Significance
Mapping neutralizing epitopes on viral entry glycoproteins allows the identification of potentially important functional regions. The structure of varicella-zoster virus (VZV) gHgL bound to two antibodies isolated from immune donors reveals a common binding site. Functional experiments demonstrate that the two antibodies neutralize VZV infection and inhibit glycoprotein gB/glycoprotein complex gHgL-mediated membrane fusion. Immunization experiments in mice demonstrate that VZV gHgL elicits potently neutralizing antibodies and confirm the key role of this antigenic site in antibody-mediated virus neutralization. This manuscript sheds light on the molecular mechanism of herpesvirus cell entry and will guide the design of subunit-based vaccines against VZV.
Keywords: VZV, gHgL, neutralizing antibodies, crystal structures, electron microscopy
Abstract
Varicella-zoster virus (VZV), of the family Alphaherpesvirinae, causes varicella in children and young adults, potentially leading to herpes zoster later in life on reactivation from latency. The conserved herpesvirus glycoprotein gB and the heterodimer gHgL mediate virion envelope fusion with cell membranes during virus entry. Naturally occurring neutralizing antibodies against herpesviruses target these entry proteins. To determine the molecular basis for VZV neutralization, crystal structures of gHgL were determined in complex with fragments of antigen binding (Fabs) from two human monoclonal antibodies, IgG-94 and IgG-RC, isolated from seropositive subjects. These structures reveal that the antibodies target the same site, composed of residues from both gH and gL, distinct from two other neutralizing epitopes identified by negative-stain electron microscopy and mutational analysis. Inhibition of gB/gHgL-mediated membrane fusion and structural comparisons with herpesvirus homologs suggest that the IgG-RC/94 epitope is in proximity to the site on VZV gHgL that activates gB. Immunization studies proved that the anti-gHgL IgG-RC/94 epitope is a critical target for antibodies that neutralize VZV. Thus, the gHgL/Fab structures delineate a site of herpesvirus vulnerability targeted by natural immunity.
Varicella-zoster virus (VZV) is an enveloped, double-stranded DNA virus of the family Alphaherpesvirinae. Primary VZV infection initiates at the respiratory mucosa. T cells are infected in regional lymph nodes and disseminate virions to the skin, causing varicella (chickenpox) (1). VZV reaches sensory ganglia by the hematogenous route or by anterograde axonal transfer from skin lesions and establishes latency in neurons (2). VZV reactivation can lead to herpes zoster (shingles), which is particularly prevalent in the elderly and in immunocompromised individuals (1, 3). An estimated one third of people in the United States will develop herpes zoster and are at risk for the chronic pain syndrome referred to as postherpetic neuralgia.
Live attenuated vaccines derived from the Oka strain are safe and effective against varicella and herpes zoster in healthy individuals but are contraindicated in most immunocompromised patients (4, 5). Subunit vaccines have the potential to be safe in these populations (6). High-titer VZV immunoglobulin given shortly after exposure decreases the incidence of varicella (7), indicating that antibodies against the VZV envelope glycoproteins can interfere with the initiation of infection and cell-to-cell spread. Thus, subunit vaccines made from one or more of these proteins could be an alternative to live attenuated VZV vaccines. Indeed in a phase 1/2 clinical trial, AS01B-adjuvanted envelope glycoprotein gE has been reported to be more immunogenic than a live attenuated VZV vaccine in healthy individuals in the age groups of 18–30 or 50–70 y. Advances are occurring in the development of this gE-based vaccine candidate against herpes zoster (6).
Similar to other herpesviruses, VZV entry into host cells is presumed to be initiated by virion attachment to the cell surface, followed by fusion between the virus envelope and the plasma membrane of the target cell. The envelope glycoproteins gB and the gHgL heterodimer (8) are highly conserved components of the herpesvirus entry machinery. VZV gB and gHgL are necessary and sufficient to induce cell fusion, which is considered a surrogate for virion envelope fusion (9, 10).
The crystal structures of the gHgL heterodimer ectodomain for herpes simplex virus 2 (HSV-2) and Epstein–Barr virus (EBV) showed that gH and gL cofold to form a tightly packed complex with no similarity to any known viral fusion proteins (11, 12). In contrast, the structures of HSV-1 and EBV gB are highly similar to the vesicular stomatitis virus membrane fusion glycoprotein G, indicating that gB is the herpesvirus fusion protein (13–15). Fluorescence bimolecular complementation studies with HSV-1 envelope glycoproteins imply that gHgL interacts with gB during membrane fusion (16, 17). This interaction was inhibited by the anti-gHgL neutralizing monoclonal antibody (mAb) LP11 (11). How gHgL facilitates the conformational changes in gB required for membrane fusion and viral entry remains elusive.
A homology model of VZV gH based on the HSV-2 gHgL structure was used to identify gH residues involved in membrane fusion (9). Incorporating gH mutations into recombinant VZV showed the importance of the gH ectodomain and cytoplasmic domain for viral replication and regulation of cell fusion in vitro and for infection of human skin xenografts in the SCID mouse model of VZV pathogenesis, as are gB mutations (9, 18, 19).
