or SV40 sT expression plasmids into 293 cells and immunoprecipitated 48 h later with anti-HA or anti-myc antibodies, followed by immunoblotting using mixed anti-MCV sT (CM8E6) and anti-SV40 sT (PAb419) antibodies. Cdc20 interaction with MCV sT was nearly eliminated in the sTmLSD mutant protein, whereas partial interaction was retained between MCV sTmLSD and myc-Cdh1 proteins. Weak interaction between SV40 sT and myc-Cdh1 only was detected. Asterisk indicates IgG heavy chain. (C) APC/C target proteins (AURKA/B, Cdc20, Skp2, Plk1, CYCA2, and claspin) are stabilized by MCV sT expression. BJ-T cells were treated with CHX (100 µg/mL) to inhibit new protein synthesis and harvested at the indicated time points. The half-lives of proteins regulated by APC/C are extended by expression of MCV sT but not empty vector or MCV sTmLSD controls. CYCD1 is not directly regulated by Cdh1, and its half-life was unchanged by MCV sT expression. A representative α-tubulin loading control is shown. Representative results are shown from three independent experiments.
MCV sT Induces mTOR-Independent δ-4E-BP1 Phosphorylation.
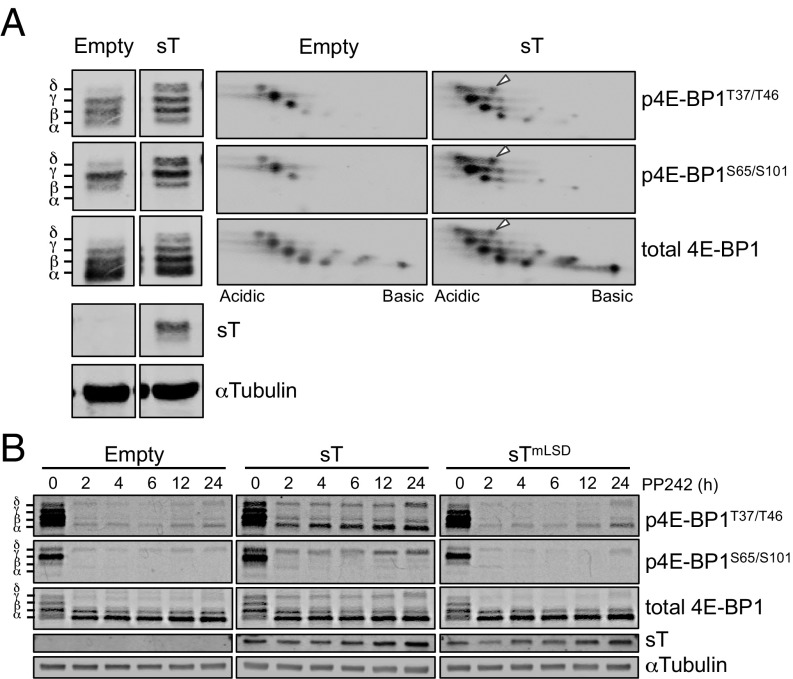
We next examined the role of MCV sT in 4E-BP1 hyperphosphorylation. 4E-BP1 hyperphosphorylation isoforms are named α through δ according to ascending molecular mass (Fig. 2A, Left) (28). Most notable was the appearance of the highest molecular mass form, δ, containing phosphorylation marks at T37/T46 and S65/S101, after transfection of the sT expression vector into 293 cells, as previously described (21). A 2D gel immunoblot (Fig. 2A, Right) aligned to the corresponding 1D SDS/PAGE immunoblot shows that during MCV sT expression, a new phosphoisoform appears at the δ position (arrows) staining for p4E-BP1T37/T46 and p4E-BP1S65/S101. MCV sT expression prolonged 4E-BP1 phosphorylation (Fig. 2B) in the presence of the mTOR inhibitor PP242 (29) compared with empty vector control and sTmLSD transfected cells, indicating that δ-4E-BP1 phosphorylation may be independent of mTOR kinase activity.
Fig. 2.
MCV sT induces cellular 4E-BP1 hyperphosphorylation. (A) MCV sT induces 4E-BP1T37/T46 and 4E-BP1S65/S101 phosphorylation. 4E-BP1 phosphospecies are named α through δ according to molecular mass. Higher molecular mass isoforms, particularly β–δ, were increased following sT expression in 293 cells and include authentic phosphorylation sites as detected by phospho-specific antibodies. A 2D gel fractionation of these same lysates (pH 3–6 isoelectric focusing/SDS/PAGE) is aligned to the 1D gel. Arrowheads indicate new 4E-BP1 isoelectric focusing spots after sT expression detected by p4E-BP1T37/T46, p4E-BP1S65/S101, and total 4E-BP1 antibodies. (B) MCV sT-induced 4E-BP1 phosphorylation is partially resistant to mTOR inhibition. The 293 cells were transfected with MCV sT, sTmLSD, or empty vector expression plasmids for 48 h; treated with the mTOR inhibitor PP242; and harvested at indicated time points. MCV sT depends on an intact LSD region to maintain PP242-resistant 4E-BP1 phosphorylation. Representative results are shown from three independent experiments.
CDK1/CYCB1 Directly Phosphorylates 4E-BP1, in the Presence and Absence of sT, to the δ Isoform During Mitosis.
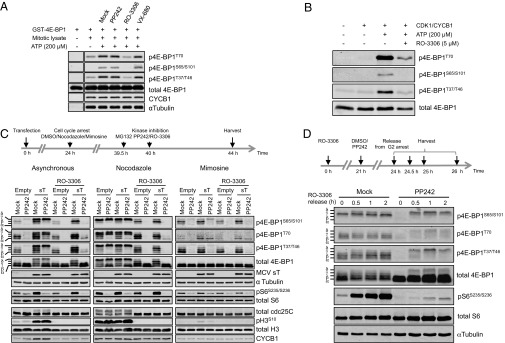
The 4E-BP1 phosphorylation is induced by microtubule assembly inhibitors such as nocodazole and paclitaxel that arrest cells in mitosis (15, 16). To assess the role of various kinases on mitotic 4E-BP1 phosphorylation, nocodazole-treated HeLa mitotic cell lysates were reacted with recombinant GST–4E-BP1 and kinase inhibitors, including PP242 (mTORC1 and mTORC2), RO-3306 (CDK1), and VX-680 (pan AURK) (Fig. 3A). GST–4E-BP1 was robustly phosphorylated at authentic sites by mitotic HeLa lysates, and this was reversed by inhibition of CDK1 but not by mTOR or AURK inhibition.