The contribution of VZV gHgL to viral entry and spread was also demonstrated by the capacity of an anti-gH mouse mAb, mAb206, to neutralize VZV in vitro and to severely impair infection of human skin xenografts (20–22). Human neutralizing mAbs generated from a library of pooled B cells obtained from naturally immune individuals also target gHgL (23). Two of these, designated IgG-24 and IgG-94, and their respective fragment of antigen-binding (Fab), had the most VZV-neutralizing activity. Recently, a VZV-neutralizing human mAb (rec-RC IgG), which recognized the gHgL heterodimer in VZV infected cells, was isolated from plasmablasts recovered from an individual who was immunized with a zoster vaccine (24).
The present study provides the first report, to our knowledge, on the structure of a herpesvirus gHgL heterodimer in complex with human mAbs and delineates the location of clinically relevant neutralizing epitopes on VZV gHgL. Structural and binding studies demonstrated that Fab-RC and Fab-94 target the same epitope formed by gH and gL residues, whereas Fab-24 binds to a distal site on gH. Immunization studies proved that the gHgL Fab-RC/-94 epitope is a critical target for neutralizing Abs against VZV. Compared with postfusion gB, gHgL elicited Abs in mice that more potently inhibited cell fusion and neutralized VZV. Vulnerability to Ab-mediated inhibition suggests the gHgL complex could be a component of an effective subunit vaccine.
Results
Binding of Human and Mouse Anti-gHgL Neutralizing mAbs to VZV gHgL.
The interactions between VZV gHgL and the Fab fragments of IgG-RC, IgG-94, and IgG-24 were studied by surface plasmon resonance (SPR). Each of the three Fabs binds to gHgL, with comparable dissociation constants (KD) in the subnanomolar range. The mouse mAb206 bound gHgL at ∼100-fold lower affinity than the others (Table S1 and Fig. S1 A and B). Additional binding experiments demonstrated that Fab-94 and Fab-24 were able to bind VZV gHgL simultaneously, whereas Fab-94 and Fab-RC competed with each other (Fig. S1C). Immunoprecipitation experiments confirmed that Fab-24 does not compete with IgG-RC for gHgL binding. Furthermore, neither Fab-RC nor Fab-24 competed with mAb206, suggesting an independent epitope for mAb206 (Fig. S1D).
Because mutations in residues 38–41 of gH abolished mAb206 binding (9), the epitopes of the four Abs were further investigated using a truncated form of gHgL (ΔNgHgL; aa 46–795 of gH) lacking the first 28 residues of the gH ectodomain (Fig. 1A and Fig. S2). The binding affinities of ΔNgHgL for Fab-RC and Fab-24 did not differ from wild-type (WT) gHgL, whereas mAb206 did not bind the truncated complex (Fig. S1 A and B). Thus, the gH N terminus is part of the mAb206 epitope that differs from the epitopes recognized by IgG-24, IgG-94, and IgG-RC.
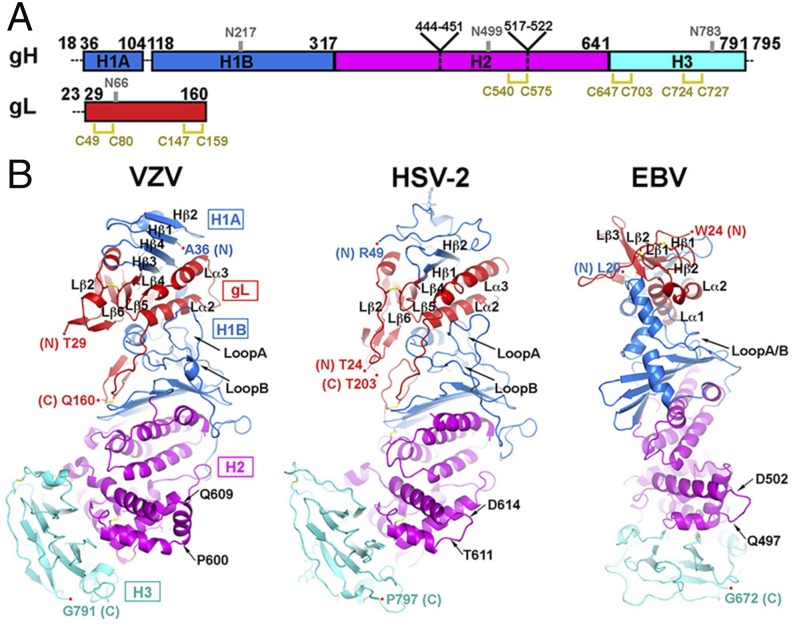
Fig. 1.
Structural comparison of the VZV gHgL with HSV and EBV gHgL. (A) Primary structure of VZV gH and gL. gH domains H1 (H1A and H1B), H2, and H3 are colored blue, magenta, and cyan, respectively. gL is colored red. Dashed lines represent regions of the construct missing in the final model. Disulfide bonds are shown in yellow. Glycosylated asparagines are labeled in gray. (B) Structure of VZV gHgL complex in comparison with those of HSV-2 and EBV. gHgL domains are colored as in A.
Structure Determination of VZV gHgL/Fab Complexes.
To delineate the molecular details of VZV gHgL and epitopes recognized by neutralizing mAbs, the gHgL/Fab-RC and gHgL/Fab-94 complexes were crystallized and their structures were determined by molecular replacement at 3.1- and 3.9-Å resolution, respectively (Fig. S2 and Table S2). Crystals could not be grown for gHgL/Fab-24 alone, and only poorly diffracting crystals were obtained for the gHgL/Fab-94/Fab-24 ternary complex (Fig. S2). Therefore, negative-stain electron microscopy (EM) reconstruction was used to identify the Fab-24 binding site.
Despite the relatively low-resolution diffraction data for the gHgL/Fab-94 complex, electron densities of Fab-94 and gHgL residues at the complex interface were clearly defined in simulated annealed omit maps, allowing unambiguous tracing and model building (Fig. S3A). In contrast, the electron densities for the Fab-94 constant domains lacked connectivity in several regions, even at the final stage of the refinement. Electron density was not observed for residues N-terminal to A36 of gH in either the gHgL/Fab-RC or the gHgL/Fab-94 structures, suggesting the gH N terminus is flexible.
VZV gHgL Has an Extended N-Terminal β-Sheet Compared with Herpesvirus Homologs.
The gHgL/Fab-RC crystal structure was used for the structural analysis of gHgL because of its higher resolution. VZV gHgL assumes a boot-like shape reminiscent of HSV-2 gHgL, but different from the straight domain packing of the EBV gHgL or pseudorabies virus (PrV) gH (Fig. 1) (11, 12, 25). Similar to other herpesviruses, VZV gL cofolds with gH. The structural core of VZV gL has a chemokine fold comprising a three-stranded β-sheet (Lβ4/Lβ5/Lβ6), two α-helices (Lα2/Lα3), and a structurally conserved disulfide bond involving residues C49 and C80 (26). VZV gL was more similar to the alphaherpesvirus HSV-2 gL (Cα rmsd: 8.7 Å) than the gammaherpesvirus EBV gL (Cα rmsd: 19.7 Å), as anticipated from homology modeling (9) (Fig. 1B).
VZV gH can be divided into three domains: H1A/B, H2, and H3, as described for HSV-2 gH (Fig. 1) (11). The H1 domain of VZV gH differs most significantly from HSV and EBV gH. The six-stranded antiparallel β-sheet in the N terminus of the HSV-2 gHgL heterodimer, with four strands from gL (Lβ2/Lβ6/Lβ5/Lβ4) and two strands from gH H1 domain (Hβ1/Hβ2 in HSV-2, Hβ3/Hβ4 in VZV gH), are extended in VZV gH by two antiparallel β-strands, Hβ1/Hβ2 (Fig. 1B). Hβ1 (aa 36–45) bridges the interaction between Hβ2 and the remainder of the hybrid β-sheet. Unlike VZV Hβ1/Hβ2, the equivalent N terminus of HSV-2 gH adopts a coil conformation. EBV gHgL forms only a five-stranded β-sheet, with two strands contributed by gH and three by gL (Fig. 1B). In addition to the eight-stranded β-sheet, VZV gH and gL also interact through multiple loops of the gH H1B domain and two conserved α-helices of gL, Lα2/Lα3. A thumb-like loop in gH (loop A; aa 285–298) lies on the surface of Lα2 and Lα3, whereas a second loop in gH (loop B; aa 154–161) and the remainder of H1B elements function as a supporting “palm” for these two gL α-helices.
The H2 domain is mostly α-helical, and the H3 domain is composed of a 10-stranded β-sandwich. The first four α-helices in H2 domain have been alternatively assigned as part of H1B domain because of their interaction with the extended β-sheet in H1B, leading to the formation of a “syntaxin-like bundle” (12, 25). The sequence of the central H2 and C-terminal H3 domains is more conserved than that of the N-terminal H1 domain across herpesvirus gHs, and this similarity is reflected in their structure. With the exception of an α-helix (aa 600–609) in the VZV H2 domain that replaces a loop in HSV-2 (aa 611–614), EBV (aa 497–502), and PrV (aa 447–451), secondary structural elements in H2 and H3 domain are well conserved (Fig. 1B). In particular, VZV and HSV-2 H2 and H3 domains are very similar, with Cα rmsd of 1.56 and 1.49 Å, respectively. This observation suggests that the functions of the H2 and H3 domains are conserved across the Herpesviridae.
Structural Characterization of Neutralizing Epitopes on VZV gHgL.
The mAbs -RC and -94 belong to independent clonal lineages (24, 27). Using IMGT V-QUEST and IMGT junction analysis on V, D, and J germline segments (28), the closest homologous germline genes were identified as IGHV3-30–3*01 F, IGHJ6*02 F, and IGHD3-10*01 F for Fab-94 heavy chain (HC) and IGKV1-12*01 F and IGKJ4*01 F for Fab-94 light chain (LC). The germline genes of Fab-RC are IGHV1-2*04 F and IGKV1-9*01 F, based on a partial nucleotide sequence of its HC and LC. Remarkably, the crystal structures of gHgL bound to Fab-RC and Fab-94 revealed that the two Fabs share a common epitope, which spans a surface composed of residues from both the gH-H1 domain and gL (Fig. 2). The buried surface of gHgL is larger in the complex with Fab-RC compared with Fab-94 (1036 and 822 Å2, respectively), consistent with the slightly higher affinity of the gHgL/Fab-RC complex (Table S1).
Fig. 2.
Fab-RC, Fab-94, and Fab-24 bind to two distinct epitopes on VZV gHgL. (A) Crystal structures of VZV gHgL bound to Fab-RC or Fab-94 presented at a side view and an inset of a top view. gH and gL are colored as in Fig. 1. The HCs and LCs of Fabs are colored in green and yellow, respectively. CH, constant domain of HC; CL, constant domain of LC; VH, variable domain of HC; VL, variable domain of LC. (B) Negative-stain EM 3D reconstruction of VZV gHgL bound to both Fab-94 and Fab-24. (C) Footprints of Fab-RC, Fab-94, and Fab-24 on the surface of VZV gHgL are outlined in yellow, green, and white dotted lines, respectively. gH-Hβ1 (aa 36–45), part of the conformational epitope of mAb206, is also outlined with a dotted line in cyan.