Fig. 3.
CDK1/CYCB1 phosphorylates 4E-BP1 during mitosis. (A) CDK1 inhibition in mitotic lysates reduces 4E-BP1 phosphorylation. Mitotic HeLa cell lysates (10 µg) enriched by nocodazole arrest were mixed with 0.2 µg GST–4E-BP1, reacted for 30 min at 30 °C in the presence or absence of 5 µM mTOR (PP242), CDK1 (RO-3306), or AURK (VX-680) kinase inhibitors and then immunoblotted with antibodies as shown. ATP-dependent 4E-BP1 phosphorylation was sensitive to CDK1 inhibitor but resistant to mTOR and AURK inhibitors. Equal loading of total 4E-BP1, CYCB1, and α-tubulin is shown. Representative results are shown from three independent experiments. (B) Recombinant CDK1/CYCB1 kinase phosphorylates GST–4E-BP1 at the known regulatory residues T70, S65/S101, and T37/T46. CDK1/CYCB1 (20 units) was mixed with bacterial-expressed GST–4E-BP1 in kinase reaction buffer for 30 min at 30 °C and immunoblotted with phospho-specific antibodies. ATP-dependent 4E-BP1 phosphorylation by CDK1/CYCB1 occurred at phospho-specific sites and was sensitive to the CDK1 active site inhibitor RO-3306. Representative results are shown from two independent experiments. (C) δ-4E-BP1 is induced during mitosis and inhibited by a CDK1 inhibitor. The 293 cells were transfected with empty vector or MCV sT and arrested for 20 h with DMSO (asynchronous), nocodazole (prometaphase), and mimosine (late G1). Cells were treated at 16 h with kinase inhibitors (5 µM PP242 or 10 µM RO-3306 + 10 µM MG132) as indicated. MCV sT induces δ-4E-BP1 in asynchronous cells sensitive to RO-3306 but not PP242. Nocodazole arrest induces similarly RO-3306–sensitive and PP242-resistant δ-4E-BP1 even in the absence of sT, whereas δ-4E-BP1 is only weakly induced by sT in mimosine-arrested cells. Markers for mitosis (pH3S10, CYCB1), a CDK1 substrate (cdc25C), and an mTORC1 downstream substrate (pS6S235/S236) showed active drug treatments. Representative results are shown from two independent experiments. (D) δ-4E-BP1 phosphorylation during mitosis occurs in the absence of active mTOR. U2OS cells were arrested at the G2/M boundary with 10 µM RO-3306 for 24 h, released by washing, and harvested at the time points shown. Cells were treated for 3 h prerelease with DMSO or 5 µM PP242. In the absence of mTOR inhibition, no δ-4E-BP1 is found at 0 h but accumulates, together with β and γ isoforms, during mitotic transit. During PP242 inhibition, δ-4E-BP1 still accumulates during mitosis, but lower molecular mass (β–γ) isoforms are reduced. Results shown are from a single experiment.
Evidence that CDK1 is responsible for δ-4E-BP1 mitotic phosphorylation was also obtained by treatment of nocodazole-arrested HeLa cells with the CDK1 inhibitor RO-3306 (Fig. S3A). δ-4E-BP1 hyperphosphorylation could not be fully restored by RO-3306/MG132 cotreatment. A technical issue in using mitotic kinase inhibitors to assess 4E-BP1 phosphorylation is the occurrence of mitotic slippage, a side effect of kinase inhibition concurrently causing enforced exit from mitosis with general loss of mitotic kinase activities (30, 31). Mitotic slippage can be prevented by simultaneous inhibition of APC/C-mediated protein degradation with the proteasome inhibitor MG132, which in effect “freezes” the mitotic phenotype. Like RO-3306, treatment of nocodazole-arrested HeLa cells with the AURK inhibitor VX-680 also eliminated δ-4E-BP1 phosphorylation (Fig. S3B). Unlike RO-3306, however, this was completely reversed by cotreatment with VX-680/MG132, suggesting that AURK inhibition effects on 4E-BP1 phosphorylation are due to mitotic slippage. Extensive in vitro phosphorylation studies also failed to reveal evidence for direct 4E-BP1 phosphorylation by purified AURKB. To confirm direct 4E-BP1 phosphorylation by CDK1/CYCB1, we generated an in vitro phosphorylation reaction using purified CDK1/CYCB1 and GST–4E-BP1 (Fig. 3B). CDK1 phosphorylation of 4E-BP1 was ATP-dependent and -inhibitable by RO-3306. CDK1 phosphorylation occurred at the previously described T70 residue (10) as well as at authentic 4E-BP1 phosphorylation sites, including T37/T46 and S65/S101, which are known to regulate 4E-BP1 binding to eIF4E.
Mitotic δ-4E-BP1 phosphorylation was also examined in nocodazole-arrested 293 cells in the presence of CDK1 and mTOR inhibitors (Fig. 3C). MG132 was added to nocodazole-arrested cells 30 min before RO-3306 treatment to prevent CDK1 inhibition-induced mitotic slippage (30). In this experiment, pH3S10+ mitotic cells comprised ∼0.9% of the total asynchronous (no cell cycle arrest) cell population (Fig. 3 C, Left and Fig. S1). MCV sT expression promotes formation of PP242-resistant δ-4E-BP1 that is lost after treatment with RO-3306. Notably, S6S235/S236 phosphorylation, a known phosphorylation mark for mTORC1 kinase activity (32, 33), is nearly ablated by PP242 but not by RO-3306. These results are consistent with sT induction of δ-4E-BP1 through CDK1 rather than mTOR kinase activity.
Distinctive 4E-BP1 phosphorylation patterns were seen during nocodazole (prometaphase) and mimosine (late G1) cell cycle arrest (Fig. 3C). During nocodazole arrest, the δ-4E-BP1 isoforms became prominent even in the absence of MCV sT expression. In contrast, δ-4E-BP1 isoforms were nearly absent under all conditions for cells arrested in G1 by mimosine. Whereas δ-4E-BP1 was resistant to mTOR inhibition, CDK1 inhibition during nocodazole mitotic arrest ablated δ-4E-BP1. These results were confirmed in HeLa cells treated with nocodazole and kinase inhibitors (Fig. S3).