The main structural components of the epitope are two loops in gH, loop A and loop B, and Lα3 of gL. Both Fab-RC and Fab-94 recognize essentially the same epitope, but assume different orientations, being rotated almost 180° (Fig. 2A). Although the HC complementarity determining region 3 (HCDR3) fit into the same cleft on gHgL, the other CDRs occupy opposite positions. Thus, the surface area of gHgL that contacts the LC CDRs (LCDRs) of Fab-RC interacts with the Fab-94 HCDRs, whereas the HCDRs of Fab-RC and the LCDRs of Fab-94 bury a similar surface of gHgL (Fig. 2A).
The Fab-24 binding site was identified using a negative-stain EM 3D reconstruction of the gHgL/Fab-94/Fab-24 ternary complex at a resolution of 15 Å. Unlike Fab-RC and Fab-94, Fab-24 contacts the gH H2 domain and part of H1 domain, and its epitope is composed of residues within amino acids 119FGFLSHP125, 171ERPFGVSLLPARPTVP186, 207TFFSAEAIIT216, 278HTVK281, and 381MGRTTEYF388 (Fig. 2 B and C and Fig. S3B). Although bound to gHgL at a distinct location from the Fab-RC/Fab-94 epitope, Fab-24 appears to interact with structural elements directly linked to loops A and B. Thus, the structural data identified the epitopes of Fab-RC, Fab-94, and Fab-24, and together with binding data (Fig. S1D), confirmed that these human mAbs do not compete with mouse mAb206 for gHgL binding (Fig. 2C).
The Fab-RC and Fab-94 Epitopes Have Common Molecular Features.
Residue W291 in the gH loop A plays a prominent role in both gHgL/Fab-RC and gHgL/Fab-94 interactions. In the gHgL/Fab-RC complex, the indole ring of W291 protrudes into a hydrophobic pocket formed by residues from LCDR1–LCDR3 and HCDR3 (Fig. 3A). The interaction is further enhanced by additional hydrophobic and charge–charge interactions around loop A, including F292 pointing toward LCDR2 and D288 forming hydrogen bonds with the main chain amides of HCDR3 to hold it in position (Table S3). In the complex with Fab-94, a backbone rearrangement of gH-loop A allows the aromatic ring of W291 and F292 to occupy a hydrophobic pocket formed by HCDRs 1–3 and LCDR3. In particular, the W291 side chain becomes sandwiched between the phenyl ring of HCDR2-Y59 and gH-F292 (Fig. 3B and Fig. S4 A and B).
Fig. 3.
Interface between gHgL and Fab-RC or Fab-94. Side chains of interface residues in the complex of (A) gHgL/Fab-RC and (B) gHgL/Fab-94 are shown as sticks. For clarity, Fab residues forming the hydrophobic pocket are selectively labeled. Coloring is as in Fig. 2.
A six-amino-acid stretch in the HCDR3 of either Fab-RC or Fab-94 interacts with a surface cleft defined on one side by loop B 156PLVW159 and on the opposing side by loop A 294LNPP297 (Fig. 3). In the gHgL/Fab-RC complex, this cleft is more open and occupied by the HCDR3 M102 side chain. The HCDR3 of Fab-94 inserts less deeply and uses the aromatic side chains of 103HYYY106 to contact residues in the upper part of the cleft (Fig. 3 and Table S3). Y103H is the only somatic mutation found in the Fab-94 variable domain that plays a role on the interface of gHgL/Fab-94 (Fig. S4C).
Fab-94 and Fab-RC both form specific interactions with the gL Lα3. E114 in the loop between Lα2 and Lα3 and R121 at the N-terminal end of Lα3 form H-bonds with Fab-94 HCDR2 S52 and N57, respectively. At the Fab-RC interface, LCDR2 R56 forms salt bridges with both D122 and D126 of Lα3. This arginine is one of the three somatic mutations (S56R) identified on the gHgL/Fab-RC interface in the variable domain nucleotide sequence of IgG-RC. The other two are S30D, which contributes to a charge–charge interaction with gH, and Y32F, which is part of the hydrophobic pocket in Fab-RC, accommodating gH-W291 (Fig. 3 and Fig. S4C and Table S3). Therefore, comparison of the two gHgL/Fab interfaces revealed a number of shared features, consistent with the competition of Fab-94 and Fab-RC for gHgL binding.
Loop A Residues 291WF292 in gH Are Critical for the RC/94 Epitope.
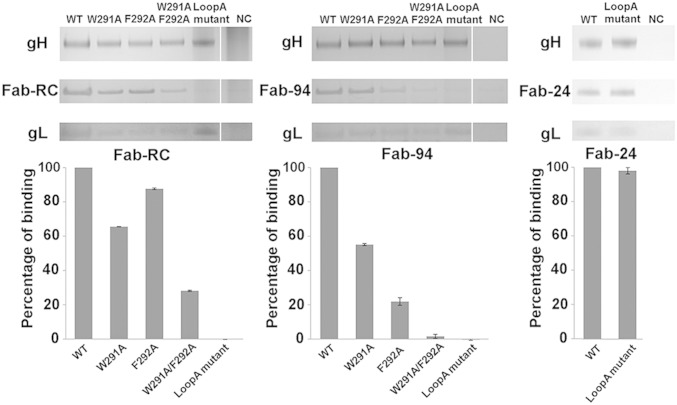
Mutations were incorporated into gH loop A to identify residues that are critical for Fab-94 and Fab-RC binding. Affinity pull-down experiments demonstrated that single and double mutations of loop A 291WF292 to alanines substantially reduced the interaction of gHgL with Fab-RC or Fab-94 (Fig. 4 and Fig. S2A). Mutation of loop A residues 288DTTWFQL294 to 288AGGAAQD294 abolished binding to both Fabs. All the mutant gH proteins formed stable complexes with gL and retained their interaction with Fab-24.
Fig. 4.
Mutational analyses of the gHgL/Fab-RC and gHgL/Fab-94 interfaces. Affinity pull-down using gHgL with mutations in gH (W291A, F292A, W291A/W292A, Loop A mutant) revealed reduced binding to Fab-RC or Fab-94. Loop A mutant (D288A/T289G/T290G/W291A/F292A/L294D) did not affect Fab24 binding.
The role of 291WF292 in Fab-RC binding was confirmed when the full-length gHgL complex was over-expressed in CHO cells. Although mature gHgL with individual substitutions W291A or F292A was immunoprecipitated by IgG-24, IgG-RC, and mAb206, the 291AA292 or loop A 288AGGAAQD294 mutants were only immunoprecipitated by IgG-24 and mAb206, but not IgG-RC (Fig. S1E).
VZV gHgL Neutralizing Abs Inhibit gB/gHgL-Mediated Membrane Fusion.
The capacity of IgG-24, IgG-94, and IgG-RC and their corresponding Fabs to interfere with cell fusion was assessed as evidence for inhibition of virus entry (9). All of the IgGs and Fabs were tested at a single concentration. Fab-24, Fab-94, and Fab-RC significantly reduced fusion by 50–75% compared with the no-Ab and Fab controls, indicating that different subdomains of gH contribute to gB/gHgL-mediated fusion (Fig. 5A). Both IgG-94 and IgG-RC had significantly increased inhibition of fusion compared with their corresponding Fabs (Fig. 5B), and they had inhibitory activities comparable to the mouse mAb206. Cross-linking of gHgL molecules or an avidity effect resulting from the bivalent nature of Ab binding could play a role in such enhanced inhibition. In contrast, the levels of IgG-24 and Fab-24 fusion inhibition were similar. These data suggest that regions of gHgL bound by IgG-24, IgG-94, and IgG-RC constitute vulnerable sites for Ab-mediated inhibition of membrane fusion.
Fig. 5.
The role of VZV gHgL epitopes in gB/gHgL-mediated cell fusion and VZV neutralization. (A and B) gHgL binding Fabs (A) and IgGs (B) inhibit VZV cell fusion. Percentage of fusion was plotted relative to that observed in the absence of the Ab (No Ab). Fab-Ctrl and IgG-Ctrl are control Fab/IgG that bind an unrelated antigen. Transfection with empty vectors (vector) with no gHgL or gB was used as a negative control. (C and D) gHgL binding Fabs (C) or IgGs (D) neutralize VZV. Error bars show standard errors of the mean (SEM). One-way ANOVA was performed to establish significant differences in fusion or VZV neutralization compared with the vector (black asterisks in A and B), No Ab (red asterisks in A, B, C, and D) or mAb206 (black asterisks in C and D). ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. (E) Sera from immunized mice neutralize VZV. One-way ANOVA was performed to establish significant differences in VZV neutralization compared with the normal mouse serum (NMS, red asterisks) or mAb206 (black asterisks). ****P < 0.0001; **P < 0.01. (F) Sera from immunized mice inhibit mediated fusion. Undiluted, 10-fold (1:10) and 100-fold (1:100) diluted sera were analyzed. Error bars show SEM. NMS was used as a control.
As expected, Fab-24, Fab-94, and Fab-RC also had neutralizing activity against VZV compared with the no-Ab and Fab controls (Fig. 5C). IgG-24 and IgG-94 had more potent neutralizing activity than their corresponding Fabs (Fig. 5D) and neutralized VZV to low or undetectable titers at 72 h postinfection. The IgG-RC also neutralized the virus and reduced titers by log10 1.5 at the concentration tested, similar to mAb206.
To evaluate whether gH residues critical for Fab-RC and Fab-94 binding play a direct role in membrane fusion, the effects of the gH 291AA292 or loop A 288AGGAAQD294 mutations were tested. Mutations in loop A did not differ from WT gH in the cell fusion assay (Fig. S5 A and B). This demonstrates that the side chains of gH 288DTTWFQL294 are not required for a functionally active conformation of gHgL to mediate membrane fusion.
Immunization with gHgL Elicits VZV Neutralizing Abs that Inhibit Membrane Fusion.
To determine whether recombinant gHgL can elicit functional Abs in vivo that recognize the epitopes mapped by Fab-RC/Fab-94 or mAb206, BALB/c mice were immunized with equimolar amounts of MF59-adjuvanted gHgL, gHgL/Fab-RC, ΔNgHgL or the gB ectodomain at two different concentrations. VZV Ab titers measured by ELISA were highest in sera collected from mice in the high-dose group at day 14 after the third immunization (Fig. S6). About tenfold more antigen-specific Abs were detected in sera from mice immunized with gB compared with gHgL in both dose groups.