To confirm these findings in the absence of chemical inhibitors, we used mechanical shake-off to isolate mitotic cells from sT-expressing BJ-T cells (Fig. S4). This maneuver enriched the mitotic cell fraction from ∼2% to ∼66% as determined by flow cytometry with propidium iodide (PI) and pH3S10 staining (Fig. S4A). Shake-off cells exclusively expressed the δ-4E-BP1 isoform, whereas adherent cells expressed only α–γ isoforms of 4E-BP1 (Fig. S4B). In vitro lambda phosphatase treatment of sT-expressing and nocodazole-arrested 293 cell lysates showed that the high-molecular-mass 4E-BP1 isoforms, including the α–γ and δ isoforms, are formed as a result of phosphorylation rather than another type of posttranslational modification (Fig. S5).
Although PP242-inhibitable mTOR kinase activity contributes to mitotic 4E-BP1 phosphorylation, particularly for lower molecular mass α and β forms (Fig. 3C), mTOR may be dispensable for mitotic 4E-BP1 hyperphosphorylation under some conditions. U2OS cells were arrested at the G2/M boundary for 24 h using 10 μM RO-3306 (31, 34) (Fig. 3D). After RO-3306 removal, cells progressed through mitosis, with most exiting mitosis 3 h after RO-3306 release. PP242 pretreatment markedly reduced pS6S235/S236 but not δ-4E-BP1, consistent with mTOR-independent phosphorylation of 4E-BP1 during mitosis. The 293 cells failed to arrest in G2 with RO-3306 and could not be examined.
The 4E-BP1 δ Isoform Is Induced in Mitosis During Normal Cell Cycling.
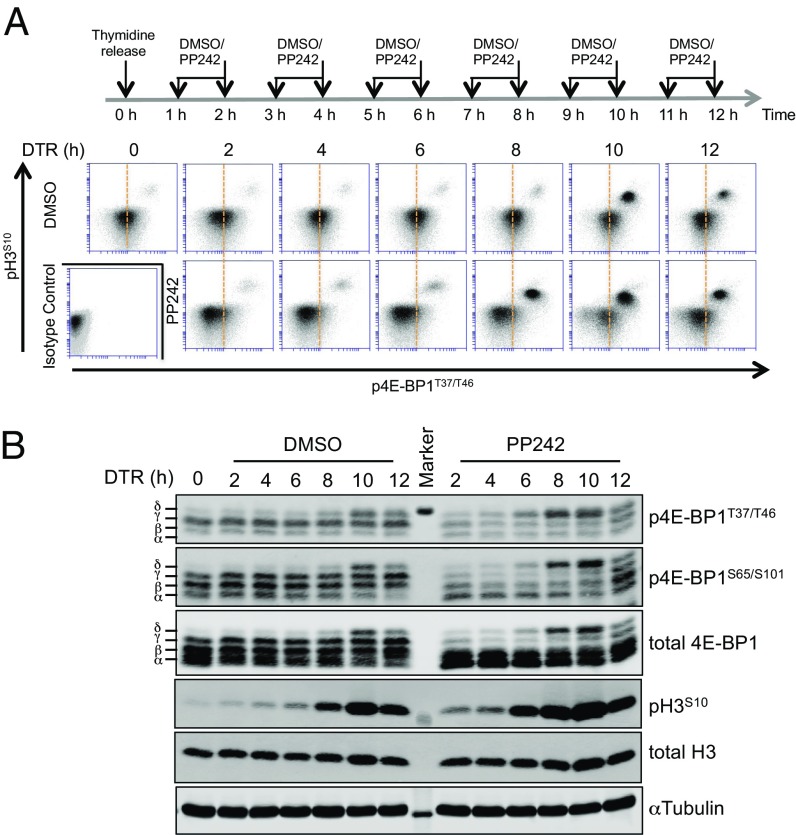
Nocodazole-arrest experiments suggest that δ-4E-BP1 accumulates during mitosis even in the absence of MCV sT expression. To confirm this in the absence of drug treatment, 293 cells were synchronized by double-thymidine block and release, harvested at sequential time points, and immunostained for pH3S10 and p4E-BP1T37/T46 (Fig. 4A). For each time point after release, cells were pretreated with PP242 or DMSO vehicle control 1 h before harvesting.
Fig. 4.
4E-BP1 is hyperphosphorylated to the δ phosphoisoform during mitosis. (A) pH3S10+ mitotic 293 cells have higher levels of p4E-BP1T37/T46+ saturation than cells in other portions of the cell cycle. Dual flow cytometry staining for pH3S10 and p4E-BP1T37/T46 was performed on 293 cells synchronized by double-thymidine block and release, which have peak mitotic entry at 10 h postrelease (see also Fig. S6). Vertical bar represents the centroid for p4E-BP1T37/T46+ fluorescence staining at time 0 h. To determine if 4E-BP1 phosphorylation depends on mTOR activity, cells were also treated 1 h before harvesting with 5 µM PP242. Mitotic cells formed an orthogonal pH3S10+/p4E-BP1T37/T46+ population having high levels of inactivated (phosphorylated) 4E-BP1 that were not dependent on mTOR activity. In contrast, interphase pH3S10– cells were largely mTOR inhibition-sensitive. PP242 treatment increases mitotic entry at 8 h postrelease. (B) PP242-resistant δ-4E-BP1 is formed during peak mitotic entry. Protein lysates were collected from cells in A and immunoblotted for p4E-BP1 and pH3S10. Representative results are shown from three independent experiments.
Flow cytometry showed peak pH3S10+ mitotic entry occurring reproducibly at 10 h, which began to diminish by 12 h after release (Fig. 4A and Fig. S6). This same pattern occurred with PP242 pretreatment, although mitotic entry was more abundant at 8 h postrelease. Unexpectedly, pH3S10+ mitotic 293 cells formed an orthogonal population with the highest per-cell saturation levels of p4E-BP1T37/T46 compared with any other stage of the cell cycle. PP242 pretreatment reduced p4E-BP1T37/T46+ staining for interphase cells at 2–8 h (note leftward shift for p4E-BP1T37/T46+ staining among pH3S10- cells) consistent with mTOR regulation of 4E-BP1. At peak mitotic entry (8–10 h postrelease), however, pH3S10+ cells were resistant to loss of p4E-BP1T37/T46+ staining with PP242 treatment.
Immunoblots performed on these same cell fractions at each time point (Fig. 4B) showed prominent α-γ 4E-BP1 phosphorylation at early time points (0–6 h), which was sensitive to mTOR inhibition. The δ isoform emerged 8–12 h after release, corresponding to maximum pH3S10+ and p4E-BP1T37/T46+ staining, and was resistant to PP242 inhibition. Similar results, but with a less abundant orthogonal pH3S10+/p4E-BP1T37/T46+ cell population, were seen in U2OS cells (Fig. S7).
CDK1/CYCB1 Activates Cap-Dependent Translation During Mitosis.
According to the existing model for 4E-BP1–regulated protein synthesis, high levels of p4E-BP1T37/T46 are predicted to promote cap-dependent translation during pH3S10+ mitosis (35). We directly examined this by using cap-binding assays for mitosis-enriched and -depleted cells and by using a flow cytometry method designed to directly measure single-cell cap-dependent protein synthesis.
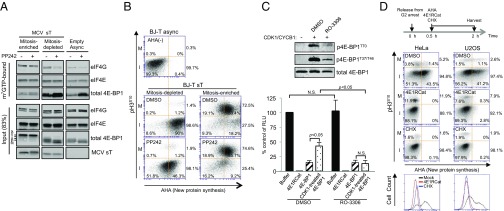
We performed m7GTP cap resin pulldown assays to assess the functional correlates of our flow cytometry and immunoblot findings. Highly enriched mitotic BJ-T cells expressing MCV sT, isolated by shake-off (nonadherent), showed m7GTP cap binding to eIF4G that was unaffected by PP242 treatment (Fig. 5A). In contrast, although interphase-enriched BJ-T cells (adherent) had comparable levels of eIF4G, eIF4G cap binding remained sensitive to PP242. Input 4E-BP1 protein from mitosis-enriched cells was almost exclusively in the δ-4E-BP1 isoform. This is consistent with mTOR-independent cap binding during mitosis and mTOR-dependent cap binding during interphase. Qualitatively similar results were found for HeLa cells using G2/M arrest enrichment and shake-off (Fig. S8). For mitosis-enriched HeLa cells, modest but reproducible reduction in eIF4G–m7GTP cap association was present with RO-3306 treatment alone but not PP242 treatment alone. Combined RO-3306 and PP242 treatment nearly eliminated eIF4G association to m7GTP. These results were confirmed by metabolic labeling using the Click-iT methionine analog l-azidohomoalanine (AHA) to measure nascent protein synthesis (36) (Fig. 5B). In this assay, cells were incubated with AHA for 90 min, in the absence or presence of PP242, in methionine-depleted medium and then subjected to mitotic shake-off. Newly synthesized protein was then labeled with Alexa Fluor 488-alkyne by the copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition (Click-iT) reaction (37) and measured by flow cytometry. Costaining for pH3S10 allowed segregation of individual cells into “mitotic” (pH3S10+) and “interphase” (pH3S10–) populations. Up to 74% of DMSO-treated mitotic cells were AHA positive in comparison with 91% of DMSO-treated interphase cells with AHA positivity. PP242 treatment reduced new protein synthesis for pH3S10– interphase BJ-T cells but had no effect on protein synthesis for pH3S10+ mitotic BJ-T cells (Fig. 5B). Similar analyses using double thymidine block and release synchronization of 293 cells, however, revealed that PP242 reduced new protein synthesis for both mitotic and interphase cells (Fig. S9), suggesting that PP242 resistance may be cell line specific.
Fig. 5.
eIF4F formation on the m7GTP cap and direct measurement of cap-dependent protein translation during mitosis (M) and interphase (I). (A) eIF4F formation on the m7GTP cap is PP242-independent for mitosis-enriched cells but PP242-sensitive for mitosis-depleted cells. BJ-T cells expressing MCV sT were harvested by mitotic shake-off to enrich for mitotic (nonadherent) and nonmitotic (adherent) populations and compared with asynchronous empty vector BJ-T cells without shake-off. Lysates were bound to m7GTP-resin, precipitated, and immunoblotted. Mitosis-enriched cell eIF4G binding, as well as 4E-BP1 binding, to the eIF4E/cap complex was unaffected by mTOR inhibition. For mitosis-depleted cells, eIF4G binding was reduced and 4E-BP1 binding was increased by mTOR inhibition. Representative results from two independent experiments are shown. (B) Nascent protein synthesis during mitosis is resistant to mTOR inhibition in BJ-T cells. BJ-T cells stably expressing sT were labeled with AHA for 45 min in methionine-depleted media, separated by mitotic shake-off as in A, reacted with Click-iT Alexa Fluor 488 alkyne after permeabilization, and new protein synthesis was measured by flow cytometry. Relative mitotic protein synthesis was determined by dividing percentage of pH3S10+-AHA+ cells by percentage of total pH3S10+ cells. Likewise, interphase protein synthesis was determined by dividing percentage of pH3S10–-AHA+ cells by percentage of total pH3S10– cells. Approximately 91% of pH3S10– adherent interphase cells showed AHA incorporation that was sensitive to mTOR inhibition. Only 74% of pH3S10+-positive mitotic cells were positive for AHA uptake, but this new protein synthesis was resistant to PP242 treatment. Baseline fluorescence was determined in asynchronous BJ-T cells without AHA incubation. I, interphase pH3S10– cells; M, mitotic pH3S10+ cells. (C) In vitro capped mRNA translation is inhibited by 4E1RCat and activated by CDK1/CYCB1. Capped and polyadenylated luciferase mRNA was generated in a T7 polymerase reaction and used to produce luciferase protein in rabbit reticulocyte lysate. The addition of 4E1RCat abolished luciferase translation, whereas addition of GST–4E-BP1 reduced translation to 15% of buffer control (averages for three independent experiments with SEM shown). When GST–4E-BP1 was phosphorylated by CDK1/CYCB1 in kinase reaction buffer, translation increased to 43% of buffer control. This effect was eliminated by RO-3306 pretreatment. Insert shows GST–4E-BP1 phosphorylation immunoblot. (D) Mitotic translation is primarily cap-dependent for HeLa and U2OS. HeLa or U2OS cells were synchronized for 24 h at the G2/M boundary with RO-3306 (10 µM), released by washing, and incubated with 25 µM of AHA for 90 min in methionine-depleted media, and then harvested 2 h after release. Harvested cells were permeabilized and reacted with Alexa Fluor 488 alkyne to measure AHA incorporation into protein. DMSO vehicle control, CHX (100 µg/mL), or 4E1RCat (50 µM) were added together with AHA 30 min after release. Vertical bar represents maximum AHA incorporation after CHX translation inhibition. Fewer mitotic (26%) than interphase (42%) HeLa cells were positive for new protein synthesis, but all cells were sensitive to 4E1RCat inhibition of cap-dependent translation. For U2OS, cell numbers positive for total mitotic and interphase translation were identical (42%), and cap-dependent translation represented 73% and 85% of mitotic and interphase translation, respectively. Bottom panel shows 4N-gated AHA positivity for treated cells, which shows that 4E1RCat inhibition (cap-dependent) is similar to CHX (total) translation inhibition. Representative results are shown for one of three independent experiments.