Mice immunized with gHgL or ΔNgHgL developed neutralizing Abs that significantly reduced cell-associated VZV titers in melanoma cells by log10 1.2 or 0.9, respectively, compared with the control mouse group (Fig. 5E). In contrast, gHgL/Fab-RC induced much lower levels of neutralizing Abs compared with the gHgL complex. These results suggest the Fab-RC epitope contributes to the induction of a significant fraction of the total VZV-neutralizing Abs that target gHgL. The mice immunized with gB did not produce neutralizing Abs even though the gB-specific Ab titers were higher than those obtained with the gHgL antigens by ELISA. It is known that recombinantly expressed ectodomain of herpesvirus gB tends to fold in the postfusion conformation, and it remains possible that a stabilized prefusion gB would elicit more potent neutralizing Abs (14, 15, 29, 30).
When pooled sera were tested in the membrane fusion assay, sera from all groups of gHgL immunized mice inhibited membrane fusion (Fig. 5F). Tenfold dilutions of gHgL, gHgL/Fab-RC, and ΔNgHgL sera retained the ability to inhibit fusion, whereas the gB sera only produced a 20% reduction in fusion at the same dilution. Inhibition of fusion was reduced significantly when all sera were tested at a 1:100 dilution. Inhibition by sera from mice given gHgL/Fab-RC indicates that the IgG-24 and mAb206 epitopes are sufficient to elicit fusion inhibitory Abs. Thus, gHgL was a more effective antigen than postfusion gB for eliciting fusion-inhibiting Abs in mice.
Discussion
The structural analysis of VZV gHgL in the present study identified epitopes targeted by mAbs that interfere with gB/gHgL-mediated membrane fusion and that have neutralizing activity against VZV. The serum Ab responses of mice given the gHgL, gHgL/Fab-RC, and ΔNgHgL immunogens demonstrated the role of the Fab-RC/Fab-94 epitopes in generating neutralizing Abs to VZV. Together, these data suggest that VZV gHgL could be used alone or in combination with other viral envelope glycoproteins, such as gE, to induce Abs that inhibit VZV infection. Antigen design strategies aimed at eliciting Abs specifically targeting the Fab-RC/Fab-94 epitope could be exploited to induce a potent neutralizing Ab response against VZV infection (31).
Inhibition of gB/gHgL-mediated membrane fusion reflects one mechanism to neutralize cell-associated VZV. Abs to gH may be internalized by VZV-infected cells (21) and might restrict VZV replication not only by inhibiting fusion/entry but also by interfering with intracellular events necessary for the production of progeny virions. These complementary neutralization mechanisms could contribute to the differing capacities of human mAbs/Fabs or sera from immunized mice to neutralize VZV compared with their inhibition of gB/gHgL-mediated fusion.
The analysis of the VZV gHgL crystal structures showed that the N-terminal 18 residues (aa 18–35) are flexible, and that this region is followed by two β-strands (Hβ1/ Hβ2) that are absent in HSV-2 gH. Deletion of residues 18–45 from the VZV gH N terminus, including the flexible N terminus and Hβ1, abrogated binding to the murine neutralizing mAb206 without affecting binding to Fab-RC or Fab-24. These data are consistent with a previous study in which substitution of residues 38LREY41 in Hβ1 to 38GRGG41 significantly reduced the binding affinity of mAb206, along with two other murine neutralizing mAbs, 258 and SG3 (9). The neutralization potency of these mAbs against a recombinant VZV mutant containing the 38GRGG41 gH substitutions was significantly decreased. In addition, the same mutant had impaired replication in human skin. Our crystal structures revealed that 38LREY41 is part of Hβ1 (aa 36–45), suggesting the 38GRGG41 mutation would destabilize the conformation of Hβ1, and therefore disrupt the packing of Hβ2. Together, these data indicate that Hβ1/Hβ2 is part of the conformational epitope of VZV gH recognized by mAb206, mAb258, and mAbSG3. It also supports the proposed role of Hβ1 and Hβ2 in VZV skin tropism and receptor binding (9). Consistent with these proposed roles, the VZV gH N-terminal region is the most divergent from HSV-2 gH.
Residues 19–38 at the extreme N terminus of the HSV-2 gH were also absent from the crystal structure of HSV-2 gHgL. This region was mapped as an epitope for the mAbs CHL17 and CHL32 that neutralized HSV-1 infection and blocked viral fusion (32). In addition, HSV-2 gH missing the N-terminal 29 residues 19–47, or the first 10 residues, 19–28, induced a low but constitutive level of fusion by gB even when gD was absent. Therefore, HSV gHgL with N-terminal truncations of gH was suggested to resemble the activated form of gHgL (33). The extreme N terminus of VZV gH could potentially keep gHgL in its inactive form through a similar mechanism. Whether or not the seemingly flexible N terminus of VZV gH plays a role together with the Fab-RC, Fab-94, and mAb206 epitopes during VZV viral entry requires further investigation.