We next generated capped, polyadenylated luciferase reporter mRNA using T7 polymerase (38, 39) and performed in vitro translation in commercial rabbit reticulocyte lysates (RRLs) to measure cap-dependent translation (Fig. 5C). Addition of 4E1RCat (40), a cap-dependent translation inhibitor that prevents eIF4F formation, virtually abolished translation. Addition of recombinant GST–4E-BP1 reduced cap-dependent translation in the reticulocyte lysates to ∼20% of buffer control (Fig. 5B). This inhibition was reduced to 45% of buffer control when GST–4E-BP1 was phosphorylated (p4E-BP1) by a CDK1/CYCB1 kinase reaction. This reversal of inhibition was antagonized by the CDK1 inhibitor RO-3306.
Measurement of cap-dependent protein synthesis during mitosis was directly determined for HeLa and U2OS cells after G2 release and synchronization using our AHA assay in cells treated with 4E1RCat (Fig. 5D). Costaining for pH3S10 allowed segregation of cells into mitotic (pH3S10+) and interphase (pH3S10–) populations. Nonspecific AHA incorporation was determined using the ribosome translation elongation inhibitor CHX (Fig. 5D, vertical lines), and new protein synthesis was reflected by AHA fluorescence above this baseline.
Like BJ-T cells, fewer (27%) mitotic HeLa cells were positive for new protein synthesis compared with interphase (46%) HeLa cells (Fig. 5D). In contrast, percentages of mitotic and interphase U2OS cells with new protein synthesis were identical (42% of mitotic and interphase cells). For both cell lines, however, nearly all new protein synthesis in both mitosis and interphase was cap-dependent and -sensitive to 4E1RCat treatment. Preliminary analyses revealed that MG132 treatment nonspecifically inhibited protein synthesis as previously reported (41), preventing us from accurately measuring the effects of CDK1 inhibition on mitotic translation under conditions that inhibit mitotic slippage. Using direct AHA uptake, however, we could confirm that nocodazole treatment specifically inhibits mitotic protein synthesis (Fig. S10).
Discussion
Tumor viruses have been central to cell biology because their oncogenes allow interrogation of specific cell proliferation and survival pathways. Among many critical findings, viral oncoproteins have been essential to the discovery of cellular oncogenes (42) and the tumor suppressor p53 (43–45), the characterization of the G1/S checkpoint (46) and the Akt-mTOR pathway (47), and identification of common innate immune and tumor suppressor signaling networks (48). MCV sT, an oncoprotein for MCC, induces mTOR-resistant 4E-BP1 hyperphosphorylation and cell transformation (21), which led us to investigate mTOR-independent 4E-BP1 signaling and cap-dependent translation in mitosis.
In addition to targeting Fbw7 (24), MCV sT inhibits APC/C E3 ligases and induces mitogenesis in sT-expressing cells. One consequence of this is increased mitotic CDK1/CYCB1 activity that is responsible for 4E-BP1 phosphorylation and δ-4E-BP1 formation. Caution is appropriate in interpreting our data, as mitotic kinase inhibition can cause mitotic slippage and exit from the mitotic phenotype. Considerable effort by our group was devoted to evaluating AURKA and AURKB as potential 4E-BP1 mitotic kinases, because AURK inhibitors (e.g., VX-680, MK-5108, and AZD-1152) also reduce 4E-BP1 hyperphosphorylation during mitosis. This was reversible, however, by cotreatment with MG132 to prevent APC/C-mediated mitotic egress, and we have no evidence that AURKs are directly responsible for 4E-BP1 phosphorylation. In contrast, there is considerable evidence from this study and others (10, 16) to indicate that CDK1/CYCB1 is a bona fide kinase for 4E-BP1.
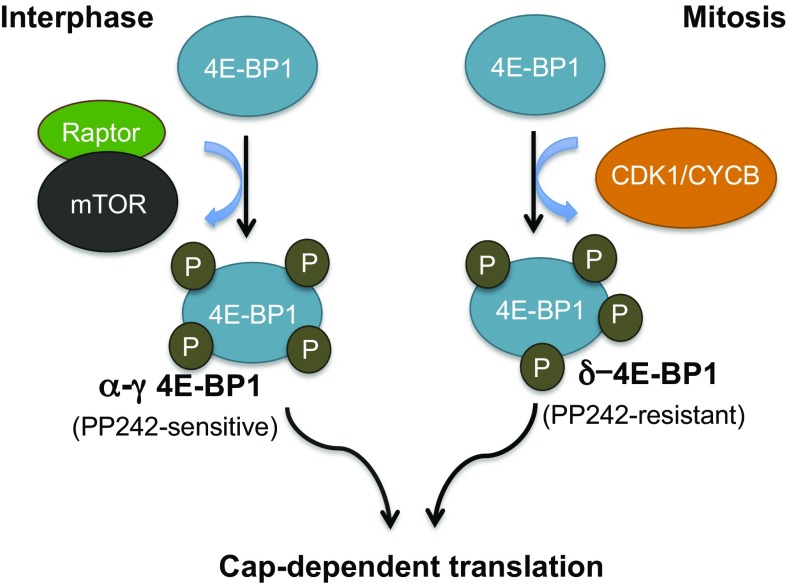
This study suggests an alternative pathway for CDK1/CYCB1-regulated cap-dependent translation during mitosis (Fig. 6). We find that mitotic 4E-BP1 is highly phosphorylated at the priming residues T37 and T46 in pH3S10+ cells, which runs counter to what would be predicted if cap-dependent translation is reduced during mitosis through an mTOR-related mechanism. The high-molecular-mass δ-4E-BP1 isoform is specific to mitosis, and our data indicate that this results from CDK1-mediated phosphorylation. Although δ-4E-BP1 can form under mitotic conditions in which mTOR is inhibited, it seems likely that mTOR cooperates with CDK1/CYCB1 to generate the mitotic δ-4E-BP1 by phosphorylating lower molecular mass α–γ isoforms that may be precursors to the δ-4E-BP1 isoform. Another limitation to our study is that we measure only 4E-BP1 phosphorylation but not δ-4E-BP1 dephosphorylation or turnover. These are likely to affect steady-state p4E-BP1 levels as well.
Fig. 6.
Model for cell cycle-dependent 4E-BP1 regulation of cap-dependent translation. Interphase 4E-BP1 is inhibited by mTORC1 kinase, whereas CDK1/CYCB1 is primarily responsible for δ-4E-BP1 inactivation during mitosis.
Our findings contrast with studies suggesting that loss of mTOR activity leads to inhibition of mitotic eIF4G cap-association and cap-dependent translation. We see cap-dependent protein translation is sustained during mitosis using a pulse flow cytometry approach. Pharmacological (4E1RCat) cap-dependent translation inhibition provides evidence that this effect is generalizable. AHA pulse labeling allows direct measurement of translation in mitotic cells, which avoids confounding issues stemming from bulk culture measurements. Although most mitotic translation was cap-dependent in all of the cell lines tested by AHA uptake, differences in relative mitotic and interphase translation were present between cell lines. Like [35S]methionine incorporation studies, AHA incorporation measurements require incubation of cells in low-methionine media.
We suspect that technical issues, which have only recently been resolved, explain differences between our studies and those of others. Measurement of mitotic protein translation (both cap-dependent and -independent) has relied on separation of mitotic and interphase cells in bulk culture, often using nocodazole-induced mitotic enrichment. We confirm that nocodazole inhibits mitotic translation for synchronized 293 cells. This has been ascribed by Coldwell et al. (15) to inhibitory phosphorylation of eIF2 and eIF4GII by nocodazole downstream to 4E-BP1 regulation. This is consistent with our findings that nocodazole both promotes δ-4E-BP1 and inhibits mitotic translation. We have not tested other mitotic-arrest compounds (e.g., paclitaxel) to determine if they have similar limitations. A second technical challenge is that mitotic cells represent a small fraction of the total cell population. Contamination with interphase cells is nearly inevitable in mitotic enrichment protocols and will dramatically alter conclusions, such as the role of mTOR in regulating 4E-BP1 during mitosis. In our experience, flow cytometry can help to resolve this dilemma by directly measuring mitotic status (pH3S10 or pMPM2 status) in cells while simultaneously determining translation regulator status, such as p4E-BP1T37/T46. Finally, newly developed classes of cap-dependent translation inhibitors such as 4E1RCat now allow direct determination of cap-dependent translation. When used in combination with AHA incorporation, direct measurement of mitotic cap-dependent translation can be determined.
Both nocodazole and PP242 are nonetheless important inhibitors to measure 4E-BP1 phosphorylation and translation during mitosis. As indicated, nocodazole does not interfere with δ-4E-BP1 formation and is useful for accentuating mitotic regulation of 4E-BP1. mTOR regulates translation through ribosomal biosynthesis as well as direct phosphorylation of translation machinery components downstream from 4E-BP1, such as eIF4B (49) and eEF2 elongation factor (50). Further, eIF4B regulation by 14-3-3σ may also play a role in later stages of mitotic protein translation and may be missed in our study of early mitosis (51). Thus, PP242 may affect mitotic translation by acting downstream to 4E-BP1. We also find evidence that in most cells mTOR typically acts in concert with CDK1/CYCB1 to promote mitotic cap-dependent translation.
Cap-dependent translation of preformed mRNAs provides rapid regulation of gene expression that may be required for short-lived cellular responses, such as transit through mitosis. These changes generally cannot be accurately measured by standard mRNA expression techniques. Mounting evidence suggests that dysregulated cap-dependent translation from aberrant PI3K–Akt–mTOR and MEF–RAF–MEK–ERK signaling contributes to cancer cell transformation (3, 52). Regardless of the contribution of activated cap-dependent translation to cancer cell transformation, such as in MCV-positive MCC, our findings point toward the possibility that combined mTOR and CDK1/CYCB1 inhibition may prove useful for cancer treatment, particularly for mTOR inhibitor-resistant cancers.
Materials and Methods
Plasmids, antibodies, primers, and standard methods are described in SI Materials and Methods.
2D Electrophoresis.
The 293 cells were lysed using lysis buffer (50 mM Tris∙HCl, pH 7.4, 0.15 M NaCl, 1% Triton X-100, 2 mM Na3VO4, 2 mM NaF) supplemented with protease inhibitors (Roche). Clarified lysates were focused using immobilized pH 3–6 gradient strips (Bio-Rad) with linear voltage ramping for 2 h at 200 V, 2 h at 500 V, and 16 h at 800 V. Focused proteins were then subjected to SDS/PAGE for 2D resolution and detected by immunoblotting.
Flow Cytometry.
The 293 and BJ-T cells were trypsinized and fixed in 70% (vol/vol) ethanol for DNA staining or in 10% (vol/vol) buffered formalin for AHA incorporation assays. Fixed cells were washed with PBS containing 1% FBS and permeabilized with 0.25% Triton X-100 for 30 min on ice. For cell cycle analysis, cells were resuspended in PI/RNase staining solution (0.05 mg/mL PI, 0.1 mg/mL RNase A in 1× PBS) and incubated for 30 min at room temperature. For phospho-histone H3S10 and phospho–4E-BP1T37/T46 analysis, cells were incubated with the corresponding fluorophore-conjugated antibodies for 2 h at room temperature.
Cell Cycle Synchronization and Mitotic Cell Enrichment.