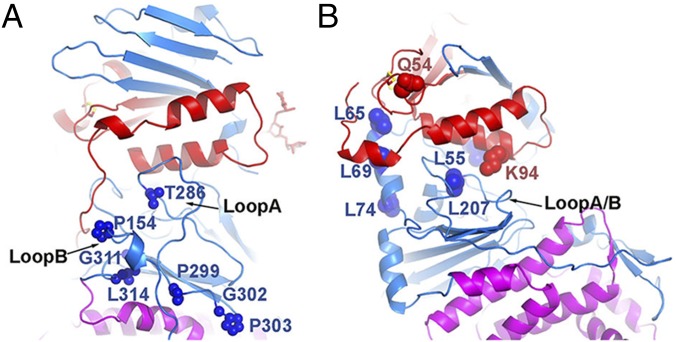
The location of the Fab-RC/94 epitope suggests this region is a critical target of acquired humoral immunity. LP11, a potent HSV-1 gHgL neutralizing mAb, blocks interactions between gB and gHgL in a bimolecular fluorescence complementation assay and inhibits the formation of syncytia in a fusion assay (11, 34). Substitutions found in the HSV-1 gH sequence encoded by LP11-resistant viruses (34) and insertions in the gH coding sequence that disrupt the LP11 epitope (35) were located in regions that corresponded to the gH loop A and loop B regions of VZV (Fig. 6A).
Fig. 6.
A conserved gB activation site on herpesvirus gHgL for membrane fusion is in proximity to the Fab-RC/94 epitope. (A) Side chains of corresponding residues mapped as part of the mAb LP11 epitope that was suggested as a gB binding site on HSV-1 gHgL are shown as spheres on VZV gHgL. (B) Side chains of residues important for EBV membrane fusion and gB activation are shown as spheres on EBV gHgL.
The structure of VZV gHgL is more distantly related to the EBV gHgL structure than that of the corresponding HSV proteins. The N-terminal half of EBV gH has a large groove next to the α-helices of EBV gL. Loops at locations analogous to VZV gH loop A and loop B are found inside this groove (Fig. 6B). Mutations of hydrophobic residues in EBV gH around this groove either reduce (L65A or L69A or L207A) or enhance (L55A/L74A) cell fusion (36, 37). L65 and L69 are also near the binding site for the EBV-neutralizing mAb E1D1 (36). In addition, two polar residues of EBV gHgL, Q54 in one of the conserved gL α-helices and K94 around the groove, are essential to specifically activate EBV-gB fusion activity (38). Similarly, polar residues in the VZV gL Lα3 are important components of the Fab-RC/94 epitope. Thus, the regions analogous to the Fab-RC/94 VZV gH epitope in EBV and HSV-2 gHgL play an important role in gB activation. Overall, these structural comparisons suggest the Fab-RC/94 epitope may be in proximity to the site on VZV gHgL that activates gB for membrane fusion.
In conclusion, this study identified a site of vulnerability in VZV gHgL recognized by neutralizing mAbs from VZV immune human subjects. Immunization studies further support that this site contributes significantly to induce potently neutralizing Abs against VZV. These findings are relevant for the design of gHgL-based VZV subunit vaccines to prevent or reduce the severity of both primary and recurrent VZV infections.
Materials and Methods
Details are provided in SI Materials and Methods. It describes construct generation, protein expression, and purification, binding analysis, X-ray and EM analysis, immunization and ELISAs, and VZV neutralization assays and cell fusion assays.
Supplementary Material
Acknowledgments
We thank Rajiv Chopra (Novartis Institutes for Biomedical Research) for help with data collection. We are grateful to Roxana Tarnita, Karen Matsuoka, and Yukti Aggarwal for excellent technical help and Zene Matsuda (Institute of Medical Science, University of Tokyo), and Marvin Sommer (Stanford University) for plasmids used in the fusion assay. Finally, we thank Enrico Malito and Matthew Bottomley for critically reading the manuscript. Work in the A.M.A. laboratory was supported by NIH Grant AI081994, and work in the C. Grose laboratory was supported by NIH Grant AI53846.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501176112/-/DCSupplemental.
References
- 1.Arvin A, Abendroth A. 2007. VZV: Immunobiology and host response. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, eds Arvin A, Campadelli-Fiume G, Mocarski E, et al. (Cambridge Univ Press, Cambridge, UK)
- 2.Zerboni L, Sen N, Oliver SL, Arvin AM. Molecular mechanisms of varicella zoster virus pathogenesis. Nat Rev Microbiol. 2014;12(3):197–210. doi: 10.1038/nrmicro3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moffat J, Ku C-C, Zerboni L, Sommer M, Arvin A. 2007. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, eds Arvin A, Campadelli-Fiume G, Mocarski E, et al. (Cambridge Univ Press, Cambridge, UK)
- 4.Takahashi M, et al. Clinical experience with Oka live varicella vaccine in Japan. Postgrad Med J. 1985;61(Suppl 4):61–67. [PubMed] [Google Scholar]
- 5.Holcomb K, Weinberg JM. A novel vaccine (Zostavax) to prevent herpes zoster and postherpetic neuralgia. J Drugs Dermatol. 2006;5(9):863–866. [PubMed] [Google Scholar]
- 6.Leroux-Roels I, et al. A phase 1/2 clinical trial evaluating safety and immunogenicity of a varicella zoster glycoprotein e subunit vaccine candidate in young and older adults. J Infect Dis. 2012;206(8):1280–1290. doi: 10.1093/infdis/jis497. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC) Updated recommendations for use of VariZIG—United States, 2013. MMWR Morb Mortal Wkly Rep. 2013;62(28):574–576. [PMC free article] [PubMed] [Google Scholar]