The 293 cells were treated with medium containing 0.5 μM nocodazole or 0.5 mM l-mimosine for 16 h to induce mitotic arrest or G1 arrest, respectively. Mitotic cells were enriched by double-thymidine block (2 mM) and release using 293 and U2OS cells or by mitotic shake-off (53) using BJ-T cells stably expressing wild-type MCV sT. To block the cell cycle at late G2, HeLa and U2OS cells were incubated in medium containing CDK1 inhibitor RO-3306 (10 μM) for 24 h. Cell cycle entry from G2 to mitosis was induced by RO-3306 washout. Cells arrested by nocodazole and released from RO-3306 arrest were treated with 10 μM of proteasome inhibitor MG132 for 30 min before kinase inhibitor treatment to retain cells in mitosis.
Kinase Inhibitors.
The following active-site kinase inhibitors were dissolved in DMSO and used for in vivo kinase inhibition and in vitro phosphorylation experiments: mTOR kinase inhibitor PP242 (Selleckchem), CDK1 kinase inhibitor RO-3306 (Calbiochem), and pan Aurora kinase inhibitor VX-680 (Selleckchem).
In Vitro Phosphorylation Assays.
Recombinant GST–4E-BP1 (0.2 μg) (SignalChem) was incubated in a 24-μL reaction containing 1× protein kinase buffer (NEB) and 20 units of recombinant CDK1/CYCB1 (NEB) or 10 μg of mitotic HeLa cell lysate, supplemented with 200 μM ATP and/or 5 μM active site kinase inhibitors, for 30 min at 30 °C. HeLa cells were arrested in mitosis by treatment with 0.5 μM nocodazole for 16 h and enriched by mechanical shake-off for lysis in nondenaturing lysis buffer (50 mM Tris∙HCl, pH 7.4, 0.15 M NaCl, 1% Triton X-100, 2 mM Na3VO4, and 2 mM NaF). The reactions were stopped by adding 5× SDS sample buffer to 1× concentration and boiling for 5 min. Reaction samples were then subjected to SDS/PAGE and immunoblotting. For in vitro protein dephosphorylation, 293 cell extracts were incubated with lambda phosphatase in protein metallophosphatase reaction buffer (NEB) supplemented with 2 mM MnCl2 for 30 min at 37 °C. Reactions were stopped by adding 2× SDS sample buffer and then subjected to SDS/PAGE and immunoblotting.
m7GTP Cap-Binding Assay.
Shake-off (mitosis-enriched) and adherent cells (mitotic-depleted) from MCV sT-transduced BJ-T cells or asynchronous BJ-T cells were lysed in buffer (50 mM Tris·HCl, pH 7.4, 0.15 M NaCl, 1% Triton X-100, 2 mM Na3VO4, and 2mM NaF) supplemented with protease inhibitors (Roche). Lysates (30 μg of total protein) were incubated with 5.0 μL of m7GTP Sepharose beads (GE Healthcare) overnight at 4 °C. Beads were collected, washed with lysis buffer, and subjected to SDS/PAGE and immunoblotting. We loaded 25 μg of total protein as input control (83%).
In Vitro mRNA Synthesis and Translation.
Capped firefly-luciferase (FLuc) reporter mRNA was synthesized by the MessageMAX T7 ARCA-Capped message transcription kit (Cell Script) using 1 μg pCD-V5-FLuc linearized by MscI as template. pCD-V5-Fluc was constructed by ligating FLuc from pGL3 to pCDNA6/V5-HisB between Hind II and XbaI. Purified RNA was polyadenylated using the A-Plus Poly (A) polymerase tailing kit (Cell Script). Translation reactions were performed in a final volume of 10 μL consisting of 7 μL of nuclease-treated RRL (Promega), 0.8 pmol of capped and polyadenylated reporter mRNAs, and an amino acid mixture (50 μM each). GST–4E-BP1 was incubated with either CDK1 or 1 μg/mL BSA in the presence of DMSO or 10 μM RO-3306 (see reaction setup for previous section). As a control, PBS alone was incubated with the same amount of either DMSO or 10 μM RO-3306. Either buffer or pretreated GST–4E-BP1 (12.4 μL) was added to the RRL reaction mixture. The prepared RRL mixture was incubated for 15 min at 30 °C. The reaction was then stopped by adding 10 μL of luciferase lysis buffer to the mixture. Translation was measured as firefly luciferase activity.
Nascent Protein Synthesis Analysis.
BJ-T sT stable cells were labeled with an azide-linked methionine analog AHA (Life Technologies) at 25 μM for 45 min in the presence or absence of PP242 (5 μM), followed by mitotic shake-off to separate mitotic cells and interphase cells. To analyze mitotic cap-dependent translation in U2OS and HeLa cells, cells were arrested at the G2/M boundary by 10 μM RO-3306 treatment for 24 h (34). After 30 min of RO-3306 removal, cells were labeled with AHA (25 μM) for 90 min in methionine-depleted DMEM (Corning Cellgro) after optimization of preexperiments. Translation inhibitors [4E1RCat (50 μM) or CHX (100 μg/mL)] or DMSO (0.1%) were added to cells with AHA. Cells were trypsinized and fixed in 10% (vol/vol) formalin for 5 min. Fixed cells were permeabilized in PBS containing 0.1% saponin and 1% FBS for 30 min at room temperature. Cells were harvested and labeled with the Alexa Fluor 488 alkyne using the Click-iT cell reaction buffer kit (Life Technologies). AHA incorporation in cells was analyzed by flow cytometry as a measure for nascent protein synthesis in interphase and mitotic cells.
Statistic Analysis.
One-sided t test was performed for densitometric analysis of m7GTP pulldown assays and two-sided t test (unequal variances) for in vitro translation assays. A P value less than 0.05 was considered to be significant.
Supplementary Material
Acknowledgments
We thank Susanne Lens and Rutger Hengeveld for access to unpublished aurora kinase analyses, and Ioanna Tzani, Gary Loughran, and John Atkins, University of Cork, for access to unpublished ribosomal profiling studies. The authors also thank Susanne Lens, Nahum Sonenberg, and Chris Bakkenist for helpful scientific suggestions; Wei Qian for technical assistance; Jing Hu for providing reagents for cap-dependent translation analysis; Nahum Sonenberg for providing 4E-BP1 knockout MEFs used in unpublished comparative studies; and Joe Zawinul and Missy Mazzoli for help in writing the manuscript. This work was supported by NIH National Cancer Institute Grants R01CA136806, CA136363, and CA170354 and American Cancer Society Professorships (to P.S.M. and Y.C.). M.S. was supported in part by University of Pittsburgh Skin Cancer Specialized Program of Research Excellence (SPORE) Grant CA12197305. Flow cytometry was performed in the University of Pittsburgh Cancer Institute (UPCI) Cytometry Facility, which is supported in part by NIH Grant P30CA047904.
Footnotes
The authors declare no conflict of interest.
See QnAs on page 5862.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505787112/-/DCSupplemental.
References
- 1.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25(48):6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh AC, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17(3):249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.She QB, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18(1):39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Proud CG. mTORC1 signaling: What we still don’t know. J Mol Cell Biol. 2011;3(4):206–220. doi: 10.1093/jmcb/mjq038. [DOI] [PubMed] [Google Scholar]
- 5.Armengol G, et al. 4E-binding protein 1: A key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67(16):7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 6.Sabatini DM. mTOR and cancer: Insights into a complex relationship. Nat Rev Cancer. 2006;6(9):729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 7.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95(4):1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gingras AC, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15(21):2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbert TP, Tee AR, Proud CG. The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J Biol Chem. 2002;277(13):11591–11596. doi: 10.1074/jbc.M110367200. [DOI] [PubMed] [Google Scholar]
- 10.Heesom KJ, Gampel A, Mellor H, Denton RM. Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1) Curr Biol. 2001;11(17):1374–1379. doi: 10.1016/s0960-9822(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 11.Shang ZF, et al. 4E-BP1 participates in maintaining spindle integrity and genomic stability via interacting with PLK1. Cell Cycle. 2012;11(18):3463–3471. doi: 10.4161/cc.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin S, et al. Glycogen synthase kinase-3β positively regulates protein synthesis and cell proliferation through the regulation of translation initiation factor 4E-binding protein 1. Oncogene. 2014;33(13):1690–1699. doi: 10.1038/onc.2013.113. [DOI] [PubMed] [Google Scholar]
- 13.Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- 14.Konrad CG. Protein synthesis and Rna synthesis during mitosis in animal cells. J Cell Biol. 1963;19:267–277. doi: 10.1083/jcb.19.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coldwell MJ, et al. Phosphorylation of eIF4GII and 4E-BP1 in response to nocodazole treatment: A reappraisal of translation initiation during mitosis. Cell Cycle. 2013;12(23):3615–3628. doi: 10.4161/cc.26588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg VL, Zimmer SG. Paclitaxel induces the phosphorylation of the eukaryotic translation initiation factor 4E-binding protein 1 through a Cdk1-dependent mechanism. Oncogene. 2005;24(30):4851–4860. doi: 10.1038/sj.onc.1208624. [DOI] [PubMed] [Google Scholar]
- 17.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrama D, Ugurel S, Becker JC. Merkel cell carcinoma: Recent insights and new treatment options. Curr Opin Oncol. 2012;24(2):141–149. doi: 10.1097/CCO.0b013e32834fc9fe. [DOI] [PubMed] [Google Scholar]
- 19.Chang Y, Moore PS. Merkel cell carcinoma: A virus-induced human cancer. Annu Rev Pathol. 2012;7:123–144. doi: 10.1146/annurev-pathol-011110-130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spurgeon ME, Lambert PF. Merkel cell polyomavirus: A newly discovered human virus with oncogenic potential. Virology. 2013;435(1):118–130. doi: 10.1016/j.virol.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121(9):3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shuda M, Chang Y, Moore PS. Merkel cell polyomavirus-positive Merkel cell carcinoma requires viral small T-antigen for cell proliferation. J Invest Dermatol. 2014;134(5):1479–1481. doi: 10.1038/jid.2013.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwun HJ, et al. Restricted protein phosphatase 2A targeting by Merkel cell polyomavirus small T antigen. J Virol. 2015;89(8):4191–4200. doi: 10.1128/JVI.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwun HJ, et al. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe. 2013;14(2):125–135. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhaegen ME, et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J Invest Dermatol 2014 doi: 10.1038/jid.2014.446. , 10.1038/jid.2014.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingras AC, et al. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 1999;13(11):1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanie T, et al. Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Mol Cell Biol. 2012;32(3):590–605. doi: 10.1128/MCB.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12(4):502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7(2):e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyler RK, Shpiro N, Marquez R, Eyers PA. VX-680 inhibits Aurora A and Aurora B kinase activity in human cells. Cell Cycle. 2007;6(22):2846–2854. doi: 10.4161/cc.6.22.4940. [DOI] [PubMed] [Google Scholar]
- 31.Vassilev LT, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103(28):10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69(7):1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 33.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441(1):1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 34.Vassilev LT. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle. 2006;5(22):2555–2556. doi: 10.4161/cc.5.22.3463. [DOI] [PubMed] [Google Scholar]
- 35.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5(4):607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 36.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci USA. 2002;99(1):19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, et al. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125(11):3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen DA, Shapiro DJ. Preparation of capped RNA transcripts using T7 RNA polymerase. Nucleic Acids Res. 1986;14(14):5936. doi: 10.1093/nar/14.14.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001;7(10):1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 40.Cencic R, et al. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci USA. 2011;108(3):1046–1051. doi: 10.1073/pnas.1011477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18(7):2603–2618. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260(5547):170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 43.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278(5701):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 44.Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17(1):43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 45.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 46.DeCaprio JA, et al. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54(2):275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 47.Chang HW, et al. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276(5320):1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 48.Moore PS, Chang Y. Antiviral activity of tumor-suppressor pathways: Clues from molecular piracy by KSHV. Trends Genet. 1998;14(4):144–150. doi: 10.1016/s0168-9525(98)01408-5. [DOI] [PubMed] [Google Scholar]
- 49.Shahbazian D, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25(12):2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, et al. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20(16):4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilker EW, et al. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446(7133):329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 52.Boussemart L, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513(7516):105–109. doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 53.Terasima T, Tolmach LJ. Growth and nucleic acid synthesis in synchronously dividing populations of HeLa cells. Exp Cell Res. 1963;30:344–362. doi: 10.1016/0014-4827(63)90306-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.