- 8.Peng T, et al. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J Virol. 1998;72(7):6092–6103. doi: 10.1128/jvi.72.7.6092-6103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vleck SE, et al. Structure-function analysis of varicella-zoster virus glycoprotein H identifies domain-specific roles for fusion and skin tropism. Proc Natl Acad Sci USA. 2011;108(45):18412–18417. doi: 10.1073/pnas.1111333108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suenaga T, et al. Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc Natl Acad Sci USA. 2010;107(2):866–871. doi: 10.1073/pnas.0913351107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdary TK, et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17(7):882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc Natl Acad Sci USA. 2010;107(52):22641–22646. doi: 10.1073/pnas.1011806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313(5784):187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 14.Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313(5784):217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 15.Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci USA. 2009;106(8):2880–2885. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atanasiu D, et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci USA. 2007;104(47):18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avitabile E, Forghieri C, Campadelli-Fiume G. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: The interaction between gB and gH/gL does not necessarily require gD. J Virol. 2009;83(20):10752–10760. doi: 10.1128/JVI.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang E, Arvin AM, Oliver SL. The cytoplasmic domain of varicella-zoster virus glycoprotein H regulates syncytia formation and skin pathogenesis. PLoS Pathog. 2014;10(5):e1004173. doi: 10.1371/journal.ppat.1004173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliver SL, et al. An immunoreceptor tyrosine-based inhibition motif in varicella-zoster virus glycoprotein B regulates cell fusion and skin pathogenesis. Proc Natl Acad Sci USA. 2013;110(5):1911–1916. doi: 10.1073/pnas.1216985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montalvo EA, Grose C. Neutralization epitope of varicella zoster virus on native viral glycoprotein gp118 (VZV glycoprotein gpIII) Virology. 1986;149(2):230–241. doi: 10.1016/0042-6822(86)90124-8. [DOI] [PubMed] [Google Scholar]
- 21.Vleck SE, et al. Anti-glycoprotein H antibody impairs the pathogenicity of varicella-zoster virus in skin xenografts in the SCID mouse model. J Virol. 2010;84(1):141–152. doi: 10.1128/JVI.01338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez JE, Moninger T, Grose C. Entry and egress of varicella virus blocked by same anti-gH monoclonal antibody. Virology. 1993;196(2):840–844. doi: 10.1006/viro.1993.1543. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki K, Akahori Y, Asano Y, Kurosawa Y, Shiraki K. Isolation of therapeutic human monoclonal antibodies for varicella-zoster virus and the effect of light chains on the neutralizing activity. J Med Virol. 2007;79(6):852–862. doi: 10.1002/jmv.20838. [DOI] [PubMed] [Google Scholar]
- 24.Birlea M, et al. Human anti-varicella-zoster virus (VZV) recombinant monoclonal antibody produced after Zostavax immunization recognizes the gH/gL complex and neutralizes VZV infection. J Virol. 2013;87(1):415–421. doi: 10.1128/JVI.02561-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Backovic M, et al. Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. Proc Natl Acad Sci USA. 2010;107(52):22635–22640. doi: 10.1073/pnas.1011507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malkowska M, Kokoszynska K, Dymecka M, Rychlewski L, Wyrwicz LS. Alphaherpesvirinae and Gammaherpesvirinae glycoprotein L and CMV UL130 originate from chemokines. Virol J. 2013;10:1. doi: 10.1186/1743-422X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akahori Y, et al. Characterization of neutralizing epitopes of varicella-zoster virus glycoprotein H. J Virol. 2009;83(4):2020–2024. doi: 10.1128/JVI.02097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brochet X, Lefranc MP, Giudicelli V. 2008. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36(Web Server issue):W503–W508.
- 29.Corey L, et al. Chiron HSV Vaccine Study Group Recombinant glycoprotein vaccine for the prevention of genital HSV-2 infection: Two randomized controlled trials. JAMA. 1999;282(4):331–340. doi: 10.1001/jama.282.4.331. [DOI] [PubMed] [Google Scholar]
- 30.Rieder F, Steininger C. Cytomegalovirus vaccine: Phase II clinical trial results. Clin Microbiol Infect. 2014;20(Suppl 5):95–102. doi: 10.1111/1469-0691.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Correia BE, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507(7491):201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cairns TM, et al. Epitope mapping of herpes simplex virus type 2 gH/gL defines distinct antigenic sites, including some associated with biological function. J Virol. 2006;80(6):2596–2608. doi: 10.1128/JVI.80.6.2596-2608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atanasiu D, et al. Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors. MBio. 2013;4(2):e00046-13. doi: 10.1128/mBio.00046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gompels UA, et al. Characterization and sequence analyses of antibody-selected antigenic variants of herpes simplex virus show a conformationally complex epitope on glycoprotein H. J Virol. 1991;65(5):2393–2401. doi: 10.1128/jvi.65.5.2393-2401.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galdiero M, et al. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J Virol. 1997;71(3):2163–2170. doi: 10.1128/jvi.71.3.2163-2170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omerović J, Lev L, Longnecker R. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J Virol. 2005;79(19):12408–12415. doi: 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Jardetzky TS, Longnecker R. The large groove found in the gH/gL structure is an important functional domain for Epstein-Barr virus fusion. J Virol. 2013;87(7):3620–3627. doi: 10.1128/JVI.03245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plate AE, Smajlović J, Jardetzky TS, Longnecker R. Functional analysis of glycoprotein L (gL) from rhesus lymphocryptovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J Virol. 2009;83(15):7678–7689. doi: 10.1128/JVI.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.